Abstract
COPASI is software used for the creation, modification, simulation and computational analysis of kinetic models in various fields. It is open-source, available for all major platforms and provides a user-friendly graphical user interface, but is also controllable via the command line and scripting languages. These are likely reasons for its wide acceptance. We begin this review with a short introduction describing the general approaches and techniques used in computational modeling in the biosciences. Next we introduce the COPASI package, and its capabilities, before looking at typical applications of COPASI in biotechnology.
Keywords: COPASI, Biotechnology, Systems Biology
1. Introduction
Computational modeling has been used in the context of biology for decades. Early studies on enzyme kinetics have already employed small computational models and their simulation (e.g. [1]). Also, in the context of population ecology and epidemiology, computational modeling has been involved for many years to describe and predict population sizes of species or to analyze the spread of diseases. Cell biological questions, e.g. the investigation of the mechanisms behind calcium oscillations (for a review see e.g. [2]) or the understanding of the regulation of glycolysis [3] are another context in which models have played a role since the 1960’s. However, despite the successes and the knowledge about the usefulness of the respective techniques, usage only started to virtually explode with the turn of the century, when systems biology developed as its own discipline [4]. This is probably due to the advances both on the experimental side (e.g. live-cell imaging, high-throughput data), as well as on the computational side (widespread availability of computers, faster machines, more and tailored algorithms). Another likely contributor to this virtual explosion in computational modeling, has been the emergence of user-friendly software, which makes model set-up and analysis much more accessible, and practical, for non-mathematicians.
Different computational methods are employed on different scales of abstraction and detail. In the areas of biochemistry and cell biology the two largest classes [4] of models are: a) stoichiometric models including constraint based genome scale models, and b) kinetic models. The former are based on static reaction schemes and used to calculate potential flux distributions mostly in metabolic networks. The latter allows for a more detailed picture and for more fine-grained analysis, but also demands more knowledge, namely about the kinetics of individual processes.
For these two classes of computational models the German Network for Bioinformatics Infrastructure, de.NBI, hosts two widely used resources. Stoichiometric models, including genome scale constraint based models, can be set-up and analyzed using CellNetAnalyzer [5], whereas COPASI [6, 7] specializes in setting up and analyzing kinetic models while also providing some basic stoichiometric analyses.
Computational models are widely employed in biotechnology. The following sections will go into some detail about this. Generally speaking, often the goal is to manipulate systems to either maximize the yield, flux, or titer of desirable end-products, or to minimize the amounts or fluxes of undesirable side-products.
Below, we will first give an overview of the current features of the COPASI software. Then we describe its application in the context of biotechnology by presenting concrete examples where COPASI was applied to systems of biotechnological interest with varying levels of complexity.
2. The COPASI Software
COPASI [6, 7, 8] is a modeling and simulation environment, jointly founded by the groups of Pedro Mendes (now at UConn Health) and Ursula Kummer (now at Heidelberg University). Currently the groups of Stefan Hoops, Jürgen Pahle, and Sven Sahle are also part of the consortium. All 3 have been former group members and are now PIs on their own, and are actively participating in development of the software. The software is platform-independent, developed as open source, and distributed available under the terms of the Artistic License 2.01, which makes it free to use for any purpose. Source and binary distributions are available from the COPASI Website (http://copasi.org) for Windows, Linux, and Mac OS X. This makes it very easy for researchers to install COPASI and get started quickly. Those who prefer it can also download the source code and compile their own specific versions of the software.
2.1. Features
Model Editing
Models are entered in (bio)chemical terms. (Bio)chemical species, located in compartments, can be transformed through reactions. The rate of each reaction is given by a kinetic rate law, that can be either selected from an included function database (filtered to apply to the selected reaction), or freely defined. Additionally, discrete events can be added to the model, that change model elements based on arbitrary expressions. From this formalism, COPASI derives the mathematical representation of the model automatically. Thus users do not have to specify the differential equations themselves. However, expert users can add additional differential equations to the model, including for compartment volumes or any other variable (i.e. not just chemical species).
Model Standards
COPASI stores the model and information what to do with it in its own specialized XML format. However, wherever possible, applicable community standards are supported. Models can be imported and exported in the Systems Biology Markup Language (SBML, [9]) format, where all major versions are supported. This enables COPASI to consume models from a variety of online model repositories (such as the BioModels Database [10]), as well as from many other software tools that support the SBML standard.
Where possible, the analysis tasks are exported to the Simulation Experiment Description Markup Language (SED-ML, [11]). Specifically, time course simulations and parameter scans can be exported to SED-ML, as SED-ML does not specify further simulation types yet.
Currently the COPASI team is extending COPASI to additionally support the COMBINE Archive [12] which will make it easier to exchange COPASI models together with experimental data.
Simulation Algorithms
At the core, COPASI supports two main formalisms for simulating the dynamics of the defined model (time course simulations). One is the traditional chemical kinetics approach of using ordinary differential equations (ODE), where the software uses the LSODAR integrator [13]. The second is the stochastic formalism [14], where individual reaction events are drawn from probability distributions, using either of the following algorithms: Gillespie’s Direct Method [15], Gibson-Bruck [16], τ-Leap, or adaptive SSA/τ-leap [17]. To aid users in adapting deterministic rate equations into their stochastic equivalent, COPASI features an option that splits up reversible reactions into their forward and backward-directional forms. Additionally there is an option to apply stochastic corrections to rate equations.
Since stochastic simulations can be rather time consuming, a third class of simulation algorithms available consist of hybrid solvers. The solvers “Hybrid (Runge-Kutta)” and “Hybrid (LSODA)” employ a dynamic partitioning strategy that treats all reactions involving species that have a particle number below a specified threshold with the stochastic approach, while the remaining reactions are treated with the differential equation approach. A new solver was recently introduced, “Hybrid (RK-45)”, which allows modelers to define this partitioning themselves, and thus control which reactions are to be simulated stochastically or deterministically.
In the future, solvers for Stochastic Differential Equations (SDEs) and Delay Differential Equations (DDEs) will also be added.
Analysis Tasks
The model evaluations through time course simulations or steady state computations at the core of COPASI support a number of additional analysis tasks.
Parameter Scan: The parameter scan task can be used to run any of the other tasks repeatedly. Frequently one needs to examine how the model behaves when some parameter(s) vary, and this task allows programming this parameter variation in an easy way. Parameter values can be either “scanned” in a given interval (i.e. their values changed through regular intervals), or sampled from a random distribution; these methods can be intermixed such that some parameters can be scanned and other sampled. This can be applied to time course simulations or steady state simulations, but also to any other task in this list. This allows for complex operations, such as multi-dimensional parameter scans, repeating stochastic time course simulations to determine the time-dependent probability distributions for each model variable, creating bifurcation plots (running the cross section task for different values of a parameter), evaluating the fitness landscape around an optimization solution, or even determining profile likelihood plots [18].
Optimization: In the optimization task users can enter an arbitrary expression of model quantities (or even results from the execution of other tasks) that should be maximized or minimized. Additionally the model elements that should be modified, where appropriate with optional constraints, are specified. COPASI implements a wide selection of local (Hooke & Jeeves [19], Levenberg-Marquardt [20], Nelder-Mead [21], Praxis [22], Steepest Descent, Truncated Newton[23]) and global optimization methods (Differential Evolution [24], Evolutionary Strategy [25], Evolutionary Programming [26], Genetic Algorithm [27], Particle Swarm [28], Scatter Search [29], Random Search, Simulated Annealing [30]), that can be used for the optimization.
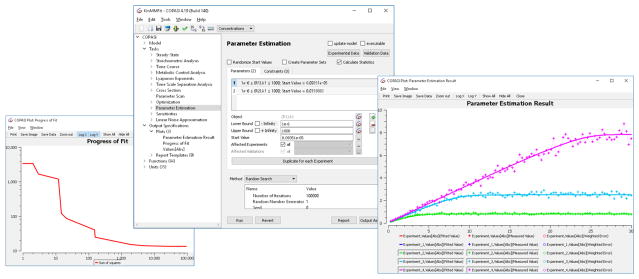
Parameter Estimation: Parameter estimation is the activity of changing the model’s parameter values in order to make the behavior of the model as close as possible to a set of experimental measurements. Parameter estimation is a special form of optimization, in which the distance between the model and the experimental data is minimized. This is achieved by changing user-selected model parameters through specific optimization algorithms (the same ones that are available for general optimization described above). COPASI supports both experimental data in the form of time courses and steady state measurements; several data sets of either type can be mixed together. Users can specify as many data sets as required. A new feature allows validation data sets to be specified as well. These experimental data sets are not used during the fitting process, but only during a validation phase to measure how robust are the model predictions. The middle window in Fig. 1 shows the definition of parameters to be modified in a parameter estimation run.
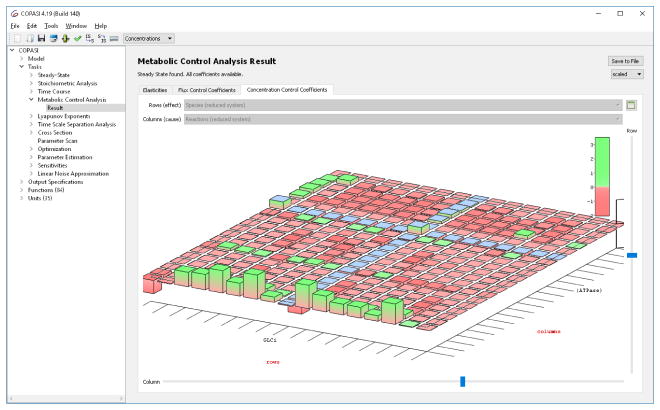
Metabolic Control Analysis: Metabolic control analysis (MCA) is a special type of sensitivity analysis that calculates how much a perturbation in the rate of a reaction affects the steady state concentrations or fluxes [31]. These are systems level coefficients and are called “control coefficients”. MCA establishes a link [32] between the control coefficients and “elasticity coefficients”, which are properties of each reaction in isolation (they are the derivatives of the kinetic rate law in terms of the reaction effectors). COPASI calculates the elasticities as well as flux and concentration control coefficients. In Fig. 2 a three dimensional representation of an MCA result is displayed.
Sensitivities: Sensitivities provide a measure of how much a selected model variable (the effect) changes when a selected parameter (the cause) is changed. COPASI includes a pre-defined set of effects and causes, but users are also free to select any specific model elements as well. Sensitivities are always calculated based on another task (selected by the user): time courses, steady states, optimization, parameter estimation, or cross sections. Second order sensitivities are also available, where the user needs to select two causes (this is essentially an estimate of a second order derivative). The control coefficients of MCA are actually a subset of the sensitivities for a steady state, however they are calculated with a different approach (using Reder’s matrix method).
Linear Noise Approximation: The Linear Noise Approximation (LNA) task estimates the covariances between species particle numbers [33, 34] without time-demanding stochastic simulations. It sets up a Lyapunov equation for the system in steady state and then solves this equation for the covariances using the Bartels-Stewart algorithm. This task can be used in a closed-loop fashion with parameter scans or optimization to efficiently find parameter sets where, e.g., the coefficient of variation of a species’ particle number is low or where the correlation between two species is high.
Time scale separation analysis: The time scale separation analysis task [35] provides information on time scales of different parts of the model (and at different times in the dynamics). This is accomplished by simulating the model for a user specified duration and analyzing the time-dependent Jacobian matrix in order to decompose species and reactions into different time scales.
Lyapunov Exponents: Lyapunov exponents provide a means to characterize the dynamical behavior of a model, including an indication if the model has chaotic behavior (sensitivity to initial conditions). COPASI implements the Wolf method [36] for calculating Lyapunov exponents from time series data. A user only has to specify how many exponents should be calculated.
Cross section: The cross section task facilitates the detection of crossings through a hyper surface in phase space. The user defines the surface by selecting a model variable and a certain value, then, when COPASI runs a simulation, the cross section task will only yield an output point when the specified value is crossed in a specified direction. In conjunction with the scan task, the cross section task makes it possible to create bifurcation diagrams (displayed in Fig. 3). A step by step tutorial to create these plots is available in our users manual2.
Figure 1.
Parameter estimation using COPASI. The left window shows a progress graph, that is updated during the parameter estimation run and shows the current objective value. While the numeric value is not of particular interest, any reduction in its value demonstrates an improvement in the fit. In the middle image the setup of parameters and the specification of the method is displayed. The right image shows a plot of the parameter estimation result. For each measured quantity the fit result is being displayed (line) along with the experimental data (points).
Figure 2.
Metabolic control analysis results displayed in a 3D widget that makes it easy to identify contributions of individual reactions to the control of concentrations or fluxes.
Figure 3.
Setting up a parameter scan over a cross section task makes it possible to create bifurcation diagrams in COPASI. Left we see a change of oscillation behavior in a calcium model [46], from simple oscillation to bursting as the number of peaks per oscillation change from one to four. Right we see the period doubling route to chaos in an abstract model [47]. In both cases a bifurcation parameter is scanned (x-axis) and the maxima of one variable of the model are plotted after the system has settled on the respective attractor (y-axis).
2.2. COPASI User Interface
Users have several options for running COPASI. The most common option is to run the interactive graphical user interface (GUI), which was written in such a way that it works natively on all operating systems. A command line version of COPASI is also available, which allows for processing simulations without a graphical interface and which is useful for running simulations offline on servers, etc. The most frequent use case involving the command line version is to prepare a COPASI file using the GUI, mark the desired task (such as parameter estimation) as executable, and then move the resulting COPASI file to a cluster environment where it is run by the command line version and the results are saved in files. Additional to these two versions of COPASI, libraries are also provided with which COPASI can be used from inside programs written in all major programming languages.
While the computations executed by the command line version and the libraries are the same as with the GUI there are two features only present when using COPASI through the graphical user interface:
Plotting Plots of several types can be used for visualization of results of all analysis tasks in COPASI. This includes basic scatter and line plots, histograms, and contour plots (a recent addition). Fig. 1 displays a combination of several plots as used during parameter estimation. Whenever results are obtained in matrix form (as is the case for stoichiometric analysis of the model, calculation of sensitivities and the like), they are displayed in a sortable table by default. However, users can also opt to have the results displayed in a three dimensional representation as is shown in Fig. 2.
Network Diagrams The GUI also has support for model diagrams. The diagrams can be created from scratch, using a force based layout algorithm, or they can be imported from other software tools using either the SBML Level 3 Layout [37] and Rendering package format or the CellDesigner [38] format. These diagrams can be used to display the result of a time course simulation (as an animation) or the decomposition of the network in elementary flux modes [39].
2.3. Support/Training
Apart from a user manual3 describing all COPASI features in detail, we regularly provide hands-on tutorials both in our own institutions, at conferences, or other venues. Additionally, online training videos can be found on our website that describe basic and advanced features of COPASI. The COPASI user forum4 is the best way to quickly get in touch with the COPASI community. Not only will the COPASI developers provide answers to questions asked, but also the other members of the forum provide answers to questions.
3. Applications in biotechnology
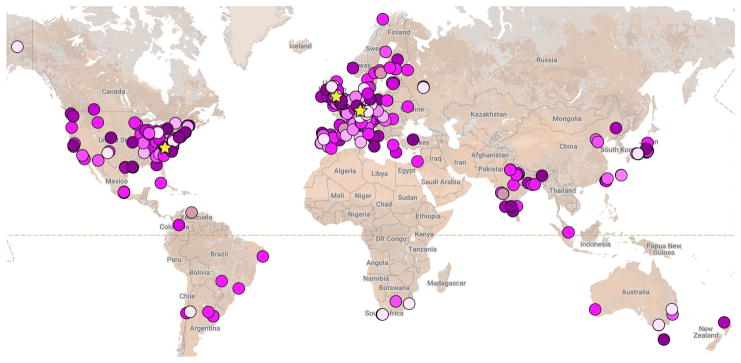
COPASI is widely used in a number of scientific fields. Although its original main purpose was for research on biochemical systems, COPASI is also used in other fields such as chemistry or ecology. We have investigated the usage of the software by carefully analyzing publications where it has been applied and classifying its research. We wanted to differentiate between citations that just mention COPASI, and actual studies that used the software for their scientific work. This analysis is ongoing and updated results can be seen at our webpage (https://www.copasi.org/Research). A visualization of the locations where these studies have been published from is displayed in Fig. 4.
Figure 4.
Manually curated world map of locations from where research has been carried out with COPASI (judged from their author’s addresses).
Here we want to focus on applications of the software in studies targeting biotechnological applications. In this context, and similar to other fields, COPASI is used for modeling and computational analysis of systems at different levels of granularity and complexity — from single enzymes to large cellular networks. In the following we want to show typical use cases that represent the most common scenarios in literature. A complete review of the literature would be far beyond the scope of this article.
At the molecular scale, the general goal is usually to determine the exact mechanism of biotechnologically relevant enzymes and their regulation, and to use this information to influence the reaction in order to e.g. increase the yield of a desired product. Pursuing this goal involves either one of two quite common scenarios. First of all, scientists use the software often to calculate steady state properties in conjunction with structural analysis and/or molecular modeling studies. Thus, Pingoud et al. [40] investigated the divalent metal ion dependence of restriction endonucleases of the EcoRI family. In their extensive study, they experimentally determined binding properties of different enzymes of this family with respect to Mg2+ and Mn2+ and to a lesser extend Ca2+ and their subsequent catalytic properties. They used the experimental results together with insights from molecular dynamics simulations to generate an extensive elementary reaction scheme for the kinetic mechanism of the enzymatic catalysis of DNA cleavage which involves binding and unbinding of different metal ions at two different binding sites. This scheme was entered as a model into COPASI and the software was used to calculate steady state distributions in different scenarios. The authors conclude that the mechanism of this class of biotechnologically relevant enzymes involves one metal ion binding site essential for the enzymatic activity, and a second one which modulates this activity.
The second common scenario on the molecular scale also involves the set-up of a model for the kinetics of an enzyme. But instead of experimentally or computationally (via molecular dynamics simulations) determined kinetic parameters it uses the parameter estimation task to estimate the values of the parameters of interest. As an example, Kashyap et al. [41] used COPASI to investigate the kinetics and mechanism of the formation of fructo-oligosaccharides using fructosyltransferase. They experimentally measured time courses and enzyme activity for various conditions. They then set up a detailed model of the elementary reactions involved in the catalytic mechanisms of this enzyme, including binding of glucose which leads to an inhibition of the enzyme. The parameters of the model were fitted with COPASI using an extensive amount of experimental measurements. In this case, the genetic algorithm was the optimization routine of choice. The result of the fitting process was statistically analyzed and the corresponding parameter values were used in simulations reproducing further experimental conditions, now no longer fitting the model but rather validating the accuracy of its predictions. Simulations were performed with the LSODA integrator of COPASI. The authors conclude how the yield of the biotechnological product, namely the fructo-oligosaccharides can be optimized with respect to the amount of substrate and enzyme.
Moving to a more complex scale, biotechnologically relevant pathways are of course also subject to similar questions, e.g. deciphering their mechanism and regulation and thus learning how to manipulate them. Therefore, also quite similar scenarios can be found as described for the molecular scale. Thus, Ma et al. [42] used COPASI to model nitrogen assimilation in E. coli. They were especially interested in understanding how this process is regulated at low ammonium concentrations. They used COPASI to set up a kinetic model describing all essential reactions participating in nitrogen assimilation in this bacterial species. Most of the parameters and concentrations had been experimentally determined. They then calculated steady-state concentrations and fluxes at different ammonium concentrations. Subsequently, they investigated the influence of the expression of AmtB (Am transport protein B) on the ability of E. coli to take up ammonium. Furthermore, the behavior of the system at ammonium impulses was studied. For this purpose, simulations with the LSODA algorithm were performed.
In similar context, but with a larger system and more unknown parameters, Raganati et al. [43] studied butanol production from different sugars by Clostridium acetobutylicum. The authors were interested in the different products and the yield in order to be able to optimize the respective fermenters. They set up an extensive model using COPASI and used parameter estimation with experimental time course data to estimate the unknown parameter values. In this instance they used the simulated annealing algorithm. It is noteworthy that — as often reported — every set of model and experimental data has individual requirements that can not be satisfied by one common single optimization routine. Rather, different algorithms are efficient for different data, without a clear pattern (i.e. there is no prescribed rule that associates algorithms with specific types of data or problem). Thus this is why COPASI offers many different optimization algorithms and allows users easily to switch from one to another. The final model was used to simulate the behavior of the system with respect to different sugars.
In contrast to the molecular level, kinetic models of whole pathways are often analyzed via metabolic control analysis (MCA). An example is the study by Trausinger et al. [44] who studied the metabolic interactions influencing carbon flux from xylose to ethanol in different yeast strains. Here, a kinetic model of this pathway was first subject to parameter estimation in COPASI (using evolutionary programming as optimization routine). The model was then studied using MCA to identify those reactions that have the largest influence on the conversion and therefore, would be the most promising targets for manipulation.
Apart from the expected usage of the software as described above, we were actually surprised not to find certain usages in biotechnological applications. Of course, they could be out there and simply not yet included in our analysis. We had expected that optimization algorithms would be more commonly used to predict conditions to optimize biotechnological strains (as some of us argued quite some time ago [45]). The optimization task in COPASI allows one to ask questions such as “What are the conditions for maximal yield?” and then to let parameters that are amenable to experimental manipulation (e.g. expression levels) be adjusted for the optimal combination. Perhaps we ought to put some more emphasis on this approach in our outreach and training efforts.
Highlights.
Reviews current features of COPASI – a software for modeling biological processes
Shows usage of the software COPASI
Represents use cases of COPASI when applied in biotechnology
Acknowledgments
Over the years, COPASI has received funding from the following funding agencies and programmes: the BMBF funded de.NBI initiative (031L0104A) and Virtual Liver Project, National Institutes of Health (NIGMS Grant: GM080219), the BBSRC (grant BB/J019259/1), BBSRC and EPSRC (grant BB/C008219/1) as well as the Klaus Tschira Foundation and the Virginia Bioinformatics Institute. We would like thank Norman Paton and Yang Cao for their collaboration. We would also like to thank all the programmers and student helpers that worked on COPASI over the years, particularly, Ralph Gauges, Joseph Dada, Ettore Murabito, Peyman Zarrineh, Natalia Simus and Irina Surovtsova. Marina Kaiser is acknowledged for literature digging. Finally, we would like to acknowledge the members of the COPASI user forum for their active support.
Footnotes
Authors are in alphabetical order.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Chance B. The reaction of catalase and cyanide. Journal of Biological Chemistry. 1949;179:1299–1309. [PubMed] [Google Scholar]
- 2.Schuster S, Marhl M, Höfer T. Modelling of simple and complex calcium oscillations. European Journal of Biochemistry. 2002;269:1333–1355. doi: 10.1046/j.0014-2956.2001.02720.x. [DOI] [PubMed] [Google Scholar]
- 3.Sel’Kov E. Self-oscillations in glycolysis. European Journal of Biochemistry. 1968;4:79–86. doi: 10.1111/j.1432-1033.1968.tb00175.x. [DOI] [PubMed] [Google Scholar]
- 4.Hübner K, Sahle S, Kummer U. Applications and trends in systems biology in biochemistry. FEBS Journal. 2011;278:2767–2857. doi: 10.1111/j.1742-4658.2011.08217.x. [DOI] [PubMed] [Google Scholar]
- 5.Klamt S, Saez-Rodriguez J, Gilles ED. Structural and functional analysis of cellular networks with CellNetAnalyzer. BMC Systems Biology. 2007;1:2. doi: 10.1186/1752-0509-1-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hoops S, Sahle S, Gauges R, Lee C, Pahle J, Simus N, Singhal M, Xu L, Mendes P, Kummer U. COPASI — a COmplex PAthway SImulator. Bioinformatics. 2006;22:3067–3074. doi: 10.1093/bioinformatics/btl485. [DOI] [PubMed] [Google Scholar]
- 7.Mendes P, Hoops S, Sahle S, Gauges R, Dada J, Kummer U. Computational modeling of biochemical networks using COPASI. In: Maly IV, editor. Systems Biology. Humana Press; Totowa, NJ: 2009. pp. 17–59. [DOI] [PubMed] [Google Scholar]
- 8.Mendes P, Messiha H, Malys N, Hoops S. Enzyme kinetics and computational modeling for systems biology. Methods in Enzymology. 2009;467:583–599. doi: 10.1016/S0076-6879(09)67022-1. [DOI] [PubMed] [Google Scholar]
- 9.Hucka M, et al. The Systems Biology Markup Language (SBML): A medium for representation and exchange of biochemical network models. Bioinformatics. 2003;19:524–531. doi: 10.1093/bioinformatics/btg015. [DOI] [PubMed] [Google Scholar]
- 10.Le Novère N, Bornstein B, Broicher A, Courtot M, Donizelli M, Dharuri H, Li L, Sauro H, Schilstra M, Shapiro B, Snoep JL, Hucka M. BioModels Database: a free, centralized database of curated, published, quantitative kinetic models of biochemical and cellular systems. Nucleic Acids Research. 2006;34:D689–D691. doi: 10.1093/nar/gkj092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Waltemath D, Adams R, Bergmann FT, Hucka M, Kolpakov F, Miller AK, Moraru II, Nickerson D, Sahle S, Snoep JL, et al. Reproducible computational biology experiments with SED-ML - the simulation experiment description markup language. BMC Systems Biology. 2011;5:1. doi: 10.1186/1752-0509-5-198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bergmann FT, Adams R, Moodie S, Cooper J, Glont M, Golebiewski M, Hucka M, Laibe C, Miller AK, Nickerson DP, et al. COMBINE archive and OMEX format: one file to share all information to reproduce a modeling project. BMC Bioinformatics. 2014;15:1. doi: 10.1186/s12859-014-0369-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Petzold L. Automatic selection of methods for solving stiff and nonstiff systems of ordinary differential equations. SIAM Journal on Scientific and Statistical Computing. 1983;4:136–148. [Google Scholar]
- 14.Pahle J. Biochemical simulations: stochastic, approximate stochastic and hybrid approaches. Briefings in Bioinformatics. 2009;10:53–64. doi: 10.1093/bib/bbn050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gillespie DT. A general method for numerically simulating the stochastic time evolution of coupled chemical reactions. Journal of Computational Physics. 1976;22:403–434. [Google Scholar]
- 16.Gibson MA, Bruck J. Efficient exact stochastic simulation of chemical systems with many species and many channels. The Journal of Physical Chemistry A. 2000;104:1876–1889. [Google Scholar]
- 17.Cao Y, Gillespie DT, Petzold LR. Efficient step size selection for the tau-leaping simulation method. The Journal of Chemical Physics. 2006;124:044109. doi: 10.1063/1.2159468. [DOI] [PubMed] [Google Scholar]
- 18.Schaber J. Easy parameter identifiability analysis with COPASI. Biosystems. 2012;110:183–185. doi: 10.1016/j.biosystems.2012.09.003. [DOI] [PubMed] [Google Scholar]
- 19.Hooke R, Jeeves TA. “direct search” solution of numerical and statistical problems. Journal of the ACM. 1961;8:212–229. [Google Scholar]
- 20.Levenberg K. A method for the solution of certain non-linear problems in least squares. Quarterly of applied mathematics. 1944;2:164–168. [Google Scholar]
- 21.Nelder JA, Mead R. A simplex method for function minimization. The Computer Journal. 1965;7:308–313. [Google Scholar]
- 22.Brent PR. A new algorithm for minimizing a function of several variables without calculating derivatives. In: Brent PR, editor. Algorithms for minimization without derivatives. Prentice-Hall, Inc; Englewood Cliffs, NJ: 1973. pp. 117–167. [Google Scholar]
- 23.Nash SG. Newton-type minimization via the lanczos method. SIAM Journal of Numerical Analysis. 1984;21:770–788. [Google Scholar]
- 24.Storn R, Price K. Differential evolution – a simple and efficient heuristic for global optimization over continuous spaces. Journal of Global Optimization. 1997;11:341–359. [Google Scholar]
- 25.Runarsson T, Yao X. Stochastic ranking for constrained evolutionary optimization. IEEE Transactions on Evolutionary Computation. 2000;4:284–294. [Google Scholar]
- 26.Fogel DB, Fogel LJ, Atmar JW. Meta-evolutionary programming. In: Chen RR, editor. 25th Asilomar conference on signals, systems & computers. IEEE Computer Society; 1992. pp. 540–545. [Google Scholar]
- 27.Michalewicz Z. Genetic algorithms + data structures = evolution programs. 3. Springer-Verlag; Berlin: 1994. [Google Scholar]
- 28.Kennedy J, Eberhart R. Particle swarm optimization. IEEE International Conference on Neural Networks; IEEE Computer Society; 1995. pp. 1942–1948. [Google Scholar]
- 29.Egea JA, Balsa-Canto E, García MSG, Banga JR. Dynamic optimization of nonlinear processes with an enhanced scatter search method. Industrial & Engineering Chemistry Research. 2009;48:4388–4401. [Google Scholar]
- 30.Corana A, Marchesi M, Martini C, Ridella S. Minimizing multimodal functions of continuous variables with the “simulated annealing” algorithm. ACM Transactions on Mathematical Software. 1987;13:262–280. [Google Scholar]
- 31.Fell DA. Understanding the Control of Metabolism. Portland Press; 1996. [Google Scholar]
- 32.Reder C. Metabolic control theory: A structural approach. 1988;135:175–201. doi: 10.1016/s0022-5193(88)80073-0. [DOI] [PubMed] [Google Scholar]
- 33.Pahle J, Challenger J, Mendes P, McKane A. Biochemical fluctuations, optimisation and the linear noise approximation. BMC Systems Biology. 2012;6:86. doi: 10.1186/1752-0509-6-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Challenger J, McKane A, Pahle J. Multi-compartment linear noise approximation. J Stat Mech. 2012;2012:P11010. [Google Scholar]
- 35.Surovtsova I, Simus N, Lorenz T, Knig A, Sahle S, Kummer U. Accessible methods for the dynamic time-scale decomposition of biochemical systems. Bioinformatics. 2009;25:2816–2823. doi: 10.1093/bioinformatics/btp451. [DOI] [PubMed] [Google Scholar]
- 36.Wolf A, Swift JB, Swinney HL, Vastano JA. Determining Lyapunov exponents from a time series. Physica D: Nonlinear Phenomena. 1985;16:285–317. [Google Scholar]
- 37.Gauges R, Rost U, Sahle S, Wengler K, Bergmann FT. The systems biology markup language (SBML) level 3 package: Layout, version 1 core. Journal of Integrative Bioinformatics. 2015;12:550–602. doi: 10.2390/biecoll-jib-2015-267. [DOI] [PubMed] [Google Scholar]
- 38.Funahashi A, Matsuoka Y, Jouraku A, Morohashi M, Kikuchi N, Kitano H. CellDesigner 3.5: a versatile modeling tool for biochemical networks. Proceedings of the IEEE. 2008;96:1254–1265. [Google Scholar]
- 39.Schuster S, Dandekar T, Fell DA. Detection of elementary flux modes in biochemical networks: a promising tool for pathway analysis and metabolic engineering. Trends in Biotechnology. 1999;17:53–60. doi: 10.1016/s0167-7799(98)01290-6. [DOI] [PubMed] [Google Scholar]
- 40.Pingoud V, Wende W, Friedhoff P, Reuter M, Alves J, Jeltsch A, Mones L, Fuxreiter M, Alfred P. On the divalent metal ion dependence of DNA cleavage by restriction endonucleases of the EcoRI family. Journal of Molecular Biology. 2009;393:140–160. doi: 10.1016/j.jmb.2009.08.011. [DOI] [PubMed] [Google Scholar]
- 41.Kashyap R, Palai T, Bhattacharya PK. Kinetics and model development for enzymatic synthesis of fructo-oligosaccharides using fructosyltransferase. Bioprocess and Biosystems Engineering. 2015;38:2417–2426. doi: 10.1007/s00449-015-1478-4. [DOI] [PubMed] [Google Scholar]
- 42.Ma H, Booger FC, Goryanin I. Modelling nitrogen assimilation of escherichia coli at low ammonium concentration. Journal of Biotechnology. 2009;144:175–183. doi: 10.1016/j.jbiotec.2009.09.003. [DOI] [PubMed] [Google Scholar]
- 43.Raganati F, Procentese A, Olivieri G, Götz P, Salatino P, Marzocchella A. Kinetic study of butanol production from various sugars by clostridium acetobutylicum using a dynamic model. Biochemical Engineering Journal. 2015;99:156–166. [Google Scholar]
- 44.Trausinger G, Gruber C, Krahulec S, Magnes C, Nidetzky B, Klimacek M. Identification of novel metabolic interactions controlling carbon flux from xylose to ethanol in natural and recombinant yeasts. Biotechnology for Biofuels. 2015;8:157. doi: 10.1186/s13068-015-0340-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mendes P, Kell DB. Non-linear optimization of biochemical pathways: applications to metabolic engineering and parameter estimation. Bioinformatics. 1998;14:869–883. doi: 10.1093/bioinformatics/14.10.869. [DOI] [PubMed] [Google Scholar]
- 46.Kummer U, Olsen LF, Dixon CJ, Green AK, Bornberg-Bauer E, Baier G. Switching from simple to complex oscillations in calcium signaling. Biophysical Journal. 2000;79:1188–1195. doi: 10.1016/S0006-3495(00)76373-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Baier G, Sahle S. Design of hyperchaotic flows. Physical Review E. 1995;51:R2712–R2714. doi: 10.1103/physreve.51.r2712. [DOI] [PubMed] [Google Scholar]