Abstract
Objective
Roseburia hominis is a flagellated gut anaerobic bacterium belonging to the Lachnospiraceae family within the Firmicutes phylum. A significant decrease of R. hominis colonization in the gut of ulcerative colitis patients has recently been demonstrated. In this work, we have investigated the mechanisms of R. hominis–host cross talk using both murine and in vitro models.
Design
The complete genome sequence of R. hominis A2-183 was determined. C3H/HeN germ-free mice were mono-colonized with R. hominis, and the host–microbe interaction was studied using histology, transcriptome analyses and FACS. Further investigations were performed in vitro and using the TLR5KO and DSS-colitis murine models.
Results
In the bacterium, R. hominis, host gut colonization upregulated genes involved in conjugation/mobilization, metabolism, motility, and chemotaxis. In the host cells, bacterial colonization upregulated genes related to antimicrobial peptides, gut barrier function, toll-like receptors (TLR) signaling, and T cell biology. CD4+CD25+FoxP3+ T cell numbers increased in the lamina propria of both mono-associated and conventional mice treated with R. hominis. Treatment with the R. hominis bacterium provided protection against DSS-induced colitis. The role of flagellin in host–bacterium interaction was also investigated.
Conclusion
Mono-association of mice with R. hominis bacteria results in specific bidirectional gene expression patterns. A set of genes thought to be important for host colonization are induced in R. hominis, while the host cells respond by strengthening gut barrier function and enhancing Treg population expansion, possibly via TLR5-flagellin signaling. Our data reveal the immunomodulatory properties of R. hominis that could be useful for the control and treatment of gut inflammation.
Keywords: Roseburia, T lymphocytes, immune tolerance, inflammatory bowel disease, flagellin, TLR5
Introduction
The human gut microbiota consists of more than 500–1,000 different phylotypes the majority of which belong to the Bacteroidetes and Firmicutes bacterial phyla (1). Successful symbiotic relationships arising from bacterial colonization of the human gut yield a wide variety of metabolic, structural, protective, and other beneficial functions. The immunological importance of the gut microbiota is also well recognized; it is particularly apparent in germ-free (GF) animals that have an impaired immune system, which, however, can be functionally reconstituted by the introduction of gut commensal bacteria (2–4).
In sharp contrast to the production of secretory intestinal IgA, which is mainly driven by microbial colonization per se (5, 6), the development and differentiation of T cells require colonization by specific commensal bacteria. Some species of clostridia, such as segmented filamentous bacteria (SFB), appear to be potent inducers of differentiation and maturation of intestinal Th1, Th17, and Treg cell lineages (7, 8). Recent studies have demonstrated that the clostridia clusters IV and XIVa and the Altered Schaedler Flora can induce de novo generation of Treg cells, while mono-colonization with Bacteroides fragilis can correct the Th1/Th2 imbalance in germ-free mice by promoting the expansion of Treg cells (4, 9, 10). The effects of commensal bacteria on T cell differentiation pathways are variable and may be influenced by a specific array of toll-like receptors (TLR) ligands associated with particular bacteria (11). For instance, the Treg-enhancing effects of B. fragilis are implemented via TLR2 signaling by polysaccharide A (12).
Dramatic changes in microbiota composition affecting the balance of symbionts and pathobionts have been documented in gastrointestinal disorders such as inflammatory bowel disease. Crohn’s disease (CD), for example, is characterized by a greater relative abundance of the Proteobacteria and a reduction of other bacteria such as the Bacteroidales and Clostridiales (13–17). A specific decrease of Roseburia spp. in patients with CD has also been noted (16, 18). Interestingly, this microbial dysbiosis is also associated with imbalances in T effector cell populations. The gut microbiota alterations in ulcerative colitis (UC) have not been characterized to the same extent. Only recently, a study of UC patients has demonstrated that Roseburia hominis, together with Faecalibacterium prausnitzii, is significantly decreased in this disease (16, 19). Both species display an inverse correlation with disease activity. Previous studies have shown anti-inflammatory effects of F. prausnitzii colonization on the host (20). Much less is known about R. hominis, which is a member of the clostridia cluster XIVa.
The complete DNA sequence and annotation of the R. hominis genome has been previously described (21). In this paper, bacterial and host transcriptome responses to R. hominis colonization were investigated. Bacterial responses included expression of genes involved in colonization of and adaptation to the environment of the murine gut, while host responses to colonization included genes of immunity and gut function.
Materials and Methods
Bacterial Cultures
Roseburia hominis A2-183T (=DSM 16839T = NCIMB 14029T) and other Roseburia species were maintained and grown on synthetic YCFA media as described before (22). Escherichia coli and Salmonella enterica strains were cultivated in LB medium. All Roseburia culture manipulations were performed in a MACS-MG-1000 anaerobic workstation (Don Whitley Scientific) under an atmosphere of 80% N2, 10% CO2 and 10% H2 at 37°C.
Animals, Experimental Design, and Sampling
Germ-free animal experiments were performed in the gnotobiotic rodent breeding facility of INRA (ANAXEM platform, Institut Micalis, INRA, Jouy-en-Josas, France). The GF C3H/HeN male mice were allocated into the control (N = 8) and treatment (N = 10) groups and caged individually. R. hominis A2-183T (=DSM 16839T = NCIMB 14029T) was grown anaerobically at 37°C in YCFA media. At days 0, 1, and 2, animals in the treatment group were given 109 colony-forming units (CFU) of R. hominis culture by gavage, while control animals were given 100 µL of YCFA medium. The ileal, ascending colonic, and cecal samples were collected at days 14 and 28. GF TLR5KO (C57BL/6 genetic background, N = 3) and C57BL/6 (N = 3) animals (Jouy-en-Josas), and conventional TLR5KO (C57BL/6 genetic background, N = 6) and Boy/J (C57BL/6 congenic, N = 6) animals (Medical Research Facility, University of Aberdeen) were gavaged with the live cultures of R. hominis to evaluate its functional importance. Boy/J (C57BL/6 B6 Cd45.1) mice were the background controls for conventional TLR5K0. They carry a CD45.1 pan leukocyte marker but are otherwise equivalent to C57BL/6 wild-type mice (23, 24). Twelve female C57BL/6 (6 weeks old) mice were used to evaluate the effect of R. hominis during DSS-induced mild colitis. Three mice were dosed daily by direct administration of a culture of R. hominis for a total of 8 days. Six mice were dosed with culture medium. Exposure to DSS was for 4 days (from day 3 to day 6). Three R. hominis-dosed and three control mice were offered sterile water containing DSS (20 g/L) over this period. On day 7, these mice were switched back to sterile water with no DSS. The mice were euthanased and dissected aseptically on day 9. Twenty-two female C57BL/6 mice (6 weeks old) were used to evaluate the effect of R. hominis during DSS-induced pathological colitis. After the acclimatization period of 7–10 days, the mice were dosed daily with 50 µL (109 CFU) of R. hominis in growth culture medium for 14 days. Control animals were given growth culture medium. From day 9, mice were given DSS (MW 50 kDa, 30 g/L) in their drinking water for 6 days. The animals were euthanized on day 15 and tissue sampling was performed as described earlier. The management and experimental procedures with animals were approved by the respective Local Ethical Review Committees.
Transcriptome Analyses
Bacterial RNA from the mouse cecum contents was isolated and labeled with dCTP-Cy3 or dCTP-Cy5 during cDNA synthesis. PCR products amplified from an R. hominis small fragment library (6,000 clones) were used to create duplicate microarrays on glass slides using a MicroGrid II TAS array-spotting robot (BioRobotics). Microarray hybridization was performed in a GeneTAC hybridization station (Genomic Solutions). Dye-swap and separate RNA purification protocols were used to avoid potential biases.
Murine RNA was extracted from the ileum and ascending colon tissues and hybridized to the GeneChip “NuGO Mouse Array” and GeneChip “Mouse Genome Array” (Affymetrix). Data analysis was performed using R1 and Bioconductor.2 The microarray data were submitted to NCBI GEO (Gene Expression Omnibus) with accession number GSE25544.
The R. hominis-specific primers 5′-CCCACTGACAGAGTATGTAATGTAC-3′ and 5′-GCACCACCTGTCACCAC-3′ were used for qPCR analyses of fecal samples to validate the efficiency of colonization. The analyses were performed using a 7500 Fast Real-Time PCR System (Applied Biosystems) with a Power SYBR Green PCR Master Mix (Applied Biosystems). RT-qPCR analyses were performed on the ileal and colonic RNA samples for host gene expression studies using a QuantiFast SYBR Green PCR Kit (Qiagen) and QuantiTect Primer Assays (Qiagen).
Immunofluorescence and Histology
FISH analyses were performed with neutral buffered, formalin-fixed gut tissue sections (2 µm thick) with 16S rRNA probes, specific for the domain of Bacteria (Cy3-labeled Eub338) and specific for the R. hominis strain A2-183 (FITC-labeled GTACATTACATACTCTGTCAGTG). Bacteria were visualized at x630 magnification. Immunolocalization of R. hominis flagellin was examined in methanol-fixed colon content smears using rabbit antisera anti-FlaA1 or anti-FlaA2 (Covalab) and Alexa donkey anti-rabbit 488 (Molecular Probes). T cell markers were examined on sequential intestinal tissue sections (8 µm) using Ly6G-FITC, CD3-FITC, isospecific IgG (BD Biosciences), and double-labeled FoxP3 Alexa Fluor 594 (Abcam) and CD3-FITC. Intestinal tissue sections (4 µm) were also stained with hematoxylin/eosin. A complete transverse cross-section of the colon from each animal was visualized at ×200 magnification. Each field of view was scored from 0 to 4 according to the method based on Ref. (25).
Cell Experiments
Caco-2 and HT29 cells were grown at 37°C in a 75% humidified atmosphere with 5% CO2 in 24-well plates. Before any treatment, cells were washed twice with Hanks’ Balanced Salt Solution and kept in DMEM supplemented with l-glutamine, selenium, and transferrin for 24 h. Epithelial cells were harvested after incubation for 2 or 4 h and then stored in RNAlater for further analyses.
Bone marrow cells were harvested from the femur and tibia of C3H/HeN and C57BL/6 mice. Bone marrow-derived dendritic cells (BMDCs) were generated by Flt3L or GM-CSF culture, stimulated with 100 ng/mL flagellin, and analyzed by flow cytometry. Supernatants were collected and tested for IL-10 and IL-12 by cytometry bead arrays (BD Bioscience).
Cells from the intestine and mesenteric lymph nodes (MLNs) were isolated as described previously (26), with minor modifications. In brief, cell suspensions were incubated with 100 U/mL of collagenase VIII (Sigma-Aldrich) in RPMI supplemented with 20% of FBS at 37oC for 20 min (MLN) or 60 min (intestine). Single cell suspensions were analyzed by flow cytometry. For mixed leukocyte reactions, naïve CD4+ T cells were purified from the major lymph nodes and spleens of OTII transgenic mice using MACs Miltenyi CD4+ T cell Isolation Kit as per manufactures instruction. Purified CD4+ T cells were co-incubated with flagellin activated Flt3L-derived BMDCs in the absence or presence of OVA323–339 peptide (1 µg/ml). After 5 days, T cell differentiation was determined by flow cytometry.
Flow Cytometry
Intestinal lamina propria cells and MLN cells were labeled with CD4-FITC, CD25-APC (eBioscience), CD8-APC-Cy7, CD3-PerCP (Biolegend), and B220-BV570 (BD Biosciences). Labeling of intracellular FoxP3 was performed after extracellular staining and fixation/permeabilization of cells. GM-CSF-derived dendritic cells were labeled with CD11b-PerCP Cy5.5 (BD Biosciences), CD11c-PE-Cy7, I-A/I-E-APC-Cy7, CD80-PE, CD86-APC, CD8-FITC, and B220-BV570 (Biolegend). Flt3L-derived dendritic cells were labeled with CD11c-PE-Cy7, CD11b- or Siglec-H-PerCP Cy5.5 (Biolegend), I-A/I-E-APC-Cy7, CD317-PE, CD40-Alexa Fluor 647, CD103-FITC, and B220-BV570. Coculture T cells were labeled with CD4-PerCP Cy5.5 (BD Biosciences), CD127-PE-Cy7, CD73-APC, CD195 (CCR5)-FITC, CD62L-BV570, and CD25-APC-Cy7 (Biolegend).
Statistical Analyses
Spot intensities on the microarrays were log-transformed and Loess normalization was applied to the microarray results. RT-qPCR data were transformed on a base-2 logarithmic scale and analyzed by one-way analysis of variance with a significance threshold of P < 0.05. Statistical significance between the treatment groups was evaluated using Student’s t-test.
More detailed protocols are available in SI Materials and Methods in Supplementary Material.
Results
R. hominis Responds to the Gut Environment by Upregulating Chemotaxis, Motility, and Mobilization Genes
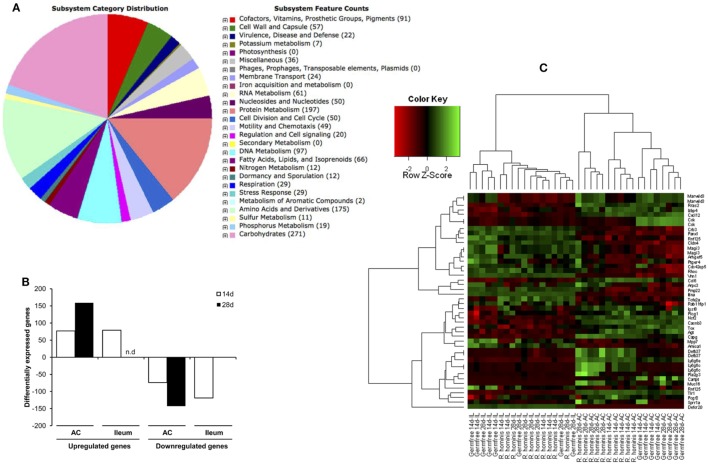
Genome analysis (Figure 1A) identified about 5% of all genes were related to chemotaxis and motility function including four different flagellin genes of which one located within the flagellar operon is associated with flagellar motility.
Figure 1.
Sequence features of Roseburia hominis genome and differentially expressed transcripts in the murine gut after mono-association with R. hominis. (A) Subsystem category distribution in the genome of R. hominis according to RAST. The largest subsystem feature counts belong to carbohydrates, amino acids and derivatives, and protein metabolism. (B) The number of differentially expressed genes in R. hominis-colonized mice (N = 5) compared to germ-free (GF) control (N = 4) identified with the Affymetrix microarray. Bar graphs represent the number of genes up- or downregulated in mice after 14 and 28 days of colonization.(C) Heatmap generated from differentially expressed genes with functional significance between GF and R. hominis-colonized mice at 14 and 28 days.
A biomass of R. hominis was given by gavage to GF C3H/HeN mice on three consecutive days. R. hominis-colonized both the ileum and colon but was found in much higher numbers in the colon, up to 1 × 1010 bacteria/g feces (Figure S1 in Supplementary Material) and was found to be closely associated with the colonic mucosa.
Differential gene expression in the bacterium in response to association with the host was investigated using the R. hominis microarray. Bacterial RNA was isolated from three different experimental conditions to distinguish between the effects of the gut environment and animal dietary components, (i) in vivo, from the cecum of mono-associated mice; (ii) in vitro, from bacteria grown in the presence of dietary components; and (iii) in vitro, from bacteria grown in culture.
Colonization of GF mice with R. hominis correlated with the increased host gene expression, which was highest in the colon, particularly at day 28 post-colonization (Figure 1B). The number of differentially expressed genes in the ileum was very low at day 28, consistent with the reduced bacterial numbers. Differential transcriptome responses in the two tissue sites and time points were illustrated by a clear separation of significant transcripts in the heatmap analysis (Figure 1C).
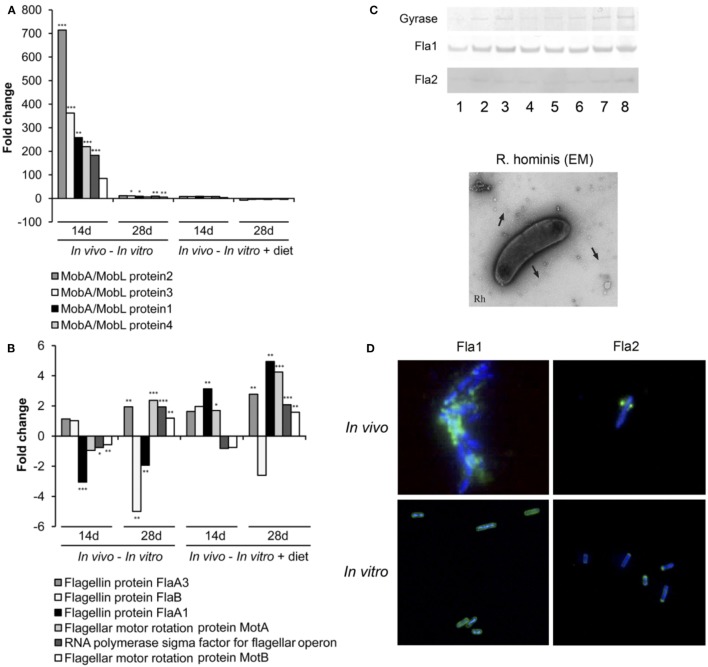
Differentially expressed genes were identified (in vivo vs. in vitro) with RT-qPCR validation performed on 42 differentially expressed genes (Tables S1 and S2 in Supplementary Material). The mobA- and mobL-like genes that are involved in conjugation/mobilization transfer showed strong upregulation in vivo (Figure 2A). Other subsystems induced by the gut environment included membrane transport, in particular magnesium transport, and motility and chemotaxis including multiple methyl-accepting chemotaxis proteins and genes of the flagellar operon (Figure 2B). R. hominis possesses multiple flagellin genes: flaA1, flaA2, flaA3, and flaB. The expression of flaA1and flaA2 in the murine gut environment was verified by Western–blot and immunocytochemistry (Figure 2C).
Figure 2.
Responses of Roseburia hominis to the gut environment. (A) RT-qPCR quantification of R. hominis transcripts involved in conjugation/mobilization transfer. R. hominis RNA samples: in vivo—isolated from mono-associated animals; in vitro—isolated from YCFA medium-grown cultures; in vitro + diet—isolated from cultures grown on YCFA medium with addition of murine chow. (B) RT-qPCR quantification of R. hominis transcripts involved in motility and chemotaxis. R. hominis RNA samples: in vivo—isolated from mono-associated animals; in vitro—isolated from YCFA medium-grown cultures; in vitro + diet—isolated from cultures grown on YCFA medium with addition of murine chow. (C) Western blot of R. hominis proteins grown in vitro in the presence of UV-irradiated standard murine chow. The membrane was immunostained with affinity-purified 1 antibody, Fla2 specific antiserum, and anti-DNA gyrase A antibody (lane 1: no diet, lane 2: 0.01 g diet/10 mL of R. hominis culture, lane 3: 0.02 g diet/10 mL, lane 4: 0.05 g diet/10 mL, lane 5: 0.1 g diet/10 mL, lane 6: 0.2 g diet/10 mL, lane 7: 0.5 g diet/10 mL, and lane 8: 1 g diet/10 mL). Electron microscopy of R. hominis showing flagella (black arrows). (D) Immunocytochemistry with FlaA1 and FlaA2 specific antisera performed on R. hominis from the luminal contents of mono-colonized mice and on R. hominis grown in vitro. Original magnification is ×1,000. RT-qPCR results are presented as fold change. Statistical significance: *P < 0.05, **P < 0.01, and ***P < 0.001.
R. hominis Affects Host Innate Signaling Pathways
Genes involved in innate immunity and gut barrier function of the host were significantly induced by R. hominis colonization [Figure 1C; NCBI GEO (Gene Expression Omnibus), accession number GSE25544]. The GO process “innate immune response” (GO:0045087) was upregulated and included the TLR-related genes Tlr5, Tlr1, and Vnn1. The upregulation of Tlr5 was of particular interest, given the presence of flagellar genes in the genome of R. hominis and the expression of the corresponding proteins in the gut of mono-colonized mice. The flagellins may have a role in this innate signaling pathway to mediate innate and adaptive immune responses. Other innate immunity genes affected by R. hominis colonization included the antimicrobial peptides Defb37, Pla2g3, Muc16, and Itln and the gut barrier function genes Sprr1a, Cldn4, Pmp22, Crb3, and Magi3. Innate immunity genes that were upregulated in the ileum in response to R. hominis included Defcr20, Pcgf2, Ltbp4, Igsf8, and Tcfe2a. We also found a negative regulation of the NF-κB pathway (GO:0043124) by R. hominis, which may contribute to the immune homeostasis by downregulating this inflammatory cascade.
R. hominis Directs T Cell Pathways
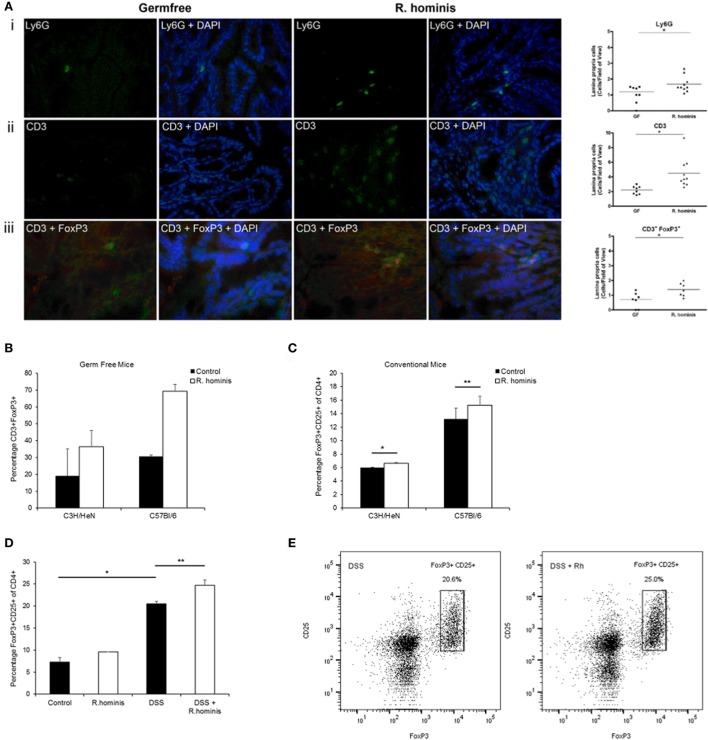
Immune response was a major pathway induced by R. hominis at day 28 in the ascending colon of mono-associated mice. The pathways significantly affected in this category were mostly involved in T cell function, including IL-10 signaling and regulation of T cell function by CTLA-4 (Table S3 in Supplementary Material). The Ly6 gene family was also induced in the ascending colon. In particular, the GPI-anchored gene product of Ly6g6c was upregulated 25-fold, and the related gene Ly6g6e was upregulated twofold at day 28. The majority of hematopoietic cells, including neutrophils and plasmacytoid dendritic cells, express one or more members of the Ly6 family. The increased presence of Ly6G+ cells in R. hominis-colonized mice was confirmed by immunocytochemistry (Figure 3A).
Figure 3.
Induction of FoxP3+ Treg cells by Roseburia hominis. (A) Immunofluorescence analysis of lamina propria cells labeled with (i) anti-Ly6G, (ii) anti-CD3, and (iii) anti-CD3/anti-FoxP3 in the ascending colon of germ-free (GF) and R. hominis-treated C3H/HeN mice. Data are shown as the number of positive cells per field of view in GF (N = 7–8) and R. hominis-treated mice (N = 8–10). Original magnification ×630. Statistical significance: *P < 0.05. (B) Immunofluorescence analysis of lamina propria cells in the ascending colon labeled with anti-CD3 and anti-FoxP3 in germ-free and R. hominis mono-colonized C3H/HeN (N = 8) and C57BL/6 (N = 3) mice. (C) Flow cytometry analysis of FoxP3+ Treg cells in the lamina propria of conventional mice treated for 14 days with R. hominis: C3H/HeN mice (*P = 0.04 between control and R. hominis treatment) and C57Bl/6 mice (**P = 0.05 between control and R. hominis treatment). (D) Flow cytometry analysis of FoxP3+ Tregs in lamina propria of FoxP3-tagged conventional mice treated for 8 days with R. hominis, with 4 days of low dose DSS treatment or untreated. Statistical significance: *P < 0.005 control versus DSS treatment and **P = 0.02 DSS treatment versus DSS+ R. hominis treatment. (E) Flow cytometry plots of FoxP3+ Treg cells in the colonic lamina propria of DSS-treated mice and DSS+ R. hominis-treated mice. Plots are representative of three experiments. The gating strategy for the FACS plots was as follows: FSC/SSC (removal of cell debris), live/dead-SSC, singlets (FSC-A/FSC-H), CD3+CD8−, and CD25+FoxP3+. The percentages were calculated as CD25+FoxP3+ cells of total CD4+ T cells in each preparation. CD4+ cells were derived from the gating strategy of CD3+CD8−.
While the pathways of T cell regulation were significantly influenced by association with R. hominis, the corresponding effects on T cell differentiation required further investigation. We therefore assessed the number of double-positive CD3+FoxP3+ cells in the colonic lamina propria of mono-associated C3H/HeN mice. A significant increase in the number of regulatory T cells (CD3+FoxP3+ cells) was detected in the germfree C3H/HeN and C56BL/6 mice colonized with R. hominis (Figures 3A,B).
Conventional C3H/HeN and C57BL/6 mice were gavaged with R. hominis for 14 days to determine the impact of the bacterium on Treg cells in the lamina propria of animals with a normal microbiota. This treatment resulted in a small but significant increase in the population of CD3+CD4+CD25+FoxP3+ T cells in the C3H/HeN (P = 0.04) and a smaller increase in the C57BL/6 mice (P = 0.05) when compared to non-colonized mice (Figure 3C). While the population of Tregs was induced in conventional mice after colonization with R. hominis under homeostatic conditions, it was unknown if this induction could be maintained under conditions of mild colitis. We therefore investigated the impact of the bacterium on Treg cells in the lamina propria of animals that were given low dose DSS. FoxP3-tagged mice of C57BL/6 background were colonized with R. hominis for 2 days before treatment with low dose of DSS for 4 days followed by 2 days of pure drinking water. DSS treatment alone significantly increased the population of CD3+CD4+CD25+FoxP3+ T cells (P = 0.02) (Figure 3D). When the animals were additionally treated with R. hominis, there was a further significant increase in the population of CD3+CD4+CD25+FoxP3+ T cells (P = 0.02) versus DSS (Figures 3D,E).
The Role of R. hominis Flagellins in Modulation of T Cell Differentiation
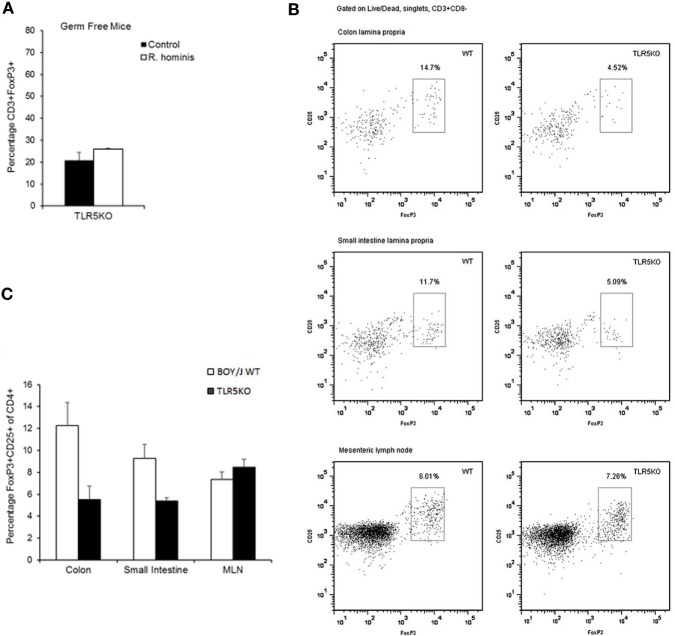
The influence of bacteria on the differentiation of T cells may reflect a specific array of TLR ligands displayed by a particular bacterium. TLR5KO (GF and conventional) and WT murine strains were gavaged with the bacterium to evaluate the functional importance of R. hominis and its flagellins. Analysis of the differentially expressed genes in TLR5KO and WT Boy/J mice colonized with R. hominis revealed that although T cell pathways were still influenced by the colonization event in TLR5KO mice, responses were more related to IL-4, IL-5, IL-6, IL-9 pathways and not to IL-10 and CTLA-4 (Table S4 in Supplementary Material). Furthermore, in contrast to mono-associated C3H/HeN and C57BL/6 animals (Figure 3B) and conventional Boy/J animals (Figures 4B,C), the numbers of double-positive CD3+FoxP3+ cells in the lamina propria of TLR5KO mice were not increased due to R. hominis treatment (Figure 4A).
Figure 4.
Modulation of T cell differentiation by Roseburia hominis flagellins. (A) Quantification of lamina propria cells in the ascending colon immunofluorescently labeled with anti-CD3 and anti-FoxP3 in germ-free (GF) and R. hominis mono-colonized TLR5KO mice (N = 2). (B) Flow cytometry plots of FoxP3+ Treg cells in the colonic and small intestinal lamina propria and mesenteric lymph nodes (MLNs) of conventional TLR5KO mice and Boy/J WT mice treated for 14 days with R. hominis. (C) Flow cytometry analysis of FoxP3+ Treg cells in conventional TLR5KO mice and Boy/J WT mice treated for 14 days with R. hominis: in the colonic lamina propria (P = 0.01 between TLR5KO and WT), in the lamina propria of the small intestine (P = 0.03 between TLR5KO and WT), and in MLNs (P = 0.33 between TLR5KO and WT).
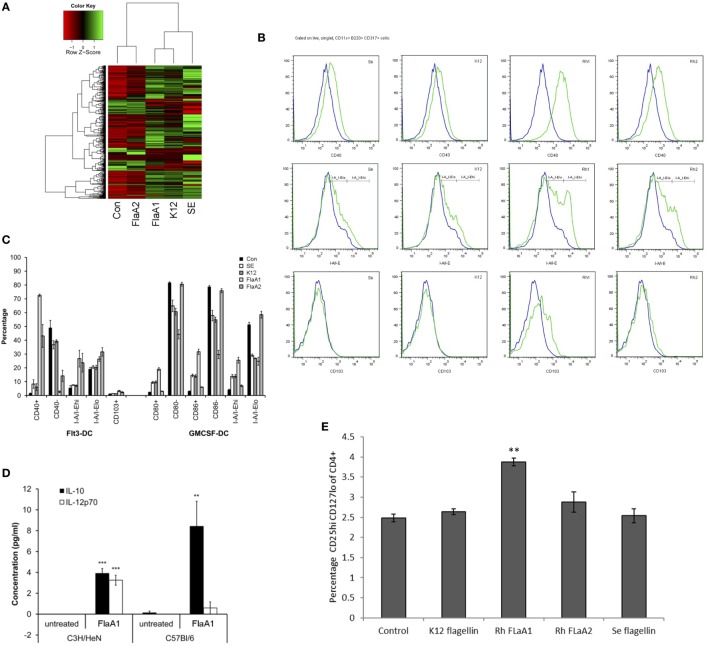
A set of flagellins from pathogenic and commensal bacteria, including recombinant R. hominis flagellins, was used to compare their effect on activation of signaling responses in IECs and BMDCs. Caco-2 cells were treated with the identical concentrations of different bacterial flagellins (Figure 5A). The flagellin of pathogenic S. enterica serotype Enteritidis (SE) induced a larger gene panel than the flagellins of commensal E. coli K12 or R. hominis FlaA1. E. coli and R. hominis induced a strong response and, with a similar gene complement between the two, formed a separate clade distinct from SE. In contrast, FlaA2 was generally neither pro-inflammatory nor did it activate the conserved gene signature (IL8, CXCL1, CXCL2, and CXCL10) that was induced by the other recombinant bacterial flagellins. In fact, the set of genes affected was closer to that of the control cells and formed a cluster separated from the other flagellins. FlaA1 from R. hominis induced different responses in Flt3L- and GM-CSF-derived BMDCs compared to SE and K12 (Figures 5B,C). In particular, FlaA1 was uniquely able to activate Flt3L-expanded DCs, with the upregulation of I-A/I-E and CD40 and production of IL-10 by BMDCs from both C3H/HeN and C57BL/6 mice. The IL-10/IL-12 ratio was particularly elevated in C57BL/6 DCs (Figure 5D), which were found to be CD103+Siglec-H+. To test whether flagellin activated BMDCs could differentially modulate naïve T cell differentiation, mixed leukocyte cultures of CD4+ OTII cells with flagellin stimulated Flt3-derived BMDC were performed. The data show that the production of IL-10 and IL-12 by the flagellin stimulated Flt3-derived BMDC ratio triggered naïve T cell differentiation toward a Tregs profile (Figure 5E). In contrast to FlaA2, FlaA1 significantly increased the Treg population in the T cell cocultures.
Figure 5.
Effects of Roseburia hominis flagellin FlaA1 on intestinal epithelial cells and murine bone marrow-derived dendritic cells. (A) Heatmap of differentially expressed genes in Caco-2 cells treated with different bacterial flagellins: from Salmonella enterica serotype Enteritidis (SE), Escherichia coli K12 (K12), and R. hominis (FlaA1 and FlaA2). (B) Expression of CD40; I-A/I-E and CD103 by CD11c+B220+CD317+ Flt3L-derived dendritic cells from conventional C3H/HeN, control (blue) and after 24 h incubation with recombinant flagellins (SE, K12, FlaA1, FlaA2; all in green) determined by flow cytometry. Histogram represents data from three experiments. (C) Frequencies of Flt3L- and GM-CSF-derived dendritic cells from conventional C3H/HeN gated on CD11c+B220+CD317+ cells and CD11c+CD11b+B220− cells treated with different recombinant flagellins (SE, K12, FlaA1, and FlaA2). Results are shown as the percentage of the total, live and singlet cells, mean ± SD from three experiments. (D) The level of IL-10 and IL-12 cytokines was measured by cytometry bead arrays in supernatants from control (unstimulated DCs; N = 3) and FlaA1-treated DCs (N = 3) derived from C3H/HeN and C57BL/6. Data are presented as mean ± SD. Statistical significance: ***P < 0.001. (E) Flagellin stimulated Flt3-derived dendritic cells were cocultured with CD4+ T cells for 5 days, and T cell were stained for the expression of T cell differentiation markers. Graph indicates the percentage of activated Treg cells. Statistical significance: **P < 0.01.
R. hominis Attenuates Colitis in DSS-Treated Mice
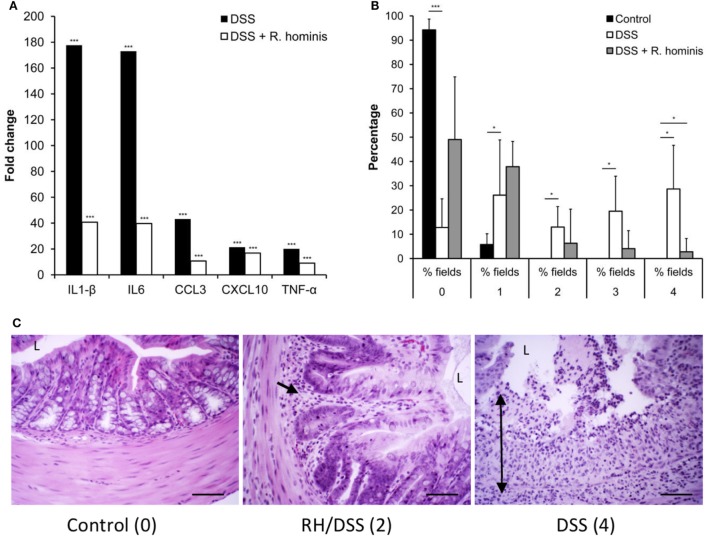
The effects of R. hominis on innate and adaptive immunity prompted us to test its therapeutic efficacy using the murine DSS model (Figure S2 in Supplementary Material; Figure 6). The treatment group was dosed daily with R. hominis (~50 μL, ca. 109 CFU) via gavage for a period of 14 days while the untreated group received the same amount of bacterial growth medium. The two groups were then given DSS in drinking water (MW 50 kDa, 30 g/L) beginning from day 8 onward. On day 15, DSS-treated mice without R. hominis addition had a strong elevation of a panel of pro-inflammatory biomarkers compared to control mice, with gene induction levels ranging from 4- to 49-fold (Figure 6A). Induction of pro-inflammatory genes was significantly lower in the R. hominis-treated mice compared to the control group, indicating a strong therapeutic benefit of oral administration of R. hominis. On day 15, severe inflammation was detected in the ascending colon of DSS-treated mice without an R. hominis supplement, while the colonic mucosa in the R. hominis-supplemented group displayed only low-level inflammation, consistent with the reduced expression of pro-inflammatory genes (Figures 6B,C). Treatment with R. hominis also alleviated the weight loss in DSS-treated mice (Figure S3 and Table S5 in Supplementary Material).
Figure 6.
Roseburia hominis attenuates inflammation in the DSS model of colitis. (A) DSS mice untreated with R. hominis (N = 8) had strong elevation of all genes compared to the control mice (N = 4), while differential gene expression was lower in R. hominis-treated animals (N = 10). RT-qPCR results obtained on day 15 are presented as a fold change compared to control. Statistical significance: ***P < 0.001. (B) Histopathology tissue scoring (25) presented as mean percentage of fields of view at a given grade. DSS treatment significantly altered all fields of view at grades 0, 2, 3, and 4. R. hominis significantly reduced the % fields of view for grade 4 pathology (P = 0.02) and increased the % fields of view for grade 0. Data obtained on day 15 are presented as mean ± SD. (C) Ascending colon (hematoxylin/eosin stained) of control, DSS+ R. hominis-treated and DSS-treated animals. Images shown are the representative fields of view for each treatment group with the allocated score in parenthesis. Single black arrow head indicates infiltrating cells, and the double headed arrow indicates transmural infiltration. L, lumen. Original magnification ×200. Scale bar represents 50 µm.
R. hominis Colonization Influences Body Composition and Expression of Satiety Genes
In the next set of experiments, we investigated metabolic changes in the host and bacterium due to host–microbe association. Mice mono-associated with R. hominis displayed significant metabolic changes compared to control GF mice. In particular, the GO processes “negative regulation of response to food” (GO:0032096), “negative regulation of appetite” (GO:0032099), and “regulation of catecholamine secretion” (GO:0050433) were all downregulated in the ascending colon after colonization by R. hominis [NCBI GEO (Gene Expression Omnibus), accession number GSE25544]. The genes involved in these processes were Agt, Cartpt, Cck, and Cxcl12, with the corresponding mRNA changes induced ranging from 2- to 12-fold. Cck, in particular, plays a major role in digestion and satiety as a hunger suppressant. Gcg expression was also downregulated at this gut site.
To establish whether the transcriptional changes of these genes have any physiological relevance in terms of body composition, analyses of dry carcass weight and lipid composition were performed. Dry carcass weights of R. hominis-associated mice were significantly heavier than those of GF animals (Figure 7A). Carcass lipid analysis showed that the total adiposity was also significantly higher in R. hominis-colonized animals at day 14 (Figure 7B).
Figure 7.
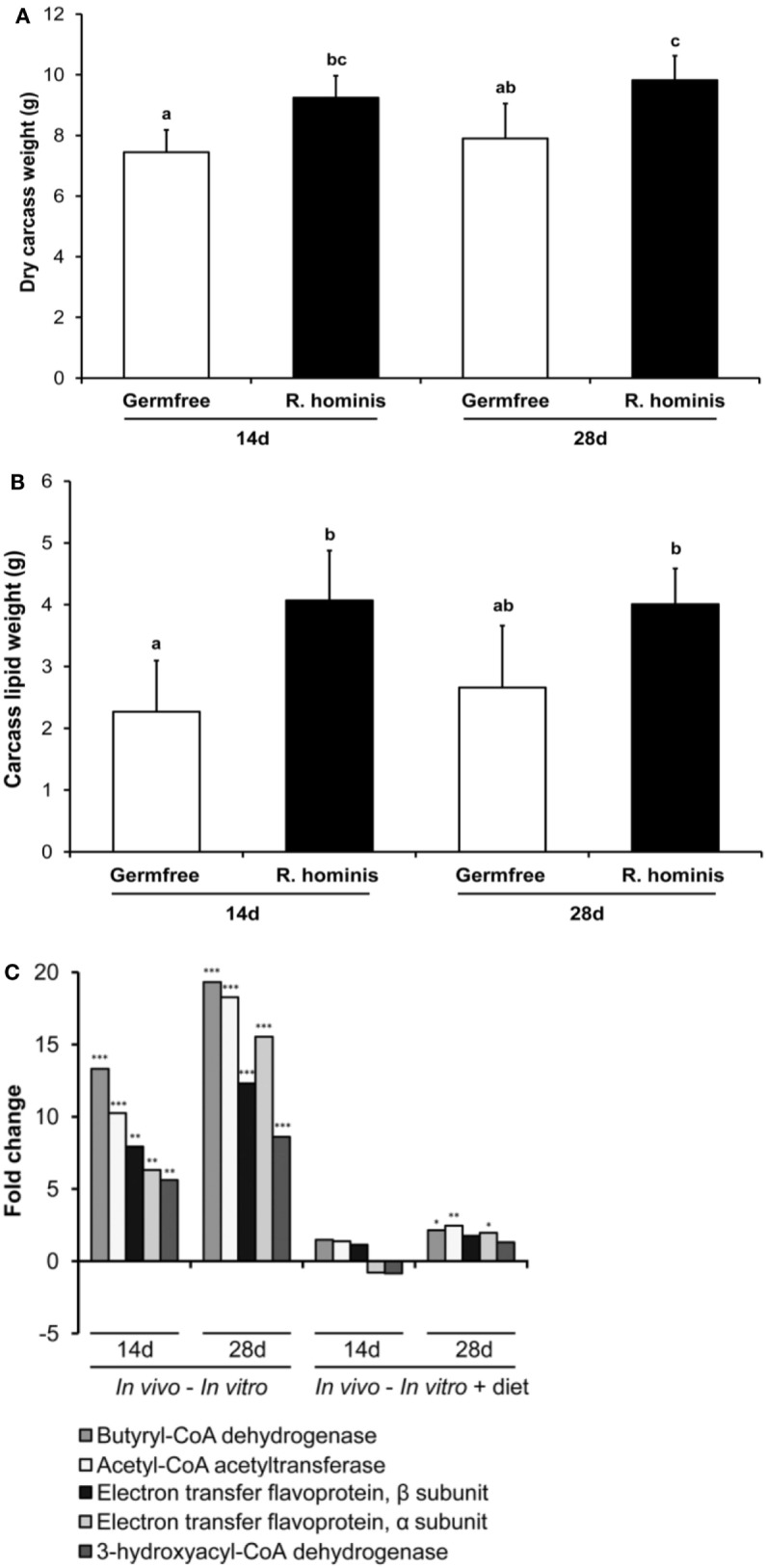
Roseburia hominis colonization influences satiety genes and body weight. The germ-free (GF) C3H/HeN male mice were allocated into the control (N = 8) and treatment (N = 10) groups. At days 0, 1, and 2, animals in the treatment group were dosed by ~109 CFU of R. hominis in YCFA medium (100 µL) by gavage, while control animals were given 100 µL of YCFA medium alone. Dry body weight and lipid content of carcasses were analyzed. (A) Weight of dry carcasses of R. hominis-associated mice was significantly heavier compared to GF animals. (B) Carcass lipid analyses showed that the total adiposity was also significantly higher in R. hominis-associated animals at day 14. Data are presented as mean ± SD. (C) RT-qPCR analysis of genes involved in butyrate metabolism.
Significant metabolic changes were also detected on the other side of the host–microbe interaction spectrum. In particular, expression of R. hominis genes involved in butyrate production was upregulated by the gut environment (Figure 7C). These were the genes encoding acetyl-CoA acetyltransferase, 3-hydroxyacyl-CoA dehydrogenase, butyryl-CoA dehydrogenase, and phosphoenolpyruvate carboxykinase. The stimulatory effect of host epithelial cells on production of acetate and butyrate by R. hominis was further confirmed in separate in vitro experiments (Figure S4 in Supplementary Material). Bacterial cells incubated with Caco-2 and HT29 cells produced significantly higher amounts of these metabolites compared to R. hominis control cells incubated under the same conditions but without the epithelial cells.
Discussion
A long-term co-evolution of host–microbe interaction has likely driven the selection of functionally important bacterial species in the gut, the majority of which are not encountered in other ecosystems. There is only limited information available at present about the contribution of individual members of the microbial community to intestinal functions, in particular to the development and regulation of mucosal immunity.
The specific functions of certain intestinal bacteria such as B. fragilis and SFB have been investigated in the mouse gut to define their individual contributions to T cell biology, and these bacteria have been shown to be potent inducers of Tregs and Th17 cells (4, 7, 8). Members of the Clostridium clusters IV and XIVa have recently received more attention. For instance, their presence in the altered Schaedler flora and the contribution of a mixed culture of Clostridium spp. to T cell differentiation has also been noted (9, 10).
We report here the successful and stable mono-association of R. hominis, a strictly anaerobic flagellated member of the Firmicutes phylum, in the gut of GF mice. The transcriptional responses of R. hominis following mono-colonization could be attributed to both the host gut environment and diet. These included chemotaxis and motility subsystems. The role of motility and the flagellar apparatus in host colonization is well understood for pathogenic bacteria but less is known about the role of flagellins in commensal bacteria. Genes encoding cell motility functions, and flagellins in particular, are generally expressed at variable and low abundance rates in the gut microbiome (27). In addition, only a subset of human gut microbiomes contains detectable flagellin genes (28). We showed that R. hominis flagellins are expressed in the mouse intestinal environment. Flagellin gene expression appears to be partly dependent on the presence of certain dietary constituents. Previous work has shown that flagellin gene expression of the related species Roseburia inulinivorans is indeed substrate dependent; its expression is higher in the presence of starch compared to glucose, inulin, or fructan (29).
The presence of R. hominis in the gut induces genes involved in promoting gut barrier function and innate immunity. Tight junctions, gap junctions, and adherens junctions operate to limit bacterial translocation to the subepithelial layer of the gut, and these functions may be promoted by intestinal bacteria (30). Both CD and UC are characterized by the loss of barrier function and the integrity of tight junctions. Interestingly, dysbiosis of the gut microbiota in UC and CD is associated with a significant reduction of R. hominis and F. prausnitzii (16, 19). Our results concerning the active contribution of R. hominis to gut barrier function support the view that its loss in patients with CD or UC may have significant consequences. Tight junction complexes can be activated by a number of other commensal and probiotic bacteria (31), potentially contributing to the amelioration of the “leaky gut” condition in these diseases.
Roseburia hominis induced the expression of genes such as Ly6g6c, as well as pathways involved in the regulation of T cells. The most affected T cell pathways included those related to IL-10, ICOS, and CTLA-4, which are all involved in the differentiation of Treg cells. Significant increases in colonic CD3+CD4+CD25+FoxP3+ cells were observed in mice either mono-colonized with R. hominis or in conventional mice fed a supplement of live R. hominis culture. Our findings complement recent reports on Treg differentiation effects by other Clostridium species (10, 32). We have shown here that the single bacterial strain R. hominis A2-183 is able to promote mucosal T cell expansion and impact T cell differentiation in mono-associated, conventional and DSS-treated mice.
Members of the Roseburia genus are some of the most prevalent motile bacterial species in the normal healthy human intestinal microbiota (33). Flagellins are potent immunomodulatory proteins: flagellated bacteria, such as R. hominis, could interact actively with the host immune system. Another important consequence of motility among the Roseburia species is their ability to penetrate the mucus layer and attach to the host gut epithelial cell surfaces (34). This property is considered to be a very desirable characteristic enhancing the probiotic potential of bacteria (35). Close proximity to the host cells allows commensal gut bacteria to exert potent physiological effects, potentially reversing metabolic disorders and controlling inflammation, gut barrier function, and gut peptide secretion.
Flagellin signaling by many pathogenic bacteria through the host’s TLR5 receptors induces a strong pro-inflammatory response, mainly driven by the activation of the NF-κB signaling pathway (36). However, the numerical prevalence of flagellated commensal bacteria makes a strong pro-inflammatory scenario seem unlikely. These bacteria are capable of flagellin/TLR5 signaling, which is important for host defense and disease protection, because deletion of TLR5 results in colitis (37). The presence of flagellin genes in the SFB genomes (38, 39) may explain the potent induction of Th17 cells by these bacteria (7, 8).
Differential expression of R. hominis flagellins in vitro has effects on IECs and BMDCs. The panel of flagellins tested affected IECs and BMDCs differently, although all the flagellin structures included the conserved Arg90 domain associated with flagellins that bind and activate TLR5 (40), which suggests that other sequence/structural properties might account for the signaling responses mediated by FlaA1 and FlaA2. Certain commensal flagellin structures may help to direct immune tolerance responses through TLR5 expressed on either CD103+DC or Treg subsets (23, 41, 42). The significance of flagellin-TLR5 signaling in Treg responses induced by R. hominis was further investigated using TLR5KO mice. We showed that R. hominis-induced CD3+CD4+CD25+FoxP3+ cells were significantly lower in TLR5KO mice, indicating that TLR5/flagellin signaling is an important mediator for the expansion of this T cell subset. The immunomodulatory effects of R. hominis were shown in DSS-treated mice. Although the DSS model of colitis is generally a T cell-independent model, we showed that R. hominis induced regulatory T cell populations in these animals. Other anti-inflammatory molecules such as butyrate may also contribute to immune tolerance and limit tissue damage (43, 44). Another effect of short-chain fatty acids produced by the bacterium is the improved food conversion efficiency. The dietary constituents such as polysaccharides that are poorly utilized by the host are degraded by the bacterium and used by the host resulting in a better energy recovery from the diet. Differential expression of satiety genes may be another factor contributing to the heavier weight and higher adiposity in R. hominis-colonized animals.
In summary, gut colonization with R. hominis caused the induction of specific subsets of genes on both the bacterial and host sides of the interaction. In this study, we have focused on signaling amongst other molecular mechanisms to explain the nature of this cross talk. TLR5/flagellin signaling could, potentially, drive the expansion of Treg cells via a TLR5-dependent mechanism, and our results suggest a potential therapeutic benefit of R. hominis in the treatment of UC, which is characterized by the loss of this bacterium from the gut (16).
Ethics Statement
The management and experimental procedures with animals were approved by the respective Local Ethical Review Committees at Institut Micalis, INRA, Jouy-en-Josas, France and Medical Research Facility, University of Aberdeen, United Kingdom.
Author Contributions
AP, IM, and RA designed research; AP, IM, AT, AL, NC-B, VG-R, KG, EL, MD, AC, EM, VF, RI, GG, and RA performed research; AP, IM, AL, GG, and RA analyzed data; AP, IM, GG, and RA wrote the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors would like to thank Andrew Gewirtz (Georgia State University, Atlanta, GA, USA) and Adam Cunningham (University of Birmingham, Edgbaston, Birmingham, UK) for generously providing the germ-free and conventional TLR5KO mice, respectively. The authors also wish to acknowledge a significant contribution to the inception of the work, experimental work, interpretation of results, and writing of the manuscript by Denise Kelly, as well as the support from RESAS (Rural and Environmental Science and Analytical Services) of the Scottish Government. EM and VF were supported by the Marie Curie Initial training network Fellowships funded by the EU (grant #215532). The authors acknowledge a generous support of Prof. Harry Flint and Dr. Sylvia Duncan in the microbiological part of this work. The skilled technical support of Gillian Campbell and Pauline Young at RINH Genomics is also gratefully acknowledged.
Footnotes
Supplementary Material
The Supplementary Material for this article can be found online at http://journal.frontiersin.org/article/10.3389/fimmu.2017.01166/full#supplementary-material.
References
- 1.Eckburg PB, Bik EM, Bernstein CN, Purdom E, Dethlefsen L, Sargent M, et al. Diversity of the human intestinal microbial flora. Science (2005) 308(5728):1635–8. 10.1126/science.1110591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Macpherson AJ, Hunziker L, McCoy K, Lamarre A. IgA responses in the intestinal mucosa against pathogenic and non-pathogenic microorganisms. Microbes Infect (2001) 3(12):1021–35. 10.1016/S1286-4579(01)01460-5 [DOI] [PubMed] [Google Scholar]
- 3.Macpherson AJ, Martinic MM, Harris N. The functions of mucosal T cells in containing the indigenous commensal flora of the intestine. Cell Mol Life Sci (2002) 59(12):2088–96. 10.1007/s000180200009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mazmanian SK, Liu CH, Tzianabos AO, Kasper DL. An immunomodulatory molecule of symbiotic bacteria directs maturation of the host immune system. Cell (2005) 122(1):107–18. 10.1016/j.cell.2005.05.007 [DOI] [PubMed] [Google Scholar]
- 5.Macpherson AJ. IgA adaptation to the presence of commensal bacteria in the intestine. Curr Top Microbiol Immunol (2006) 308:117–36. 10.1007/3-540-30657-9_5 [DOI] [PubMed] [Google Scholar]
- 6.Chung H, Kasper DL. Microbiota-stimulated immune mechanisms to maintain gut homeostasis. Curr Opin Immunol (2010) 22(4):455–60. 10.1016/j.coi.2010.06.008 [DOI] [PubMed] [Google Scholar]
- 7.Gaboriau-Routhiau V, Rakotobe S, Lecuyer E, Mulder I, Lan A, Bridonneau C, et al. The key role of segmented filamentous bacteria in the coordinated maturation of gut helper T cell responses. Immunity (2009) 31(4):677–89. 10.1016/j.immuni.2009.08.020 [DOI] [PubMed] [Google Scholar]
- 8.Ivanov II, Atarashi K, Manel N, Brodie EL, Shima T, Karaoz U, et al. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell (2009) 139(3):485–98. 10.1016/j.cell.2009.09.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Geuking MB, Cahenzli J, Lawson MA, Ng DC, Slack E, Hapfelmeier S, et al. Intestinal bacterial colonization induces mutualistic regulatory T cell responses. Immunity (2011) 34(5):794–806. 10.1016/j.immuni.2011.03.021 [DOI] [PubMed] [Google Scholar]
- 10.Atarashi K, Tanoue T, Shima T, Imaoka A, Kuwahara T, Momose Y, et al. Induction of colonic regulatory T cells by indigenous Clostridium species. Science (2011) 331(6015):337–41. 10.1126/science.1198469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nutsch KM, Hsieh CS. T cell tolerance and immunity to commensal bacteria. Curr Opin Immunol (2012) 24(4):385–91. 10.1016/j.coi.2012.04.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Round JL, Lee SM, Li J, Tran G, Jabri B, Chatila TA, et al. The toll-like receptor 2 pathway establishes colonization by a commensal of the human microbiota. Science (2011) 332(6032):974–7. 10.1126/science.1206095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Frank DN, St Amand AL, Feldman RA, Boedeker EC, Harpaz N, Pace NR. Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc Natl Acad Sci U S A (2007) 104(34):13780–5. 10.1073/pnas.0706625104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Scanlan PD, Shanahan F, Marchesi JR. Culture-independent analysis of desulfovibrios in the human distal colon of healthy, colorectal cancer and polypectomized individuals. FEMS Microbiol Ecol (2009) 69(2):213–21. 10.1111/j.1574-6941.2009.00709.x [DOI] [PubMed] [Google Scholar]
- 15.Kang S, Denman SE, Morrison M, Yu Z, Dore J, Leclerc M, et al. Dysbiosis of fecal microbiota in Crohn’s disease patients as revealed by a custom phylogenetic microarray. Inflamm Bowel Dis (2010) 16(12):2034–42. 10.1002/ibd.21319 [DOI] [PubMed] [Google Scholar]
- 16.Machiels K, Joossens M, Sabino J, De Preter V, Arijs I, Eeckhaut V, et al. A decrease of the butyrate-producing species Roseburia hominis and Faecalibacterium prausnitzii defines dysbiosis in patients with ulcerative colitis. Gut (2014) 63(8):1275–83. 10.1136/gutjnl-2013-304833 [DOI] [PubMed] [Google Scholar]
- 17.Gevers D, Kugathasan S, Denson LA, Vazquez-Baeza Y, Van Treuren W, Ren B, et al. The treatment-naive microbiome in new-onset Crohn’s disease. Cell Host Microbe (2014) 15(3):382–92. 10.1016/j.chom.2014.02.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hedin CR, McCarthy NE, Louis P, Farquharson FM, McCartney S, Taylor K, et al. Altered intestinal microbiota and blood T cell phenotype are shared by patients with Crohn’s disease and their unaffected siblings. Gut (2014) 63(10):1578–86. 10.1136/gutjnl-2013-306226 [DOI] [PubMed] [Google Scholar]
- 19.Tilg H, Danese S. Roseburia hominis: a novel guilty player in ulcerative colitis pathogenesis? Gut (2014) 63(8):1204–5. 10.1136/gutjnl-2013-305799 [DOI] [PubMed] [Google Scholar]
- 20.Sokol H, Pigneur B, Watterlot L, Lakhdari O, Bermudez-Humaran LG, Gratadoux JJ, et al. Faecalibacterium prausnitzii is an anti-inflammatory commensal bacterium identified by gut microbiota analysis of Crohn’s disease patients. Proc Natl Acad Sci U S A (2008) 105(43):16731–6. 10.1073/pnas.0804812105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Travis AJ, Kelly D, Flint HJ, Aminov RI. Complete genome sequence of the human gut symbiont Roseburia hominis. Genome Announc (2015) 3(6):e1286–1215. 10.1128/genomeA.01286-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Duncan SH, Aminov RI, Scott KP, Louis P, Stanton TB, Flint HJ. Proposal of Roseburia faecis sp. nov., Roseburia hominis sp. nov. and Roseburia inulinivorans sp. nov., based on isolates from human faeces. Int J Syst Evol Microbiol (2006) 56(Pt 10):2437–41. 10.1099/ijs.0.64098-0 [DOI] [PubMed] [Google Scholar]
- 23.Flores-Langarica A, Marshall JL, Hitchcock J, Cook C, Jobanputra J, Bobat S, et al. Systemic flagellin immunization stimulates mucosal CD103+ dendritic cells and drives Foxp3+ regulatory T cell and IgA responses in the mesenteric lymph node. J Immunol (2012) 189(12):5745–54. 10.4049/jimmunol.1202283 [DOI] [PubMed] [Google Scholar]
- 24.Zhan T, Cao C, Li L, Gu N, Civin CI, Zhan X. MIM regulates the trafficking of bone marrow cells via modulating surface expression of CXCR4. Leukemia (2016) 30(6):1327–34. 10.1038/leu.2016.39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Berg DJ, Davidson N, Kuhn R, Muller W, Menon S, Holland G, et al. Enterocolitis and colon cancer in interleukin-10-deficient mice are associated with aberrant cytokine production and CD4(+) TH1-like responses. J Clin Invest (1996) 98(4):1010–20. 10.1172/JCI118861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Monteleone I, Platt AM, Jaensson E, Agace WW, Mowat AM. IL-10-dependent partial refractoriness to toll-like receptor stimulation modulates gut mucosal dendritic cell function. Eur J Immunol (2008) 38(6):1533–47. 10.1002/eji.200737909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Neville BA, Sheridan PO, Harris HM, Coughlan S, Flint HJ, Duncan SH, et al. Pro-inflammatory flagellin proteins of prevalent motile commensal bacteria are variably abundant in the intestinal microbiome of elderly humans. PLoS One (2013) 8(7):e68919. 10.1371/journal.pone.0068919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Qin J, Li R, Raes J, Arumugam M, Burgdorf KS, Manichanh C, et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature (2010) 464(7285):59–65. 10.1038/nature08821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Scott KP, Martin JC, Chassard C, Clerget M, Potrykus J, Campbell G, et al. Substrate-driven gene expression in Roseburia inulinivorans: importance of inducible enzymes in the utilization of inulin and starch. Proc Natl Acad Sci U S A (2011) 108(Suppl 1):4672–9. 10.1073/pnas.1000091107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ukena SN, Singh A, Dringenberg U, Engelhardt R, Seidler U, Hansen W, et al. Probiotic Escherichia coli Nissle 1917 inhibits leaky gut by enhancing mucosal integrity. PLoS One (2007) 2(12):e1308. 10.1371/journal.pone.0001308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ulluwishewa D, Anderson RC, McNabb WC, Moughan PJ, Wells JM, Roy NC. Regulation of tight junction permeability by intestinal bacteria and dietary components. J Nutr (2011) 141(5):769–76. 10.3945/jn.110.135657 [DOI] [PubMed] [Google Scholar]
- 32.Narushima S, Sugiura Y, Oshima K, Atarashi K, Hattori M, Suematsu M, et al. Characterization of the 17 strains of regulatory T cell-inducing human-derived Clostridia. Gut Microbes (2014) 5(3):333–9. 10.4161/gmic.28572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Aminov RI, Walker AW, Duncan SH, Harmsen HJ, Welling GW, Flint HJ. Molecular diversity, cultivation, and improved detection by fluorescent in situ hybridization of a dominant group of human gut bacteria related to Roseburia spp. or Eubacterium rectale. Appl Environ Microbiol (2006) 72(9):6371–6. 10.1128/AEM.00701-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Van den Abbeele P, Belzer C, Goossens M, Kleerebezem M, De Vos WM, Thas O, et al. Butyrate-producing Clostridium cluster XIVa species specifically colonize mucins in an in vitro gut model. ISME J (2013) 7(5):949–61. 10.1038/ismej.2012.158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Juge N. Microbial adhesins to gastrointestinal mucus. Trends Microbiol (2012) 20:30–9. 10.1016/j.tim.2011.10.001 [DOI] [PubMed] [Google Scholar]
- 36.Choi YJ, Im E, Chung HK, Pothoulakis C, Rhee SH. TRIF mediates toll-like receptor 5-induced signaling in intestinal epithelial cells. J Biol Chem (2010) 285(48):37570–8. 10.1074/jbc.M110.158394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Singh V, Yeoh BS, Carvalho F, Gewirtz AT, Vijay-Kumar M. Proneness of TLR5-deficient mice to develop colitis is microbiota dependent. Gut Microbes (2015) 6(4):279–83. 10.1080/19490976.2015.1060390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Prakash T, Oshima K, Morita H, Fukuda S, Imaoka A, Kumar N, et al. Complete genome sequences of rat and mouse segmented filamentous bacteria, a potent inducer of Th17 cell differentiation. Cell Host Microbe (2011) 10(3):273–84. 10.1016/j.chom.2011.08.007 [DOI] [PubMed] [Google Scholar]
- 39.Sczesnak A, Segata N, Qin X, Gevers D, Petrosino JF, Huttenhower C, et al. The genome of Th17 cell-inducing segmented filamentous bacteria reveals extensive auxotrophy and adaptations to the intestinal environment. Cell Host Microbe (2011) 10(3):260–72. 10.1016/j.chom.2011.08.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yoon SI, Kurnasov O, Natarajan V, Hong M, Gudkov AV, Osterman AL, et al. Structural basis of TLR5-flagellin recognition and signaling. Science (2012) 335(6070):859–64. 10.1126/science.1215584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Crellin NK, Garcia RV, Hadisfar O, Allan SE, Steiner TS, Levings MK. Human CD4+ T cells express TLR5 and its ligand flagellin enhances the suppressive capacity and expression of FOXP3 in CD4+CD25+ T regulatory cells. J Immunol (2005) 175(12):8051–9. 10.4049/jimmunol.175.12.8051 [DOI] [PubMed] [Google Scholar]
- 42.Kinnebrew MA, Buffie CG, Diehl GE, Zenewicz LA, Leiner I, Hohl TM, et al. Interleukin 23 production by intestinal CD103(+)CD11b(+) dendritic cells in response to bacterial flagellin enhances mucosal innate immune defense. Immunity (2012) 36(2):276–87. 10.1016/j.immuni.2011.12.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Smith PM, Howitt MR, Panikov N, Michaud M, Gallini CA, Bohlooly-Y M, et al. The microbial metabolites, short-chain fatty acids, regulate colonic Treg cell homeostasis. Science (2013) 341(6145):569–73. 10.1126/science.1241165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Furusawa Y, Obata Y, Fukuda S, Endo TA, Nakato G, Takahashi D, et al. Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells. Nature (2013) 504(7480):446–50. 10.1038/nature12721 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.