Abstract
Background and Aims
Proteomics holds promise for individualizing cancer treatment. We analyzed to what extent the proteomic landscape of human colorectal cancer (CRC) is maintained in established CRC cell lines and the utility of proteomics for predicting therapeutic responses.
Methods
Proteomic and transcriptomic analyses were performed on 44 CRC cell lines, compared against primary CRCs (n=95) and normal tissues (n=60), and integrated with genomic and drug sensitivity data.
Results
Cell lines mirrored the proteomic aberrations of primary tumors, in particular for intrinsic programs. Tumor relationships of protein expression with DNA copy number aberrations and signatures of post-transcriptional regulation were recapitulated in cell lines. The five proteomic subtypes previously identified in tumors were represented among cell lines. Nonetheless, systematic differences between cell line and tumor proteomes were apparent, attributable to stroma, extrinsic signaling and growth conditions. Contribution of tumor stroma obscured signatures of DNA mismatch repair identified in cell lines with a hypermutation phenotype. Global proteomic data showed improved utility for predicting both known drug-target relationships and overall drug sensitivity as compared to genomic or transcriptomic measurements. Inhibition of targetable proteins associated with drug responses further identified corresponding synergistic or antagonistic drug combinations. Our data provide evidence for CRC proteomic subtype-specific drug responses.
Conclusions
Proteomes of established CRC cell line are representative of primary tumors. Proteomic data tend to exhibit improved prediction of drug sensitivity as compared to genomic and transcriptomic profiles. Our integrative proteogenomic analysis highlights the potential of proteome profiling to inform personalized cancer medicine.
Keywords: colorectal cancer, cell lines, proteomics, drug sensitivity
INTRODUCTION
Studies of the genomic and transcriptomic landscapes of human colorectal cancer (CRC), have been instrumental in advancing our understanding of disease biology and the identification of clinically actionable aberrations 1–3. While the major genomic and transcriptomic hallmarks and subtypes of CRC have been defined 4, 5, these explain only part of tumor clinical heterogeneity. The next challenge is to gain a detailed understanding of the dynamic protein pathways involved in normal and disease states, and we have recently characterized the proteome of primary CRCs from patients participating in The Cancer Genome Atlas (TCGA) project, identifying five major proteomic subtypes (Clinical Proteomic Tumor Analysis Consortium (CPTAC) 6). From a therapeutic perspective, most drug targets are proteins rather than nucleic acids, and we have shown that DNA- or mRNA-level measurements are poor predictors of protein abundance 6.
Cancer cell lines are the most commonly utilized model systems in tumor biology and therapy development. Large cancer cell line-based projects, such as NCI-60 7, Cancer Cell Line Encyclopedia (CCLE) 2 and Genomics of Drug Sensitivity in Cancer (GDSC) 3, have used molecularly heterogeneous cancer cell lines to identify stratification biomarkers and signatures for precision medicine. Nonetheless, controversy remains whether cell lines provide an appropriate representation of primary tumors, given the lack of organismal context, different growth conditions, and selection or acquisition of additional aberrations in vitro. Genomic analyses indicate that established cancer cell lines are suitable molecular proxies for primary tumors in many cancer types 2, yet findings at the transcriptomic level have been variable, with data for hepatocellular carcinoma 8 and colorectal cancer (CRC) 9 indicating similarity between cell lines and primary tumors, whilst data for breast cancer suggest pronounced differences 10. Although some global proteomics data sets for cancer cell lines are available 11, 12 there exists no large-scale proteomic study comparing cell lines with primary tumors. It remains unknown whether cancer cell lines are representative of primary tumors at the proteome level and to what extent molecular programs and proteogenomic relationships are maintained in vitro. The relative utility of proteomic data as a predictor of anti-cancer drug responses in comparison to genomic and transcriptomic modalities has not been systematically investigated.
Here, we generated proteomic and transcriptomic data for a panel of 44 human CRC cell lines previously characterized at the genomic level 13. We performed a comprehensive integrative proteogenomic analysis across these 44 cell lines and 95 CRCs and 60 normal tissue biopsies analyzed in our CPTAC project 6 to systematically evaluate cell lines as tumor models. We further integrated cell line proteogenomic data with drug sensitivity measurements to assess the utility of different types of omics data for predicting therapeutic responses and to connect tumor proteomic subtypes to drug sensitivity.
MATERIALS AND METHODS
CRC cell lines and primary tumors
A total of 44 CRC cell lines were studied (Supplementary Table 1, Supplementary Methods). In addition, we retrieved previously published genomic, transcriptomic and proteomic data on 95 primary tumor specimens from 90 CRC patients and proteomics data from 60 normal colon biopsies from 30 patients from our original CPTAC study 6, as well as RNA-Seq data for 48 normal colon and rectum samples deposited by the TCGA (Supplementary Table 2–3).
LC/MS-MS
The protein extraction and tryptic digestion of the frozen cell line pellets were performed as previously described for TCGA CRC specimens 6 (Supplementary Methods). Raw data for the cell lines, database search results, and the two versions of assemblies can be found at the Mass spectrometry Interactive Virtual Environment (MassIVE, ftp://massive.ucsd.edu/MSV000080374).
Transcriptome sequencing
RNA samples from CRC cell lines were extracted from pellets collected at the same time as the samples for proteomics analysis and sequenced to a depth of at least 28 million reads. Reads were subsequently aligned to human genome build Hg19 using Tophat (Supplementary Methods).
SNP microarray analysis
SNP array data on 38 cell lines from our cohort have been published previously 13. SNP array assays on the additional DiFi, GEO, IS1, IS2, IS3 and V9P cells were performed at the Australian Genome Research Facility (AGRF) using CytoSNP-850K v1.1 and processed using OncoSNP v2.18 suite (Supplementary Methods).
Exome-capture sequencing
Whole exome mutation data on 35 CRC cell lines from our cohort have been published previously 13. Additionally, DIFI, GEO, IS1, IS2, IS3, LIM1863, LIM2537, V9P and VAC05 cells were sequenced using the Nextera Rapid Capture Expanded Exome Enrichment Kit (Illumina) on an Illumina HiSeq 2000 System at the AGRF. Sequence alignment and calling of SNVs and INDEL in the absence of matched normal tissue were performed using a hybrid of the GATK Germline and Somatic Best Practice Variant Detection Protocols (Supplementary Methods).
Variant peptide identification and analysis
To identify variant peptides, we derived customized protein sequence databases from matched WES and RNA-Seq data and then performed database searches using these databases for individual samples (Supplementary Methods).
VOOM/LIMMA analysis
Voom/limma analyses were performed using Limma and edgeR R packages, and method sensitivity and specificity for spectral count data were validated using the spike-in data set generated by the 2015 study of the Proteome Informatics Research Group (iPRG) of the Association of Biomolecular Resource Facilities (ABRF) (Supplementary Fig. 1, Supplementary Methods).
Online databases
The Human Protein Atlas, tumor stroma markers, KEGG pathways and GDSC (Genomics of Drug Sensitivity in Cancer) drug sensitivity data were downloaded from online resources (Supplementary Methods).
Correlation analysis
Spearman’s correlation analysis of steady state mRNA and protein abundance, mRNA and protein variation, and relative mRNA-protein abundances required additional normalization steps that are outlined in Supplementary Methods.
Pathway signature identification
To assess whether genes in a given KEGG pathway had differing expression in tumors or cell lines relative to normal colorectal tissue, we modelled the protein or mRNA expression levels (cpm values for quantifiable genes) of pathway members using a linear mixed-effects model (lme4 R package) (Supplementary Methods).
Comparison of the impact of copy number alteration on protein abundance for cell lines and tumors
Evaluation of the association between copy number alteration and protein or mRNA levels were carried out using voom/limma analysis utilizing robust linear regression for gene-level log R ratios against protein or RNA-Seq expression levels (Supplementary Methods).
Comparison of the effect of candidate oncogene-targeting shRNAs on the proliferation of colon cancer cell lines
The shRNA gene level data was downloaded from the Achilles project website (https://portals.broadinstitute.org/achilles/datasets/5/download) and contained eight colon cancer cell lines overlapping with our 44 cell lines. We calculated the Spearman’s correlation between shRNA score and log-transformed DNA copy number data across eight cell lines for each candidate oncogene (Supplementary Methods).
Drug sensitivity studies
Oxaliplatin (Cat# S1224), erlotinib (Cat# S7786) and regorafenib (Cat# S1178) were purchased from Selleck Chemicals. 5-fluorouracil (Cat# F6627) was obtained from Sigma. Cells were seeded into 384-well plates with compounds added to the cells in quadruplicate for 72hr. Cell viability was determined using CellTiter Glo 2 (Supplementary Methods). For drug combination screening in HCT116 cells, 123 drugs were accessed from Compounds Australia, Griffith University, Australia (Supplementary Tables 4–5).
Comparison of omic modalities for prediction of drug sensitivity
Assessment of the utility of proteomic data for drug sensitivity prediction relative to mutation, DNA copy number, and mRNA expression data was undertaken using random forests and five-fold cross-validation for 5-fluorouracil, erlotinib, oxaliplatin, regorafenib and SN-38 over our panel of 44 CRC cell lines (Supplementary Methods).
Cell line proteomic and CMS subtype predictions
To assign cell lines to our previously identified proteomic subtypes 6, the R package pamr (http://CRAN.R-project.org/package=pamr) was used to apply our predefined signature genes from our CPTAC CRC tumor study to the cell line proteomic data (Supplementary Methods).
To assign CMS subtypes to cell lines and a dataset of 5 matched primary and metastatic tumor pairs (deposited in NCBI GEO: GSE90814), we used the CMSclassifier package in R (https://github.com/Sage-Bionetworks/CMSclassifier). (Supplementary Methods).
RESULTS
Proteomic analysis of CRC cell lines
We performed liquid chromatography-tandem mass spectrometry (LC-MS/MS) based shotgun proteomic analyses on 44 established CRC cell lines (Supplementary Table 1, Supplementary Fig. 2), identifying a total of 10,643 distinct peptides (2,548,082 spectra) in an assembly of 7,796 protein groups with a protein-level False Discovery Rate (FDR) of 4% (Supplementary Table 6). To capture protein variants, we further searched customized protein sequence databases derived from matched whole exome sequencing (WES) and RNA-Seq data (Supplementary Tables 7–8). Out of 111,022 non-synonymous SNVs from RNA-Seq and WES data, we observed 1,702 unique variants at the proteomic level including 276 somatic variants reported in the TCGA/COSMIC databases and 952 germline variants listed in the Single Nucleotide Polymorphism Database (dbSNP) (Supplementary data, Supplementary Table 9, Supplementary Fig. 3–4). The sparse detection of non-synonymous SNVs by peptide sequencing is consistent with our previous findings in primary tumors 6, reflecting the partial protein-coding sequence coverage achievable with the current proteomic technology.
Protein inventory concordance between cell line, tumor and normal samples
The cell line proteomic analysis was performed on the same platform previously used for the analysis of the TCGA tumors (n=95) and normal tissues (n=60) in our CPTAC project 6, and analysis of quality control samples across both projects demonstrated high platform stability (Supplementary Fig. 5). To determine the overlap between protein inventories of CRC cell line, tumor and normal colon samples, proteomic data were integrated into a joint assembly of 9,101 protein groups (Supplementary Table 2). The protein inventory of cell lines was highly similar to those from tumor and normal tissues, exhibiting 98.0% and 90.9% overlap, respectively (Supplementary Fig. 6a). 103, 42 and 20 proteins were detected exclusively in cell line, tumor and normal samples, but most of these were low abundance proteins at the threshold of detection (Supplementary Fig. 6b–6d). Notably, proteome analysis detected 48% of the 18,178 protein-coding genes identified in corresponding mRNA datasets including the 44 matched cell line samples, 87 matched tumor samples, and 48 normal samples (Supplementary Table 3), with similar representation of the major Gene Ontology (GO) categories (Supplementary Fig. 7). In the following analyses, we only used robustly quantifiable proteins, i.e., proteins with a spectral count per million (CPM) >20 in ≥20% of samples.
Contribution of stroma components to tumor proteomes
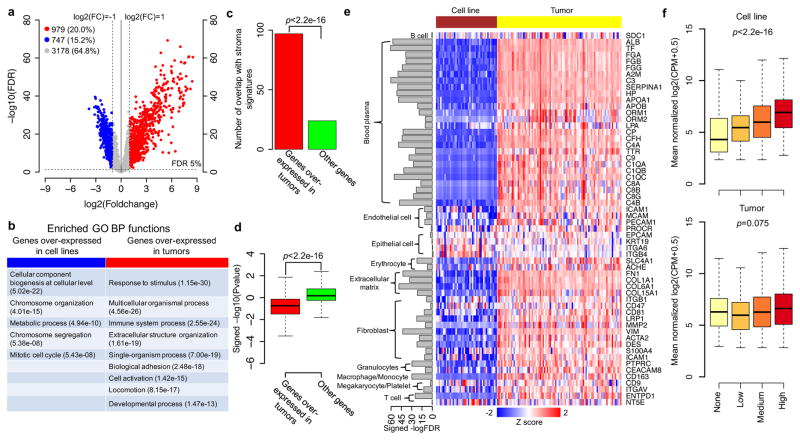
Among the 4,904 quantifiable proteins from the CRC tumor and cell line proteomes, 747 (15.2%) exhibited significantly higher levels in the cell lines, whereas 979 (20.0%) displayed higher levels in the tumors (FDR<5% and fold-change >2, voom/limma, Figure 1a, Supplementary Table 10). Using GO enrichment analysis in WebGestalt 14, cell line-overexpressed proteins were enriched for cell growth and proliferation-related biological processes, such as metabolism and cell cycle, as anticipated for a comparison between in vitro cultured cell lines in log phase growth and primary tumor cells in vivo (Figure 1b, Supplementary Table 11a and Supplementary Fig. 8a). In contrast, tumor-overexpressed proteins were enriched for processes related to immune response, extracellular matrix, and response to extrinsic stimuli (Figure 1b, Supplementary Table 11b, and Supplementary Fig. 7b). The latter proteins also significantly overlapped with previously published cancer-associated fibroblast, leukocyte, or endothelial cell signatures 15 (p<2.2e-16, hypergeometric test, Figure 1c), indicating a substantial contribution of stroma to the tumor proteomes. Indeed, protein abundance for 82.3% of the tumor-overexpressed genes showed a negative correlation with tumor purity scores (ABSOLUTE algorithm 16), in contrast to 38.4% among other genes (p<2.2e-16, two-sided Wilcoxon rank sum test, Figure 1d and Supplementary Table 12). We also compared the mRNA profiles of cell lines and tumors and obtained similar results (Supplementary Table 13–15 and Supplementary Fig. 9–10).
Figure 1. Comparison of protein abundances between CRC cell lines and tumors.
(a) Volcano plot indicating proteins overexpressed in cell lines (blue) or tumors (red) (FDR<5% and fold change>2); other genes are colored in grey. (b) The GO Biological Processes (BP) enriched for proteins overexpressed in cell lines (blue) or tumors (red) identified using WebGestalt 14. (c) Overlap of stroma signatures with genes overexpressed in tumors versus other genes. p value for hypergeometric test. (d) Distributions of the signed -log10 p values (voom/limma) of the associations between protein abundance and tumor purity score for genes overexpressed in tumors versus other genes. p value for Wilcox rank sum test. (e) Heatmap of tumor stroma and epithelial protein marker expression in tumors and cell lines. The bar plot to the left of the heatmap represents the signed -log10 FDR (voom/limma) comparing protein abundances of tumor and cell line samples. (f) Box plots comparing protein abundance measurements for cell lines and tumors against tumor-cell specific IHC scores defined by the Human Protein Atlas. p values for Jonckheere’s trend test.
To characterize which components of the tumor stroma contributed to the tumor-overexpressed gene signature, we interrogated our tumor and cell line data for the expression of relevant stroma markers. Protein markers for blood plasma, extracellular matrix, endothelial cells, erythrocytes, fibroblasts, granulocytes, macrophages/monocytes, megakaryocytes/platelets and T cells were generally overexpressed (FDR<0.05 and fold-change>2, voom/limma) in the CRC samples as compared to the cell lines (Figure 1e, Supplementary Table 16). Analysis of RNA-Seq data additionally identified overexpression for markers of B lymphocytes and natural killer cells (Supplementary Table 17). Markers of the various tumor stroma components identified in the proteomics or RNA-Seq based analyses were verified by immunohistochemistry (IHC) data from the Human Protein Atlas (HPA)17 (Supplementary Fig. 11). In contrast, IHC supported epithelial cell markers (EPCAM, KRT19, ITGA6, ITGB4; Supplementary Fig. 12), displayed similar expression levels (fold-change<2) in the cell lines and tumors (Figure 1e, Supplementary Fig. 9e).
To examine the impact of “contaminating” stroma on tumor proteome profiles, we intersected cell line and tumor data with the tumor-cell specific IHC expression scores from the HPA. Protein abundance measurements in tumor specimens showed only a weak concordance with corresponding IHC expression scores (p=0.075, Jonckheere’s trend test), while cell line data exhibited a high level of concordance (p<2.2e-16) (Figure 1f).
Cell line proteomes reveal intrinsic biology of the hypermutation phenotype
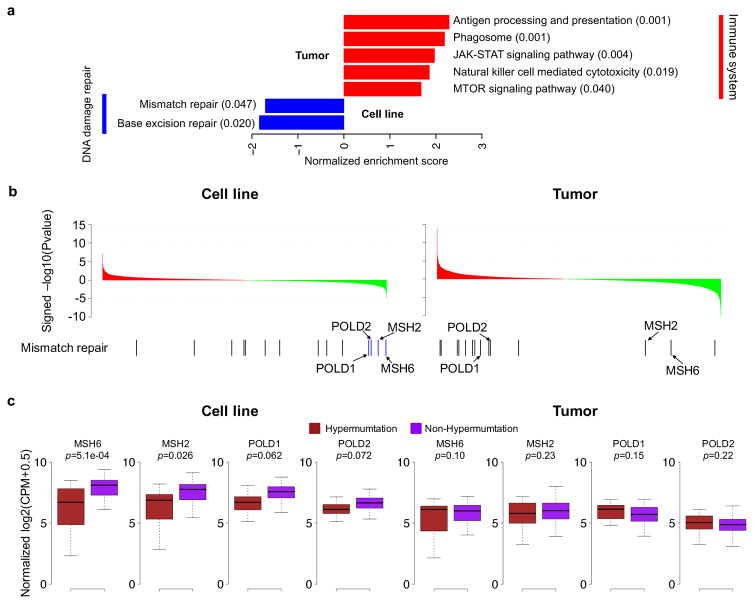
To compare the utility of proteomic data from cell lines against that of tumor samples to elucidate cell-intrinsic molecular mechanisms, we investigated the protein profiles associated with the well-characterized hypermutation phenotype (both cohorts included 19 hypermutated cases; Supplementary Table 1 and Supplementary Fig. 2). Using differential protein expression analysis followed by gene set enrichment analysis (GSEA) (Supplementary Tables 18–20), the DNA mismatch repair pathway was found to be significantly underexpressed in hypermutated cell lines compared with non-hypermutated cell lines (FDR=0.047), but this was not observed in tumors (Figure 2a). Genes contributing to the statistical significance in the cell line data (blue bars, GSEA leading edge, Figure 2b) included the mismatch repair proteins MSH2 and MSH6, as well as two subunits of DNA polymerase delta, POLD1 and POLD2 (Figure 2b and 2c). Loss of MSH2 and MSH6 expression are diagnostic of defective DNA mismatch repair, in particular for CRC associated with Lynch syndrome 18, and loss of POLD1 proof-reading function by somatic mutation in the exonuclease domain is implicated in causing tumor hypermutation phenotypes 19. In contrast, tumor data associated the hypermutation phenotype with strong immune system signatures (Figure 2a), consistent with documented high levels of lymphocyte infiltration in hypermutated cases 20. These results were replicated when examining mRNA-expression (Supplementary Table 21–23, Supplementary Fig. 13). Notably, MLH1 protein, loss of which underlies most hypermutated sporadic CRCs 18, was not detected in the proteomics data but was observed in the RNA-Seq data, with a greater dynamic signal range in cell lines relative to tumor samples (p=6.85e-05, Levene’s test).
Figure 2. Pathways associated with the hypermutation phenotype in CRC cell lines and tumors.
(a) GSEA enrichment scores for significant KEGG pathways in cell lines and tumors. Red and blue bars represent the positively and negatively enriched pathways, respectively. The numbers in the parentheses represent the enriched FDR of the pathways. (b) Genes sorted by differential expression between hypermutated and non-hypermutated samples. Red and green represent overexpression in hypermutated and non-hypermutated samples, respectively. Bars in the bottom panel represent genes annotated to the mismatch repair pathway with blue bars indicating the leading-edge genes reported by GSEA. (c) Comparison of protein abundance between hypermutated and non-hypermutated samples for the leading-edge genes identified from the cell line data.
Tumor pathway signatures of post-transcriptional regulation are maintained in cell lines
We previously reported that mRNA and protein levels are only modestly correlated in the TCGA CRC cohort suggesting a major impact of post-transcriptional regulation 6, although omics analyses in tumor samples were performed on different specimen sections. To evaluate the relative contributions from biological and specimen variability, we compared mRNA-protein correlations in tumors samples with those from cell lines.
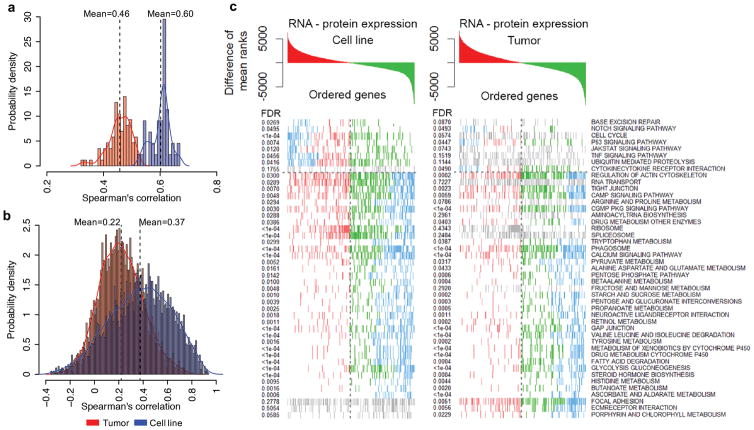
The average Spearman’s correlation between steady-state mRNA and protein abundance within individual samples across genes was 0.60 for cell lines, compared to 0.46 for tumors (Figure 3a); the average correlation across samples within genes was 0.37 for cell lines, compared to 0.22 for tumors (Figure 3b). These results indicate that the tumor-based analyses likely have underestimated the protein-mRNA correlations, and emphasize the necessity of performing mRNA and protein measurements on the same tissue sample. Nevertheless, even for the cell line data, mRNA measurements remained poor predictors of protein abundance variations for many genes.
Figure 3. Comparison of the correlations between mRNA and protein abundance in tumor and cell line data.
(a) Correlations between steady state mRNA and protein abundance across genes within individual samples. (b) Correlations between mRNA and protein variation across cell line or tumor samples for each gene. (c) GSEA KEGG enrichment for average differences in mRNA-protein ranks across genes in both the cell line and tumor data. Genes colored in red are ranked higher in RNA, genes in green ranked higher in proteomics and blue are the leading-edge GSEA genes.
To investigate whether tumor signatures of post-transcriptional regulation at the biological pathway level were maintained in cell lines, we performed GSEA KEGG enrichment analysis on the average rank-differences between mRNA and protein expression (see Methods, Figure 3c and Supplementary Table 24). Tumors and cell lines exhibited high concordance for putative post-transcriptionally modulated pathways, with 66.7% of significant pathways overlapping between these cohorts (p<2.2e-16, hypergeometric test). Post-transcriptional up-regulation of protein expression was observed in both cohorts for 28 processes including 20 metabolic pathways, cAMP, cGMP signaling and cell adhesion functions. Only two pathways, p53 and Notch, showed evidence of coordinated post-transcriptional down-regulation.
Tumor intrinsic protein expression and pathway signatures are retained in CRC cell lines
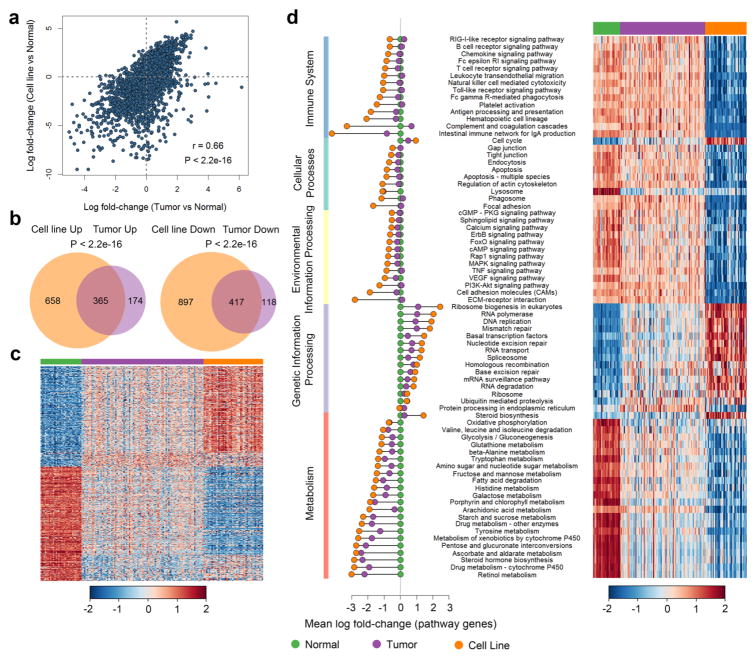
To investigate to what extent proteome dysregulation in primary tumors was recapitulated in CRC cell lines, we compared protein abundances from cell line and tumor samples against those from normal samples. Cell lines and tumors exhibited a high correlation of expression changes relative to normal tissue (Spearman’s correlation=0.66, p<2.2e-16; Figure 4a, Supplementary Table 25), with significant overlap between up-regulated and down-regulated proteins (FDR<0.05, fold change>2, voom/limma) identified for each group (p<2.2e-16, Fisher’s exact test, Figure 4b). Nonetheless, expression in tumor samples tended to lie between that for normal tissues and cell lines, observed for 82.2% of the overlapping dysregulated proteins (p<2.2e-16, proportion test, Figure 4c), consistent with tumor samples representing an admixed population of neoplastic and normal cell types. Similar results were obtained when considering mRNA expression (Supplementary Fig. 14a–14b, Supplementary Table 26).
Figure 4. Comparison of cell lines and tumors to normal tissues based on protein abundance data.
(a) Correlation of protein expression changes for cell line and tumor relative to normal tissue. (b) Overlap between up-regulated and down-regulated proteins (FDR<0.05, fold change>2) relative to normal. (c) Heat map showing protein expression in normal, tumor and cell line samples. (d) Coordinated protein expression changes within KEGG pathways determined using a linear mixed-effects model. Mean log fold change as compared to normal and heatmap of pathway expression shown for normal, tumor and cell line samples.
To gain a more detailed understanding of the level of conservation between cell lines and tumors at the level of protein pathways, we tested for coordinated protein expression changes within KEGG pathways as compared to normal tissue. Overall, changes in pathway expression were highly concordant between cell lines and tumors as observed at the individual protein level (Spearman’s correlation=0.69, p<2.2e-16; Supplementary Fig. 15a, Supplementary Table 27). In particular, significant “intrinsic” pathways (FDR<0.05 for either group comparisons; left panel of Figure 4d) related to genetic information processing and metabolism showed a high consistency of protein expression between tumors (purple points) and cell lines (orange points) compared to normal tissues (green points), with tumor pathway expression levels generally between cell lines and normal tissues. However, for “extrinsic” and stroma-related pathways including environmental information processing, cellular and immune-system related processes, tumors were more similar to normal tissues, while expression in cell lines was markedly decreased. These global protein and pathway category patterns were recapitulated for RNA-Seq data (Supplementary Fig. 14c, Supplementary Fig. 15b, Supplementary Table 28).
Influence of copy number aberrations on protein abundance across cell lines and tumors
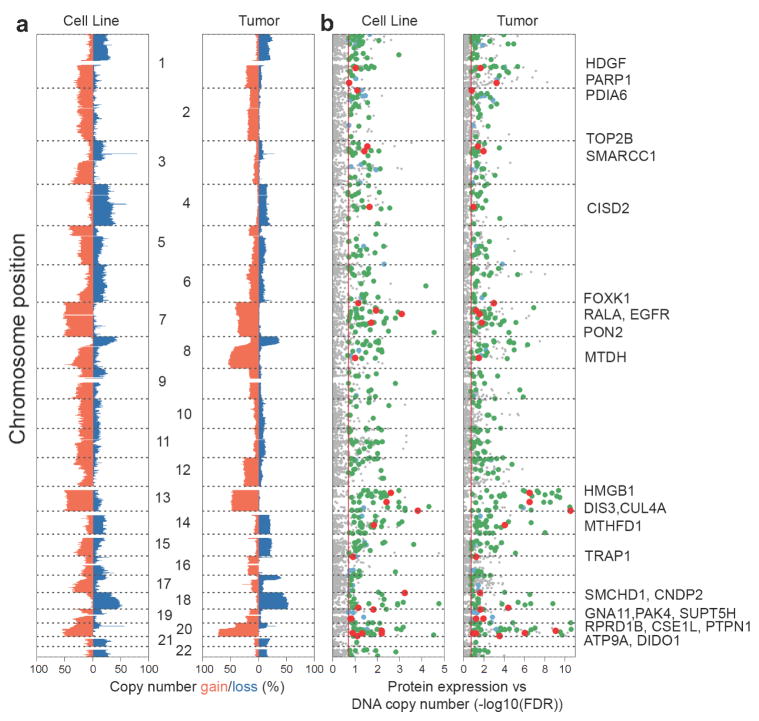
While the impact of DNA copy number on mRNA expression is well established 4, our previous analysis of TCGA tumors suggested that this impact is less apparent with respect to protein expression 6. To compare the effect of DNA copy number aberrations on gene expression between tumors and CRC cell lines, we retrieved DNA copy number states from matched SNP array data. DNA copy-number spectra in cell lines closely resembled those seen in tumors, with the most commonly gained chromosome arms including chromosome 7, 8q, 13, and 20q, and the most common deleted regions including 8p, 17p, and 18q (% gain = red bars, % loss = blue bars in Figure 5a). Overall, 989 proteins in CRC cell lines and 1524 proteins in tumors were associated with DNA copy-number changes (FDR<0.2, voom/limma, see Methods), with strengths of associations tracking with the respective frequencies of DNA copy number loss or gain (Figure 5b, Supplementary Table 29–30). As expected, similar but more pronounced results were found when analyzing associations between DNA copy number aberrations and mRNA expression (Supplementary Table 31–32). 438 protein-DNA measurement relationships were detected across both tumors and cell lines (p<2.2e-16 for overlap, hypergeometric test; large points in Figure 5b), 90.0% of which also were detected at the mRNA level (green/red points). Among these proteins, 26 are known or proposed cancer genes (red points in Figure 5b, Supplementary Table 33). Proteins identified in regions of gain included the established EGFR oncogene 21, and candidates such as FOXK1, a forkhead transcription factor, and CNDP2, an activator of MAPK pathways. Increased expression of FOXK1 has been shown to promote CRC invasion and metastasis 22, and up-regulation of CNDP2 to facilitate colon cancer proliferation 23. In regions of loss, we identified several putative tumor suppressors, including MTHFD1, a 1-tetrahydrofolate synthase. MTHFD1 deficiency has been shown to increase intestinal tumor incidence, number and burden in transgenic mouse models 24. Using shRNA knockdown data from the Achilles project 25, we further validated six oncogene candidates (USP39, PARP1, EGFR, DLD, SRI and IDH3B) as “essential” genes in CRC (Supplementary Fig. 16).
Figure 5. Proteome alterations associated with copy-number aberrations.
(a) DNA copy-number spectra (% gain = red bars, % loss = blue bars, relative to ploidy) in cell lines and tumors. (b) Strengths of association for protein expression with corresponding DNA copy-number changes (-log10(FDR)). Grey = not significantly associated with copy number alterations, blue = significant across proteomics cell line and tumor data only, green = significant for both proteomics and mRNA expression across cell line and tumor, red = candidate tumor suppressor and oncogenes.
Proteomics data better predicts CRC drug sensitivity
To evaluate the relative utility of proteomics data as a marker of drug responses in comparison to mutation, DNA copy number, and mRNA expression data, we retrieved response profiles for 210 drugs from the GDSC database which comprised 18 cell lines from our CRC cell line panel.
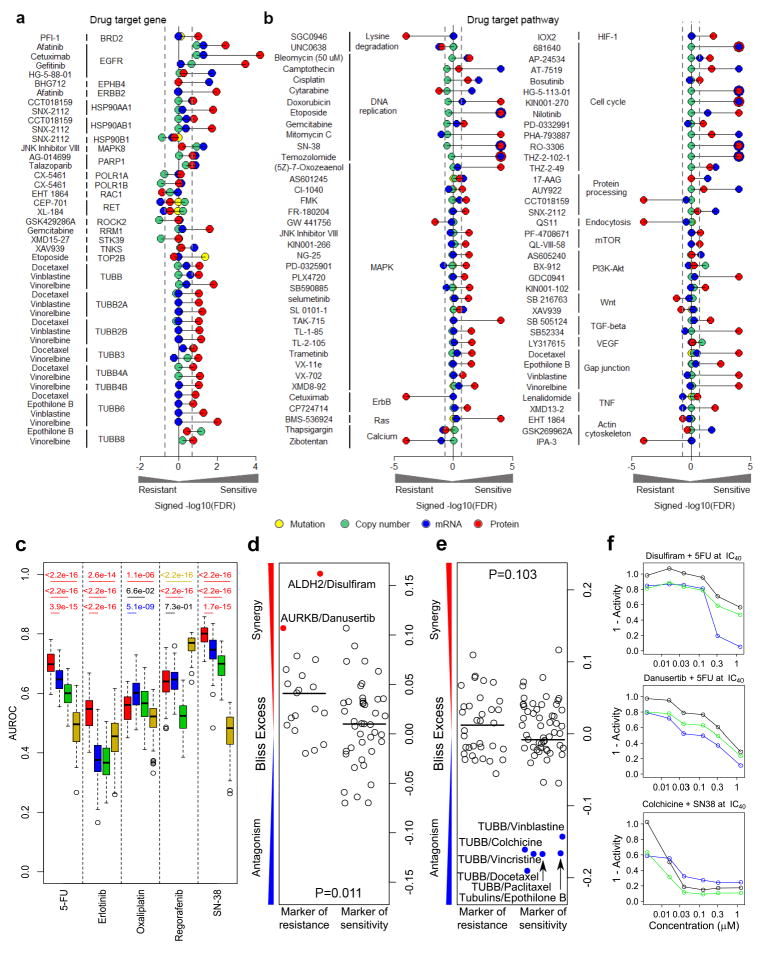
Considering 191 known drug-target gene associations quantifiable at the protein level (Supplementary Table 34), proteomics data identified 16.2% of the relationships (FDR <0.2), as compared to only 6.3% for mRNA, 5.3% for DNA copy number and 1.9% for mutation data (Supplementary Table 35). Among the significant drug-target gene pairs detected at the protein level were multiple associations for EGF receptor family members (afatinib, cetuximab, gefitinib), heat shock protein 90 (CCT018159, SNX-2112) and β-tubulin family members (docetaxel, epothilone B, vinblastine, vinorelbine) (Figure 6a). Among pairwise comparisons in which at least one omics modality showed a discernable association (FDR <0.2), proteomic data showed greater correlations with drug responses than mRNA and DNA copy number data for 77.1% (27/35, p=0.001) and 81.6% (31/38, p=9.53e-05) of cases, respectively. Mutation data could not formally be evaluated for this latter comparison as only two quantifiable cases were significant in the overlap with the proteomics data.
Figure 6. Proteomics data utility for predicting therapeutic responses.
(a, b) Associations of proteomic, mutation, DNA copy number and mRNA data with (a) established drug-target associations and (b) drug-pathway associations. Associations are shown for drug-target gene associations quantifiable at the protein level and significant in at least one of the four modalities as signed -log10(FDR) values from voom/limma and GSEA analyses, respectively. (c) Comparison of the utility of four omic modalities to predict drug sensitivity for 5-fluoruracil (5-FU), erlotinib, oxaliplatin, regorafenib and SN-38: proteomic data (red); RNA-Seq data (blue); CNA data (green); and exome mutation data (yellow). For each drug-omic modality combination, area under the receiver operating characteristic curve (AUROCs) were generated from 100 times of 5-fold cross-validations. The two-sided Wilcoxon rank sum test was used to compare the performance between protein-based models and models based on other omics data types. For each comparison, the p value is colored based on the color of the omic data type with significantly better performance. (d–e) Pharmacological targeting of proteins associated with resistance or sensitivity to (d) 5-FU or (e) SN-38. Bliss excess values are shown for drug combinations with 5-FU (at IC30 concentration) and SN-38 (at IC40 concentration) in HCT116 cells. The protein targets were restricted to those with FDR< 0.2 from the relevant voom/limma calculation; drugs are detailed in Supplementary Tables 4–5. p-values for Student’s t-test. (f) Dose-response plots for selected compounds alone (black), with either a 5-FU or SN-38 (blue), or the predicted response under the assumption of Bliss independence for the two compounds (green). Bliss synergy = blue line below green line; Bliss antagonism = blue line above green line.
Extending our association analyses to known drug-pathway relationships, proteomics data again identified more relationships (52.8%) than mRNA (25.2%), DNA copy number (1.6%) and mutation data (0%) (Figure 6b, Supplementary Table 36). The KEGG DNA replication (e.g. mitomycin C, SN-38), MAPK (e.g. TAK-715, trametinib) and PI3K-Akt (GDC0941, KIN001-102) pathways were among the significant drug-pathway pairs detected at the protein level (Figure 6b). Similarly, for pairwise comparisons, proteomic data showed greater correlations with drug response than mRNA, DNA copy number and mutation data for 74.3% (55/74, p=2.36e-05), 97.0% (65/67, p=1.80e-14) and 100% (62/62, p=1.65e-13) of respective cases. In addition to the established drug-target relationships, responses for many drugs were correlated with protein signatures reflective of cell doubling rate (Supplementary Fig. 17–18, Supplementary Tables 37–38).
To more formally assess the utility of proteomic data for prediction of drug sensitivity relative to mRNA expression, DNA copy number and mutation data, we evaluated predictive models using random forests and five-fold cross-validation. Given the limited number of CRC cell lines with matched GDSC data, we screened our 44 CRC cell lines panel against four major drugs used in the treatment of human CRC, 5-fluoruracil (5-FU), oxaliplatin, SN-38 and regorafenib. In addition, we tested the small molecule inhibitor erlotinib as a proxy for anti-EGFR antibody therapeutics (Supplementary Table 39). Significant correlations were observed between GDSC and our drug sensitivity data for two overlapping drugs, 5-FU and SN-38 (Supplementary Fig. 19).
As shown in Fig 6c, proteomics data demonstrated better performance for predicting sensitivity to 5-FU, SN-38, erlotinib, regorafenib and oxaliplatin in 11 out of 15 pair-wise comparisons against other modalities. Notably, proteomics data displayed a striking advantage for 5-FU, SN-38 and erlotinib. For regorafenib and oxaliplatin, only mutation data (yellow) and mRNA data (blue) outperformed proteomics data, respectively. In general, proteomics data thus provides an improved ability to predict the drug sensitivity of the CRC cell lines.
Proteins associated with drug sensitivity may be functionally implicated in determining drug responses. Pharmacological inhibition of targetable proteins contributing to drug resistance may synergize with baseline treatment, whereas inhibition of proteins conferring sensitivity may be antagonistic. To test this contention, we assembled 60 and 92 drugs whose inhibitory profiles included targetable protein implicated in responses to 5-FU or SN-38 (the active metabolite of irinotecan) (FDR <0.2 in GDSC, 48 and 56 targets), respectively, two mainstay treatments for CRC (Supplementary Tables 4–5). Dose-response curves for the inhibitor panel were determined for HCT116 colon cancer cells in the presence or absence of 5-FU or SN-38 at IC30/40 concentrations, and drug combinations evaluated for evidence of synergy or antagonism based on excess over the Bliss (EOB) independence model. For both 5-FU and SN-38 treatment, EOBs tended to differ between drugs targeting protein markers of resistance as compared to markers of sensitivity (5-FU, p=0.011, SN-38, p=0.103, t-test), with the expected propensities to synergy or antagonism (Figure 6d–e, Supplementary Tables 4–5). For 5-FU treatment, inhibition with disulfiram, an efficacious ALDH inhibitor (incl. ALDH1 and ALDH2), was the top synergistic combination detected (Figure 6f). ALDH is a family of enzymes that play a key role in the metabolism of aldehydes and have been shown to oxidize and inactivate several prominent chemotherapeutic drugs 26. ALDH activity has been associated with colon cancer resistance to irradiation and 5-FU 27. Accordingly, disulfiram has previously been shown to potentiate gemcitabine and 5-FU treatment in colon cancer cells 28, 29.
Danusertib, an inhibitor against for Aurora A/B/C was identified as another synergistic compound with 5-FU (Figure 6f), and multiple inhibitors of Aurora kinases have been evaluated for the treatment of CRC in combination with 5-FU, with several in clinical trials 30. Consistent with our findings, several studies have indicated that overexpression of Aurora kinases has a role in chemo- and radiotherapy resistance of CRC 31, 32.
For SN-38, combination with multiple tubulin inhibitors showed evidence of antagonism (Figure 6f). It has previously been reported that a primary mechanism of tubulin inhibitor resistance is simultaneous administration of a compound that inhibits cell cycle progression at the G2-M phase, the main phase of action of SN-38 33.
Cell lines connect proteomic subtypes to drug sensitivity
Colorectal tumors can be classified into five proteomic subtypes that are largely distinct from the established transcriptomic subtypes 6. Using a PAM prediction model trained on the primary tumor samples (Supplementary Fig. 20), 40 cell lines were assigned to a proteomic subtype with a prediction probability of >0.8 (Supplementary Fig. 21), with representative cell lines identified for all five subtype classes (A–E) (Figure 7a, Supplementary Table 1). Cell lines were further categorized into transcriptomic subtypes using the CMSclassifier algorithm 5. Subtypes CMS1, CMS2 and CMS3 were identified among cell lines, but subtype CMS4 was not assigned (Supplementary Fig. 22a, Supplementary Table 1). The failure to detect CMS4-assigned cell lines may be coincidental given our limited cohort size, or perhaps reflect the observation that this subtype signature is largely dominated by signals from tumor stroma 15, 34.
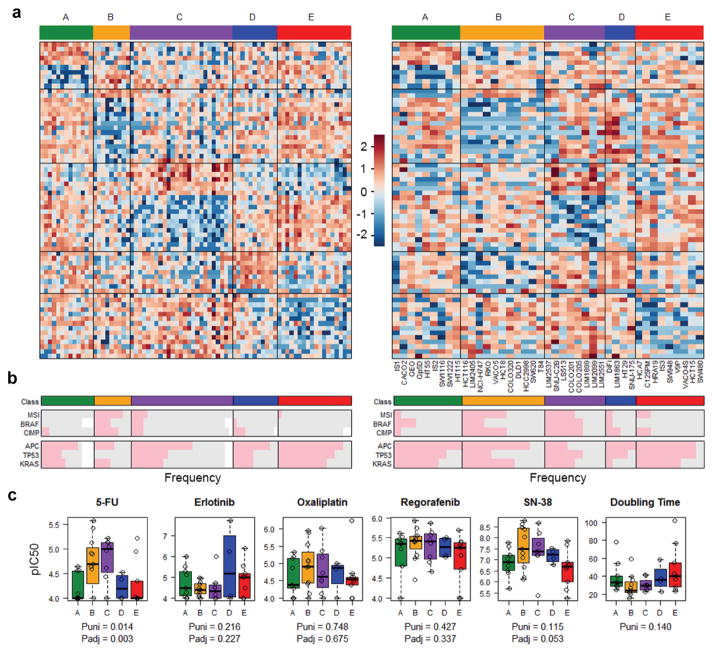
Figure 7. Concordance of proteomic CRC subtypes in cell lines and tumors.
(a) Heatmap of protein abundances indicating proteomic subtypes for tumors (left panel) and cell lines (right panel). Samples are arranged along the X axis and genes are arranged along the Y axis. Increased expression (red) and decreased expression (blue) relative to the mean-centered and scaled expression of the gene (normalized CPM) across the samples. (b) Representation of genomic hallmarks among proteomic subtypes. (c) Drug responses of proteomic subtypes to 5-fluoruracil (5-FU), erlotinib, oxaliplatin, regorafenib and SN-38 treatment, and relationships with cell doubling time. Puni (univariate) is the P-value obtained from univariate ANOVA, and Padj (adjusted) is the P-value from two-way ANOVA adjusting for cell doubling time.
Comparing cell lines and tumors, proteomic and CMS subtypes were associated with similar distributions of genomic hallmarks across the cohorts, including MSI and CIMP status and mutations in BRAF, APC, TP53 and KRAS (Figure 7b, Supplementary Fig. 22b). Interestingly, analysis of paired cell lines derived from the same tumor or primary tumor and metastatic derivatives identified some discordant assignments for proteomic subtypes. Discordances were also observed for transcriptomic subtypes, suggesting that proteomic and transcriptomic subtypes may represent transient states, with tumors adopting different subtypes with clonal evolution (Supplementary Data). Consistent with this suggestion, mutational differences were evident between paired cell lines at the genetic level (Supplementary Data). The transient nature of expression-based subtypes was further supported by microarray analysis for 5 matched primary tumors and liver metastases identifying discordant CMS subtypes for 3 of these pairs (Supplementary Table 40).
To evaluate the potential value of tumor proteomic subtypes to predict drug response, we analysed the 5-FU, oxaliplatin, SN-38, regorafenib and erlotinib data for our 44 CRC cell line panel. GDSC data were not evaluated, due to the small cohort size. Although the number of cell lines in each subgroup were limited, proteomics subtypes were significantly associated with response to 5-FU, with subtype C exhibiting the greatest sensitivity (univariate p=0.014, ANOVA, Figure 7c). The association of proteomic subtypes with 5-FU response remained significant when adjusting for cell doubling time (adjusted p=0.003 ANOVA), which itself was directly related to 5-FU sensitivity (adjusted p=0.0007, ANOVA), or when the analysis was limited to microsatellite stable cases (univariate p=0.031, adjusted p=0.009, ANOVA, Supplementary Fig. 23). In contrast, transcriptomic subtypes showed no significant associations with drug response (Supplementary Fig. 22c). The mechanism underlying the increased sensitivity of proteomics subtype C to 5-FU remains to be elucidated, but may be related to differences in 5-FU metabolic activation, detoxification or drug export 35, 36. These results further underscore the potential of proteomic data for drug response prediction, in line with our protein signature and pathway analyses.
DISCUSSION
Our global proteomic characterization firstly demonstrates that CRC cell line proteomes maintain the major cell-intrinsic molecular programs, proteogenomic relationships and proteomic subtypes observed in primary tumors, highlighting the utility of cell lines as models for tumor biology, biomarker discovery and therapeutics. Most proteome aberrations and intrinsic pathway signatures (e.g. genetic information processing and metabolism) showed concordant differences in both cell lines and tumors as compared to normal tissues. Relationships between protein expression and somatic DNA copy number changes in primary tumors were recapitulated in cell lines, identifying both established (EGFR) and candidate cancer genes (e.g. FOXK1, CNDP2 and MTHFD1) 22–24. Integration of proteomic and transcriptomic data indicates that tumor post-transcriptional regulation at the biological pathway level is maintained in cell lines. The five proteomic subtypes previously identified for primary tumors were represented among cell lines and showed similar distributions of established genomic hallmarks. Notably, some heterogeneity in proteomic subtype assignments was observed between paired cell lines, as for transcriptomic subtypes, suggesting that expression-based subtype signatures may represent transient states.
Nonetheless, systematic differences between cell line and tumor proteomes were apparent, with major changes attributable to tumor stroma, extrinsic signaling and different growth conditions. Because of the significant contribution of the tumor stroma, the anticipated signatures of DNA mismatch repair and DNA proofreading polymerases identified in cell lines with a hypermutation phenotype were not detectable in the primary tumors. Instead, the proteomes of hypermutated tumors were characterized by signatures of immune infiltrates that are typically associated with such cases 20. Overall, protein abundance measurements in cell lines showed a higher concordance with tumor-cell specific IHC expression measurements than did proteome profiles of admixed tumor specimens. Together, these findings underscore both the value and limitation of cell line models for unraveling tumor biology.
Multiple studies have explored genomic and transcriptomic markers for drug sensitivity in cancer cell lines 1, 3, but data on the proteome remain limited 11. Our comparison of omics modalities for the identification of known drug-target gene or pathway relationships in CRC cell lines demonstrates the potential of global proteomic data to predict therapeutic responses. Consistent with our observation that DNA and mRNA measurements are poor predictors of protein abundance, protein level data outperformed mRNA, DNA copy number and mutation data in 11 out of 15 pairwise comparisons for five evaluated standard therapies (5-fluoruracil, SN-38, erlotinib a proxy for anti-EGFR antibody therapy, regorafenib, oxaliplatin). Furthermore, proteomic data more closely predicted known drug-target relationships, both at the individual gene and the target pathway levels. Pharmacological inhibition of targetable proteins associated with CRC cell line resistance or sensitivity to standard chemotherapies (5-FU and SN-38) identified markers that may be functionally implicated in determining drug responses, exhibiting synergy or antagonism in combination treatments, respectively. In addition, our data suggest that tumor proteomic subtypes may be useful predictors of drug responses, warranting further investigation in expanded studies. A caveat to our analysis is that we could not validate proteome-drug sensitivity relationships in our cohort of TCGA primary cancers due to insufficient cases with single-agent treatment and outcome data.
In summary, our integrative analysis demonstrates the utility of CRC cell lines as representative models of primary tumors at the proteome level, and highlights the potential of global proteomic data to inform personalized cancer medicine. Our data provide a rich resource for the scientific community and are available in public repositories and for interrogation via customized online research tools.
Supplementary Material
Acknowledgments
GRANT SUPPORT
This study was supported by the Ludwig Institute for Cancer Research, a NHMRC Project Grant (APP1079362), a Cancer Council Victoria Grant-in-Aid (APP1060964) and the Victorian Government’s Operational Infrastructure Support Program, by National Cancer Institute (NCI) CPTAC awards U24CA159988 and U24CA210954, by NCI SPORE award P50CA095103, and by contract 15X038 from Leidos Biomedical Research, Inc. We also thank the support from NCI-Funded Special Programs of Research Excellence in GI Cancer. This research was supported by a Victorian Life Sciences Computation Initiative (VLSCI) grant number [VR0310, VR0311] on its Peak Computing Facility at the University of Melbourne, an initiative of the Victorian Government, Australia. O.M.S. is a NHMRC R.D. Wright Biomedical Fellow (APP1062226). B.Z. is Cancer Prevention & Research Institutes of Texas (CPRIT RR160027) Scholar and McNair Medical Institute Scholar
We thank Doreen Agyapomaa for the preparation of compound screening plates and Eugene Kapp for providing access to the spike-in data set generated by the 2015 study of the Proteome Informatics Research Group (iPRG) of the Association of Biomolecular Resource Facilities (ABRF). We also thank the support from NCI-Funded Special Programs of Research Excellence in GI Cancer.
Footnotes
CONFLICT OF INTEREST STATEMENT.
No conflict of interest or competing financial interests to disclose for all authors.
AUTHOR CONTRIBUTIONS
Conceptualization: AWB, BZ, DCL, OMS, RJCC, RJCS
Methodology: BZ, DCL, DM, JW, OMS, XW
Software: CGL, CY, DM, JW, MCC, RJCS, RNJ, SV, XC, XW, YB, ZS
Validation: BZ, DM, JW, OMS, XW
Formal Analysis: BZ, DM, JW, OMS, RNJ, XW
Investigation: HJ, JW (Weinstock), KJL, KL, LJZ, SL
Resources: AWB, BZ, DCL, JM, OMS, ZS
Data curation: BZ, DM, JW, MCC, OMS, RJCS, XW
Writing – Original Drafts: BZ, DM, JW, OMS, XW
Write – Review & Editing: All authors
Visualization: BZ, DM, JW, OMS, XW
Supervision: BZ, OMS
Project Administration: AWB, BZ, DCL, OMS
Funding Acquisition: AWB, BZ, DCL, OMS, RJCC
References
- 1.Garnett MJ, Edelman EJ, Heidorn SJ, et al. Systematic identification of genomic markers of drug sensitivity in cancer cells. Nature. 2012;483:570–5. doi: 10.1038/nature11005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barretina J, Caponigro G, Stransky N, et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature. 2012;483:603–7. doi: 10.1038/nature11003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Iorio F, Knijnenburg TA, Vis DJ, et al. A Landscape of Pharmacogenomic Interactions in Cancer. Cell. 2016;166:740–54. doi: 10.1016/j.cell.2016.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cancer Genome Atlas N. Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487:330–7. doi: 10.1038/nature11252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Guinney J, Dienstmann R, Wang X, et al. The consensus molecular subtypes of colorectal cancer. Nat Med. 2015;21:1350–6. doi: 10.1038/nm.3967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang B, Wang J, Wang X, et al. Proteogenomic characterization of human colon and rectal cancer. Nature. 2014;513:382–7. doi: 10.1038/nature13438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shoemaker RH. The NCI60 human tumour cell line anticancer drug screen. Nat Rev Cancer. 2006;6:813–23. doi: 10.1038/nrc1951. [DOI] [PubMed] [Google Scholar]
- 8.Chen B, Sirota M, Fan-Minogue H, et al. Relating hepatocellular carcinoma tumor samples and cell lines using gene expression data in translational research. BMC Med Genomics. 2015;8(Suppl 2):S5. doi: 10.1186/1755-8794-8-S2-S5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Medico E, Russo M, Picco G, et al. The molecular landscape of colorectal cancer cell lines unveils clinically actionable kinase targets. Nat Commun. 2015;6:7002. doi: 10.1038/ncomms8002. [DOI] [PubMed] [Google Scholar]
- 10.Vincent KM, Findlay SD, Postovit LM. Assessing breast cancer cell lines as tumour models by comparison of mRNA expression profiles. Breast Cancer Res. 2015;17:114. doi: 10.1186/s13058-015-0613-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lawrence RT, Perez EM, Hernandez D, et al. The proteomic landscape of triple-negative breast cancer. Cell Rep. 2015;11:630–44. doi: 10.1016/j.celrep.2015.03.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gholami AM, Hahne H, Wu Z, et al. Global proteome analysis of the NCI-60 cell line panel. Cell Rep. 2013;4:609–20. doi: 10.1016/j.celrep.2013.07.018. [DOI] [PubMed] [Google Scholar]
- 13.Mouradov D, Sloggett C, Jorissen RN, et al. Colorectal cancer cell lines are representative models of the main molecular subtypes of primary cancer. Cancer Res. 2014;74:3238–47. doi: 10.1158/0008-5472.CAN-14-0013. [DOI] [PubMed] [Google Scholar]
- 14.Wang J, Duncan D, Shi Z, et al. WEB-based GEne SeT AnaLysis Toolkit (WebGestalt): update 2013. Nucleic Acids Res. 2013;41:W77–83. doi: 10.1093/nar/gkt439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Isella C, Terrasi A, Bellomo SE, et al. Stromal contribution to the colorectal cancer transcriptome. Nat Genet. 2015;47:312–9. doi: 10.1038/ng.3224. [DOI] [PubMed] [Google Scholar]
- 16.Aran D, Sirota M, Butte AJ. Systematic pan-cancer analysis of tumour purity. Nat Commun. 2015;6:8971. doi: 10.1038/ncomms9971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Uhlen M, Oksvold P, Fagerberg L, et al. Towards a knowledge-based Human Protein Atlas. Nat Biotechnol. 2010;28:1248–50. doi: 10.1038/nbt1210-1248. [DOI] [PubMed] [Google Scholar]
- 18.Poulogiannis G, Frayling IM, Arends MJ. DNA mismatch repair deficiency in sporadic colorectal cancer and Lynch syndrome. Histopathology. 2010;56:167–79. doi: 10.1111/j.1365-2559.2009.03392.x. [DOI] [PubMed] [Google Scholar]
- 19.Briggs S, Tomlinson I. Germline and somatic polymerase epsilon and delta mutations define a new class of hypermutated colorectal and endometrial cancers. J Pathol. 2013;230:148–53. doi: 10.1002/path.4185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Smyrk TC, Watson P, Kaul K, et al. Tumor-infiltrating lymphocytes are a marker for microsatellite instability in colorectal carcinoma. Cancer. 2001;91:2417–22. [PubMed] [Google Scholar]
- 21.Radinsky R, Risin S, Fan D, et al. Level and function of epidermal growth factor receptor predict the metastatic potential of human colon carcinoma cells. Clin Cancer Res. 1995;1:19–31. [PubMed] [Google Scholar]
- 22.Wu Y, Peng Y, Wu M, et al. Oncogene FOXK1 enhances invasion of colorectal carcinoma by inducing epithelial-mesenchymal transition. Oncotarget. 2016 doi: 10.18632/oncotarget.9457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xue C, Zhang Z, Yu H, et al. Up-regulation of CNDP2 facilitates the proliferation of colon cancer. BMC Gastroenterol. 2014;14:96. doi: 10.1186/1471-230X-14-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.MacFarlane AJ, Perry CA, McEntee MF, et al. Mthfd1 is a modifier of chemically induced intestinal carcinogenesis. Carcinogenesis. 2011;32:427–33. doi: 10.1093/carcin/bgq270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cheung HW, Cowley GS, Weir BA, et al. Systematic investigation of genetic vulnerabilities across cancer cell lines reveals lineage-specific dependencies in ovarian cancer. Proc Natl Acad Sci U S A. 2011;108:12372–7. doi: 10.1073/pnas.1109363108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Moreb JS, Ucar-Bilyeu DA, Khan A. Use of retinoic acid/aldehyde dehydrogenase pathway as potential targeted therapy against cancer stem cells. Cancer Chemother Pharmacol. 2017;79:295–301. doi: 10.1007/s00280-016-3213-5. [DOI] [PubMed] [Google Scholar]
- 27.Bellamkonda K, Sime W, Sjolander A. The impact of inflammatory lipid mediators on colon cancer-initiating cells. Mol Carcinog. 2015;54:1315–27. doi: 10.1002/mc.22207. [DOI] [PubMed] [Google Scholar]
- 28.Guo X, Xu B, Pandey S, et al. Disulfiram/copper complex inhibiting NFkappaB activity and potentiating cytotoxic effect of gemcitabine on colon and breast cancer cell lines. Cancer Lett. 2010;290:104–13. doi: 10.1016/j.canlet.2009.09.002. [DOI] [PubMed] [Google Scholar]
- 29.Wang W, McLeod HL, Cassidy J. Disulfiram-mediated inhibition of NF-kappaB activity enhances cytotoxicity of 5-fluorouracil in human colorectal cancer cell lines. Int J Cancer. 2003;104:504–11. doi: 10.1002/ijc.10972. [DOI] [PubMed] [Google Scholar]
- 30.Bavetsias V, Linardopoulos S. Aurora Kinase Inhibitors: Current Status and Outlook. Front Oncol. 2015;5:278. doi: 10.3389/fonc.2015.00278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cammareri P, Scopelliti A, Todaro M, et al. Aurora-a is essential for the tumorigenic capacity and chemoresistance of colorectal cancer stem cells. Cancer Res. 2010;70:4655–65. doi: 10.1158/0008-5472.CAN-09-3953. [DOI] [PubMed] [Google Scholar]
- 32.Wu X, Liu W, Cao Q, et al. Inhibition of Aurora B by CCT137690 sensitizes colorectal cells to radiotherapy. J Exp Clin Cancer Res. 2014;33:13. doi: 10.1186/1756-9966-33-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ehrhardt H, Pannert L, Pfeiffer S, et al. Enhanced anti-tumour effects of Vinca alkaloids given separately from cytostatic therapies. Br J Pharmacol. 2013;168:1558–69. doi: 10.1111/bph.12068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Calon A, Lonardo E, Berenguer-Llergo A, et al. Stromal gene expression defines poor-prognosis subtypes in colorectal cancer. Nat Genet. 2015;47:320–9. doi: 10.1038/ng.3225. [DOI] [PubMed] [Google Scholar]
- 35.Longley DB, Harkin DP, Johnston PG. 5-fluorouracil: mechanisms of action and clinical strategies. Nat Rev Cancer. 2003;3:330–8. doi: 10.1038/nrc1074. [DOI] [PubMed] [Google Scholar]
- 36.Li H, Zhu F, Boardman LA, et al. Aspirin Prevents Colorectal Cancer by Normalizing EGFR Expression. EBioMedicine. 2015;2:447–455. doi: 10.1016/j.ebiom.2015.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.