Abstract
Topoisomerase 2 (TOP2) DNA transactions are essential for life, and proceed via formation of the TOP2 cleavage complex (TOP2cc), a covalent enzyme-DNA reaction intermediate that is vulnerable to trapping by potent anticancer TOP2 drugs. How genotoxic TOP2 DNA-protein crosslinks are resolved is unclear. Here, we show that the SUMO ligase ZATT (ZNF451) is a multifunctional DNA repair factor that controls cellular responses to TOP2 damage. ZATT binding to TOP2cc facilitates a proteasome-independent Tyrosyl-DNA phosphodiesterase 2 (TDP2) hydrolase activity on stalled TOP2cc. The ZATT SUMO ligase activity further promotes TDP2 interactions with SUMOylated TOP2, regulating efficient TDP2 recruitment through a "split-SIM" SUMO2 engagement platform. These findings uncover a ZATT–TDP2 catalyzed and SUMO2-modulated pathway for direct resolution of TOP2cc.
Topoisomerase 2 (TOP2α and TOP2β in mammalian cells) regulates DNA topology through the production of a transient DNA double strand breaks (1). The key intermediate in the TOP2 reaction is the covalent protein-DNA TOP2 cleavage complex (TOP2cc), a protein-DNA crosslink that forms between the active site TOP2 tyrosine and the 5′-terminus of the incised DNA duplex (2) (Fig. 1A). TOP2 is critical for facilitating DNA processes such as replication and transcription (3–5). However, it can be trapped on DNA by poisons, including front-line anti-cancer drugs such as etoposide, or by binding to existing DNA damage. Poisoning results in stable protein–blocked DNA breaks that are impediments to elongating RNA and DNA polymerases and cause cell death (1–4, 6, 7).
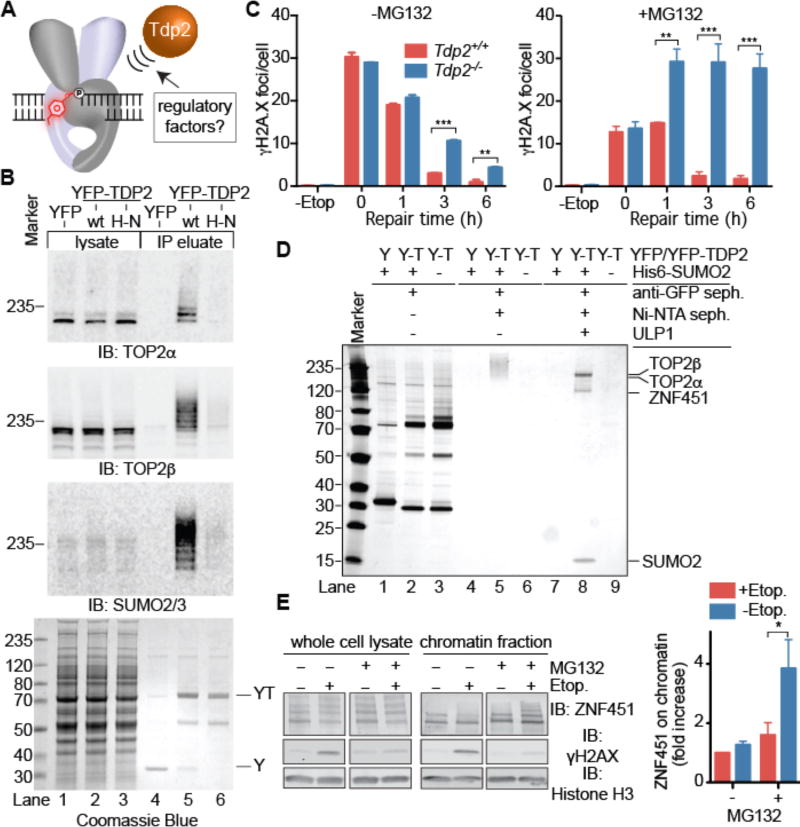
Figure 1. TDP2 binds SUMOylated TOP2 and ZNF451.
(A) Cellular mechanisms that regulate TDP2-catalyzed phosphotyrosyl bond hydrolysis in poisoned TOP2cc are unknown. (B) Immunoblotting of soluble cell lysates and IPs from cells expressing YFP, YFP-TDP2, or YFP-TDP2H351N (H–N). (C) Resolution of γH2AX foci in MEFs after etoposide exposure (20 µM). Average ±s.e.m.; n=3; ** p<0.01, *** p<0.001 (two-way ANOVA with Bonferroni post-test). (D) Silver-stained SDS-PAGE of YFP–TDP2 (Y-T)–associated SUMO2–modified proteins isolated as in fig. S2A. (E) Immunoblotting of ZNF451 (upper panel) in whole-cell extracts (WCE) or chromatin fraction. Quantification (right) of ZNF451 levels in chromatin. Average ± s.e.m.; N=3; *p<0.05 (two-way ANOVA with Bonferroni post-test).
Vertebrate Tyrosyl-DNA phosphodiesterase 2 (TDP2, also known as VpG unlinkase, TTRAP, EAPII) directly resolves the protein-DNA linkages (5′-phosphotyrosyl) characteristic of TOP2-induced DSBs (7–11). In this context, TDP2 modulates cellular (7, 10) and organismal (12) survival following TOP2 targeting anticancer drug treatments, and TDP2 inhibitors hold promise for chemotherapy (13, 14). A critical question in TOP2 biology is how TDP2 accesses the TOP2-DNA phosphotyrosyl chemical bond which is protected within the TOP2 protein shell (15, 16) (Fig. 1A). As etoposide treatment can trigger TOP2 degradation by the proteasome (17–19), it is hypothesized that TDP2 processes TOP2-DNA oligopeptides after TOP2cc proteolytic degradation (20). However, the molecular basis for regulation, coordination, and control of TOP2cc metabolism remains enigmatic.
To identify modulators of TDP2 dependent TOP2cc repair, we stably expressed YFP-tagged TDP2 in HEK293F cells, purified TDP2-containing protein complexes using anti-GFP/YFP single domain camelid nanobody (sdAb) resin, and identified co-purifying proteins by LC-MS/MS (fig. S1, A–C). We observed robust enrichment of TOP2α, TOP2β, and Small Ubiquitin-like Modifier 2 (SUMO2), but not SUMO1 peptides in YFPTDP2 immunoprecipitates (IP) (tables S1 and S2). Western blotting revealed that TDP2 interacts with a ladder of intact (non-proteolysed) TOP2α and TOP2β (Fig. 1B, lane 5) which is post-translationally modified with SUMO2 (fig. S1D, lane 4).
TOP2 is conjugated with SUMO2 during mitosis or in response to TOP2 poisons (21–23), and we find etoposide treatment prior to TDP2 IP increased the amount of high-molecular weight SUMO2 and the extent of modification of TOP2α and TOP2β (fig. S1E, lanes 8–12). Intriguingly, IP conducted with a catalytically inactive variant of TDP2H351N (8) was nearly devoid of SUMO2-TOP2 (Fig. 1B, lane 6), suggesting TDP2 catalysis is required to liberate intact SUMOylated TOP2 from the insoluble chromatin fraction. This prompted us to evaluate the untested link between the proteasome and TDP2 in repair of poisoned TOP2cc by monitoring the resolution of phospho-histone H2AX (γH2AX) foci following etoposide treatment. As reported previously, Tdp2 knockout (Tdp2−/−) mouse embryonic fibroblasts (MEFs) display delayed resolution of etoposide-induced γH2AX foci compared to Tdp2+/+cells (11), and this was severely impaired by proteasome inhibition with MG132 (Fig. 1C, S1F). These data are consistent with a TDP2 SUMO2-dependent TOP2cc resolution mechanism that acts independently of, or parallel to proteasome-mediated TOP2cc repair.
To identify factors that regulate SUMO2 and TDP2 dependent TOP2cc repair, we conducted tandem affinity purifications (TAP) from HEK293F cells expressing YFP-TDP2 and His6-tagged SUMO2 (Fig. 1D and S2A). TAP samples contained a ladder of SUMO2 modified proteins (Fig. 1D, lanes 5). ULP1 SUMO2 protease treatment of this ladder uncovered four proteins (Fig. 1D, lanes 8), TOP2α, TOP2β, SUMO2, and ZNF451 — a prototypical member of a recently identified class of SUMO2 E3/E4 ligases (fig. S2B) (24, 25). Similar to TDP2, IP samples of endogenous ZNF451 or GFP-ZNF451 are enriched with SUMO2, TOP2α, and TOP2β (tables S1 and S3, fig. S2C and S2D), suggesting that these proteins form a functional complex in cells. Furthermore, recombinant ZNF451 binds to TOP2α and TOP2β (fig. S2E). ZNF451 is recruited to the cellular chromatin fraction following etoposide treatment when the proteasome is inhibited (Fig. 1E), but not by ionizing radiation induced DSBs (fig. S2F). These data indicate ZNF451 directly binds to TOP2, and is recruited to chromatin following TOP2 poisoning.
TDP2 is unable to hydrolyze intact, recombinant Saccharomyces cerevisiae TOP2 DNA-protein crosslinks (ScTOP2cc) in vitro but the activity is enabled by heat denaturation of ScTOP2cc (20), suggesting that TOP2cc resolution could be regulated by proteasome-independent mechanisms. Given the direct binding of ZNF451 to TOP2 and TDP2 (figs. S2D and S3A), we hypothesized that ZNF451 could regulate the activity of TDP2 on TOP2cc. To test this, we generated reconstituted TOP2cc via reaction of TOP2α or TOP2β with a suicide oligonucleotide substrate (20) (Fig. 2A), and assayed for TDP2-dependent TOP2cc resolution (Fig. 2B and fig. S3B–D). We found mammalian TOP2cc is generally refractory to direct resolution by TDP2, except at high concentrations of TDP2 where we observed liberation of a small amount of 15nt DNA product diagnostic of TDP2-catalyzed hydrolysis of TOP2cc (Fig. 2B, lanes 7 and 8, and fig. S3B). Strikingly, TDP2 is >1000-fold more active on TOP2cc (α and β) in the presence of ZNF451, with nanomolar concentrations of ZNF451 and TDP2 being sufficient to catalyze TOP2cc hydrolysis (Fig. 2B and figS3, B–E). Neither ZNF451 alone nor a catalytically impaired TDP2H351N mutant supported this reaction (fig. S3F, lanes 4 and 6) indicating that ZNF451 stimulates TOP2cc hydrolysis catalyzed by TDP2. Probing of the TOP2cc structure suggests that ZNF451 alters the conformation of TOP2cc to facilitate the TDP2 direct resolution reaction (fig. S3G).
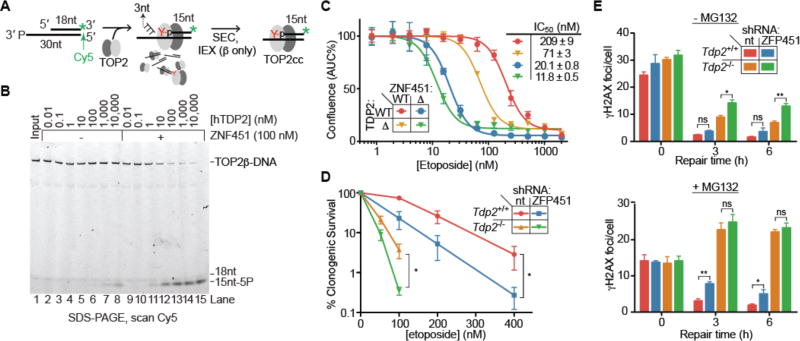
Figure 2. ZNF451 promotes phosphotyrosyl bond hydrolysis by TDP2.
(A) Synthesis and purification of stalled TOP2cc. (B) TOP2βcc (0.2 nM) hydrolysis by TDP2 is enhanced by ZNF451. Representative gel from 3 experiments. (C) Cellular proliferation of HEK293F or CRISPR knockout cells measured by area-under-the curve (AUC) of cell confluency after 6 days growth with etoposide. Average ± s.d., N=4. (D) Clonogenic survival of MEF cells in the indicated concentrations of etoposide. nt = non-targeting. Average ± s.e.m.; N≥4; *p≤0.05 (F-test, log values quadratic regression). (E) Resolution of DSBs marked by γH2AX foci in MEF cells post-etoposide exposure in the presence or absence of MG132. Average ±s.e.m.; N=3; ns p≥0.05, *p<0.05, ** p<0.01 (two-way ANOVA with Bonferroni post-test).
To assess the contribution of ZNF451 to TOP2cc repair in mammalian cells we examined cellular sensitivity of ΔZNF451, ΔTDP2, and ΔTDP2/ΔZNF451 double knockout HEK293F cell lines to etoposide (Fig. 2C and fig. S4, A and B). ZNF451 deletion conferred an even more severe etoposide sensitivity than TDP2 deletion (Fig. 2C). Similarly, shRNA-mediated ZNF451 knockdown sensitized HEK293F cells to etoposide, while GFP-ZNF451 overexpression decreased etoposide sensitivity, indicating that ZNF451 expression directly correlates with etoposide resistance in a dose dependent manner (fig. S4, C and D). This effect was specific to TOP2 drugs, as ZNF451 knockdown did not affect sensitivity to camptothecin, methyl methanesulfonate, or Zeocin (fig. S4D). TDP2 deletion further increased etoposide sensitivity in ΔZNF451, but to a lesser degree than in wild-type cells, suggesting both collaborative and unique functions of these proteins in the response to TOP2 damage.
Knockdown of the murine ZNF451 homolog (ZFP451) also decreased cell survival following etoposide treatment, in both wild-type and Tdp2−/− transformed MEFs (Fig. 2D and fig. S4E). Although ZFP451 depletion alone did not confer a significant defect in γH2AX foci resolution, it caused a delay in the repair kinetics when combined with Tdp2 deletion (Tdp2−/−) or proteasome inhibition (Fig. 2E). In line with protein-protein interaction results, ZFP451 and TDP2 appear to act in the same proteasome independent TOP2cc repair pathway, as ZFP451 depletion in Tdp2−/− cells did not further impair resolution of etoposide induced γH2AX foci in MG132 treated cells. These effects were not the result of global impairment of DSB repair by proteasome inhibition, as MG132 did not cause major defects in γH2AX foci resolution following ionizing radiation treatment (fig. S4F). Overall, our results identify ZNF451 as a component in the cellular response to TOP2-induced damage that operates through both TDP2-dependent and -independent mechanisms.
ZNF451 is a SUMO2 E3/E4 ligase (24, 25), so we examined TOP2 SUMOylation in vitro using reactions containing SUMO E1 (Sae2/Aos2), E2 (Ubc9), SUMO2, and full-length ZNF451. ZNF451 exhibited robust auto-SUMOylation and catalyzed poly-SUMOylation of recombinant TOP2α (Fig 3A, lanes 4–8). ZNF451 further displayed a marked preference for SUMOylating TOP2cc over TOP2 (Fig. 3B, and fig. S5, A–C), suggesting TOP2cc is a preferred and specific target for ZNF451 SUMO2 ligase activity. In HEK293F cells, steady-state levels of TOP2α (Fig. 3, C and D) and TOP2β (fig. S5, D and E) SUMOylation with SUMO2 are largely dependent on ZNF451 (Fig. 3c, lanes 7 and 8), as is etoposide induced stimulation of TOP2 SUMOylation (Fig. 3C, lanes 9–10). Intriguingly, ZNF451-dependent TOP2 SUMOylation is also triggered by treatment with ICRF- 193, a drug that induces TOP2 clamping on DNA (Fig. 3C, lanes 11,12). These results indicate that ZNF451 regulates TOP2 SUMOylation following treatment with drugs that perturb the TOP2 reaction cycle by distinct mechanisms.
Figure 3. ZNF451 preferentially SUMOylates TOP2cc and enhances TDP2 activity.
(A) Recombinant TOP2α was incubated with SUMO E1, E2, and increasing concentrations of SUMO2 in the presence or absence of ZNF451. (B) Reactions with 2 nM TOP2αcc (DNA-Cy5 label) and 75 nM free TOP2α were incubated with E1, E2, SUMO2, and the indicated concentration of ZNF451, then separated by SDS-PAGE. TOP2cc SUMOylation is monitored by scanning for the Cy5 label, while free TOP2β sumoylation is monitored by western blotting. (C) Immunoblot of lysates and Ni-NTA purified samples from cells expressing His6-SUMO2 pretreated with the indicated drugs for 20 mins. (D) Quantification of SUMO2-TOP2α signal from lanes 7–12 in panel C. N=3; Average ± s.d.; *=p<0.05, **=p<0.001 (2-tailed t-test). (E) Representative image (top) and quantification (bottom) of TOP2β or SUMO2 covalently bound to genomic DNA in MEFs following 1 h etoposide treatment and recovery in the presence or absence of 20 µM MG132. Average ± s.e.m.; N≥7; *p<0.05 (two-way ANOVA with Bonferroni post-test).
TOP2 SUMOylation by ZNF451 also increased the efficiency of ZNF451-TDP2 TOP2cc hydrolysis by an additional ~75 % (fig. S5F). Thus, we tested the role of SUMOylation in TDP2-catalyzed removal of TOP2cc with an In vivo Complexes of Enzyme (ICE) assay that monitors TOP2β and SUMO2-modified TOP2β DNA-protein crosslink removal from chromatin (11) (Fig.3E, Supplementary text, Fig. S6). Following TOP2 poisoning and etoposide removal, the turnover of the SUMO2 modified TOP2β fraction was specifically delayed in Tdp2−/− cells, but only when the proteasome was inhibited (Fig. 3E, fig. S6E). In vitro treatment of TOP2cc from ICE samples with recombinant wild type TDP2 also showed enhanced removal of the SUMOylated TOP2βcc fraction compared to total TOP2β (fig. S6F), consistent with a model where TOP2cc SUMOylation promotes TDP2– catalyzed TOP2cc direct resolution.
SUMOylation enhances protein-protein associations in the DNA damage response (26,27), and TDP2 binds SUMO2 (but not SUMO1) (28). Thus, covalent labeling of TOP2cc with SUMO2 may recruit TDP2 to poisoned TOP2cc. Maltose Binding Protein (MBP) pulldowns map the SUMO2 binding region of TDP2 to the catalytic domain (aa 108–362, TDP2cat) (fig. S7A), which lacks a canonical SUMO interaction motif (SIM) (28). To define the molecular basis for this non-canonical SUMO interaction we crystallized and determined X-ray structures of mouse TDP2cat as binary mTDP2cat-SUMO2 and ternary mTDP2cat-SUMO2-DNA complexes (Fig. 4A and fig. S7B, table S4). SUMO2 binds distal to the TDP2 catalytic center through five SUMO binding elements, SB1–SB5 (Fig. 4, A and B). We confirmed that this overall architecture of mTDP2cat-SUMO2 is consistent with that in solution using small-angle X-ray scattering (fig. S7, C–H). The core of the SUMO2-TDP2 interface is composed of a "split-SIM" structure, distinct from well-characterized SUMO-SIM interfaces. In a triangular configuration, two β–strands (β0 of SB1, and C-terminal β14 from SB5) engage the SUMO2 β–sheet (Fig. 4B). A prominent feature of this interface is the insertion of the TDP2 C-terminal Leu370 into a SUMO2 hydrophobic pocket, where the terminal carboxylate of Leu370 forms a salt bridge with SUMO2 Lys42 (Fig. 4, B and C). Loops SB2, SB3, and SB4 augment this split-SIM core, which comprises an interface (~800 Å2) that is larger than typical SUMO-SIM interfaces (500–600 Å2) (fig. S7, I and J). Accordingly, the dissociation constant (Kd) for the TDP2-SUMO2 interaction (fig. S7, K and L) is 880 nM, placing TDP2 amongst the stronger SUMObinding proteins (28).
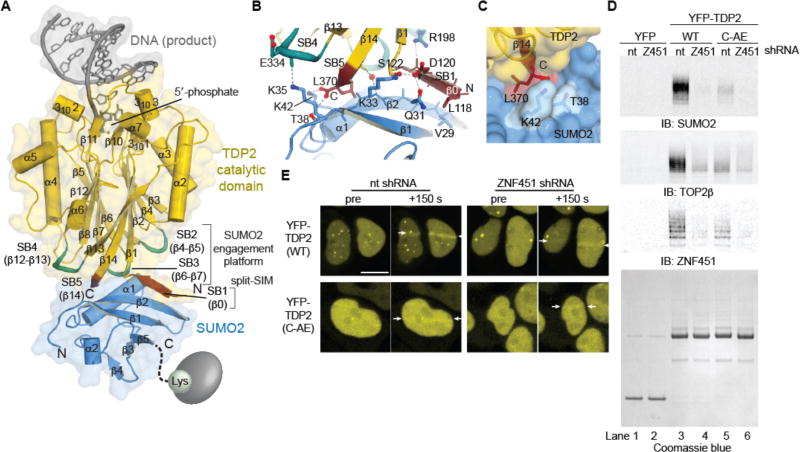
Figure 4. Structure of SUMO2–mTDP2cat reveals molecular basis for recruitment of TDP2 by SUMOylation.
(A) Structure of the DNA/mTDP2cat/SUMO2 complex. SUMO2 binds the TDP2 catalytic domain with a split– SIM (red) and three loops (turquoise) distal from the DNA binding site. (B) A “β–triangle” is formed by strands β0 and β14 of mTDP2cat and β2 of SUMO2. (C) The C–terminus of TDP2 fits in a pocket on SUMO2. (D) Immunoblotting of IPs from cells co-expressing YFP, YFP-TDP2, or YFP-TDP2 C-AE with shRNAs. (E) HEK293F cells expressing YFP-TDP2 (wt or C-AE mutant) and a non–targeting (nt) or ZNF451-targeting shRNA control were microirradiated with a UV laser between the 2 white arrows at t=0. Scale bar, 10 µm.
To probe functions of the TDP2-SUMO2 interaction we mutated β0 to encode proline substitutions (TDP2PQ), and/or extended the buried C-terminal Leu370 by two residues (TDP2C-AE) to create steric blocks to SUMO2 engagement (Figs. 4C and S7M). In HEK293 cells, YFP-TDP2 C-AE, PQ, or C-AE/PQ double mutants fail to colocalize with SUMO2 (fig. S8A). In vitro, TDP2C-AE blocked SUMO2 binding in size-exclusion chromatography and when monitored by NMR, yet did not impair TDP2 phosphotyrosylase activity (fig. S8, BE). YFP-TDP2 association with SUMO2-TOP2 was impaired by the C-AE SUMO-binding mutation, ZNF451 knockdown, or by a combination of these defects, demonstrating the TDP2-SUMO2 interface is important for engagement of SUMOylated TOP2 (Fig. 4D).
In comparison to strong phenotypes observed with ZNF451 and TDP2 depletion (Fig. 2, C and D), we found that overexpressed TDP2C-AE complements Tdp2−/− MEFs in clonogenic survival and γH2AX repair assays following etoposide treatment (fig. S8, F and G). We reasoned that SUMOylation of TOP2 by ZNF451 may thus act secondarily to it's enhancement of TOP2cc hydrolase activity, with SUMOylation acting to direct TDP2 to stalled cleavage complexes. We thus examined the kinetics of TDP2 recruitment to TOP2 DNA damage generated by UV microirradiation (10, 29, 30). YFP-TDP2 accumulates within 150 s following UV treatment (fig. S8H), and both ZNF451 knockdown and the C-AE mutation impair TDP2 mobilization to DNA damage (Figs. 4E and S8I). TDP2C-AE mutation also delayed, but did not ablate removal of the SUMOylated fraction of TOP2 from ICE purified TOP2cc (fig. S8J). Altogether, these results indicate that the TDP2 interaction with TOP2cc is enhanced by interactions between TDP2 and SUMO2, and dependent on ZNF451 SUMO2 ligase. Furthermore, TDP2 directly binds TOP2 and ZNF451 in vitro (figs. S3A and fig. S7A), so this array of interactions with SUMO2, TOP2 and ZNF451 likely facilitates recruitment of TDP2 to SUMO2-TOP2cc in cells.
The ZNF451-TDP2 modulated TOP2cc direct resolution pathway (Fig. S9) may contribute to tumor adaptation during chemotherapy with TOP2 poisons, and thus constitutes a new potential target for chemotherapeutic intervention. In addition to TDP2-related functions, ZNF451 displays TDP2-independent effects on the cellular response to TOP2 poisoning that will require further investigation. It will be important in future work to elucidate the mechanics of ZNF451-TDP2 influence on TOP2 and its potential for modulating TOP2 regulation of genome dynamics and transcription (4, 12, 31, 32). Given ZNF451 association with TDP2- mediated TOP2 repair, we propose renaming it to ZATT (Zinc finger protein Associated with TDP2 and TOP2) to appropriately reflect these cellular functions.
Supplementary Material
Acknowledgments
We thank NIEHS staff and core facilities: AM, JK, and LP (X-ray crystallography), NM and MW (Viral Vector), CM and BP (Protein Expression), AJ and JT (Fluorescence Microscopy Imaging Center), AA and KJ (Mass Spectrometry Research and Support Group), and MS and CB (Flow Cytometry), and NG (NIEHS). Funding was provided by US NIH Intramural Research Program grants 1Z01ES102765 to RSW, 1ZIAES050111- 26 to RL, and ZES102488-09 to JW, and grants from the Spanish and Andalusian Government (SAF2010-21017, SAF2013-47343-P, SAF2014-55532-R, CVI-7948, FEDER funds) to FCL and (BES-2015-071672) to AHR, the ERC (ERC-CoG-2014-647359) to FCL, and the University of Seville to JAL (PIF-2011). The APS SERCAT beamline is supported by the US DOE OBES, (W-31-109-Eng-38). The ALS is operated by LBNL on behalf of the DOE OBES and the IDAT program, supported by DOE OBER and NIH project MINOS (R01GM105404). CABIMER is supported by the Andalusian Government. Coordinates were deposited in the RCSB Protein Data Bank under 5TVP for TDP2–SUMO2–DNA and 5TVQ for TDP2–SUMO2.
Footnotes
Author contributions: Conceptualization, MS, FC, and RSW; Methodology, MS, JW, GM, FCL, and RSW; Investigation, MS, JL, AHR, LB, JW, AMC, GM; Writing – original draft, MS, FCL, RSW; Writing – Reviewing and editing, MS, JL, JW, GM, FCL, RSW; Funding Acquisition FCL, RL, and RSW; Supervision, RL, FCL, and RSW.
The authors declare no competing financial interests.
Materials and Methods
References
- 1.Pommier Y. Drugging topoisomerases: lessons and challenges. ACS Chem Biol. 2013;8:82–95. doi: 10.1021/cb300648v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ashour ME, Atteya R, El-Khamisy SF. Topoisomerase-mediated chromosomal break repair: an emerging player in many games. Nature reviews. Cancer. 2015;15:137–151. doi: 10.1038/nrc3892. [DOI] [PubMed] [Google Scholar]
- 3.Ju BG, et al. A topoisomerase IIbeta-mediated dsDNA break required for regulated transcription. Science. 2006;312:1798–1802. doi: 10.1126/science.1127196. [DOI] [PubMed] [Google Scholar]
- 4.Madabhushi R, et al. Activity-Induced DNA Breaks Govern the Expression of Neuronal Early-Response Genes. Cell. 2015;161:1592–1605. doi: 10.1016/j.cell.2015.05.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tammaro M, Barr P, Ricci B, Yan H. Replication-dependent and transcription-dependent mechanisms of DNA double-strand break induction by the topoisomerase 2-targeting drug etoposide. PLoS One. 2013;8:e79202. doi: 10.1371/journal.pone.0079202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Deweese JE, Osheroff N. The DNA cleavage reaction of topoisomerase II: wolf in sheep's clothing. Nucleic Acids Res. 2009;37:738–748. doi: 10.1093/nar/gkn937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schellenberg MJ, et al. Reversal of DNA damage induced Topoisomerase 2 DNA-protein crosslinks by Tdp2. Nucleic Acids Res. 2016;44:3829–3844. doi: 10.1093/nar/gkw228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schellenberg MJ, et al. Mechanism of repair of 5'-topoisomerase II-DNA adducts by mammalian tyrosyl-DNA phosphodiesterase 2. Nat Struct Mol Biol. 2012;19:1363–1371. doi: 10.1038/nsmb.2418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shi K, et al. Structural basis for recognition of 5'-phosphotyrosine adducts by Tdp2. Nat Struct Mol Biol. 2012;19:1372–1377. doi: 10.1038/nsmb.2423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cortes Ledesma F, El Khamisy SF, Zuma MC, Osborn K, Caldecott KW. A human 5'-tyrosyl DNA phosphodiesterase that repairs topoisomerase-mediated DNA damage. Nature. 2009;461:674–678. doi: 10.1038/nature08444. [DOI] [PubMed] [Google Scholar]
- 11.Alvarez-Quilon A, et al. ATM specifically mediates repair of double-strand breaks with blocked DNA ends. Nat Commun. 2014;5:3347. doi: 10.1038/ncomms4347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gomez-Herreros F, et al. TDP2 protects transcription from abortive topoisomerase activity and is required for normal neural function. Nat Genet. 2014;46:516–521. doi: 10.1038/ng.2929. [DOI] [PubMed] [Google Scholar]
- 13.Marchand C, et al. Deazaflavin Inhibitors of Tyrosyl-DNA Phosphodiesterase 2 (TDP2) Specific for the Human Enzyme and Active against Cellular TDP2. ACS Chem Biol. 2016;11:1925–1933. doi: 10.1021/acschembio.5b01047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hornyak P, et al. Mode of action of DNA-competitive small molecule inhibitors of tyrosyl DNA phosphodiesterase 2. Biochem J. 2016;473:1869–1879. doi: 10.1042/BCJ20160180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schmidt BH, Burgin AB, Deweese JE, Osheroff N, Berger JM. A novel and unified two-metal mechanism for DNA cleavage by type II and IA topoisomerases. Nature. 465:641–644. doi: 10.1038/nature08974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wu CC, et al. Structural basis of type II topoisomerase inhibition by the anticancer drug etoposide. Science. 2011;333:459–462. doi: 10.1126/science.1204117. [DOI] [PubMed] [Google Scholar]
- 17.Mao Y, Desai SD, Ting CY, Hwang J, Liu LF. 26 S proteasome-mediated degradation of topoisomerase II cleavable complexes. J Biol Chem. 2001;276:40652–40658. doi: 10.1074/jbc.M104009200. [DOI] [PubMed] [Google Scholar]
- 18.Zhang A, et al. A protease pathway for the repair of topoisomerase II-DNA covalent complexes. J Biol Chem. 2006;281:35997–36003. doi: 10.1074/jbc.M604149200. [DOI] [PubMed] [Google Scholar]
- 19.Ban Y, Ho CW, Lin RK, Lyu YL, Liu LF. Activation of a Novel Ubiquitin-Independent Proteasome Pathway when RNA Polymerase II Encounters a Protein Roadblock. Mol Cell Biol. 2013;33:4008–4016. doi: 10.1128/MCB.00403-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gao R, et al. Proteolytic Degradation of Topoisomerase II (Top2) Enables the Processing of Top2.DNA and Top2.RNA Covalent Complexes by Tyrosyl-DNA-Phosphodiesterase 2 (TDP2) J Biol Chem. 2014;289:17960–17969. doi: 10.1074/jbc.M114.565374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Azuma Y, Arnaoutov A, Dasso M. SUMO-2/3 regulates topoisomerase II in mitosis. J Cell Biol. 2003;163:477–487. doi: 10.1083/jcb.200304088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ryu H, Furuta M, Kirkpatrick D, Gygi SP, Azuma Y. PIASy-dependent SUMOylation regulates DNA topoisomerase IIalpha activity. J Cell Biol. 2010;191:783–794. doi: 10.1083/jcb.201004033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Agostinho M, et al. Conjugation of human topoisomerase 2 alpha with small ubiquitin-like modifiers 2/3 in response to topoisomerase inhibitors: cell cycle stage and chromosome domain specificity. Cancer Res. 2008;68:2409–2418. doi: 10.1158/0008-5472.CAN-07-2092. [DOI] [PubMed] [Google Scholar]
- 24.Cappadocia L, Pichler A, Lima CD. Structural basis for catalytic activation by the human ZNF451 SUMO E3 ligase. Nat Struct Mol Biol. 2015;22:968–975. doi: 10.1038/nsmb.3116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Eisenhardt N, et al. A new vertebrate SUMO enzyme family reveals insights into SUMO-chain assembly. Nat Struct Mol Biol. 2015;22:959–967. doi: 10.1038/nsmb.3114. [DOI] [PubMed] [Google Scholar]
- 26.Jackson SP, Durocher D. Regulation of DNA damage responses by ubiquitin and SUMO. Mol Cell. 2013;49:795–807. doi: 10.1016/j.molcel.2013.01.017. [DOI] [PubMed] [Google Scholar]
- 27.Gareau JR, Lima CD. The SUMO pathway: emerging mechanisms that shape specificity, conjugation and recognition. Nat Rev Mol Cell Biol. 2010;11:861–871. doi: 10.1038/nrm3011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hecker CM, Rabiller M, Haglund K, Bayer P, Dikic I. Specification of SUMO1- and SUMO2-interacting motifs. J Biol Chem. 2006;281:16117–16127. doi: 10.1074/jbc.M512757200. [DOI] [PubMed] [Google Scholar]
- 29.Hung F, Luo D, Sauve DM, Muller MT, Roberge M. Characterization of topoisomerase II-DNA interaction and identification of a DNA-binding domain by ultraviolet laser crosslinking. FEBS Lett. 1996;380:127–132. doi: 10.1016/0014-5793(96)00035-x. [DOI] [PubMed] [Google Scholar]
- 30.Mielke C, Christensen MO, Barthelmes HU, Boege F. Enhanced processing of UVA-irradiated DNA by human topoisomerase II in living cells. J Biol Chem. 2004;279:20559–20562. doi: 10.1074/jbc.C400032200. [DOI] [PubMed] [Google Scholar]
- 31.Canela A, et al. Genome Organization Drives Chromosome Fragility. Cell. 2017;170:507–521. e518. doi: 10.1016/j.cell.2017.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Karvonen U, Jaaskelainen T, Rytinki M, Kaikkonen S, Palvimo JJ. ZNF451 is a novel PML body-and SUMO-associated transcriptional coregulator. J Mol Biol. 2008;382:585–600. doi: 10.1016/j.jmb.2008.07.016. [DOI] [PubMed] [Google Scholar]
- 33.Salmon P, Trono D. Production and titration of lentiviral vectors. Curr Protoc Neurosci. 2006;Chapter 4(Unit 4):21. doi: 10.1002/0471142301.ns0421s37. [DOI] [PubMed] [Google Scholar]
- 34.Mandraju RK, Kannapiran P, Kondapi AK. Distinct roles of Topoisomerase II isoforms: DNA damage accelerating alpha, double strand break repair promoting beta. Arch Biochem Biophys. 2008;470:27–34. doi: 10.1016/j.abb.2007.10.017. [DOI] [PubMed] [Google Scholar]
- 35.Kirchhofer A, et al. Modulation of protein properties in living cells using nanobodies. Nat Struct Mol Biol. 2010;17:133–138. doi: 10.1038/nsmb.1727. [DOI] [PubMed] [Google Scholar]
- 36.Rothbauer U, et al. A versatile nanotrap for biochemical and functional studies with fluorescent fusion proteins. Mol Cell Proteomics. 2008;7:282–289. doi: 10.1074/mcp.M700342-MCP200. [DOI] [PubMed] [Google Scholar]
- 37.Hutchins JR, et al. Systematic analysis of human protein complexes identifies chromosome segregation proteins. Science. 2010;328:593–599. doi: 10.1126/science.1181348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Uuskula-Reimand L, et al. Topoisomerase II beta interacts with cohesin and CTCF at topological domain borders. Genome Biol. 2016;17:182. doi: 10.1186/s13059-016-1043-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xing M, et al. Interactome analysis identifies a new paralogue of XRCC4 in non-homologous end joining DNA repair pathway. Nat Commun. 2015;6:6233. doi: 10.1038/ncomms7233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ochi T, et al. DNA repair. PAXX, a paralog of XRCC4 and XLF, interacts with Ku to promote DNA double-strand break repair. Science. 2015;347:185–188. doi: 10.1126/science.1261971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nitiss JL, Soans E, Rogojina A, Seth A, Mishina M. Topoisomerase assays. Curr Protoc Pharmacol. 2012;Chapter 3(Unit 3):3. doi: 10.1002/0471141755.ph0303s57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Otwinowski Z, Minor W. Processing of X-ray Diffraction Data Collected in Oscillation Mode. Methods in Enzymology. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 43.McCoy AJ, et al. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Huang WC, Ko TP, Li SS, Wang AH. Crystal structures of the human SUMO-2 protein at 1.6 A and 1.2 A resolution: implication on the functional differences of SUMO proteins. Eur J Biochem. 2004;271:4114–4122. doi: 10.1111/j.1432-1033.2004.04349.x. [DOI] [PubMed] [Google Scholar]
- 45.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Adams PD, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta crystallographica. Section D, Biological crystallography. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Classen S, et al. Implementation and performance of SIBYLS: a dual endstation small-angle X-ray scattering and macromolecular crystallography beamline at the Advanced Light Source. J Appl Crystallogr. 2013;46:1–13. doi: 10.1107/S0021889812048698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dyer KN, et al. High-throughput SAXS for the characterization of biomolecules in solution: a practical approach. Methods Mol Biol. 2014;1091:245–258. doi: 10.1007/978-1-62703-691-7_18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schneidman-Duhovny D, Hammel M, Sali A. FoXS: a web server for rapid computation and fitting of SAXS profiles. Nucleic Acids Res. 2010;38:W540–544. doi: 10.1093/nar/gkq461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schneidman-Duhovny D, Hammel M, Tainer JA, Sali A. Accurate SAXS profile computation and its assessment by contrast variation experiments. Biophys J. 2013;105:962–974. doi: 10.1016/j.bpj.2013.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kitahara R, et al. Basic folded and low-populated locally disordered conformers of SUMO-2 characterized by NMR spectroscopy at varying pressures. Biochemistry. 2008;47:30–39. doi: 10.1021/bi7014458. [DOI] [PubMed] [Google Scholar]
- 52.Logan TM, Olejniczak ET, Xu RX, Fesik SW. A general method for assigning NMR spectra of denatured proteins using 3D HC(CO)NH-TOCSY triple resonance experiments. J Biomol NMR. 1993;3:225–231. doi: 10.1007/BF00178264. [DOI] [PubMed] [Google Scholar]
- 53.Delaglio F, et al. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 54.Williamson MP. Using chemical shift perturbation to characterise ligand binding. Prog Nucl Magn Reson Spectrosc. 2013;73:1–16. doi: 10.1016/j.pnmrs.2013.02.001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.