Abstract
We present Omni-ATAC, an improved ATAC-seq protocol for chromatin accessibility profiling that works across multiple applications with substantial improvement of signal-to-background ratio and information content. The Omni-ATAC protocol generates chromatin accessibility profiles from archival frozen tissue samples and 50-μm sections, revealing the activities of disease-associated DNA elements in distinct human brain structures. The Omni-ATAC protocol enables the interrogation of personal regulomes in tissue context and translational studies.
The mapping of regulatory landscapes that control gene expression and cell state has become a widespread area of interest. Recent methodological advances, such as the advent of the assay for transposase-accessible chromatin by sequencing1 (ATAC-seq) and the application of DNase hypersensitivity sequencing (DNase-seq) to low cell numbers2, have enabled the generation of high-fidelity chromatin accessibility profiles for a variety of cell types3–9. However, certain cell types and tissues require individualized protocol optimizations10,11, making data difficult to compare across multiple studies. To this end, we report an improved, broadly applicable ATAC-seq protocol, called Omni-ATAC, that is suitable for diverse cell lines, tissue types, and archival frozen samples while simultaneously improving data quality across all cell types and cell contexts tested (Supplementary Protocol 1).
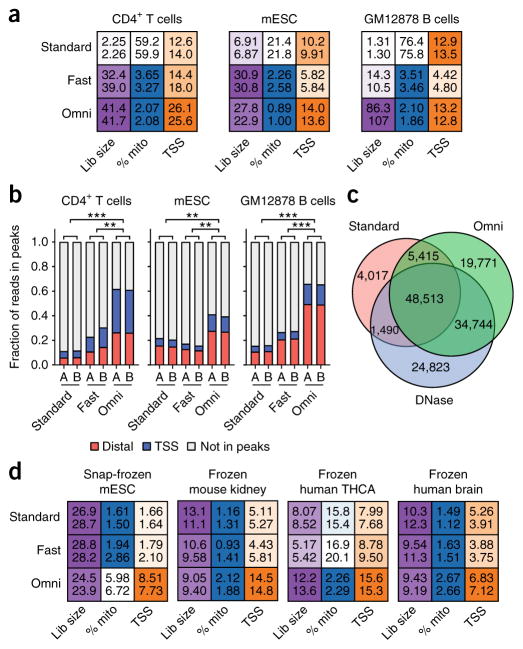
Systematic protocol alterations lead to stepwise improvements in ATAC-seq data quality while maintaining the simplicity of the standard ATAC-seq protocol (Supplementary Fig. 1a). These improvements include (i) the use of multiple detergents (such as NP40, Tween-20, and digitonin) to improve permeabilization across a wide array of cell types and to remove mitochondria from the transposition reaction, (ii) a post-lysis wash step using Tween-20 to further remove mitochondria and to increase the complexity of the library, and (iii) the use of phosphate-buffered saline (PBS) in the transposition reaction to increase the signal-to-background ratio (Supplementary Fig. 1a and Supplementary Note). The ATAC-seq data generated using the Omni-ATAC protocol are consistent with previously published standard ATAC-seq1 (R = 0.73), Fast-ATAC11 (R = 0.88), and DNase-seq12 (R = 0.72) measurements in GM12878 B cells and CD4+ T cells (Supplementary Figs. 1b–i and 2a–f). However, as compared to the standard ATAC-seq protocol, the Omni-ATAC protocol lowers sequencing costs by generating 13-fold fewer sequencing reads that map to mitochondrial DNA, and it improves data quality by yielding threefold higher percentage of reads that map to peaks of chromatin accessibility and by yielding a 15-fold greater number of unique fragments per input cell (median values from n = 14 cell types or contexts; all values determined from 5 million random aligned de-duplicated reads; Fig. 1a, Supplementary Fig. 3a, and Supplementary Table 1). Of the sequencing reads that map to known peaks, the Omni-ATAC protocol generated a higher percentage of both reads that mapped to promoters (defined as sequences within 500 bp of a transcriptional start site (TSS)) and reads that mapped to distal elements (defined as sequences that are more than 500 bp away from a TSS), as compared to those generated by the standard ATAC-seq, Fast-ATAC, and DNase-seq methodologies (Fig. 1b and Supplementary Fig. 3b). Because there is more information per sequencing read, the Omni-ATAC protocol identifies as many or more peaks with consistently higher significance at constant sequencing depth than previously published standard ATAC-seq, Fast-ATAC, and DNase-seq methodologies (Supplementary Fig. 3c–e). Of the peaks identified by at least two methods, 53.8% are identified by all methods, 38.5% are not identified by the standard ATAC-seq method, 6.0% are not identified by DNase-seq, and 1.7% are not identified by the Omni-ATAC protocol (Fig. 1c). This demonstrated that both the Omni-ATAC protocol and DNase-seq were able to identify a substantial number of peaks that were neither identified nor showed robust signal using the standard ATAC-seq protocol (Supplementary Fig. 3f–h). Stronger signals could clearly be observed in sequencing tracks that were derived from equivalent numbers of nonduplicate, aligned reads (Supplementary Fig. 3i–l), demonstrating that the Omni-ATAC protocol produces accessibility data with a substantially higher signal-to-background ratio than alternative tested methods.
Figure 1.
Comparison of the Omni-ATAC protocol to standard ATAC-seq or Fast-ATAC. (a) Heat-map-based representation of ATAC-seq quality control metrics, including estimated library size (“Lib size”; purple), percentage of reads mapping to mitochondrial DNA (“% mito”; blue), and enrichment of signal at TSSs (orange) (Online Methods). Deeper color is used to depict the most desirable value of each statistic and ranges following a linear scale starting at 0 (white) and ending at the maximum value for the given cell type. In the case of the percentage of reads that map to mitochondrial DNA (% mito), the color scale starts at the maximum value (white) and ends at 5% (blue). All values were determined from 5 million random aligned reads. Two technical replicates are shown per sample as numeric values within each box. (b) Fraction of reads in peaks that map to TSSs (±500 bp of TSS) and distal elements (>500 bp from TSS) from libraries generated in this study, using the standard ATAC-seq method (“Standard”), the Fast-ATAC-seq method (“Fast”), or the Omni-ATAC protocol (“Omni”). Each bar represents a single technical replicate. **P < 0.01; ***P < 0.001 by two-tailed unpaired Student’s t-test comparing the fraction of reads in peaks to reads outside of peaks. All values were determined from 5 million random aligned de-duplicated reads. mESC, mouse embryonic stem cell. (c) Overlap of GM12878 peaks called from 60 million reads using standard ATAC-seq, DNase-seq, and Omni-ATAC. All input reads were trimmed to equal lengths (36 bp) before alignment. Data represent the mean of three individual downsampling replicates. (d) Heat-map-based representation of ATAC-seq quality control metrics, including estimated library size, percentage of reads mapping to mitochondrial DNA, and enrichment of signal at TSSs, as shown in a. Deeper color is used to depict the most desirable value of each statistic. All values were determined from 5 million random aligned reads. Two technical replicates are shown per sample.
The Omni-ATAC protocol also improves chromatin accessibility measurements from very small numbers of cells. Previous publications have demonstrated the applicability of standard ATAC-seq to as few as 500 cells1,6. ATAC-seq using the Omni-ATAC protocol in 500 GM12878 B cells led to a significant increase in the signal-to-background ratio and the fraction of reads in peaks, as compared to those in previously published data1 (P < 0.001; Supplementary Fig. 4a–g and Supplementary Table 1). Moreover, ATAC-seq using the Omni-ATAC protocol in 500 GM12878 cells identified more known accessible chromatin regions (Supplementary Fig. 4h) and showed a greater correlation with libraries that were generated from 50,000 cells than using standard ATAC-seq (Supplementary Fig. 4i,j). We also note that the Omni-ATAC protocol has the potential to reduce reaction costs by enabling the use of less amounts of transposase enzyme (Supplementary Fig. 5a–e, Supplementary Table 1, and Supplementary Note).
In addition to improving data quality in previously studied cell types, the Omni-ATAC protocol enabled the generation of robust chromatin accessibility data from cell types and cell contexts that previously proved difficult to assay. For example, standard ATAC-seq and Fast-ATAC generally perform poorly on snap-frozen pellets, requiring the transposition reaction to be performed on fresh cells. This constraint has prevented the broad application of ATAC-seq to banked frozen pellets. However, the Omni-ATAC protocol allowed for the generation of high signal-to-background chromatin accessibility profiles from snap-frozen pellets of just 50,000 cells (Fig. 1d and Supplementary Table 1). Similarly, although both standard ATAC-seq and Fast-ATAC perform poorly on primary human keratinocytes, yielding data with low signal-to-background ratios and a low fraction of reads in peaks, the Omni-ATAC protocol allowed for the generation of high-quality, information-rich chromatin accessibility data under a single consistent protocol (Supplementary Table 1). Overall, the Omni-ATAC protocol simplifies laboratory workflows and enables data acquisition from biomaterials previously deemed unusable for native chromatin accessibility profiling.
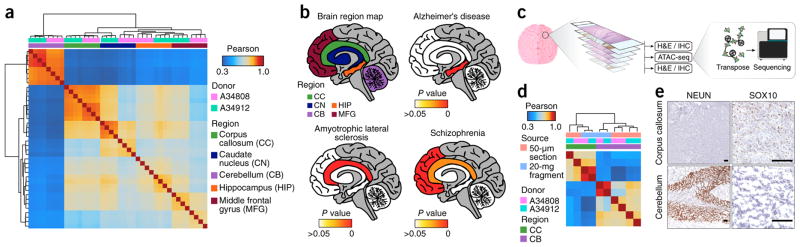
We sought to generate high-quality chromatin accessibility profiles from clinically relevant frozen tissues, such as brain, in which clinical specimens are acquired from rapid autopsy and preserved by snap-freezing, or cancer, in which patients’ tissue samples are often banked as snap-frozen fragments. We first isolated nuclei from 20 mg of frozen tissue via Dounce homogenization followed by density gradient centrifugation (Supplementary Protocol 2 and Supplementary Fig. 6a,b). The Omni-ATAC protocol provided improved signal-to-background ratios and overall data quality in frozen tissues from individuals with thyroid cancer (THCA), frozen post-mortem human brain samples, and a diverse array of frozen mouse tissues, including colon, heart, liver, lung, spleen, and kidney (Fig. 1d and Supplementary Table 1). We then applied the Omni-ATAC protocol to study diverse macro-dissected human brain regions, including the cerebellum, caudate nucleus, corpus callosum, middle frontal gyrus, and hippocampus from two donors (Supplementary Table 2). Comparison of the five brain regions showed strong intra-region correlation across technical replicates and biological donors (Fig. 2a and Supplementary Fig. 6c), which allowed for delineation of region-specific signatures of differentially accessible chromatin and differential transcription factor motif usage that correlated with known brain-specific transcriptional drivers13 (Supplementary Fig. 6d,e).
Figure 2.
Omni-ATAC in defined regions of the post-mortem human brain. (a) Pearson correlation heat map showing sample-by-sample unsupervised clustering on all peaks identified across all regions. Biological donor and brain region are indicated by the color across the top. CC, corpus callosum; CN, caudate nucleus; CB, cerebellum; HIP, hippocampus; MFG, middle frontal gyrus. All regions are represented by four technical replicates per donor. (b) Significance of enrichment of disease-specific GWAS polymorphisms in the uniquely accessible regions of five different brain regions (shown in the top left image) from individuals with the indicated diseases. The empirical P value is depicted colorimetrically with reference to association-based permutations of the GWAS SNPs. Nonsignificant empirical P values are represented in white. (c) Schematic of the experimental strategy for the generation of chromatin accessibility profiles from 50-μm frozen tissue sections with adjacent relevant histological staining. (d) Pearson correlation heat map showing sample-by-sample unsupervised clustering on all peaks. Data derived from the 20-mg tissue fragments represent the mean of four technical replicates from a single biological donor. Data derived from the 50-μm tissue sections show individual technical replicates. (e) Representative images (n = 3) showing histology and anti-NEUN (left) or anti-SOX10 (right) staining of 5-μm frozen sections from the corpus callosum (top) and cerebellum (bottom) immediately adjacent to the 50-μm section used for ATAC-seq shown in Supplementary Figure 8. Scale bars, 100 μm. SOX10 staining is shown at a higher resolution due to a more diffuse staining pattern.
These chromatin accessibility profiles also enabled interpretation of the results of genome-wide association studies (GWAS) that have mapped putative brain-disease-relevant single-nucleotide polymorphisms (SNPs) to noncoding regions (Fig. 2b, Supplementary Fig. 7a–c, and Supplementary Table 3). For example, the hippocampus, a region that has key roles in memory formation and exhibits atrophy in individuals with Alzheimer’s disease14, showed the strongest enrichment for Alzheimer’s- disease-related GWAS SNPs (Fig. 2b and Supplementary Table 3). Similarly, the corpus callosum, a region that is consistently involved in amyotrophic lateral sclerosis (ALS)15, showed significant enrichment for ALS-related GWAS SNPs16 (Fig. 2b, Supplementary Fig. 8a, and Supplementary Table 3).
Given the potential applicability of epigenomic information to clinical diagnosis and prognostication, we developed a methodological framework to combine routine histopathology with submillimeter-precision ATAC-seq. This approach enables collection of multiple thin 5-μm tissue sections for pathology that are immediately adjacent to a single 50-μm tissue section that is used for ATAC-seq Fig. 2c. On 50-μm frozen tissue sections from human brain regions, the Omni-ATAC protocol generated chromatin accessibility profiles comparable to those generated from bulk tissue (Fig. 2d, Supplementary Fig. 8b–f, and Supplementary Table 1). These chromatin accessibility profiles correlated well with adjacent histopathological staining. Regions with high glial cell abundance by SOX10 immunohistochemistry showed increased accessibility near glial-specific genes such as OLIG2 (Fig. 2e, Supplementary Fig. 8b,c,g, and Supplementary Fig. 9a–d). Similarly, regions with high neuronal cell abundance by NEUN immunohistochemistry and Nissl staining showed increased accessibility near neuron-specific genes such as NEUROD1 (Fig. 2e, Supplementary Fig. 8d,g, and Supplementary Fig. 9e–h). Thus, the Omni-ATAC protocol enables the application of epigenomics to clinically relevant specimens, paving the way for assays and diagnostics that leverage the highly informative and cell-type-specific signals of the open chromatin landscape.
Altogether, our data demonstrate that the Omni-ATAC protocol provides a robust, broadly applicable platform for the generation of high-quality and information-rich chromatin- accessibility data. By enabling profiling in a wider array of cell types and cell contexts, we believe the Omni-ATAC protocol will make chromatin accessibility landscapes more universally comparable, thereby facilitating the use of ATAC-seq in difficult cell lines, rare primary cells, and clinically relevant frozen tissues.
ONLINE METHODS
Code availability
All custom code used in this work is available upon request.
Publicly available data used in this work
GM12878 standard ATAC-seq data was obtained as raw fastq files from GEO GSE47753. GM12878 DNase-seq data was obtained as unfiltered and filtered alignments from ENCODE ENCSR000EMT. mESC ES-14 DNase-seq data was obtained as filtered alignments from ENCODE ENCSR000CMW.
Genome annotations
All human data is aligned and annotated for the hg19 reference genome. All mouse data is aligned and annotated for the mm10 reference genome.
Sequencing
All deep sequencing was performed using 2 × 75-bp reads on an Illumina HiSeq4000 instrument that was purchased with funds from the NIH under award number S10OD018220 to the Stanford Functional Genomics Facility. Prior to sequencing on a HiSeq4000 instrument, pooled ATAC-seq libraries were purified using PAGE gel size selection (for fragments >100 bp) to remove excess primers (<100 bp). All low-depth sequencing was performed using 2 × 75-bp reads on an Illumina MiSeq instrument.
Sample acquisition and patient consent
Primary blood cells, primary brain tissue, primary thyroid cancer tissue, and primary keratinocytes were acquired with written and informed consent through Stanford Institutional Review Board (IRB) protocols 27804, 29259, 11977, and 35324, respectively. Human donor sample sizes were chosen to provide sufficient confidence to validate methodological conclusions of the applicability of Omni-ATAC. All animal studies were performed in compliance with Stanford University IACUC and APLAC regulations.
Omni-ATAC protocol
See Protocol Exchange17 or Supplementary Protocols 1 and 2 for a detailed protocol. Cells grown in tissue culture were pretreated with 200 U/ml DNase (Worthington) for 30 min at 37 °C to remove free-floating DNA and to digest DNA from dead cells. This medium was then washed out, and the cells were resuspended in cold PBS. For primary human T cells, cells were sorted using a Becton Dickinson FACS Aria II instrument based on the expression of CD45, CD3, and CD4, as described previously11. After the cells were counted, 50,000 cells were resuspended in 1 ml of cold ATAC-seq resuspension buffer (RSB; 10 mM Tris-HCl pH 7.4, 10 mM NaCl, and 3 mM MgCl2 in water). Cells were centrifuged at 500 r.c.f. for 5 min in a pre-chilled (4 °C) fixed-angle centrifuge. After centrifugation, 900 μl of supernatant was aspirated, which left 100 μl of supernatant. This remaining 100 μl of supernatant was carefully aspirated by pipetting with a P200 pipette tip to avoid the cell pellet. Cell pellets were then resuspended in 50 μl of ATAC-seq RSB containing 0.1% NP40, 0.1% Tween-20, and 0.01% digitonin by pipetting up and down three times. This cell lysis reaction was incubated on ice for 3 min. After lysis, 1 ml of ATAC-seq RSB containing 0.1% Tween-20 (without NP40 or digitonin) was added, and the tubes were inverted to mix. Nuclei were then centrifuged for 10 min at 500 r.c.f. in a pre-chilled (4 °C) fixed-angle centrifuge. Supernatant was removed with two pipetting steps, as described before, and nuclei were resuspended in 50 μl of transposition mix (25 μl 2× TD buffer (recipe in Supplementary Protocol 1) 2.5 μl transposase26 (100 nM final), 16.5 μl PBS, 0.5 μl 1% digitonin, 0.5 μl 10% Tween-20, and 5 μl water) by pipetting up and down six times. Transposition reactions were incubated at 37 °C for 30 min in a thermomixer with shaking at 1,000 r.p.m. Reactions were cleaned up with Zymo DNA Clean and Concentrator 5 columns. The remainder of the ATAC-seq library preparation was performed as described previously18. All libraries were amplified with a target concentration of 20 μl at 4 nM, which is equivalent to 80 femtomoles of product. Minor protocol modifications were used for Omni-ATAC in frozen tissues and in limiting cell numbers. These modifications are outlined in the corresponding Online Methods sections.
Other ATAC-seq methodologies
Standard ATAC-seq and Fast-ATAC were performed as described previously1,11 without additional modifications.
ATAC-seq of frozen cell pellets with the Omni-ATAC protocol
Frozen cell pellets of 50,000 cells were directly transposed using the Omni-ATAC transposition mix without additional lysis and wash steps. However, depending on the cell type and application, it may improve data quality to thaw the cell pellet in Omni-ATAC lysis buffer, wash the cells, and then transpose them.
ATAC-seq with the Omni-ATAC protocol for 500 cells
GM12878 cells were counted five times using a manual hemocytometer. The mean cell count was used to resuspend the cells to a concentration of 500 cells per 100 μl by the addition of PBS. From this diluted cell mixture, 100 μl (500 cells) were deposited into a 0.5-ml DNA LoBind tube (Eppendorf #022431005) containing 400 μl of cold ATAC-seq RSB. This was done to simulate a work-flow involving FACS sorting. These tubes were centrifuged at 500 r.c.f. for 10 min in a pre-chilled (4 °C) fixed-angle centrifuge with 0.6-ml tube adapters. All of the supernatant was removed using the two pipetting steps described above, first by removing 400 μl with a P1000 pipette tip followed by removal of the remaining volume with a P200 pipette tip. We note that a gradual but constant removal of supernatant is crucial and that the final supernatant removal step should be completed in a single motion to avoid disrupting the cell pellet. After supernatant removal, lysis and transposition were performed simultaneously to avoid cell loss, and the total reaction volume was reduced for the same reason. As such, 10 μl of transposition mix (3.3 μl PBS, 1.15 μl water, 5 μl 2× TD Buffer, 0.25 μl 1:10 diluted Tn5 enzyme26, 0.1 μl 1% digitonin, 0.1 μl 10% Tween-20, and 0.1 μl 10% NP40) was added directly to the invisible cell pellet, and the pellet was resuspended by pipetting up and down six times. The transposition reaction was incubated at 37 °C for 30 min in a thermomixer with shaking at 1,000 r.p.m. Note that Tn5 should be diluted in 1× TD Buffer (for example, 5 μl 2× TD Buffer, 4 μl of water, 1 μl Tn5).
ATAC-seq using the Omni-ATAC protocol on nuclei isolated from frozen tissues
See Supplementary Protocol 2 for a detailed protocol. This protocol is highly similar to the INTACT method19 and either protocol can be used for the isolation of nuclei with equivalent results. All of the steps were carried out at 4 °C. A frozen tissue fragment ~20 mg was placed into a pre-chilled 2-ml Dounce homogenizer containing 2 ml of cold 1× homogenization buffer (320 mM sucrose, 0.1 mM EDTA, 0.1% NP40, 5 mM CaCl2, 3 mM Mg(Ac)2, 10 mM Tris pH 7.8, 1× protease inhibitors (Roche, cOmplete), and 167 μM β-mercaptoethanol, in water). Tissue was homogenized with approximately ten strokes with the loose ‘A’ pestle, followed by 20 strokes with the tight ‘B’ pestle. Connective tissue and residual debris were precleared by filtration through an 80-μm nylon mesh filter followed by centrifugation for 1 min at 100 r.c.f. While avoiding the pelleted debris, 400 μl was transferred to a pre-chilled 2-ml round bottom Lo-Bind Eppendorf tube. An equal volume (400 μl) of a 50% iodixanol solution (50% iodixanol in 1× homogenization buffer) was added and mixed by pipetting to make a final concentration of 25% iodixanol. 600 μl of a 29% iodixanol solution (29% iodixanol in 1× homogenization buffer containing 480 mM sucrose) was layered underneath the 25% iodixanol mixture. A clearly defined interface should be visible. In a similar fashion, 600 μl of a 35% iodixanol solution (35% iodixanol in 1× homogenization containing 480 mM sucrose) was layered underneath the 29% iodixanol solution. Again, a clearly defined interface should be visible between all three layers. In a swinging-bucket centrifuge, nuclei were centrifuged for 20 min at 3,000 r.c.f. After centrifugation, the nuclei were present at the interface of the 29% and 35% iodixanol solutions. This band with the nuclei was collected in a 300 μl volume and transferred to a pre-chilled tube. Nuclei were counted after addition of trypan blue, which stains all nuclei due to membrane permeabilization from freezing. 50,000 counted nuclei were then transferred to a tube containing 1 ml of ATAC-seq RSB with 0.1% Tween-20. Nuclei were pelleted by centrifugation at 500 r.c.f. for 10 min in a pre-chilled (4 °C) fixed-angle centrifuge. Supernatant was removed using the two pipetting steps described above. Because the nuclei were already permeabilized, no lysis step was performed, and the transposition mix (25 μl 2× TD buffer, 2.5 μl transposase (100 nM final), 16.5 μl PBS, 0.5 μl 1% digitonin, 0.5 μl 10% Tween-20, 5 μl water) was added directly to the nuclear pellet and mixed by pipetting up and down six times. Transposition reactions were incubated at 37 °C for 30 min in a thermomixer with shaking at 1,000 r.p.m. Reactions were cleaned up with Zymo DNA Clean and Concentrator 5 columns. The remainder of the ATAC-seq library preparation was performed as described previously18.
ATAC-seq using the Omni-ATAC protocol on thin, frozen tissue sections
Omni-ATAC on thin, frozen tissue sections was performed using the same protocol as described for the 20-mg tissue fragments described above, with one modification. To prevent sample loss, 50-μm sections were prepared in a 2-ml Dounce homogenizer containing 500 μl of 1× homogenization buffer. We determined that, despite some bubble formation, the quality of nuclei recovered from homogenization in a 2-ml Dounce with 500 μl is superior to the quality of nuclei recovered when smaller Dounce homogenizers were used.
ATAC-seq data analysis
ATAC-seq data analysis used the following tools and versions: Samtools v1.3, Picard v2.2.1, Bowtie2 v2.2.8, macs2 v2.1.0.20150731, and bedtools v2.23.0. First, Nextera adaptor sequences were trimmed from the reads by using a custom Python script. These reads were aligned to a reference genome using bowtie2, with standard parameters and a maximum fragment length of 2,000. Picard was then used to remove duplicate reads. These de-duplicated reads were then filtered for high quality (MAPQ ≥ 30), nonmitochondrial chromosome, non-Y chromosome, and properly paired (samtools flag 0 × 2) reads.
ATAC-seq library quality control (QC) statistics
Library size was determined from a subsample of 5 million aligned reads before any filtration. Subsampling to 5 million reads was used because current tools to estimate library size are very sensitive to the input read depth. In this way, because the library size estimates were obtained from the same number of input reads, our library size estimates were comparable across assays and cell types, but they may have been an underestimate of the actual library complexity, as only 5 million reads were used. The percentage of reads that aligned to mitochondrial DNA and the enrichment of TSSs were also determined from the same 5-million-read subset or aligned reads. TSS enrichment was determined using hg19 RefSeq TSSs. Enrichment was calculated by counting transposition events in 1-bp bins in the regions ±2,000 bp surrounding all TSSs. The value of each bin was then normalized by dividing by the mean value of the first 200 single-base-pair bins. In this way, the signal from bases –2,000 to –1,800 was used to represent the ‘background’ signal. For the low-depth libraries presented in Supplementary Table 1, only 100,000 aligned reads were used for these metrics.
Footprinting analysis
Meta-footprints were generated for CCCTC-binding factor (CTCF) using pyDNase, a tool based on the Wellington algorithm20. CTCF motif occurrences were filtered for those sites that overlapped with an ATAC-seq peak with a peak score (−log10(P value)) >50. The resulting high-confidence CTCF motif set was used as input to pyDNase dnase_average_profile.py using the –c (stranded) and –A (accounts for Tn5 cut-site offset) options. Meta-footprints were generated from all of the available filtered reads.
Fraction of reads in peaks (FRIP)
The FRIP was determined by using a subsample of 5 million aligned de-duplicated reads before any filtration. For FRIP calculations, called peaks were marked as ‘distal’ if they showed no overlap with ±500 bp from annotated TSSs. Reads that overlapped with regions ±500 bp from TSSs were binned as ‘TSS’ reads. Reads that overlapped with distal peaks were binned as ‘distal’ reads. Reads that did not overlap with either one of these regions were labeled as ‘not in peaks’. Overlap of reads with genomic regions was determined using bedtools intersect, with standard parameters. Reads that mapped to mitochondrial DNA were categorically binned as ‘not in peaks’. For the low-depth libraries presented in Supplementary Table 1, only 100,000 aligned reads were used for these metrics. Peak files used for FRIP calculations are outlined in Supplementary Table 1.
Peak calling and peak scores
All peak calling was performed with macs2 using ‘macs2 callpeak–nomodel–nolambda–keep-dup all–call-summits’. For simulations of peaks called per input read, aligned and de-duplicated BAM files were used without any additional filtering.
Peak overlap and Venn diagrams
For peak overlap of DNase, standard ATAC-seq, and Omni-ATAC-seq, peaks were called using fully processed filtered and merged BAM files that represented the union of all available replicates. These individual peak sets were concatenated, and a union peak set was made as described previously11. Briefly, overlapping peaks that were called in the different data sets were resolved by retaining the peak with the higher macs2 score. In this way, we generated a non-overlapping union peak set containing all of the peaks that were called in the data from all three assays. This union peak set was then intersected individually with each of the peak sets for DNase-seq, standard ATAC-seq, and Omni-ATAC-seq. Each individual intersection represented the total peaks called in each individual assay. These peak sets were then intersected with each other to determine the overlap of peaks called. All intersections were performed using bedtools21 with either the ‘-v’ (unique) and the ‘-u’ (shared) options.
Sequencing tracks
All sequencing tracks were made using the Washington University Epigenome Browser. Sequencing-coverage tracks that were used to compare DNase, standard ATAC-seq, and Omni-ATAC-seq data were generated by subsampling 60 million reads from an aligned and de-duplicated BAM file that had not been additionally filtered. These equal-depth BAM files were then converted to bigwig for visualization. For comparisons involving DNase-seq, all ATAC-seq reads were trimmed to 36 bp to match the single-end 36-bp sequencing reads used in DNase before alignment. The y-axis scale for all sequencing tracks was set to range from 0 to the maximum height among the three data sets. In this way, the heights of the tracks were comparable across techniques, as they were derived from the same number of equal-length input reads. Sequencing tracks that were used to compare 500-cell Omni-ATAC to 500-cell standard ATAC-seq data in GM12878 cells were not normalized. In these visualizations, all pass-filter reads were used to generate sequencing tracks under the assumption that these libraries were sequenced to near-full depth. This assumption is necessary due to differences in the library sizes of the Omni-ATAC and standard ATAC-seq 500-cell libraries. Sequencing tracks related to frozen human brain tissue were all normalized by the total number of reads in the peaks.
Genome-wide association study (GWAS) enrichment
To test for enrichment of GWAS variants in our region-specific uniquely accessible peak sets (Supplementary Fig. 6d), we used all GWAS data sets in the GRASP database (n = 178)22. The GWAS SNPs were pruned to contain no variants in linkage disequilibrium by keeping the most significant P value where there were multiple linked variants for the same trait. We kept only GWAS with at least 900 SNPs, after pruning, in the analysis for sufficient quality to calculate an enrichment. The set of pruned SNPs was then expanded to all linked variants with r2 ≥ 0.8 for individuals of European descent for all further analysis.
We performed a rank-based enrichment of GWAS variants in each set of upregulated differential peaks, and we merged broad and narrow peaks of each brain region. We segmented each GWAS study into bins that represented decreasing tiers of significance. We set a minimum bin size of 50 and filled the first bin with the 50 most significantly associated variants for each study. We then filled the next bins with 2×50, 4×50 and 8×50 variants and then segmented the remaining variants into bins at the four quartiles of the remaining P value distribution. We used the pruned set of SNPs for setting the bin thresholds. We then computed the rank fold-change enrichment of ATAC peaks across the segmented GWAS23. For each bin, we computed the fraction of GWAS variants that was less than or equal to the bin’s P value threshold that overlapped the peaks for each brain region. We calculated the fold-change enrichment by dividing this fraction by the fraction of all GWAS variants of any significance level overlapping our regions. Baseline enrichment of 1 indicates no change from the base rate of overlap of all significant and nonsignificant variants in the study. An enrichment <1 meant that the most significant variants were depleted relative to the baseline value, and any value >1 indicated significant enrichment of variants. To compute the significance of these enrichments, we permuted the P value associated with each GWAS SNP in the study 200 times and recomputed the enrichment relative to the baseline value. The empirical P value indicated the number of permuted studies where the true study had a greater enrichment for the most significant bin of GWAS hits.
Correlation analyses
Pearson correlation heat maps were generated using variance-stabilizing transformed read counts via DESeq2 (ref. 24). Briefly, read counts were determined for all called peaks using bedtools multicov and then quantile-normalized and rounded to the nearest read count. This normalized count matrix was input to DESeq2. Plots of sample-by-sample correlation at all peaks were generated based on the quantile-normalized read counts.
ATAC-seq peak and transcription factor (TF) analyses
Brain-region-unique peaks were identified using quantile-normalized read counts. Briefly, peaks were identified as unique if they were found to be more than two s.d. away from the mean of all other brain regions. TF deviation analysis was performed as described previously25. Enrichment of TF motifs in ATAC-seq peak regions was also performed using the hypergeometric optimization of motif enrichment (Homer) algorithm, with standard parameters.
Histology and immunohistochemistry
Histology and immunohistochemistry were performed according to the manufacturers’ protocols. Anti-NEUN (ab177487) and anti-SOX10 (ab212843) were purchased from Abcam.
Transposase production
All of the transposase used in this work was produced and prepared as described previously26.
Cell lines
All of the cell lines used in this study were purchased from the ATCC or DSMZ. Where possible, cell lines were validated by comparison to published sequencing data or by in-house genotyping with comparison to the Cancer Cell Line Encyclopedia. Cell lines were tested for mycoplasma contamination upon receipt and periodically thereafter, but not before each experiment.
Statistical analysis
All statistical tests performed are included in the figure legends where relevant.
Data availability
All raw sequencing files generated in this work are available through the Sequencing Read Archive via BioProject PRJNA380283 or SRA SRP103230.
Supplementary Material
Supplementary Figure 1. Comparison of technical replicate consistency between and across the standard ATAC-seq method, DNase-seq, and Omni-ATAC.
(a) Heatmap-based representation of ATAC-seq quality control metrics including library size (purple), percent of reads mapping to mitochondrial DNA (blue), and enrichment of signal at TSSs (orange) for each optimization made to produce the Omni-ATAC protocol. Deeper color is used to depict the most desirable value of each statistic. All values were determined from 100,000 random aligned reads. Data represents the average of two technical replicates per square. (b–d) Correlation of technical replicates from GM12878 ATAC-seq data generated using (b) the Omni-ATAC protocol, (c) the standard ATAC-seq protocol, and (d) DNase-seq. Data for standard ATAC-seq and DNase-seq were obtained from publicly available data. Each dot represents an individual peak. Peaks analyzed were derived from a union peak set using data from all three methods. Only 50,000 random peaks with more than 5 reads are shown. R value indicated at the top of each plot. (e,f) Correlation of GM12878 ATAC-seq data generated using (e) Omni-ATAC protocol and (f) the standard ATAC-seq protocol with DNase-seq as in (b–d). (g) Pearson correlation heatmap showing sample by sample unsupervised clustering on all peaks identified across all technical replicates from each method. Each sample represents an individual technical replicate. (h,i) CTCF meta-footprint across the top 20,000 ATAC-seq peaks containing a CTCF motif in data from GM12878 cells derived using (h) the standard ATAC-seq protocol and (i) the Omni-ATAC protocol. Data for standard ATAC-seq method derived from published data1.
Supplementary Figure 2. Comparison of Omni-ATAC to the Fast-ATAC method in primary human T cells.
(a–c) Correlation of technical replicates from primary human CD4+ T cells cells using (a) the Omni-ATAC protocol, (b) the Fast-ATAC protocol, and (c) the standard ATAC-seq protocol. All data shown was generated in this study from the same sample of T cells. Each dot represents an individual peak. Peaks analyzed were derived from a union peak set using data from all three methods. Only 50,000 random peaks with more than 5 reads are shown. R value indicated at the top of each plot. (d,e) Correlation of ATAC-seq data generated using the Omni-ATAC protocol with data generated using (d) the Fast-ATAC protocol or (e) the standard ATAC-seq protocol in CD4+ T cells as in (a–c). (f) Pearson correlation heatmap showing sample by sample unsupervised clustering on all peaks identified across all technical replicates of CD4+ T cells from each method.
Supplementary Figure 3. Comparison of Omni-ATAC to standard ATAC-seq and DNase-seq at the level of peaks.
(a) The number of PCR cycles required to amplify ATAC-seq libraries to an absolute yield of 80 femtomoles (20 ul at 4 nM) using the standard ATAC-seq protocol, the Fast-ATAC protocol, and the Omni-ATAC protocol. N=28 for each box chart, representing 2 technical replicates from 14 distinct cell types and cell contexts (Supplementary Table 1). (b) Fraction of reads in peaks mapping to TSSs (+/− 500 bp of TSS) and distal elements (>500 bp from TSS) from GM12878 libraries generated in this study and from public repositories using the standard ATAC-seq protocol (“Standard”), the Fast-ATAC-seq protocol (“Fast”), the Omni-ATAC-seq protocol (“Omni-ATAC”), and DNase-seq11 (“DNase”). Each bar represents the mean of 2 technical replicates with the exception of the previously published ATAC-seq data which had 4 technical replicates. Error bars represent standard deviation. **p < 0.01, ***p < 0.001 by two-tailed unpaired students t-test comparing the fraction of reads in peaks to reads outside of peaks. All values were determined from 5 million random aligned de-duplicated reads. (c) Histogram comparing the distribution of macs2 peak scores (−log10(pvalue of peak call)) from peaks called from 20 million randomly selected aligned de-duplicated reads in GM12878 cells as in (d). (d,e) In silico simulation of peak calling from variable read depth using data derived from (d) GM12878 cells with the standard ATAC-seq protocol, DNase-seq, and the Omni-ATAC protocol or (e) CD4+ T cells with the standard ATAC-seq protocol, the Fast-ATAC protocol, and the Omni-ATAC protocol. Input reads were randomly selected from a pool of properly aligned, non-duplicate reads. All input reads were trimmed to equal length (36 bp) prior to alignment to match DNase-seq data. GM12878 data from the standard ATAC-seq protocol and DNase-seq were obtained from publicly available sources. All data for CD4+ T cells was generated from a single donor. Data points represent the mean of three individual down-sampled technical replicates. (f–h) Signal at peaks called as “unique” in the analysis presented in Figure 1c for (f) the standard ATAC-seq protocol, (g) the Omni-ATAC protocol, and (h) DNase-seq. Peak score is calculated in the same way as the TSS enrichment score but for a different set of specified peaks. (i–l) Input read normalized sequencing tracks at a control locus (i,j) and a B cell-related locus (k,l) from GM12878 cells derived using standard ATAC-seq, DNase-seq, and Omni-ATAC. In each case, 60 million properly aligned, non-duplicate reads were used to allow for direct comparison. All tracks in each panel are shown on the same y-axis scale. Tracks shown in (j,l) depict the same regions shown in (i,k) but with a reduced y-axis scale to show the difference in signal-to-background in data generated using the Omni-ATAC protocol compared to the standard ATAC-seq protocol and DNase-seq. Peaks that pass above the 10 read y-axis limit are truncated at y=10. All input reads were trimmed to equal length (36 bp) prior to alignment to match DNase-seq. Regions shown represent (i,j) chr19:36,097,939–36,272,939, and (k,l) chr9:36,373,966–37,494,500.
Supplementary Figure 4. Omni-ATAC outperforms the standard ATAC-seq method in situations requiring low cell numbers.
(a,b) Sequencing tracks of ATAC-seq data at (a) a control locus and (b) the PAX5 locus derived from the standard ATAC-seq protocol and the Omni-ATAC protocol using 500 GM12878 cells. No normalization was performed. All available filtered reads are shown under the assumption that the libraries have been sequenced to relatively complete depth. Each track represents an individual technical replicate. Data for standard ATAC-seq from Buenrostro et al. 20131. Region shown represents (a) chr19:36,097,939–36,272,939 or (b) chr9:36,373,966–37,494,500. (c) The total raw reads, filtered reads, and the percent duplication (how completely the libraries have been sequenced) are shown for each library to the right. (d,e) Sequencing tracks shown in (a,b) but with a y-axis scale of 5 to show the difference in signal-to-background in data derived using the Omni-ATAC protocol compared to the standard ATAC-seq protocol. Peaks that pass above the 5 read y-axis limit are truncated at y=5. Each track represents an individual technical replicate. Region shown represents (d) chr19:36,097,939–36,272,939 or (e) chr9:36,373,966–37,494,500. (f) Fraction of reads in peaks mapping to TSSs (+/− 500 bp of TSS) and distal elements (>500 bp from TSS) from the libraries shown in (a–e). Each bar represents the mean of 3 technical replicates. Error bars represent standard deviation. ***p<0.001 by two-tailed unpaired students t-test. Data for standard ATAC-seq from Buenrostro et al. 20131. (g) Metaplot showing enrichment of reads at TSSs in data derived from 500 GM12878 cells using Omni-ATAC (N=3) compared to published data using standard ATAC-seq1 (N=3). (h) Overlap of 500 cell GM12878 peaks called from all available reads using the standard ATAC-seq protocol and the Omni-ATAC protocol. Numbers represent the mean of three individual technical replicates in each case. Overlap was performed with a union set of GM12878 peaks identified by DNase-seq, standard ATAC-seq, and Omni-ATAC. Data for standard ATAC-seq from Buenrostro et al. 20131. (i) Correlation of Omni-ATAC data from 500 GM12878 cells with Omni-ATAC data from 50,000 GM12878 cells. Each dot represents an individual peak. Peaks analyzed were derived from a union peak set using data from the standard ATAC-seq method, DNase-seq, and Omni-ATAC. Only 50,000 random peaks with more than 5 reads are shown. R value indicated at the top of the plot. (j) Pearson correlation heatmap showing sample by sample unsupervised clustering on all peaks identified across all technical replicates of 500 and 50,000 GM12878 cells. Data for standard ATAC-seq for both 500 cell and 50,000 cell GM12878 data from Buenrostro et al. 20131. DNase-seq was performed on an unknown number of cells.
Supplementary Figure 5. Omni-ATAC enables the use of less amounts of transposase without decreasing data quality.
(a) Heatmap-based representation of ATAC-seq quality control metrics including library size, percent of reads mapping to mitochondrial DNA, and enrichment of signal at TSSs. Deeper color is used to depict the most desirable value of each statistic. All values were determined from 5 million random aligned reads. Data represents two technical replicates per square. (b–d) Correlation of Omni-ATAC data generated with varying amounts of transposase enzyme. Technical replicates using (b) 100% Tn5 transposase input and (c) 20% transposase input are shown alongside (d) the correlation of 100% Tn5 transposase input with 20% Tn5 transposase input. Peaks analyzed were derived from DNase-seq data. Only 50,000 random peaks with more than 5 reads are shown. R value indicated at the top of each plot. (e) Fraction of reads in peaks mapping to TSSs (+/− 500 bp of TSS) and distal elements (>500 bp from TSS) for Omni-ATAC data performed with various amounts of Tn5 transposase input. Peak set used was derived from DNase-seq data. Each bar represents the mean of 2 technical replicates. Error bars represent standard deviation. NS = not significant.
Supplementary Figure 6. Application of Omni-ATAC to frozen post-mortem human brain samples yields robust data.
(a) Representative phase contrast image of nuclei isolated from frozen post-mortem human brain. Scale bar represents 10 μm. Experiment was repeated 5 times with 5 total images collected. (b) Representative DAPI staining of the nuclei shown in (a). Scale bar represents 10 μm. (c) Three-dimensional principal component analysis showing the first 3 principal components from ATAC-seq data generated with the Omni-ATAC protocol in 5 different brain regions from 2 different biological donors, each with 2 technical replicates. Different brain regions are indicated by color. CC = corpus callosum; CN = caudate nucleus; CB = cerebellum; HIP = hippocampus; MFG = middle frontal gyrus. (d) Differentially-accessible peaks defined as being at least 2 standard deviations away from the mean of all other regions. Transcription factors whose binding motifs are identified as enriched using the Hypergeometric Optimization of Motif Enrichment (Homer) algorithm in the accessible chromatin regions of each brain region are shown to the right. (e) Transcription factor deviation of all technical replicates and biological donors. TFs enriched in each cluster are shown to the right. Color scale represents the minimum and maximum of each row. Each row represents an individual TF motif. Each column represents an individual replicate of ATAC-seq using the Omni-ATAC protocol.
Supplementary Figure 7. GWAS analysis of ATAC-seq data from different brain regions provides key insights into the ontogeny of various brain diseases.
(a) Box plot of the minimum empirical p value from all brain regions for each GWAS SNP set assayed. *p < 0.05 by two-tailed unpaired students t-test. (b) Significance of enrichment of disease-specific GWAS polymorphisms in the uniquely-accessible regions of the 5 different brain regions shown in Fig. 2b. The empirical p value is depicted colorimetrically with reference to association-based permutations of the GWAS SNPs. (c) Heatmap representation of the enrichment (left) and significance (right) of the enrichment of GWAS SNPs in the uniquely-accessible regions of each brain region. For enrichment, 0 values are represented colorimetrically the same as a log2(enrichment) of -3. Regions without significant associations are shown as white in the p value heatmap.
Supplementary Figure 8 The Omni-ATAC protocol enables generation of robust ATAC-seq data from 50-μm-thick frozen tissue sections.
(a) Depth-normalized sequencing tracks surrounding amyotrophic lateral sclerosis disease-associated SNP rs7175096. Tracks were normalized according to the total number of reads in peaks. Each individual replicate (N=4 technical replicates for each of 2 donors per tissue fragment, N=1 technical replicate for each of 2 donors for corpus callosum sections, and N=2 technical replicates for each of 2 donors for cerebellum sections) is displayed with 25% transparency to show consistency among technical replicates and biological donors. Region shown represents chr15:92,390,775–92,470,760. Data derived from 20 mg tissue fragments and 50 μm tissue sections are represented by a cartoon image to the right. (b–d) Depth normalized sequencing tracks from all replicates from all regions comparing ATAC-seq data generated using the Omni-ATAC protocol in 50 μm tissue sections and larger 20 mg tissue fragments. Each individual replicate (N=4 technical replicates for each of 2 donors per tissue fragment, N=1 technical replicate for each of 2 donors for corpus callosum sections, and N=2 technical replicates for each of 2 donors for cerebellum sections) is displayed with 25% transparency to show consistency among technical replicates and biological donors. Regions shown represent (c) chr2:182,378,211–182,585,768, (d) chr21:34,391,306–34,455,298, and (e) chr22:38,347,790–38,398,955. (e–f) Correlation of ATAC-seq data from 50 μm sections of (f) corpus callosum or (g) cerebellum with ATAC-seq data generated from a nearby 20 mg fragment of the same brain region from the same donor. Each dot represents an individual peak from a union set of peaks called from all technical replicates and biological donors of data derived from tissue fragments from the same brain region. Only 50,000 random peaks with more than 5 reads are shown. (g) Histology and immunohistochemistry of 5 μm frozen sections from cerebellum and corpus callosum immediately adjacent to the 50 μm section used for ATAC-seq shown in Figure 2d–e. Stains from left to right: hematoxylin and eosin (H&E), Nissl. Scale bar represents 100 μm in each image.
Supplementary Figure 9. Histology of tissue immediately adjacent to a 50-μm section used for ATAC-seq.
(a,e) Hematoxylin and eosin staining of corpus callosum and cerebellum. Scale bar represents 1 mm. (b,f) Nissl staining of corpus callosum and cerebellum. Scale bar represents 1 mm. (c,g) Anti-NEUN immunohistochemistry of corpus callosum and cerebellum. Scale bar represents 1 mm. (d,h) Anti-SOX10 immunohistochemistry of corpus callosum and cerebellum. Scale bar represents 100 μm. All images represent same tissue shown in Figure 2d.
Supplementary Table 2 - Clinical characteristics of brain donors
Acknowledgments
We thank the Stanford Alzheimer’s Disease Research Center (NIH P50 AG047366; to V. Henderson), the Pacific Udall Center for Excellence in Parkinson’s Disease Research (NIH P50 NS062684; T.J.M.), and their participants for donating samples for research. We also thank J. Coller and X. Ji for sequencing assistance, E. Plowey and D. Channappa for tissue preparation, and P. Chu and A. Grewall for histology assistance. This work was supported by a grant from the Leukemia & Lymphoma Society Career Development Program (M.R.C.), US National Institutes of Health (NIH) training grant R25CA180993 (M.R.C.), NIH grants P50-HG007735 (H.Y.C. and W.J.G.), UM1HG00943 (W.J.G.), and U19AI057266 (W.J.G.), National Institute on aging grant RF1 AG053959 (T.J.M.), the Rita Allen Foundation (W.J.G.), the Human Frontier Science Program (W.J.G.), the National Science Foundation Graduate Research Fellowship Program (A.E.T.), and a US Department of Defense National Defense Science and Engineering Graduate (NDSEG) Fellowship (N.A.S.-A.).
Footnotes
Note: Any Supplementary Information and Source Data files are available in the online version of the paper.
AUTHOR CONTRIBUTIONS
M.R.C. and H.Y.C. conceived the project; M.R.C., E.G.H., and A.T.S. performed all of the experiments, with help from M.R.M. and A.C.C.; S.W.C. produced the Tn5 transposase complex that was used in all of the experiments; P.G.G. performed all of the GWAS analysis, with guidance and supervision from A. Kundaje; M.R.C. and A.J.R. performed all other data analysis; M.R.C., A.E.T., S.V., and N.A.S.-A., developed methods for the isolation of nuclei from frozen tissues; B.W., A. Kathiria, V.I.R., and W.J.G. provided protocol expertise and recommendations; K.S.M. and T.J.M. oversaw all brain tissue acquisition and processing; M.K. and L.A.O. oversaw acquisition and processing of thyroid cancer tissue; A.J.R. and P.A.K. oversaw acquisition and processing of primary foreskin keratinocytes; and M.R.C., E.G.H., W.J.G., and H.Y.C. wrote the manuscript with input from all authors.
COMPETING FINANCIAL INTERESTS
The authors declare competing financial interests: details are available in the online version of the paper.
A Life Sciences Reporting Summary is available.
References
- 1.Buenrostro JD, Giresi PG, Zaba LC, Chang HY, Greenleaf WJ. Nat Methods. 2013;10:1213–1218. doi: 10.1038/nmeth.2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jin W, et al. Nature. 2015;528:142–146. doi: 10.1038/nature15740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Roadmap Epigenetics Consortium. et al. Nature. 2015;518:317–330. [Google Scholar]
- 4.Koues OI, et al. Cell. 2016;165:1134–1146. doi: 10.1016/j.cell.2016.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shih HY, et al. Cell. 2016;165:1120–1133. doi: 10.1016/j.cell.2016.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wu J, et al. Nature. 2016;534:652–657. doi: 10.1038/nature18606. [DOI] [PubMed] [Google Scholar]
- 7.Gray LT, et al. eLife. 2017;6:e21883. doi: 10.7554/eLife.21883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Scott LJ, et al. Nat Commun. 2016;7:11764. doi: 10.1038/ncomms11764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xu J, et al. Nat Genet. 2017;49:377–386. doi: 10.1038/ng.3769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bao X, et al. Genome Biol. 2015;16:284. doi: 10.1186/s13059-015-0840-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Corces MR, et al. Nat Genet. 2016;48:1193–1203. doi: 10.1038/ng.3646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.The ENCODE Project Consortium. Nature. 2012;489:57–74. [Google Scholar]
- 13.Konopka G, et al. Neuron. 2012;75:601–617. doi: 10.1016/j.neuron.2012.05.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Henneman WJP, et al. Neurology. 2009;72:999–1007. doi: 10.1212/01.wnl.0000344568.09360.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Filippini N, et al. Neurology. 2010;75:1645–1652. doi: 10.1212/WNL.0b013e3181fb84d1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jones AR, et al. Neurobiol Aging. 2013;34:2234.e1–2234.e7. doi: 10.1016/j.neurobiolaging.2013.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Corces MR, et al. Protocol Exchange. 2017 http://dx.doi.org/10.1038/protex.2017.096.
- 18.Buenrostro JD, Wu B, Chang HY, Greenleaf WJ. Curr Protoc Mol Biol. 2015;109:21.29.1–21.29.9. doi: 10.1002/0471142727.mb2129s109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mo A, et al. Neuron. 2015;86:1369–1384. doi: 10.1016/j.neuron.2015.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Piper J, et al. Nucleic Acids Res. 2013;41:e201. doi: 10.1093/nar/gkt850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Quinlan AR, Hall IM. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Leslie R, O’Donnell CJ, Johnson AD. Bioinformatics. 2014;30:i185–i194. doi: 10.1093/bioinformatics/btu273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maurano MT, et al. Science. 2012;337:1190–1195. doi: 10.1126/science.1222794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Love MI, Huber W, Anders S. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schep AN, Wu B, Buenrostro JD, Greenleaf WJ. bioRxiv. 2017:110346. [Google Scholar]
- 26.Picelli S, et al. Genome Res. 2014;24:2033–2040. doi: 10.1101/gr.177881.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1. Comparison of technical replicate consistency between and across the standard ATAC-seq method, DNase-seq, and Omni-ATAC.
(a) Heatmap-based representation of ATAC-seq quality control metrics including library size (purple), percent of reads mapping to mitochondrial DNA (blue), and enrichment of signal at TSSs (orange) for each optimization made to produce the Omni-ATAC protocol. Deeper color is used to depict the most desirable value of each statistic. All values were determined from 100,000 random aligned reads. Data represents the average of two technical replicates per square. (b–d) Correlation of technical replicates from GM12878 ATAC-seq data generated using (b) the Omni-ATAC protocol, (c) the standard ATAC-seq protocol, and (d) DNase-seq. Data for standard ATAC-seq and DNase-seq were obtained from publicly available data. Each dot represents an individual peak. Peaks analyzed were derived from a union peak set using data from all three methods. Only 50,000 random peaks with more than 5 reads are shown. R value indicated at the top of each plot. (e,f) Correlation of GM12878 ATAC-seq data generated using (e) Omni-ATAC protocol and (f) the standard ATAC-seq protocol with DNase-seq as in (b–d). (g) Pearson correlation heatmap showing sample by sample unsupervised clustering on all peaks identified across all technical replicates from each method. Each sample represents an individual technical replicate. (h,i) CTCF meta-footprint across the top 20,000 ATAC-seq peaks containing a CTCF motif in data from GM12878 cells derived using (h) the standard ATAC-seq protocol and (i) the Omni-ATAC protocol. Data for standard ATAC-seq method derived from published data1.
Supplementary Figure 2. Comparison of Omni-ATAC to the Fast-ATAC method in primary human T cells.
(a–c) Correlation of technical replicates from primary human CD4+ T cells cells using (a) the Omni-ATAC protocol, (b) the Fast-ATAC protocol, and (c) the standard ATAC-seq protocol. All data shown was generated in this study from the same sample of T cells. Each dot represents an individual peak. Peaks analyzed were derived from a union peak set using data from all three methods. Only 50,000 random peaks with more than 5 reads are shown. R value indicated at the top of each plot. (d,e) Correlation of ATAC-seq data generated using the Omni-ATAC protocol with data generated using (d) the Fast-ATAC protocol or (e) the standard ATAC-seq protocol in CD4+ T cells as in (a–c). (f) Pearson correlation heatmap showing sample by sample unsupervised clustering on all peaks identified across all technical replicates of CD4+ T cells from each method.
Supplementary Figure 3. Comparison of Omni-ATAC to standard ATAC-seq and DNase-seq at the level of peaks.
(a) The number of PCR cycles required to amplify ATAC-seq libraries to an absolute yield of 80 femtomoles (20 ul at 4 nM) using the standard ATAC-seq protocol, the Fast-ATAC protocol, and the Omni-ATAC protocol. N=28 for each box chart, representing 2 technical replicates from 14 distinct cell types and cell contexts (Supplementary Table 1). (b) Fraction of reads in peaks mapping to TSSs (+/− 500 bp of TSS) and distal elements (>500 bp from TSS) from GM12878 libraries generated in this study and from public repositories using the standard ATAC-seq protocol (“Standard”), the Fast-ATAC-seq protocol (“Fast”), the Omni-ATAC-seq protocol (“Omni-ATAC”), and DNase-seq11 (“DNase”). Each bar represents the mean of 2 technical replicates with the exception of the previously published ATAC-seq data which had 4 technical replicates. Error bars represent standard deviation. **p < 0.01, ***p < 0.001 by two-tailed unpaired students t-test comparing the fraction of reads in peaks to reads outside of peaks. All values were determined from 5 million random aligned de-duplicated reads. (c) Histogram comparing the distribution of macs2 peak scores (−log10(pvalue of peak call)) from peaks called from 20 million randomly selected aligned de-duplicated reads in GM12878 cells as in (d). (d,e) In silico simulation of peak calling from variable read depth using data derived from (d) GM12878 cells with the standard ATAC-seq protocol, DNase-seq, and the Omni-ATAC protocol or (e) CD4+ T cells with the standard ATAC-seq protocol, the Fast-ATAC protocol, and the Omni-ATAC protocol. Input reads were randomly selected from a pool of properly aligned, non-duplicate reads. All input reads were trimmed to equal length (36 bp) prior to alignment to match DNase-seq data. GM12878 data from the standard ATAC-seq protocol and DNase-seq were obtained from publicly available sources. All data for CD4+ T cells was generated from a single donor. Data points represent the mean of three individual down-sampled technical replicates. (f–h) Signal at peaks called as “unique” in the analysis presented in Figure 1c for (f) the standard ATAC-seq protocol, (g) the Omni-ATAC protocol, and (h) DNase-seq. Peak score is calculated in the same way as the TSS enrichment score but for a different set of specified peaks. (i–l) Input read normalized sequencing tracks at a control locus (i,j) and a B cell-related locus (k,l) from GM12878 cells derived using standard ATAC-seq, DNase-seq, and Omni-ATAC. In each case, 60 million properly aligned, non-duplicate reads were used to allow for direct comparison. All tracks in each panel are shown on the same y-axis scale. Tracks shown in (j,l) depict the same regions shown in (i,k) but with a reduced y-axis scale to show the difference in signal-to-background in data generated using the Omni-ATAC protocol compared to the standard ATAC-seq protocol and DNase-seq. Peaks that pass above the 10 read y-axis limit are truncated at y=10. All input reads were trimmed to equal length (36 bp) prior to alignment to match DNase-seq. Regions shown represent (i,j) chr19:36,097,939–36,272,939, and (k,l) chr9:36,373,966–37,494,500.
Supplementary Figure 4. Omni-ATAC outperforms the standard ATAC-seq method in situations requiring low cell numbers.
(a,b) Sequencing tracks of ATAC-seq data at (a) a control locus and (b) the PAX5 locus derived from the standard ATAC-seq protocol and the Omni-ATAC protocol using 500 GM12878 cells. No normalization was performed. All available filtered reads are shown under the assumption that the libraries have been sequenced to relatively complete depth. Each track represents an individual technical replicate. Data for standard ATAC-seq from Buenrostro et al. 20131. Region shown represents (a) chr19:36,097,939–36,272,939 or (b) chr9:36,373,966–37,494,500. (c) The total raw reads, filtered reads, and the percent duplication (how completely the libraries have been sequenced) are shown for each library to the right. (d,e) Sequencing tracks shown in (a,b) but with a y-axis scale of 5 to show the difference in signal-to-background in data derived using the Omni-ATAC protocol compared to the standard ATAC-seq protocol. Peaks that pass above the 5 read y-axis limit are truncated at y=5. Each track represents an individual technical replicate. Region shown represents (d) chr19:36,097,939–36,272,939 or (e) chr9:36,373,966–37,494,500. (f) Fraction of reads in peaks mapping to TSSs (+/− 500 bp of TSS) and distal elements (>500 bp from TSS) from the libraries shown in (a–e). Each bar represents the mean of 3 technical replicates. Error bars represent standard deviation. ***p<0.001 by two-tailed unpaired students t-test. Data for standard ATAC-seq from Buenrostro et al. 20131. (g) Metaplot showing enrichment of reads at TSSs in data derived from 500 GM12878 cells using Omni-ATAC (N=3) compared to published data using standard ATAC-seq1 (N=3). (h) Overlap of 500 cell GM12878 peaks called from all available reads using the standard ATAC-seq protocol and the Omni-ATAC protocol. Numbers represent the mean of three individual technical replicates in each case. Overlap was performed with a union set of GM12878 peaks identified by DNase-seq, standard ATAC-seq, and Omni-ATAC. Data for standard ATAC-seq from Buenrostro et al. 20131. (i) Correlation of Omni-ATAC data from 500 GM12878 cells with Omni-ATAC data from 50,000 GM12878 cells. Each dot represents an individual peak. Peaks analyzed were derived from a union peak set using data from the standard ATAC-seq method, DNase-seq, and Omni-ATAC. Only 50,000 random peaks with more than 5 reads are shown. R value indicated at the top of the plot. (j) Pearson correlation heatmap showing sample by sample unsupervised clustering on all peaks identified across all technical replicates of 500 and 50,000 GM12878 cells. Data for standard ATAC-seq for both 500 cell and 50,000 cell GM12878 data from Buenrostro et al. 20131. DNase-seq was performed on an unknown number of cells.
Supplementary Figure 5. Omni-ATAC enables the use of less amounts of transposase without decreasing data quality.
(a) Heatmap-based representation of ATAC-seq quality control metrics including library size, percent of reads mapping to mitochondrial DNA, and enrichment of signal at TSSs. Deeper color is used to depict the most desirable value of each statistic. All values were determined from 5 million random aligned reads. Data represents two technical replicates per square. (b–d) Correlation of Omni-ATAC data generated with varying amounts of transposase enzyme. Technical replicates using (b) 100% Tn5 transposase input and (c) 20% transposase input are shown alongside (d) the correlation of 100% Tn5 transposase input with 20% Tn5 transposase input. Peaks analyzed were derived from DNase-seq data. Only 50,000 random peaks with more than 5 reads are shown. R value indicated at the top of each plot. (e) Fraction of reads in peaks mapping to TSSs (+/− 500 bp of TSS) and distal elements (>500 bp from TSS) for Omni-ATAC data performed with various amounts of Tn5 transposase input. Peak set used was derived from DNase-seq data. Each bar represents the mean of 2 technical replicates. Error bars represent standard deviation. NS = not significant.
Supplementary Figure 6. Application of Omni-ATAC to frozen post-mortem human brain samples yields robust data.
(a) Representative phase contrast image of nuclei isolated from frozen post-mortem human brain. Scale bar represents 10 μm. Experiment was repeated 5 times with 5 total images collected. (b) Representative DAPI staining of the nuclei shown in (a). Scale bar represents 10 μm. (c) Three-dimensional principal component analysis showing the first 3 principal components from ATAC-seq data generated with the Omni-ATAC protocol in 5 different brain regions from 2 different biological donors, each with 2 technical replicates. Different brain regions are indicated by color. CC = corpus callosum; CN = caudate nucleus; CB = cerebellum; HIP = hippocampus; MFG = middle frontal gyrus. (d) Differentially-accessible peaks defined as being at least 2 standard deviations away from the mean of all other regions. Transcription factors whose binding motifs are identified as enriched using the Hypergeometric Optimization of Motif Enrichment (Homer) algorithm in the accessible chromatin regions of each brain region are shown to the right. (e) Transcription factor deviation of all technical replicates and biological donors. TFs enriched in each cluster are shown to the right. Color scale represents the minimum and maximum of each row. Each row represents an individual TF motif. Each column represents an individual replicate of ATAC-seq using the Omni-ATAC protocol.
Supplementary Figure 7. GWAS analysis of ATAC-seq data from different brain regions provides key insights into the ontogeny of various brain diseases.
(a) Box plot of the minimum empirical p value from all brain regions for each GWAS SNP set assayed. *p < 0.05 by two-tailed unpaired students t-test. (b) Significance of enrichment of disease-specific GWAS polymorphisms in the uniquely-accessible regions of the 5 different brain regions shown in Fig. 2b. The empirical p value is depicted colorimetrically with reference to association-based permutations of the GWAS SNPs. (c) Heatmap representation of the enrichment (left) and significance (right) of the enrichment of GWAS SNPs in the uniquely-accessible regions of each brain region. For enrichment, 0 values are represented colorimetrically the same as a log2(enrichment) of -3. Regions without significant associations are shown as white in the p value heatmap.
Supplementary Figure 8 The Omni-ATAC protocol enables generation of robust ATAC-seq data from 50-μm-thick frozen tissue sections.
(a) Depth-normalized sequencing tracks surrounding amyotrophic lateral sclerosis disease-associated SNP rs7175096. Tracks were normalized according to the total number of reads in peaks. Each individual replicate (N=4 technical replicates for each of 2 donors per tissue fragment, N=1 technical replicate for each of 2 donors for corpus callosum sections, and N=2 technical replicates for each of 2 donors for cerebellum sections) is displayed with 25% transparency to show consistency among technical replicates and biological donors. Region shown represents chr15:92,390,775–92,470,760. Data derived from 20 mg tissue fragments and 50 μm tissue sections are represented by a cartoon image to the right. (b–d) Depth normalized sequencing tracks from all replicates from all regions comparing ATAC-seq data generated using the Omni-ATAC protocol in 50 μm tissue sections and larger 20 mg tissue fragments. Each individual replicate (N=4 technical replicates for each of 2 donors per tissue fragment, N=1 technical replicate for each of 2 donors for corpus callosum sections, and N=2 technical replicates for each of 2 donors for cerebellum sections) is displayed with 25% transparency to show consistency among technical replicates and biological donors. Regions shown represent (c) chr2:182,378,211–182,585,768, (d) chr21:34,391,306–34,455,298, and (e) chr22:38,347,790–38,398,955. (e–f) Correlation of ATAC-seq data from 50 μm sections of (f) corpus callosum or (g) cerebellum with ATAC-seq data generated from a nearby 20 mg fragment of the same brain region from the same donor. Each dot represents an individual peak from a union set of peaks called from all technical replicates and biological donors of data derived from tissue fragments from the same brain region. Only 50,000 random peaks with more than 5 reads are shown. (g) Histology and immunohistochemistry of 5 μm frozen sections from cerebellum and corpus callosum immediately adjacent to the 50 μm section used for ATAC-seq shown in Figure 2d–e. Stains from left to right: hematoxylin and eosin (H&E), Nissl. Scale bar represents 100 μm in each image.
Supplementary Figure 9. Histology of tissue immediately adjacent to a 50-μm section used for ATAC-seq.
(a,e) Hematoxylin and eosin staining of corpus callosum and cerebellum. Scale bar represents 1 mm. (b,f) Nissl staining of corpus callosum and cerebellum. Scale bar represents 1 mm. (c,g) Anti-NEUN immunohistochemistry of corpus callosum and cerebellum. Scale bar represents 1 mm. (d,h) Anti-SOX10 immunohistochemistry of corpus callosum and cerebellum. Scale bar represents 100 μm. All images represent same tissue shown in Figure 2d.
Supplementary Table 2 - Clinical characteristics of brain donors
Data Availability Statement
All raw sequencing files generated in this work are available through the Sequencing Read Archive via BioProject PRJNA380283 or SRA SRP103230.