Abstract
Racemates often have lower solubility than enantiopure compounds, and mixing of enantiomers can enhance aggregation propensity of peptides. Amyloid β (Aβ) 42 is an aggregation-prone peptide, believed to play a key role in Alzheimer’s Disease. Soluble Aβ42 aggregation intermediates (oligomers) have emerged as particularly neurotoxic. We hypothesized that addition of mirror image (D-) Aβ42 should reduce the concentration of toxic oligomers formed by natural (L-) Aβ42. We synthesized L- and D-Aβ42 and found their equimolar mixing to lead to accelerated fibril formation. Confocal microscopy with fluorescently labeled analogs of the enantiomers showed their co-localization in racemic fibrils. Reflecting enhanced fibril formation propensity, racemic Aβ42 was less prone to form soluble oligomers. This resulted in protection of cells from toxicity of L-Aβ42 at concentrations ranging up to 50 µM. In summary, mixing of Aβ42 enantiomers induces accelerated formation of non-toxic fibrils.
Keywords: D-peptides, Racemate, Amyloid β, Aggregation, Toxic oligomers
COMMUNICATION
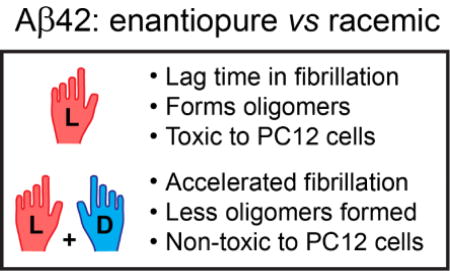
We synthesized both enantiomers of Aβ42 and found their mixing to induce drastic acceleration of fibrillization. When made from fluorescently labelled analogues, racemic fibrils showed high degree of co-localization of the enantiomers. The racemate showed a reduced propensity to yield oligomeric aggregation intermediates, which, remarkably, resulted in inhibition of toxicity of the natural L-Aβ42 enantiomer in the racemate.
Enantiomers and racemates of the same molecule may exhibit drastically different properties upon aggregation, which can lead to pronounced differences in both structure and reactivity of the resultant molecular assemblies.[1] Pauling and Corey in 1953 proposed that racemic peptide mixtures should be able to form stable structures with alternating L- and D-amino acid-derived peptide units. The authors referred to this arrangement as the „rippled sheet“ configuration.[2a] Recent studies have shown that such heterochiral interfaces can indeed be formed from biologically relevant peptides.[2b] Mixing of the enantiomers of certain intrinsically disordered peptides was also found to lead to the formation of peptidic frameworks with enhanced stability.[3]
Amyloid beta (Aβ) is a hydrophobic, intrinsically disordered, aggregation-prone peptide that is believed to play a pivotal role in Alzheimer’s disease.[4] Aβ is produced as a cleavage product of the transmembrane protein APP and can range from 36 to 43 amino acids in length.[4a] The 40 amino acid long variant (Aβ40) is the most abundant form of the peptide, but its 42 amino acid long analogue (Aβ42) is substantially more neurotoxic, which has been attributed to the higher aggregation propensity of the latter.[5] Although Aβ42 fibrils were initially believed to be the disease culprit, intermediates of aggregation (commonly referred to as diffusible oligomers) have been recognized as significantly more harmful.[5b] Aβ peptides can form diverse fibrillary aggregates,[6] and cases exist, where Aβ42 fibrils were found to be non-toxic.[7]
Racemates often have lower solubility than their enantiopure counterparts.[1a] Because Aβ42 fibrils are believed to act as (potentially protective) reservoirs that scavenge the toxic oligomers,[8] we hypothesized that mixing of the Aβ42 enantiomers should accelerate fibril formation and attenuate toxicity of the native L-peptide.
We synthesized the natural L-Aβ42 as well as its mirror image, i.e., the D-enantiomer (see Figure S1 and associated SI for preparative procedures). As expected, the two peptides had reciprocal circular dichroism (CD) spectra,[9] and equimolar mixing of L- and D-Aβ42 led to disappearance of the CD signal (Figure S2). We employed the Thioflavin T (ThT) assay to measure kinetics of fibril formation of the two enantiomers of Aβ42 and their racemic mixture. In agreement with previous reports,[9b] both enantiopure compounds exhibited a sigmoidal fibril growth profile.
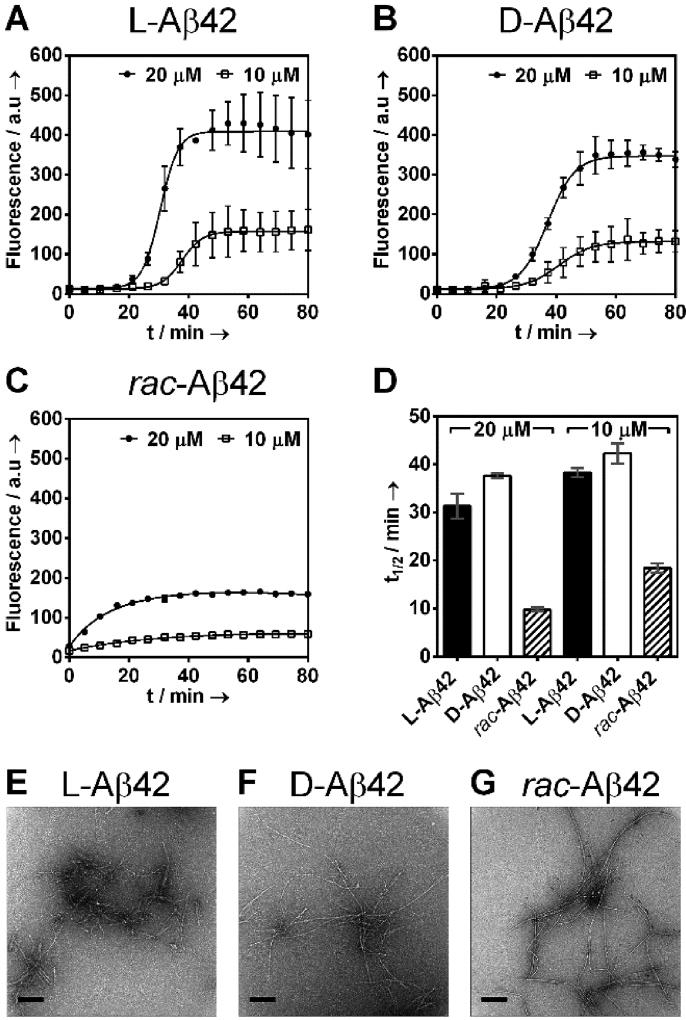
Both enantiomers had a lag phase of 20–30 min (Figure 1A,B). Consistent with recent reports, longer incubation times were needed at lower concentrations.[10] The racemic mixture showed a qualitatively different fibril formation profile that was devoid of any lag phase (Figure 1C). The induction period in the enantiopure cases was followed by rapid increase in fluorescence, diagnostic for fibril formation. Time required to reach half of the maximum fluorescence (t1/2) was comparable for the two enantiomers, but substantially shorter with theracemate, exhibiting a 3 to 4-fold acceleration of fibril formation (31.4 ± 2.6 min, L-Aβ42, 20 µM; 37.6 ± 0.5 min, D-Aβ42, 20 µM; 9.8 ± 0.4 min, rac-Aβ42, 20 µM). Racemic Aβ42 also exhibited lower final fluorescence (~2-fold reduction at 20 µM total concentration, see Figure 1). Consistent trends were observed at lower concentrations (Figure 1A–D), whereas the racemic mixture prepared from 20 µM each L- and D-Aβ42 (40 µM total) resulted in fibril formation that was too rapid to monitor (Figure S3)a Fibrils formed in the ThT experiments were readily observed via TEM, both for the two enantiopure samples as well as the racemate (Figure 1E–G shows representative images and SI appendix A displays all TEM images obtained, 100 per condition).
Figure 1.
Aggregation kinetics of (A) L-Aβ42 (10, 20 µM), (B) D-Aβ42 (10, 20 µM) and (C) rac-Aβ42 (20, 10 µM total concentration) monitored by Thioflavin T (ThT, 20 µM) fluorescence at 37 °C in PBS (pH 7.4) in presence of 0.02% NaN3. Racemic Aβ42 was prepared by mixing equal amounts of L-Aβ42 and D-Aβ42 in 20 mM NaOH before diluting with PBS. Each data point is an average of three replicates with error bars representing the standard deviations. (D) Half times (t½) of Aβ42 fibril formation; t½ is defined as time required to reach half the maximum fluorescence intensity measured in ThT assays (A–C). See SI for more details. Transmission electron microscopy (TEM) of the fibrils of (E) L-Aβ42, (F) D-Aβ42 and (G) racemic Aβ42. Samples were taken directly from the ThT assay (20 µM) at the endpoint of the experiment. All scale bars are 200 nm. See SI Appendix A for further TEM images.
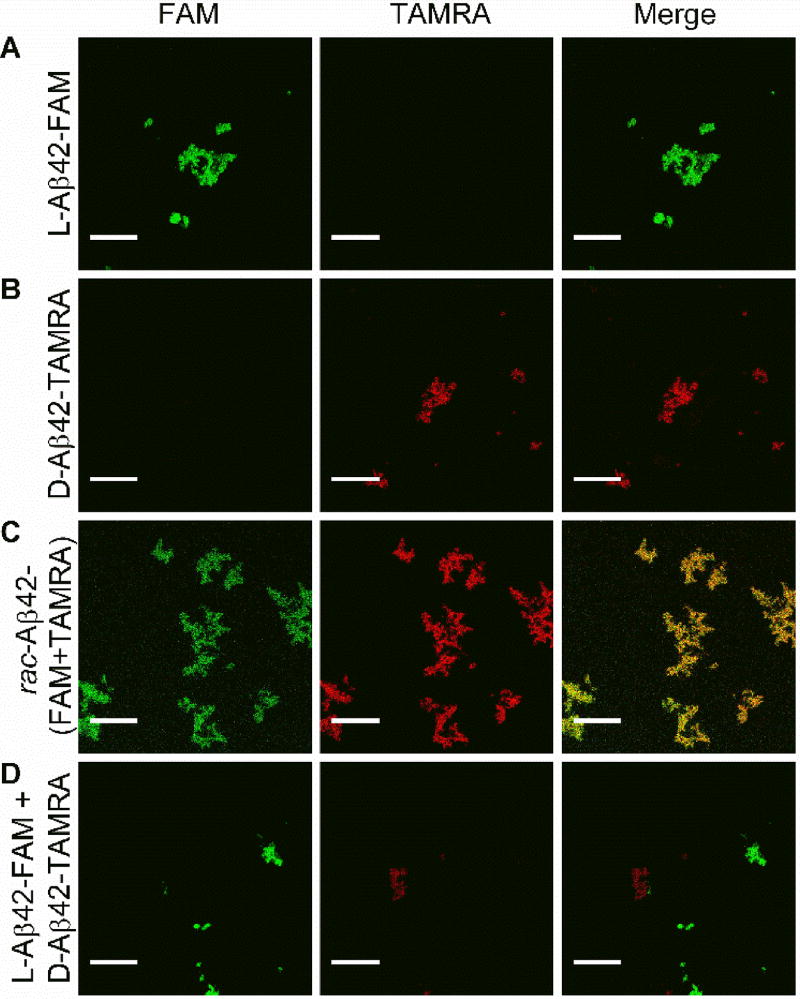
Results from ThT experiments suggested that fibrils obtained from rac-Aβ42 were distinct from those formed by enantiopure materials. To better understand the composition of the racemic fibrils, we made fluorescently labeled analogs of the two Aβ42 enantiomers. L-Aβ42 was N-terminally labeled with 5(6)-Carboxyfluorescein (denoted as L-Aβ42-FAM) and D-Aβ42 was N-terminally labeled with 5(6)-Carboxytetramethylrhodamine (denoted as D-Aβ42-TAMRA). Chemical modifications were performed as described in the SI, (Figure S6). The use of conditions employed in ThT experiments was found to yield fibrils of the fluorescently labeled peptides (Figure S7). Two-channel confocal imaging was performed (Channel 1, FAM: excitation at 476 nm, emission over 484–514 nm; Channel 2, TAMRA: excitation at 543 nm; emission over 630–690 nm). Under those conditions, we found the L-Aβ42-FAM fluorescence to be detectable exclusively via Channel 1 (Figure 2A), whereas D-Aβ42-TAMRA was seen via Channel 2 only (Figure 2B). Importantly, fibrils grown from an equimolar (i.e., “racemic”) mixture of L-Aβ42-FAM and D-Aβ42-TAMRA were fluorescent in both channels (Figure 2C), and the two signals had high degree of co-localization (Pearson Correlation Coefficient, PCC = 0.93; see SI for details). On the other hand, when mature fibrils that had been grown from either L-Aβ42-FAM or D-Aβ42-TAMRA were subsequently mixed, we found that these mixtures were fluorescently active either via Channel 1 (L-Aβ42-FAM) or Channel 2 (D-Aβ42-TAMRA), with very low signal co-localization (Figure 2D; PCC = 0.03).
Figure 2.
Two-channel confocal microscopy imaging (Channel 1, left panel, monitors FAM: excitation at 476 nm, emission over 484–514 nm; Channel 2, middle panel, monitors TAMRA: excitation at 543 nm; emission over 630–690 nm; right panel: merging of Channels 1 and 2 allows to probe for co-localization of the fluorescent labels). L-Aβ42-FAM fibrils were fluorescentl in Channel 1, but not Channel 2 (A), whereas D-Aβ42-TAMRA fibrils were active via Channel 2 only (B). Fibrils grown from an equimolar mixture of L-Aβ42-FAM and D-Aβ42-TAMRA were fluorescently active in both Channel 1 and 2, with robust signal co-localization (C). In a control experiment (D), fibrils grown from either L-Aβ42-FAM or D-Aβ42-TAMRA were subsequently mixed and were fluorescently active either in Channel 1 or Channel 2, with very low co-localization. Scalebar: 20 µm.
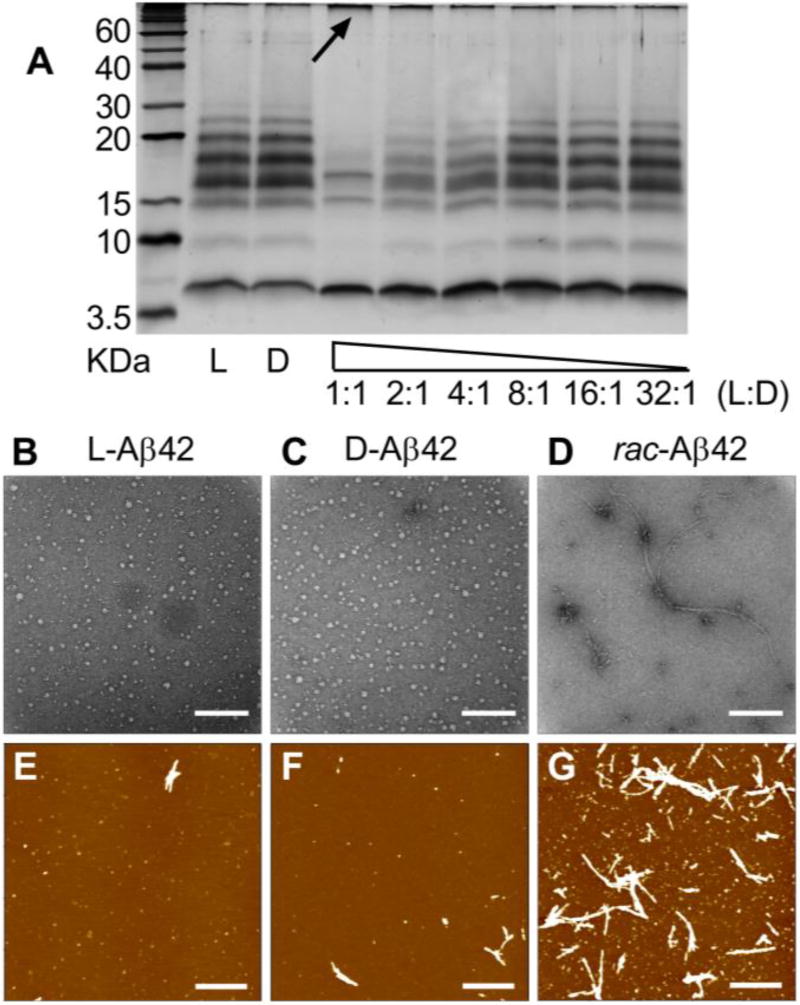
The acceleration of fibril formation from racemic Aβ42 (Figure 1) and the co-localization of L- and D-Aβ42 in fibrils grown from the racemic mixture (Figure 2C) prompted us to investigate earlier stages of aggregation. To measure the abundance of oligomers in solution, we conducted PICUP experiments.[5c] Performed with either enantiopure Aβ42 (L- or D-, 50 µM) or their racemic mixture (50 µM L- and 50 µM D-), the experiment revealed suppression of oligomer formation with rac-Aβ42 (Figure 3A), which was accompanied by an increased formation of a high molecular weight band (see arrow in Figure 3A). Indicating increased formation of larger aggregates, this is consistent with the enhancement of fibril formation upon mixing of the two enantiomers (Figure 1). The oligomer distribution obtained from L-Aβ42 was consistent with previous reports by us and others,[5c,12] and D-Aβ42 showed a virtually identical oligomerization pattern, as expected for a mirror-image peptide. The suppression of oligomer formation from L-Aβ42 showed dose dependence upon addition of sub-stoichiometric amounts of D-Aβ42 above a 4:1 ratio.
Figure 3.
A) A PICUP gel of L- and D-Aβ42 (50 µM), as well as various mixtures of the two enantiomers. In mixtures, L-Aβ42 was always kept at 50 µM, and D-Aβ42 covered the range between 50 µM and 1.56 µM, as indicated. (B–D) representative TEM images for L-, D- and rac-Aβ42 aggregation intermediates (Scalebar: 200 nm). Samples were prepared as described in SI Appendix B; method adopted from ref. 13. (E–G) representative AFM images for L-, D- and rac-Aβ42 aggregation intermediates, sample preparation same as TEM (Scalebar: 1000 nm); see SI for further TEM and AFM images.
We explored the inhibition of oligomer formation further by conducting TEM experiments on aggregation intermediates, following recently published protocols (see SI Appendix B).[13] These experiments revealed that both L- and D-Aβ42 were forming spherical oligomers with diameters of 12.9 ± 3.7 nm and 13.6 ± 4.1 nm, respectively (Figure 3B,C), which is consistent with literature values (see SI Appendix B for further images and histogram analysis of oligomer size distribution).[13] With rac-Aβ42, we observed a more heterogeneous mixture of aggregation intermediates, which frequently contained advanced (fibrillary) aggregates. Atomic Force Microscopy (AFM) confirmed the presence of these aggregation intermediates (Figure 3E–G; see Figure S8 for further images). From AFM imaging, both L- and D- Aβ42 were more likely to form oligomers and only few fibrils. (Figure 3E,F) By comparison, rac-Aβ42 produced more fibrils with high aspect (Figure 3G). Oligomers obtained from enantiopure (L- or D-) Aβ42 were disc-shaped and had an average height of ~0.2–0.3 nm, which is in agreement with previous reports.[13] With rac-Aβ42, on the other hand, the average height of the oligomers was higher at 0.4 nm and the average height of the high aspect ratio fibers was ~3 nm, reflecting the enhanced aggregation propensity upon mixing of the two enantiomers. Overall, the results from AFM imaging were in qualitative agreement with TEM.
Taken together, our findings on Aβ42 aggregation intermediates (Figure 3) were consistent with the observation that mixing of the enantiomers accelerates fibril formation (Figure 1) and that the enantiomers co-localize in fibrils grown from rac-Aβ42 (Figure 2).
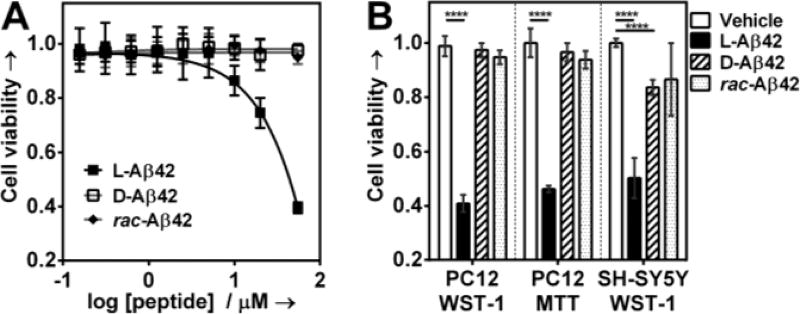
Diffusible Aβ42 oligomers have been recognized as particularly toxic,[5] and we found their concentration to be substantially reduced in the racemate. Hence, we hypothesized that rac-Aβ42 should exhibit reduced cytotoxicity in comparison to (natural) L-Aβ42. Incubation of PC12 neuron-like cells with 20 µM L-Aβ42 reduced cell viability (measured via WST-1 assay, see SI for details) by ~25 %, which is in agreement with our previous findings (Figure 4A).[12] The D-enantiomer was non-toxic under those conditions. The addition of one equivalent D-Aβ42 to L-Aβ42 prior to dosing the PC12 resulted in full suppression of toxicity (final concentration of L-Aβ42 in the racemate was 20 µM). Because cytotoxicity observed with L-Aβ42 at 20 µM was modest, we also conducted the experiment at 50 µM. At that concentration, L-Aβ42 reduced PC12 viability by 60 %, whereas D-Aβ42 again showed no toxicity. Remarkably, in the racemic mixture (50 µM L-Aβ42 and 50 µM D-Aβ42) the toxicity of L-Aβ42 was completely suppressed. We also measured partial toxicity reduction against PC12 cells by WST-1 in the mixture of 50 µM L- / 25 µM D-Aβ42, but not 50 µM L- / 12.5 µM D-Aβ42 (Figure S9).
Figure 4.
A) Cellular viability of the PC12 adhesive cell line in response to dosing in of varied concentrations of enantiopure (L- or D-) or racemic Aβ42 B) Cell viability (PC12 or SH-SY5Y) in response to treatment with either enantiopure (L- or D-; 50 µM in both cases) or rac-Aβ42 (50 µM L-Aβ42 and 50 µM D-Aβ42). Cells were plated and allowed to adhere for 24 h. Peptides were then added and cells incubated for further 72 h. Cell viability was determined using the cell proliferation reagent WST-1 or MTT, as shown. Data are represented as mean ± s.d, (****P < 0.0001, calculated by unpaired t-test).
Consistent observations were made when the MTT cytotoxicity assay was employed in place of WST-1 (Figure 4B and Figure S10), and when the SH-SY5Y cell line was used instead of PC12 (Figure 4B). We did note marginaltoxicity of D-Aβ42 against SH-SY5Y cells (~14% viability reduction). We are aware of two independent studies that compared L- and D-Aβ42, with apparently contradictory results.[9] The investigation by Cribbs et al. reported comparable toxicity for both L- and D-Aβ42,[9a] whereas the study by Ciccotosto et al. found D-Aβ42 to be non-toxic.[9b] The first study used hippocampal neurons, while the second used cortical neurons. The different cell model systems chosen in those studies may underlie the apparent contradiction. We also measured LDH release induced by L- D- and racemic Aβ42 in PC12 cells. We found L-Aβ42 to induce ~40 % maximum LDH release, whereas with D- or rac-Aβ42 it was less than 10 % (Figure S11). The amount of LDH released upon exposing PC12 cells to D- or rac-Aβ42 is comparable to that recently reported for scrambled Aβ42, which the authors also found to be non-toxic against rat brain endothelial cells by MTT.[14]
Examples of enhancement of Aβ42 fibrillization as a strategy to protect from toxicity are scarce.[7] Tailored D-peptides have been previously employed to attenuate Aβ42 aggregation,[15] but not to promote fibril formation. Our approach to induce non-toxic fibril formation through addition of mirror-image Aβ42 (i.e., “Chiral Inactivation”) is based on fundamentals of molecular stereochemistry, and we were able to recapitulate the trend of accelerated fibril formation upon enantiomer mixing with the Aβ40 system (Figure S4). Of relevance to our work, Kar et al. found recently that mirror-image (D-)polyglutamine can recruit L-polyglutamine and lead to formation of potentially toxic inclusion bodies in cell-based assays.[16] We therefore note that, although there appears to be some generality of stereochemical aspects underlying the formation of homo- and heterochiral aggregates from intrinsically disordered peptides, biological consequences will be specific to the system under study.
In summary, we find that racemic Aβ42 forms fibrils substantially more rapidly than enantiopure, and that the fluorescently labeled derivatives of the two enantiomers co-localize in racemic fibrils. Acceleration of fibrillization is accompanied by a suppression of oligomer formation with racemic Aβ42. These changes in aggregation properties lead to inhibition of toxicity against PC12 and SH-SY5Y cells.
Supplementary Material
Acknowledgments
JAR thanks UCSC for flexible start-up funds, special research grants and the Hellman Foundation for a Fellowship. We acknowledge the NIHS10OD016246-01A1 award for the purchase of the JASCO J1500 CD. MR and XZ acknowlege Office of Naval Research Award - N000141410724 and Award N000141612507 (DURIP) for the purchase of the AFM.
Footnotes
We performed the corresponding ThT fibril formation experiments with the Aβ40 isoform and observed accelerated fibril formation for the racemic mixture there as well (Figure S4). Our racemic fibril formation experiment has to be distinguished from the seeding experiment performed by Esler et al.,11 where pre-formed fibrils of L-Aβ40 were found to seed stereospecifically the fibrillization of the L- (but not D-) enantiomer. We found their results to qualitatively hold true for the Aβ42 system as well (Figure S5).
References
- 1.Jacques J, Collet A, Wilen SH. Enantiomers, Racemates and Resolutions. Wiley; New York: 1981. [Google Scholar]; b) Weissbuch I, Illos RA, Bolbach G, Lahav M. Acc. Chem. Res. 2009;42:1128–1140. doi: 10.1021/ar900033k. [DOI] [PubMed] [Google Scholar]; c) Raskatov JA, Thompson AL, Cowley AR, Claridge TDW, Brown JM. Chem. Sci. 2013;4:3140–3147. [Google Scholar]; d) Soai K, Kawasaki T, Matsumoto A. Acc. Chem. Res. 2014;47:3643–3654. doi: 10.1021/ar5003208. [DOI] [PubMed] [Google Scholar]
- 2.a) Pauling L, Corey RB. Proc. Natl. Acad. Sci. USA. 1953;39:253–256. doi: 10.1073/pnas.39.4.253. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Torbeev V, Grogg M, Ruiz J, Boehringer R, Schirer A, Hellwig P, Jeschke G, Hilvert D. J. Pept. Sci. 2016;22:290–304. doi: 10.1002/psc.2861. [DOI] [PubMed] [Google Scholar]
- 3.a) Nagy KJ, Giano MC, Jin A, Pochan DJ, Schneider JP. J. Am. Chem. Soc. 2011;133:14975–3977. doi: 10.1021/ja206742m. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Swanekamp RJ, DiMaio JT, Bowerman CJ, Nilsson BL. J. Am. Chem. Soc. 2012;134:5556–5559. doi: 10.1021/ja301642c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.a) Querfurth HW, LaFerla FM. N. Engl. J. Med. 2010;362:329–344. doi: 10.1056/NEJMra0909142. [DOI] [PubMed] [Google Scholar]; b) Chiti F, Dobson CM. Annu. Rev. Biochem. 2006;75:333–366. doi: 10.1146/annurev.biochem.75.101304.123901. [DOI] [PubMed] [Google Scholar]
- 5.a) Gong YS, Chang L, Viola KL, Lacor PN, Lambert MP, Finch CE, Krafft GA, Klein WL. Proc. Natl. Acad. Sci. USA. 2003;100:10417–10422. doi: 10.1073/pnas.1834302100. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Haass C, Selkoe DJ. Nat. Rev. Mol. Cell. Biol. 2007;8:101–112. doi: 10.1038/nrm2101. [DOI] [PubMed] [Google Scholar]; c) Bitan G, Kirkitadze MD, Lomakin A, Vollers SS, Benedek GB, Teplow DB. Proc. Natl. Acad. Sci. USA. 2003;100:330–335. doi: 10.1073/pnas.222681699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.a) Schmidt M, Rohou A, Lasker K, Yadav JK, Schiene-Fischer C, Fandrich M, Grigorieff N. Proc. Natl. Acad. Sci. USA. 2015;112:11858–11863. doi: 10.1073/pnas.1503455112. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Xiao Y, Ma B, McElheny D, Parthasarathy S, Long F, Hoshi M, Nussinov R, Ishii Y. Nat. Struct. Mol. Biol. 2015;22:499–505. doi: 10.1038/nsmb.2991. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Colvin MT, Silvers R, Ni QZ, Can TV, Sergeyev I, Rosay M, Donovan KJ, Michael B, Wall J, Linse S, Griffin RG. J. Am. Chem. Soc. 2016;138:9663–9674. doi: 10.1021/jacs.6b05129. [DOI] [PMC free article] [PubMed] [Google Scholar]; d) Wälti MA, Ravotti F, Arai H, Glabe CG, Wall JS, Bockmann A, Guntert P, Meier BH, Riek R. Proc. Natl. Acad. Sci. USA. 2016;113:E4976–4984. doi: 10.1073/pnas.1600749113. [DOI] [PMC free article] [PubMed] [Google Scholar]; e) Fändrich M, Meinhardt J, Grigorieff N. Prion. 2009;3:89–93. doi: 10.4161/pri.3.2.8859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.a) Ghanta J, Shen CL, Kiessling LL, Murphy RM. J. Biol. Chem. 1996;271:29525–29528. doi: 10.1074/jbc.271.47.29525. [DOI] [PubMed] [Google Scholar]; b) Bieschke J, Herbst M, Wiglenda T, Friedrich RP, Boeddrich A, Schiele F, Kleckers D, Lopez del Amo JM, Gruning BA, Wang Q, Schmidt MR, Lurz R, Anwyl R, Schnoegl S, Fandrich M, Frank RF, Reif B, Gunther S, Walsh DM, Wanker EE. Nat. Chem. Biol. 2011;8:93–101. doi: 10.1038/nchembio.719. [DOI] [PubMed] [Google Scholar]
- 8.a) Jiang L, Liu C, Leibly D, Landau M, Zhao M, Hughes MP, Eisenberg DS. eLife. 2013;2:e00857. doi: 10.7554/eLife.00857. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Selkoe DJ, Hardy J. EMBO Mol. Med. 2016;8:595–608. doi: 10.15252/emmm.201606210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.a) Cribbs DH, Pike CJ, Weinstein SL, Velazquez P, Cotman CW. J. Biol. Chem. 1997;272:7431–7436. doi: 10.1074/jbc.272.11.7431. [DOI] [PubMed] [Google Scholar]; b) Ciccotosto GD, Tew DJ, Drew SC, Smith DG, Johanssen T, Lal V, Lau TL, Perez K, Curtain CC, Wade JD, Separovic F, Masters CL, Smith JP, Barnham KJ, Cappai R. Neurobiol. Aging. 2011;32:235–248. doi: 10.1016/j.neurobiolaging.2009.02.018. [DOI] [PubMed] [Google Scholar]
- 10.Meisl G, Kirkegaard JB, Arosio P, Michaels TC, Vendruscolo M, Dobson CM, Linse S, Knowles TP. Nat. Protoc. 2016;11:252–272. doi: 10.1038/nprot.2016.010. [DOI] [PubMed] [Google Scholar]
- 11.Esler WP, Stimson ER, Fishman JR, Ghilardi JR, Vinters HV, Mantyh PW, Maggio JE. Biopolymers. 1999;49:505–514. doi: 10.1002/(SICI)1097-0282(199905)49:6<505::AID-BIP8>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- 12.Warner CJ, Dutta S, Foley AR, Raskatov JA. Chem. Eur. J. 2016;22:11967–11970. doi: 10.1002/chem.201601763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ahmed M, Davis J, Aucoin D, Sato T, Ahuja S, Aimoto S, Elliott JI, Van Nostrand WE, Smith SO. Nat. Struct. Mol. Biol. 2010;17:561–567. doi: 10.1038/nsmb.1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Deli MA, Veszelka S, Csiszar B, Toth A, Kittel A, Csete M, Sipos A, Szalai A, Fulop L, Penke B, Abraham CS, Niwa M. J. Alzheimers Dis. 2010;22:777–794. doi: 10.3233/JAD-2010-100759. [DOI] [PubMed] [Google Scholar]
- 15.a) Wiesehan K, Stohr J, Nagel-Steger L, van Groen T, Riesner D, Willbold D. Protein Eng. Des. Sel. 2008;21:241–246. doi: 10.1093/protein/gzm054. [DOI] [PubMed] [Google Scholar]; b) Sievers SA, Karanicolas J, Chang HW, Zhao A, Jiang L, Zirafi O, Stevens JT, Munch J, Baker D, Eisenberg D. Nature. 2011;475:96–100. doi: 10.1038/nature10154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kar K, Arduini I, Drombosky KW, van der Wel PCA, Wetzel R. J. Mol. Biol. 2014;426:816–829. doi: 10.1016/j.jmb.2013.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.