Abstract
Single nucleus RNA-seq (sNuc-seq) profiles RNA from tissues that are preserved or cannot be dissociated, but does not provide the throughput required to analyse many cells from complex tissues. Here, we develop DroNc-seq, massively parallel sNuc-Seq with droplet technology. We profile 39,111 nuclei from mouse and human archived brain samples to demonstrate sensitive, efficient and unbiased classification of cell types, paving the way for systematic charting of cell atlases.
Single cell RNA-seq (scRNA-seq) has become instrumental for interrogating cell types, dynamic states and functional processes in complex tissues1,2. However, current protocols require preparation of single cell suspension from fresh tissue, a major roadblock in many applications, including handling of clinical samples, archived materials, and tissues that cannot be readily dissociated. The necessary harsh enzymatic dissociation is particularly problematic for brain tissue because it harms the integrity of neurons and their RNA, biases data in favour of recovery of some cell types, and works only on samples from young organisms, precluding, for example, analyzing those obtained from deceased patients with neurodegenerative disorders. To address this challenge, we3 and others4–6 developed single nucleus RNA-seq (snRNA-seq) for analysis of RNA in single nuclei from fresh, frozen or lightly fixed tissues. snRNA-seq methods such as sNuc-Seq3, Div-Seq3, and others4,5 can handle minute samples of complex tissues that cannot be successfully dissociated, providing access to archived samples, such as fresh-frozen or lightly fixed samples. However, these methods either rely on sorting nuclei by FACS into plates (96 or 384 wells)3,5 or on C1 microfluidics4, neither of which are scalable, precluding profiling tens of thousands of nuclei (needed for human brain tissue) or large numbers of samples (e.g., tumor biopsies from patients). Conversely, massively parallel scRNA-seq methods, such as Drop-seq7 and related methods8–10 can be readily applied at scale11 in a cost-effective manner12, but require intact single cell suspension as input.
Here, we addressed this challenge by developing DroNc-seq (Fig. 1a), a massively parallel single nucleus RNA-seq method that combines the advantages of sNuc-Seq and Drop-seq to profile nuclei at low cost and high throughput. We modified Drop-seq7 to accommodate the relatively lower amount of RNA in nuclei compared to cells, including a modified microfluidic design and changes in the nuclei isolation protocol (Supplementary Fig. 1, Supplementary Table 1, Supplementary Data 1, Methods).
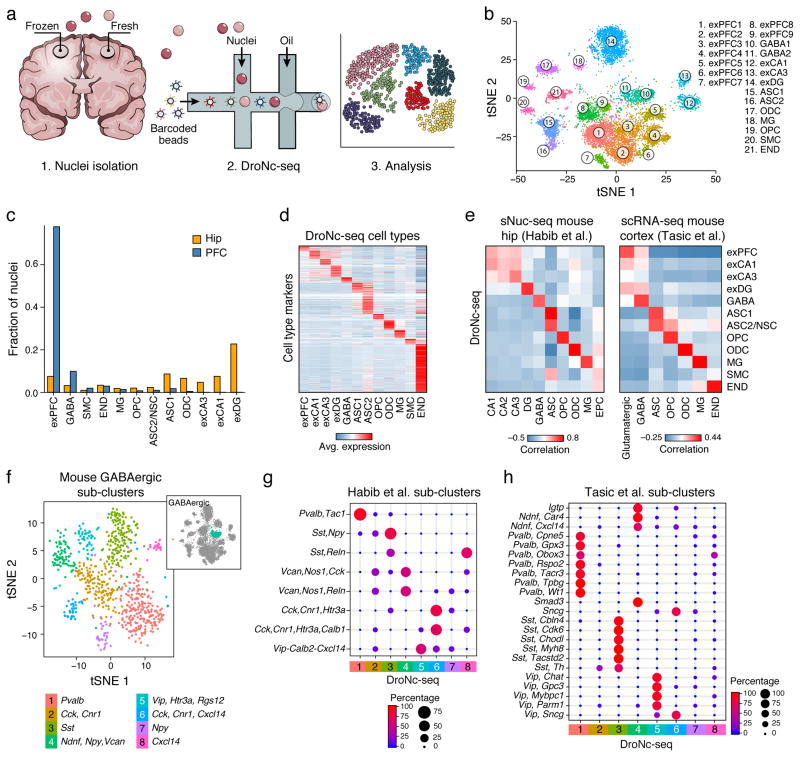
Figure 1. DroNc-seq: Massively parallel single nucleus RNA-Seq.
(a) Overview. (b) DroNc-seq of adult frozen mouse hippocampus and prefrontal cortex. A two-dimensional tSNE plot of 13,133 DroNc-seq nuclei profiles (>10,000 reads and >200 genes per nucleus) from hippocampus (hip; 4 samples) and prefrontal cortex (PFC; 4 samples). Nuclei (dots) are colored by cluster membership and labelled post hoc by cell types and anatomical distinctions (exPFC=glutamatergic neurons from the PFC, exCA1/3=pyramidal neurons from the Hip CA region, GABA=GABAergic interneurons, exDG=granule neurons from the Hip dentate gyrus region, ASC=astrocytes, NSC=neuronal stem cells, MG=microglia, ODC=oligodendrocytes, OPC=oligodendrocyte precursor cells, NSC=neuronal stem cells, SMC=smooth muscle cells, END= endothelial cells). Clusters are grouped by cell types as in Supplementary Fig. 3a. Flagged clusters (Supplementary Fig. 3b and Supplementary Table 3, Methods) were removed. (c) Fraction of nuclei from each brain region associated with each cell type. Cell types are defined as in Supplementary Fig. 3a and sorted from left by types enriched in PFC vs. Hip. (d) Cell type signatures. The average expression of differentially expressed signature genes (rows, Methods) in each DroNc-seq mouse brain cell subset (columns). (e) DroNc-seq cell-type expression signatures in the mouse brain agree with previous studies. Pairwise correlations of the average expression (Methods) for the genes in each cell-type signature defined by DroNc-seq and in cell-types defined by sNuc-Seq in the mouse hippocampus3 (left) and scRNA-seq in the visual cortex17 (right). (f) Sub-sets of mouse GABAergic neurons. tSNE embedding of 816 DroNc-seq nuclei profiles from the GABAergic neurons cluster (Clusters 10–11 in Fig. 1b; inset, blue), color coded by sub-cluster membership. (g,h) Congruence of GABAergic neurons sub-clusters defined here (from j) with subsets defined from nuclei profiles in the mouse hippocampus3 (g) and single cell profiles in the mouse visual cortex17 (h). Dot plot shows the proportion of cells in each cluster defined by the other two datasets that were classified to each DroNc-seq cluster using a multi-class random forest classifier (as in11, Methods) trained on the DroNc-seq sub-clusters.
We used DroNc-seq to robustly generate high quality expression profiles of nuclei from a mouse cell line (3T3, 5,636 nuclei), adult frozen mouse brain tissue (19,561), and archived frozen adult human post-mortem tissue (19,550 nuclei). DroNc-seq (for samples sequenced at 160,000 reads per nucleus, Methods) detected on average 3,295 genes (4,643 transcripts) for 3T3 nuclei, 2,731 genes (3,653 transcripts) for mouse brain, and 1,683 genes (2,187 transcripts) for human brain (Supplementary Fig. 2). Using down-sampling, we estimate that 19,000–26,000 of transcriptome-mapped reads per nucleus are required for saturation (Supplementary Fig. 2f,g).
To assess the throughput and sensitivity of DroNc-seq, we profiled 3T3 cells at the single cell (with Drop-seq) and single nucleus (with DroNc-seq) levels, followed by deep sequencing (~160,000 reads per nucleus/cell). Both Drop-seq and DroNc-seq yielded high-quality libraries, detecting an average of 5,134 and 3,295 genes for cells and nuclei, respectively (Supplementary Fig. 2b,c). DroNc-seq had similar throughput to Drop-seq with efficiencies of 78% (for 3T3 nuclei), 89% (mouse brain), and 95% (human brain) (1,003, 1,251 and 1,333 high-quality nuclei per library out of 1,400 expected nuclei, given our loading parameters, for cell lines, mouse and human brain respectively), compared to 72% high-quality cells per library (1,444 nuclei out of 2,000 expected) (Methods). Notably, libraries were sampled from a larger pool of 20,000 STAMPs (Single Transcriptome Associated Micro Particles7) produced from a given input of nuclei, which can be re-sampled multiple times if a user wishes to sequence additional nuclei from the same input (Methods).
The average expression profile of single nuclei correlated well with the average profile of single cells (Pearson r=0.87, Supplementary Fig. 2d). Those genes with significantly higher expression in nuclei (e.g., lncRNAs Malat1 and Meg3) or cells (mitochondrial genes Mt-nd1/2/4) (Supplementary Fig. 2d) were consistent with known distinct enrichment in these compartments (Supplementary Table 2). In both methods over 84% of reads align to the genome (in a representative example), but in cells 75.2% of these genomic reads map to exons and 9.1% map to introns, whereas in nuclei 46.2% of genomic reads map to exons and 41.8% to introns (Supplementary Fig. 2e), reflecting the enrichment of nascent transcripts in the nucleus3,13–16. To allow comparison with previous studies, we used only exonic reads subsequently, although intronic reads can be leveraged in future13.
Clustering11 of 13,313 nuclei profiled from frozen adult mouse hippocampus (n=4 mice) and prefrontal cortex (PFC, n=4) (sequenced at low depth of >10,000 reads and >200 genes detected per nucleus) with an average of 1,810 genes in neurons and 1,077 in non-neuronal cells (Methods) revealed groups of nuclei corresponding to known cell types (e.g., GABAergic neurons) and to anatomically distinct brain regions or sub-regions (e.g., CA1, CA3 within the hippocampus; Fig. 1b,c, Supplementary Fig. 3,4 and Supplementary Table 3). Each had a distinct expression signature (Fig. 1d, Supplementary Table 4) and was supported by nuclei from all mice (Supplementary Fig. 5a). GABAergic neurons of the same class but from different brain regions (and different samples) group together, as do non-neuronal cells (Fig. 1b,c, Supplementary Fig. 5). Among non-neuronal cells, different glia cell-types, including astrocytes, microglia, oligodendrocytes, and oligodendrocyte precursor cells (OPC), readily partitioned into separate clusters (Fig. 1b), despite their relatively low RNA levels and correspondingly lower numbers of detected genes (Supplementary Fig. 5c,d). Finally, despite the lower number of genes detected per nucleus in this setting, the cell types and their signatures from DroNc-seq are comparable to those obtained previously with sNuc-Seq of mouse hippocampus3 and scRNA-seq of the visual cortex17 (Fig. 1e, Methods).
We also captured finer distinctions between closely related cells, congruent with earlier, lower-throughput studies. For example, we distinguished eight sub-sets of GABAergic neurons (Fig. 1f, Supplementary Fig. 6a,b), each expressing a unique combination of canonical marker genes and signatures (Supplementary Fig. 6c,d, Supplementary Table 5). To determine the congruence between cell subtypes obtained from DroNc-seq and those in previous datasets, we trained a multi-class random forest classifier11 (Methods) on the DroNc-seq GABAergic sub-clusters and used it to map GABAergic neuronal cells17 or nuclei3 from other datasets (Fig. 1g,h, Methods). Despite the different brain regions, experimental methods, and lower number of genes detected, the DroNc-seq sub-clusters matched with a nearly one-to-one mapping to sub-clusters defined by sNuc-Seq3 on the hippocampus, and with a satisfactory match to sets of fine-resolution sub-clusters defined by scRNA-seq of a distinct brain region, the visual cortex17 (Fig. 1g,h, Supplementary Fig. 6).
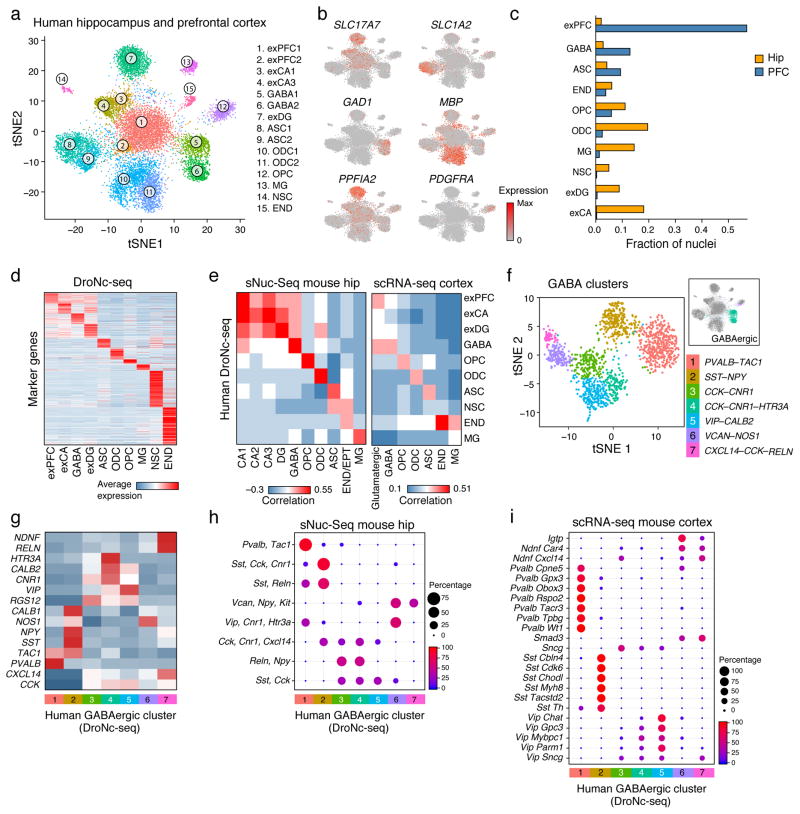
To demonstrate the utility of DroNc-seq on archived human tissue, we profiled seven frozen post-mortem samples of human hippocampus and PFC from five adults (40–65 years old), archived for 3.5–5.5 years by the GTEx project18 (Supplementary Table 6). Our analysis of 14,963 nuclei (sequenced to low depth at >10,000 reads per nucleus; with an average of 1,238 genes in neurons and 607 in non-neuronal cells, Fig. 2a–d, Supplementary Fig. 7) revealed distinct nuclei clusters corresponding to known cell types (labelled by cluster in Fig. 2a, and by major cell-type in Supplementary Fig. 7a, Supplementary Table 7). Although the human archived samples varied in quality, DroNc-seq yielded high-quality libraries of both neurons and glia cells from each sample (Supplementary Fig. 7c,d). By analyzing a large number of cells, we were able to recover rare cell types, such as cluster 14 (Fig. 2a), consisting only of hippocampal cells, and likely comprised of neural stem cells, based on expression of marker genes (Supplementary Fig. 7f).
Figure 2. DroNc-seq distinguishes cell types and signatures in adult post-mortem human brain tissue.
(a) Cell-type clusters. tSNE embedding of 14,963 DroNc-seq nuclei profiles (each with >10,000 reads and >200 genes) from adult frozen human hippocampus (Hip, 4 samples) and prefrontal cortex (PFC, 3 samples) from five donors. Nuclei are color-coded by cluster membership and clusters are labeled post-hoc (abbreviations as in Fig. 1b). (b) Marker genes. Shown is the same plot as in (a) but with nuclei colored by the expression level of known cell-type marker genes. (SLC17A7 – excitatory neurons, GAD1 – GABAergic neurons, PPFIA2 – exDG, SLC1A2 – ASC, MBP – ODC, PDGFRA – OPC). (c) Fraction of nuclei from each brain region associated with each cell type. Cell types are defined as in Supplementary Fig. 7a and sorted from left by types enriched in PFC vs. Hip. (d) Cell type expression signatures. The average expression of differentially expressed signature genes (Methods, rows) in each DroNc-seq human brain cell subset (columns; defined as in Supplementary Fig. 7a). (e) DroNc-seq cell-type expression signatures in the human brain agree with previous mouse datasets. Pairwise correlations of the average expression (Methods) for the genes in each cell-type signature defined by DroNc-seq (rows) and cell-types defined by sNuc-Seq in the mouse hippocampus3 (left, columns) and scRNA-seq in the visual cortex17 (right, columns). (f–i) GABAergic neurons sub-clusters. (f) tSNE embedding of 1,500 DroNc-seq nuclei profiles from the GABAergic neurons cluster (clusters 5–6 in Fig. 2a; inset), color coded by sub-cluster membership. (g) Average expression of canonical GABAergic marker genes (rows) in each of the nuclei sub-clusters (columns) defined in (f). (h,i) Mapping of human GABAergic neurons sub-cluster defined here (columns, from f) to subsets defined from nuclei profiles in the mouse hippocampus3 (h) and single cell profiles in the mouse visual cortex17 (i) (rows). Dot plot shows the proportion of cells in each cluster defined by the other two datasets that were classified to each DroNc-seq cluster (as in Fig. 1k,l, Methods).
The cell-type specific gene signatures we determined for each human cell-type cluster (Fig. 2d, Supplementary Table 8) agreed well with previously defined signatures in mouse hippocampus3 and cortex17 (Fig. 2e), and highlighted specific pathways (Supplementary Fig. 7e). Moreover, we captured finer distinctions between closely related cells (Fig. 2f and Supplementary Fig. 8–10). These include: subtypes of CA pyramidal neurons, reflecting anatomical distinctions within the hippocampus (Supplementary Fig. 8); subtypes of glutamatergic neurons in the PFC expressing unique cortical layer marker genes, such as RORB (layer 4–54,17) (Supplementary Fig. 9, Supplementary Table 9); and subtypes of GABAergic neurons (Fig. 2f, Supplementary Fig. 10a–c), each associated with a distinct combination of canonical markers and signatures (Fig. 2g, Supplementary Fig. 10d–e, Supplementary Table 9), as previously reported3,4,17,19. Notably, we found good congruence between our GABAergic sub-clusters and those previously defined3,4,17 in mouse and human (Fig. 2h,i, Supplementary Fig. 11, and Supplementary Table 9) using a classifier trained on one dataset and tested on the other (Methods). Human GABAergic sub-clusters mapped well to previously defined clusters in the mouse hippocampus3 (sNuc-Seq, Fig. 2h), mouse visual cortex17 (scRNA-seq, Fig. 2i), and human cortex4 (snRNA-seq, Supplementary Fig. 11), including the same assignment of canonical marker genes to each cluster (e.g. PVALB, SST, and VIP), despite the different species, experimental methods, and brain regions used in each study, as well as the lower number of genes detected in DroNc-seq.
In conclusion, DroNc-seq is a massively-parallel single nucleus RNA-seq method that is robust, cost-effective, and easy to use. DroNc-seq profiling from mouse and human frozen archived brain tissues successfully identified cell types and subtypes, rare cells, expression signatures, and activated pathways. Classifications and signatures derived from DroNc-seq profiles were congruent with prior studies in human and mouse (despite the lower number of detected genes per nucleus), but were derived with significantly improved throughput and cost. Moreover, DroNc-seq readily identified rare cell types without the need for enrichment. Nuclei grouped primarily by cell type and not by individual, indicating that cell-type signatures are largely consistent across individuals. Future studies with larger numbers of individuals should assess inter-individual variations, which may increase with aging and pathological conditions20. DroNc-seq opens the way to systematic single nucleus analysis of complex tissues that are inherently challenging to dissociate or already archived, helping create vital atlases of human tissues and clinical samples.
ONLINE METHODS
See Protocol Exchange21 for a step-by-step protocol for DroNc-seq.
EXPERIMENTAL PROCEDURES
Microfluidic device design and fabrication
Microfluidic devices were designed using AutoCAD (AutoDESK, USA), tested using COMSOL Multiphysics as well as empirically, and fabricated using soft lithographic techniques22 (Supplementary Data 1). The devices were tested on a Drop-seq setup, using bare beads (Tosoh, Japan, Cat # HW-65s) in Drop-Seq Lysis Buffer (DLB7; 10 ml stock consists of 4 ml of nuclease-free H2O, 3 ml 20% Ficoll PM−400 (Sigma, Cat # F5415-50ML), 100 μl 20% Sarkosyl (Teknova, Cat # S3377), 400 μl 0.5M EDTA (Life Technologies), 2 ml 1M Tris pH 7.5 (Sigma), and 500 μl 1M DTT (Teknova, Cat # D9750), where the DTT is added fresh) and 1x PBS, to optimize flow and bead occupancy parameters in drops. Droplet generation was assessed under a microscope in real time using a fast camera (Photron, Model # SA5), and later by sampling the emulsion using a disposable hemocytometer (Life Technologies, Cat # 22-600-100) to check droplet integrity, size, and bead occupancy. The device design is provided as a Supplementary File 1 and Supplementary Fig. 1a. The unit in the CAD provided is 1 unit = 1 μm; channel depth on device is 75 μm.
Cell culture
3T3 and HEK293 cells were prepared as described7. TF1 cells were cultured according to ATCC’s instructions. For DroNc-seq, cells were washed once with PBS, scraped with 2 ml nuclease- and protease-free Nuclei EZ lysis or EZ PREP buffer (Sigma, Cat # EZ PREP NUC-101) and processed as tissues, described below.
Dissection of mouse hippocampus and pre-frontal cortex (PFC)
Microdissections of mouse hippocampus and PFC were performed under stereomicroscope3. Dissected sub-regions were flash frozen on dry ice and stored at −80°C until processed for nuclei isolation. To validate DroNc-seq for fixed tissue (Supplementary Fig. 1e), sub-regions were placed in ice-cold RNAlater (ThermoFisher Scientific, Cat # AM7020), stored at 4°C overnight, after which RNAlater was removed and samples were stored at −80°C until processing.
Human hippocampus and PFC samples
Human hippocampus and PFC samples were obtained from the Genotype-Tissue Expression (GTEx) project. Samples were originally collected from recently deceased, non-diseased donors18,23. For this study, we selected samples of frozen hippocampus and PFC from five male donors, aged 40–65 (including three samples of PFC and four samples of hippocampus). We used RNA quality from tissues as a proxy for tissue quality, and selected tissues with RNA Integrity Number (RIN) values of 6.9 or higher (average RIN was 7.3). Average post-mortem ischemic interval for tissues was 12.4 hours (Supplementary Table 6).
Nuclei isolation
Nuclei were isolated with EZ PREP buffer (Sigma, Cat #NUC-101). Tissue samples cut into pieces < 0.5 cm or cell pellets were homogenized using a glass dounce tissue grinder (Sigma, Cat #D8938) (25 times with pastel A, and 25 times with pastel B) in 2 ml of ice-cold EZ PREP and incubated on ice for 5 minutes, with additional 2 ml ice-cold EZ PREP. Nuclei were centrifuged at 500 x g for 5 minutes at 4°C, washed with 4 ml ice-cold EZ PREP and incubated on ice for 5 minutes. After centrifugation, the nuclei were washed in 4 ml Nuclei Suspension Buffer (NSB; consisting of 1x PBS, 0.01% BSA and 0.1% RNAse inhibitor (Clontech, Cat #2313A)). Isolated nuclei were resuspended in 2 ml NSB, filtered through a 35 μm cell strainer (Corning, Cat # 352235) and counted. A final concentration of 300,000 nuclei/ml was used for DroNc-seq experiments.
For comparison experiments of nuclei isolation protocols (Supplementary Fig. 1c,d), nuclei were also isolated using the sucrose gradient centrifugation method described for sNuc-Seq3. The nuclei isolation protocol used here is more efficient than the gradient centrifugation based method and does not require ultra-centrifugation. This reduced processing time and minimized RNA degradation, facilitating processing of multiple samples.
Co-encapsulation of nuclei and barcode beads
10 μl of the single nuclei suspension in NSB (described above) was stained with DAPI (Fisher, Cat # D1306), loaded on a hemocytometer, and checked under microscope to ensure that nuclei were adequately isolated into singletons. The nuclei were suspended in NSB at ~300,000 nuclei/ml. Using ~75 um droplets, loading concentration of 300,000 nuclei/ml, and ~4.5 million drops/ml, amounts to a Poisson loading parameter, λ ~ 300,000/4,500,000= 0.07).
Barcoded beads (Chemgenes, Cat # Macosko-2011-10) were prepared as in Ref 7. Because the DroNc-seq microfluidic device has narrow channels (~70 μm), they are likely to clog from large beads, compared to Drop-seq. We therefore size-selected beads < 40 μm diameter using a strainer (PluriSelect, Cat # 43-50040-03); in our experience, these smaller beads comprise roughly 55% of the purchased bead pool. The barcoded beads were suspended in DLB (described above) and counted at 1:1 dilution in 20% PEG solution using a hemocytometer (VWR, Cat # 22-600-102)7, at concentrations between 325,000 and 350,000 per ml.
The nuclei and barcoded bead suspension were loaded7 and flown at 1.5 ml/hr each, along with carrier oil (BioRad Sciences, Cat # 186-4006) at 16 ml/hr, to co-encapsulate single nuclei and beads in ~75 μm drops (vol. ~ 200 pl) at 4,500 drops/sec and double Poisson loading concentrations. The smaller droplet volume in DroNc-seq results in higher mRNA concentration in drops (> 5x) compared to 125 μm drops in Drop-seq.
The theoretical Poisson loading concentration at 1/10 bead and nuclei occupancy for devices with channels 70 μm wide and 75 μm deep is ~520,000/ml, and 100 μm depth (also tested) is 340,000/ml. We tested bead and cell loading at this and other concentrations using species-mixing experiments7 (e.g., Supplementary Fig. 1f and Supplementary Table 1) and ease of bead flow as metrics and found that beads at 350,000/ml and nuclei at 300,000/ml concentrations performed best, in terms of low human-mouse doublet rate and fewer clogging events during droplet generation. At the nuclei loading concentrations used, the occurrence of one or more nuclei in a drop follows a Poisson distribution, P(x) = λx e−λ/x!, where λ = Poisson parameter, and x=2 for doublet estimation. As a theoretical lower bound, increasing nuclei concentration will increase doublet rate as λ2 e−λ/2; e.g., if nuclei loading is increased by 10%, the probability of getting two nuclei in a drop will increase from 0.21% to 0.25%. However, the probability of getting two or more nuclei in a drop, i.e., doublets, triplets, etc., all of which would be indistinguishable in species-mixing experiments, is P(x ≥ 2; λ = 0.07) = 0.5%. In practice, nuclei that stick together or cellular debris could also contribute to doublets or doublet-like phenomena. Empirical doublet rates in experiments ranged from ~1% (mouse tissue; clustering analysis) to ~5% (species-mixing).
For nuclei experiments on human and mouse tissue, 75 μm DroNc-seq devices were used, except for where 125 μm Drop-seq device was used for comparison (Supplementary Fig. 1b). Note that for 3T3 nuclei, both 125 μm Drop-seq and 75 μm DroNc-seq devices yielded similar results, while 3T3 cells profiled by Drop-seq had better efficiency and complexity.
Droplet breaking, washes, and reverse transcription (RT)
Microfluidic emulsion was collected into 50 ml Falcon tubes for ~22 min each, and left at room temperature for up to 45 min before breaking drops7 and performing RT7.
Post RT wash, exonuclease I treatment, PCR, and library preparation
Post RT, each barcoded bead had cDNA barcoded with the bead’s unique barcode (BC) bound onto it, also referred to as a STAMP7. STAMPs from multiple collections of a given sample were pooled at this point, resuspended in 1 mL H2O, and a 10 μl aliquot of the suspension was mixed with 10 μl of 20% PEG solution and counted. Aliquots of 5,000 beads were amplified7 using the following PCR steps: 95°C for 3 min; then 4 cycles of: 98°C for 20 sec, 65°C for 45 sec, 72°C for 3 min; then X cycles of: 98°C for 20 sec, 67°C for 20 sec, 72°C for 3 min; and finally, 72°C for 5 min, where X was adjusted according to sample quality. STAMPs from mouse tissue were amplified for X=10 cycles and PCR products were pooled in batches of 4 wells or 16 wells. STAMPS from human tissue were amplified for X=10 or 12 cycles. Human PCR products were pooled in batches of 4 wells (X=12) or 16 wells (X=10). Supernatants from each well were combined in a 1.5 ml Eppendorf tube and cleaned with 0.6X SPRI beads (Ampure XP, Beckman Coulter, Cat # A63881).
Notably, the number of PCR wells from a DroNc-seq run depends on the number of STAMPs obtained. A user may access the STAMPs in different ways, depending on the number of nuclei they wish to sequence. One would either access the pool one time or more, each time taking only a portion of the STAMPs to generate a library, and repeating the process if more is desired. For mouse and human brain, it was optimal to use 5,000 STAMPs in each PCR reaction and then pool 4 PCR wells together for library preparation, which is expected to yield 1,400 nuclei profiles based on our loading and flow parameters. Depending on the desired number of reads per nucleus and sequencing yield, one can pool higher numbers of PCR wells in a single Illumina Nextera™ library, as demonstrated here using 16–32 wells for libraries used in the clustering analysis of mouse and human brain tissue.
Purified cDNA was quantified7 and 550 pg of each sample was fragmented, tagged, and amplified in each Nextera reaction7.
Sequencing
The libraries were sequenced at 2.2 pM (mouse, 16 wells pool), 2.7 pM (mouse, 4 wells pool), and 2.3 pM (human) on an Illumina NextSeq 500. We used NextSeq 75 cycle v3 kits to sequence 20bp and 64bp paired-end reads, with Custom Read1 primer7. The sequencing cluster density and percent passing filter number from different experiments varied according to the quality of nuclei samples used, but were optimized around cluster density of 220 and 90% passing filter.
COMPUTATIONAL DATA ANALYSIS
Preprocessing of DroNc-seq data
Read filtering and alignment
Paired-end sequence reads were processed mostly as previously described7,11. Briefly, the left read was used to infer both the cell of origin, based on the first 12 bases (the Nucleus Barcode or NB), and the molecule of origin, based on the next 8 bases (Unique Molecular Index or UMI). Reads were first filtered by quality score, and the right mate of each read pair was trimmed and aligned to the genome (mouse mm10 UCSC, human hg19 UCSC) using STAR v2.4.0a24. Reads mapping to exonic regions of genes as per the mouse UCSC genome (version mm10) or the human UCSC genome (version hg19) were recorded.
Digital gene expression
Nucleus (cell) barcodes that represent genuine nuclei RNA libraries rather than technical and sequencing errors were distinguished as previously described7,11 as true or “core” nucleus barcodes. Briefly, barcodes were first filtered based on a minimum number of transcripts associated with them and then barcodes were checked for synthesis errors and collapsed to core barcodes if they were within an edit distance of 1. To account for amplification bias, gene counts were collapsed within each sample, using UMI sequences (within an edit distance of 1, substitutions only), as previously described7,11. The expression count (or number of transcripts) for a given gene in a given nucleus was determined by counting unique UMIs, and compiled into a digital gene expression (DGE) matrix. The DGE matrix was scaled by total UMI counts, multiplied by the mean number of transcripts (calculated for each dataset separately) and the values were log transformed. To reduce the effects of library quality and complexity on cluster identity, a linear model was used to regress out effects of the number of transcripts and genes detected per nucleus (using the ‘RegressOut’ function in the Seurat software package).
Gene detection and quality controls
Additional filtering of the expression matrix
Nuclei with less than 200 detected genes and less than 10,000 usable reads were filtered out. We note that, as for scRNA-seq, depending on the cell-type in question; the cut-off may need to be set on a case-by-case basis, based on the characteristic RNA content of the cell type. A gene is considered detected in a cell if it has at least two unique UMIs (transcripts) associated with it. For each analysis, genes were removed that were detected in less than 10 nuclei. After filtering, the number of cells and nuclei were as follows: (1) 1,710 cells from the 3T3 single cell libraries (collected by Drop-seq) across two replicates; (2) 5,636 3T3 nuclei across 6 replicates; (3) 19,561 nuclei from the mouse brain (4 PFC samples and 4 hippocampus samples from 4 mice used for cell type analysis, and an additional 8 cortical samples from 4 mice used for quality control experiments); and (4) 19,550 nuclei from the human brain (3 PFC samples and 4 hippocampus samples from 5 donors). Clusters and cell-type classification were robust for different gene detection thresholds. The above threshold was used in all the clustering analyses. For the quality control experiments (specifically, testing the performance with RNA Later, different nuclei isolation protocols, different microfluidic devices; Supplementary Fig. 1), at least 20,000 usable reads per nucleus were required (the number of reads at which we estimated sample saturation; Supplementary Fig. 2f,g). For the assessment of the complexity and sensitivity of DroNc-seq at least 80,000 usable reads per nucleus were required; this analysis was performed only with the samples sequenced deeply to an average of 160,000 reads per nucleus, as required for saturation analysis.
QC metrics
A list of quality metrics was obtained for all DroNc-seq datasets using Samtools (http://samtools.sourceforge.net/), Picard Tools (http://broadinstitute.github.io/picard/), and in-house scripts. For each single-nucleus profile, we calculated the total number of reads mapped to coding regions and UTRs, number of genes detected per nucleus, and the percentage of the total number of reads assigned to nucleus barcode that were from: (1) coding regions, (2) UTRs, (3) intronic regions, (4) intergenic regions, (5) ribosomal RNA (rRNA), and (6) transcripts derived from the mitochondrial genome.
Comparison of Drop-Seq (cells) and DroNc-seq (nuclei)
We compared DroNc-seq (nuclei) and Drop-seq (cells) using several measures. (1) We compared the capture rate efficiency of DroNc-seq and Drop-seq in libraries derived from pooling four PCR wells, followed by sequencing to an average depth of 160,000 usable reads per nucleus/cell. The efficiency is defined as the percent of nuclei actually observed out of the proportion expected per library, given the Poisson loading of 0.07 for DroNc-seq and 0.1 for Drop-seq. For example, at 100% efficiency, a DroNc-seq pool of 20,000 beads is expected to contain 1,400 nuclei (2,000 cells in Drop-seq). On average, we observed 87% efficiency for DroNc-seq (78%, 89%, and 95% efficiency for cell lines, mouse brain, and human brain tissue, respectively) and 72% for Drop-seq on cell lines. (2) We compared the means and the distributions of the number of genes and transcripts detected for all cells and nuclei that pass our quality filter (Supplementary Figure 2b–c). (3) We compared the expression profiles of nuclei and cells (3T3 cell line) by computing the average expression for each gene (average log transformed UMI counts) in each replicate, and then the Pearson correlation coefficients between technical replicates of cells or nuclei (all have r=0.99+/−stdev=0.0023), and between nuclei and cells (r=0.81+/−stdev= 0.0024). (Supplementary Figure 2d) (4) We tested for genes differentially expressed between cells and nuclei (3T3 cell lines) after pooling technical replicates. We defined differentially expressed genes using Student’s t-test, requiring FDR < 0.001, log-ratio > 1 and an average expression across all nuclei or cell samples log(UMI count) > 3. We found only two genes up-regulated in the nuclei (lincRNAs Malat1 and Meg3), and 57 genes up regulated in cells, including many mitochondrial RNAs and ribosomal protein RNAs (known to be stable and thus enriched in cells compared to nuclei13,14) (Supplementary Table 2). (5) We compared the fraction of the total number of reads that were mapped to (1) coding regions, (2) UTRs, (3) intronic regions, (4) intergenic regions, and (5) ribosomal RNA (as described above) (Supplementary Figure 2e).
PCA, clustering, and tSNE visualization
Finding variable genes
To select highly variable genes, we fit a relationship between mean counts and coefficient of variation using a Gamma distribution on the data from all the genes19,25, and ranked genes by the extent of excess variation as a function of their mean expression (using a threshold of at least 0.2 difference in the coefficient of variation between the empirical and the expected and a minimal mean transcript count of 0.005).
Dimensionality reduction using PCA
We used a DGE matrix consisting only of variable genes as defined above, scaled and log transformed, and then reduced its dimensions with principal components analysis (PCA). We used the fast ‘rpca’ function in R (package ‘rsvd’), and chose the most significant principal components (or PCs) based on the largest eigen value gap3 (separately for each dataset) to use as input in downstream analysis.
Graph clustering
We partitioned the profiles into clusters of transcriptionally similar nuclei using the top significant PCs as an input to a graph based clustering algorithm, as previously described11. Briefly, in the first step, we compute a k-nearest neighbor (k-NN) graph, connected each nucleus to its k-nearest neighbors (based on Euclidean distance, using the ‘nng’ function of the ‘igraph’ package in R). We next used the k-NN graph as an input to the Infomap algorithm26, which decomposes an input graph into modules using the ‘cluster_infomap’ function in R). The clustering results were visualized by coloring a tSNE27 2D map post hoc (below). We used k=100 for clustering of each full dataset, and k=80 for the human brain subset clustering (Fig. 2f, Supplementary Fig. 8–9).
Sub-clustering
To identify subtypes of cells, the same analyses were performed as described above but on a specific subset of nuclei (one or few of the major clusters; as described in the main text) to partition it to sub-clusters.
tSNE visualization
We generated a two-dimensional (2D) non-linear embedding of the nuclei profiles using tSNE. The scores along the top significant PCs estimated above were used as input to the algorithm (using the ‘Rtsne’ package, with a maximum of 2,000 iterations, disabling the initial PCA step and setting the perplexity parameter to 100 for detection of the major clusters and 60 for sub-clusters). Since tSNE can produce different visualizations in different runs, we used these coordinates only for visualization and not to identify cell clusters. Interestingly, we can associate nuclei with a distinct known cell type even for those nuclei with as few as 100 genes detected, suggesting that the cell-type identity in the brain can be encoded by a small set of genes, easily detected with shallow sequencing, as previously observed in other systems11.
To visualize the expression of known marker genes (e.g., subtypes of GABAergic neurons in the hippocampus and cortex3,19) or genes found to be up-regulated, we visualized the average expression of the markers across each cluster or cell type as violin plots, and visualized the distribution of the expression across cells in the tSNE space by color coding the dots based on expression levels.
Testing for batch and technical effects
To rule out the possibility that the resulting clusters are driven by batch or other technical effects, we examined the distribution of samples within each cluster and the distribution of the number of genes detected across clusters (as a measure of nuclei quality). Overall, the nuclei separated into distinct point clouds in tSNE space that were not driven by batch; each cluster/cloud was an admixture of cells from all technical and biological replicates, with variable numbers of genes. Related to the number of genes, we note that there is a distinct biological difference in cell size (and expected RNA content) between neuronal and glial cells in the brain.
Transcript and gene saturation analysis
To assess the extent of saturation and required read depth of the DroNc-seq libraries, we used nuclei libraries from a mouse cell line (3T3), mouse brain tissue, and human brain tissue (cortex) each sequenced to an average read depth of 160,000 reads per nucleus. We removed nuclei with less than either 200 genes detected or 10,000 reads. We performed saturation analyses for transcripts (UMI) and genes for each nucleus separately by sub-sampling reads with replacement across the range of reads for that nucleus (from 0.02 to 0.98 of the total read counts within a given nucleus or cell, in 0.02 increments). For each subsampling, we calculated the number of reads and transcripts detected. This sampling procedure was repeated 10 times and the mean values were reported. Saturation limits for UMI/genes were estimated by nonlinear fitting of the following saturation function to all points generated by the sampling procedure:
Cluster annotation, filtering, differential expression, and pathway analysis
Major cell-type clusters were identified by using a set of known cell-type marker genes from the literature, as previously described3,19. In addition, we identified signatures of up-regulated genes for each cluster (Supplementary Tables 4,5,8 and 9), which we used to further validate the identity of the cluster by matching these signatures with canonical cell-type marker genes and by testing for enriched pathways. Differentially expressed signatures were calculated using a binomial likelihood ratio test28 to find genes that are up-regulated within each cluster compared to the rest of the nuclei in the dataset, with FDR 0.01 and requiring that these genes are expressed in at least 20% of nuclei in the given cluster and have a minimum difference of 20% in the fraction of nuclei in which they are detected. The differential expression signatures were tested for enriched pathways and gene sets using a hypergeometric test (FDR < 0.01). Pathways were taken from the MSigDB/GSEA resource (combining data from Hallmark pathways, REACTOME, KEGG, GO and BIOCARTA)29.
We flagged problematic clusters to be disregarded in downstream analysis by any one of three criteria: (1) clusters that had dubious quality of nuclei, in which the nuclei associated mainly with one sample did not associate with specific cell type markers. (2) clusters with nuclei expressing both overlapping markers of two different cell types and having a relatively higher number of transcripts, indicating they might be nuclei doublets; or (3) clusters expressing markers of neighboring brain regions that might be a result of non-specific tissue dissection (such as genes enriched in the choroid plexus, Supplementary Fig. 3b). Several small clusters in the human and mouse brain were discarded from downstream analysis (as annotated in Supplementary Tables 3 and 7, and in Supplementary Fig. 3b).
Cell types were defined by combining clusters of all subtypes (e.g. the GABAergic sub-clusters were combined into one group of GABAergic neurons), which was used in the downstream analysis for testing the number of genes and transcripts in each cell type, defining cell-type specific expression signatures, sub-clustering, and comparing cell-type signatures to previous datasets.
Comparison of DroNc-seq data to previous datasets
Comparison of cell-type signatures
Cell-type specific expression patterns were compared to signatures previously defined in several relevant datasets by calculating the pairwise Pearson correlations coefficients between each pair of cell types in the other dataset and DroNc-seq datasets for the same set of genes. First, we compared to average cell-type specific signatures from sNuc-Seq analysis in the mouse hippocampus3 (Supplementary Tables in Ref. 3). Second, we compared to the single cell RNA-seq dataset of the mouse visual cortex (Tasic et al.17), using the previously defined cell-type annotations and expression values per cell (from GEO dataset GSE71585 and Ref. 17). Average log transformed TPM counts, FPKM counts, or scaled UMI counts were used to generate the mouse hippocampus3, mouse visual cortex17, and DroNc-seq signatures, respectively.
Comparison of mouse and human GABAergic sub-clusters to previously defined sub-clusters in mouse brain
To determine the congruence of cell subtypes between the DroNc-seq analyses to other neural datasets, we adopted an approach which we previously described in an analysis of retinal neurons11. Briefly, we trained a multi-class random forest classifier30 on the clusters defined on the DroNc-seq data separately for human and mouse GABAergic neurons. In each case, we used the most variable genes (approximately 700–2,000 genes across datasets, as described above) to build a classifier on 60% of the data (training set). For each dataset, the classifier was tested on the remaining 40% of the data that was not used for training (test set) to obtain an estimate of the classification accuracy. Nuclei in the test set mapped to their correct classes at a rate of 93% for the human GABAergic neurons and 91% for the mouse GABAergic neurons (expected accuracy based on random assignment was 12.5%). These classifiers were then used to map cells or nuclei in other datasets including single nucleus RNA-seq in the mouse hippocampus brain region3 and single cell RNA-seq in the mouse visual cortex17.
Comparison of human GABAergic sub-clusters to previously defined sub-clusters in human brain
To determine the congruence of neuron subtypes between DroNc-Seq analysis of hippocampus and PFC and previous analyses of human visual cortex (Lake et al.4), we used the classifier previously defined in Lake et al.4 that includes a set of signature genes at each point along a decision tree leading to the classification of eight GABAergic subtypes. To classify the DroNc-seq nuclei profiles, at each branch point in the tree, we scored each nucleus profile using the left and right gene signatures, by the average expression level of all signature genes per nucleus (log transformed UMI counts centered around the mean value), and assigned the tested nucleus by the higher score.
RNA in situ hybridization data
RNA in situ hybridization (ISH) images for marker genes was taken from the Allen Institute Brain Atlas31.
Data Availability
Raw human sequencing data is available at dbGaP, under accession phs000424.v8.p1, and expression tables are available at http://www.gtexportal.org/home/datasets. Raw and processed mouse sequencing data is available at https://portals.broadinstitute.org/single_cell and at the Gene Expression Omnibus (GEO) database.
Supplementary Material
Acknowledgments
We thank Rhiannon Macare, Assaf Rotem, Christoph Muus and Eugene Drokhlyansky for helpful discussions, Talia Habib for babysitting, Timothy Tickle and Asma Bankapur for technical support, Leslie Gaffney and Ania Hupalowska for help with graphics. Work was supported by Klarman Cell Observatory, NIMH grant U01MH105960, NCI grant 1R33CA202820-1 (to A.R.), and Koch Institute Support (core) grant P30-CA14051 from the NCI. Microfluidic devices were fabricated at the Center for Nanoscale Systems, Harvard University, member of NNIN, supported by NSF award no. 1541959. N.H. is a Howard Hughes Medical Institute (HHMI) Fellow of The Helen Hay Whitney Foundation. A.R. is an HHMI Investigator, a member of Scientific Advisory Boards for Thermo Fisher Scientific, Syros Pharmaceuticals and Driver Genomics. F.Z. is a New York Stem Cell Foundation-Robertson Investigator. F.Z. is supported by NIMH (5DP1-MH100706 and 1R01-MH110049), NSF, HHMI, the New York Stem Cell, Simons, Paul G. Allen Family, and Vallee Foundations; and James and Patricia Poitras, Robert Metcalfe, and David Cheng. D.A.W. thanks NSF DMR-1420570, NSF DMR-1310266 and NIH P01HL120839 grants for their support. NH is a HHMI fellow for Helen Hey Whitney Foundation. GTEx is supported by the Common Fund of the Office of the Director of NIH, through Contract HHSN268201000029C (to K.A LDACC, Broad Institute).
Footnotes
Author Contributions
N.H., I.A.D., A.B., O.R., F.Z. and A.R. conceived the study. A.R. and N.H. devised analyses. N.H., K.S., M.H., and F.A., analyzed the data. A.B. designed and fabricated the microfluidics device. D.A.W. devised the microfluidics design. A.B. I.A.D., N.H., and T.B. designed and conducted the experiments. S.R.C. provided the mice brain tissue. E.G., and K.A., provided the human brain tissue, N.H., I.A.D., A.B., and A.R., wrote the paper with input from all of the authors.
Competing Financial Interests Statement
N.H., A.B., I.A.D., D.A.W., F.Z. and A.R. are co-inventors on international patent application PCT/US16/59239 of Broad Institute, Harvard and MIT, relating to inventions of methods of this manuscript.
References
- 1.Wagner A, Regev A, Yosef N. Revealing the vectors of cellular identity with single-cell genomics. Nat Biotechnol. 2016;34:1145–1160. doi: 10.1038/nbt.3711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tanay A, Regev A. Scaling single-cell genomics from phenomenology to mechanism. Nature. 2017;541:331–338. doi: 10.1038/nature21350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Habib N, et al. Div-Seq: Single-nucleus RNA-Seq reveals dynamics of rare adult newborn neurons. Science. 2016;353:925–928. doi: 10.1126/science.aad7038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lake BB, et al. Neuronal subtypes and diversity revealed by single-nucleus RNA sequencing of the human brain. Science. 2016;352:1586–1590. doi: 10.1126/science.aaf1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lacar B, et al. Nuclear RNA-seq of single neurons reveals molecular signatures of activation. Nat Commun. 2016;7:11022. doi: 10.1038/ncomms11022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Grindberg RV, et al. RNA-sequencing from single nuclei. Proc Natl Acad Sci U S A. 2013;110:19802–19807. doi: 10.1073/pnas.1319700110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Macosko EZ, et al. Highly Parallel Genome-wide Expression Profiling of Individual Cells Using Nanoliter Droplets. Cell. 2015;161:1202–1214. doi: 10.1016/j.cell.2015.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dixit A, et al. Perturb-Seq: Dissecting Molecular Circuits with Scalable Single-Cell RNA Profiling of Pooled Genetic Screens. Cell. 2016;167:1853–1866. e1817. doi: 10.1016/j.cell.2016.11.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Adamson B, et al. A Multiplexed Single-Cell CRISPR Screening Platform Enables Systematic Dissection of the Unfolded Protein Response. Cell. 2016;167:1867–1882 e1821. doi: 10.1016/j.cell.2016.11.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Klein AM, et al. Droplet barcoding for single-cell transcriptomics applied to embryonic stem cells. Cell. 2015;161:1187–1201. doi: 10.1016/j.cell.2015.04.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shekhar K, et al. Comprehensive Classification of Retinal Bipolar Neurons by Single-Cell Transcriptomics. Cell. 2016;166:1308–1323 e1330. doi: 10.1016/j.cell.2016.07.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ziegenhain C, et al. Comparative Analysis of Single-Cell RNA Sequencing Methods. Mol Cell. 2017;65:631–643 e634. doi: 10.1016/j.molcel.2017.01.023. [DOI] [PubMed] [Google Scholar]
- 13.Rabani M, et al. Metabolic labeling of RNA uncovers principles of RNA production and degradation dynamics in mammalian cells. Nat Biotechnol. 2011;29:436–442. doi: 10.1038/nbt.1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rabani M, et al. High-resolution sequencing and modeling identifies distinct dynamic RNA regulatory strategies. Cell. 2014;159:1698–1710. doi: 10.1016/j.cell.2014.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schwanhausser B, et al. Global quantification of mammalian gene expression control. Nature. 2011;473:337–342. doi: 10.1038/nature10098. [DOI] [PubMed] [Google Scholar]
- 16.Cheadle C, et al. Control of gene expression during T cell activation: alternate regulation of mRNA transcription and mRNA stability. BMC Genomics. 2005;6:75. doi: 10.1186/1471-2164-6-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tasic B, et al. Adult mouse cortical cell taxonomy revealed by single cell transcriptomics. Nat Neurosci. 2016;19:335–346. doi: 10.1038/nn.4216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Consortium GTHuman genomics. The Genotype-Tissue Expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science. 2015;348:648–660. doi: 10.1126/science.1262110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zeisel A, et al. Brain structure Cell types in the mouse cortex and hippocampus revealed by single-cell RNA-seq. Science. 2015;347:1138–1142. doi: 10.1126/science.aaa1934. [DOI] [PubMed] [Google Scholar]
- 20.Tirosh I, et al. Dissecting the multicellular ecosystem of metastatic melanoma by single-cell RNA-seq. Science. 2016;352:189–196. doi: 10.1126/science.aad0501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Habib N, Avraham-Davidi I, Basu A, Burks T, Zhang F, Regev A. Protocl: Massively-parallel single nucleus RNA-seq with DroNc-seq. Protocol Exchange. 2017 doi: 10.1038/nmeth.4407. http://dx.doi.org/ [DOI] [PMC free article] [PubMed]
- 22.McDonald JC, et al. Fabrication of microfluidic systems in poly(dimethylsiloxane) Electrophoresis. 2000;21:27–40. doi: 10.1002/(SICI)1522-2683(20000101)21:1<27::AID-ELPS27>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 23.Carithers LJ, et al. A Novel Approach to High-Quality Postmortem Tissue Procurement: The GTEx Project. Biopreserv Biobank. 2015;13:311–319. doi: 10.1089/bio.2015.0032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dobin A, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brennecke P, et al. Accounting for technical noise in single-cell RNA-seq experiments. Nat Methods. 2013;10:1093–1095. doi: 10.1038/nmeth.2645. [DOI] [PubMed] [Google Scholar]
- 26.Rosvall M, Bergstrom CT. Maps of random walks on complex networks reveal community structure. Proc Natl Acad Sci U S A. 2008;105:1118–1123. doi: 10.1073/pnas.0706851105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.van der Maaten L, Hinton G. Visualizing Data using t-SNE. J Mach Learn Res. 2008;9:2579–2605. [Google Scholar]
- 28.McDavid A, et al. Data exploration, quality control and testing in single-cell qPCR-based gene expression experiments. Bioinformatics. 2013;29:461–467. doi: 10.1093/bioinformatics/bts714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Subramanian A, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Breiman L. Random Forests. Machine Learning. 2001;45:5. doi: 10.1023/A:1010933404324. [DOI] [Google Scholar]
- 31.Lein ES, et al. Genome-wide atlas of gene expression in the adult mouse brain. Nature. 2007;445:168–176. doi: 10.1038/nature05453. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw human sequencing data is available at dbGaP, under accession phs000424.v8.p1, and expression tables are available at http://www.gtexportal.org/home/datasets. Raw and processed mouse sequencing data is available at https://portals.broadinstitute.org/single_cell and at the Gene Expression Omnibus (GEO) database.