Abstract
Hedgehog (Hh) signaling regulates cell fate and self-renewal in development and cancer. Canonical Hh signaling is mediated by Hh ligand binding to the receptor Patched (Ptch), which in turn activates Gli-mediated transcription through Smoothened (Smo), the molecular target of the Hh pathway inhibitors used as cancer therapeutics. Small cell lung cancer (SCLC) is a common, aggressive malignancy with universally poor prognosis. Although preclinical studies have shown that Hh inhibitors block the self-renewal capacity of SCLC cells, the lack of activating pathway mutations have cast doubt over the significance of these observations. In particular, the existence of autocrine, ligand-dependent Hh signaling in SCLC has been disputed. In a conditional Tp53;Rb1 mutant mouse model of SCLC, we now demonstrate a requirement for the Hh ligand Sonic Hedgehog (Shh) for the progression of SCLC. Conversely, we show that conditional Shh overexpression activates canonical Hh signaling in SCLC cells, and markedly accelerates tumor progression. When compared to mouse SCLC tumors expressing an activating, ligand-independent Smo mutant, tumors overexpressing Shh exhibited marked chromosomal instability and Smoothened-independent upregulation of Cyclin B1, a putative non-canonical arm of the Hh pathway. In turn, we show that overexpression of Cyclin B1 induces chromosomal instability in mouse embryonic fibroblasts lacking both Tp53 and Rb1. These results provide strong support for an autocrine, ligand-dependent model of Hh signaling in SCLC pathogenesis, and reveal a novel role for non-canonical Hh signaling through the induction of chromosomal instability.
Keywords: Hedgehog, Lung Cancer, Mouse Models, Tumor Progression
Introduction
The Hedgehog (Hh) pathway regulates cell fate and self-renewal in development.1,2 In mammals, the pathway is activated by three lipid modified Hh proteins, Sonic (Shh), Indian (Ihh) and Desert (Dhh) Hedgehogs, which bind to the receptor Patched (Ptch1), which in turn acts to constitutively inhibit Smoothened (Smo), the molecular target of the small molecule Hh inhibitors in clinical use for basal cell carcinoma and medulloblastoma.1,2 Canonical Hh signaling is mediated via Smo activation, which induces stabilization and activation of the Gli family of latent zinc finger transcription factors.1 Transcription of Gli target genes as a consequence of Smo activation such as Gli1, cMyc and Ccnd1 then transduce the major cellular effects of canonical Hh signaling.1,2 In contrast, non-canonical Hh signaling can occur through Ptch acting as a dependence receptor independent of Smo through the regulation of Cyclin B1 and Caspase 9.1,2
Mutations in either PTCH1 or SMO result in aberrant Hh pathway activation in medulloblastoma and basal cell carcinoma.3 Although clinical trials of small molecule inhibitors of Smo have clearly shown that these tumors are addicted to aberrant Hh signaling, results in tumors that lack activating mutations have been disappointing.4 Expression of Shh ligand is frequently seen in SCLC, and preclinical studies have shown that self-renewal of SCLC cells can be inhibited by targeting Smo with small molecules, siRNA, or by conditional genetic deletion.3,5,6 One model that might explain Hh pathway activation in SCLC is autocrine signaling through the overexpression of Shh ligand.5,6 In the present study, we sought to definitively resolve the role of ligand-dependent Hh signaling in SCLC pathogenesis using a well-described conditional genetic mouse model of SCLC by determining the effects of Shh gain or loss-of-function on the tumor phenotype.
Results and Discussion
Shh is necessary and sufficient for the progression of SCLC
To better define the importance of ligand-dependent Hh signaling in SCLC, we employed a well-characterized mouse model in which conditional loxP knockout alleles of both Tp53 and Rb1 can be specifically deleted in the airway epithelium.6,7 Following the inhalation of a Cre expressing adenoviral vector (AdCre), mice carrying both p53lox/lox and Rblox/lox alleles (hereafter p53RbKO mice) develop SCLC within 9 months.6,7 To manipulate Shh expression in this model, we crossed these animals with either gain-of-function or loss-of-function Shh alleles in order to observe the effects on SCLC initiation and progression in vivo.
The conditional loss-of-function allele harbors loxP sites introduced into the endogenous Shh locus flanking exon 2, with Cre-mediated recombination resulting in a frameshift mutation.8 These mice were crossed in order to generate p53lox/lox × Rblox/lox × Shhlox/lox triple homozygotes (hereafter p53RbKOShhKO mice). The gain-of-function allele contains a transgenic lox-EGFP-STOP-lox-Shh expression cassette driven by a constitutive promoter, allowing for Cre-mediated overexpression of mouse Shh protein9 when crossed into the p53RbKO background (hereafter p53RbKOShhTg mice). Cohorts were treated with a single dose of a recombinant adenoviral vector expressing Cre recombinase (AdCre),10 observed for 9 months, and then sacrificed (Figure 1a). As a quality control for AdCre delivery, mice carrying a lox-STOP-lox-βGal cassette knocked into the Rosa locus11 were included in parallel to monitor the efficacy of airway epithelial Cre-mediated recombination (Supplementary Figure 1a).
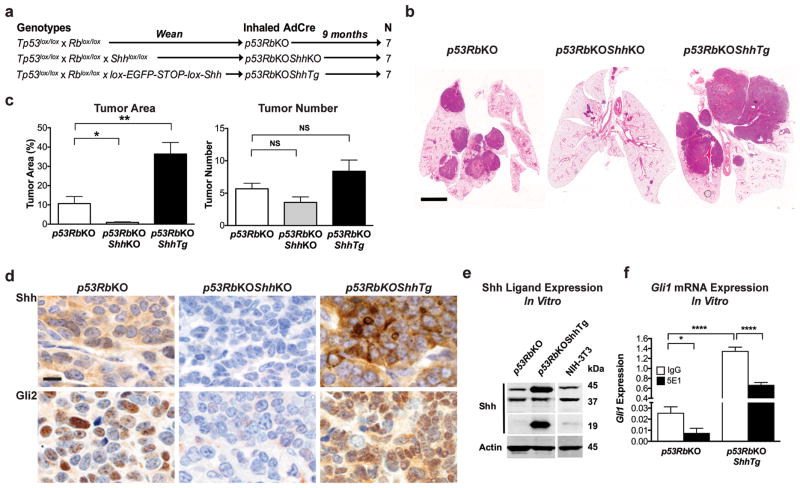
Figure 1.
Shh drives tumor progression in a mouse model of SCLC. (a) A schematic summarizing mouse cohorts and treatments. Animal experiments were conducted with the approval of the Monash University Animal Ethics Committee on a pure C57Bl6 background. Mice were obtained from Jackson Laboratories with the exception of the conditional lox-STOP-lox-Shh transgenic line9. Genotyping was performed according to Jackson Laboratory protocols, or as described9. Mice were anesthetized with Avertin and then administered 5×108 PFU Ad5CMVCre virus (University of Iowa) by intranasal inhalation at 8 weeks of age. The sample size was chosen based on published studies using this SCLC mouse model.6 (b) Representative photomicrograph images of hematoxylin and eosin stained sections of whole lungs from mice with genotypes indicated, administered AdCre by intranasal inhalation and aged for 9 months. Scale bar, 4mm. (c) Quantification of tumor area and tumor number. n = 7, data are shown as mean ± SEM. *P < 0.05, **P < 0.01, one-way ANOVA with Bonferroni correction. Tumor burden was determined by a blinded observer on each section using Imagescope software (Leica Biosystems) by calculating the total surface area of tumours as a ratio of the lung surface area in the section (Supplementary Figure S1b). (d) Representative photomicrograph images of tumor sections stained for Shh (Santa Cruz sc-9024, 1:200,34) and Gli2 (Abcam ab7195, 1:200,12,13) in lung tumour sections from the same animals shown in Figure 1. Immunoperoxidase signal is shown in brown, counterstained with hematoxylin. Scale bar, 5μm. Immunohistochemistry was performed using epitope retrieval in 0.01M Citrate Buffer (pH 6.0) and the Vectastain Elite ABC HRP Kit (Vector Laboratories) (e) Western blot analysis of Shh (Santa Cruz sc-9024, 1:100034) and actin expression in mouse SCLC cell lines derived from tumors with genotypes indicated. Mouse SCLC cell lines were grown in Advanced RPMI 1640 supplemented with 1% newborn calf serum, Penicillin-Streptomycin and Glutamax (Life Technologies). All cell lines were mycoplasma tested every 6 months. (f) Quantitative real-time RT-PCR analysis of Gli1 mRNA expression in mouse SCLC cell lines, normalised to expression of β2-microglobulin. n = 4, mean ± SEM. ****P < 0.0001, *P < 0.0001, one-way ANOVA with Bonferroni correction. RNA was extracted using RNeasy Mini Kit (Qiagen) and DNase treated on column. PCR was carried out using an ABI Prism HT-7900 (Applied Biosystems). Power SYBR Green PCR Mastermix (Life Technologies) was used according to the manufacturer’s instructions. Primer sequences: Gli1 Fwd: 5′-gaggttgggatgaagaagca-3′; Gli1 Rev: 5′-cttgtggtggagtcattgga-3′; β2m Fwd: 5′-cctggtctttctggtgcttg-3′; β2m Rev: 5′-ttcagtatgttcggcttccc-3′.
Lungs from wild type (WT), ShhKO and ShhTg mice treated with inhaled AdCre that did not carry the conditional Tp53 and Rb alleles were macroscopically and histologically normal (data not shown). In keeping with previous observations,6 p53RbKO mice developed multiple SCLC tumours 9 months after AdCre administration (Figure 1b). Strikingly, p53RbKOShhKO mice developed a similar number of tumors, most of which failed to progress beyond 1–2 mm in size (Figure 1b,c and Supplementary Figure 1b,c), and could not be propagated in cell culture. By contrast, p53RbKOShhTg animals developed much larger tumors, at a similar frequency to their p53RbKO counterparts (Figure 1b,c). Cell lines from both genotypes were generated (Supplementary Figure 1d), and expressed the pulmonary epithelial marker Thyroid Transcription Factor 1 (Ttf1), and the neuroendocrine markers Calcitonin Gene Related Peptide (Cgrp) and Cd56 (Supplementary Figure 1e). These data suggest that although deletion of Tp53 and Rb1 in the airway epithelium can initiate SCLC, Shh is required for a fully penetrant malignant phenotype.
Shh overexpression induces canonical Hh pathway activation in SCLC cells
Immunohistochemical analysis of Shh ligand expression revealed the absence or overexpression of Shh in p53RbKOShhKO or p53RbKOShhTg tumors respectively (Figure 1d). Staining for nuclear expression of Gli2, an indirect measure of canonical Hh signaling commonly used in mouse development and cancer models12,13 revealed heterogeneous expression in p53RbKO tumors that contrasted with absent expression in p53RbKOShhKO tumors, and widespread strong nuclear expression in p53RbKOShhTg tumors (Figure 1d). To detect the presence of cell-autonomous, ligand-dependent Hh signaling, we confirmed Shh overexpression in cells derived from p53RbKOShhTg tumors (Figure 1e), as well as overexpression of Gli1, commonly used in mouse cell culture models to measure canonical Hh signaling.1,2 Treatment with 5E1, a monoclonal antibody that inhibits binding of Hh ligands to the Ptch1 receptor14 attenuated Gli1 expression in both p53RbKO SCLC and p53RbKOShhTg SCLC (Figure 1f). Although we cannot exclude a functional contribution of stromal Shh signaling in this model in vivo, these data show that transgenic overexpression of Shh in SCLC can trigger ligand-dependent, canonical Hh signaling in a similar way to that seen in response to mutational activation of Smo6.
Shh overexpression results in highly proliferative tumors with large cell features
Since the SmoM2 allele directly activates canonical Hh signaling, one would predict that overexpression of Shh would phenocopy the effect of cell autonomous Smo activation in SCLC.6 In order to compare the effects of conditional overexpression of Shh with cell autonomous activation of Smo, we re-analysed previously described SCLC samples from p53RbKO mice carrying a conditional active SmoM2 allele (hereafter p53RbKOSmoM2)6, and compared these results with tumors obtained from the experiments shown in Figure 1.
As expected, tumors from p53RbKO animals developed SCLC with typical histologic features6,7. (Figure 3a). Tumors from the p53RbShhKO cohort were characterized by less nuclear pleomorphism and scant cytoplasm (Figure 2a). By contrast, p53RbKOShhTg and p53RbKOSmoM2 tumors were highly proliferative, confirmed by quantitative immunohistochemical analysis of Pcna expression (Figure 2a,b). In tumours from all three genotypes, ubiquitous expression of Ttf1 confirmed that the tumors were of pulmonary origin (Supplementary Figure 2). In all three genotypes, heterogeneous expression of Cgrp, and Cd56 was also consistent with a neuroendocrine phenotype (Supplementary Figure 2). Unexpectedly, although p53RbKOShhTg tumors clearly possess a growth advantage, an increase in TUNEL staining suggested an increase spontaneous apoptosis compared to tumors from the other genotypes (Figure 2a,c).
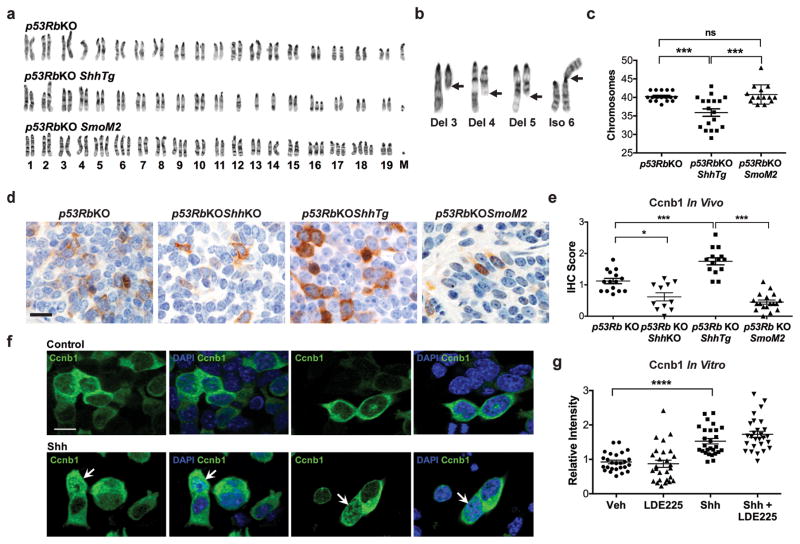
Figure 3.
(a) Examples of karyotypes in cell lines derived from p53RbKO, p53RbKOShhTg and p53RbKOSmoM2 mouse SCLC tumors. Unblinded karyotyping was performed as described.37 (b) Examples of chromosomal abnormalities in a p53RbKOShhTg SCLC cell line. Del, deletion; Iso, isochromosome. (c) Karyotype analysis shown as total chromosome number in cell lines derived from p53RbKO, p53RbKOShhTg and p53RbKOSmoM2 tumors, 3–4 cell lines, 4–5 mitoses per cell line. **P < 0.001; ***P < 0.0001, one-way ANOVA with Bonferroni correction. (d) Representative photomicrograph images of SCLC tumor sections stained for Ccnb1, (Cell Signaling 4138S, 1:200,38) from mice with the genotypes indicated. Immunoperoxidase staining is in brown, counterstained with hematoxylin in blue. Scale bar, 10μm. (e) Quantitative assessment of Ccnb1 staining in the same experiment depicted in Figure 3d, data plotted as mean ± SEM. p53RbKO, 22 tumors from 7 animals; p53RbKOShhKO 11 tumors from 7 animals; p53RbKOShhTg, 27 tumors from 7 animals p53RbKOShhTg, 27 tumors from 7 animals. *P < 0.05, ***P < 0.001, one-way ANOVA with Bonferroni correction. (f) Examples of immunofluorescence images of p53RbKO SCLC cells treated with vehicle of Sonic Hedgehog (Shh, 1μg/ml). Cells were stained for Ccnb1 (green), and a DNA counterstained (DAPI, blue). Scale bar, 2μm. (g) Quantitative analysis of nuclear Ccnb1 staining in p53RbKO SCLC cells treated with vehicle, the Smo antagonist LDE225 (400nM), recombinant Sonic Hedgehog (Shh, 1μg/ml), or in combination. n = 5, mean ± SEM. **P <0.01, ***P < 0.001 one-way ANOVA with Bonferroni correction. Cells were fixed in 4% paraformaldehyde, permeabilized with 0.5% Triton X-100 and stained as described39. FIJI-ImageJ and Adobe Photoshop CC were used for image handling. The nuclear fluorescence intensity was calculated using FIJI-ImageJ (v1.51c) as previously described.39
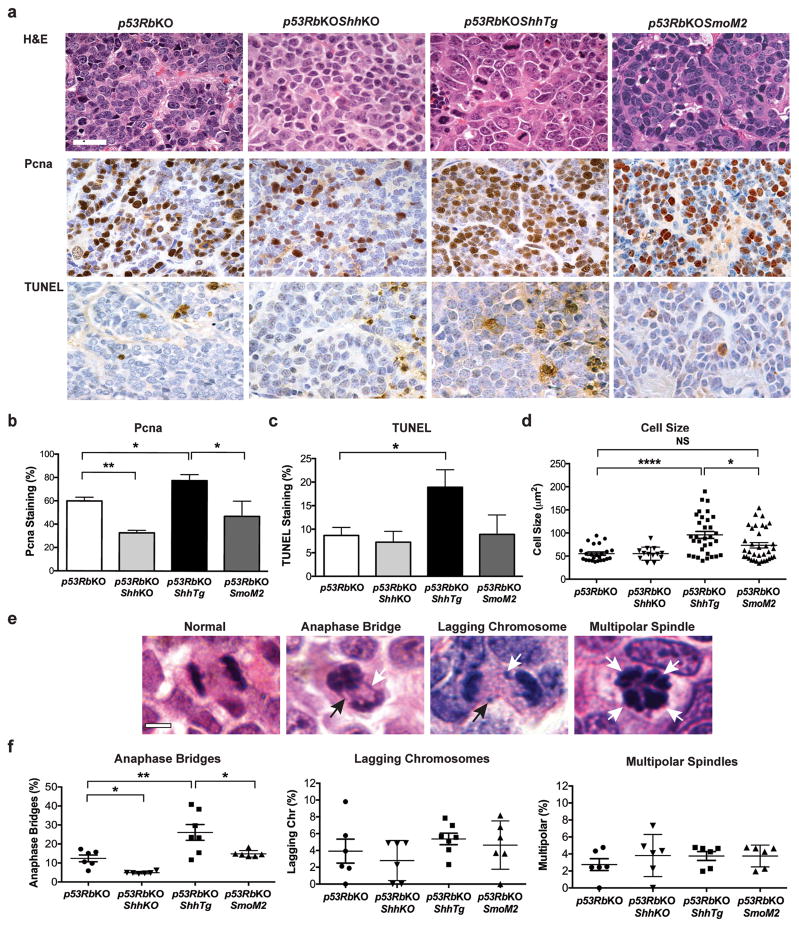
Figure 2.
Effects of gain or loss of function Shh or gain of function Smo alleles on the SCLC phenotype in a conditional Tp53/Rb1 knockout mouse model. (a) Representative photomicrographs of formalin fixed, paraffin-embedded sections of mouse SCLC tumors stained with hematoxylin and eosin (H&E), immunostained for Proliferating Cell Nuclear Antigen (Pcna) Dako M0879, 1:1200,35 or with Terminal deoxynucleotidyl transferase dUTP nick end labeling (TUNEL) (ApopTag system, Merck-Millipore). Scale bar, 50μm. (b, c) Quantitative assessment of Pcna and TUNEL staining, data plotted as mean ± SEM. p53RbKO, 22 tumors from 4 animals; p53RbKOShhKO 14 tumors from 5 animals; p53RbKOShhTg, 27 tumors from 5 animals; p53RbKOSmoM2 34 tumors from 5 animals. **P < 0.01, *P < 0.05, one-way ANOVA with Bonferroni correction. Staining was analyzed by an independent, blinded observer using the QuickScore method.36 (d) Quantitative assessment of cell size from tumors from the same experiment. Data plotted as mean ± SEM. ****P < 0.0001, *P < 0.05, one-way ANOVA with Bonferroni correction. Cell size was measured using Aperio ImageScope software by a blinded independent observer. (e) Hematoxylin and eosin stained sections mouse SCLC showing examples of anaphase abnormalities. Scale bar, 2μm. (f) Quantitative assessment of anaphase bridges, lagging chromosomes and multipolar spindles by a blinded observer in tumors from mice with the genotypes indicated. Data shown as mean ± SEM. n = 3–7 animals, 40 anaphases counted per animal. *P < 0.05, **P < 0.01 one-way ANOVA with Bonferroni correction.
Although p53RbKOShhTg and p53RbKOSmoM2 tumors both exhibited a neuroendocrine phenotype, histopathologic analysis also revealed a striking large cell phenotype in p53RbKOShhTg tumors (Figure 2a,d and Supplementary Figure 3a). By contrast, cell size in p53RbKO and p53RbKOSmoM2 tumors were more typical of SCLC (Figure 2a,d and Supplementary Figure 3a). These data show that although Shh overexpression can induce canonical Hh signaling through Smo in SCLC cells, and that the accelerated SCLC phenotype is in keeping with that seen in response to Smo activation,6 the large cell phenotype and spontaneous apoptosis conferred by overexpression of Shh was not seen in p53RbKOSmoM2 tumors. This suggested that Shh overexpression may play an additional role beyond the activation of Smo and Gli-mediated transcription.
Shh overexpression drives chromosomal instability and segmental aneuploidy in SCLC
Histologic analysis of SCLC tumors revealed an increase in the prevalence of anaphase bridges as well as bizarre mitotic figures, pleomorphic nuclei and giant cell formation in p53RbKOShhTg when compared to tumors from the other genotypes (Figure 2e,f and Supplementary Figure 3b). All of these features are characteristic of chromosomal instability.15 Other markers of chromosomal instability, such as lagging chromosomes and multipolar mitoses were similar across all four genotypes (Figure 2f).
Chromosomal instability is typically associated with segmental or numerical aneuploidy in cancer cells16. To confirm this in our SCLC model, we performed karyotype analysis on a series of newly derived SCLC lines from p53RbKO and p53RbKOShhTg tumors, as well established cell lines from p53RbKOSmoM2 tumors.6 Cells derived from p53RbKO tumors contained a near-diploid chromosomal content, whereas p53RbKOShhTg tumors exhibited numerical aneuploidy characterised by chromosomal loss, as well as frequent segmental chromosomal defects typically seen in association with anaphase bridge formation17 (Figure 3a,b,c). In contrast, cells derived from p53RbKOSmoM2 tumors lacked segmental defects, and were characterized by whole chromosome gain, more in keeping with errors in mitotic segregation17 (Figure 3a,b,c). These data suggest that Shh overexpression induces a pattern of chromosomal instability distinct from that resulting from ligand-independent constitutive activation of Smo.
Shh overexpression activates Ccnb1 signaling in SCLC independent of Smo
We first considered whether two well-characterized transcriptional targets of canonical Hh signaling, Myc and Cyclin D1 (Ccnd1)1,2 might explain the chromosomal instability phenotype seen in p53RbKOShhTg tumors. In particular, Ccnd1 was an attractive candidate due to its reported ability to induce chromosomal instability in the absence of Rb18. As shown in Supplementary Figure 4, p53RbKOShhTg and p53RbKOSmoM2 tumours showed no consistent differences in the expression of either Myc or Ccnd1 protein when comparing SCLC tumors across all genotypes. Given these data, we next asked whether non-canonical, Smo-independent Hh signaling could explain the chromosomal instability phenotype induced by overexpression of Shh.
Independent of Smo, Ptch has been shown to act as a dependence receptor by constitutively activating Caspase 9.19 Since Shh overexpression induced an increase in TUNEL staining in our SCLC model, we dismissed this as a potential mechanism to explain our findings. We next focused on Cyclin B1 (Ccnb1) as a non-canonical Hh pathway target that might explain the chromosomal instability seen in p53RbKOShhTg tumors. Evidence from several groups suggests that Ptch acts as cell cycle “gate keeper” by inhibiting the nuclear localization of Ccnb1, and that inhibition of Ptch through a variety of ways can lead to activation of Ccnb1 signaling independent of Smo.20,21 Moreover, transgenic overexpression of Ccnb1 in vivo induces cancer that is associated with chromosomal instability characterized by the formation of anaphase bridges.22,23
To test this idea, we analyzed Ccnb1 expression and localization in SCLC tumors from all four genotypes. As shown in Figure 3d and e, an increase in nuclear expression of Ccnb1 was seen in p53RbKOShhTg SCLC tumors compared with both p53RbKO and p53RbKOSmoM2 tumors. By contrast, SCLC tumors in p53RbShhKO mice expressed lower levels of Ccnb1. To confirm this result in vitro, we treated p53RbKO SCLC cells with recombinant Shh ligand in culture. This induced nuclear expression of Ccnb1, an effect that was not inhibited by treatment with the Smo antagonist LDE225 (Figure 3f,g). These data suggest that inhibition of Ptch by Shh is able to induce nuclear localisation of Ccnb1 that mirrors the effect of Ptch inactivation20, independent of Smo signaling, consistent with activation of non-canonical Hh signaling.
Ccnb1 overexpression induces chromosomal instability in cells lacking both Tp53 and Rb1
To determine whether aberrant Ccnb1 signaling could induce chromosomal instability in the absence of both Tp53 and Rb1, we employed a mouse embryonic fibroblast model in which the conditional mouse knockout lines described in Figure 1a were crossed with transgenic mice carrying a tamoxifen-inducible Cre recombinase expression cassette knocked into the Rosa26 locus24. In preliminary experiments, we found that MEFs provide significant advantages over cultured mouse SCLC cells with respect to adherent morphology and reproducible visualization of anaphase architecture (data not shown). This allowed us to directly test the relationship between activation of Ccnb1 and the generation of chromosomal instability in cells lacking Tp53 and Rb. To do this, we used an overexpression approach in order to induce aberrant Ccnb1 signaling.22,23
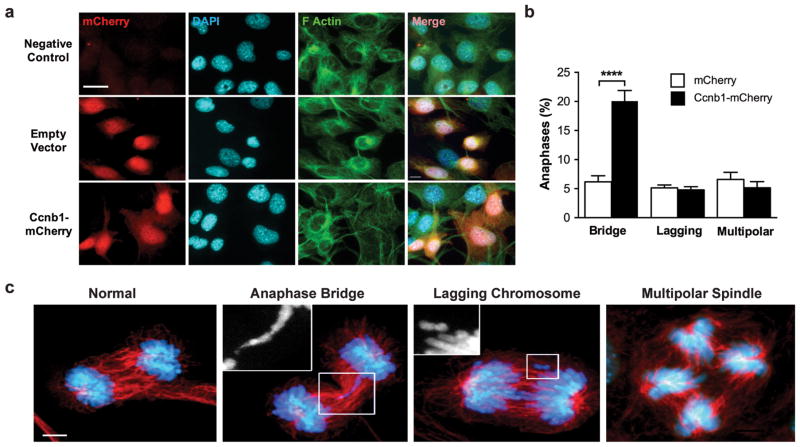
Mouse embryonic fibroblasts (MEFs) generated from p53RbKOEsrCre embryos were treated with tamoxifen and then allowed to spontaneously immortalize (data not shown). Using lentiviral transduction, we generated stable p53RbKOEsrCre MEF lines expressing either mCherry, or a Ccnb1-mCherry bicistronic expression vector (Figure 4a). Quantitative analysis revealed an increase in the prevalence of anaphase bridges, but not lagging chromosomes or multipolar mitoses in response to overexpression of Ccnb1 (Figure 4b,c). These data replicate the effects on anaphase morphology of Shh overexpression in the mouse SCLC model, and are consistent with the effects of Ccnb1 overexpression in a transgenic mouse tumor model 22,23. These results suggest that upregulation of Ccnb1 can induce chromosomal instability in addition the deregulation of the cell cycle induced by the combined loss of both Tp53 and Rb1.
Figure 4.
Cyclin B1 overexpression induces chromosomal instability in mouse embryonic fibroblast cells (MEFs) lacking both Tp53 and Rb. Mouse embryonic fibroblast p53RbKO cell lines were generated and grown as described.40 All cell lines were mycoplasma tested every 6 months. (a) Fluorescence images of mCherry expression in p53RbKO MEFs transduced with an mCherry expressing empty vector control (transduction efficiency 75%), or mCherry-Ccnb1 (transduction efficiency 60%). Cells were counterstained with DAPI and for F-Actin. Scale bar, 10 μm. Plasmid constructs (GeneCopoeia) containing mouse Ccnb1-mCherry (pReceiver-LV213-mCCNB1-IRES2_mCherry-IRES-Puro), or mCherry control (pReceiver-LV213-IRES2-mCherry-IRES-Puro) were used to prepare lentiviral particles by transfecting HEK293T cells with Lipofectamine3000 (Life Technologies) and the packaging plasmids pMDLg/pRRE, pRSV-Rev and pMD2.G. Culture supernatants were collected after 48 h, filtered through 0.45 um and used to transduce MEF p53RbKO cells with 8 μg/mL polybrene. 72 hours after transduction, cells were selected with 2 ug/ml puromycin to create stable populations. (b) Quantitative assessment of aberrant anaphases in p53RbKO MEFs transduced with lentiviruses expressing either empty vector mCherry or Cyclin B1-mCherry. N=4 independent experiments, mean ± SEM. ****P < 0.001, one-way ANOVA with Bonferroni correction. (c) Representative confocal immunofluorescence images showing examples of anaphase bridge formation (highlighted), lagging chromosomes (highlighted) and a multipolar spindle. Tubulin (custom produced in mouse, 1:50038) is shown in red, DNA stained with DAPI (blue). Images were captured using a 63× lens on a Leica DM6000 SP8 Confocal microscope in 0.3 μm z-sections displayed as 2D maximum projections generated with FIJI (v8.1). Anaphase and telophase cells were identified on the presence of clearly separated and condensed chromosomes and the elongation of the mitotic spindle and establishment of the mid-spindle.
We have identified a definitive role for Shh ligand in the progression of SCLC in vivo using a genetically modified mouse model. This is in stark contrast to the effect of deleting Shh in pancreatic cancer precursor cells, which results in more aggressive tumors unrestrained by a stromal reaction dependent on epithelial-stromal Hh signaling25. Since activating mutations in the Hh pathway do not occur in SCLC, our findings support an important role of ligand-dependent Hh signaling in SCLC pathogenesis in vivo, and suggest that targeting Shh with monoclonal antibodies may represent a potential therapeutic opportunity.
The large cell features and chromosomal instability seen in p53RbKOShhTg tumors was unexpected. Although the conditional p53RbKO mouse model of SCLC accurately recapitulates the human disease in many respects, the slow development of tumors (9–12 months) and lack of chromosomal instability suggest that factors that cooperate with the loss of Tp53 and Rb1 contribute to the tumor biology of this disease in humans26,27. Although these differences may reflect the potent mutagenic effects of tobacco exposure seen in human SCLC, with recent evidence suggesting that in the genetic mouse model, cooperating genetic events can have a major impact on tumor progression28–30. Since chromosomal instability is now recognized as a critical event in tumor progression, chemoresistance, clonal heterogeneity, transcriptional and metabolic deregulation29,31–33, our results reveal a novel role for Shh signaling in the progression of SCLC.
Supplementary Material
Acknowledgments
Financial Support: National Health and Medical Research Council of Australia (GNT1048669): VG, JEC, DNW
The Victorian Cancer Agency (TS10-01): VG, JEC, DNW
The Petre Foundation: DNW
Victorian Government’s Operational Infrastructure Support Program: DNW
National Institutes of Health (NCI CA201513): JS
Cancer Institute of NSW Fellowship (10/FRL/3-02) and the Patricia Helen Guest Fellowship: AB
This work was supported by the National Health and Medical Research Council of Australia (GNT1048669), The Victorian Cancer Agency (TS10-01), The Petre Foundation, the Victorian Government’s Operational Infrastructure Support Program, and the National Institutes of Health (NCI CA201513). The contents of this manuscript are solely the responsibility of the participating institutions and individual authors, and do not reflect the views of these funding agencies.
Footnotes
Conflict of Interest: DNW is a co-inventor on a patent related to aspects of this work.
Supplementary Information accompanies this paper on the Oncogene website (www.nature.com/Oncogene)
References
- 1.Briscoe J, Thérond PP. The mechanisms of Hedgehog signalling and its roles in development and disease. Nat Rev Mol Cell Biol. 2013;14:418–431. doi: 10.1038/nrm3598. [DOI] [PubMed] [Google Scholar]
- 2.Pak E, Segal RA. Hedgehog Signal Transduction: Key Players, Oncogenic Drivers, and Cancer Therapy. Dev Cell. 2016;38:333–344. doi: 10.1016/j.devcel.2016.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Marini KD, Payne BJ, Watkins DN, Martelotto LG. Mechanisms of Hedgehog signalling in cancer. Growth Factors. 2011;29:221–234. doi: 10.3109/08977194.2011.610756. [DOI] [PubMed] [Google Scholar]
- 4.Amakye D, Jagani Z, Dorsch M. Unraveling the therapeutic potential of the Hedgehog pathway in cancer. Nat Med. 2013;19:1410–1422. doi: 10.1038/nm.3389. [DOI] [PubMed] [Google Scholar]
- 5.Watkins DN, Berman DM, Burkholder SG, Wang B, Beachy PA, Baylin SB. Hedgehog signalling within airway epithelial progenitors and in small-cell lung cancer. Nature. 2003;422:313–317. doi: 10.1038/nature01493. [DOI] [PubMed] [Google Scholar]
- 6.Park K-S, Martelotto LG, Peifer M, Sos ML, Karnezis AN, Mahjoub MR, et al. A crucial requirement for Hedgehog signaling in small cell lung cancer. Nat Med. 2011;17:1504–1508. doi: 10.1038/nm.2473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Meuwissen R, Linn SC, Linnoila RI, Zevenhoven J, Mooi WJ, Berns A. Induction of small cell lung cancer by somatic inactivation of both Trp53 and Rb1 in a conditional mouse model. Cancer Cell. 2003;4:181–189. doi: 10.1016/s1535-6108(03)00220-4. [DOI] [PubMed] [Google Scholar]
- 8.Lewis PM, Dunn MP, McMahon JA, Logan M, Martin JF, St-Jacques B, et al. Cholesterol modification of sonic hedgehog is required for long-range signaling activity and effective modulation of signaling by Ptc1. Cell. 2001;105:599–612. doi: 10.1016/s0092-8674(01)00369-5. [DOI] [PubMed] [Google Scholar]
- 9.Wang DH, Clemons NJ, Miyashita T, Dupuy AJ, Zhang W, Szczepny A, et al. Aberrant epithelial-mesenchymal Hedgehog signaling characterizes Barrett’s metaplasia. Gastroenterology. 2010;138:1810–1822. doi: 10.1053/j.gastro.2010.01.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.DuPage M, Dooley AL, Jacks T. Conditional mouse lung cancer models using adenoviral or lentiviral delivery of Cre recombinase. Nat Protocol. 2009;4:1064–1072. doi: 10.1038/nprot.2009.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Soriano P. Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat Genet. 1999;21:70–71. doi: 10.1038/5007. [DOI] [PubMed] [Google Scholar]
- 12.Wong SY, Seol AD, So P-L, Ermilov AN, Bichakjian CK, Epstein EH, et al. Primary cilia can both mediate and suppress Hedgehog pathway-dependent tumorigenesis. Nat Med. 2009;15:1055–1061. doi: 10.1038/nm.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Youssef KK, Van Keymeulen A, Lapouge G, Beck B, Michaux C, Achouri Y, et al. Identification of the cell lineage at the origin of basal cell carcinoma. Nat Cell Biol. 2010;12:299–305. doi: 10.1038/ncb2031. [DOI] [PubMed] [Google Scholar]
- 14.Ericson J, Morton S, Kawakami A, Roelink H, Jessell TM. Two critical periods of Sonic Hedgehog signaling required for the specification of motor neuron identity. Cell. 1996;87:661–673. doi: 10.1016/s0092-8674(00)81386-0. [DOI] [PubMed] [Google Scholar]
- 15.Geigl JB, Obenauf AC, Schwarzbraun T, Speicher MR, Speicher MR. Defining ‘chromosomal instability’. Trends Genet. 2008;24:64–69. doi: 10.1016/j.tig.2007.11.006. [DOI] [PubMed] [Google Scholar]
- 16.Heng HH, Bremer SW, Stevens JB, Horne SD, Liu G, Abdallah BY, et al. Chromosomal instability (CIN): what it is and why it is crucial to cancer evolution. Cancer Metastasis Rev. 2013;32:325–340. doi: 10.1007/s10555-013-9427-7. [DOI] [PubMed] [Google Scholar]
- 17.Pfau SJ, Amon A. Chromosomal instability and aneuploidy in cancer: from yeast to man. EMBO Reports. 2012;13:515–527. doi: 10.1038/embor.2012.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Casimiro MC, Di Sante G, Crosariol M, Loro E, Dampier W, Ertel A, et al. Kinase-independent role of cyclin D1 in chromosomal instability and mammary tumorigenesis. Oncotarget. 2015;6:8525–8538. doi: 10.18632/oncotarget.3267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fombonne J, Bissey P-A, Guix C, Sadoul R, Thibert C, Mehlen P. Patched dependence receptor triggers apoptosis through ubiquitination of caspase-9. Proc Natl Acad Sci USA. 2012;109:10510–10515. doi: 10.1073/pnas.1200094109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Adolphe C, Hetherington R, Ellis T, Wainwright B. Patched1 Functions as a Gatekeeper by Promoting Cell Cycle Progression. Cancer Res. 2006;66:2081–2088. doi: 10.1158/0008-5472.CAN-05-2146. [DOI] [PubMed] [Google Scholar]
- 21.Barnes EA, Kong M, Ollendorff V, Donoghue DJ. Patched1 interacts with cyclin B1 to regulate cell cycle progression. EMBO J. 2001;20:2214–2223. doi: 10.1093/emboj/20.9.2214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.de Cárcer G, Malumbres M. A centrosomal route for cancer genome instability. Nat Cell Biol. 2014;16:504–506. doi: 10.1038/ncb2978. [DOI] [PubMed] [Google Scholar]
- 23.Nam H-J, van Deursen JM. Cyclin B2 and p53 control proper timing of centrosome separation. Nat Cell Biol. 2014;16:538–549. doi: 10.1038/ncb2952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Badea TC, Wang Y, Nathans J. A noninvasive genetic/pharmacologic strategy for visualizing cell morphology and clonal relationships in the mouse. J Neurosci. 2003;23:2314–2322. doi: 10.1523/JNEUROSCI.23-06-02314.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rhim AD, Oberstein PE, Thomas DH, Mirek ET, Palermo CF, Sastra SA, et al. Stromal elements act to restrain, rather than support, pancreatic ductal adenocarcinoma. Cancer Cell. 2014;25:735–747. doi: 10.1016/j.ccr.2014.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gazdar AF, Savage TK, Johnson JE, Berns A, Sage J, Linnoila RI, et al. The Comparative Pathology of Genetically Engineered Mouse Models for Neuroendocrine Carcinomas of the Lung. J Thorac Oncol. 2015;10:553–564. doi: 10.1097/JTO.0000000000000459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gazdar AF, Hirsch FR, Minna JD. From Mice to Men and Back: An Assessment of Preclinical Model Systems for the Study of Lung Cancers. J Thorac Oncol. 2016;11:287–299. doi: 10.1016/j.jtho.2015.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cui M, Augert A, Rongione M, Conkrite K, Parazzoli S, Nikitin AY, et al. PTEN is a potent suppressor of small cell lung cancer. Mol Cancer Res. 2014;12:654–659. doi: 10.1158/1541-7786.MCR-13-0554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McFadden D, Papagiannakopoulos T, Taylor-Weiner A, Stewart C, Carter SL, Cibulskis K, et al. Genetic and Clonal Dissection of Murine Small Cell Lung Carcinoma Progression by Genome Sequencing. Cell. 2014;156:1298–1311. doi: 10.1016/j.cell.2014.02.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wu N, Jia D, Ibrahim AH, Bachurski CJ, Gronostajski RM, MacPherson D. NFIB overexpression cooperates with Rb/p53 deletion to promote small cell lung cancer. Oncotarget. 2016 doi: 10.18632/oncotarget.11583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shaukat Z, Liu D, Choo A, Hussain R, O’Keefe L, Richards R, et al. Chromosomal instability causes sensitivity to metabolic stress. Oncogene. 2015;34:4044–4055. doi: 10.1038/onc.2014.344. [DOI] [PubMed] [Google Scholar]
- 32.Stevens JB, Horne SD, Abdallah BY, Ye CJ, Heng HH. Chromosomal instability and transcriptome dynamics in cancer. Cancer Metastasis Rev. 2013;32:391–402. doi: 10.1007/s10555-013-9428-6. [DOI] [PubMed] [Google Scholar]
- 33.Sheltzer JM. A transcriptional and metabolic signature of primary aneuploidy is present in chromosomally unstable cancer cells and informs clinical prognosis. Cancer Res. 2013;73:6401–6412. doi: 10.1158/0008-5472.CAN-13-0749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.O’Toole SA, Machalek DA, Shearer RF, Millar EK, Millar EKA, Nair R, et al. Hedgehog Overexpression Is Associated with Stromal Interactions and Predicts for Poor Outcome in Breast Cancer. Cancer Res. 2011;71:4002–4014. doi: 10.1158/0008-5472.CAN-10-3738. [DOI] [PubMed] [Google Scholar]
- 35.Roos G, Landberg G, Huff JP, Houghten R, Takasaki Y, Tan EM. Analysis of the epitopes of proliferating cell nuclear antigen recognized by monoclonal antibodies. Lab Invest. 1996;993(68):204–210. [PubMed] [Google Scholar]
- 36.Detre S, Saclani Jotti G, Dowsett M. A ‘quickscore’ method for immunohistochemical semiquantitation: validation for oestrogen receptor in breast carcinomas. Journal of Clinical Pathology. 1995;48:876–878. doi: 10.1136/jcp.48.9.876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nagy A, Gertsenstein M, Vintersten K, Behringer R. Karyotyping mouse cells. CSH Protoc. 2008;2008 doi: 10.1101/pdb.prot4706. pdb.prot4706. [DOI] [PubMed] [Google Scholar]
- 38.McCloy RA, Rogers S, Caldon CE, Lorca T, Castro A, Burgess A. Partial inhibition of Cdk1 in G 2 phase overrides the SAC and decouples mitotic events. Cell Cycle. 2014;13:1400–1412. doi: 10.4161/cc.28401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rogers S, Fey D, McCloy RA, Parker BL, Mitchell NJ, Payne RJ, et al. PP1 initiates the dephosphorylation of MASTL, triggering mitotic exit and bistability in human cells. J Cell Sci. 2016;129:1340–1354. doi: 10.1242/jcs.179754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Robles-Oteiza C, Taylor S, Yates T, Cicchini M, Lauderback B, Cashman CR, Burds AA, Winslow MM, Jacks T, Feldser DM. Recombinase-based conditional and reversible gene regulation via XTR alleles. Nat Commun. 2015;6:8783. doi: 10.1038/ncomms9783. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.