Abstract
Spinal muscular atrophy (SMA), a prominent genetic disease of infant mortality, is caused by low levels of survival motor neuron (SMN) protein owing to deletions or mutations of the SMN1 gene. SMN2, a nearly identical copy of SMN1 present in humans, cannot compensate for the loss of SMN1 due to predominant skipping of exon 7 during pre-mRNA splicing. With the recent FDA approval of nusinersen (Spinraza™), the potential for correction of SMN2 exon 7 splicing as a SMA therapy has been affirmed. Nusinersen is an antisense oligonucleotide that targets intronic splicing silencer N1 (ISS-N1) discovered in 2004 at the University of Massachusetts Medical School. ISS-N1 has emerged as the model target for testing the therapeutic efficacy of antisense oligonucleotides using different chemistries as well as different mouse models of SMA. Here we provide a historical account of events that led to the discovery of ISS-N1 and describe the impact of independent validations that raised the profile of ISS-N1 as one of the most potent antisense targets for the treatment of a genetic disease. Recent approval of nusinersen provides a much-needed boost for antisense technology that is just beginning to realize its potential. Beyond treating SMA, the ISS-N1 target offers myriad potentials for perfecting various aspects of the nucleic-acid-based technology for the amelioration of the countless number of pathological conditions.
Introduction
Spinal muscular atrophy (SMA) is a prominent and devastating genetic cause of infant mortality with an incidence of ~1 in 10,000 live births.1,2 SMA results from low levels of survival motor neuron (SMN) protein due to deletions of or mutations in the SMN1 gene.3,4 SMN is an essential protein involved in diverse functions including snRNP assembly, snoRNP assembly, DNA repair, transcription, telomerase biogenesis, translation, RNA trafficking, selenoprotein synthesis, stress granule formation and cell signaling.5 While motor neurons and neuromuscular junctions are the first to be affected in SMA,6 other tissues including the cardiovascular system, lung, bone, intestine, liver, pancreas, spleen and testis are also impacted by low levels of SMN.7–18 The disease spectrum of SMA, divided into four types (I through IV) is broad and ranges from infant mortality to adult onset.1 In general, SMA severity correlates with SMN2 copy number: the lower the SMN2 copy number, the lower the SMN and the more severe the disease.1 Lorson and Hahnen, working in the Androphy laboratory at Tufts Medical School, found that SMN2, a nearly identical copy of SMN1, fails to compensate for the loss of SMN1 due to defective splicing.19 The skipping of SMN2 exon 7 is attributed to a silent c.840C>T transition that corresponds to the 6th position of the 54-nucleotide (54-nt)-long exon 7.19,20 Since this silent mutation does not affect the protein sequence, correction of SMN2 exon 7 splicing has the potential to restore the fully functional SMN protein.21,22 The SMN2 exon 7-skipped mRNA itself leads to the production of SMNΔ7, a partially stable protein.23–26
Pre-mRNA splicing is a complex process involving multiple RNA-protein, protein-protein and RNA-RNA interactions unique for the removal of each individual intron.27 Several cis-elements and transacting factors have been implicated in regulation of SMN exon 7 splicing. Many recent reviews describe the mechanism of splicing regulation of SMN exon 7.28–30 The generation and nature of various splice variants of SMN have been described elsewhere.31–34 Here we provide background information pertaining to the discovery of the RNA regulatory element that produced nusinersen (Spinraza™), the first US Food and Drug Administration (FDA)-approved drug for SMA.35 Several investigations had demonstrated that C6U weakens the 3′-splice site (3′ss) of exon 7 due to its close proximity to the 3′ss.36–38 However, a subsequent analysis using live cells expressing an exon 7 splicing cassette revealed that a weak 5′ss is also a limiting factor for inclusion of exon 7.39 In vivo selection is an unbiased approach in which the significance of every nucleotide at each exonic position is tested employing a large pool of unique molecules (>1012) with random mutations.40,41 Surprisingly, the results of in vivo selection revealed that an adenosine residue at the last position (A54) of exon 7 has a more detrimental effect on SMN2 exon 7 splicing than the natural C6U substitution known to cause SMN2 exon 7 skipping.39 In agreement with the results of in vivo selection, an A54G substitution restored exon 7 inclusion even in the absence of positive cis-elements considered to be important for inclusion of exon 7 in both SMN1 and SMN2.39 The strong stimulatory effect of A54G on SMN2 exon 7 splicing was attributed to several factors, including the disruption of an inhibitory structure that sequesters the 5′ss of exon 7 and/or lengthening of the RNA:RNA duplex formed between U1 snRNA and the 5′ss of exon 7.39,40 Both of these mechanisms would result in enhanced recruitment of U1 snRNP at the 5′ss of exon 7. Indeed, a follow up study confirmed the presence of an inhibitory terminal stem-loop structure (TSL2) that facilitates SMN2 exon 7 skipping by sequestering the 5′ss of this exon.42 It was also confirmed that increasing the size of the duplex formed between U1 snRNA and the 5′ss of exon 7 has a strong stimulatory effect on SMN2 exon 7 inclusion.42
These insights into the processing of the exon 7 SMN2 mRNA brought immediate attention to the role of the 5′ss in regulation of SMN exon 7 splicing. Subsequent studies in the Singh and Androphy labs at the University of Massachusetts Medical School (UMMS) in Worcester, MA, USA discovered Intronic Splicing Silencer N1 (ISS-N1) as the major inhibitory element in SMN exon 7 splicing regulation.35,43,44 Here we describe how the discovery of ISS-N1 transformed our understanding of SMN exon 7 splicing regulation and paved the way for an antisense-oligonucleotide (ASO)-based therapy for SMA. Nusinersen (Spinraza™), the recently FDA-approved drug for SMA, is an ISS-N1 targeting ASO (Figure 1). We will present what lessons can be learnt from the successes of Spinraza™ for future ASO-based therapies of SMA and other diseases.
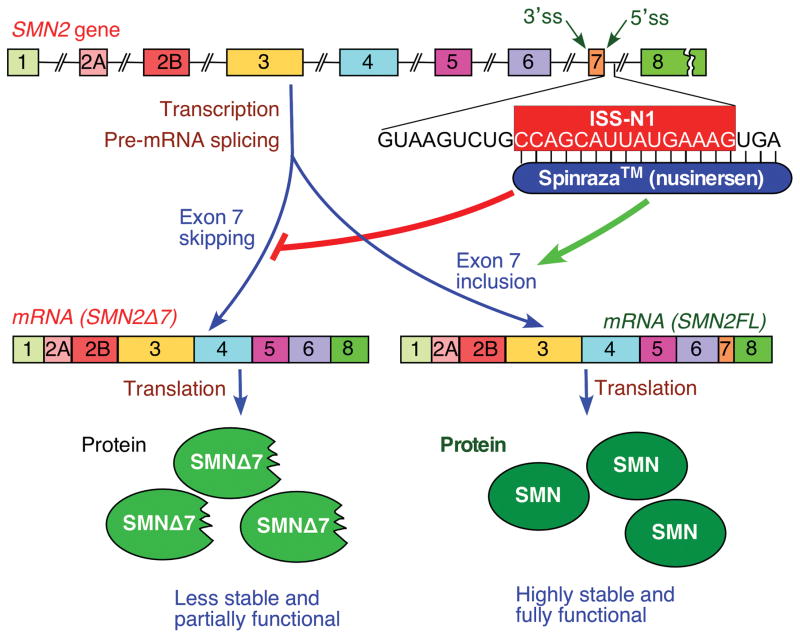
Figure 1.
Diagrammatic representation of SMN2 gene and Spinraza mode of action. SMN2 exons are represented by colored boxes, while introns are shown as broken lines. Intronic sequence immediately down stream of exon 7 is given. ISS-N1 region within this sequence is highlighted in pink box. Positions to which Spinraza anneals are indicated. SMN2 pre-mRNA splicing results in exon 7-included (SMN2FL) and exon 7-skipped (SMN2Δ7) transcripts, translation of which leads to production of the full length functional SMN protein and a truncated less stable isoform, respectively. Targeting of ISS-N1 by Spriranza prevents exon 7 skipping and as a consequence increases levels of the full length SMN.
Defining features of the ISS-N1 target
ISS-N1 is 15-nt long motif located immediately downstream of the 5′ss of exon 7. Deletion of ISS-N1 fully restored inclusion of SMN2 exon 7.43 Interestingly, the effect of ISS-N1 deletion was similar to what was observed with A54G mutation, since positive cis-elements became dispensable for exon 7 inclusion when ISS-N1 was deleted.39,43 Hence, Baralle and colleagues termed ISS-N1 as the master checkpoint of SMN2 exon 7 splicing.45 Consistent with the strong inhibitory effect of ISS-N1, an ASO complementary to and thereby blocking ISS-N1 fully restored SMN2 exon 7 inclusion in cultured fibroblasts derived from the SMA patient.43 The ASOs employed in this study carried phosphorothioate backbone and the 2′O-methyl (2′OMe ) modifications in all sugar residues. The stimulatory effect of the ISS-N1-targeting ASO was robust even at the very low concentration of 5 nM, suggesting that the ISS-N1 target is structurally accessible.43 Demonstrating the target specificity, two mismatch mutations within the ISS-N1 target as well as in an ASO that targeted it, completely abolished the stimulatory effect on splicing of exon 7.43 Additional validation of specificity came from an antisense walk in which the effect of three control ASOs that annealed to sequences downstream of ISS-N1 were evaluated. As expected, none of these control ASOs had any appreciable effect on SMN2 exon 7 splicing.43 These observations presented a rare example of a highly specific ASO with desired high activity at a record low nanomolar concentration. Equally significant was the target location within an intron, since an ASO annealing to an intronic sequence cannot have an adverse effect on mRNA export and protein translation. Consistent with the splicing correction, the ISS-N1-targeting ASO increased SMN protein in human SMA derived fibroblasts at the low nanomolar concentration.43 Overall, these findings were highly instructive in suggesting that the devastating consequences of exon 7 skipping could be fully overcome by abrogating the activity of an intronic inhibitory element. For commercial exploitation and drug development, UMMS filed for intellectual property rights with the US Patent Office in 2004 and subsequently secured a series of patents on the ISS-N1 target.35,44
At the time the report on the ISS-N1 target was published in 2006, Ionis Pharmaceuticals (previously ISIS Pharmaceuticals, Carlsbad, CA, USA), in collaboration with the Krainer group (Cold Spring Harbor Laboratory, Long Island, NY, USA), was working to develop an ASO-based therapy for SMA. In 2007 Ionis Pharmaceuticals presented the results of screening of a large number of ASOs that targeted exon 7.46 The ASOs used in these experiments carried phosphorothioate backbone and the 2′-O-methoxyethyl (MOE) modifications in all sugar residues. While the findings of this study validated the outcome of in vivo selection of exon 7, none of the ASOs identified emerged as a contender for therapeutic application. Hoping to find a better lead candidate, Ionis Pharmaceuticals continued with another MOE ASO library screen, this time targeting intronic sequences upstream and downstream of exon 7 including the ISS-N1 region.47 This study was particularly significant since it allowed a side-by-side comparison of an ISS-N1-targeting ASO with a likely novel lead ASO. The ISS-N1-targeting ASO (ASO 10–27) emerged as the best ASO in this study.47 Thus, ISS-N1 became the first antisense target for SMA therapy to be independently validated by employing an ASO with different chemistry.
Encouraged by these findings, the Krainer laboratory in collaboration with Ionis Pharmaceuticals and others, performed a series of in vivo studies with the ISS-N1-targeting ASO (ASO 10–27).48–50 The results of these studies showed its unprecedented therapeutic efficacy, including the record lifespan extension benefits in the severe SMA models (described below). While in vivo studies on the ISS-N1 target were still underway, Ionis Pharmaceuticals achieved another milestone in 2010 by securing an exclusive license from UMMS to develop SMA drug based on the ISS-N1 target.35 These advances put Ionis Pharmaceuticals in 2011 at the forefront of launching a clinical trial of an ISS-N1-targeting drug (ISIS-SMNRX) for the treatment of SMA. Subsequent years also witnessed a series of in vivo studies employing phosphorodiamidate morpholino oligonucleotides (PMOs) targeting ISS-N1.13,15,51–54 These studies provided additional independent validations of the efficacy of ISS-N1-targeting ASOs.
Mechanism of Action of ISS-N1-targeting ASOs
The mechanism by which an ISS-N1-targeting ASO stimulates SMN2 exon 7 inclusion appears to be complex. Sequences downstream of ISS-N1 harbor TIA1 binding sites.55 Overexpression of TIA1 restores inclusion of SMN2 exon 7, suggesting that factor(s) interacting with ISS-N1 interfere with the recruitment of this protein.55 Owing to the presence of two hnRNP A1/A2 motifs spanning the last 14 positions of the 15-nt long ISS-N1, it has been proposed that the inhibitory effect of ISS-N1 is due to interaction of ISS-N1 with hnRNP A1/A2.47 ISS-N1 overlaps with an 8-nt long GC-rich sequence (GCRS) spanning from the 7th to 14th positions of intron 7.56 Sequestration of GCRS by an 8-mer 2′OMe ASO also restores SMN2 exon 7 inclusion. The GCRS-targeting ASO partially sequestered only one of the two putative hnRNP A1/A2 motifs and yet this ASO produced the robust stimulatory effect on SMN2 exon 7 splicing. These surveys suggest that ISS-N1 may be a composite landing site for several splicing factors.
The most compelling evidence against the role of hnRNP A1/A2 in the negative effect of ISS-N1 came from a systematic study employing two 14-mer 2′OMe ASOs called L14 and F14.57 F14 and L14 bind to the first and the last fourteen nucleotides of the 15-nt long ISS-N1, respectively. As expected, F14 restored SMN2 exon 7 inclusion. Surprisingly, L14 (which sequestered both hnRNP A1/A2 sites but not the 1st position of ISS-N1) strongly inhibited SMN2 exon 7 splicing.57 Similar results were observed with two 14-mer ASOs with different chemistry, namely locked nucleic acid, suggesting that chemistry is not the cause of the L14-induced skipping of SMN2 exon 7.57 These observations ruled out the role of hnRNP A1/A2 as the sole regulator(s) associated with the negative effect of ISS-N1 and called for a unique mechanism in which sequestration of the first position of ISS-N1 is necessary for the stimulatory effect of an ISS-N1 targeting ASO.
The opposite effect of F14 and L14 on exon 7 splicing was investigated in a recent study where the 1st position of ISS-N1 was shown to be involved in the formation of a unique long-distance interaction (LDI) with a deep intronic sequence termed ISS-N2 (Figure 2).58 In particular, an internal stem formed by a LDI (ISTL1) sequesters the 1st position of ISS-N1 in addition to sequestering a portion of the 5′ss of exon 7.59 An ISS-N1-targeting ASO was found to disrupt ISTL1 and increase the accessibility of the 5′ss of exon 7 for the recruitment of U1 snRNP.59 Supporting the inhibitory role of ISTL1, an ISS-N2-targeting ASO that disrupted ISTL1, also produced a strong stimulatory effect on SMN2 exon 7 splicing.58 Hence, the stimulatory effect of an ISS-N1-targeting ASO is at least in part due to the breaking of the inhibitory structure, ISTL1. It is also likely that the sequestration of ISS-N1 promotes recruitment of TIA1 at sequences downstream of ISS-N1.59 In turn, an enhanced binding of TIA1 is likely to promote recruitment of U1 snRNP to the 5′ss of exon 7 causing its inclusion similarly as recently described.59,60 Longer morpholino ASOs that target ISS-N1 as well as downstream sequences also stimulated SMN2 exon 7 splicing.52 These results suggest that the recruitment of TIA1 downstream of ISS-N1 may not be critical once the ISS-N1 site is sequestered and ISTL1 is disrupted. However, longer ASOs may bring additional changes within the intron 7 structure and these changes may obviate the requirement for TIA1 in SMN exon 7 splicing regulation.
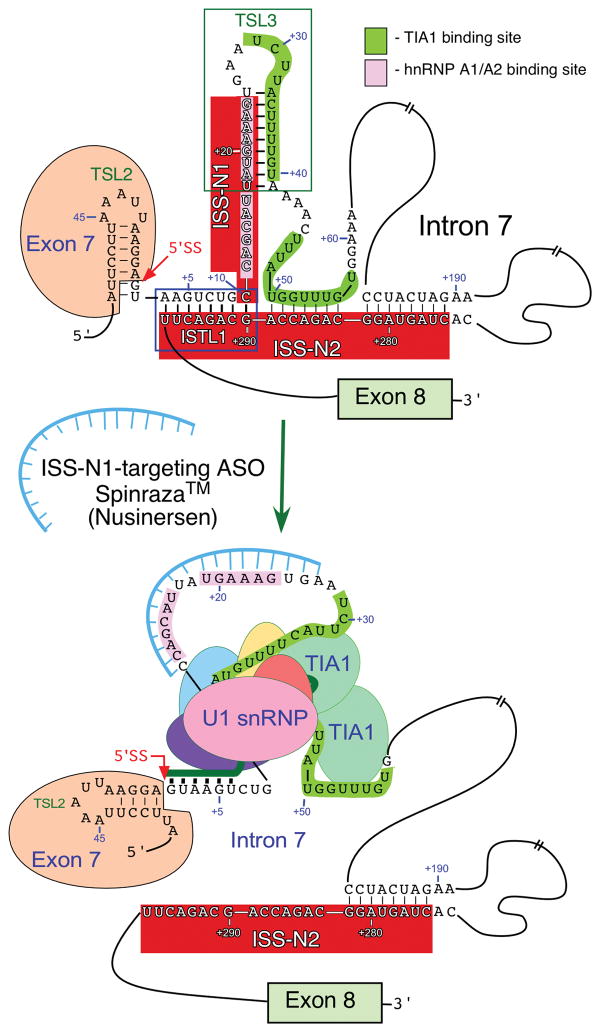
Figure 2.
Mechanism of exon 7 splicing correction by an ISS-N1 targeting ASO. Only relevant portions of exon 7 and intron 7 are shown. Exonic and intronic sequences are presented in the context of experimentally derived structures.42,58 Neutral and positive numbering start from the first position of exon 7 and the first position of intron 7, respectively. Splicing regulatory cis-elements and structures, such as the 5′ ss of exon 7, ISS-N1, ISS-N2, TSL2 and ISTL1 are highlighted. Binding sites for hnRNPA1/A2 and TIA1 are indicated. Annealing positions of the ASO and U1 snRNA are shown. Targeting of ISS-N1 by ASO causes structural rearrangements, such as disruption of TSL3 and ISTL1 and blocks the binding sites of hnRNP A1/A2. As the result TIA1 binding sites become accessible and the recruitment of U1 snRNP to the 5′ ss of exon 7 is increased. Abbreviations: ASO, antisense oligonucleotide; ISS, intronic splicing silencer; ISTL, internal stem-loop structure; ss, splice site; TSL, terminal stem-loop structure.
In vivo studies with ISS-N1-targeting ASOs
ISS-N1-targeting ASOs have shown remarkable efficacy in ameliorating the phenotype of several SMA mouse models when administered soon after birth (Table 1).13,15,48–54,61–72 Initially, the 2′OMe ISS-N1-targeting ASO43 increased SMN protein in the central nervous system (CNS) and improved motor function in the Δ7 mouse.61 This finding led to studies from Krainer and colleagues, and independently from the Burghes (The University of Ohio School of Medicine, Columbus, OH, USA) and Muntoni groups (University College London, London, UK), to further explore the efficacy of the ISS-N1 inhibitor, albeit they used contrasting delivery strategies and ASOs of different lengths and chemistries.48,50–53 The Krainer group first published results using an 18-mer MOE ASO (targeted to intron 7 nucleotides 10 to 27) in Taiwanese SMA mice, which can be bred to express only the human SMN2 transcript.48,50 Remarkably, in these mice subcutaneous (SC) administration of the MOE ASO extended median lifespan 25-fold and improved neuromuscular junction development.50 This survival benefit remains one of the greatest increases observed for all SMA treatments.21 Interestingly, MOE ASO delivery directly to the CNS provided relatively little survival benefit.50 Comparatively, the Burghes group observed that a single 27, 54 or 81 μg intracerebroventricular (ICV) dose of a 20-mer ISS-N1-targeting PMO (targeted to intron 7 nucleotides 10 to 29) increased median lifespan nearly 7-fold non-dose dependently in the relatively less severe Δ7 mouse model, which is transgenic for human SMN2 plus the SMNΔ7 cDNA.51 It remains unclear which ASO chemistry, length and route of delivery would provide the most efficacious treatment strategy for SMA. ICV injection is relatively invasive, but provides targeted ASO delivery to the CNS. Since the blood brain barrier (BBB) in neonatal mice is leaky, ASO delivered systemically could enter the CNS. However, BBB leakiness is much less prominent in humans and systemic administration (SC or intravascular) would most likely require a high dose to effectively distribute the ASO in the human CNS. High doses could potentially produce toxic effects especially in the liver and kidneys. The Krainer laboratory aimed to determine whether SMN restoration in the CNS is required to rescue the SMA phenotype in Taiwanese mice.67 SC delivery of an 18-mer ISS-N1-targeting MOE ASO accompanied with an ICV administration of a decoy MOE ASO complementary to the ISS-N1 targeting ASO mitigated the SMA phenotype similarly to SC 18-mer ISS-N1-targeting MOE ASO administration alone.67 Thus, a SMN increase in peripheral organs rather than strictly in the CNS appears important to the therapeutic outcome. This finding is not entirely surprising, since a host of recent studies have revealed peripheral organ defects in mouse models of SMA.5 Overall, regardless of their chemistry and length, ISS-N1-targeting ASOs consistently show the greatest efficacy in improving the phenotype of SMA mouse models.21 Collectively, these findings confirm the potency of the ISS-N1 target in stimulating SMN2 exon 7 inclusion and for the treatment of SMA. Because of this potency, the ISS-N1 target can be useful in investigating and perfecting emerging antisense technologies, including chemical modifications that could target the ASO to specific organs. For instance, Wood and colleagues have recently reported promising CNS delivery of systemically-administered peptide-conjugated PMOs.70,71 However, the pharmacokinetics and tolerability of these conjugated ASOs remains to be further examined in non-human primates. Nevertheless, these modifications could be particularly informative for other genetic diseases in which nucleic-acid-based treatment may be beneficial.
Table 1.
In vivo studies using ISS-N1-targeting ASOs
| Chemistry (Length) | Treatment Details | Outcome Measures | Ref |
|---|---|---|---|
| Studies in Δ7 mouse model | |||
| PS 2′OMe (20) | ICV P1, P3, P5, P7, P10 (1 μg) | Increased SMN protein in the CNS; improved motor function; increased body weight | 61 |
| PS MOE (18) | ICV P0 (4 μg) | Median survival increased 16 d to 26 d; improved muscle and NMJ development | 49 |
| PS 2OMe (21a) | ICV P1, P3, P5 (1 or 10 μg) | Median survival increased ~10 d to ~20 d; improved motor function | 62 |
| PMO (20) | ICV P0 (27, 45 or 81 μg) | Median survival increased 15 d to ~100 d; improved motor function; increased body weight | 51 |
| PMO (20, 22 and 25) | ICV P0 (0.5, 1, 2, 4 or 6 mM) | 25-mer dose-dependently increased median survival 15 d to 37–126 d | 53 |
| PMO (20) | ICV P1 (40 μg) | Age-dependent normalization of CMAP and MUNE | 63 |
| PMO and Dendrimeric octoguanadine (Vivo)-PMO (25) | ICV P0, P3 (2 nM) | Median survival increased 17 d to 23 d | 64 |
| SC P0, P3 (10, 12 or 24 nM) | Median survival increased 17 d to ~27 d | ||
| ICV P0 (2, 5, 10 or 12 nM) and SC P0, P3 (2, 5, 10 or 12 nM) | Median survival dose-dependently increased 17 d to between 40–120 d; combination of highest doses improved NMJ maturation and motor function | ||
| PMO (20) | ICV P0–P2, P4 or P6 (40 μg) | Treatment at P2 improved CMAP, MUNE and EIM measures when examined at P12 | 65 |
| PMO (20) | ICV P1 (40 μg) | Normalization of several SMA biomarkers | 66 |
| Studies in Taiwanese type I mouse model | |||
| PS MOE (18) | ICV P1 (20 μg) | Median survival increased 10 d to 16 d | 50 |
| SC P0–1, P2–3 (40, 50, 80 or 160 μg/g) | Median survival dose-dependently increased 10 d to between 84–248 d; improved NMJ maturation, motor function; motor neuron protection | ||
| ICV P1 (20 μg) and SC P0–1, P2–3 (50 μg/g) | Median survival increased 10 d to 173 d | ||
| PMO (18 and 25) | ICV P1 (20 or 40 μg/g) | Median lifespan increased 9.5 d to 12 d or 32 d (18-mer) or 43 d or 85.5 d (25-mer) | 52 |
| PMO (25) | IV P0 (40 μg/g) | Median lifespan increased 9.5 d to >230 d | |
| PMO (25) | IV P0 (40 μg/g) and IP or SC P3 (40 μg/g) | Median survival increased 9.5 d to 93.5 d | |
| Vivo-PMO (25) | IV P0 10 μg/g and IP P3 10 μg/g | Median survival was 16 d | |
| PMO (25) | ICV P0 (10, 20 or 40 μg/g) | Median survival increased 9.5 d to 22 d (10 μg/g), 24 d (20 μg/g) or 212 d (40 μg/g) | 54 |
| SC P0 (10, 20 or 40 μg/g) | Median survival increased 9.5 d to 22 d (10 μg/g), 58 d (20 μg/g) or 261 d (40 μg/g); improved NMJ and muscle maturation | ||
| SC P0 (10 μg/g), SC P5 and every 2 weeks (10 μg/g) | Survival increased 9.5 d to >150 d; improved muscle and NMJ maturation | ||
| PS MOE (18) | SC P0, P2 (120 mg/kg per day) | Median survival increased 10 d to 237 d; improved NMJ maturation and motor function; motor neuron protection | 67 |
| PS MOE (18) with decoy ASO | SC P0, P2 (120 mg/kg per day); ICV decoy ASO P0 (10 μg), P20 (20 μg) | Median survival increased 10 d to 212 d; improved NMJ maturation and motor function and protected motor neurons | |
| PMO (20) | SC P0 (80 μg/g) | Mean lifespan increased from 7.7 d to 19.7 d; FDB-2 muscle protection | 68 |
| PMO (25) | SC P0 (40 μg/g) | Normalized intestine development | 13 |
| PMO (25) | SC P0 (40 μg/g) | Normalized select miRNA levels in SC, muscle, serum | 69 |
| PMO and Pip6a-conjugated PMO (20) | IV P0 (10 μg/g) | Median survival increased 12 d to 167 d (Pip6a-PMO); no effect with unconjugated PMO | 70 |
| IV P0, P2 (5 or 10 μg/g each day) | Median survival increased 12 d to 57 d (unconjugated) or 283 d (5 μg/g Pip6a-conjugated) or 457 (10 μg/g Pip6a-conjugated) days; improved weight gain, NMJ maturation, motor function | ||
| PMO (25) | SC P0 (40 μg/g) | Normalized liver development | 15 |
| PMO and branched ApoE derivative-conjugated (20) | IV P0, P2 (10 mg/kg each day) | Median survival increased 10.5 d to 29 (unconjugated) or 78 d (ApoE-PMO) | 71 |
| Studies in Taiwanese type III mouse model | |||
| PS MOE (18) | ICV infusion for 7 days in adult mice (10, 25, 50, 100 or 150 μg per day) | Increased SMN2 exon 7 inclusion and SMN protein in the CNS | 48 |
| ICV E15 (2.5, 5, 10 or 20 μg) | Dose-dependent increase in tail length and delayed tail necrosis | ||
| PMO (25) | IV P0 (20 μg/g) | Tail necrosis onset delayed from 3 to 8 weeks | 52 |
| Vivo-PMO (25) | IV P0 (10 μg/g) | Tail necrosis onset delayed from 3 weeks to 9 months | |
| PS MOE (18) | IP P0, P2 (40, 80 or 120 mg/kg) | Prevented tail necrosis, but tails significant shorter than control mice | 67 |
| Study in Burgheron mouse model | |||
| PS MOE (20) | IP P10, P12 (80 μg/g) | Significantly extended lifespan; prevented tail necrosis; improved NMJ maturation | 72 |
| IP P25,P27 (80 μg/g) | Significantly extended lifespan | ||
Examined and ISS-N1-targeting ASO as well as ISS-N1-targeting ASO conjugated to sequence to recruit either SF2/ASF or hTra2β1
Abbreviations: 2′OMe, 2′-O-methyl; CMAP, compound muscle action potential; d, days; EIM, electrical impedance myography; ICV, intracerebroventricular; IP, intraperitoneal; IV, intravascular; MOE, 2′-O-methoxyethyl; MUNE, motor unit number estimation; NMJ, neuromuscular junction; P, postnatal day; PMO, phosphorodiamidate morpholino oligonucleotide; PS, phosphorothioate; SC, subcutaneous
ISS-N1-targeting ASOs also provide a powerful tool to further understand SMA disease mechanisms. Using ISS-N1-targeting ASOs allows researchers to examine the temporal impact of increasing SMN protein. For example, recent studies show impaired intestine and liver development in Taiwanese mice.13,15 SC delivery of a 25-mer PMO targeted to ISS-N1 normalized the development of these organs indicating that SMN is required for their development.12,14 ISS-N1-targeting ASOs can also validate biomarkers that could be examined in SMA patients in clinical trials. Namely, electrophysiological abnormalities and serum biomarkers in the Δ7 mouse are partially corrected with administration of a 20-mer ISS-N1-targeting PMO.63,65,66 Finally, these ISS-N1-targeting ASOs may also develop new models of the disease. The Wirth group generated an intermediate SMA mouse model through SC administration of suboptimal doses of the 18-mer ISS-N1-targeting MOE ASO in severe Taiwanese mice.73 This new model allowed for examination of the benefit of the SMA genetic modifier plastin3.73 Intermediate models of the disease would also allow one to capture potential defects in organ development that may not be observed in severe models with early postnatal lethality.
Therapeutic development
Based on the strong pre-clinical data on the efficacy of ISS-N1, Ionis Pharmaceuticals commenced clinical trials with nusinersen, an 18-mer ASO with their proprietary MOE chemistry. Given the neuromuscular nature of SMA, nusinersen was delivered intrathecally through a lumbar puncture for these clinical trials.74–76 Lumbar puncture effectively distributed nusinersen throughout the CNS and did not produce adverse effects beyond what had been previously reported for this procedure.75 In the phase I study, nusinersen was well-tolerated and led to an increase in SMN protein in the cerebrospinal fluid at 9–14 months post treatment.74 In addition, the highest dose (9 mg) increased motor function up to 9–14 months post treatment as assessed by the Hammersmith Functional Motor Score Expanded (HFMSE). The HFSME score increase was particularly promising since it constituted what would be considered a clinically meaningful outcome and led to commencement of larger phase 2 and 3 clinical trials.74 A phase 2 clinical trial enrolled severe type I SMA infants with symptom onset between 3 weeks and 4 months.76 Multiple doses of nusinersen were well-tolerated and the majority of the adverse effects were mild. Nusinersen was broadly distributed throughout the CNS, increased full-length SMN2 transcript and SMN protein and most infants exhibited improved motor function as well as survival and electrophysiological function.76 The promising results from these clinical trials led to a placebo-controlled double-blinded phase III clinical trials. While the results have yet to be published, the trials were terminated early because the primary endpoint at interim analysis was met;77 FDA approved Spinraza™ on December 23, 2016. Hence, Spinraza™ became the first FDA-approved drug for SMA as well as the first antisense drug to treat the major population of a genetic disease through splicing correction.
Conclusions and Future directions
Correction of SMN2 exon 7 splicing has long been considered as the most efficacious therapeutic option for most SMA patients who retain at least one copy of the SMN2 allele. The specificity with which an oligonucleotide corrects splicing remains unmatched. The major obstacle in the way of an effective ASO-based therapy of SMA is the delivery of the oligonucleotide across the BBB. Other concerns include the stability and the tolerability of the oligonucleotide. With the recent FDA approval of Spinraza™, much of the skepticism of the oligonucleotide-based therapy of SMA has been put to the rest. Animal studies underscore that an early restoration of SMN is key to achieving the maximum therapeutic benefit. The availability of Spinraza™ provides SMA patients with their first opportunity to receive a drug that could increase SMN levels. This development puts SMA among a handful of rare diseases with at least one FDA-approved drug. This is also a great win for nucleic-acid therapeutics, which holds the promise for the treatment of rare and orphan diseases.
Despite the anticipated enthusiasm among the SMA community and caregivers, it is understandable that Spinraza™ may not act with equal efficacy in all SMA patients. This is due to a variety of reasons including a limited number of SMN2 alleles, delayed age of drug administration and the presence or absence of other disease modifying factors. There may be some instances in which Spinraza™ may be completely unusable due to an acute immune response. These concerns call for a continued progress towards developing alternative therapies for SMA. Fortunately, all oligonucleotide chemistries incorporated into ISS-N1-targeting ASOs have shown promising results. These independent validations of target efficacy suggest bright prospects for developing advanced ASO-based SMA therapies. We now know that low levels of SMN affect most tissues. The launch of Spinraza™ has invigorated the field of nucleic acid-based therapeutics to further translate the potential use of ASOs targeting other regulatory elements in SMN2, and modifications that enable penetration into all tissues when delivered employing noninvasive procedures. Recent reports have also validated the efficacy of additional antisense targets, including Element 1 in SMN2 intron 678 and ISS-N2 in SMN2 intron 779 in SMA mouse models. In addition, the potential for dual-masking ASOs that simultaneously sequester two targets could increase the repertoire of ASOs for SMA therapy.80 Given the spectrum of the SMA phenotype, it will be useful to have multiple ASO targets so as to treat best treat patients who may not respond to specific ASOs.
Success of an ASO-based therapy for SMA has implications for other diseases in which nucleic acid-based therapy remains an option. Progress thus far exhibits a classic example of smooth transition from fundamental discovery of the ISS-N1 target to independent pre-clinical validations of ISS-N1-targeting ASOs to independent clinical trials of Spinraza™. The coming years will be critical for evaluating the long-term efficacy of Spinraza™ in large cohorts comprised of the appropriate age-matched patients. However, there remains a concern of drug affordability as the high cost of Spinraza™ might limit the number of patients who might otherwise benefit from this drug. Irrespective of these concerns, there is every reason to believe that tomorrow’s SMA patients will have more options thanks in part to the new insights brought forth from the basic investigations into the regulatory elements that control SMN2 exon 7 alternative splicing.
Acknowledgments
The nonprofit organization Cure SMA (formerly Families of SMA) supported the initial studies in the laboratories of Drs. Ravindra Singh and Elliot Androphy that led to the discovery of the ISS-N1 target at University Massachusetts Medical School. RNS is supported by US National Institutes of Health NIH R01 NS055925, Iowa Center of Advanced Neurotoxicology (ICAN) and Salsbury Endowment at Iowa State University. EJA is grateful for support from NIH grants R01 NS040275 and R01 NS0682284 and Cure SMA.
Footnotes
Financial & competing interests disclosure
The ISS-N1 target (US7838657) was discovered in the Singh laboratory at UMass Medical School (MA, USA). Inventors, including RN Singh, NN Singh, EJA and UMASS Medical School, are currently benefiting from licensing of the ISS-N1 target to Ionis Pharmaceuticals. Iowa State University holds intellectual property rights on GCRS and ISS-N2 targets. Therefore, inventors including RN Singh, NN Singh and Iowa State University could potentially benefit from any future commercial exploitation of GCRS and ISS-N2 targets.
References
- 1.Awano T, Kim J-K, Monani UR. Spinal muscular atrophy: journeying from bench to bedside. Neurotherapeutics. 2014;11:786–795. doi: 10.1007/s13311-014-0293-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sugarman EA, Nagan N, Zhu H, Akmaev VR, Zhou Z, Rohlfs EM, et al. Pan-ethnic carrier screening and prenatal diagnosis for spinal muscular atrophy: clinical laboratory analysis of >72,400 specimens. Eur J Hum Genet. 2012;20:27–32. doi: 10.1038/ejhg.2011.134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lefebvre S, Bürglen L, Reboullet S, Clermont O, Burlet P, Viollet L, et al. Identification and characterization of a spinal muscular atrophy-determining gene. Cell. 1995;80:155–165. doi: 10.1016/0092-8674(95)90460-3. [DOI] [PubMed] [Google Scholar]
- 4.Wirth B. An update of the mutation spectrum of the survival motor neuron gene (SMN1) in autosomal recessive spinal muscular atrophy (SMA) Hum Mutat. 2000;15:228–237. doi: 10.1002/(SICI)1098-1004(200003)15:3<228::AID-HUMU3>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 5.Singh RN, Howell MD, Ottesen EW, Singh NN. Diverse role of survival motor neuron protein. Biochim Biophys Acta. 2017;1860:299–315. doi: 10.1016/j.bbagrm.2016.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ahmad S, Bhatia K, Kannan A, Gangwani L. Molecular Mechanisms of Neurodegeneration in Spinal Muscular Atrophy. J Exp Neurosci. 2016;10:39–49. doi: 10.4137/JEN.S33122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shababi M, Habibi J, Yang HT, Vale SM, Sewell WA, Lorson CL. Cardiac defects contribute to the pathology of spinal muscular atrophy models. Hum Mol Genet. 2010;19:4059–4071. doi: 10.1093/hmg/ddq329. [DOI] [PubMed] [Google Scholar]
- 8.Heier CR, Satta R, Lutz C, DiDonato CJ. Arrhythmia and cardiac defects are a feature of spinal muscular atrophy model mice. Hum Mol Genet. 2010;19:3906–3918. doi: 10.1093/hmg/ddq330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schreml J, Riessland M, Paterno M, Garbes L, Roßbach K, Ackermann B, et al. Severe SMA mice show organ impairment that cannot be rescued by therapy with the HDACi JNJ-26481585. Eur J Hum Genet. 2013;21:643–652. doi: 10.1038/ejhg.2012.222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Michaud M, Arnoux T, Bielli S, Durand E, Rotrou Y, Jablonka S, et al. Neuromuscular defects and breathing disorders in a new mouse model of spinal muscular atrophy. Neurobiol Dis. 2010;38:125–135. doi: 10.1016/j.nbd.2010.01.006. [DOI] [PubMed] [Google Scholar]
- 11.Shanmugarajan S, Tsuruga E, Swoboda KJ, Maria BL, Ries WL, Reddy SV. Bone loss in survival motor neuron (Smn(−/−) SMN2) genetic mouse model of spinal muscular atrophy. J Pathol. 2009;219:52–60. doi: 10.1002/path.2566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gombash SE, Cowley CJ, Fitzgerald JA, Iyer CC, Fried D, McGovern VL, et al. SMN deficiency disrupts gastrointestinal and enteric nervous system function in mice. Hum Mol Genet. 2015;24:3847–3860. doi: 10.1093/hmg/ddv127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sintusek P, Catapano F, Angkathunkayul N, Marrosu E, Parson SH, Morgan JE, et al. Histopathological Defects in Intestine in Severe Spinal Muscular Atrophy Mice Are Improved by Systemic Antisense Oligonucleotide Treatment. PLoS ONE. 2016;11:e0155032. doi: 10.1371/journal.pone.0155032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vitte JM, Davoult B, Roblot N, Mayer M, Joshi V, Courageot S, et al. Deletion of murine Smn exon 7 directed to liver leads to severe defect of liver development associated with iron overload. Am J Pathol. 2004;165:1731–1741. doi: 10.1016/S0002-9440(10)63428-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Szunyogova E, Zhou H, Maxwell GK, Powis RA, Francesco M, Gillingwater TH, et al. Survival Motor Neuron (SMN) protein is required for normal mouse liver development. Sci Rep. 2016;6:34635. doi: 10.1038/srep34635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bowerman M, Swoboda KJ, Michalski J-P, Wang G-S, Reeks C, Beauvais A, et al. Glucose metabolism and pancreatic defects in spinal muscular atrophy. Ann Neurol. 2012;72:256–268. doi: 10.1002/ana.23582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Thomson AK, Somers E, Powis RA, Shorrock HK, Murphy K, Swoboda KJ, et al. Survival of motor neurone protein is required for normal postnatal development of the spleen. J Anat. 2016 doi: 10.1111/joa.12546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ottesen EW, Howell MD, Singh NN, Seo J, Whitley EM, Singh RN. Severe impairment of male reproductive organ development in a low SMN expressing mouse model of spinal muscular atrophy. Sci Rep. 2016;6:20193. doi: 10.1038/srep20193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lorson CL, Hahnen E, Androphy EJ, Wirth B. A single nucleotide in the SMN gene regulates splicing and is responsible for spinal muscular atrophy. Proc Natl Acad Sci USA. 1999;96:6307–6311. doi: 10.1073/pnas.96.11.6307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Monani UR, Lorson CL, Parsons DW, Prior TW, Androphy EJ, Burghes AH, et al. A single nucleotide difference that alters splicing patterns distinguishes the SMA gene SMN1 from the copy gene SMN2. Hum Mol Genet. 1999;8:1177–1183. doi: 10.1093/hmg/8.7.1177. [DOI] [PubMed] [Google Scholar]
- 21.Seo J, Howell MD, Singh NN, Singh RN. Spinal muscular atrophy: An update on therapeutic progress. Biochim Biophys Acta. 2013;1832:2180–2190. doi: 10.1016/j.bbadis.2013.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Howell MD, Singh NN, Singh RN. Advances in therapeutic development for spinal muscular atrophy. Future Med Chem. 2014;6:1081–1099. doi: 10.4155/fmc.14.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lorson CL, Androphy EJ. An exonic enhancer is required for inclusion of an essential exon in the SMA-determining gene SMN. Hum Mol Genet. 2000;9:259–265. doi: 10.1093/hmg/9.2.259. [DOI] [PubMed] [Google Scholar]
- 24.Vitte J, Fassier C, Tiziano FD, Dalard C, Soave S, Roblot N, et al. Refined characterization of the expression and stability of the SMN gene products. Am J Pathol. 2007;171:1269–1280. doi: 10.2353/ajpath.2007.070399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Burnett BG, Muñoz E, Tandon A, Kwon DY, Sumner CJ, Fischbeck KH. Regulation of SMN protein stability. Mol Cell Biol. 2009;29:1107–1115. doi: 10.1128/MCB.01262-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cho S, Dreyfuss G. A degron created by SMN2 exon 7 skipping is a principal contributor to spinal muscular atrophy severity. Genes Dev. 2010;24:438–442. doi: 10.1101/gad.1884910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee Y, Rio DC. Mechanisms and Regulation of Alternative Pre-mRNA Splicing. Annu Rev Biochem. 2015;84:291–323. doi: 10.1146/annurev-biochem-060614-034316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Singh NN, Singh RN. Alternative splicing in spinal muscular atrophy underscores the role of an intron definition model. RNA Biol. 2011;8:600–606. doi: 10.4161/rna.8.4.16224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Singh NN, Lee BM, Singh RN. Splicing regulation in spinal muscular atrophy by an RNA structure formed by long-distance interactions. Ann N Y Acad Sci. 2015;1341:176–187. doi: 10.1111/nyas.12727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Singh NN, Howell MD, Singh RN. Transcriptional and Splicing Regulation of Spinal Muscular Atrophy Genes. In: Sumner CJ, Paushkin S, Ko C-P, editors. Spinal Muscular Atrophy. Academic Press; 2017. pp. 75–97. [Google Scholar]
- 31.Setola V, Terao M, Locatelli D, Bassanini S, Garattini E, Battaglia G. Axonal-SMN (a-SMN), a protein isoform of the survival motor neuron gene, is specifically involved in axonogenesis. Proc Natl Acad Sci USA. 2007;104:1959–1964. doi: 10.1073/pnas.0610660104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Singh NN, Seo J, Rahn SJ, Singh RN. A multi-exon-skipping detection assay reveals surprising diversity of splice isoforms of spinal muscular atrophy genes. PLoS ONE. 2012;7:e49595. doi: 10.1371/journal.pone.0049595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Seo J, Singh NN, Ottesen EW, Sivanesan S, Shishimorova M, Singh RN. Oxidative Stress Triggers Body-Wide Skipping of Multiple Exons of the Spinal Muscular Atrophy Gene. PLoS ONE. 2016;11:e0154390. doi: 10.1371/journal.pone.0154390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Seo J, Singh NN, Ottesen EW, Lee BM, Singh RN. A novel human-specific splice isoform alters the critical C-terminus of Survival Motor Neuron protein. Sci Rep. 2016;6:30778. doi: 10.1038/srep30778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ottesen EW. ISS-N1 makes the first FDA-approved drug for spinal muscular atrophy. Translat Neurosci. 2017;8:1–6. doi: 10.1515/tnsci-2017-0001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Singh NN, Androphy EJ, Singh RN. An extended inhibitory context causes skipping of exon 7 of SMN2 in spinal muscular atrophy. Biochem Biophys Res Commun. 2004;315:381–388. doi: 10.1016/j.bbrc.2004.01.067. [DOI] [PubMed] [Google Scholar]
- 37.Singh NN, Androphy EJ, Singh RN. The regulation and regulatory activities of alternative splicing of the SMN gene. Crit Rev Eukaryot Gene Expr. 2004;14:271–285. doi: 10.1615/critreveukaryotgeneexpr.v14.i4.30. [DOI] [PubMed] [Google Scholar]
- 38.Singh RN. Evolving concepts on human SMN pre-mRNA splicing. RNA Biol. 2007;4:7–10. doi: 10.4161/rna.4.1.4535. [DOI] [PubMed] [Google Scholar]
- 39.Singh NN, Androphy EJ, Singh RN. In vivo selection reveals combinatorial controls that define a critical exon in the spinal muscular atrophy genes. RNA. 2004;10:1291–1305. doi: 10.1261/rna.7580704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Singh RN. Unfolding the mystery of alternative splicing through a unique method of in vivo selection. Front Biosci. 2007;12:3263–3272. doi: 10.2741/2310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Singh RN, Singh NN. Functional Analysis of Large Exonic Sequences Through Iterative In Vivo Selection. In: Stamm S, Smith CWJ, Lührmann R, editors. Alternative pre-mRNA Splicing: Theory and Protocols. Vol. 2012. Wiley-VCH Verlag GmbH & Co; KGaA, Weinheim, Germany: 2012. pp. 200–209. [Google Scholar]
- 42.Singh NN, Singh RN, Androphy EJ. Modulating role of RNA structure in alternative splicing of a critical exon in the spinal muscular atrophy genes. Nucleic Acids Res. 2007;35:371–389. doi: 10.1093/nar/gkl1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Singh NK, Singh NN, Androphy EJ, Singh RN. Splicing of a critical exon of human Survival Motor Neuron is regulated by a unique silencer element located in the last intron. Mol Cell Biol. 2006;26:1333–1346. doi: 10.1128/MCB.26.4.1333-1346.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Singh RN, Singh NN, Singh NK, Androphy EJ. Spinal muscular atrophy (SMA) treatment via targeting of SMN2 splice site inhibitory sequences. US7838657. US patent publication. 2010 (Also published as US8110560, US8586559, US9476042, US20070292408, US20100087511, US20120165394, US20140066492)
- 45.Buratti E, Baralle M, Baralle FE. Defective splicing, disease and therapy: searching for master checkpoints in exon definition. Nucleic Acids Res. 2006;34:3494–3510. doi: 10.1093/nar/gkl498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hua Y, Vickers TA, Baker BF, Bennett CF, Krainer AR. Enhancement of SMN2 exon 7 inclusion by antisense oligonucleotides targeting the exon. PLoS Biol. 2007;5:e73. doi: 10.1371/journal.pbio.0050073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hua Y, Vickers TA, Okunola HL, Bennett CF, Krainer AR. Antisense masking of an hnRNP A1/A2 intronic splicing silencer corrects SMN2 splicing in transgenic mice. Am J Hum Genet. 2008;82:834–848. doi: 10.1016/j.ajhg.2008.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hua Y, Sahashi K, Hung G, Rigo F, Passini MA, Bennett CF, et al. Antisense correction of SMN2 splicing in the CNS rescues necrosis in a type III SMA mouse model. Genes Dev. 2010;24:1634–1644. doi: 10.1101/gad.1941310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Passini MA, Bu J, Richards AM, Kinnecom C, Sardi SP, Stanek LM, et al. Antisense oligonucleotides delivered to the mouse CNS ameliorate symptoms of severe spinal muscular atrophy. Sci Transl Med. 2011;3:72ra18. doi: 10.1126/scitranslmed.3001777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hua Y, Sahashi K, Rigo F, Hung G, Horev G, Bennett CF, et al. Peripheral SMN restoration is essential for long-term rescue of a severe spinal muscular atrophy mouse model. Nature. 2011;478:123–126. doi: 10.1038/nature10485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Porensky PN, Mitrpant C, McGovern VL, Bevan AK, Foust KD, Kaspar BK, et al. A single administration of morpholino antisense oligomer rescues spinal muscular atrophy in mouse. Hum Mol Genet. 2012;21:1625–1638. doi: 10.1093/hmg/ddr600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhou H, Janghra N, Mitrpant C, Dickinson RL, Anthony K, Price L, et al. A Novel Morpholino Oligomer Targeting ISS-N1 Improves Rescue of Severe Spinal Muscular Atrophy Transgenic Mice. Hum Gene Ther. 2013;24:331–342. doi: 10.1089/hum.2012.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mitrpant C, Porensky P, Zhou H, Price L, Muntoni F, Fletcher S, et al. Improved antisense oligonucleotide design to suppress aberrant SMN2 gene transcript processing: towards a treatment for spinal muscular atrophy. PLOS ONE. 2013;8:e62114. doi: 10.1371/journal.pone.0062114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhou H, Meng J, Marrosu E, Janghra N, Morgan J, Muntoni F. Repeated low doses of morpholino antisense oligomer: an intermediate mouse model of spinal muscular atrophy to explore the window of therapeutic response. Hum Mol Genet. 2015;24:6265–6277. doi: 10.1093/hmg/ddv329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Singh NN, Seo J, Ottesen EW, Shishimorova M, Bhattacharya D, Singh RN. TIA1 prevents skipping of a critical exon associated with spinal muscular atrophy. Mol Cell Biol. 2011;31:935–954. doi: 10.1128/MCB.00945-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Singh NN, Shishimorova M, Cao LC, Gangwani L, Singh RN. A short antisense oligonucleotide masking a unique intronic motif prevents skipping of a critical exon in spinal muscular atrophy. RNA Biol. 2009;6:341–350. doi: 10.4161/rna.6.3.8723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Singh NN, Hollinger K, Bhattacharya D, Singh RN. An antisense microwalk reveals critical role of an intronic position linked to a unique long-distance interaction in pre-mRNA splicing. RNA. 2010;16:1167–1181. doi: 10.1261/rna.2154310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Singh NN, Lawler MN, Ottesen EW, Upreti D, Kaczynski JR, Singh RN. An intronic structure enabled by a long-distance interaction serves as a novel target for splicing correction in spinal muscular atrophy. Nucleic Acids Res. 2013;41:8144–8165. doi: 10.1093/nar/gkt609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Singh NN, Lee BM, DiDonato CJ, Singh RN. Mechanistic principles of antisense targets for the treatment of spinal muscular atrophy. Future Med Chem. 2015;7:1793–1808. doi: 10.4155/fmc.15.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rogalska ME, Tajnik M, Licastro D, Bussani E, Camparini L, Mattioli C, et al. Therapeutic activity of modified U1 core spliceosomal particles. Nat Commun. 2016;7:11168. doi: 10.1038/ncomms11168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Williams JH, Schray RC, Patterson CA, Ayitey SO, Tallent MK, Lutz GJ. Oligonucleotide-mediated survival of motor neuron protein expression in CNS improves phenotype in a mouse model of spinal muscular atrophy. J Neurosci. 2009;29:7633–7638. doi: 10.1523/JNEUROSCI.0950-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Osman EY, Yen P-F, Lorson CL. Bifunctional RNAs targeting the intronic splicing silencer N1 increase SMN levels and reduce disease severity in an animal model of spinal muscular atrophy. Mol Ther. 2012;20:119–126. doi: 10.1038/mt.2011.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Arnold WD, Porensky PN, McGovern VL, Iyer CC, Duque S, Li X, et al. Electrophysiological Biomarkers in Spinal Muscular Atrophy: Preclinical Proof of Concept. Ann Clin Transl Neurol. 2014;1:34–44. doi: 10.1002/acn3.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Nizzardo M, Simone C, Salani S, Ruepp M-D, Rizzo F, Ruggieri M, et al. Effect of combined systemic and local morpholino treatment on the spinal muscular atrophy Δ7 mouse model phenotype. Clin Ther. 2014;36:340–356. e5. doi: 10.1016/j.clinthera.2014.02.004. [DOI] [PubMed] [Google Scholar]
- 65.Arnold W, McGovern VL, Sanchez B, Li J, Corlett KM, Kolb SJ, et al. The neuromuscular impact of symptomatic SMN restoration in a mouse model of spinal muscular atrophy. Neurobiol Dis. 2016;87:116–123. doi: 10.1016/j.nbd.2015.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Arnold WD, Duque S, Iyer CC, Zaworski P, McGovern VL, Taylor SJ, et al. Normalization of Patient-Identified Plasma Biomarkers in SMNΔ7 Mice following Postnatal SMN Restoration. PLoS ONE. 2016;11:e0167077. doi: 10.1371/journal.pone.0167077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hua Y, Liu YH, Sahashi K, Rigo F, Bennett CF, Krainer AR. Motor neuron cell-nonautonomous rescue of spinal muscular atrophy phenotypes in mild and severe transgenic mouse models. Genes Dev. 2015;29:288–297. doi: 10.1101/gad.256644.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lin T-L, Chen T-H, Hsu Y-Y, Cheng Y-H, Juang B-T, Jong Y-J. Selective Neuromuscular Denervation in Taiwanese Severe SMA Mouse Can Be Reversed by Morpholino Antisense Oligonucleotides. PLoS ONE. 2016;11:e0154723. doi: 10.1371/journal.pone.0154723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Catapano F, Zaharieva I, Scoto M, Marrosu E, Morgan J, Muntoni F, et al. Altered Levels of MicroRNA-9, -206, and -132 in Spinal Muscular Atrophy and Their Response to Antisense Oligonucleotide Therapy. Mol Ther Nucleic Acids. 2016;5:e331. doi: 10.1038/mtna.2016.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hammond SM, Hazell G, Shabanpoor F, Saleh AF, Bowerman M, Sleigh JN, et al. Systemic peptide-mediated oligonucleotide therapy improves long-term survival in spinal muscular atrophy. Proc Natl Acad Sci USA. 2016;113:10962–10967. doi: 10.1073/pnas.1605731113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Shabanpoor F, Hammond SM, Abendroth F, Hazell G, Wood MJA, Gait MJ. Identification of a Peptide for Systemic Brain Delivery of a Morpholino Oligonucleotide in Mouse Models of Spinal Muscular Atrophy. Nucleic Acid Ther. 2017 doi: 10.1089/nat.2016.0652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Bogdanik LP, Osborne MA, Davis C, Martin WP, Austin A, Rigo F, et al. Systemic, postsymptomatic antisense oligonucleotide rescues motor unit maturation delay in a new mouse model for type II/III spinal muscular atrophy. Proc Natl Acad Sci USA. 2015;112:E5863–5872. doi: 10.1073/pnas.1509758112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hosseinibarkooie S, Peters M, Torres-Benito L, Rastetter RH, Hupperich K, Hoffmann A, et al. The Power of Human Protective Modifiers: PLS3 and CORO1C Unravel Impaired Endocytosis in Spinal Muscular Atrophy and Rescue SMA Phenotype. Am J Hum Genet. 2016;99:647–665. doi: 10.1016/j.ajhg.2016.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Chiriboga CA, Swoboda KJ, Darras BT, Iannaccone ST, Montes J, De Vivo DC, et al. Results from a phase 1 study of nusinersen (ISIS-SMN(Rx)) in children with spinal muscular atrophy. Neurology. 2016;86:890–897. doi: 10.1212/WNL.0000000000002445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Haché M, Swoboda KJ, Sethna N, Farrow-Gillespie A, Khandji A, Xia S, et al. Intrathecal Injections in Children With Spinal Muscular Atrophy: Nusinersen Clinical Trial Experience. J Child Neurol. 2016;31:899–906. doi: 10.1177/0883073815627882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Finkel RS, Chiriboga CA, Vajsar J, Day JW, Montes J, De Vivo DC, et al. Treatment of infantile-onset spinal muscular atrophy with nusinersen: a phase 2, open-label, dose-escalation study. Lancet. 2016;388:3017–3026. doi: 10.1016/S0140-6736(16)31408-8. [DOI] [PubMed] [Google Scholar]
- 77.Garber K. Big win possible for Ionis/Biogen antisense drug in muscular atrophy. Nat Biotechnol. 2016;34:1002–1003. doi: 10.1038/nbt1016-1002. [DOI] [PubMed] [Google Scholar]
- 78.Osman EY, Miller MR, Robbins KL, Lombardi AM, Atkinson AK, Brehm AJ, et al. Morpholino antisense oligonucleotides targeting intronic repressor Element1 improve phenotype in SMA mouse models. Hum Mol Genet. 2014;23:4832–4845. doi: 10.1093/hmg/ddu198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Howell MD, Ottesen EW, Singh NN, Anderson RL, Singh RN. Gender-Specific Amelioration of SMA Phenotype upon Disruption of a Deep Intronic Structure by an Oligonucleotide. Mol Ther. 2017 doi: 10.1016/j.ymthe.2017.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Pao PW, Wee KB, Yee WC, Pramono ZAD, Dwipramono ZA. Dual masking of specific negative splicing regulatory elements resulted in maximal exon 7 inclusion of SMN2 gene. Mol Ther. 2014;22:854–861. doi: 10.1038/mt.2013.276. [DOI] [PMC free article] [PubMed] [Google Scholar]