Abstract
Rationale
Duchenne muscular dystrophy (DMD) is a severe inherited form of muscular dystrophy caused by mutations in the reading frame of the dystrophin gene disrupting its protein expression. Dystrophic cardiomyopathy is a leading cause of death in DMD patients and currently no effective treatment exists to halt its progression. Recent advancement in genome editing technologies offers a promising therapeutic approach in restoring dystrophin protein expression. However, the impact of this approach on DMD cardiac function has yet to be evaluated. Therefore, we assessed the therapeutic efficacy of CRISPR (clustered regularly interspaced short palindromic repeats)-mediated genome editing on dystrophin expression and cardiac function in mdx/Utr+/− mice after a single systemic delivery of recombinant adeno-associated virus (AAV).
Objective
To examine the efficiency and physiological impact of CRISPR-mediated genome editing on cardiac dystrophin expression and function in dystrophic mice.
Methods and Results
Here we packaged SaCas9/gRNA constructs into an AAV vector and systemically delivered them to mdx/Utr+/− neonates. We showed that CRIPSR-mediated genome editing efficiently excised the mutant exon 23 in dystrophic mice and immunofluorescence data supported the restoration of dystrophin protein expression in dystrophic cardiac muscles to a level approaching 40%. Moreover, there was a noted restoration in the architecture of cardiac muscle fibers and a reduction in the extent of fibrosis in dystrophin deficient hearts. The contractility of cardiac papillary muscles was also restored in CRISPR-edited cardiac muscles compared to untreated controls. Furthermore, our targeted deep sequencing results confirmed that our AAV-CRISPR-Cas9 strategy was very efficient in deleting the ~23 kb of intervening genomic sequences.
Conclusions
This study provides evidence for using CRISPR-based genome editing as a potential therapeutic approach for restoring dystrophic cardiomyopathy structurally and functionally.
Keywords: AAV, cardiomyopathy, CRISPR, dystrophin, genome editing, muscular dystrophy, gene therapy, Duchenne muscular dystrophy cardiomyopathy
Subject Terms: Gene Therapy, Cardiomyopathy
INTRODUCTION
Duchenne muscular dystrophy (DMD) is the most common form of muscular dystrophy affecting approximately one in 3500 live male births1. DMD is caused by mutations in the dystrophin gene (DMD) that result in the absence of dystrophin2, a critical component of the dystrophin-glycoprotein complex (DGC) that integrates cytoskeleton with the extracellular matrix (ECM). As a consequence, progressive muscle degeneration occurs resulting in the development of cardiac dysfunction and respiratory muscle weakness that ultimately leads to premature death3. Notably, cardiomyopathy develops in >90% of DMD patients and represents the leading cause of death in DMD patients4, 5. Current treatments for DMD-associated cardiomyopathy include glucocorticoids in conjunction with either angiotensin-converting enzyme (ACE) inhibitors or beta-blockers; however, these treatments focus on preserving residual muscle strength and extending lifespan not the prevention of disease pathogenesis3.
Genome editing provides an exciting avenue for the treatment of severe forms of human disease such as DMD. In fact, recent work illustrates the potential power of in vivo CRISPR (Clustered regularly interspaced short palindromic repeats)-mediated genome editing for DMD treatment6–12. However, to date the impact of this approach on DMD cardiac muscle function has not been evaluated. We previously showed SpCas9/gRNA can restore dystrophin reading frame in skeletal muscle of mdx mice. SpCas9/gRNA constructs were packaged into an adenovirus and delivered locally and systemically demonstrating the efficiency of the SpCas9/gRNA in restoring the expression of dystrophin protein in mdx heart as detected by immunofluorescence and immunoblotting. However, adenovirus is highly antigenic, has limited translational ability and was only able to transduce a limited layer of the heart mainly pericardial which limits its therapeutic potential. Therefore, we attempted to package our SaCas9/gRNA constructs into an adeno-associated virus (AAV) serotype rh.74 and we delivered it to mdx/Utr+/− mice systemically via a retro-orbital approach. Our results not only showed restoration of the dystrophin protein expression but also demonstrated the restoration of the cardiac myofiber architecture and reduction of the fibrotic area within the heart was also observed. Furthermore, functional improvement of the contractility of papillary muscles was evident in the AAV-treated mice versus the untreated control mdx/Utr+/− mice.
METHODS
Detailed methods can be found in the Online Data Supplement.
RESULTS
Systemic adenovirus-Cas9/gRNAs delivery in mdx mice restores dystrophin expression in the pericardial muscles
The mdx mouse has been used extensively as a mouse model for DMD. It carries a point mutation at exon 23 resulting in the disruption of dystrophin expression in both skeletal and cardiac muscles13. We previously demonstrated that in-frame deletion of the genomic DNA covering exons 21, 22 and 23 restored functional dystrophin expression in skeletal muscles of mdx mice using intramuscular Ad-Cas9/gRNAs delivery6. Here, we attempted to deliver the Ad-Cas9/gRNAs into mdx neonates (postnatal day 1–3) and test the hypothesis that CRISPR-gene editing could restore cardiac dystrophin expression. The adenovirus carries a GFP cassette to illustrate transduction into the heart in mdx mice. Cardiac dystrophin expression was restored in the Ad-Cas9/gRNAs treated mdx mice, as evidenced by immunofluorescence, PCR and immunoblotting (Online Figures I and II). These studies demonstrate that CRISPR-mediated genome editing allows excision of the mutant exon in dystrophin-deficient mice and restoration of dystrophin expression in the heart muscle. However, consistent with previous report14, adenovirus has limited transduction efficiency to the heart and it only targets peripheral myocardium as shown in Online Figure III. Therefore, we subsequently tested systemic delivery of CRISPR/Cas9 using adeno-associated virus (AAV) in dystrophic mice.
Comparison of SpCas9 and SaCas9 for editing the dystrophin gene
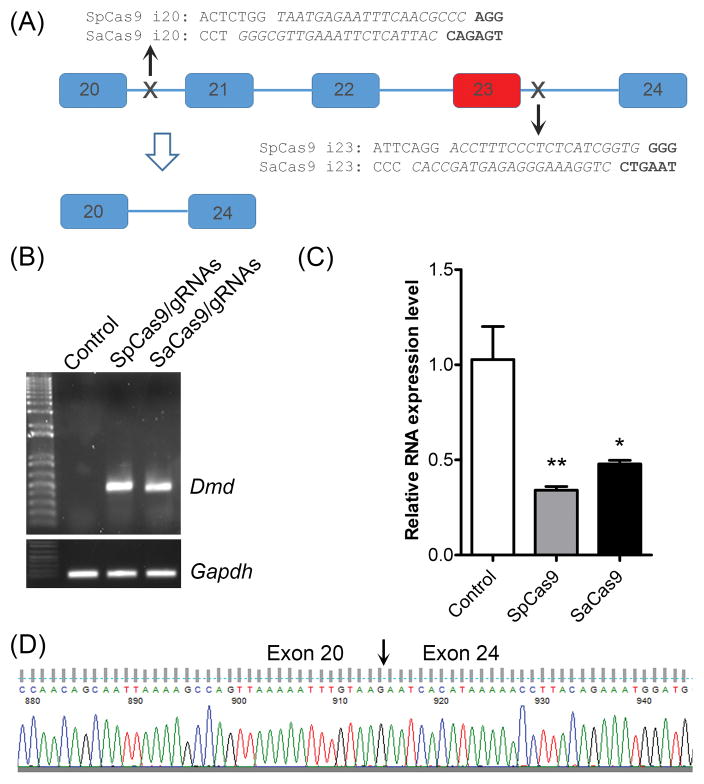
In the Ad-CRISPR system, we used Streptococcus pyogenes Cas9 (SpCas9), whose size is very close to the packaging limit of the AAV. Therefore, we sought to employ a smaller version of Cas9 from Staphylococcus aureus (SaCas9) for efficiently packaging into the AAV vector. For comparison, we designed the gRNAs for SaCas9 to target the same regions as for SpCas9 (Fig. 1A). When electroporated into C2C12 cells, both SpCas9/gRNAs and SaCas9/gRNAs induced efficient gene editing as evidenced by PCR analysis of the genomic DNA. A 500-bp PCR product as predicted could be amplified from both Cas9-treated samples, but not in the control cells (Fig. 1B). To quantitatively determine the relative efficiency of SpCas9 versus SaCas9, we designed a real-time RT-PCR primer pair in Exon 22, a region that should be deleted in correctly edited cells. Greater reduction in the PCR indicates higher editing efficiency. The transfected cells with SpCas9/gRNAs showed ~70% reduction of the WT transcript carrying Exon 22 (Fig. 1C). Although slightly less efficient than SpCas9/gRNAs, SaCas9/gRNAs transfection also resulted in over 50% reduction of the WT transcript (Fig. 1C). Moreover, DNA sequencing confirmed that the 500-bp transcript in Fig. 1B amplified from C2C12 cells treated with gRNA/SaCas9 were formed due to successful deletion of exons 21–23 (Fig. 1D). We then package the SaCas9/gRNAs into AAVrh74, a serotype shown to be nonpathogenic and mediate effective transgene expression in striated muscles15–18.
Figure 1. Comparison of SpCas9/gRNAs and SaCas9/gRNAs for deletion of exon 23 of the mouse Dmd gene in C2C12 cells.
(A) Diagram showing the genomic locus of mouse Dmd around exon 23. The gRNA targeting sites and sequences for both SpCas9 and SaCas9 were labeled. The mutant exon 23 is highlighted in red. (B) PCR analysis of genomic DNA extracted from C2C12 cells treated without or with SpCas9/gRNAs or SaCas9/gRNAs. (C) Quantitative RT-PCR analysis of the dystrophin expression in C2C12 cells treated without or with SpCas9/gRNAs or SaCas9/gRNAs. (D) DNA sequencing analysis of RT-PCR product of SaCas9-gRNA transduced C2C12 cells. *P<0.05 and **P<0.01. All data are representative of a minimum of three experiments.
Systemic delivery of AAV-SaCas9/gRNAs restores dystrophin protein expression in cardiac muscle
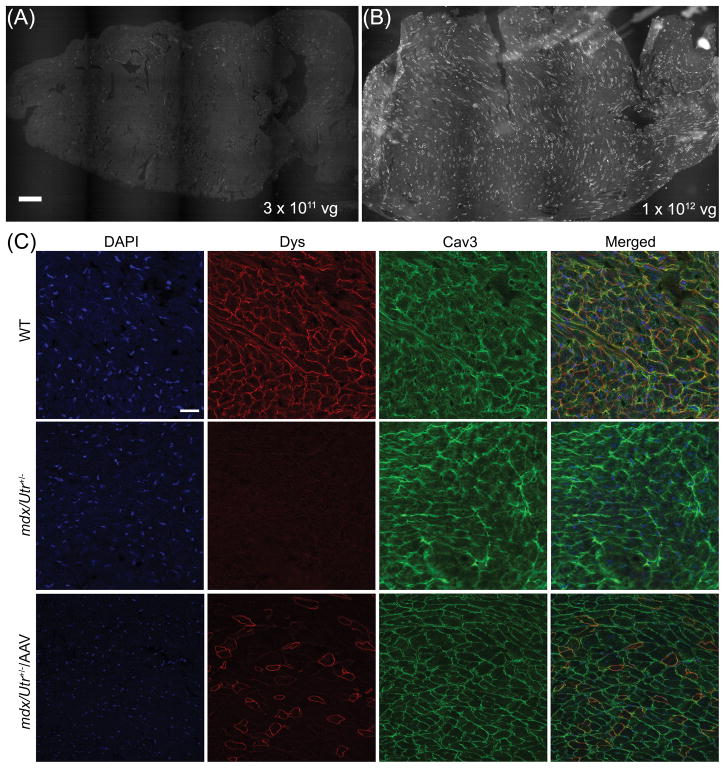
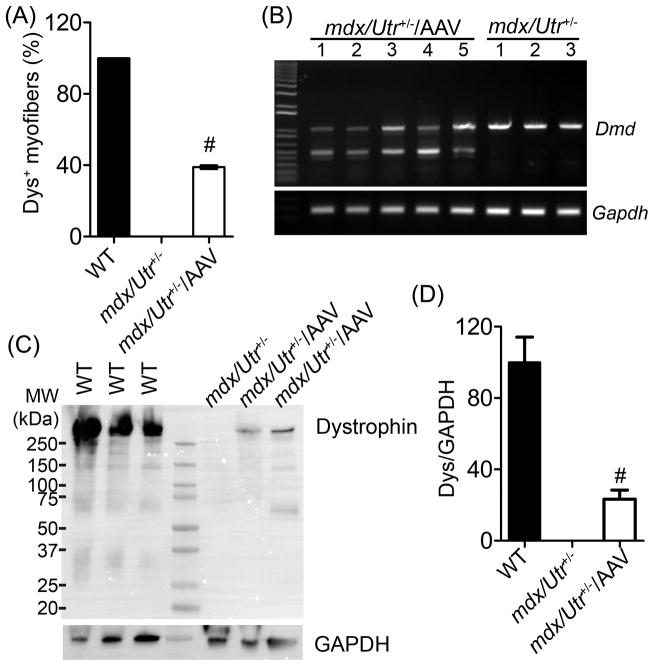
For the AAV study, we chose the mdx/Utr+/− mice because these animals exhibit a more severe phenotype and mimic human DMD patients better than the mdx mice due to the reduced compensation from the dystrophin homologue utrophin19. In particular, cardiomyopathy develops earlier in these mice and is much more severe, making the mdx/Utr+/− mice an excellent model to study potential cardiac therapeutics20. Systemic delivery of AAV-SaCas9/gRNAs (low dose, 3 × 1011 or high dose, 1 × 1012 vg/mouse) was performed in (day 3) mdx/Utr+/− neonates. At 10 weeks after treatment, the heart muscles of the animals were analyzed for dystrophin expression by immunofluorescence staining. The low dose treatment group showed scattered dystrophin-positive fibers (Fig. 2A) while the high dose treatment restored dystrophin expression widely in many cardiomyocytes across the entire heart section of mdx/Utr+/− mice (Fig. 2B). At a higher magnification, dystrophin was found to be correctly localized at the sarcolemma of the cardiomyocytes (Fig. 2C). For the high dose treatment group, we found that about 40% (39.0 ± 0.4 %, N=4) of cardiomyocytes were dystrophin positive (Fig. 3A).
Figure 2. Immunofluorescence staining of the heart sections from mdx/Utr+/− mice at 10 weeks following AAV-SaCas9/gRNAs treatment.
(A, B) Immunofluorescence images of dystrophin in the entire heart sections of mdx/Utr+/− mice treated with 3 × 1011 (A) or 1 × 1012 (B) vg AAV-SaCas9/gRNA systemically (N=4 per group). Scale bar: 500 μm. (C) Immunofluorescence images of dystrophin (red) and caveolin-3 (green) in the heart sections of WT and mdx/Utr+/− mice treated with or without 1 × 1012 vg AAV-SaCas9/gRNA systemically. Scale bar: 50 μm.
Figure 3. Restoration of dystrophin in the cardiomyocytes of mdx/Utr+/− mice.
(A) Measurement of dystrophin-positive cardiomyocytes (% of total caveolin-3-positive cardiomyocytes; mean ± SD) in mdx/Utr+/− heart cryosections treated with or without 1 × 1012 vg AAV-SaCas9/gRNAs (N=4 per group). (B) RT-PCR analysis of heart tissues from mdx/Utr+/− mice treated with (N=5) or without (N=3) 1 × 1012 vg AAV-SaCas9/gRNA. Genome editing is expected to produce a smaller (~500 bp) dystrophin band. (C) Immunoblotting of heart lysates from WT, mdx/Utr+/− and mdx/Utr+/− with AAV-SaCas9/gRNAs using anti-dystrophin and anti-GAPDH antibodies. (D) Quantitative analysis of dystrophin expression by immunoblotting of heart lysates from wild-type (WT), mdx/Utr+/− and AAV treated mdx/Utr+/− (N=3 per group). # p < 0.01.
RT-PCR analysis of the hearts showed that the edited smaller dystrophin transcript was expressed in all AAV-treated mdx/Utr+/− samples (Fig. 3B). Immunoblotting also confirmed the expression of dystrophin protein in the AAV-SaCas9/gRNAs treated mdx/Utr+/− hearts (Fig. 3C). Densitometry analysis of the dystrophin bands showed it was restored to 23.3 ± 5.1 % of the WT (wild-type) level (Fig. 3D).
To assess the non-homologous end joining (NHEJ) events occurred at the gRNA target sites, we performed targeted deep sequencing of PCR amplicons from both gRNA annealing sites (left, amplicon A; right, amplicon B; Online Figure IV and Sequences). In addition, we performed targeted deep sequencing of the amplicon spanning across the junction connecting the left and right sides. This amplicon represents the deletion of ~ 23 kb from chr. X due to CRISPR-Cas9-guided NHEJ in vivo (amplicon C). We used both Illumina (Online Figure IV) and Ion Torrent platforms (data not shown). Very few insertions, deletions or base substitutions were detected in amplicon A, and only slightly more were detected in amplicon B (Online Figure IV A&B). By contrast, most sequences from amplicon C showed that the end joining between targets A and B was almost always perfect, deleting ~ 23 kb of intervening genomic sequences, just as we had designed (Online Figure IV C&D). We found that 76% of all targeted sequences from amplicon C showed precise joining due to CRISPR-Cas9, and 86% of these sequences revealed no or only very short (< 5 nt) insertions or deletions at the desired recombinant junction site (Online Figure IV D). In addition, very low numbers of individual mutations at one target site or the other were detected alone. This suggested that the gRNAs at A and at B did not typically operate alone, but instead coordinated together in deleting the intervening genomic sequences. Very similar results were obtained using both the Illumina MiSeq and Ion Torrent platforms. These results confirmed that our CRISPR-Cas9 recombination strategy was very highly effective in deleting the ~23 kb of intervening genomic sequences.
Furthermore, to test if CRISPR-mediated genome editing could restore dystrophin expression in the adult heart of dystrophic mice, we administered AAV-SaCas9/gRNAs (1 × 1012 vg/mouse) into mdx/Utr+/− mice at the age of 16 weeks by tail vein injection. As shown in Online Figure V, measurable restoration of dystrophin protein was also detected using immunofluorescence seven days after the treatment.
Overall, this is highly encouraging that a single systemic treatment with AAV-SaCas9/gRNAs restored the expression in nearly half of the cardiomyocytes across the entire heart of dystrophin-deficient mice. Therefore, we further examined whether the observed levels of dystrophin restoration results in pathological and/or functional improvements in the cardiac muscles of mdx/Utr+/− mice.
Dystrophin restoration in AAV-CRISPR/SaCas9 treated mdx/Utr+/− mice resulted in decreased cardiac fibrosis and improved contractility of papillary muscles
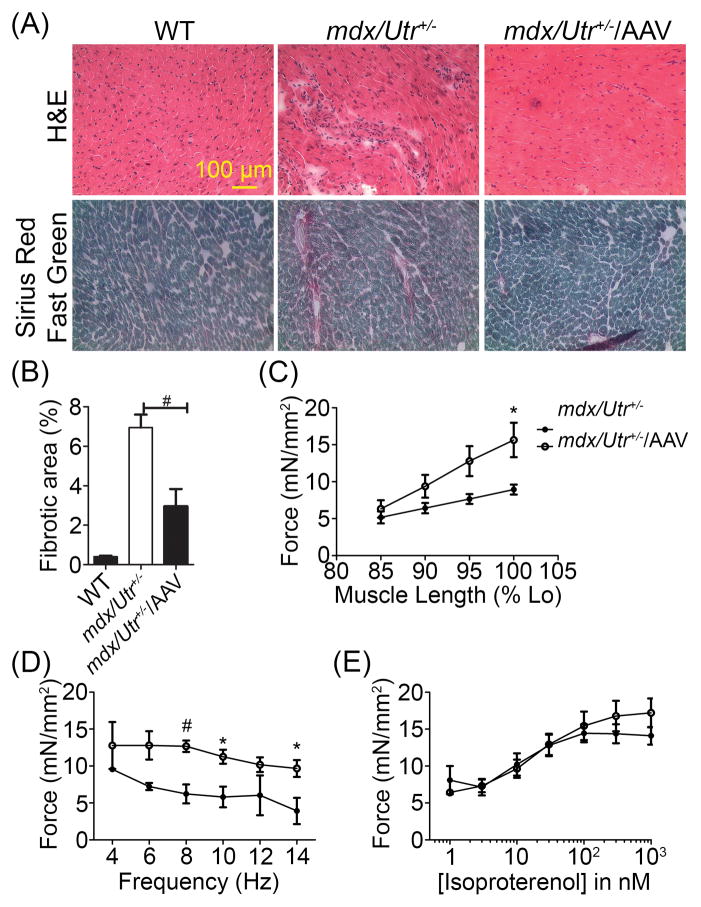
Histological analysis was performed to determine whether the restored cardiac dystrophin expression also resulted in reduced cardiac pathology. H&E staining illustrated cardiac muscle architecture and Sirius red/fast green staining illustrated the amount of fibrosis present in the heart. Reduced fibrosis was visually noticed in the AAV-SaCas9/gRNAs injected mice (Fig. 4A). By measuring the fibrotic area, we found that AAV-CRISPR/Cas9 treated mice had significantly reduced fibrotic area over the total heart cross section when compared with untreated controls (Fig. 4B).
Figure 4. Functional restoration of cardiac contractility after in vivo genome editing.
(A) Upper lane: H&E stained WT and mdx/Utr+/− heart cryosections with or without AAV-SaCas9/gRNAs. Lower lane: Sirius Red/Fast Green stained WT and mdx/Utr+/− heart cryosections with or without AAV-SaCas9/gRNAs (N≥3 per group). (B) Quantitative analysis of fibrotic area from Sirius Red/Fast Green stained mdx/Utr+/− heart cryosections with or without AAV-SaCas9/gRNAs (N≥3 per group). (C) Length-dependent activation of heart contractility evaluated by recording the contractile parameters at 85, 90, 95, and 100% of optimal length (N=5 for untreated mdx/Utr+/− and 6 for AAV-mdx/Utr+/−). (D) The force-frequency response measured at optimal length by adjusting the stimulation frequency from 4 to 6, 8, 10, 12 and 14 Hz (N=4 for untreated mdx/Utr+/− and 6 for AAV-mdx/Utr+/−). (E) β-adrenergic responsiveness in mdx/Utr+/− mice with or without SaCas9/gRNA-AAV (N=5 for untreated mdx/Utr+/− and 6 for AAV-mdx/Utr+/−). # p ≤ 0.01 and * p ≤ 0.05.
To determine whether the gene editing treatment also improves cardiac function, we studied the contractility of the isolated papillary muscle at 10 weeks21. Three main mechanisms of cardiac contractile regulation were assessed as previously described22, 23, including length dependent activation, frequency-dependent activation, and beta-adrenergic stimulation. The length-dependent activation was evaluated by recording the contractile parameters at 85, 90, 95, and 100% of optimal muscle length. A statistically significant increase in force at 100% optimal muscle length was observed in the mdx/Utr+/− AAV treated hearts compared to the untreated hearts (Fig. 4C). The force-frequency response was measured at optimal length by adjusting the stimulation frequency from 4 to 6, 8, 10, 12 and 14 Hz, with 2–3 minute stabilization time between each frequency. Results also showed a statistically significant increase in force at 8, 10 and 14 Hz in the AAV treated hearts vs. mdx/Utr+/− controls (Fig. 4D). Finally, after allowing the muscle to stabilize for 10 minutes at 4 Hz, the β-adrenergic responsiveness was assessed by measuring the contractile parameters with the addition of isoproterenol at semi-log steps between 1 nM and 1 μM. Data demonstrated no significant difference in the β-adrenergic responsiveness in the AAV treated group versus the untreated controls to the increase in isoproterenol concentration (Fig. 4E).
DISCUSSION
DMD is one of the most common genetic diseases and DMD patients lose their life prematurely due to respiratory or cardiac complications5, 24. Therefore, an effective therapy for DMD needs to address the complications in both the skeletal muscle and heart. Previous studies from others and us showed that CRISPR-based genome editing effectively restored dystrophin expression in the skeletal muscle of a postnatal mouse model of DMD6–12. In the present study, we presented data to demonstrate that AAV-mediated systemic delivery of CRISPR reagents leads to cardiac dystrophin expression and functional improvement, further highlighting the therapeutic promise of genome editing therapy for DMD.
Several therapeutic strategies have been explored to treat the primary cause of DMD by restoring dystrophin expression, such as mini-dystrophin cDNA, exon skipping and genome editing. Both mini-dystrophin cDNA and exon skipping have shown therapeutic potential in limiting DMD progression, whereas both strategies have their limitations. The mini-dystrophins are potentially less functional and more immunogenic due to the large deletions and/or altered protein structure25, while exon skipping showed only transient restoration of dystrophin26–28, requiring frequently repeated treatments. A more revolutionary approach was recently brought to the DMD therapeutic arena, which takes advantage of the CRISPR/Cas9 genome editing technology. CRISPR/Cas9 was originally discovered in bacteria as a defense mechanism whereby the Cas9 enzyme cleaves foreign nucleic acids29. The CRISPR/Cas9 system has been modified for genome editing as it can precisely cleave a specific target sequence based on the gRNA supplied29, 30. Previous work8 performed by Long et al. demonstrated that injecting Cas9, gRNA and a single-stranded oligodeoxynucleotide (ssODN) donor template into the one-cell embryo of mdx mice resulted in correction of the dystrophin mutation in the offspring through homologous recombination8. However, this technique has limited translational abilities. A subsequent study10 by Ousterout et al. in 2015 used CRISPR/Cas9 and gRNAs to correct specific mutations in myoblasts from DMD patients. More recently, our laboratory demonstrated for the first time that CRISPR/Cas9 could restore the expression of dystrophin protein and DGC components in postnatal mdx mice6. Functional improvements in the edited muscles were observed, as skeletal muscle was protected from damage during downhill treadmill running. Soon after, three other laboratories published their studies that employed AAV to deliver the CRISPR/Cas9 and gRNAs either systemically or intramuscularly into mdx mice7, 9, 11. Earlier this year, a study by Bengtsson et al.12 demonstrated a restoration of dystrophin up to 70% in the skeletal muscles with increased force generation following local muscular delivery. Moreover, systemic administration of their AAV vectors resulted in dystrophin expression in both skeletal and cardiac muscles. All these studies demonstrated some levels of dystrophin restoration in skeletal and cardiac muscles. However, functional assessment of the edited hearts has not been explored. Therefore, the present study is the first to demonstrate that a single AAV injection through a retro-orbital approach could restore dystrophin expression in nearly 40% cardiac muscle fibers across the entire heart and demonstrate a significant functional improvement in cardiac contractility in the mdx/Utr+/− mouse model.
The major advantage of the CRISPR-based genome editing therapy for DMD is that such a treatment would be permanent. However, there are also several concerns for moving this strategy forward into clinical trials. First, the most often used CRISPR/Cas9 systems (SpCas9 or SaCas9) are well known for their off-target activities. The off-target activities resulting from the non-targeted cleavage in vivo could be potentially dangerous, in particular with the long-lasting expression mediated by AAV delivery. In this regards, the recently reported high-fidelity Cas9 engineering is encouraging31, 32. Second, Cas9 protein may activate cellular immune response as a recent study reported that expression of SpCas9 in adult tibialis anterior muscle evoked host immune response regardless of delivery method33. Interestingly, this same study33 did not observe significant cytolytic activities resulting from the AAV-CRISPR/Cas9 delivery even though it activates the host immunity. Nevertheless, there is little concern of immune response in the present study as we injected the AAV into pups at the age of day 1–3 when the immune system has not been fully developed. Finally, the delivery technology is still the bottleneck and awaiting major breakthroughs as for all other gene therapy approaches. Currently, AAV is one of the most widely used delivery vehicles for gene therapy. However, the delivery efficiency, packaging capacity, tissue specificity, immune activating potential, temporal control, and dosage control are all of concern and needed to be taken into consideration for engineering more efficient delivery tools in the future.
In summary, our data showed the clear evidence of restoration of cardiac muscle function in genetically edited dystrophin deficient hearts of postnatal mice. In addition, our targeted deep sequencing results confirmed that our AAV-CRISPR-Cas9 strategy was very efficient in deleting the ~23 kb of intervening genomic sequences. Although future studies are required to deal with many associated issues such as off-target activities, immunological responses, delivery efficiency, delivery to adult mice, temporal control and tissue specificity, our current study together with previous studies suggest that in vivo gene editing holds a great promise as a “permanent” therapy for DMD.
Supplementary Material
NOVELTY AND SIGNIFICANCE.
What Is Known?
Duchenne muscular dystrophy (DMD) is a genetic disease with cardiac manifestations, caused by disrupted expression of dystrophin.
Currently, there is no existing treatment for this devastating disease, and patients are offered only supportive treatment.
What New Information Does This Article Contribute?
Dystrophin gene editing can restore protein expression in a substantial population of heart muscle cells in mdx/Utr+/− mice.
Genome editing-mediated dystrophin restoration resulted in functional improvement of the dystrophic heart.
DMD is the most common form of muscular dystrophy that affects 1:3500 live male births. As the disease progresses, life threatening cardiac complications can occur and most patients die in early adulthood. There is no current treatment for the disease and no cure to stop its progression to heart damage. Here we showed that genome editing efficiently corrected the genetic defect in a mouse model of DMD and restored dystrophin protein expression in the heart muscles to nearly 40%. There was also a noted improvement in the architecture of heart muscle fibers, pathological changes and contractile strength. Our study demonstrates that genome editing can restore dystrophin expression and heart function in dystrophic mice after a single treatment. This study holds great promise for the treatment of DMD patients.
Acknowledgments
We thank Yeh Siang Lau for technical assistance. We also thank the Viral Vector Core at the Nationwide Children’s hospital for their technical assistance in producing the AAV.
SOURCES OF FUNDING
R.H. is supported by US National Institutes of Health grants (R01HL116546 and R01 AR064241). M.E.R. is a recipient of the NIH T32 postdoctoral fellowship (T32HL0980391). The Genomics Shared Resource of the OSU Comprehensive Cancer Center is supported in part by NCI grant number P30CA016058.
Nonstandard Abbreviations and Acronyms
- AAV
Adeno-associated virus
- ACE
Angiotensin-converting enzyme
- Ad
Adenovirus
- CRISPR
Clustered regularly interspaced short palindromic repeats
- DGC
Dystrophin-glycoprotein complex
- DMD
Duchenne muscular dystrophy
- ECM
Extracellular matric
- Gapdh
Glyceraldehyde 3-phosphate dehydrogenase
- NGS
Next generation sequencing
- SaCas9
Cas9 from Staphylococcus aureus
- SpCas9
Cas9 from Streptococcus pyogenes
- ssODN
Single-stranded oligodeoxynucleotide
Footnotes
DISCLOSURES
None.
References
- 1.Emery AE. Population frequencies of inherited neuromuscular diseases--a world survey. Neuromuscul Disord. 1991;1:19–29. doi: 10.1016/0960-8966(91)90039-u. [DOI] [PubMed] [Google Scholar]
- 2.Hoffman EP, Brown RH, Kunkel LM. Dystrophin: The protein product of the duchene muscular dystrophy locus. 1987. Biotechnology. 1992;24:457–466. [PubMed] [Google Scholar]
- 3.Bushby K, Finkel R, Birnkrant DJ, Case LE, Clemens PR, Cripe L, Kaul A, Kinnett K, McDonald C, Pandya S, Poysky J, Shapiro F, Tomezsko J Constantin C, Group DMDCCW. Diagnosis and management of duchenne muscular dystrophy, part 1: Diagnosis, and pharmacological and psychosocial management. Lancet Neurol. 9:77–93. doi: 10.1016/S1474-4422(09)70271-6. [DOI] [PubMed] [Google Scholar]
- 4.Moriuchi T, Kagawa N, Mukoyama M, Hizawa K. Autopsy analyses of the muscular dystrophies. Tokushima J Exp Med. 1993;40:83–93. [PubMed] [Google Scholar]
- 5.McNally EM. New approaches in the therapy of cardiomyopathy in muscular dystrophy. Annual review of medicine. 2007;58:75–88. doi: 10.1146/annurev.med.58.011706.144703. [DOI] [PubMed] [Google Scholar]
- 6.Xu L, Park KH, Zhao L, Xu J, El Refaey M, Gao Y, Zhu H, Ma J, Han R. Crispr-mediated genome editing restores dystrophin expression and function in mdx mice. Mol Ther. 2015;24:564–569. doi: 10.1038/mt.2015.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Long C, Amoasii L, Mireault AA, McAnally JR, Li H, Sanchez-Ortiz E, Bhattacharyya S, Shelton JM, Bassel-Duby R, Olson EN. Postnatal genome editing partially restores dystrophin expression in a mouse model of muscular dystrophy. Science. 2016;351:400–403. doi: 10.1126/science.aad5725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Long CZ, McAnally JR, Shelton JM, Mireault AA, Bassel-Duby R, Olson EN. Prevention of muscular dystrophy in mice by crispr/cas9-mediated editing of germline DNA. Science. 2014;345:1184–1188. doi: 10.1126/science.1254445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nelson CE, Hakim CH, Ousterout DG, Thakore PI, Moreb EA, Castellanos Rivera RM, Madhavan S, Pan X, Ran FA, Yan WX, Asokan A, Zhang F, Duan D, Gersbach CA. In vivo genome editing improves muscle function in a mouse model of duchenne muscular dystrophy. Science. 2016;351:403–407. doi: 10.1126/science.aad5143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ousterout DGK, AM, Thakore PI, Majoros WH, Reddy TE, Gersbach CA. Multiplex crispr/cas9-based genome editing for correction of dystrophin mutations that cause duchenne muscular dystrophy. Nat Commun. 2015;6:6244. doi: 10.1038/ncomms7244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tabebordbar M, Zhu K, Cheng JK, Chew WL, Widrick JJ, Yan WX, Maesner C, Wu EY, Xiao R, Ran FA, Cong L, Zhang F, Vandenberghe LH, Church GM, Wagers AJ. In vivo gene editing in dystrophic mouse muscle and muscle stem cells. Science. 2016;351:407–411. doi: 10.1126/science.aad5177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bengtsson NE, Hall JK, Odom GL, Phelps MP, Andrus CR, Hawkins RD, Hauschka SD, Chamberlain JR, Chamberlain JS. Muscle-specific crispr/cas9 dystrophin gene editing ameliorates pathophysiology in a mouse model for duchenne muscular dystrophy. Nat Commun. 2017;8:14454. doi: 10.1038/ncomms14454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sicinski P, Geng Y, Ryder-Cook AS, Barnard EA, Darlison MG, Barnard PJ. The molecular basis of muscular dystrophy in the mdx mouse: A point mutation. Science. 1989;244:1578–1580. doi: 10.1126/science.2662404. [DOI] [PubMed] [Google Scholar]
- 14.Rincon MY, VandenDriessche T, Chuah MK. Gene therapy for cardiovascular disease: Advances in vector development, targeting, and delivery for clinical translation. Cardiovasc Res. 2015;108:4–20. doi: 10.1093/cvr/cvv205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pozsgai ER, Griffin DA, Heller KN, Mendell JR, Rodino-Klapac LR. Beta-sarcoglycan gene transfer decreases fibrosis and restores force in lgmd2e mice. Gene Ther. 1038;23:57–66. doi: 10.1038/gt.2015.80. [DOI] [PubMed] [Google Scholar]
- 16.Rodino-Klapac LR, Janssen PM, Shontz KM, Canan B, Montgomery CL, Griffin D, Heller K, Schmelzer L, Handy C, Clark KR, Sahenk Z, Mendell JR, Kaspar BK. Micro-dystrophin and follistatin co-delivery restores muscle function in aged dmd model. Hum Mol Genet. 1093;22:4929–4937. doi: 10.1093/hmg/ddt342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Duncan FJ, Naughton BJ, Zaraspe K, Murrey DA, Meadows AS, Clark KR, Newsom DE, White P, Fu H, McCarty DM. Broad functional correction of molecular impairments by systemic delivery of scaavrh74-hsgsh gene delivery in mps iiia mice. Mol Ther. 2015;23:638–647. doi: 10.1038/mt.2015.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chicoine LG, Rodino-Klapac LR, Shao G, Xu R, Bremer WG, Camboni M, Golden B, Montgomery CL, Shontz K, Heller KN, Griffin DA, Lewis S, Coley BD, Walker CM, Clark KR, Sahenk Z, Mendell JR, Martin PT. Vascular delivery of raavrh74.Mck.Galgt2 to the gastrocnemius muscle of the rhesus macaque stimulates the expression of dystrophin and laminin alpha2 surrogates. Mol Ther. 2014;22:713–724. doi: 10.1038/mt.2013.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhou L, Rafael-Fortney JA, Huang P, Zhao XS, Cheng G, Zhou X, Kaminski HJ, Liu L, Ransohoff RM. Haploinsufficiency of utrophin gene worsens skeletal muscle inflammation and fibrosis in mdx mice. J Neurol Sci. 2008;264:106–111. doi: 10.1016/j.jns.2007.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rafael JA, Tinsley JM, Potter AC, Deconinck AE, Davies KE. Skeletal muscle-specific expression of a utrophin transgene rescues utrophin-dystrophin deficient mice. Nat Genet. 1998;19:79–82. doi: 10.1038/ng0598-79. [DOI] [PubMed] [Google Scholar]
- 21.Delfin DA, Xu Y, Peterson JM, Guttridge DC, Rafael-Fortney JA, Janssen PM. Improvement of cardiac contractile function by peptide-based inhibition of nf-kappab in the utrophin/dystrophin-deficient murine model of muscular dystrophy. Journal of translational medicine. 2011;9:68. doi: 10.1186/1479-5876-9-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Biesiadecki BJ, Davis JP, Ziolo MT, Janssen PM. Tri-modal regulation of cardiac muscle relaxation; intracellular calcium decline, thin filament deactivation, and cross-bridge cycling kinetics. Biophys Rev. 6:273–289. doi: 10.1007/s12551-014-0143-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Janssen PM. Kinetics of cardiac muscle contraction and relaxation are linked and determined by properties of the cardiac sarcomere. Am J Physiol Heart Circ Physiol. 299:H1092–1099. doi: 10.1152/ajpheart.00417.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McNally EM. Cardiomyopathy in muscular dystrophy: When to treat? JAMA cardiology. 2016 doi: 10.1001/jamacardio.2016.4910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mendell JR, Campbell K, Rodino-Klapac L, Sahenk Z, Shilling C, Lewis S, Bowles D, Gray S, Li C, Galloway G, Malik V, Coley B, Clark KR, Li J, Xiao X, Samulski J, McPhee SW, Samulski RJ, Walker CM. Dystrophin immunity in duchenne’s muscular dystrophy. N Engl J Med. 2010;363:1429–1437. doi: 10.1056/NEJMoa1000228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Voit T, Topaloglu H, Straub V, Muntoni F, Deconinck N, Campion G, De Kimpe SJ, Eagle M, Guglieri M, Hood S, Liefaard L, Lourbakos A, Morgan A, Nakielny J, Quarcoo N, Ricotti V, Rolfe K, Servais L, Wardell C, Wilson R, Wright P, Kraus JE. Safety and efficacy of drisapersen for the treatment of duchenne muscular dystrophy (demand ii): An exploratory, randomised, placebo-controlled phase 2 study. The Lancet. Neurology. 2014;13:987–996. doi: 10.1016/S1474-4422(14)70195-4. [DOI] [PubMed] [Google Scholar]
- 27.Unger EF, Califf RM. Regarding eteplirsen for the treatment of duchenne muscular dystrophy. Ann Neurol. 2016 doi: 10.1002/ana.24842. [DOI] [PubMed] [Google Scholar]
- 28.Mendell JR, Rodino-Klapac LR, Sahenk Z, Roush K, Bird L, Lowes LP, Alfano L, Gomez AM, Lewis S, Kota J, Malik V, Shontz K, Walker CM, Flanigan KM, Corridore M, Kean JR, Allen HD, Shilling C, Melia KR, Sazani P, Saoud JB, Kaye EM Eteplirsen Study G. Eteplirsen for the treatment of duchenne muscular dystrophy. Ann Neurol. 2013;74:637–647. doi: 10.1002/ana.23982. [DOI] [PubMed] [Google Scholar]
- 29.Mali P, Yang L, Esvelt KM, Aach J, Guell M, DiCarlo JE, Norville JE, Church GM. Rna-guided human genome engineering via cas9. Science. 339:823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shalem O, Sanjana NE, Hartenian E, Shi X, Scott DA, Mikkelsen TS, Heckl D, Ebert BL, Root DE, Doench JG, Zhang F. Genome-scale crispr-cas9 knockout screening in human cells. Science. 343:84–87. doi: 10.1126/science.1247005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kleinstiver BP, Pattanayak V, Prew MS, Tsai SQ, Nguyen NT, Zheng Z, Joung JK. High-fidelity crispr-cas9 nucleases with no detectable genome-wide off-target effects. Nature. 2016;529:490–495. doi: 10.1038/nature16526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Slaymaker IM, Gao L, Zetsche B, Scott DA, Yan WX, Zhang F. Rationally engineered cas9 nucleases with improved specificity. Science. 2016;351:84–88. doi: 10.1126/science.aad5227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chew WL, Tabebordbar M, Cheng JK, Mali P, Wu EY, Ng AH, Zhu K, Wagers AJ, Church GM. A multifunctional aav-crispr-cas9 and its host response. Nat Methods. 2016;13:868–874. doi: 10.1038/nmeth.3993. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.