Abstract
Immune tolerance hinders the potentially destructive responses of lymphocytes to host tissues. Tolerance is regulated at the stage of immature B cell development (central tolerance) by clonal deletion, involving apoptosis, and by receptor editing, which reprogrammes the specificity of B cells through secondary recombination of antibody genes. Recent mechanistic studies have begun to elucidate how these divergent mechanisms are controlled. Single-cell antibody cloning has revealed defects of B cell central tolerance in human autoimmune diseases and in several human immunodeficiency diseases caused by single gene mutations, which indicates the relevance of B cell tolerance to disease and suggests possible genetic pathways that regulate tolerance.
B cells have an essential role in host defence via the production of the antibody response to microorganisms. Individuals lacking B cells fail to produce any antibodies and are prone to serious infectious disease. Each B cell carries a unique receptor for antigen (the B cell receptor (BCR)) that is composed of the membrane-bound form of its antibody. Upon antigen recognition by the membrane-bound receptor, reactive B cells proliferate to increase their numbers and differentiate to secrete their specific antibody as one of five immunoglobulin classes: IgM, IgD, IgG, IgA or IgE. In collaboration with CD4+ T follicular helper (TFH) cells and other cell types, activated B cells can also undergo somatic mutation of the variable portion of the expressed antibody genes to alter and improve antigen specificity and affinity. High-affinity antibodies provide protection against many types of infection, as well as immunity in response to vaccination. However, antibodies that have ‘inappropriate’ specificities for host tissue can be pathogenic and are diagnostic of many autoimmune or rheumatological diseases, such as systemic lupus erythematosus (SLE), rheumatoid arthritis and insulin-dependent type 1 diabetes. Therapeutic depletion of B cells is often beneficial in diseases of this kind, probably because it reduces antigen presentation to autoreactive T cells as well as the production of harmful autoantibodies.
Under normal circumstances, autoreactive B cells are regulated in several ways to decrease their frequency in the B cell repertoire, their affinity for self-tissue or their functionality. These immune tolerance mechanisms function at various stages of B cell development. Central tolerance refers to the regulatory mechanisms that occur at the early stages of B cell development in the bone marrow, when B cells carry a surface antigen receptor of the IgM class but are not fully mature. Later developmental stages of B cells take place mainly in the spleen, lymph nodes and other tissues, where B cells co-express IgM and IgD, acquire the capacity to be fully activated, and are able to respond productively with T cells and antigen to produce high-affinity antibodies. Tolerance mechanisms that occur at these later developmental stages are referred to as peripheral tolerance. Although mechanisms of peripheral tolerance — such as the induction of anergy, antigen receptor desensitization or tolerance to antigens that co-engage sialic acid-binding immunoglobulin-like lectin (Siglec) inhibitory receptors1–4 — regulate the survival and activation of B cells after they exit the bone marrow, none of these can be considered as fail-safe mechanisms; most of the mechanisms of peripheral tolerance are reversible because of the potential need for mature B cells to respond to viruses and microorganisms that may carry similar epitopes to self-antigens5. Therefore, central tolerance has a key role in reducing the frequency of autoreactive cells in the naive, pre-immune B cell repertoire.
A novel aspect of central tolerance that has attracted recent research attention is the mechanism of receptor editing, which permits ongoing immunoglobulin gene recombination to modify the specificity of B cells carrying autoreactive antigen receptors. At the same time, receptor editing contributes to immune diversity by promoting the use of antibody genes that initially rearrange inefficiently. Apoptosis resulting from the recognition of self-antigens also has a major role in central tolerance in both B cells and T cells, as cells at early developmental stages are particularly sensitive to this form of cell death. Defects in these tolerance processes have been implicated in the pathogenesis of autoimmune diseases and in certain immunodeficiency disorders. Here, I discuss the processes that regulate autoreactive B cells as they emerge in the bone marrow and the dysregulation of these processes in disease states, based on studies in mouse models and humans. In particular, I describe how antigen receptor signalling in B cell development regulates the nature of the receptor itself, aiding in receptor selection and correction, to eliminate autoreactivity by reprogramming the antigen receptor genes. This discussion requires a brief review of B cell development, BCR signalling and V(D)J recombination. I also review several recent studies that assess central tolerance mechanisms in mouse and human B cells, with particular emphasis on the effects of single gene defects (TABLE 1). This Review is not intended to cover studies that exclusively focus on mature, peripheral B cells, although such studies are occasionally mentioned when they help to formulate ideas of relevance to immature B cells.
Table 1.
Genes and proteins implicated in B cell central tolerance and receptor editing
| Gene | Model | Comment | Refs |
|---|---|---|---|
| ADA | Human | Deficiency hinders tolerance | 133 |
| AICDA | Human and mouse | Deficiency hinders tolerance | 141,142 |
| Bcl2 | Mouse | Enforced expression hinders tolerance | 16,101 |
| Bim (also known as Bcl2l11) | Mouse | Deficiency hinders tolerance | 99,160 |
| Blnk | Mouse | Deficiency hinders editing | 71 |
| BTK | Human and mouse | Deficiency hinders tolerance | 176–178 |
| Cbl and Cblb | Mouse | Deficiency promotes autoimmunity | 155 |
| CD19 | Human and mouse | Deficiency hinders tolerance | 137,179,180 |
| Cd45 | Mouse | Deficiency hinders tolerance | 136,181 |
| CIN85 (also known as SH3KBP1) | Human cell line | Knockdown promotes B cell survival | 182 |
| Fkbp11 | Mouse | Overexpression hinders tolerance | 183 |
| Gadd45a | Mouse | Deficiency hinders editing | 160 |
| Hcls1 (encodes HS1) | Mouse cell line | Deficiency hinders apoptosis | 184 |
| IRAK4 | Human | Deficiency hinders tolerance | 139 |
| Irf4 | Mouse | Deficiency hinders editing | 82 |
| Itpkb | Mouse | Deficiency hinders tolerance | 185 |
| mir-148a | Mouse | Overexpression hinders deletion | 160 |
| miR-17~92 | Mouse | Overexpression hinders deletion | 63,161 |
| Myc | Mouse | Knockdown hinders positive selection | 63 |
| MYD88 | Human | Deficiency hinders tolerance | 139 |
| Nfkb (also known as Rela) | Mouse | Inhibition hinders tolerance | 79,83 |
| Nras | Mouse | Constitutive activity drives positive selection | 186 |
| Pik3cd (encodes p110δ) | Mouse | Deficiency hinders positive selection | 187 |
| Pik3ca (encodes p110α) | Mouse | Constitutive activity drives positive selection | 60 |
| Pik3r1 (encodes p85α) | Mouse | Deficiency hinders positive selection | 68 |
| Plcg2 | Mouse | Deficiency hinders editing | 188 |
| Prkcd (encodes PKCδ) | Mouse | Deficiency hinders deletion | 104 |
| Pten | Mouse | Deficiency hinders tolerance | 61,63,160 |
| PTPN22 T | Human | Polymorphism hinders tolerance | 129 |
| Raf1 | Mouse | Constitutive activity suppresses editing | 185 |
| RAG1 | Human | Partial deficiency hinders tolerance | 134 |
| Rasgrp1 | Mouse | Deficiency hinders tolerance | 189 |
| Rasgrp1 or Rasgrp3 | Mouse | Deficiency hinders apoptosis | 104 |
| Syk | Mouse | Deficiency hinders positive selection | 190,191 |
| Syk and Zap70 | Mouse | Double deficiency hinders positive selection | 54 |
| TACI (also known as TNFRSF13B) | Human | Mutation hinders tolerance Mutation hinders tolerance | 135 |
| UNC93B | Human | Deficiency hinders tolerance | 139 |
ADA, adenosine deaminase; AICDA, activation-induced cytidine deaminase; Bcl2, B cell leukaemia/lymphoma 2; Blnk, B cell linker; BTK, Bruton’s tyrosine kinase; CIN85, CBL-interacting protein of 85 kDa; Fkbp11, FK506-binding protein 11; Gadd45a, growth arrest and DNA-damage-inducible 45 alpha; Hcls1, haematopoietic cell-specific Lyn substrate 1; IRAK4, interleukin-1 receptor-associated kinase 4; Irf4, interferon regulatory factor 4; Itpkb, inositol-1,4,5-trisphosphate 3-kinase B; mir-148a, microRNA-148a; miR-17~92, microRNA cluster 17~92; MYD88, myeloid differentiation primary response 88; Nfkb, nuclear factor kappaB; Pik3cd, phosphoinositide 3-kinase catalytic delta polypeptide; Pik3ca, phosphoinositide 3-kinase catalytic alpha polypeptide; Pik3r1, phosphoinositide 3-kinase regulatory subunit polypeptide 1; Plcg2, phospholipase C gamma 2; Prkcd, protein kinase C delta; Pten, phosphatase and tensin homologue; PTPN22 T, R620W allele of protein tyrosine phosphatase non-receptor type 22; RAG1, recombination-activating gene 1; Rasgrp1, RAS guanyl-releasing protein 1; Syk, spleen tyrosine kinase; Zap70, zeta-chain-associated protein of 70 kDa; TACI, transmembrane activator and CAML interactor
B cell development
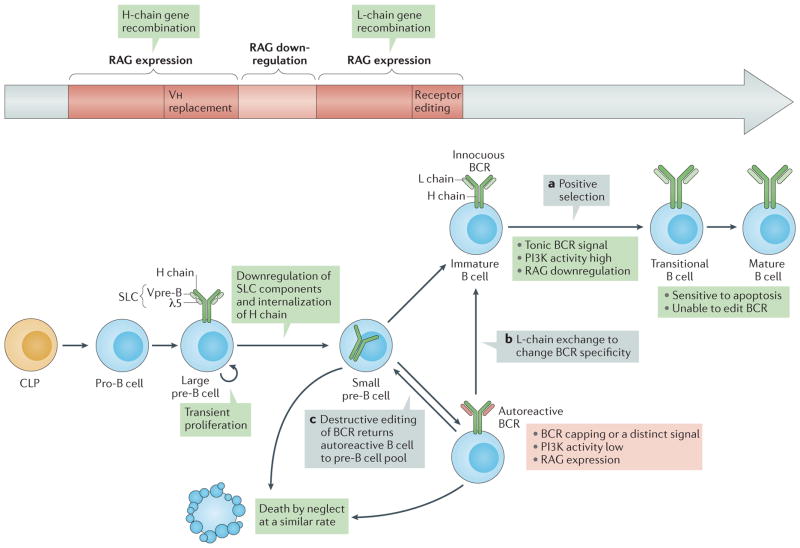
B cells are generated in the bone marrow from progenitor cells committed to the B cell lineage (known as pro-B cells). B cell development is a complex process that involves several differentiation steps (FIG. 1) controlled by a network of transcription factors interacting with environmental cues6. Each pro-B cell undergoes independent rearrangement and assembly of diverse variable (V), diversity (D) and joining (J) gene segments of the immunoglobulin heavy (H)-chain locus7 (FIG. 2). Rearrangement of the H-chain locus creates in each B cell a variable exon with a unique sequence upstream of the immunoglobulin constant (C)-region exons, and drives the expression of H-chain protein. Sensing this maturation step, the cells (now, by definition, pre-B cells (FIG. 1)) then proliferate, exit the cell cycle and differentiate to commence immunoglobulin light (L)-chain gene recombination. This involves the assembly of V elements to J elements in one of two L-chain loci, κ or λ, that provide a different downstream C region (FIG. 2). When a B cell expresses L-chain protein, it pairs with the previously rearranged H chain and is expressed as membrane immunoglobulin on the cell surface.
Figure 1. B cell development.
Pro-B cells in the bone marrow, which are derived from common lymphoid progenitor (CLP) cells, initiate heavy (H)-chain gene rearrangement through expression of recombination-activating genes (RAG1 and RAG2; collectively referred to here as RAG) and epigenetic modifications of the H-chain loci that promote accessibility6. Productive H-chain gene assembly leads to the association of IgM H-chain (μ-chain) protein with surrogate light-chain (SLC) components λ5 (also known as IGLL1) and Vpre-B, and surface expression of the pre-B cell receptor (pre-BCR) in large pre-B cells162,163. Pro-B cells can also undergo H-chain variable region (Vh) replacement reactions, whereby Vh elements recombine to a conserved heptamer within the Vh element of an already rearranged VDJ exon164. Spontaneous, antigen-independent triggering of the pre-BCR promotes progression to the large pre-B cell stage, which involves downregulation of RAG expression and transient proliferation. Differentiation to the small pre-B cell stage follows; at this stage, SLC components are downregulated, RAG is re-expressed and RAG activity is redirected to the L-chain genes163. L chains that pair with H chains trigger tonic BCR signalling, which promotes positive selection when the BCR is non-autoreactive (part a) or receptor editing (parts b and c) when the BCR is autoreactive or if tonic signalling is impaired. Editing can lead to exchange of one functional L chain for another, which can render the BCR innocuous and allow developmental progression (part b), or to secondary rearrangements that prevent L-chain expression (part c), such as out-of-frame joins that destroy the original L-chain gene but fail to replace it, which returns the cell to the pre-B cell compartment25,69. Cells that go through positive selection enter the transitional B cell stage, at which stage B cells seem to be extremely sensitive to apoptosis while losing the ability to edit the BCR97,98. Small pre-B cells and editing B cells have a similar turnover rate, which I interpret to indicate that they die ‘by neglect’ owing to prolonged insufficiency of phosphoinositide 3-kinase (PI3K)–AKT activity.
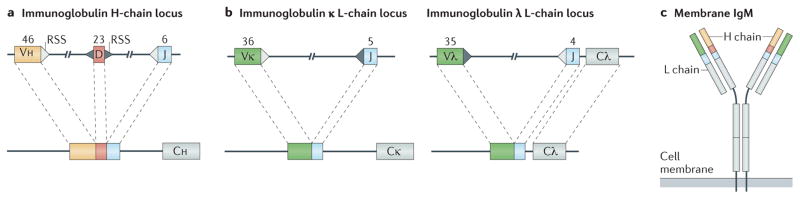
Figure 2. Antibody gene assembly by DNA recombination between gene segments.
The variable part of antibody genes is composed of variable (V), diversity (D) and joining (J) elements on the locus encoding the heavy (H) chain (part a), and V and J elements on each of two loci encoding L chains, λ and κ7 (part b). Triangles show the recombination signal sequences (RSSs) adjacent to the gene segments. Numbers above each element indicate the estimated sums of the indicated gene elements arranged along the human immunoglobulin loci165–168. In addition to the coding element shown, each V region has its own upstream promoter, leader exon and intron (not shown). Each V, D and J element can recombine as shown to generate many combinations of elements within a locus. On the H-chain locus, D-to-J rearrangements occur first, followed by V-to-DJ recombination7. The cartoon of membrane IgM protein (part c) shows the approximate placement of amino acid residues encoded by each element. Also shown is the transmembrane region of IgM and the short cytoplasmic tail, which has the sequence Lys-Val-Lys. C, constant region.
At this immature B cell developmental stage, B cells express the membrane isoform of IgM, which is a dimer of IgM H chain (μ-chain) associated with L chain (FIG. 2). Transport of IgM to the plasma membrane and antigen recognition require the formation of a complex of the membrane form of IgM with CD79A–CD79B (also known as Igα–Igβ)8. CD79A and CD79B are transmembrane proteins that carry on their cytoplasmic tails tyrosine phosphorylation sites within immunoreceptor tyrosine-based activation motifs (ITAMs), which are important docking sites in signal transduction9. This complex is the BCR (FIG. 3). On naive B cells, single BCRs are probably present in equilibrium with dimers or higher-order complexes10.
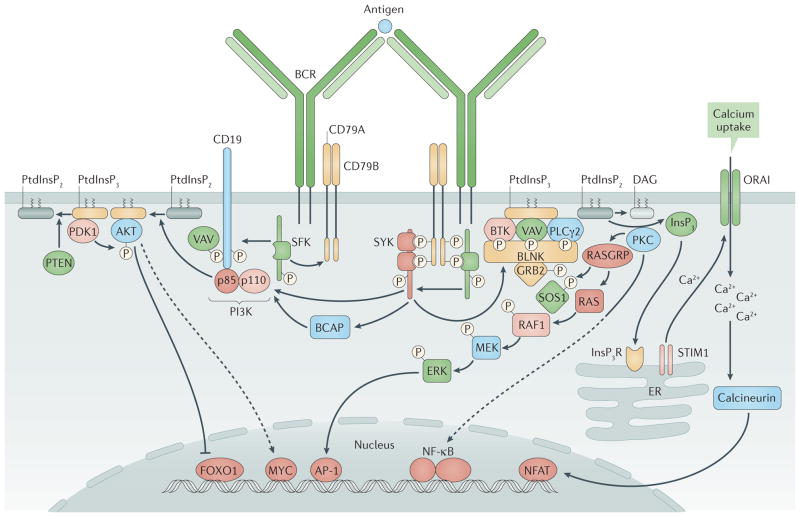
Figure 3. B cell receptor signalling.
In the assembled B cell receptor (BCR), CD79A–CD79B is weakly bound by Src family kinases (SFKs). Antigen binding promotes tyrosine phosphorylation of CD79A and CD79B on their immunoreceptor tyrosine-based activation motifs (ITAMs)9,169. Phosphorylated ITAMs recruit spleen tyrosine kinase (SYK) and upregulate its kinase activity52,170. CD19 functions as a BCR co-receptor, leading to tyrosine phosphorylation within YXXM motifs in the cytoplasmic tail of CD19 that recruit the p85 regulatory subunit of phosphoinositide 3-kinase (PI3K)53,55,171 (left). PI3K activation mediated through this pathway or by the adaptor protein B cell adaptor for PI3K (BCAP)55 mediates phosphorylation of phosphatidylinositol-4,5-bisphosphate (PtdInsP2), generating phosphatidylinositol-3,4,5-trisphosphate (PtdInsP3), which recruits AKT, 3-phosphoinositide-dependent protein kinase 1 (PDK1), Bruton’s tyrosine kinase (BTK) and other enzymes that are essential for signal propagation172. In a distinct pathway (right), B cell linker protein (BLNK)173 functions as a scaffold and substrate for SYK- and SFK-mediated phosphorylation, promoting the recruitment of phospholipase Cγ2 (PLCγ2), BTK, VAV guanine nucleotide exchange factor proteins174 and growth factor receptor-bound protein 2 (GRB2). Activated PLCγ2 hydrolyses PtdInsP2 to diacylglycerol (DAG) and inositol-1,4,5-trisphosphate (InsP3), which promotes Ca2+ mobilization through the InsP3 receptor (InsP3R) and opening of the plasma membrane Ca2+ channel ORAI. DAG recruits protein kinase C (PKC) isoforms and RAS guanyl-releasing proteins (RASGRPs). During positive selection of immature B cells, AKT activity suppresses forkhead box protein O1 (FOXO1) nuclear localization, which turns off recombination-activating gene (RAG) expression, and increases production of the transcription factor MYC, which (together with AKT) promotes cell survival. Another BCR-triggered pathway involves the guanine nucleotide exchange factor son of sevenless 1 (SOS1), the small GTPase RAS, the serine kinase RAF1, MAPK/ERK kinase (MEK) and extracellular signal-regulated kinases (ERKs). Ca2+ mobilization through InsP3R and ORAI promotes the activation of calcineurin and the nuclear localization of nuclear factor of activated T cells (NFAT). PKC activation promotes activation of nuclear factor−κB (NF−κB). PI3K and its downstream activities seem to be promoted in immature B cells by the unligated BCR, whereas BCR ligation activates the BLNK pathway transiently. This leads to BCR internalization and reduced signalling through both pathways, and drives RAG expression, developmental arrest and cell starvation. AP-1, activator protein 1; ER, endoplasmic reticulum; PTEN, phosphatase and tensin homologue; STIM1, stromal interaction molecule 1. Adapted with permission from REF. 175, F1000Research.
At this stage of immature B cell development, the cell-surface antibody can bind antigens. In the bone marrow microenvironment in which immature B cells emerge, antigens that engage the BCR will almost always be self-antigens, which makes regulation at this stage essential. Ligation of the BCR by self-antigens promotes signalling that triggers regulatory processes to reduce self-reactivity. These processes are collectively known as central tolerance, and the mechanisms by which they are achieved are the main focus of this Review.
A brief history of B cell tolerance
In 1905, Morgenroth and Ehrlich found that haemolytic antibodies could be produced by immunizing animals with red blood cells from individuals of the same species. They coined the term ‘horror autotoxicus’ to illustrate the potential of the antibody response to destroy host tissue, and they suggested that mechanisms must be in place to prevent this in healthy individuals. The phenomenon of B cell tolerance to self-antigens was first demonstrated in the 1970s and 1980s using elegant functional assays to quantify the numbers and responses of rare antigen-reactive B cells that were tracked based on their ability to secrete specific antibodies (see REF. 5 for a review of these early studies). However, these findings were met with considerable scepticism, in part because self-reactive B cells and antibodies could often be found in normal individuals11. In addition, when tolerance could be measured, it was difficult to distinguish between the loss of self-reactive cells or their functional inactivation or suppression.
Transgenic mouse technology developed some years later provided the means to more readily visualize and assess the fate of B cells in vivo12. Transgene-driven expression of immunoglobulin H chains or L chains suppresses endogenous immunoglobulin gene rearrangement and expression, so B cells with the antigen specificity encoded by the transgenes are produced in large numbers. In some models, B cells that were programmed by the transgenes to be autoreactive were functionally inactivated (rendered anergic) when they developed in the presence of autoantigen, but not in its absence4. As initially described4, anergic B cells had reduced expression of IgM on the cell surface, starting at the immature B cell stage. In other transgenic models, autoreactive B cells were eliminated from the secondary immune tissues of the spleen and lymph nodes, but were detectable in the bone marrow as immature B cells carrying low levels of surface receptor, which is consistent with the central clonal deletion of autoreactive cells occurring at the primary site of B cell generation13–16.
At the time these experiments were carried out (in the late 1980s and early 1990s), tolerance was presumed to be achieved either by a form of cell destruction or through functional inactivation5. Studies using transgenic mice expressing anti-DNA antibodies, which are associated with SLE, found that both deletion and anergy of autoreactive B cells could occur depending on the specificity or affinity of the BCR for self-antigen17,18. These findings led to the notion that the molecular presentation of the autoantigen determines the mode of tolerance, with multimeric or membrane-bound antigens promoting B cell deletion and monomeric soluble antigens leading to B cell anergy. These interpretations were strongly influenced by the success of the clonal selection hypothesis5, the central paradigm in the field, which posited that the immune system is controlled at the level of cells carrying distinct, clonally distributed antigen receptors. Notably, it was thought that cells could be regulated only through control of their growth, responsiveness, survival or death.
This view changed radically over the ensuing years when it was found that central tolerance could occur by receptor editing, a process in which ongoing L-chain gene recombination (FIG. 1) alters B cell specificity and rescues autoreactive cells from deletion through the replacement of one immunoglobulin L chain by another19–22. Such genetic reprogramming uncouples the fate of the B cell from that of its receptor and is more properly referred to as receptor selection. Antibody genes encoded by conventional transgenes (which are randomly inserted into the genome) cannot be efficiently edited because antibody gene recombination must occur in cis no more than 2–3 million base pairs away from immunoglobulin gene segments carrying recombination signals. The development of transgenic mice in which the antibody L-chain transgenes are targeted to the physiological immunoglobulin locus revealed the efficiency of receptor editing23–29. The notion of anergy as a mechanism of B cell tolerance also changed when it was shown that anergic B cells in the peripheral lymphoid organs had a reduced lifespan when in competition with non-autoreactive B cells, such that over time the anergic cells were effectively deleted30,31.
These features, and related studies, led to a developmental model of tolerance in which autoreactivity early in B cell development results primarily in receptor editing, whereas later in B cell development, autoreactivity is negatively regulated by decreasing the viability and functionality of cells21. Several additional checkpoints of various kinds have now been identified that limit B cell reactivity, survival or function at later stages of development32. As mentioned above, here I do not cover these later stages of B cell development and focus solely on recent advances in our understanding of central tolerance. However, to understand these processes, one must delve into how BCR signalling controls not just receptor editing and central tolerance but also B cell development in general.
BCR signalling in B cell quality control
V(D)J recombination is inherently random and error-prone, generating diversity, but often assembling genes that are non-productive or that encode potentially autoreactive antibodies. Moreover, although each B cell has the genetic potential to simultaneously express two different H chains and four or more different L chains (two κ and two λ L chains in humans; two κ and four λ L chains in mice), a B cell usually expresses a unique H-chain–L-chain pair (known as allelic exclusion). Maintaining a unique specificity of each B cell is thought to be important for robust recognition of antigens and for the specificity of the antibody response33. Signalling through the BCR (FIG. 3) controls B cell development to favour the expression on each B cell of a single H-chain–L-chain pair that is of limited self-reactivity. Several feedback mechanisms in pre-B cell development select for cells expressing exactly one functional H chain by sensing productive rearrangement, inhibiting further H-chain gene rearrangements and promoting progression in development34–41, thereby preventing the expression of two different H chains as well as preventing the survival of those cells lacking any H chain. Functional H-chain expression in pro-B cells is required for developmental progression, induction of transient proliferation and the subsequent redirection of V(D)J recombination to the L-chain loci. Similarly, functional H-chain–L-chain expression provides signals that are required for full B cell maturation, cessation of V(D)J recombination, migration to the peripheral lymphoid tissues and cell survival21.
When small pre-B cells first express productive immunoglobulin L-chain gene rearrangements and produce L chains that can assemble with H chains to form surface IgM, they by definition become B cells that can detect antigens for the first time. At this point in development, a B cell has three options (FIG. 1). First, it can undergo positive selection, which involves ceasing V(D)J recombination and initiating maturation and migration to the secondary lymphoid organs. Second, it can ignore the expression of its BCR if the positive selection signal is insufficient, for example if the H-chain–L-chain pairing is inefficient or its expression is too low, thus continuing ongoing L-chain rearrangements. Third, it can undergo receptor editing or apoptosis if the BCR is ligated by antigen present in the bone marrow (and hence, presumably, self-antigen). Ongoing immunoglobulin gene rearrangement can alter BCR specificity, as outlined in FIG. 4. These checkpoints provide a form of molecular quality control, preventing the development of B cells that fail to express a functional H-chain–L-chain pair and limiting the numbers of B cells that carry two different H chains or have autoreactive H-chain–L-chain combinations. These checkpoints thus rely on the ability of the BCR to signal in different ways. In its unligated form, the BCR seems to transmit a tonic signal that is important for B cell survival and developmental maturation42. The nature of the tonic signal has been best characterized in naive splenic B cells, in which the BCR can function as a signalling hub required for in vivo survival and can also facilitate signalling through Toll-like receptors (TLRs)43–46. In newly formed B cells, antigen recognition leads to distinct signals through the BCR that promote receptor editing and apoptosis21. Cells that fail to assemble any functional L chain or that inactivate their L-chain gene in the process of receptor editing lack all BCR signals and fail to mature47.
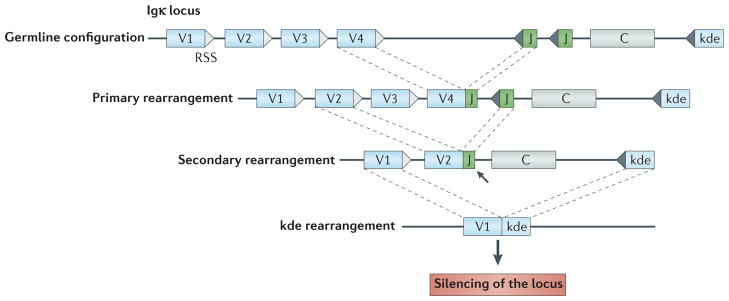
Figure 4. Recombination events associated with receptor editing on the immunoglobulin κ locus can silence or alter gene expression.
The top line of the figure shows the germline configuration of gene segments in the human immunoglobulin κ (Igκ) locus, with variable (V) and joining (J) elements flanked by recombination signal sequences (RSSs). Also shown is the κ deleting element (kde). The first rearrangement on a locus is known as the primary rearrangement. Secondary rearrangement can silence the primary rearrangement; for example, in this case, V2 rearrangement to a downstream J element replaces the V4–J join. Note that some Vκ genes rearrange by inversion rather than excision of intervening DNA (not shown). When they are in-frame, secondary rearrangements can replace one functional V region with another. However, like all immunoglobulin gene rearrangements, these rearrangements are often non-functional and so can silence κ light-chain protein expression rather than replace it. Out-of-frame primary rearrangements can similarly be displaced by secondary rearrangements. Even loci that have ‘used up’ the J elements (arrow) can silence the locus by subsequent kde recombination (bottom line). The kde has no coding function, but behaves as a non-functional, but rearrangeable, J element, leading to deletion of the constant (C) region and silencing of the locus. Such rearrangements are often found in cells expressing λ light chains. The λ locus in humans has a similar organization, although a kde-like element has not been identified.
The simplest hypothesis for how such a mechanism might work is a ‘Goldilocks’ model in which only BCR signalling of intermediate strength promotes positive selection. Small pre-B cells that lack a signal owing to the absence of cell-surface BCR persist in a default state of developmental arrest, with ongoing L-chain gene recombination in an attempt to produce a functional BCR, but having a limited lifespan of ~3 days in which to achieve this. By contrast, positive selection would require a special signal (discussed later in this section) transmitted by an adequate level of unligated BCR on the plasma membrane to promote developmental progression. Finally, newly formed, autoreactive B cells encountering antigen will at first signal excessively and then rapidly down regulate surface IgM, leading to a reduced steady-state BCR signal that mimics that of the pre-B cell and accordingly yields ongoing L-chain recombination, which in this context can change BCR specificity. In such a model, B cells carrying BCRs that are incapable of positive selection (owing to lack of surface expression or to autoreactivity) would share with pre-B cells a short lifespan of ~3 days. Within this time frame, ongoing L-chain recombination can rescue both pre-B cells and non-selected B cells by generating new BCRs that promote positive selection. Consistent with this model, underexpression of surface IgM and of many BCR signalling components promotes ongoing L-chain gene recombination and receptor editing even when the BCR itself is non-autoreactive48. Induced deletion of surface IgM on B cells at this stage similarly promotes ongoing L-chain recombination49.
Positive selection of innocuous B cells
How does the unligated BCR induce positive selection? Many components of the BCR signalling complex are preassembled before antigen stimulation50. The unligated BCR promotes phosphoinositide 3-kinase (PI3K) signalling in immature B cells through tonic tyrosine phosphorylation of the adaptor proteins CD19 and/or B cell adaptor for PI3K (BCAP) by Src family kinases (SFKs)51–53 and, possibly, SYK and ζ-chain associated protein of 70 kDa (ZAP70)54, and the subsequent recruitment of PI3K through its p85 regulatory subunit55 (FIG. 3). In splenic B cells, SYK may be specifically excluded from unligated BCRs56. It is unclear whether this is also true in immature B cells undergoing positive selection, but if so, it might provide a biochemical mechanism to distinguish ligated versus unligated BCRs. Downstream of PI3K activity, the expression of recombination-activating genes (RAG1 and RAG2; collectively referred to here as RAG) is negatively regulated by phosphatidylinositol-3,4,5-trisphosphate (PtdIns(3,4,5)P3)-mediated activation of AKT, which leads to phosphorylation of the forkhead box protein O1 (FOXO1) transcription factor and its sequestration from the nucleus57–59. This pathway also promotes B cell survival60,61. PI3K activity is opposed by phosphatase and tensin homologue (PTEN), a phosphatase that catalyses the reverse reaction and is particularly abundant in immature B cells61,62. Defects in positive selection caused by CD19 deficiency are substantially overcome by mutation of PTEN62,63. A second, possibly related, pathway implicated in positive selection involves a low level of RAS activation. Defects in positive selection caused by BCR underexpression can be compensated for by the expression of active RAS, which promotes B cell developmental progression and the upregulation of maturation markers, and indirectly promotes PI3K activity and the downregulation of RAG1 and RAG2 expression64. The RAS–extracellular signal-regulated kinase (ERK) pathway may also regulate the essential B cell transcription factor E2A (also known as TCF3)65. E2A is required for RAG expression and L-chain gene accessibility, and its effective levels are increased in pre-B cells and immature B cells6,66,67.
Negative selection and receptor editing
Next I consider the mechanisms of negative selection through which antigen encounter leads to tolerance. Consistent with the Goldilocks model, upon BCR ligation there is downregulation of cell-surface expression of the BCR (BOX 1), leading to greatly reduced PI3K signalling, nuclear localization of FOXO1, increased RAG expression and receptor editing68–70. The developmental arrest of autoreactive B cells can be reversed by editing to swap an autoreactive receptor for an innocuous one or by the withdrawal of autoantigen15,19. However, in opposition to the Goldilocks model, the ligated BCR is believed to signal in a manner that is distinct in some ways from BCR underexpression, in that the BCR-associated kinases are more strongly activated by the ligated BCR. This leads, through the activation of B cell linker protein (BLNK), to the suppression of AKT activity, which in turn releases FOXO1 to promote RAG gene expression and receptor editing59,71. (It is unclear whether immature B cell fate differs in the context of BCR ligation versus under expression, although the preferences of L-chain V-to-J rearrangements could be affected through effects on locus accessibility.) This mechanism of negative selection leading to receptor editing is analogous to the signals that promote primary L-chain gene rearrangements in pre-B cells, with that checkpoint being regulated by the pre-BCR. Because the pre-BCR spontaneously signals owing to self-aggregation and/or interactions with invariant host ligands, it is presumed to have parallels to an autoreactive BCR72. In immature B cells, BCR signalling is opposed not only by the protein and lipid phosphatase activities of enzymes such as SHP1 (also known as PTPN6) and PTEN73,74 but also by arginine methylation of CD79A. In the absence of a conserved arginine in CD79A, B cell activation is enhanced and development is impaired75. In an independent feedback loop, the RAG-mediated double-stranded DNA breaks that occur during immunoglobulin gene rearrangement activate serine-protein kinase ATM and nuclear factor-κB (NF-κB), which upregulate the transcription factor SPIC, thereby inhibiting the expression of SYK and BLNK and potentially providing a mechanism to limit the rate of L-chain rearrangement and the number of alleles accessible76,77. In chicken DT40 cells, BLNK also has a role in BCR capping78, which downregulates BCR levels and presumably also reduces tonic signalling by BCR depletion. BCR ligation also activates NF-κB signalling, which has been implicated in receptor editing by promoting the accessibility of L-chain loci to RAG1 and RAG2 through induction of interferon regulatory factor 4 (IRF4); in promoting RAG expression; and in extending the survival of cells through the induction of the serine/threonine protein kinase PIM2 (REFS 79–83). Overall, therefore, the signals that promote receptor editing in autoreactive B cells seem to be similar to those that initiate L-chain recombination in pre-B cells, but additional signals may distinguish self-antigen ligation of the BCR in immature B cells from those signals driving ongoing immunoglobulin gene recombination in pre-B cells.
Box 1. B cell receptor internalization triggered by antigen.
Internalization of the B cell receptor (BCR) in immature B cells is poorly understood. Most of what we know comes from studies of cell lines or mature B cells, in which BCR internalization is important for the delivery of antigen to endocytic vesicles for presentation in association with MHC class II molecules. Upon antigen recognition, the BCR rapidly associates with lipid rafts146, leading to clathrin phosphorylation and hence clathrin-mediated internalization147. Internalization is mediated by interactions between the endocytic clathrin adaptor AP2 and motifs in CD79A–CD79B148,149. CD79A–CD79B and Src family kinases (SFKs) facilitate internalization of the BCR, whereas spleen tyrosine kinase (SYK) seems to be dispensible150–154. Mice with a B cell-specific deficiency of the E3 ubiquitin ligases CBL and CBLB have a significant defect in internalization of cell-surface IgM and develop a lupus-like disease155. CBL and CBLB may be recruited to the BCR by the CBL-interacting protein of 85 kDa (CIN85; also known as SH3KBP1), which is constitutively associated with B cell linker protein (BLNK)156. However, CIN85-deficient mice have no defect in BCR internalization, possibly owing to the function of the related protein CD2AP157. The adaptor BAM32 (also known as DAPP1) is recruited to the membrane by SH2 domain and pleckstrin homology domain interactions, where it also contributes significantly to BCR internalization151. BAM32 internalization is associated with mitogen-activated protein kinase activation151,158, whereas CIN85 internalization is associated with nuclear factor-κB (NF-κB) activation157. Despite these connections between BCR activation, signalling and internalization, BCRs whose immunoreceptor tyrosine-based activation motifs (ITAMs) are phosphorylated seem to be preferentially retained on the cell surface, whereas internalized BCRs lack phosphorylation159. Thus, much more research needs to be carried out to understand how BCR internalization occurs biochemically and how it fits into the overall picture of central tolerance.
Implications and limitations of editing
Receptor editing seems to occur in a significant fraction of developing B cells29,84,85, but the proportion of B cells that edit as a result of self-reactivity, as opposed to inefficient positive selection, is difficult to measure. One study estimated that ~20% of immature B cells have downregulated expression of the BCR and are actively undergoing tolerance-induced editing69.
Receptor editing is directional, as ongoing L-chain gene recombination is not entirely random. Rearrangements in each B cell tend to progress sequentially over time from upstream to downstream Jκ elements on the κ L-chain loci, before continuing on λ L-chain loci22,23,86–88 (reviewed in REF. 21). There is probably also a progression from efficiently rearranged genes to less efficiently rearranged genes89. Thus, receptor editing contributes to the diversification of the antibody repertoire and promotes λ L-chain usage. λ L chains may have a specialized role in editing because of their distinct physiochemical properties, including in the complementarity-determining region (CDR)–L3 loop formed by V–J joining90, and in some studies λ chains are particularly effective at quenching autoreactivity19,91,92. Editing also seems to take place on both alleles (and on both κ and λ L chains), and so can sometimes lead to the co-expression of two L chains and the escape of partly self-reactive B cells from the bone marrow48,86,93.
Apoptosis in central deletion
Several lines of evidence have highlighted the efficiency of receptor editing in central tolerance21,26, which allows several attempts at editing to occur in a limited time. B cells that are experimentally prevented from editing or that have H chains with features that cannot be efficiently corrected by exchanging L chains eventually undergo an apoptotic process13,16,69,94–96 (FIG. 1). Apoptosis in this population of B cells and in the newly formed B cells that transition from bone marrow to the spleen is thought to be largely driven by the proapoptotic protein BIM and can be inhibited by enforced expression of its anti-apoptotic binding partners, such as B cell leukaemia/lymphoma 2 (BCL-2)97–100. Enforced BCL-2 expression facilitates receptor editing, which is consistent with the notion that receptor editing is limited by BIM-dependent cell death101. BIM expression is driven by FOXO family proteins102; in B cells, this probably involves FOXO1 together with E2A6, and so BIM expression is presumably high in pre-B cells and editing B cells and is decreased by PI3K-mediated activation of AKT in positively selected B cells.
As newly formed B cells are more sensitive to antigen-induced apoptosis than are mature B cells5, it has been of interest to understand the differences in their signalling biochemistry. BCR ligation in immature bone marrow B cells leads to robust tyrosine phosphorylation and Ca2+ mobilization, despite reduced steady-state levels of inositol-1,4,5-trisphosphate formation, compared with more mature B cell stages103. These differences are probably the result of distinct levels of various BCR-coupled signalling proteins103,104 and they correlate with the increased sensitivity to apoptosis of immature B cells.
Similar to control by receptor editing, the apoptotic control of central tolerance is incomplete. Not all self-antigens are accessible in native form to B cells in the bone marrow. Antigens that are normally expressed in peripheral tissues might conceivably be presented to bone marrow B cells by antigen transport or ectopic expression, for example, although there is little evidence to support this. Some types of polyreactive B cell clearly fail to induce receptor editing or deletion, such as the major population of B1 cells in mice that are reactive to phosphatidylcholine105. It is unclear how B cells with such specificities avoid negative selection, but presumably the low affinities of their BCRs for antigen, or the low accessibility or epitope density of their ligands, have a role. B cells recognizing certain self-antigens may engage activating or inhibitory co-receptors on B cells, such as Siglecs or complement receptors, which could potentially up- or down-modulate central tolerance to certain classes of antigens2,106.
Superantigen screens for tolerance genes
Many mouse studies have identified defects in B cell central tolerance, in terms of either receptor editing or clonal deletion, in spontaneous models of SLE107,108, Sjogren syndrome109 and type 1 diabetes110, although there is as yet no real consensus as to the mechanisms involved. As editing can both destroy and create certain autoantibody specificities, insufficient as well as excessive editing may be deleterious111,112. Defects in apoptosis may also have a role. To assess the factors involved in central tolerance more systematically, investigators have begun screening for mutations that promote the escape of B cells from tolerance. One way in which this has been approached is to set up an in vivo selection system in which all normal B cells are negatively selected owing to engineered self-reactivity, such that tolerance leads to the loss of these B cells. As mentioned above, transgenic mice carrying autoreactive BCRs or superantigens reactive to the BCR are useful tools with which to study B cell tolerance in vivo. In IgM macroself mice, which carry a superantigen designed to bind to the IgM C region, central tolerance without the option of escape by receptor editing leads to a developmental block at the immature B cell stage69. This powerful example of in vivo negative selection facilitates testing of the effects of mutations or gene overexpression in B cells on abro-gating central deletion. This approach has been used successfully to identify microRNAs (miRNAs), the overexpression of which hinders tolerance induction, which in turn facilitates the identification of miRNA target genes that control tolerance (BOX 2).
Box 2. The role of miRNAs in B cell central tolerance.
Recent studies have investigated the role of microRNAs (miRNAs) in the regulation of B cell tolerance. Remarkably, in IgM macroself mice, B cell-restricted overexpression of either the miRNA cluster miR-17~92 or the individual miRNA miR-148a led to almost complete escape of B cells from central tolerance160,161. miR-17~92 is a cluster of six miRNAs that includes miR-19a and miR-19b-1, which share the same seed region and mRNA targets. These two miRNAs seem to promote the breach in B cell tolerance by downregulating expression of phosphatase and tensin homologue (PTEN)161. It was independently found that miR-17~92 overexpression could substantially overcome the developmental defects of CD19-deficient immature B cells63, which suggests that a threshold exists at which phosphoinositide 3-kinase (PI3K) activity promotes expression of the transcription factor MYC, which drives miR-17~92 expression and in turn downregulates PTEN expression, thus tipping the balance of B cell signalling to promote survival and positive selection. The effect of miR-148a on B cell tolerance was independently identified in a functional screen of a retroviral library of 113 miRNAs by transduction of haematopoietic stem cells followed by transfer into IgM macroself mice160. Of these 113 miRNAs, miR-148a led to the most potent escape from central tolerance. Identified targets of miR-148a included growth arrest and DNA-damage-inducible 45 alpha (Gadd45a), Bim and Pten, each of which was found to inhibit B cell deletion in the bone marrow when knocked out. Moreover, tolerance was less efficient in B cells with heterozygous deficiency of each one of these genes alone, indicating that their protein products make a quantitative contribution to tolerance that is probably under tight control. The precise mechanism by which GADD45α regulates tolerance is unclear, but it is believed to promote apoptosis by facilitating the localization of BIM to mitochondria160. Interestingly, GADD45α, like the recombination-activating genes (RAGs) and BIM itself, is a target of forkhead box protein O (FOXO) transcription factors102. These studies show that it may be possible to define a key set of genes that regulate immune tolerance. Using similar techniques in different cell types at various stages of their development, it might be possible to map and organize the genes into pathways with the potential to provide insight into tolerance in health and disease. TABLE 1 shows some of the genes that are implicated in B cell central tolerance in various studies of gene overexpression or loss of function in humans and mice.
Humanized mice
Most of the studies of normal immune tolerance discussed so far have been carried out in mice. It is important for our medical understanding in this regard to verify that human cells behave similarly to mouse cells (notwithstanding that most scientists do not doubt that this is the case). The heterogeneity of human genetics and environment presents a major challenge, which has inspired some ingenious approaches, such as the study of humanized mice. A recent study has combined the use of humanized mice with the macro self antigen approach to challenge human immune cells experimentally. Central tolerance of human B cells was thus studied directly in vivo using severe combined immunodeficient (SCID) mice expressing a super antigen reactive to human κ L chains113 that were reconstituted at birth with human haematopoietic stem cells. Negative selection was documented in terms of markedly reduced numbers of B cells expressing κ L chain in the spleen, whereas numbers of B cells expressing λ L chain were increased. The developing B cells of the bone marrow showed signs of receptor editing, including BCR downregulation, increased RAG gene expression and increased secondary L-chain gene recombination events, such as the use of downstream Jκ gene segments and L-chain λ-locus excision circles. This study not only showed that developing human B cells use receptor editing as a mechanism of B cell central tolerance but also suggested that among normal human donors there is genetically determined variability in the efficiency of tolerance induction. Defects in B cell central tolerance have been associated with autoimmune disease in mice and humans, including rheumatoid arthritis114, SLE107,108,111,115,116 and type 1 diabetes110,114. The exciting possibility of this type of experimental system is that it may allow for the identification, under controlled conditions, of fundamental differences in central tolerance between normal and autoimmune donors.
Central tolerance in human B cells
Single-cell antibody cloning
In another fruitful approach to studying B cell tolerance in humans, several recent studies have quantified autoreactive B cells in normal and diseased individuals to detect the elimination of self-reactive B cells in the healthy state or the breaching of tolerance checkpoints in disease. Serological analysis has established that certain autoantibody profiles are diagnostic for, and sometimes predictive of, diseases such as SLE, rheumatoid arthritis and type 1 diabetes117. For many years, researchers have attempted to identify and enumerate autoreactive B cells using techniques such as Epstein–Barr virus transformation118, tracking the usage of V genes that are biased to be autoreactive119, and flow cytometry or cloning approaches120,121. However, most recent studies have used the approach of cloning and expressing antibody genes obtained from single human B cells at different stages of their development.
This robust protocol involves the sorting of single B cells, cloning of their individual antibody H-chain and L-chain cDNAs, expression of the antibody in mammalian cells, and the analysis of antibody sequence and reactivity85. Self-reactivity is assessed in this method by antibody binding to lysate from human epithelial type 2 (HEp-2) cells that is adhered to microwell plates. Polyreactivity is defined as the ability of a particular antibody, in a particular concentration range, to bind to more than one of the following ligands: DNA, insulin or lipopolysaccharide. In some cases, antibodies are tested by immunofluorescence assay for antinuclear specificity. Although the number of B cells that can be analysed using the cloning and expression approach is limited, the frequency of random B cells that score positive for autoreactivity is surprisingly high. The frequency of autoreactive B cells starts at 50–80% in early-stage B cells of the bone marrow, then gradually decreases with developmental progression to ~10% in mature naive B cells of normal adults114,122. The loss of autoreactive B cells with increasing developmental maturity has been ascribed to central tolerance, whereas increased levels of reactivity have been associated with defective negative selection, either by reduced receptor editing or impaired apoptosis.
Use of this assay, primarily by the laboratory of Eric Meffre, has identified significant differences between healthy and immunologically diseased humans in a remarkable number of contexts114. When used to assess small samples of the immune repertoire in human autoimmune disease, statistically significant increases in the numbers of polyreactive or autoreactive B cells were observed in patients with SLE116, rheumatoid arthritis123, type 1 diabetes124, Sjogren syndrome125 and multiple sclerosis126, which is consistent with the possibility that these diseases are caused in part by defects in B cell central tolerance. However, the autoreactive antibodies that are detected using this assay are usually non-mutated and hence are unlike the affinity-matured, often highly specific, autoantibodies that are characteristic of autoimmune disease, such as those that bind DNA, chromatin or small nuclear ribonucleoproteins127. Polyreactive B cells of certain specificities are believed to be beneficial, such as those reactive to bacteria or apoptotic cells, whereas others may be potentially pathogenic11. It is currently unclear if the autoantibodies that are diagnostic of autoimmune disease are derived from non-autoreactive cells that subsequently mutate during a T cell-dependent response128, or are selected from the relatively rare escape or recruitment of high-affinity precursors. In any case, the correlations between the disease states mentioned above and defects in B cell central tolerance are striking.
One candidate gene contributing to increased levels of pre-immune autoreactive or polyreactive B cells in humans is the R620W variant of protein tyro sine phosphatase non-receptor type 22 (PTPN22)129 (also known as the PTPN22 T allele), which is a risk factor for multiple autoimmune diseases130. This PTPN22 variant affects antigen receptor signalling130, which suggests that intrinsic alterations in B cell signalling thresholds may lead to defects in central tolerance. Consistent with this possibility, mice carrying a mutation in Ptpn22 (R619W) analogous to the disease-associated human PTPN22 allele (R620W) had a B cell-autonomous defect that could promote autoimmunity and was associated with the protection of immature B cells from apoptosis131. Moreover, in mice engrafted with human haematopoietic stem cells, intrinsic expression of the R620W allele was sufficient to impair B cell central tolerance132.
Tolerance defects in immunodeficiency
The human antibody cloning assay has similarly revealed defects in the removal of autoreactive B cells in primary immunodeficiency diseases114, such as in patients with X-linked agammaglobulinaemia (XLA) (caused by deficiency in Bruton’s tyrosine kinase (BTK)), with deficiencies in interleukin-1 receptor-associated kinase 4 (IRAK4), myeloid differentiation primary response protein 88 (MYD88), UNC93B and adenosine deaminase133, and even with a hypomorphic mutation in RAG1, which is required for antigen receptor gene assembly134. Many patients with common variable immunodeficiency (CVID) seem to have dysregulated B cell central tolerance owing to null or missense mutations in the gene encoding trans membrane activator and CAML interactor (TACI; also known as TNFRSF13B), which is a receptor for the B cell survival cytokines APRIL (a proliferation-inducing ligand) and BAFF (B cell-activating factor), but which also interacts with TLR7 and TLR9 (REF. 135), which are nucleic acid sensors that induce an inflammatory response.
Several possibilities have been suggested to explain these findings. In the case of mutations that impair BCR signalling, B cell negative selection through editing or central clonal deletion may simply be incomplete. Alternatively or in addition, B cells with impaired BCR signalling and weakly autoreactive antigen receptors might not only be permitted to survive by escaping negative selection, but also be preferentially selected, as was demonstrated in the case of CD45-deficient mice136. Such effects may also explain the high levels of autoantibodies in patients with CVID who lack the B cell-restricted signalling molecule CD19 (REF. 137).
Indirect effects may also have a role in regulating B cell tolerance. For example, B cell lymphopaenia often leads to increased levels of the B cell survival factor BAFF as an indirect result of its reduced consumption114. Increased BAFF levels have been shown in mice to promote the survival and activation of autoreactive B cells even in the absence of T cells138. Altered microbial burden and attendant inflammatory processes might also have a role in altering B cell tolerance thresholds through changes to the level of BAFF or other cytokines. It is unclear how deficiencies in IRAK4, MYD88 or UNC93B hinder B cell tolerance as these proteins are involved in TLR signalling in B cells and other cells, and also have other roles in innate immunity and inflammatory cytokine responses139. However, studies from mice have implicated TLR9 signalling in promoting the death of B cells with anti-DNA specificity140. Perhaps most remarkably, activation-induced cytidine deaminase (AICDA; also known as AID) — which is required for somatic mutation and class-switch recombination in mature B cell responses — has been implicated in promoting B cell central tolerance141,142. Because humans who retain mutational activity while being deficient in H-chain class switching (owing to carboxy-terminal AICDA truncation, CD40 ligand (CD40L) deficiency or the lack of UNG) lack comparable defects in tolerance, these data support the counterintuitive concept that the point mutagenesis activity of AICDA in developing B cells might have a role in promoting the removal of autoreactive cells141. It has been suggested that DNA lesions caused by AICDA activity promote cell death through a p53-dependent pathway141.
Central tolerance as a barrier to immunity
In addition to limiting self-reactivity, B cell tolerance might also hinder immune reactions to foreign antigens to which reactivity might be desirable, such as vaccines. This possibility has been raised in connection with the difficulty in neutralizing HIV, as some of the rare, highly mutated broadly neutralizing antibodies for HIV that have been identified from patients so far also have reactivity to self-antigens143. The implication of this finding is that some conserved viral epitopes might be selected to mimic host antigens as a way to hinder the antibody response. Indeed, self-antigens that cross-react with the broadly neutralizing HIV gp41-reactive antibodies 2F5 and 4E10 have recently been identified; studies of immunoglobulin knock-in mice carrying these antibody specificities showed that the B cells were eliminated by central tolerance (reviewed in REF. 144). It is unclear whether the ability of some patients infected with HIV to produce antibodies such as 2F5 and 4E10 indicates that there is relaxed immune tolerance during chronic HIV infection or if it represents an autoimmune-prone feature of the rare donors from which these antibodies arise. Transiently relieving central tolerance in the context of vaccination might be a useful approach to raising antibodies in animals or perhaps in anticancer therapy, but given the current state of our knowledge regarding the mechanisms of central tolerance, together with the strict safety standards applicable to any product for use in otherwise healthy individuals, it is unlikely that such an approach would be feasible for use in human vaccines.
Future directions
Defects in central tolerance are clearly associated with autoimmune disease, although many of the details need to be better understood to assess definitively the extent to which such defects contribute to disease and to define the extent to which they are B cell intrinsic. The immunodeficiency diseases that are associated with dysregulation of central tolerance provide a new set of puzzles to be solved mechanistically, as many of the findings are surprising and often counterintuitive. The immune tolerance field is now entering the genomic era, in which the challenge before us is to understand all of the molecular components involved, starting with key genes whose absence promotes a breach of tolerance, and to verify their functions in signalling networks in human cells. The scale of future studies involving single-cell sequencing of the expressed antibody genes from human B cells is likely to expand markedly with advances in technology, such as droplet-based methods to enable high-throughput paired H-chain–L-chain gene sequencing145, with further potential to provide insights into the mechanisms of tolerance escape and B cell maturation in human disease. Much still needs to be learned about BCR signalling and how it can be manipulated therapeutically. This field is expanding rapidly as its importance is becoming clearer in the understanding and treatment of B cell neoplasms. Finally, it seems possible that a fuller understanding of B cell central tolerance might allow manipulations to transiently relax immune tolerance in situations in which it might be advantageous, such as in vaccine protocols for cancer or the prevention of infectious diseases.
Acknowledgments
The author thanks current and former members of his laboratory, The Scripps Research Institute Department of Immunology and Microbial Science, and the following funding sources: US Department of Health & Human Services, National Institutes of Health (NIH) grants R37AI059714, R01AI128836 and R01AI073148.
Glossary
- CD4+ T follicular helper (TFH) cells
CD4+ T cells that are specialized to interact with B cells to promote and maintain the germinal centre reaction.
- Systemic lupus erythematosus (SLE)
A prototypical autoimmune disease characterized by B cell hyperactivity, autoantibody production and a range of symptoms that can include vasculitis, arthritis, glomerulonephritis, fever and neurological abnormalities.
- Anergy
Functional impairment of B or T cells resulting from prolonged signalling, usually caused by self-recognition.
- Receptor editing
A process whereby ongoing antibody gene recombination promotes a change in the specificity or expression of the antigen receptor of a B cell; receptor editing is usually associated with central tolerance.
- Immunoreceptor tyrosine-based activation motifs (ITAMs)
Conserved motifs (YXXL/IX(6–8)YXXL/I) on the cytoplasmic tails of CD79A–CD79B, CD3 proteins and other immune system proteins that become phosphorylated upon receptor ligation and recruit SYK and ZAP70.
- Allelic exclusion
The phenomenon of expression of only one of two alleles despite the genetic potential to express both. In normal B cells, expression of both alleles of the heavy chain occurs at a frequency of ~0.0001%, and the frequency of light-chain double expression is ~1%.
- Positive selection
In the context of B cell development, this refers to the process whereby immature B cells progress from a programme of active light chain gene recombination to a more mature phenotype, which is associated with the cessation of recombination and migration from bone marrow.
- Tonic signal
The weak signal emanating from a receptor in the absence of its ligand.
- Recombination-activating genes
RAG1 and RAG2 have non-redundant roles in initiating V(D)J recombination at recombination signal sequences through their DNA binding and nuclease activities.
- Pre-BCR (Pre-B cell receptor)
The complex formed when an immunoglobulin heavy chain pairs with the surrogate light-chain components λ5 and Vpre-B, allowing surface expression of a complex including CD79A–CD79B and other signalling components.
- BCR capping (B cell receptor capping)
The process whereby antigen receptors on B cells redistribute and aggregate on the plasma membrane after antigen recognition.
- IgM macroself mice
Transgenic mice expressing a custom-designed superantigen reactive to the constant region of IgM, derived from a fusion protein of single-chain antibody fused to a part of the IgG extracellular region and transmembrane and cytoplasmic regions of an MHC class I molecule.
- MicroRNAs (mi RNAs)
Short (~22 bp) RNAs that function by downregulating mRNA translation or stability and that have important roles in many cellular pathways.
- Humanized mice
Severe combined immunodeficient (SCID) mice transplanted with haematopoietic cells from humans. These mice are useful for carrying out experiments on human immune cells in vivo.
- λ-Locus excision circles
Intervening DNA removed and religated to itself by V-to-J rearrangement in the immunoglobulin λ locus.
Footnotes
Competing interests statement
The author declares no competing interests.
References
- 1.Duong BH, et al. Decoration of T-independent antigen with ligands for CD22 and Siglec-G can suppress immunity and induce B cell tolerance in vivo. J Exp Med. 2010;207:173–174. doi: 10.1084/jem.20091873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Macauley MS, et al. Antigenic liposomes displaying CD22 ligands induce antigen-specific B cell apoptosis. J Clin Invest. 2013;123:3074–3083. doi: 10.1172/JCI69187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sabouri Z, et al. Redemption of autoantibodies on anergic B cells by variable-region glycosylation and mutation away from self-reactivity. Proc Natl Acad Sci USA. 2014;111:E2567–E2575. doi: 10.1073/pnas.1406974111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Goodnow CC, et al. Altered immunoglobulin expression and functional silencing of self-reactive B lymphocytes in transgenic mice. Nature. 1988;334:676–682. doi: 10.1038/334676a0. A monumental paper that uses immunoglobulin transgenic mice to show that clonal anergy is a mechanism of B cell tolerance. [DOI] [PubMed] [Google Scholar]
- 5.Klinman NR. The “clonal selection hypothesis” and current concepts of B cell tolerance. Immunity. 1996;5:189–195. doi: 10.1016/s1074-7613(00)80314-3. [DOI] [PubMed] [Google Scholar]
- 6.Lin YC, et al. A global network of transcription factors, involving E2A, EBF1 and Foxo1, that orchestrates B cell fate. Nat Immunol. 2010;11:635–643. doi: 10.1038/ni.1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tonegawa S. Somatic generation of antibody diversity. Nature. 1983;302:575–581. doi: 10.1038/302575a0. [DOI] [PubMed] [Google Scholar]
- 8.Hombach J, Tsubata T, Leclercq L, Stappert H, Reth M. Molecular components of the B-cell antigen receptor complex of the IgM class. Nature. 1990;343:760–762. doi: 10.1038/343760a0. [DOI] [PubMed] [Google Scholar]
- 9.Reth M. Antigen receptors on B lymphocytes. Annu Rev Immunol. 1992;10:97–121. doi: 10.1146/annurev.iy.10.040192.000525. [DOI] [PubMed] [Google Scholar]
- 10.Yang J, Reth M. Oligomeric organization of the B-cell antigen receptor on resting cells. Nature. 2010;467:465–469. doi: 10.1038/nature09357. [DOI] [PubMed] [Google Scholar]
- 11.Gunti S, Notkins AL. Polyreactive antibodies: function and quantification. J Infect Dis. 2015;212(Suppl 1):S42–S46. doi: 10.1093/infdis/jiu512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Storb U. Transgenic mice with immunoglobulin genes. Annu Rev Immunol. 1987;5:151–174. doi: 10.1146/annurev.iy.05.040187.001055. [DOI] [PubMed] [Google Scholar]
- 13.Nemazee DA, Burki K. Clonal deletion of B lymphocytes in a transgenic mouse bearing anti-MHC class I antibody genes. Nature. 1989;337:562–566. doi: 10.1038/337562a0. The first evidence for clonal deletion in B cell central tolerance. [DOI] [PubMed] [Google Scholar]
- 14.Nemazee D, Buerki K. Clonal deletion of autoreactive B lymphocytes in bone marrow chimeras. Proc Natl Acad Sci USA. 1989;86:8039–8043. doi: 10.1073/pnas.86.20.8039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hartley SB, et al. Elimination from peripheral lymphoid tissues of self-reactive B lymphocytes recognizing membrane-bound antigens. Nature. 1991;353:765–769. doi: 10.1038/353765a0. [DOI] [PubMed] [Google Scholar]
- 16.Hartley SB, et al. Elimination of self-reactive B lymphocytes proceeds in two stages: arrested development and cell death. Cell. 1993;72:325–335. doi: 10.1016/0092-8674(93)90111-3. [DOI] [PubMed] [Google Scholar]
- 17.Erikson J, et al. Expression of anti-DNA immunoglobulin transgenes in non-autoimmune mice. Nature. 1991;349:331–334. doi: 10.1038/349331a0. The first evidence for negative regulation of B cells reactive to antigens that are relevant to autoimmune disease. [DOI] [PubMed] [Google Scholar]
- 18.Fields ML, Erikson J. The regulation of lupus-associated autoantibodies: immunoglobulin transgenic models. Curr Opin Immunol. 2003;15:709–717. doi: 10.1016/j.coi.2003.09.016. [DOI] [PubMed] [Google Scholar]
- 19.Tiegs SL, Russell DM, Nemazee D. Receptor editing in self-reactive bone marrow B cells. J Exp Med. 1993;177:1009–1020. doi: 10.1084/jem.177.4.1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gay D, Saunders T, Camper S, Weigert M. Receptor editing: an approach by autoreactive B cells to escape tolerance. J Exp Med. 1993;177:999–1008. doi: 10.1084/jem.177.4.999. References 19 and 20 provide the first evidence for receptor editing in B cells. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nemazee D. Receptor editing in lymphocyte development and central tolerance. Nat Rev Immunol. 2006;6:728–740. doi: 10.1038/nri1939. [DOI] [PubMed] [Google Scholar]
- 22.Radic MZ, Erikson J, Litwin S, Weigert M. B lymphocytes may escape tolerance by revising their antigen receptors. J Exp Med. 1993;177:1165–1173. doi: 10.1084/jem.177.4.1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Luning Prak E, Weigert M. Light chain replacement: a new model for antibody gene rearrangement. J Exp Med. 1995;182:541–548. doi: 10.1084/jem.182.2.541. The first targeted transgenic model for immunoglobulin L chain shows that secondary rearrangements can edit a functional L chain gene in the physiological context. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen C, Prak EL, Weigert M. Editing disease-associated autoantibodies. Immunity. 1997;6:97–105. doi: 10.1016/s1074-7613(00)80673-1. [DOI] [PubMed] [Google Scholar]
- 25.Pelanda R, et al. Receptor editing in a transgenic mouse model: site, efficiency, and role in B cell tolerance and antibody diversification. Immunity. 1997;7:765–775. doi: 10.1016/s1074-7613(00)80395-7. [DOI] [PubMed] [Google Scholar]
- 26.Halverson R, Torres RM, Pelanda R. Receptor editing is the main mechanism of B cell tolerance toward membrane antigens. Nat Immunol. 2004;5:645–650. doi: 10.1038/ni1076. This paper provides strong evidence for the remarkable efficiency of receptor editing in rescuing autoreactive B cells. [DOI] [PubMed] [Google Scholar]
- 27.Pewzner-Jung Y, et al. B cell deletion, anergy, and receptor editing in “knock in” mice targeted with a germline-encoded or somatically mutated anti-DNA heavy chain. J Immunol. 1998;161:4634–4645. [PubMed] [Google Scholar]
- 28.Hippen KL, et al. In vivo assessment of the relative contributions of deletion, anergy, and editing to B cell self-tolerance. J Immunol. 2005;175:909–916. doi: 10.4049/jimmunol.175.2.909. [DOI] [PubMed] [Google Scholar]
- 29.Casellas R, et al. Contribution of receptor editing to the antibody repertoire. Science. 2001;291:1541–1544. doi: 10.1126/science.1056600. [DOI] [PubMed] [Google Scholar]
- 30.Fulcher DA, Basten A. Reduced life span of anergic self-reactive B cells in a double-transgenic model. J Exp Med. 1994;179:125–134. doi: 10.1084/jem.179.1.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cyster JG, Hartley SB, Goodnow CC. Competition for follicular niches excludes self-reactive cells from the recirculating B-cell repertoire. Nature. 1994;371:389–395. doi: 10.1038/371389a0. [DOI] [PubMed] [Google Scholar]
- 32.Goodnow CC, Vinuesa CG, Randall KL, Mackay F, Brink R. Control systems and decision making for antibody production. Nat Immunol. 2010;11:681–688. doi: 10.1038/ni.1900. [DOI] [PubMed] [Google Scholar]
- 33.Schlissel M. Allelic exclusion of immunoglobulin gene rearrangement and expression: why and how? Semin Immunol. 2002;14:207–212. doi: 10.1016/s1044-5323(02)00044-1. [DOI] [PubMed] [Google Scholar]
- 34.Nussenzweig MC, et al. Allelic exclusion in transgenic mice that express the membrane form of immunoglobulin mu. Science. 1987;236:816–819. doi: 10.1126/science.3107126. [DOI] [PubMed] [Google Scholar]
- 35.Barreto V, Cumano A. Frequency and characterization of phenotypic Ig heavy chain allelically included IgM-expressing B cells in mice. J Immunol. 2000;164:893–899. doi: 10.4049/jimmunol.164.2.893. [DOI] [PubMed] [Google Scholar]
- 36.Lutz J, et al. Pro-B cells sense productive immunoglobulin heavy chain rearrangement irrespective of polypeptide production. Proc Natl Acad Sci USA. 2011;108:10644–10649. doi: 10.1073/pnas.1019224108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hewitt SL, et al. RAG-1 and ATM coordinate monoallelic recombination and nuclear positioning of immunoglobulin loci. Nat Immunol. 2009;10:655–664. doi: 10.1038/ni.1735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Herzog S, Reth M, Jumaa H. Regulation of B-cell proliferation and differentiation by pre-B-cell receptor signalling. Nat Rev Immunol. 2009;9:195–205. doi: 10.1038/nri2491. [DOI] [PubMed] [Google Scholar]
- 39.Alt FW, et al. Ordered rearrangement of immunoglobulin heavy chain variable region segments. EMBO J. 1984;3:1209–1219. doi: 10.1002/j.1460-2075.1984.tb01955.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Loffert D, Ehlich A, Muller W, Rajewsky K. Surrogate light chain expression is required to establish immunoglobulin heavy chain allelic exclusion during early B cell development. Immunity. 1996;4:133–144. doi: 10.1016/s1074-7613(00)80678-0. [DOI] [PubMed] [Google Scholar]
- 41.Shimizu T, Mundt C, Licence S, Melchers F, Martensson IL. VpreB1/VpreB2/lambda 5 triple-deficient mice show impaired B cell development but functional allelic exclusion of the IgH locus. J Immunol. 2002;168:6286–6293. doi: 10.4049/jimmunol.168.12.6286. [DOI] [PubMed] [Google Scholar]
- 42.Monroe JG. ITAM-mediated tonic signalling through pre-BCR and BCR complexes. Nat Rev Immunol. 2006;6:283–294. doi: 10.1038/nri1808. [DOI] [PubMed] [Google Scholar]
- 43.Lam KP, Kuhn R, Rajewsky K. In vivo ablation of surface immunoglobulin on mature B cells by inducible gene targeting results in rapid cell death. Cell. 1997;90:1073–1083. doi: 10.1016/s0092-8674(00)80373-6. [DOI] [PubMed] [Google Scholar]
- 44.Kraus M, et al. Interference with immunoglobulin (Ig) α immunoreceptor tyrosine-based activation motif (ITAM) phosphorylation modulates or blocks B cell development, depending on the availability of an Igβ cytoplasmic tail. J Exp Med. 2001;194:455–469. doi: 10.1084/jem.194.4.455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Otipoby KL, et al. The B-cell antigen receptor integrates adaptive and innate immune signals. Proc Natl Acad Sci USA. 2015;112:12145–12150. doi: 10.1073/pnas.1516428112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schweighoffer E, et al. The BAFF receptor transduces survival signals by co-opting the B cell receptor signaling pathway. Immunity. 2013;38:475–488. doi: 10.1016/j.immuni.2012.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zou X, et al. Block in development at the pre-B-II to immature B cell stage in mice without Igκ and Igλ light chain. J Immunol. 2003;170:1354–1361. doi: 10.4049/jimmunol.170.3.1354. [DOI] [PubMed] [Google Scholar]
- 48.Pelanda R. Dual immunoglobulin light chain B cells: Trojan horses of autoimmunity? Curr Opin Immunol. 2014;27:53–59. doi: 10.1016/j.coi.2014.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tze LE, et al. Basal immunoglobulin signaling actively maintains developmental stage in immature B cells. PLoS Biol. 2005;3:e82. doi: 10.1371/journal.pbio.0030082. This paper shows that loss of BCR expression in immature B cells leads to RAG expression and L-chain gene rearrangement, which supports a role for tonic signaling in B cell maturation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang Y, Wienands J, Zurn C, Reth M. Induction of the antigen receptor expression on B lymphocytes results in rapid competence for signaling of SLP-65 and Syk. EMBO J. 1998;17:7304–7310. doi: 10.1093/emboj/17.24.7304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Saijo K, et al. Essential role of Src-family protein tyrosine kinases in NF-κB activation during B cell development. Nat Immunol. 2003;4:274–279. doi: 10.1038/ni893. [DOI] [PubMed] [Google Scholar]
- 52.Mukherjee S, et al. Monovalent and multivalent ligation of the B cell receptor exhibit differential dependence upon Syk and Src family kinases. Sci Signal. 2013;6:ra1. doi: 10.1126/scisignal.2003220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fujimoto M, et al. CD19 regulates Src family protein tyrosine kinase activation in B lymphocytes through processive amplification. Immunity. 2000;13:47–57. doi: 10.1016/s1074-7613(00)00007-8. [DOI] [PubMed] [Google Scholar]
- 54.Schweighoffer E, Vanes L, Mathiot A, Nakamura T, Tybulewicz VL. Unexpected requirement for ZAP-70 in pre-B cell development and allelic exclusion. Immunity. 2003;18:523–533. doi: 10.1016/s1074-7613(03)00082-7. [DOI] [PubMed] [Google Scholar]
- 55.Aiba Y, Kameyama M, Yamazaki T, Tedder TF, Kurosaki T. Regulation of B-cell development by BCAP and CD19 through their binding to phosphoinositide 3-kinase. Blood. 2008;111:1497–1503. doi: 10.1182/blood-2007-08-109769. [DOI] [PubMed] [Google Scholar]
- 56.Klasener K, Maity PC, Hobeika E, Yang J, Reth M. B cell activation involves nanoscale receptor reorganizations and inside-out signaling by Syk. eLife. 2014;3:e02069. doi: 10.7554/eLife.02069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Amin RH, Schlissel MS. Foxo1 directly regulates the transcription of recombination-activating genes during B cell development. Nat Immunol. 2008;9:613–622. doi: 10.1038/ni.1612. This paper shows a central role for FOXO1 in the regulation of RAG1 and RAG2 expression in B cells. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lazorchak AS, et al. Sin1-mTORC2 suppresses rag and il7r gene expression through Akt2 in B cells. Mol Cell. 2010;39:433–443. doi: 10.1016/j.molcel.2010.07.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Herzog S, et al. SLP-65 regulates immunoglobulin light chain gene recombination through the PI(3) K-PKB-Foxo pathway. Nat Immunol. 2008;9:623–631. doi: 10.1038/ni.1616. [DOI] [PubMed] [Google Scholar]
- 60.Srinivasan L, et al. PI3 kinase signals BCR-dependent mature B cell survival. Cell. 2009;139:573–586. doi: 10.1016/j.cell.2009.08.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cheng S, et al. BCR-mediated apoptosis associated with negative selection of immature B cells is selectively dependent on Pten. Cell Res. 2009;19:196–207. doi: 10.1038/cr.2008.284. [DOI] [PubMed] [Google Scholar]
- 62.Anzelon AN, Wu H, Rickert R. C Pten inactivation alters peripheral B lymphocyte fate and reconstitutes CD19 function. Nat Immunol. 2003;4:287–294. doi: 10.1038/ni892. [DOI] [PubMed] [Google Scholar]
- 63.Benhamou D, et al. A c-Myc/miR17-92/Pten axis controls PI3K-mediated positive and negative selection in B cell development and reconstitutes CD19 deficiency. Cell Rep. 2016;16:419–431. doi: 10.1016/j.celrep.2016.05.084. [DOI] [PubMed] [Google Scholar]
- 64.Rowland SL, DePersis CL, Torres RM, Pelanda R. Ras activation of Erk restores impaired tonic BCR signaling and rescues immature B cell differentiation. J Exp Med. 2010;207:607–621. doi: 10.1084/jem.20091673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Novak R, Jacob E, Haimovich J, Avni O, Melamed D. The MAPK/ERK and PI3K pathways additively coordinate the transcription of recombination-activating genes in B lineage cells. J Immunol. 2010;185:3239–3247. doi: 10.4049/jimmunol.1001430. [DOI] [PubMed] [Google Scholar]
- 66.Quong MW, et al. Receptor editing and marginal zone B cell development are regulated by the helix-loop-helix protein, E2A. J Exp Med. 2004;199:1101–1112. doi: 10.1084/jem.20031180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Beck K, Peak MM, Ota T, Nemazee D, Murre C. Distinct roles for E12 and E47 in B cell specification and the sequential rearrangement of immunoglobulin light chain loci. J Exp Med. 2009;206:2271–2284. doi: 10.1084/jem.20090756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Verkoczy L, et al. Basal B cell receptor-directed phosphatidylinositol 3-kinase signaling turns off RAGs and promotes B cell-positive selection. J Immunol. 2007;178:6332–6341. doi: 10.4049/jimmunol.178.10.6332. This paper, together with reference 187, shows a key role for PI3K in downregulating RAG expression in newly formed B cells carrying innocuous BCRs. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Duong BH, et al. Negative selection by IgM superantigen defines a B cell central tolerance compartment and reveals mutations allowing escape. J Immunol. 2011;187:5596–5605. doi: 10.4049/jimmunol.1102479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Schram BR, et al. B cell receptor basal signaling regulates antigen-induced Ig light chain rearrangements. J Immunol. 2008;180:4728–4741. doi: 10.4049/jimmunol.180.7.4728. [DOI] [PubMed] [Google Scholar]
- 71.Hayashi K, Nojima T, Goitsuka R, Kitamura D. Impaired receptor editing in the primary B cell repertoire of BASH-deficient mice. J Immunol. 2004;173:5980–5988. doi: 10.4049/jimmunol.173.10.5980. [DOI] [PubMed] [Google Scholar]
- 72.Kohler F, et al. Autoreactive B cell receptors mimic autonomous pre-B cell receptor signaling and induce proliferation of early B cells. Immunity. 2008;29:912–921. doi: 10.1016/j.immuni.2008.10.013. [DOI] [PubMed] [Google Scholar]
- 73.Bounab Y, Getahun A, Cambier JC, Daeron M. Phosphatase regulation of immunoreceptor signaling in T cells, B cells and mast cells. Curr Opin Immunol. 2013;25:313–320. doi: 10.1016/j.coi.2013.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Cornall RJ, et al. Polygenic autoimmune traits: Lyn, CD22, and SHP-1 are limiting elements of a biochemical pathway regulating BCR signaling and selection. Immunity. 1998;8:497–508. doi: 10.1016/s1074-7613(00)80554-3. [DOI] [PubMed] [Google Scholar]
- 75.Infantino S, et al. Arginine methylation of the B cell antigen receptor promotes differentiation. J Exp Med. 2010;207:711–719. doi: 10.1084/jem.20091303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Bednarski JJ, et al. RAG-mediated DNA double-strand breaks activate a cell type-specific checkpoint to inhibit pre-B cell receptor signals. J Exp Med. 2016;213:209–223. doi: 10.1084/jem.20151048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Steinel NC, et al. The ataxia telangiectasia mutated kinase controls Igκ allelic exclusion by inhibiting secondary Vκ-to-Jκ rearrangements. J Exp Med. 2013;210:233–239. doi: 10.1084/jem.20121605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Imamura Y, et al. BLNK binds active H-Ras to promote B cell receptor-mediated capping and ERK activation. J Biol Chem. 2009;284:9804–9813. doi: 10.1074/jbc.M809051200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Cadera EJ, et al. NF-κB activity marks cells engaged in receptor editing. J Exp Med. 2009;206:1803–1816. doi: 10.1084/jem.20082815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Derudder E, et al. Development of immunoglobulin lambda-chain-positive B cells, but not editing of immunoglobulin kappa-chain, depends on NF-κB signals. Nat Immunol. 2009;10:647–654. doi: 10.1038/ni.1732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Johnson K, et al. Regulation of immunoglobulin lightchain recombination by the transcription factor IRF-4 and the attenuation of interleukin-7 signaling. Immunity. 2008;28:335–345. doi: 10.1016/j.immuni.2007.12.019. [DOI] [PubMed] [Google Scholar]
- 82.Pathak S, Ma S, Trinh L, Lu R. A role for interferon regulatory factor 4 in receptor editing. Mol Cell Biol. 2008;28:2815–2824. doi: 10.1128/MCB.01946-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Verkoczy L, et al. A role for nuclear factor kappa B/rel transcription factors in the regulation of the recombinase activator genes. Immunity. 2005;22:519–531. doi: 10.1016/j.immuni.2005.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Retter MW, Nemazee D. Receptor editing occurs frequently during normal B cell development. J Exp Med. 1998;188:1231–1238. doi: 10.1084/jem.188.7.1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wardemann H, et al. Predominant autoantibody production by early human B cell precursors. Science. 2003;301:1374–1377. doi: 10.1126/science.1086907. This paper shows that polyreactive B cells are present in humans at high frequency early in development and that this frequency is lower in more mature B cells. [DOI] [PubMed] [Google Scholar]
- 86.Luning Prak E, Trounstine M, Huszar D, Weigert M. Light chain editing in kappa-deficient animals: a potential mechanism of B cell tolerance. J Exp Med. 1994;180:1805–1815. doi: 10.1084/jem.180.5.1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Constantinescu A, Schlissel MS. Changes in locus-specific V(D)J recombinase activity induced by immunoglobulin gene products during B cell development. J Exp Med. 1997;185:609–620. doi: 10.1084/jem.185.4.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Arakawa H, Shimizu T, Takeda S. Re-evaluation of the probabilities for productive arrangements on the kappa and lambda loci. Int Immunol. 1996;8:91–99. doi: 10.1093/intimm/8.1.91. [DOI] [PubMed] [Google Scholar]
- 89.Aoki-Ota M, Torkamani A, Ota T, Schork N, Nemazee D. Skewed primary igkappa repertoire and v-j joining in C57BL/6 mice: implications for recombination accessibility and receptor editing. J Immunol. 2012;188:2305–2315. doi: 10.4049/jimmunol.1103484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Townsend CL, et al. Significant differences in physicochemical properties of human immunoglobulin kappa and lambda CDR3 regions. Front Immunol. 2016;7:388. doi: 10.3389/fimmu.2016.00388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Pelanda R, Schaal S, Torres RM, Rajewsky K. A prematurely expressed Igκ transgene, but not VκJκ gene segment targeted into the Igκ locus, can rescue B cell development in λ5-deficient mice. Immunity. 1996;5:229–239. doi: 10.1016/s1074-7613(00)80318-0. [DOI] [PubMed] [Google Scholar]
- 92.Wardemann H, Hammersen J, Nussenzweig MC. Human autoantibody silencing by immunoglobulin light chains. J Exp Med. 2004;200:191–199. doi: 10.1084/jem.20040818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Li Y, Li H, Weigert M. Autoreactive B cells in the marginal zone that express dual receptors. J Exp Med. 2002;195:181–188. doi: 10.1084/jem.20011453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Spanopoulou E, et al. Functional immunoglobulin transgenes guide ordered B-cell differentiation in Rag-1-deficient mice. Genes Dev. 1994;8:1030–1042. doi: 10.1101/gad.8.9.1030. [DOI] [PubMed] [Google Scholar]
- 95.Chen C, et al. The site and stage of anti-DNA B-cell deletion. Nature. 1995;373:252–255. doi: 10.1038/373252a0. The first paper to study an antibody gene knock-in model carrying both H-chain and L-chain genes with defined specificity (anti-chromatin or anti-DNA); the study shows not only that editing is nearly complete, but that it is focused on the L-chain rather than the H-chain locus. [DOI] [PubMed] [Google Scholar]
- 96.Vela JL, Aït-Azzouzene D, Duong BH, Ota T, Nemazee D. Rearrangement of mouse immunoglobulin kappa deleting element recombining sequence promotes immune tolerance and lambda B cell production. Immunity. 2008;28:161–170. doi: 10.1016/j.immuni.2007.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Melamed D, Benschop RJ, Cambier JC, Nemazee D. Developmental regulation of B lymphocyte immune tolerance compartmentalizes clonal selection from receptor selection. Cell. 1998;92:173–182. doi: 10.1016/s0092-8674(00)80912-5. [DOI] [PubMed] [Google Scholar]
- 98.Carsetti R, Kohler G, Lamers MC. Transitional B cells are the target of negative selection in the B cell compartment. J Exp Med. 1995;181:2129–2140. doi: 10.1084/jem.181.6.2129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Enders A, et al. Loss of the pro-apoptotic BH3-only Bcl-2 family member Bim inhibits BCR stimulation-induced apoptosis and deletion of autoreactive B cells. J Exp Med. 2003;198:1119–1126. doi: 10.1084/jem.20030411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Fang W, et al. Self-reactive B lymphocytes overexpressing Bcl-xL escape negative selection and are tolerized by clonal anergy and receptor editing. Immunity. 1998;9:35–45. doi: 10.1016/s1074-7613(00)80586-5. [DOI] [PubMed] [Google Scholar]
- 101.Lang J, et al. Enforced Bcl-2 expression inhibits antigen-mediated clonal elimination of peripheral B cells in an antigen dose-dependent manner and promotes receptor editing in autoreactive, immature B cells. J Exp Med. 1997;186:1513–1522. doi: 10.1084/jem.186.9.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Calnan DR, Brunet A. The FoxO code. Oncogene. 2008;27:2276–2288. doi: 10.1038/onc.2008.21. [DOI] [PubMed] [Google Scholar]
- 103.Benschop RJ, Brandl E, Chan AC, Cambier JC. Unique signaling properties of B cell antigen receptor in mature and immature B cells: implications for tolerance and activation. J Immunol. 2001;167:4172–4179. doi: 10.4049/jimmunol.167.8.4172. [DOI] [PubMed] [Google Scholar]
- 104.Limnander A, et al. STIM1, PKC-δ and RasGRP set a threshold for proapoptotic Erk signaling during B cell development. Nat Immunol. 2011;12:425–433. doi: 10.1038/ni.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Chumley MJ, et al. A VH11Vκ9 B cell antigen receptor drives generation of CD5+ B cells both in vivo and in vitro. J Immunol. 2000;164:4586–4593. doi: 10.4049/jimmunol.164.9.4586. [DOI] [PubMed] [Google Scholar]
- 106.Dempsey PW, Allison ME, Akkaraju S, Goodnow CC, Fearon DT. C3d of complement as a molecular adjuvant: bridging innate and acquired immunity. Science. 1996;271:348–350. doi: 10.1126/science.271.5247.348. [DOI] [PubMed] [Google Scholar]
- 107.Liu Y, et al. Lupus susceptibility genes may breach tolerance to DNA by impairing receptor editing of nuclear antigen-reactive B cells. J Immunol. 2007;179:1340–1352. doi: 10.4049/jimmunol.179.2.1340. [DOI] [PubMed] [Google Scholar]
- 108.Lamoureux JL, et al. Reduced receptor editing in lupus-prone MRL/lpr mice. J Exp Med. 2007;204:2853–2864. doi: 10.1084/jem.20071268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Meng W, et al. B-Cell tolerance defects in the B6.Aec1/2 mouse model of Sjögren’s syndrome. J Clin Immunol. 2012;32:551–564. doi: 10.1007/s10875-012-9663-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Henry-Bonami RA, et al. B lymphocyte “original sin” in the bone marrow enhances islet autoreactivity in type 1 diabetes-prone nonobese diabetic mice. J Immunol. 2013;190:5992–6003. doi: 10.4049/jimmunol.1201359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Yachimovich-Cohen N, Fischel R, Bachar N, Yarkoni Y, Eilat D. Autoimmune NZB/NZW F1 mice utilize B cell receptor editing for generating high-affinity anti-dsDNA autoantibodies from low-affinity precursors. Eur J Immunol. 2003;33:2469–2478. doi: 10.1002/eji.200324025. [DOI] [PubMed] [Google Scholar]
- 112.Verkoczy LK, Martensson AS, Nemazee D. The scope of receptor editing and its association with autoimmunity. Curr Opin Immunol. 2004;16:808–814. doi: 10.1016/j.coi.2004.09.017. [DOI] [PubMed] [Google Scholar]
- 113.Lang J, et al. Receptor editing and genetic variability in human autoreactive B cells. J Exp Med. 2016;213:93–108. doi: 10.1084/jem.20151039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Meffre E. The establishment of early B cell tolerance in humans: lessons from primary immunodeficiency diseases. Ann NY Acad Sci. 2011;1246:1–10. doi: 10.1111/j.1749-6632.2011.06347.x. This paper reviews B cell repertoire analyses of tolerance using single-cell antibody cloning from normal donors and patients with primary immunodeficiencies. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Clark AG, et al. Regulation of basement membrane-reactive B cells in BXSB, (NZBxNZW)F1, NZB, and MRL/lpr lupus mice. Autoimmunity. 2013;46:188–204. doi: 10.3109/08916934.2012.746671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Yurasov S, et al. Defective B cell tolerance checkpoints in systemic lupus erythematosus. J Exp Med. 2005;201:703–711. doi: 10.1084/jem.20042251. This paper shows major defects in B cell negative selection in patients with SLE. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Robinson WH, Mao R. Biomarkers to guide clinical therapeutics in rheumatology? Curr Opin Rheumatol. 2016;28:168–175. doi: 10.1097/BOR.0000000000000250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Winger L, Winger C, Shastry P, Russell A, Longenecker M. Efficient generation in vitro, from human peripheral blood cells, of monoclonal Epstein-Barr virus transformants producing specific antibody to a variety of antigens without prior deliberate immunization. Proc Natl Acad Sci USA. 1983;80:4484–4488. doi: 10.1073/pnas.80.14.4484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Foreman AL, Van de Water J, Gougeon ML, Gershwin ME. B cells in autoimmune diseases: insights from analyses of immunoglobulin variable (Ig V) gene usage. Autoimmun Rev. 2007;6:387–401. doi: 10.1016/j.autrev.2006.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Malkiel S, et al. Checkpoints for autoreactive B cells in the peripheral blood of lupus patients assessed by flow cytometry. Arthritis Rheumatol. 2016;68:2210–2220. doi: 10.1002/art.39710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Corti D, Sallusto F, Lanzavecchia A. High throughput cellular screens to interrogate the human T and B cell repertoires. Curr Opin Immunol. 2011;23:430–435. doi: 10.1016/j.coi.2011.04.006. [DOI] [PubMed] [Google Scholar]
- 122.Wardemann H, Nussenzweig MC. B-Cell self-tolerance in humans. Adv Immunol. 2007;95:83–110. doi: 10.1016/S0065-2776(07)95003-8. [DOI] [PubMed] [Google Scholar]
- 123.Samuels J, Ng YS, Coupillaud C, Paget D, Meffre E. Impaired early B cell tolerance in patients with rheumatoid arthritis. J Exp Med. 2005;201:1659–1667. doi: 10.1084/jem.20042321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Chamberlain N, et al. Rituximab does not reset defective early B cell tolerance checkpoints. J Clin Invest. 2016;126:282–287. doi: 10.1172/JCI83840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Saadoun D, et al. Expansion of autoreactive unresponsive CD21-/low B cells in Sjögren’s syndrome-associated lymphoproliferation. Arthritis Rheum. 2013;65:1085–1096. doi: 10.1002/art.37828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Kinnunen T, et al. Specific peripheral B cell tolerance defects in patients with multiple sclerosis. J Clin Invest. 2013;123:2737–2741. doi: 10.1172/JCI68775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Radic MZ, Weigert M. Genetic and structural evidence for antigen selection of anti-DNA antibodies. Annu Rev Immunol. 1994;12:487–520. doi: 10.1146/annurev.iy.12.040194.002415. [DOI] [PubMed] [Google Scholar]
- 128.Mietzner B, et al. Autoreactive IgG memory antibodies in patients with systemic lupus erythematosus arise from nonreactive and polyreactive precursors. Proc Natl Acad Sci USA. 2008;105:9727–9732. doi: 10.1073/pnas.0803644105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Menard L, et al. The PTPN22 allele encoding an R620W variant interferes with the removal of developing autoreactive B cells in humans. J Clin Invest. 2011;121:3635–3644. doi: 10.1172/JCI45790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Stanford SM, Bottini N. PTPN22: the archetypal non-HLA autoimmunity gene. Nat Rev Rheumatol. 2014;10:602–611. doi: 10.1038/nrrheum.2014.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Dai X, et al. A disease-associated PTPN22 variant promotes systemic autoimmunity in murine models. J Clin Invest. 2013;123:2024–2036. doi: 10.1172/JCI66963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Schickel JN, et al. PTPN22 inhibition resets defective human central B cell tolerance. Sci Immunol. 2016;1:aaf7153. doi: 10.1126/sciimmunol.aaf7153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Sauer AV, et al. Defective B cell tolerance in adenosine deaminase deficiency is corrected by gene therapy. J Clin Invest. 2012;122:2141–2152. doi: 10.1172/JCI61788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Walter JE, et al. Expansion of immunoglobulin-secreting cells and defects in B cell tolerance in Rag-dependent immunodeficiency. J Exp Med. 2010;207:1541–1554. doi: 10.1084/jem.20091927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Romberg N, et al. CVID-associated TACI mutations affect autoreactive B cell selection and activation. J Clin Invest. 2013;123:4283–4293. doi: 10.1172/JCI69854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Cyster JG, et al. Regulation of B-lymphocyte negative and positive selection by tyrosine phosphatase CD45. Nature. 1996;381:325–328. doi: 10.1038/381325a0. [DOI] [PubMed] [Google Scholar]
- 137.Vince N, et al. Defects in the CD19 complex predispose to glomerulonephritis, as well as IgG1 subclass deficiency. J Allergy Clin Immunol. 2011;127:538–541. doi: 10.1016/j.jaci.2010.10.019. [DOI] [PubMed] [Google Scholar]
- 138.Mackay F, Schneider P. Cracking the BAFF code. Nat Rev Immunol. 2009;9:491–502. doi: 10.1038/nri2572. [DOI] [PubMed] [Google Scholar]
- 139.Isnardi I, et al. IRAK-4- and MyD88-dependent pathways are essential for the removal of developing autoreactive B cells in humans. Immunity. 2008;29:746–757. doi: 10.1016/j.immuni.2008.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Nickerson KM, et al. TLR9 promotes tolerance by restricting survival of anergic anti-DNA B cells, yet is also required for their activation. J Immunol. 2013;190:1447–1456. doi: 10.4049/jimmunol.1202115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Cantaert T, et al. Activation-induced cytidine deaminase expression in human B cell precursors is essential for central B cell tolerance. Immunity. 2015;43:884–895. doi: 10.1016/j.immuni.2015.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Kuraoka M, et al. Activation-induced cytidine deaminase mediates central tolerance in B cells. Proc Natl Acad Sci USA. 2011;108:11560–11565. doi: 10.1073/pnas.1102571108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Haynes BF, et al. Cardiolipin polyspecific autoreactivity in two broadly neutralizing HIV-1 antibodies. Science. 2005;308:1906–1908. doi: 10.1126/science.1111781. [DOI] [PubMed] [Google Scholar]
- 144.Verkoczy L, Kelsoe G, Haynes BF. HIV-1 envelope gp41 broadly neutralizing antibodies: hurdles for vaccine development. PLoS Pathog. 2014;10:e1004073. doi: 10.1371/journal.ppat.1004073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.DeKosky BJ, et al. In-depth determination and analysis of the human paired heavy- and light-chain antibody repertoire. Nat Med. 2015;21:86–91. doi: 10.1038/nm.3743. [DOI] [PubMed] [Google Scholar]
- 146.Weintraub BC, et al. Entry of B cell receptor into signaling domains is inhibited in tolerant B cells. J Exp Med. 2000;191:1443–1448. doi: 10.1084/jem.191.8.1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Stoddart A, et al. Lipid rafts unite signaling cascades with clathrin to regulate BCR internalization. Immunity. 2002;17:451–462. doi: 10.1016/s1074-7613(02)00416-8. [DOI] [PubMed] [Google Scholar]
- 148.Busman-Sahay K, Drake L, Sitaram A, Marks M, Drake JR. Cis and trans regulatory mechanisms control AP2-mediated B cell receptor endocytosis via select tyrosine-based motifs. PLoS ONE. 2013;8:e54938. doi: 10.1371/journal.pone.0054938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Davis RE, et al. Chronic active B-cell-receptor signalling in diffuse large B-cell lymphoma. Nature. 2010;463:88–92. doi: 10.1038/nature08638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Patel KJ, Neuberger MS. Antigen presentation by the B cell antigen receptor is driven by the alpha/beta sheath and occurs independently of its cytoplasmic tyrosines. Cell. 1993;74:939–946. doi: 10.1016/0092-8674(93)90473-4. [DOI] [PubMed] [Google Scholar]
- 151.Niiro H, et al. The B lymphocyte adaptor molecule of 32 kilodaltons (Bam32) regulates B cell antigen receptor internalization. J Immunol. 2004;173:5601–5609. doi: 10.4049/jimmunol.173.9.5601. [DOI] [PubMed] [Google Scholar]
- 152.Ma H, et al. Visualization of Syk-antigen receptor interactions using green fluorescent protein: differential roles for Syk and Lyn in the regulation of receptor capping and internalization. J Immunol. 2001;166:1507–1516. doi: 10.4049/jimmunol.166.3.1507. [DOI] [PubMed] [Google Scholar]
- 153.Gazumyan A, Reichlin A, Nussenzweig MC. Igβ tyrosine residues contribute to the control of B cell receptor signaling by regulating receptor internalization. J Exp Med. 2006;203:1785–1794. doi: 10.1084/jem.20060221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Cornall RJ, Cheng AM, Pawson T, Goodnow CC. Role of Syk in B-cell development and antigen-receptor signaling. Proc Natl Acad Sci USA. 2000;97:1713–1718. doi: 10.1073/pnas.97.4.1713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Kitaura Y, et al. Control of the B cell-intrinsic tolerance programs by ubiquitin ligases Cbl and Cbl-b. Immunity. 2007;26:567–578. doi: 10.1016/j.immuni.2007.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Oellerich T, et al. The B-cell antigen receptor signals through a preformed transducer module of SLP65 and CIN85. EMBO J. 2011;30:3620–3634. doi: 10.1038/emboj.2011.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Kometani K, et al. CIN85 drives B cell responses by linking BCR signals to the canonical NF-κB pathway. J Exp Med. 2011;208:1447–1457. doi: 10.1084/jem.20102665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Han A, Saijo K, Mecklenbrauker I, Tarakhovsky A, Nussenzweig MC. Bam32 links the B cell receptor to ERK and JNK and mediates B cell proliferation but not survival. Immunity. 2003;19:621–632. doi: 10.1016/s1074-7613(03)00275-9. [DOI] [PubMed] [Google Scholar]
- 159.Hou P, et al. B cell antigen receptor signaling and internalization are mutually exclusive events. PLoS Biol. 2006;4:e200. doi: 10.1371/journal.pbio.0040200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Gonzalez-Martin A, et al. The microRNA miR-148a functions as a critical regulator of B cell tolerance and autoimmunity. Nat Immunol. 2016;17:433–440. doi: 10.1038/ni.3385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Lai M, et al. Regulation of B-cell development and tolerance by different members of the miR-17 approximately 92 family microRNAs. Nat Commun. 2016;7:12207. doi: 10.1038/ncomms12207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Melchers F. The pre-B-cell receptor: selector of fitting immunoglobulin heavy chains for the B-cell repertoire. Nat Rev Immunol. 2005;5:578–584. doi: 10.1038/nri1649. [DOI] [PubMed] [Google Scholar]
- 163.Ochiai K, et al. A self-reinforcing regulatory network triggered by limiting IL-7 activates pre-BCR signaling and differentiation. Nat Immunol. 2012;13:300–307. doi: 10.1038/ni.2210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Sun A, et al. VH replacement in primary immunoglobulin repertoire diversification. Proc Natl Acad Sci USA. 2015;112:E458–E466. doi: 10.1073/pnas.1418001112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Matsuda F, et al. The complete nucleotide sequence of the human immunoglobulin heavy chain variable region locus. J Exp Med. 1998;188:2151–2162. doi: 10.1084/jem.188.11.2151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Brensing-Kuppers J, Zocher I, Thiebe R, Zachau HG. The human immunoglobulin κ locus on yeast artificial chromosomes (YACs) Gene. 1997;191:173–181. doi: 10.1016/s0378-1119(97)00056-5. [DOI] [PubMed] [Google Scholar]
- 167.Frippiat JP, et al. Organization of the human immunoglobulin lambda light-chain locus on chromosome 22q11.2. Hum Mol Genet. 1995;4:983–991. doi: 10.1093/hmg/4.6.983. [DOI] [PubMed] [Google Scholar]
- 168.Hieter PA, Hollis GF, Korsmeyer SJ, Waldmann TA, Leder P. Clustered arrangement of immunoglobulin lambda constant region genes in man. Nature. 1981;294:536–540. doi: 10.1038/294536a0. [DOI] [PubMed] [Google Scholar]
- 169.Ubelhart R, et al. Responsiveness of B cells is regulated by the hinge region of IgD. Nat Immunol. 2015;16:534–543. doi: 10.1038/ni.3141. [DOI] [PubMed] [Google Scholar]
- 170.Rolli V, et al. Amplification of B cell antigen receptor signaling by a Syk/ITAM positive feedback loop. Mol Cell. 2002;10:1057–1069. doi: 10.1016/s1097-2765(02)00739-6. [DOI] [PubMed] [Google Scholar]
- 171.Wang Y, et al. The physiologic role of CD19 cytoplasmic tyrosines. Immunity. 2002;17:501–514. doi: 10.1016/s1074-7613(02)00426-0. [DOI] [PubMed] [Google Scholar]
- 172.Okkenhaug K. Signaling by the phosphoinositide 3-kinase family in immune cells. Annu Rev Immunol. 2013;31:675–704. doi: 10.1146/annurev-immunol-032712-095946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Wienands J, et al. SLP-65: a new signaling component in B lymphocytes which requires expression of the antigen receptor for phosphorylation. J Exp Med. 1998;188:791–795. doi: 10.1084/jem.188.4.791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Rickert RC. New insights into pre-BCR and BCR signalling with relevance to B cell malignancies. Nat Rev Immunol. 2013;13:578–591. doi: 10.1038/nri3487. [DOI] [PubMed] [Google Scholar]
- 175.Packard TA, Cambier JC. B lymphocyte antigen receptor signaling: initiation, amplification, and regulation. F1000Prime Rep. 2013;5:40. doi: 10.12703/P5-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Ng YS, Wardemann H, Chelnis J, Cunningham-Rundles C, Meffre E. Bruton’s tyrosine kinase is essential for human B cell tolerance. J Exp Med. 2004;200:927–934. doi: 10.1084/jem.20040920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Dingjan GM, et al. Bruton’s tyrosine kinase regulates the activation of gene rearrangements at the λ light chain locus in precursor B cells in the mouse. J Exp Med. 2001;193:1169–1178. doi: 10.1084/jem.193.10.1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Middendorp S, Hendriks RW. Cellular maturation defects in Bruton’s tyrosine kinase-deficient immature B cells are amplified by premature B cell receptor expression and reduced by receptor editing. J Immunol. 2004;172:1371–1379. doi: 10.4049/jimmunol.172.3.1371. [DOI] [PubMed] [Google Scholar]
- 179.Buhl AM, Cambier JC. Co-receptor and accessory regulation of B-cell antigen receptor signal transduction. Immunol Rev. 1997;160:127–138. doi: 10.1111/j.1600-065x.1997.tb01033.x. [DOI] [PubMed] [Google Scholar]
- 180.Keren Z, Diamant E, Ostrovsky O, Bengal E, Melamed D. Modification of ligand-independent B cell receptor tonic signals activates receptor editing in immature B lymphocytes. J Biol Chem. 2004;279:13418–13424. doi: 10.1074/jbc.M311970200. [DOI] [PubMed] [Google Scholar]
- 181.Shivtiel S, Leider N, Melamed D. Receptor editing in CD45-deficient immature B cells. Eur J Immunol. 2002;32:2264–2273. doi: 10.1002/1521-4141(200208)32:8<2264::AID-IMMU2264>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 182.Niiro H, et al. CIN85 is required for Cbl-mediated regulation of antigen receptor signaling in human B cells. Blood. 2012;119:2263–2273. doi: 10.1182/blood-2011-04-351965. [DOI] [PubMed] [Google Scholar]
- 183.Ruer-Laventie J, et al. Overexpression of Fkbp11, a feature of lupus B cells, leads to B cell tolerance breakdown and initiates plasma cell differentiation. Immun Inflamm Dis. 2015;3:265–279. doi: 10.1002/iid3.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Yamanashi Y, et al. Role of tyrosine phosphorylation of HS1 in B cell antigen receptor-mediated apoptosis. J Exp Med. 1997;185:1387–1392. doi: 10.1084/jem.185.7.1387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Ramsey LB, Vegoe AL, Miller AT, Cooke MP, Farrar MA. Tonic BCR signaling represses receptor editing via Raf- and calcium-dependent signaling pathways. Immunol Lett. 2011;135:74–77. doi: 10.1016/j.imlet.2010.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Teodorovic LS, et al. Activation of Ras overcomes B-cell tolerance to promote differentiation of autoreactive B cells and production of autoantibodies. Proc Natl Acad Sci USA. 2014;111:E2797–E2806. doi: 10.1073/pnas.1402159111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Llorian M, Stamataki Z, Hill S, Turner M, Martensson IL. The PI3K p110δ is required for down-regulation of RAG expression in immature B cells. J Immunol. 2007;178:1981–1985. doi: 10.4049/jimmunol.178.4.1981. This paper shows the importance of the PI3K pathway in downregulating RAG expression in immature B cells, and defines p110δ as a key player in positive selection. [DOI] [PubMed] [Google Scholar]
- 188.Bai L, et al. Phospholipase Cγ2 contributes to light-chain gene activation and receptor editing. Mol Cell Biol. 2007;27:5957–5967. doi: 10.1128/MCB.02273-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Bartlett A, Buhlmann JE, Stone J, Lim B, Barrington RA. Multiple checkpoint breach of B cell tolerance in Rasgrp1-deficient mice. J Immunol. 2013;191:3605–3613. doi: 10.4049/jimmunol.1202892. [DOI] [PubMed] [Google Scholar]
- 190.Cheng AM, et al. Syk tyrosine kinase required for mouse viability and B-cell development. Nature. 1995;378:303–306. doi: 10.1038/378303a0. [DOI] [PubMed] [Google Scholar]
- 191.Turner M, et al. Syk tyrosine kinase is required for the positive selection of immature B cells into the recirculating B cell pool. J Exp Med. 1997;186:2013–2021. doi: 10.1084/jem.186.12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]