Abstract
Reactive oxygen species, both endogenous and exogenous, can damage nucleobases of RNA and DNA. Among the nucleobases, guanine has the lowest redox potential making it a major target of oxidation. Although, RNA is more prone to oxidation than DNA, oxidation of guanine in RNA has been studied to a significantly lesser extent. One of the reasons for this is that many tools that were previously developed to study oxidation of DNA cannot be used on RNA. In the current study, the lack of a method for seeking sites of modification in RNA where oxidation occurs is addressed. For this purpose, reverse transcription of RNA containing major products of guanine oxidation was used. Extension of a DNA primer annealed to an RNA template containing 8-oxo-7,8-dihydroguanine (OG), 5-guanidinohydantoin (Gh), or the R and S diastereomers of spiroiminodihydantoin (Sp) was studied under standing start conditions. SuperScript III reverse transcriptase is capable of bypassing these lesions in RNA inserting predominantly A opposite OG, predominantly G opposite Gh, and almost an equal mixture of A and G opposite the Sp diastereomers. These data should allow RNA sequencing of guanine oxidation products by following characteristic mutation signatures formed by the reverse transcriptase during primer elongation past G oxidation sites in the template RNA strand.
Keywords: Oxidative damage; RNA sequencing; Reverse transcription; 8-Oxo-7, 8-dihydroguanine; Guanidinohydantoin; Spiroiminodihydantoin
Introduction
The nucleic acids DNA and RNA are prone to oxidative damage from reactive oxygen species (ROS) that are formed during metabolism or induced by exogenous sources, such as UV radiation, ionizing radiation, or environmental toxins.1–4 Exposure to these damaging agents may result in modifications of nucleobases, strand breaks, or cross-links with other molecules present in the cell.5–9 Oxidation of DNA has been an area of intense study for over two decades because it can lead to irreversible mutations in the genetic code that result in cancer and numerous genetic diseases.10–12
Oxidative damage to RNA has received much less attention. The likely reasons for this include the many challenges of working with the inherently less chemically stable RNA and the assumption that oxidation of RNA does not significantly disturb normal cell functions due to turnover of RNA molecules in the cell.13 While the latter may be true for lower organisms with predominantly short-lived RNA,14 the average half-life of mRNA in human cells is ~10 hours while for the long-lived tRNA and rRNA this value can reach several days.15 On average mammalian cells contain at least as much RNA as DNA13, 16 and RNA itself is 2–25 times17–19 more susceptible to oxidation by ROS in vivo than DNA. Therefore, RNA oxidation can present a substantial challenge for cell survival, and multiple studies have linked it to development and progression of cancer and neurodegenerative diseases.20–29 Recent work has discovered pathways of surveillance, sequestration, and, in some cases, repair of RNA damage.13, 30–34
Among the nucleobases, guanine has the lowest redox potential (1.29 V vs. NHE) that makes it the major target for oxidizing agents.35 Thus, one of the most abundant lesions is a product of its 2-electron oxidation, 8-oxo-7,8-dihydroguanine (OG).36 The redox potential of OG is even lower than that of guanine (0.74 V vs. NHE) making these sites predisposed to further oxidation35 to yield two hydantoin lesions, 5-guanidinohydantoin (Gh) and spiroiminodihydantoin (Sp) (Scheme 1).37, 38 OG has been found in both cellular DNA and RNA, while Gh and Sp have thus far been characterized only in cellular DNA and in RNA oligomers mimicking the tRNA anticodon loop or short single strands of RNA.18, 39, 40 The latter lesion, Sp, exists as a pair of enantiomers (R-Sp and S-Sp) as the free base, which form a pair of diastereomers once attached to the chiral ribose or 2′-deoxyribose components in nucleic acids.41
Scheme 1.
Pathways of Guanine Oxidation.
Mapping positions of the oxidation sites in DNA has been used for achieving a deeper understanding of guanine susceptibility to oxidation and composition of the products on the sequence and structural context.36, 42–46 However, methods that have been successfully used for finding where oxidation events occur in DNA cannot be directly applied to RNA.40 Techniques developed for mapping nucleotide modifications in DNA or RNA could be broken into 3 major categories.
Category one includes methods that induce a strand break directly in the studied sequence by an enzyme or a chemical agent (e.g., Maxam-Gilbert chemistry, RNA digestion with specific RNAses, or base excision from the DNA strand by repair enzymes) with further analysis of products by PAGE or CE. We have previously tested the possibility of using simple cleavage of oxidized bases in RNA by hot aniline treatment and discovered that most products of guanine oxidation in RNA are more resistant to cleavage under standard conditions than the same lesions in DNA when treated with hot piperidine.40 In addition, RNA itself is more susceptible than DNA to position independent backbone cleavage,47 which results in a high background resulting in a limited practical use of chemical methods for locating lesion sites.
Category two comprises mass spectroscopic techniques developed for characterization of post-transcriptional modifications in RNA (e.g. simple analysis of MS/MS fragmentation of oligonucleotides48 or more complex LC-MS/MS methods developed by the McCloskey laboratory49–51). Although, these methods can possibly be highly useful for mapping guanine oxidation products, they have never been used for that purpose. Optimization of this type of analysis for detection of oxidized lesions could be quite challenging considering that oxidized lesions are likely to be more randomly distributed than localized post-transcriptional modifications.
The third category consists of primer extension assays utilizing either incorporation of natural (e.g. bisulfite sequencing of m5C, or SHAPE-MaP52) or unnatural bases (e.g. insertion of artificial nucleotides opposite m5C or O6-BnG53–56) opposite modified nucleotides or arrest of the polymerase activity, if it cannot efficiently insert a base opposite the modification site (e.g. adenosine methylation by DMS or older versions of SHAPE probing57, 58). Currently, there are no known artificial bases that can be inserted opposite OG or the hydantoin lesions with the required specificity to map exclusively these sites.59 At the same time, multiple studies have described insertion of canonical nucleobases opposite these lesions in DNA.60–65 The polymerases studied insert A or C opposite OG62, 63, 65 and A or G opposite Gh and Sp,60, 61 with the ratio highly dependent on the polymerase. Among the aforementioned papers, only one was focused on polymerase insertion fidelity opposite OG by RNA-dependent DNA polymerase62 called reverse transcriptase (RT). In that study, insertion opposite OG in a DNA (not RNA) template strand by HIV1 RT was interrogated finding insertion of A and C in a 14:1 ratio.62
Thus, there are so far no conclusive reports on how reverse transcriptase enzymes behave when they encounter OG, Gh, or Sp in a RNA template. The current paper focuses on testing whether commercially available reverse transcriptase enzymes can insert canonical nucleobases opposite OG, Gh, or Sp when located in a RNA template. For the potential use of reverse transcription as a method for detection of oxidized guanine lesions, it should either result in termination of polymerization at the lesion sites or insertion of any base other than C to allow discrimination from unoxidized G. Considering that several types of RNA (18S rRNA and tRNA in eukaryotes66) contain post-transcriptional modifications, such as m1G or m22G, that also result in polymerase arrest,67 the latter option would be preferable for application of this lesion sequencing approach on biological samples. Overall this study provides a foundation for development of a method for mapping OG, Gh, or Sp in RNA templates using reverse transcription to induce mutations that can be tracked after next-generation sequencing.
Materials and Methods
Oligomer synthesis
DNA and RNA oligomers were synthesized by the core facilities at the University of Utah using solid-phase synthesis following standard protocols. The RNA templates containing OG were synthesized using the commercially available rOG phosphoramidite (ChemGenes, Wilmington, MA). The RNA strands were synthesized with a 3′ dT to maximize the solid-phase synthesis yield of the modified RNA strands; the added dT will not impact the polymerase extension studies. All oligonucleotides were purified via analytical ion-exchange HPLC and dialyzed against ddH2O.
Synthesis of the Sp and Gh hydantoins into the RNA strands was achieved utilizing the RNA strand with rOG incorporated via its phosphoramidite at the site of the modification. Synthesis of Gh was conducted by dissolving 1 nmole of rOG-containing RNA into 50 μL of ddH2O. The sample was placed on ice for 10 min followed by a bolus addition of 12 equivalents of Na2IrCl6, and the reaction was allowed to progress for 30 min. The rGh-containing strands were purified from the reaction mixture by ion-exchange HPLC on a DNAPac PA100 column (250 × 4.6 mm). The HPLC mobile phases consisted of A = 1.5 M NaOAc (pH 7.0) in 9:1 ddH2O:MeCN and B = 9:1 ddH2O:MeCN while running a linear gradient from 25% B to 100% B over 30 min with a flow rate of 1 mL/min while monitoring the absorbance at 260 nm. The RNA strands containing diastereomers of rGh were purified and studied as a mixture because the epimers readily interconvert.68 The purified strands were dialyzed against ddH2O for 24 h while changing the ddH2O every 6 h to remove the purification salts. The rSp-containing strands were synthesized by placing 1 nmole of purified rOG-containing RNA into 50 μL of 20 mM NaPi (pH 7.4) buffer with 100 mM NaCl at 22 °C followed by a bolus addition of 12 equivalents of Na2IrCl6. The reaction was allowed to progress for 30 min followed by workup via the same method outlined for the rGh-containing RNA strands. The diastereomers of rSp were individually purified for the polymerase studies. The absolute configurations for the Sp diastereomers have been determined in DNA strands and nucleosides but not in RNA strands or nucleosides41; therefore, we validated the absolute configurations for the Sp diastereomers in RNA strands and nucleosides to find identical results between the two polymers (Figure S1). The purity of the hydantoins in the RNA oligomers studied was determined by ion-exchange HPLC (Figure S2) and the product identities were established by ESI-MS (OG-1 calcd = 6608.0, expt = 6608.1; Gh-1 calcd = 6598.0, expt = 6598.5; S-Sp-1 calcd = 6624.0, expt = 6624.8; R-Sp-1 calcd = 6624.0, expt = 6624.5).
Labeling of the DNA primer
To monitor progression of primer extension by reverse transcriptases via polyacrylamide gel electrophoresis (PAGE), the DNA primer was 5′ end-labeled with 32P following a procedure adopted from the literature69 using T4 polynucleotide kinase (New England Biolabs, Ipswich, MA.) and [γ-32P] ATP (PerkinElmer, Waltham, MA.).
Polymerase nucleotide insertion and extension efficiency assays
The following enzymes were used: SuperScript III (200 U/μL, Invitrogen, Carlsbad, CA), AMV RT (25 U/μL, New England Biolabs, Ipswich, MA.), MMLV RT (200 U/μL, New England Biolabs, Ipswich, MA.), ProtoScript II (200 U/μL, New England Biolabs, Ipswich, MA.), and Omniscript (4 U/μL, Qiagen, Hilden, Germany). Before all primer extension assays, samples were annealed by heating 10–14 μL of aqueous solution containing 0.44 pmol (22 nM in a final volume of 20 μL) of RNA template and 0.4 pmol (20 nM in 20 μL) DNA primer including ~30,000 cpm of 32P-labeled strand to 95 °C for 5 min, then incubating them at 55 °C for another 5 min followed by cooling at 15 °C for 10 min. Upon completion of annealing, stock solutions of reaction buffer (50 mM Tris-HCl, 75 mM KCl (75 mM KAc for AMV RT), 3 mM MgCl2 (8 mM MgAc2 for AMV RT), pH 8.3 in 20 μL, 1× commercial reaction buffer for Omniscript), DTT (10 mM in 20 μL and the commercially defined concentration of DTT in the buffer for Omniscript), dNTPs were added to the reaction mixture to bring the volume to 18 μL. Then 2 μL of stock solution containing one of the reverse transcriptases in 50% glycerol was added to give a final volume of 20 μL (final concentrations of dNTPs and enzymes are provided in the next paragraph). Reaction mixtures were incubated for 30 min at 37 °C, and then to quench the reaction, the mixture was diluted with an equal volume of 2× gel loading buffer (8 M urea, 0.01% xylene cyanole, 0.01% bromphenol blue, ×1 TBE buffer) and heated to 95 °C for 10 min. About 15 μL of the resulting solution were analyzed on 20% denaturing PAGE. Gels were stored with a storage phosphor screen for 12–18 hours which was then scanned using a phosphoimager. The resulting images were analyzed using ImageJ2 software.70, 71 For alignment of the gel lane pixel density plots, they were rescaled along the y-axis to normalize intensities and translated along the x-axis without rescaling to align the peaks corresponding to the unextended primer.
Final concentrations of enzymes and triphosphates for polymerase nucleotide insertion studies were as follows: for templates containing OG or G – 50 μM dATP, dCTP, dGTP, or dTTP, 3U SuperScript III, 0.4 U AMV RT, 2 U MMLV RT, 5U ProtoScript II, 0.3 U Omniscript in 20 μL; for Gh-1 template – 100 μM individual dNTPs, 4U SuperScript III in 20 μL; for S-Sp-1 template - 200 μM individual dNTPs, 40U SuperScript III in 20 μL; for R-Sp-1 template - 200 μM individual dNTPs, 20U SuperScript III in 20 μL. Final concentrations for full extension efficiency study were 100U of SuperScript III in 20 μL and 500 or 200 μM of each dNTP (dATP, dCTP, dGTP, and dTTP). Final concentrations for comparing extension efficiency past OG-A and OG-C were 6U SuperScript III in 20 μL and 100 μM dATP for lanes 1 (A), 2 (AT), 7 (AC), 8 (ACT), 200 μM dATP for lane 5 (2A), 100 μM dCTP for lanes 3 (C), 4 (CT), 7 (AC), 8 (ACT), 200 μM dCTP for lane 6 (2C), 100 μM dTTP for lanes 2 (AT), 4 (CT), 8 (ACT). Final concentrations for comparing extension efficiency with Gh-1 template were 8U SuperScript III in 20 μL and 200 μM dATP for lanes 1 (A), 2 (AT), 7 (AG), 8 (AGT), 400 μM dATP for lane 5 (2A), 200 μM dGTP for lanes 3 (G), 4 (GT), 7 (AG), 8 (AGT), 400 μM dGTP for lane 6 (2G), 200 μM dTTP for lanes 2 (AT), 4 (GT), 8 (AGT). Final enzyme concentrations for reactions using S-Sp-1 and R-Sp-1 templates were 80U and 40U SuperScript III in 20 μL correspondingly; concentrations of triphosphates were the same as described for Gh-1 template.
Steady-state kinetics
A procedure analogous to the one described in the previous section was used with the following changes. Instead of 30,000 cpm of 32P-labeled strand, 100,000 cpm was added. After annealing, only reaction buffer and DTT stock solutions were added, and then the samples were pre-incubated for 1 min at 37 °C, followed by addition of 2 μL of SuperScript III stock solution to give 1U (OG), 4U (Gh), 10U (R-Sp), or 20U (S-Sp) in 20 μL followed by pre-incubation for another 1 min at 37 °C and then addition of 2 μL of stock solution of dATP, dCTP, or dGTP of varied concentrations to initiate the reaction. Next 5-μL aliquots were taken in regular time intervals for the first 2–2.5 min of reaction, rapidly mixed into an equal volume of the loading buffer and heated to 95 °C for 10 min. The diluted stock solutions of triphosphates and enzymes were prepared fresh and used the same day. Samples were analyzed on a denaturing PAGE gel the same way as described above. Intensities of the bands corresponding to unextended primer (P) and the primer extended by one base (P+1) were quantified using ImageJ2 software70, 71 and converted into concentration of P+1 strand using equation 1,61, 72 where CP+1 is the concentration of P+1 strand, Ct is the total concentration of radiolabeled primer, and IP and IP+1 are intensities of P and P+1 bands on the gel. No significant accumulation of higher order bands was observed. Initial reaction velocities were extracted from fitting the data in coordinates CP+1 (nM) vs. time (min) to a linear regression curve as a slope of the fitted line (Figure S4). Each experiment was repeated at least 3 times. Conversion of Vmax to kcat was achieved using specific activity and the molecular weight of SuperScript III provided by Invitrogen (410,000 U/mg, 78 kDa).
| (1) |
Results and Discussion
Reverse transcriptases
Reverse transcriptases are polymerase type enzymes that synthesize a complementary DNA (cDNA) based on an RNA template. While enzymes of this type have been discovered in prokaryotes and yeast,73, 74 the majority of them have retroviral origins. Most commercially available reverse transcriptase enzymes originate either from Avian Myeloblastosis Virus (AMV) RT or Moloney Murine Leukemia Virus (MMLV) RT. The primary purpose of reverse transcription in contemporary research is sequencing and quantification of mRNA via RT-PCR and RT-qPCR. Engineered versions of these enzymes have been created by multiple vendors to improve their stability, sensitivity, and reverse transcription yield (Table 1).75, 76 In the current study, we screened insertion profiles of 5 RT enzymes (underlined in Table 1) opposite OG and G in a RNA template. Because similar insertion profiles have been observed for all reverse transcriptases used, only one of them, SuperScript III, was picked for further experiments. SuperScript III was chosen over other enzymes due to it being one of the polymerases of choice for next-generation sequencing of RNA.58, 75–78
Table 1.
Selection of commercially available reverse transcriptases.
| Original RT | Enzyme concentration | |
|---|---|---|
|
| ||
| AMV RT | AMV RT | 25 U/μL |
| MMLV RT | MMLV RT | 200 U/μL |
| ProtoScript II® | MMLV RT | 200 U/μL |
| Omniscript® | undisclosed | 4U/μL |
| Sensiscript® | undisclosed | undisclosed |
| ImProm-II® | undisclosed | undisclosed |
| ThermoScript® | AMV RT | 15 U/μL |
| Superscript II® | MMLV RT | 200 U/μL |
| Superscript III® | MMLV RT | 200 U/μL |
| Superscript IV® | MMLV RT | 200 U/μL |
| TGIRT® | Group II intron RT | 200 U/μL |
Polymerase nucleotide insertion studies
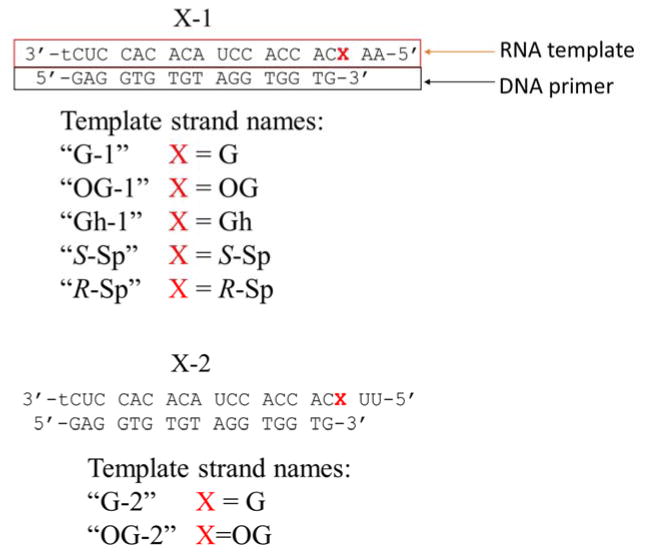
To study the behavior of RT enzymes encountering products of guanine oxidation in RNA, we designed two RNA-DNA hybrid duplexes (Figure 1). Both of them employ the same DNA primer and are suited for studying reverse transcription under standing start conditions with the only difference between the two being two nucleotides on the 5′ end following guanine or its oxidation products. To perform an initial evaluation of reverse transcription as a way of detecting oxidized lesions, we studied insertion profiles opposite G and OG in both RNA templates for 5 RT enzymes: SuperScript III, AMV RT, MMLV RT, Omniscript, and ProtoScript II. To do so, we ran reverse transcription reactions with A, G, C, or T triphosphates in separate tubes for a fixed amount of time and resolved reaction products on a polyacrylamide gel. Although all these RTs have identical unit definitions (amount of an enzyme incorporating 1 nmol dTTP in 50 μL in 10 min at 37 ºC using poly(rA)18 • poly(dT)12–18 duplex and 500 μM dTTP), their activities are standardized under different conditions (0.1–0.4 mM primer-template duplex, 3–6 mM MgCl2, 40–75 mM KCl, 1–10 mM DTT, 0–0.1 mg/mL BSA; no data is available on Omniscript). Additionally, there are slight variations in the reaction buffers supplied with AMV and the other enzymes that were used for the reactions (75 mM KOAc instead of 75 mM KCl and 8mM MgOAc2 instead of 3 mM MgCl2; no data are available on Omniscript reaction buffer composition). Thus, we found it necessary to individually adjust the concentration of each enzyme to achieve ~50% insertion of dCTP (50 μM) opposite OG in the OG-1 strand. Results of the interrogation of nucleotide insertion profiles of the reverse transcriptases studied are presented in Figure 2, Figure S3, and in Table S1. On the gels, the lowest band corresponds to the unextended primer (P) and all bands above it correspond to a primer extended by 1 (P+1), 2 (P+2), or 3 (P+3) nucleotides. Although some reverse transcriptases showed a preference for one sequence context over another (e.g., SuperScript III gave more primer extension product for the OG-1 template than for the OG-2 template), all examined enzymes demonstrated matching nucleotide insertion profiles inserting C or A opposite OG and C or T opposite G when only one of the dNTPs was present. Considering that the assayed reverse transcriptases demonstrated similar patterns of nucleotide insertion for both RNA templates, we limited further studies to using only SuperScript III and template number 1 (Figure 1).
Figure 1.
RNA-DNA hybrid duplexes studied.
Figure 2. Nucleotide insertion profiles opposite G or OG in the templates.
Insertion of A, G, C, or T in the reaction mixture with only one of the dNTPs present by SuperScript III (A) and Omniscript (B) reverse transcriptases for G-1, OG-1, G-2, and OG-2 templates. (C) Structures of OG-C, OG-A, G-C, and G-T base pairs.
Before examining which nucleotides are inserted opposite Gh and diastereomers of Sp, we again made adjustments to concentrations of the triphosphates and the enzyme to achieve similar reactivities. Based on the adjusted parameters, the rate of polymerization for the RNA templates containing products of guanine oxidation decreased in the following order OG>Gh≫R-Sp>S-Sp. All three hydantoin lesions showed similar base pairing preferences leading to insertion of A or G (Figure 3). Insertion of A or G opposite Gh, S-Sp, and R-Sp when only one of the triphosphates was present was similar to insertion of A or C opposite OG in perfect agreement with what was previously reported for insertion opposite these lesions in DNA.60–63, 65 These results show that reverse transcriptases are capable of inserting canonical A, G, or C bases opposite OG, Gh, S-Sp, and R-Sp in an RNA template.
Figure 3. Nucleotide insertion assays for Gh and Sp.
A - Insertion of A, G, C, or T in the reaction mixture with only one of the dNTPs present by SuperScript III reverse transcriptase for Gh-1, S-Sp-1, and R-Sp-1 templates. B - Structures of the base pairs between G or A and Gh or Sp are based on previous literature reports.79, 80
Steady-state kinetics
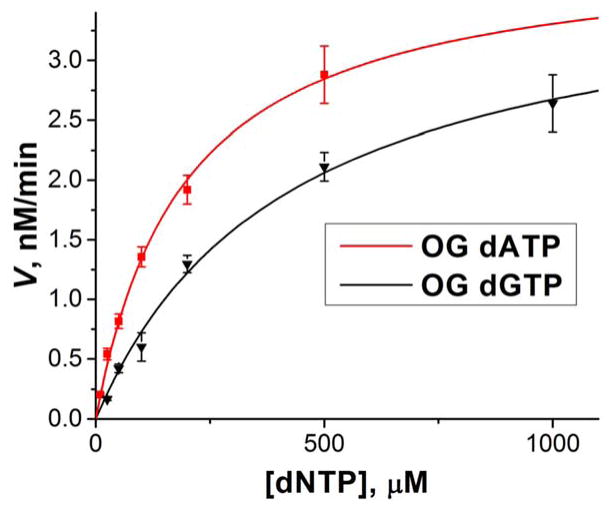
To examine SuperScript III behavior when it encounters oxidized lesions in more detail, we studied the kinetics of insertion of A or C opposite OG and A or G opposite Gh, S-Sp, or R-Sp in the template RNA strand. Additionally, insertion of C opposite G in the template was studied for a comparison that served as a control for SuperScript III kinetics when encountering canonical bases. Initial velocities of first base insertions were derived in units of nM/min and plotted against concentration of triphosphate used in each case. The data were then fitted to the Michaelis-Menten equation (2) to determine the steady-state kinetics parameters Vmax and KM. Vmax was then divided by the total enzyme concentration to calculate kcat according to the equation 3. Michaelis-Menten curves and calculated kinetics parameters are shown in Figure 4, Figure S5, and Table 2. The error bars on the graphs and in the table are representative of 68% confidence intervals.
Figure 4.
Michaelis-Menten plot for insertion of A or G opposite OG.
Table 2.
Steady-state kinetic parameters.
| Michaelis-Menten parameters | V/Et at 500 μM dNTP, min−1 | ||||
|---|---|---|---|---|---|
| Kcat, min−1 | KM, μM | Kcat/KM*103 | |||
| OG-1 | dCTP | 2.4 ± 0.2 | 450 ± 60 | 5.3 | 1.3 |
|
| |||||
| dATP | 2.5 ± 0.1 | 190 ± 20 | 13 | 1.8 | |
|
| |||||
| Gh-1 | dGTP | 0.55 ± 0.03 | 80 ± 15 | 7 | 0.47 |
|
| |||||
| dATP | 0.69 ± 0.08 | 600 ± 100 | 1 | 0.31 | |
|
| |||||
| S-Sp-1 | dGTP | 0.056 ± 0.002 | 200 ± 20 | 0.29 | 0.041 |
|
| |||||
| dATP | 0.071 ± 0.006 | 600 ± 100 | 0.12 | 0.032 | |
|
| |||||
| R-Sp-1 | dGTP | 0.12 ± 0.01 | 210 ± 50 | 0.57 | 0.085 |
|
| |||||
| dATP | 0.196 ± 0.003 | 1040 ± 30 | 0.19 | 0.064 | |
|
| |||||
| G | dCTP | 2.7 ± 0.2 | 0.028 ± 0.005 | 96,000 | 2.7 |
all error bars represent 68% confidence interval
| (2) |
| (3) |
Three main parameters derived from Michaelis-Menten curve are kcat, KM, and their ratio kcat/KM. The catalytic rate constant or turnover number kcat indicates the maximum number of reactions a single enzyme can catalyze per unit of time. In the case of reverse transcription, it is a combination of the rate of catalysis (condensation between 3′ end of the primer strand and a triphosphate being inserted opposite a studied base) and the rate of dissociation between the primer-template complex and the enzyme after the primer extension. For polymerases, kcat is normally defined by the rate of the slower dissociation step,62–64, 81, 82 thus it was not surprising that we observed very close values with overlapping confidence intervals for formation of OG-A (2.5 ± 0.1 min−1), OG-C (2.4 ± 0.2 min−1), and G-C (2.7 ± 0.2 min−1) base pairs. Similarities between turnover numbers for incorporation of a base opposite G and OG have been previously reported for translesion DNA polymerase η and HIV-1 RT by the Guengerich laboratory.62, 64 Unlike OG, the rate of phosphodiester bond formation apparently was affected by the presence of hydantoin lesions in the template strongly enough to result in lower kcat values for Gh-A (0.69 ± 0.08 min−1), Gh-G (0.55 ± 0.03 min−1), S-Sp-A (0.071 ± 0.006 min−1), S-Sp-G (0.056 ± 0.002 min−1), R-Sp-A (0.196 ± 0.003 min−1), and R-Sp-G (0.12 ± 0.01 min−1) base pairs. The kcat values for templates containing different products of guanine oxidation follow the same rule as reactivities determined for the nucleotide insertion assays: OG>Gh>R-Sp>S-Sp (Figure 2 and Table S1). Interestingly, in all cases, the kcat values for insertion of A were higher than for the insertion of C.
For polymerases, the Michaelis-Menten constant KM indicates how well an enzyme utilizes the substrate, but it cannot be directly linked to the dNTP dissociation constant.64, 82 The calculated KM values for insertion of C opposite G were more than 3 orders of magnitude higher than for the oxidized lesions showing that SuperScript III has a strong preference for this conventional Watson-Crick base pair. When we compared KM for insertion of A, C, or G opposite each product of guanine oxidation, in the case of OG we observed a significantly higher (2-fold) value for the OG-C base pair than for the OG-A base pair, while for Gh, S-Sp, and R-Sp, substantially higher values for Gh-A (6-fold), S-Sp-A (3-fold), and R-Sp-A (5-fold) were observed compared to the base pairs with G. From the kcat and KM values measured, we could calculate their ratio, kcat/KM, the catalytic efficiency of the enzyme that is proportional to a rate of dNTP insertion when the concentration of dNTP approaches 0. Catalytic efficiency is commonly used to determine enzyme preference for one dNTP over another as a substrate. Based on the determined kcat/KM values, SuperScript III has a 2.5-fold preference for inserting A over C opposite OG, a 6-fold preference for inserting G over A opposite Gh, a 2.4-fold preference for inserting G over A opposite S-Sp, and a 3-fold preference for inserting G over A opposite R-Sp.
Having the sequencing for oxidative damage to G in RNA as an ultimate goal, we used the steady-state kinetic parameters to estimate the ratio of nucleotides inserted opposite each studied base. This information would allow us to know if during cDNA synthesis SuperScript III would provide characteristic mutations that could be analyzed in order to locate sites of G oxidation. The results are shown in Table 2. The estimated insertion ratios were: for OG 1:1.4 C to A; for Gh 1:1.5 A to G; for both Sp diastereomers 1:1.3 A to G. Overall these data suggest that OG and the hydantoin lesions in RNA can be detected by sequencing cDNA created upon reverse transcription of RNA containing these products of guanine oxidation. While the three hydantoin lesions Gh, S-Sp, and R-Sp are likely not to be distinguishable by this method due to the similar sequencing signals observed in these studies, they should be easily separable from G and OG. Furthermore, knowing that a hydantoin is present at a given site in RNA would be a significant advancement in our knowledge of oxidative modification of RNA in cells. For the purpose of sequencing, a higher fraction of A insertion than C opposite OG is preferable because it allows an easier differentiation between unoxidized G and OG that is essential to increase the method sensitivity. Overall, based on the determined kinetic parameters, 3 different sequencing signals are expected in which G provides ~100% C insertion, OG yields A insertion at the modified site with ~60% efficiency, and the hydantoins yield insertion of a mixture of A and G with a modest preference for G.
Extension efficiency studies
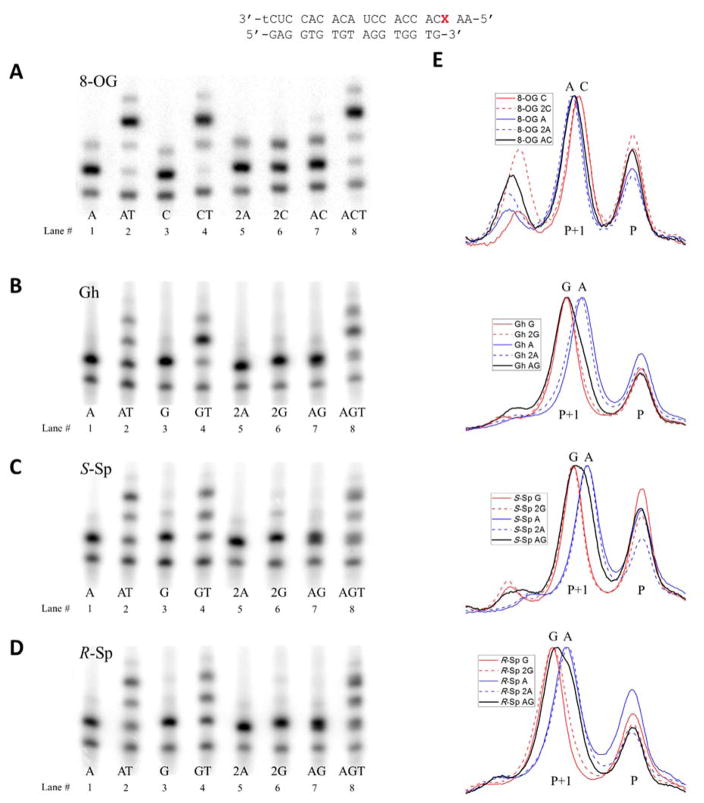
After discovering which bases are inserted opposite OG, Gh, S-Sp, and R-Sp, we wanted to investigate whether the extension efficiency is affected by what nucleotide is inserted opposite the lesions. To test this we performed primer extension assays using different combinations of dATP, dTTP, and dCTP (for OG) or dGTP (for the hydantoins). The reactions were conducted at twice higher concentrations of enzyme (6 U for OG, 8 U for Gh, 80 U for S-Sp, and 40 U for R-Sp in 20 μL) and increased concentrations of the triphosphates (100–200 μM for OG and 200–400 μM for Gh and Sp), compared to the nucleotide insertion studies; the reason for these changes was to drive the reaction closer to complete base insertion opposite the lesion site and be able to analyze products of primer extension past the site. The results of these studies are presented in Figure 5, Table S2, and Figure S6. On the gels, lanes 2 (AT) and 4 (CT or GT) correspond to reactions containing one of the triphosphates that can be efficiently inserted opposite an oxidized lesion (A and G or C) and dTTP sufficient for full extension of the primer. Thus, these studies can be used to discover which of the base pairs is more disruptive to the duplex structure and causes more polymerase inhibition. Lanes 1 (A) and 3 (C or G) serve as a control of the basal level of insertion of the first base under the same reaction conditions. Lane 8 (ACT or AGT) corresponds to reactions containing both triphosphates of the nucleotides that can be inserted opposite OG, Gh, or Sp, and dTTP shows how the efficiency of extension past the studied base is affected by the presence of both possible base pairs in the ratio at which they are inserted by reverse transcriptase. Lane 7 serves as a control study of the extent to which the first base is inserted under the same conditions. Lanes 5 and 6 serve as additional controls for insertion levels of the first base under twice higher concentrations of triphosphates corresponding to a sum of concentrations of dATP and dCTP or dGTP for lane 7. If efficiency of insertion of the first base in lane 7 is higher than in lanes 1 and 3, lanes 5 and 6 should show whether it is achievable simply by matching total dNTP concentrations.
Figure 5. Efficiency of extension past different base pairs.
A–D – polyacrylamide gels showing results of primer extension in presence of one (lanes 1, 3, 5, and 6), two (lanes 2, 4, and 7), or three (lane 8) different triphosphates aimed at comparing bypass efficiencies for base pairs between OG, Gh, S-Sp, and R-Sp and A, C or G. Samples in lanes 5 and 6 marked 2N contained a doubled concentration of corresponding dNTP compared to lanes 1 and 3. E - aligned gel lane plots showing separation between cDNA strands containing different bases inserted opposite OG, Gh, S-Sp, and R-Sp.
From the gels, it is evident that there is almost no difference in the level of polymerase blocking by the OG-A and OG-C base pairs (Figure 5A). Although, the presence of an OG-C base pair results in slightly more efficient insertion of the first T (CT lane) directly following C, insertion of the second T is less efficient than in the presence of an OG-A base pair (AT lane) leading to a similar amount of full extension product for both base pairs. Expectedly, a mixture of OG-A and OG-C base pairs leads to a very similar level of polymerase blocking (lane 8). Thus, there is practically no difference between OG-A and OG-C base pairs when it comes to how efficiently reverse transcription proceeds after them, meaning that the ratio at which they are inserted opposite OG should match the A:C ratio in a full-length cDNA.
For the template containing Gh, the presence of a Gh-G base pair (GT lane) leads to significantly more efficient insertion of the first T than the Gh-A base pair, but the polymerase struggles to insert the next base (Figure 5B and Table S2). In the case of a Gh-A base pair (AT lane) the most polymerase blockage is observed right after insertion of A. The presence of both dATP and dGTP in the reaction mixture (AGT lane) leads to polymerase arrest primarily after insertion of the T directly following the position of the Gh-N base pair. This hints at predominant insertion of G that aligns with the kinetic data for A and G insertion. Overall, insertion of G and A led to formation of comparable amounts of fully extended primer indicating that extension efficiency should not affect the A:G ratio in the fully extended cDNA.
Templates with R-Sp or S-Sp showed similar behavior with more efficient bypass of Sp-A base pairs in the AT lane than Sp-G base pairs in the GT lane (Figure 5C, D, and Table S2). An intermediate amount of polymerase blockage was observed for the case when both dATP and dGTP were present (AGT lane) for both Sp diastereomers indicating that both bases are inserted in this case. Due to a lower amount of polymerase blockage caused by the Sp-A bypass, it is likely that the fraction of this base pair in the fully extended cDNA would be higher than the ~45% estimate from the insertion kinetic studies.
There are two other noteworthy details about the gels on Figure 5. First, in all cases apart from the extension product of the expected length (P+3 for lanes 2, 4, and 8; P+1 for lanes 1, 3, 5, 6, and 7), a product overextended by one nucleotide was also observed. This is especially prominent in the case of the OG-1 template and can be attributed to strong template-independent polymerase activity of reverse transcriptase enzymes.83, 84 Second, the P+1 and P+3 bands are significantly broader in lanes 7 and 8 when both triphosphates are present. This is especially prominent for the Sp diastereomers and is caused by the dependence of electrophoretic mobility on nucleotide composition (C(fastest)>A>T>G).65, 85 Thus, insertion of a mixture of two nucleotides opposite OG, Gh, or Sp leads to a mixture of two cDNA strands with slightly different electrophoretic mobilities resulting in either broadened or split bands on the polyacrylamide gel. This can be used to get a rough estimate of composition of bases inserted opposite each studied lesion when both dNTPs are present. To do so, we generated pixel density plots for lanes 1, 3, 5, 6, and 7 and aligned them to match the position of the peak corresponding to the primer (P) (Figure 5E). It is only possible to characterize insertion of A and C or G qualitatively from this data due to the presence of the overextended peak P+2 and overlap between peaks corresponding to insertion of different bases. What could be said based on these plots is that A is predominantly inserted opposite OG, G is predominantly inserted opposite Gh, almost a 1:1 ratio of A and G are inserted opposite S-Sp, and a slightly higher fraction of G than A is inserted opposite the R-Sp isomer. Considering that Sp does not form stable base pairs with any natural nucleotides,61 insertion of comparable amounts of A and G opposite the hydantoin most likely can be attributed to fitting in the SuperScript III active site due to a similar size and minor groove binding pattern of Sp to C or T,86–88 rather than to efficient hydrogen-bonding with A or G. The ratio of A and C incorporated opposite OG by the polymerase can significantly deviate from the one calculated based on kinetic parameters in the presence of multiple triphosphates.64 Thus we tested whether this was true for SuperScript III using a gel assay (Figure S7 and S8). Since only qualitative characterization of the insertion rate was possible we attempted to adjust concentrations to achieve close to a 1:1 insertion ratio and compared it to the one expected from the reaction rates calculated at those concentrations (Table S3). In all four cases insertion of a higher fraction of A over C or G than expected from the kcat and KM was observed. Expected insertion ratios were within one standard deviation (SD) for Gh and R-Sp, two SD for S-Sp, and 4 SD for OG. Additionally, insertion ratios at 500 μM concentration of competing triphosphates were studied showing ~1:1 insertion of A and G opposite both Sp diastereomers, almost exclusive insertion of A opposite OG and predominant insertion of G opposite Gh (Figure S7 and S8).
The other question that we wanted to ask was how efficiently reverse transcriptases can fully extend the primer when a G oxidation product is located in the template strand. For this purpose, we ran RT reactions with RNA templates containing OG, Gh, the Sp diastereomers, or G, using the latter study as a control, in conditions close to those recommended for SuperScript III by the manufacturer of the polymerase (500 μM and 200 μM mixture of all four dNTPs and 100 U of enzyme in 20 μL). Products of the extension reactions were resolved on polyacrylamide gels (Figure 6). As expected, RNA templates containing G gave the highest amount of full extension product (~80% with 500 μM dNTPs), while the presence of OG, Gh, or R-Sp in the RNA template resulted in a decreased amount of fully extended primer (~60%), and the S-Sp-1 template gave the least amount of full extension product (~40%). Although, all tested RNA templates gave full extension product, it is apparent that OG, Gh, and Sp cause some level of polymerase inhibition even after the primer is extended past their position, most likely either due to the more dynamic nature of the base pairs they form or negative steric interactions with the flanking bases.89, 90 Interestingly, the highest level of inhibition was caused by S-Sp that was also found to be the most blocking in the polymerase nucleotide insertion studies (Figure 2). Analysis similar to the one presented in Figure 5E was done on fully extended primers (Figure 6B). Results for OG-1 (predominant insertion of A) and Gh-1 (predominant insertion of G) templates were comparable to the ones obtained for the insertion ratios of the first nucleotides in the presence of multiple dNTPs (Figure 5, S7, and S8). For both diastereomers of Sp, we observed a minor increase in the fraction of fully extended primer containing A inserted opposite Sp compared to the primer extended by just one nucleotide. This led to full-length cDNA containing close to a 1:1 ratio of A and G for the R-Sp-1 template and a higher fraction of A for the S-Sp template. Most likely this result was caused by a higher efficiency of bypassing the Sp-A base pair than the Sp-G base pair by the SuperScript III, as highlighted before.
Figure 6. Efficiency of complete primer extension.
A - polyacrylamide gel showing efficiency of synthesis of full-length cDNA based on the templates containing G, OG, Gh, S-Sp, or R-Sp at two different concentrations of dNTP mixture. B - aligned gel lane plots for different templates for reactions containing 500 and 200 μM dNTP mixture.
Conclusions
In the current study, we investigated the behavior of reverse transcriptases when they encounter products of guanine oxidation, OG, Gh, S-Sp, and R-Sp, in an RNA template under standing start conditions. Insertion of A or C opposite OG and A or G opposite the hydantoin lesions was observed that correlated with the previous reports for DNA containing these bases.37, 61, 64 We also report steady-state kinetic parameters for single nucleotide insertion opposite the studied lesions in the 5′-AXC-3′ sequence context by the SuperScript III reverse transcriptase. Results of this assay indicate a preference for insertion of A opposite OG and G opposite the Gh and Sp diastereomers. We discovered that all these lesions can be bypassed by the SuperScript III under the standard conditions used for RNA sequencing with sufficient efficiency to yield full-length cDNAs containing A or C inserted opposite OG with predominance of A, A or G inserted opposite Gh with predominance of G, A or G inserted opposite S-Sp with minor preference for A, and an equal amount of A or G inserted opposite R-Sp. These data indicate that mapping positions of OG, Gh, and Sp in RNA is potentially achievable with the use of reverse transcription to induce characteristic mutations upon cDNA synthesis. Sequencing of the cDNA strand should allow separate detection of unoxidized G as a non-mutagenic C, OG as a C to A mutation (G to T in the complementary stand), Gh as a C to G mutation (G to C in the complementary stand), and the Sp diastereomers as an ~1:1 mixture of C to A and C to G mutations (G to T and G to C in the complementary strand). Posttranscriptional modifications of G and I that disrupt their base pairing preference for C (m1G, m2G, m22G, and m1I) give different sequencing signals (preferential insertion of T91, 92) and could be removed with dealkylating enzymes,67, 93 therefore, they should not interfere with the reverse transcription results for the oxidized guanine lesions. Relatively efficient bypass of all lesions studied also permits amplification of cDNA via PCR (RT-PCR) that allows interrogation of much smaller quantities of RNA.
Supplementary Material
Acknowledgments
We appreciate the support of the National Institute of General Medical Sciences (R01 GM093099) for this work. The oligonucleotides were provided by the DNA/Peptide core facility at the University of Utah that is supported in part by a NCI Cancer Support Grant (P30 CA042014).
Footnotes
Conflict of interest
The authors declare no competing financial interest.
Additional information on characterization of rSp diastereomers, Michaelis-Menten plots, gel images and gel quantification data for polymerase nucleotide insertion and extension assays not included in this paper can be found in the supporting information. The supporting information is available free of charge via the Internet at http://pubs.acs.org/.
References
- 1.Lindahl T. Instability and decay of the primary structure of DNA. Nature. 1993;362:709–715. doi: 10.1038/362709a0. [DOI] [PubMed] [Google Scholar]
- 2.Loft S, Poulsen HE. Cancer risk and oxidative DNA damage in man. J Mol Med. 1996;74:297–312. doi: 10.1007/BF00207507. [DOI] [PubMed] [Google Scholar]
- 3.De Bont R, van Larebeke N. Endogenous DNA damage in humans: a review of quantitative data. Mutagenesis. 2004;19:169–185. doi: 10.1093/mutage/geh025. [DOI] [PubMed] [Google Scholar]
- 4.Wogan GN, Hecht SS, Felton JS, Conney AH, Loeb LA. Environmental and chemical carcinogenesis. Semin Cancer Biol. 2004;14:473–486. doi: 10.1016/j.semcancer.2004.06.010. [DOI] [PubMed] [Google Scholar]
- 5.Noll DM, Mason TM, Miller PS. Formation and repair of interstrand cross-links in DNA. Chem Rev. 2006;106:277–301. doi: 10.1021/cr040478b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Oleinick NL, Chiu SM, Ramakrishnan N, Xue LY. The formation, identification, and significance of DNA-protein cross-links in mammalian cells. Br J Cancer Suppl. 1987;8:135–140. [PMC free article] [PubMed] [Google Scholar]
- 7.Johansen ME, Muller JG, Xu X, Burrows CJ. Oxidatively induced DNA-protein cross-linking between single-stranded binding protein and oligodeoxynucleotides containing 8-oxo-7,8-dihydro-2′-deoxyguanosine. Biochemistry. 2005;44:5660–5671. doi: 10.1021/bi047580n. [DOI] [PubMed] [Google Scholar]
- 8.Kundu LM, Linne U, Marahiel M, Carell T. RNA is more UV resistant than DNA: the formation of UV-induced DNA lesions is strongly sequence and conformation dependent. Chem Eur J. 2004;10:5697–5705. doi: 10.1002/chem.200305731. [DOI] [PubMed] [Google Scholar]
- 9.Kong Q, Lin CL. Oxidative damage to RNA: mechanisms, consequences, and diseases. Cell Mol Life Sci. 2010;67:1817–1829. doi: 10.1007/s00018-010-0277-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ames BN, Shigenaga MK, Hagen TM. Oxidants, antioxidants, and the degenerative diseases of aging. Proc Natl Acad Sci USA. 1993;90:7915–7922. doi: 10.1073/pnas.90.17.7915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Finkel T, Holbrook NJ. Oxidants, oxidative stress and the biology of ageing. Nature. 2000;408:239–247. doi: 10.1038/35041687. [DOI] [PubMed] [Google Scholar]
- 12.Cooke MS, Evans MD, Dizdaroglu M, Lunec J. Oxidative DNA damage: mechanisms, mutation, and disease. FASEB J. 2003;17:1195–1214. doi: 10.1096/fj.02-0752rev. [DOI] [PubMed] [Google Scholar]
- 13.Li Z, Wu J, Deleo CJ. RNA damage and surveillance under oxidative stress. IUBMB Life. 2006;58:581–588. doi: 10.1080/15216540600946456. [DOI] [PubMed] [Google Scholar]
- 14.Bernstein JA, Khodursky AB, Lin PH, Lin-Chao S, Cohen SN. Global analysis of mRNA decay and abundance in Escherichia coli at single-gene resolution using two-color fluorescent DNA microarrays. Proc Natl Acad Sci USA. 2002;99:9697–9702. doi: 10.1073/pnas.112318199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Defoiche J, Zhang Y, Lagneaux L, Pettengell R, Hegedus A, Willems L, Macallan DC. Measurement of ribosomal RNA turnover in vivo by use of deuterium-labeled glucose. Clin Chem. 2009;55:1824–1833. doi: 10.1373/clinchem.2008.119446. [DOI] [PubMed] [Google Scholar]
- 16.Schmidt EE, Schibler U. Cell size regulation, a mechanism that controls cellular RNA accumulation: consequences on regulation of the ubiquitous transcription factors Oct1 and NF-Y and the liver-enriched transcription factor DBP. J Cell Biol. 1995;128:467–483. doi: 10.1083/jcb.128.4.467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hofer T, Badouard C, Bajak E, Ravanat JL, Mattsson A, Cotgreave IA. Hydrogen peroxide causes greater oxidation in cellular RNA than in DNA. Biol Chem. 2005;386:333–337. doi: 10.1515/BC.2005.040. [DOI] [PubMed] [Google Scholar]
- 18.Mangerich A, Knutson CG, Parry NM, Muthupalani S, Ye W, Prestwich E, Cui L, McFaline JL, Mobley M, Ge Z, Taghizadeh K, Wishnok JS, Wogan GN, Fox JG, Tannenbaum SR, Dedon PC. Infection-induced colitis in mice causes dynamic and tissue-specific changes in stress response and DNA damage leading to colon cancer. Proc Natl Acad Sci USA. 2012;109:E1820–1829. doi: 10.1073/pnas.1207829109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu M, Gong X, Alluri RK, Wu J, Sablo T, Li Z. Characterization of RNA damage under oxidative stress in Escherichia coli. Biol Chem. 2012;393:123–132. doi: 10.1515/hsz-2011-0247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Saikia M, Jobava R, Parisien M, Putnam A, Krokowski D, Gao XH, Guan BJ, Yuan Y, Jankowsky E, Feng Z, Hu GF, Pusztai-Carey M, Gorla M, Sepuri NB, Pan T, Hatzoglou M. Angiogenin-cleaved tRNA halves interact with cytochrome c, protecting cells from apoptosis during osmotic stress. Mol Cell Biol. 2014;34:2450–2463. doi: 10.1128/MCB.00136-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mishima E, Inoue C, Saigusa D, Inoue R, Ito K, Suzuki Y, Jinno D, Tsukui Y, Akamatsu Y, Araki M, Araki K, Shimizu R, Shinke H, Suzuki T, Takeuchi Y, Shima H, Akiyama Y, Toyohara T, Suzuki C, Saiki Y, Tominaga T, Miyagi S, Kawagisihi N, Soga T, Ohkubo T, Yamamura K, Imai Y, Masuda S, Sabbisetti V, Ichimura T, Mount DB, Bonventre JV, Ito S, Tomioka Y, Itoh K, Abe T. Conformational change in transfer RNA is an early indicator of acute cellular damage. J Am Soc Nephrol. 2014:2316–2326. doi: 10.1681/ASN.2013091001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mleczko AM, Celichowski P, Bakowska-Zywicka K. Ex-translational function of tRNAs and their fragments in cancer. Acta Biochim Pol. 2014;61:211–216. [PubMed] [Google Scholar]
- 23.Lee YS, Shibata Y, Malhotra A, Dutta A. A novel class of small RNAs: tRNA-derived RNA fragments (tRFs) Genes Dev. 2009;23:2639–2649. doi: 10.1101/gad.1837609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thompson DM, Lu C, Green PJ, Parker R. tRNA cleavage is a conserved response to oxidative stress in eukaryotes. RNA. 2008;14:2095–2103. doi: 10.1261/rna.1232808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nawrot B, Sochacka E, Duchler M. tRNA structural and functional changes induced by oxidative stress. Cell Mol Life Sci. 2011;68:4023–4032. doi: 10.1007/s00018-011-0773-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Moreira PI, Nunomura A, Nakamura M, Takeda A, Shenk JC, Aliev G, Smith MA, Perry G. Nucleic acid oxidation in Alzheimer disease. Free Radic Biol Med. 2008;44:1493–1505. doi: 10.1016/j.freeradbiomed.2008.01.002. [DOI] [PubMed] [Google Scholar]
- 27.Poulsen HE, Specht E, Broedbaek K, Henriksen T, Ellervik C, Mandrup-Poulsen T, Tonnesen M, Nielsen PE, Andersen HU, Weimann A. RNA modifications by oxidation: a novel disease mechanism? Free Radic Biol Med. 2012;52:1353–1361. doi: 10.1016/j.freeradbiomed.2012.01.009. [DOI] [PubMed] [Google Scholar]
- 28.Castellani RJ, Nunomura A, Rolston RK, Moreira PI, Takeda A, Perry G, Smith MA. Sublethal RNA oxidation as a mechanism for neurodegenerative disease. Int J Mol Sci. 2008;9:789–806. doi: 10.3390/ijms9050789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Che Y, Wang JF, Shao L, Young T. Oxidative damage to RNA but not DNA in the hippocampus of patients with major mental illness. J Psychiatry Neurosci. 2010;35:296–302. doi: 10.1503/jpn.090083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fujikawa K, Kamiya H, Yakushiji H, Nakabeppu Y, Kasai H. Human MTH1 protein hydrolyzes the oxidized ribonucleotide, 2-hydroxy-ATP. Nucleic Acids Res. 2001;29:449–454. doi: 10.1093/nar/29.2.449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wu J, Li Z. Human polynucleotide phosphorylase reduces oxidative RNA damage and protects HeLa cell against oxidative stress. Biochem Biophys Res Commun. 2008;372:288–292. doi: 10.1016/j.bbrc.2008.05.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ishibashi T, Hayakawa H, Ito R, Miyazawa M, Yamagata Y, Sekiguchi M. Mammalian enzymes for preventing transcriptional errors caused by oxidative damage. Nucleic Acids Res. 2005;33:3779–3784. doi: 10.1093/nar/gki682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bellacosa A, Moss EG. RNA repair: damage control. Curr Biol. 2003;13:R482–484. doi: 10.1016/s0960-9822(03)00408-1. [DOI] [PubMed] [Google Scholar]
- 34.Wurtmann EJ, Wolin SL. RNA under attack: cellular handling of RNA damage. Crit Rev Biochem Mol Biol. 2009;44:34–49. doi: 10.1080/10409230802594043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Steenken S, Jovanovic SV, Bietti M, Bernhard K. The trap depth (in DNA) of 8-oxo-7,8-dihydro-2 ′deoxyguanosine as derived from electron-transfer equilibria in aqueous solution. J Am Chem Soc. 2000;122:2373–2374. [Google Scholar]
- 36.Neeley WL, Essigmann JM. Mechanisms of formation, genotoxicity, and mutation of guanine oxidation products. Chem Res Toxicol. 2006;19:491–505. doi: 10.1021/tx0600043. [DOI] [PubMed] [Google Scholar]
- 37.Henderson PT, Delaney JC, Muller JG, Neeley WL, Tannenbaum SR, Burrows CJ, Essigmann JM. The hydantoin lesions formed from oxidation of 7,8-dihydro-8-oxoguanine are potent sources of replication errors in vivo. Biochemistry. 2003;42:9257–9262. doi: 10.1021/bi0347252. [DOI] [PubMed] [Google Scholar]
- 38.McKibbin PL, Fleming AM, Towheed MA, Van Houten B, Burrows CJ, David SS. Repair of hydantoin lesions and their amine adducts in DNA by base and nucleotide excision repair. J Am Chem Soc. 2013;135:13851–13861. doi: 10.1021/ja4059469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tomaszewska-Antczak A, Guga P, Nawrot B, Pratviel G. Guanosine in a single stranded region of anticodon stem-loop tRNA models is prone to oxidatively generated damage resulting in dehydroguanidinohydantoin and spiroiminodihydantoin lesions. Chem Eur J. 2015;21:6381–6385. doi: 10.1002/chem.201406409. [DOI] [PubMed] [Google Scholar]
- 40.Fleming AM, Alshykhly O, Zhu J, Muller JG, Burrows CJ. Rates of chemical cleavage of DNA and RNA oligomers containing guanine oxidation products. Chem Res Toxicol. 2015;28:1292–1300. doi: 10.1021/acs.chemrestox.5b00096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fleming AM, Orendt AM, He Y, Zhu J, Dukor RK, Burrows CJ. Reconciliation of chemical, enzymatic, spectroscopic and computational data to assign the absolute configuration of the DNA base lesion spiroiminodihydantoin. J Am Chem Soc. 2013;135:18191–18204. doi: 10.1021/ja409254z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Delaney S, Jarem DA, Volle CB, Yennie CJ. Chemical and biological consequences of oxidatively damaged guanine in DNA. Free Radic Res. 2012;46:420–441. doi: 10.3109/10715762.2011.653968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pratviel G, Meunier B. Guanine oxidation: one- and two-electron reactions. Chem Eur J. 2006;12:6018–6030. doi: 10.1002/chem.200600539. [DOI] [PubMed] [Google Scholar]
- 44.Gimisis T, Cismas C. Isolation, characterization, and independent synthesis of guanine oxidation products. Eur J Org Chem. 2006:1351–1378. [Google Scholar]
- 45.Cadet J, Wagner JR, Shafirovich V, Geacintov NE. One-electron oxidation reactions of purine and pyrimidine bases in cellular DNA. Int J Radiat Biol. 2014;90:423–432. doi: 10.3109/09553002.2013.877176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Burrows CJ, Muller JG. Oxidative nucleobase modifications leading to strand scission. Chem Rev. 1998;98:1109–1152. doi: 10.1021/cr960421s. [DOI] [PubMed] [Google Scholar]
- 47.Wells SE, Hughes JMX, Igel AH, Ares M. Use of dimethyl sulfate to probe RNA structure in vivo. Methods Enzymol. 2000;318:479–493. doi: 10.1016/s0076-6879(00)18071-1. [DOI] [PubMed] [Google Scholar]
- 48.Ross RL, Cao X, Limbach PA. Mapping Post-Transcriptional Modifications onto Transfer Ribonucleic Acid Sequences by Liquid Chromatography Tandem Mass Spectrometry. Biomolecules. 2017;7:E21. [Google Scholar]
- 49.Wagner TM, Nair V, Guymon R, Pomerantz SC, Crain PF, Davis DR, McCloskey JA. A novel method for sequence placement of modified nucleotides in mixtures of transfer RNA. Nucleic Acids Symp Ser (Oxf) 2004:263–264. doi: 10.1093/nass/48.1.263. [DOI] [PubMed] [Google Scholar]
- 50.Kowalak JA, Pomerantz SC, Crain PF, McCloskey JA. A novel method for the determination of post-transcriptional modification in RNA by mass spectrometry. Nucleic Acids Res. 1993;21:4577–4585. doi: 10.1093/nar/21.19.4577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Guymon R, Pomerantz SC, Ison JN, Crain PF, McCloskey JA. Post-transcriptional modifications in the small subunit ribosomal RNA from Thermotoga maritima, including presence of a novel modified cytidine. RNA. 2007;13:396–403. doi: 10.1261/rna.361607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Siegfried NA, Busan S, Rice GM, Nelson JA, Weeks KM. RNA motif discovery by SHAPE and mutational profiling (SHAPE-MaP) Nat Methods. 2014;11:959–965. doi: 10.1038/nmeth.3029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.von Watzdorf J, Marx A. 6-Substituted 2-aminopurine-2′-deoxyribonucleoside 5′-triphosphates that trace cytosine methylation. ChemBioChem. 2016;17:1532–1540. doi: 10.1002/cbic.201600245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.von Watzdorf J, Leitner K, Marx A. Modified nucleotides for discrimination between cytosine and the epigenetic marker 5-methylcytosine. Angew Chem Int Ed Engl. 2016;55:3229–3232. doi: 10.1002/anie.201511520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wyss LA, Nilforoushan A, Eichenseher F, Suter U, Blatter N, Marx A, Sturla SJ. Specific incorporation of an artificial nucleotide opposite a mutagenic DNA adduct by a DNA polymerase. J Am Chem Soc. 2015;137:30–33. doi: 10.1021/ja5100542. [DOI] [PubMed] [Google Scholar]
- 56.Gahlon HL, Schweizer WB, Sturla SJ. Tolerance of base pair size and shape in postlesion DNA synthesis. J Am Chem Soc. 2013;135:6384–6387. doi: 10.1021/ja311434s. [DOI] [PubMed] [Google Scholar]
- 57.Merino EJ, Wilkinson KA, Coughlan JL, Weeks KM. RNA structure analysis at single nucleotide resolution by selective 2 ′-hydroxyl acylation and primer extension (SHAPE) J Am Chem Soc. 2005;127:4223–4231. doi: 10.1021/ja043822v. [DOI] [PubMed] [Google Scholar]
- 58.Loughrey D, Watters KE, Settle AH, Lucks JB. SHAPE-Seq 2.0: systematic optimization and extension of high-throughput chemical probing of RNA secondary structure with next generation sequencing. Nucleic Acids Res. 2014;42:e165. doi: 10.1093/nar/gku909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Dahlmann HA, Vaidyanathan VG, Sturla SJ. Investigating the biochemical impact of DNA damage with structure-based probes: abasic sites, photodimers, alkylation adducts, and oxidative lesions. Biochemistry. 2009;48:9347–9359. doi: 10.1021/bi901059k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kornyushyna O, Burrows CJ. Effect of the oxidized guanosine lesions spiroiminodihydantoin and guanidinohydantoin on proofreading by Escherichia coli DNA polymerase I (Klenow fragment) in different sequence contexts. Biochemistry. 2003;42:13008–13018. doi: 10.1021/bi0350755. [DOI] [PubMed] [Google Scholar]
- 61.Kornyushyna O, Berges AM, Muller JG, Burrows CJ. In vitro nucleotide misinsertion opposite the oxidized guanosine lesions spiroiminodihydantoin and guanidinohydantoin and DNA synthesis past the lesions using Escherichia coli DNA polymerase I (Klenow fragment) Biochemistry. 2002;41:15304–15314. doi: 10.1021/bi0264925. [DOI] [PubMed] [Google Scholar]
- 62.Furge LL, Guengerich FP. Analysis of nucleotide insertion and extension at 8-oxo-7,8-dihydroguanine by replicative T7 polymerase exo(-) and human immunodeficiency virus-1 reverse transcriptase using steady-state and pre-steady-state kinetics. Biochemistry. 1997;36:6475–6487. doi: 10.1021/bi9627267. [DOI] [PubMed] [Google Scholar]
- 63.Lowe LG, Guengerich FP. Steady-state and pre-steady-state kinetic analysis of dNTP insertion opposite 8-oxo-7,8-dihydroguanine by Escherichia coli polymerases I exo- and II exo. Biochemistry. 1996;35:9840–9849. doi: 10.1021/bi960485x. [DOI] [PubMed] [Google Scholar]
- 64.Xue Q, Zhong M, Liu B, Tang Y, Wei Z, Guengerich FP, Zhang H. Kinetic analysis of bypass of 7,8-dihydro-8-oxo-2′-deoxyguanosine by the catalytic core of yeast DNA polymerase eta. Biochimie. 2016;121:161–169. doi: 10.1016/j.biochi.2015.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hsu GW, Ober M, Carell T, Beese LS. Error-prone replication of oxidatively damaged DNA by a high-fidelity DNA polymerase. Nature. 2004;431:217–221. doi: 10.1038/nature02908. [DOI] [PubMed] [Google Scholar]
- 66.Cantara WA, Crain PF, Rozenski J, McCloskey JA, Harris KA, Zhang X, Vendeix FA, Fabris D, Agris PF. The RNA modification database, RNAMDB: 2011 update. Nucleic Acids Res. 2011;39:D195–201. doi: 10.1093/nar/gkq1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zheng G, Qin Y, Clark WC, Dai Q, Yi C, He C, Lambowitz AM, Pan T. Efficient and quantitative high-throughput tRNA sequencing. Nat Methods. 2015;12:835–837. doi: 10.1038/nmeth.3478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ye Y, Muller JG, Luo W, Mayne CL, Shallop AJ, Jones RA, Burrows CJ. Formation of 13C-, 15N-, and 18O-labeled guanidinohydantoin from guanosine oxidation with singlet oxygen. Implications for structure and mechanism. J Am Ceram Soc. 2003;125:13926–13927. doi: 10.1021/ja0378660. [DOI] [PubMed] [Google Scholar]
- 69.Fleming AM, Muller JG, Dlouhy AC, Burrows CJ. Structural context effects in the oxidation of 8-oxo-7,8-dihydro-2′-deoxyguanosine to hydantoin products: electrostatics, base stacking, and base pairing. J Am Chem Soc. 2012;134:15091–15102. doi: 10.1021/ja306077b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Schindelin J, Rueden CT, Hiner MC, Eliceiri KW. The ImageJ ecosystem: An open platform for biomedical image analysis. Mol Reprod Dev. 2015;82:518–529. doi: 10.1002/mrd.22489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Morales JC, Kool ET. Importance of terminal base pair hydrogen-bonding in 3′-end proofreading by the Klenow fragment of DNA polymerase I. Biochemistry. 2000;39:2626–2632. doi: 10.1021/bi992173a. [DOI] [PubMed] [Google Scholar]
- 73.Boutabout M, Wilhelm M, Wilhelm FX. DNA synthesis fidelity by the reverse transcriptase of the yeast retrotransposon Ty1. Nucleic Acids Res. 2001;29:2217–2222. doi: 10.1093/nar/29.11.2217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Mohr S, Ghanem E, Smith W, Sheeter D, Qin Y, King O, Polioudakis D, Iyer VR, Hunicke-Smith S, Swamy S, Kuersten S, Lambowitz AM. Thermostable group II intron reverse transcriptase fusion proteins and their use in cDNA synthesis and next-generation RNA sequencing. RNA. 2013;19:958–970. doi: 10.1261/rna.039743.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Stahlberg A, Kubista M, Pfaffl M. Comparison of reverse transcriptases in gene expression analysis. Clin Chem. 2004;50:1678–1680. doi: 10.1373/clinchem.2004.035469. [DOI] [PubMed] [Google Scholar]
- 76.Okello JBA, Rodriguez L, Poinar D, Bos K, Okwi AL, Bimenya GS, Sewankambo NK, Henry KR, Kuch M, Poinar HN. Quantitative assessment of the sensitivity of various commercial reverse transcriptases based on armored HIV RNA. Plos One. 2010;5:e13931. doi: 10.1371/journal.pone.0013931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Di Giallonardo F, Zagordi O, Duport Y, Leemann C, Joos B, Kunzli-Gontarczyk M, Bruggmann R, Beerenwinkel N, Gunthard HF, Metzner KJ. Next-generation sequencing of HIV-1 RNA genomes: determination of error rates and minimizing artificial recombination. Plos One. 2013;8:e74249. doi: 10.1371/journal.pone.0074249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.He L, Sok D, Azadnia P, Hsueh J, Landais E, Simek M, Koff WC, Poignard P, Burton DR, Zhu J. Toward a more accurate view of human B-cell repertoire by next-generation sequencing, unbiased repertoire capture and single-molecule barcoding. Sci Rep. 2014;4:6778. doi: 10.1038/srep06778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Jia L, Shafirovich V, Shapiro R, Geacintov NE, Broyde S. Structural and thermodynamic features of spiroiminodihydantoin damaged DNA duplexes. Biochemistry. 2005;44:13342–13353. doi: 10.1021/bi050790v. [DOI] [PubMed] [Google Scholar]
- 80.Zhao X, Muller JG, Halasyam M, David SS, Burrows CJ. In vitro ligation of oligodeoxynucleotides containing C8-oxidized purine lesions using bacteriophage T4 DNA ligase. Biochemistry. 2007;46:3734–3744. doi: 10.1021/bi062214k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Gardner AF, Joyce CM, Jack WE. Comparative kinetics of nucleotide analog incorporation by vent DNA polymerase. J Biol Chem. 2004;279:11834–11842. doi: 10.1074/jbc.M308286200. [DOI] [PubMed] [Google Scholar]
- 82.Einolf HJ, Guengerich FP. Kinetic analysis of nucleotide incorporation by mammalian DNA polymerase delta. J Biol Chem. 2000;275:16316–16322. doi: 10.1074/jbc.M001291200. [DOI] [PubMed] [Google Scholar]
- 83.Ohtsubo Y, Nagata Y, Tsuda M. Efficient N-tailing of blunt DNA ends by Moloney murine leukemia virus reverse transcriptase. Sci Rep. 2017;7:41769. doi: 10.1038/srep41769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Bibillo A, Eickbush TH. End-to-end template jumping by the reverse transcriptase encoded by the R2 retrotransposon. J Biol Chem. 2004;279:14945–14953. doi: 10.1074/jbc.M310450200. [DOI] [PubMed] [Google Scholar]
- 85.Frank R, Koster H. DNA chain length markers and the influence of base composition on electrophoretic mobility of oligodeoxyribonucleotides in polyacrylamide-gels. Nucleic Acids Res. 1979;6:2069–2087. doi: 10.1093/nar/6.6.2069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Hottin A, Marx A. Structural Insights into the Processing of Nucleobase-Modified Nucleotides by DNA Polymerases. Acc Chem Res. 2016;49:418–427. doi: 10.1021/acs.accounts.5b00544. [DOI] [PubMed] [Google Scholar]
- 87.Ludmann S, Marx A. Getting it Right: How DNA Polymerases Select the Right Nucleotide. Chimia. 2016;70:203–206. doi: 10.2533/chimia.2016.203. [DOI] [PubMed] [Google Scholar]
- 88.Marx A, Summerer D. Molecular insights into error-prone DNA replication and error-free lesion bypass. ChemBioChem. 2002;3:405–407. doi: 10.1002/1439-7633(20020503)3:5<405::AID-CBIC405>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 89.Ye Y. PhD dissertation. Department of Chemistry, University of Utah; Salt Lake City, UT: 2007. From nucleosides and nucleotides to DNA: synthesis, enzymatic study and structural analysis of oxidized lesions beyond 8-oxo-purine. [Google Scholar]
- 90.Khutsishvili I, Zhang N, Marky LA, Crean C, Patel DJ, Geacintov NE, Shafirovich V. Thermodynamic profiles and nuclear magnetic resonance studies of oligonucleotide duplexes containing single diastereomeric spiroiminodihydantoin lesions. Biochemistry. 2013;52:1354–1363. doi: 10.1021/bi301566v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Delaney JC, Essigmann JM. Mutagenesis, genotoxicity, and repair of 1-methyladenine, 3-alkylcytosines, 1-methylguanine, and 3-methylthymine in alkB Escherichia coli. Proc Natl Acad Sci USA. 2004;101:14051–14056. doi: 10.1073/pnas.0403489101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Ryvkin P, Leung YY, Silverman IM, Childress M, Valladares O, Dragomir I, Gregory BD, Wang LS. HAMR: high-throughput annotation of modified ribonucleotides. RNA. 2013;19:1684–1692. doi: 10.1261/rna.036806.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Dai Q, Zheng G, Schwartz MH, Clark WC, Pan T. Selective enzymatic demethylation of N2, N2-dimethylguanosine in RNA and its application in high-throughput tRNA sequencing. Angew Chem Int Ed Engl. 2017;56:5017–5020. doi: 10.1002/anie.201700537. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.