Abstract
We first review fundamental insights into anti-ranavirus immunity learned with the Xenopus laevis/ranavirus FV3 model that are generally applicable to ectothermic vertebrates. We then further investigate FV3 genes involved in immune evasion. Focusing on FV3 knockout (KO) mutants defective for a putative viral caspase activation and recruitment domain-containing (CARD)-like protein (Δ64R-FV3), a β-hydroxysteroid dehydrogenase homolog (Δ52L-FV3), and an immediate-early 18kDa protein (FV3-Δ18K), we assessed the involvement of these viral genes in replication, dissemination and interaction with peritoneal macrophages in tadpole and adult frogs. Our results substantiate the role of 64R and 52L as critical immune evasion genes, promoting persistence and dissemination in the host by counteracting type III IFN in tadpoles and type I IFN in adult frogs. Comparably, the substantial accumulation of genome copy numbers and exacerbation of type I and III IFN gene expression responses but deficient release of infectious virus suggests that 18K is a viral regulatory gene.
Keywords: Amphibian, ranavirus, macrophages, virulence genes
Introduction
Over the last 50 years, infections caused by ranaviruses (Iridoviridae) in most part of the world have markedly increased in prevalence as well as in the range of species infected (Chinchar et al., 2009; Duffus et al., 2015; Kik et al., 2011; Kolby et al., 2014; Kolby et al., 2015; Price et al., 2014). Emerging infectious diseases caused by ranavirus are not only alarming for biodiversity and aquaculture, but also poses fundamental issues related to the evolution of host/pathogen interactions (Chen and Robert, 2012; Collins; Daszak et al., 1999; Gray et al., 2009; Robert and Gregory Chinchar, 2012). With regards to host immune response, it is startling that ranaviruses are capable of crossing species barriers among a wide range of ectothermic vertebrates including species from different classes of vertebrates ranging from amphibians to fish and reptile species. Such a promiscuous infectious ability suggests that these pathogens possess potent immune evasion mechanisms (Johnson et al., 2008; Mao et al., 1999; Robert and Jancovich, 2016). Furthermore, although some host species are highly susceptible to ranavirus, others are relatively resistant and can serve as asymptomatic carriers that disseminate infectious virus (Hoverman et al.; Robert et al., 2007; Teacher et al., 2009). This implies the involvement of host specific factors that determine the outcome of infection.
In mammals, the understanding of host immune responses to viruses has been by large derived from the mouse model (review in (Mahalingam et al., 2000; Panchanathan et al., 2008)). To a similar extent, the African clawed frog, Xenopus laevis, has provided and still serves as an instrumental model to gain insights into amphibian host-ranavirus pathogen interaction and immune evasion. In a relative short period of time, the experimental platform using X. laevis and the ranavirus Frog Virus 3 (FV3) has permitted to characterize critical mechanisms of host immune defenses to ranavirus. This body of work can serve as a foundation for anti-ranaviral immunity in amphibian and more generally cold blooded vertebrates (reviewed in (Chen and Robert, 2011; Chinchar and Waltzek, 2014). This model system takes advantage of the extensive characterization of the immune system of Xenopus and the availability immunological reagents such as antibodies developed for X. laevis (Robert and Ohta, 2009) as well as the large genetic and genomic resources (Xenbase), which includes the full annotated genome sequence of X. laevis (Session et al., 2016) and sister species X. tropicalis (Hellsten Uffe, 2009). We provide first here an updated review of what has been learned about immunity to ranavirus using the Xenopus laevis/ranavirus FV3 model system, before focusing on three putative ranavirus virulence genes.
1. Amphibian adaptive immune responses to ranavirus pathogens
As a preamble, studies in Xenopus have revealed a remarkable conservation at the functional level in host immune response against ranavirus, not only with DNA viruses in fish (Cuesta and Tafalla, 2009; Somamoto et al., 2014), but also with DNA viruses in mammals (Panchanathan et al., 2008). Thus, although species-specific variations and adaptations are expected, antiviral mechanisms are fundamentally conserved among cold blooded and warm-blooded vertebrates. In amphibians as in all jawed vertebrates, control and clearance of viral infection requires an efficient and timely collaboration and integration of both innate and adaptive arms of the immune system.
1.1. Adult frogs
Studies in Xenopus have demonstrated that upon a primary infection, adult frogs develop an active adaptive CD8+ T cell response. This response is characterized by an expansion of CD8+ T cells that peak at 6 days post-infection in the spleen and by corresponding increased infiltration of CD8+ T cell effectors in the kidney at the height of viral replication (Morales and Robert, 2007). These CD8+ T cells are required for subsequent viral clearance that typically occurs within 2 weeks following infection and adult host survival (Robert et al., 2005). Although, direct evidence of an anti-FV3 CD4+ T helper cell response is lacking, the role of CD4+ T cells in Xenopus antiviral response can be inferred by an expansion and infiltration of T cells recognized by the pan T cell marker CD5 that are CD8 negative, and by the production of effective IgY anti-FV3 antibodies able to inactivate FV3 in vitro (Chinchar and Waltzek, 2014; Maniero et al., 2006; Robert et al., 2005). The production of IgY antibodies requires T cell help (likely CD4+) for the isotype switch from IgM (Blomberg et al., 1980). Interestingly, as in the case of anti-pox immune response in mammals (Panchanathan et al., 2006, 2008), detectable potent IgY antibody response to FV3 in Xenopus only occurs during a secondary FV3 infection, despite the induced expression of the B cell specific activation-induced cytidine deaminase (AID) mediating isotype switch as early as 3 days following primary FV3 infection (Marr et al., 2007). Notably, B cell memory established during the primary infection, could be detected for at least 6 months after this primary infection (Maniero et al., 2006). This suggests that adult frogs surviving a primary ranavirus infection can remain resistant to a subsequent infection for long time. Indication of immunological memory leading to increased survival and/or improved viral clearance has been reported in turtles (Hausmann et al., 2015). Evidence documenting an active adaptive immune response against ranavirus infection is also emerging from recent transcriptome studies in the non-model amphibian Rana temporari species (Price et al., 2015). Of further interest in this case, is the lack of differential expression for many immunologically-related genes upon ranavirus infection, which is perhaps due to the use of metamorphic animals. In Xenopus, it is well established that it take 4 to 6 weeks after the metamorphic completion for full recovery of immune functions (Robert and Ohta, 2009).
1.2. Tadpoles
Xenopus as other anuran species is characterized by a larval stage (tadpole) during which the immune system is generally considered to be more immature than adults, with a weaker antibody response, a poor isotype switch from IgM to IgY and weaker adaptive T cell response (Robert and Ohta, 2009). Consistent with this, tadpoles are usually more susceptible to ranavirus infection (Bayley et al., 2013; Landsberg et al., 2013; Reeve et al., 2013). In X. laevis, tadpoles are typically unable to control FV3 infection and most of them succumb. However, death from FV3 infection occurs gradually over 1 to 2 months, which is quite a long time (Gantress et al., 2003; Robert et al., 2005). In addition, we have noted that death is not always correlated with a high viral load (Grayfer et al., 2015). In fact, compared to adult frogs, viral loads in tadpole tissues including the kidney are significantly lower than adults even at early stages of infection, which suggest a distinct host-pathogen interaction between FV3 and the immune system of either tadpole or adult frogs. As such, it is important to remember that, despite some weaknesses, tadpoles are not just immunologically ignorant or deficient but rather have a distinct set of immune responses adapted for their life stage (Robert and Ohta, 2009). This, in turn, would imply that differences in selective pressure have lead FV3 and other ranaviruses to adapt their infection strategies for adult and larval stages. Indeed, adult and larval frogs have different ecological niches and have, therefore, evolved different approaches to fighting viral pathogens.
1.3. Importance of innate-like T cells in amphibian host defenses against ranavirus
Besides a minor fraction of conventional CD8+ T cells and MHC class II-restricted T cells (likely CD4+ T helper), the tadpole adaptive immune system is dominated by six subsets of innate-like (i)T cells representing about 80% of CD8 negative and CD8low lymphocytes (Edholm et al., 2013; Robert and Edholm, 2014). These iT cells express a very limited or invariant T cell receptor repertoire and require non-polymorphic MHC-like molecules rather than classical MHC class I molecules for their development and function (Edholm et al., 2013). Of particular relevance for ranavirus immunity, we found that one iT cell subset expressing the invariant rearrangement Vα6-Jα1.43 restricted by the MHC class I-like molecule XNC10 is critical for anti-FV3 response in tadpoles (Edholm et al., 2013). The loss-of-function of the mhc1b10.L gene encoding the XNC10 molecule established by combining RNA interference with transgenesis, abrogates the development of Vα6 iT cells. This XNC10 loss-of-function markedly increases tadpole susceptibility to FV3 infection, resulting in increased viral replication and high lethality at early stage of infection (Edholm et al., 2013). Vα6 iT cells are also important in adult host response as evidenced by the delay in antiviral response, increased viral load and kidney damage (Edholm et al., 2015). However, the mature adult immune system is still sufficient to ultimately control the viral infection and clear FV3.
2. Amphibian innate immune responses to ranavirus
Although the innate arm of the immune system is often considered as ancillary, only assisting the adaptive arm of the immune system, its central role in host antiviral resistance has become appreciated. In adult Xenopus, FV3 infections rapidly (as early as 1 day post-infection) induce potent type I interferon (IFN) response and interferon response factors (Mx1, Mx2) as well as pro-inflammatory or inflammatory-associated (TNFα, IFNγ, IL-1β) responses in parallel with the recruitment of activated macrophages to the site of infection (De Jesus Andino et al., 2012; Grayfer et al., 2014a, 2015). In contrast, the induction of pro-inflammatory immune gene expression (TNFα, IFNγ, IL-1β) by FV3 in tadpoles is delayed and of lower magnitude compared to adult frogs (De Jesus Andino et al., 2012). In addition, there is a poor recruitment of granulocyte-colony stimulating factor (G-CSF) receptor (G-CSFR) expressing granulocytes into tadpole kidneys, consistent with the weak inflammatory response to FV3 (Koubourli et al., 2017). However, it would be too simplistic to conclude that tadpole antiviral immunity is inefficient.
2.1. Interferon response
Type I IFN has clearly potent antiviral activity as demonstrated by its ability to protect for FV3 infection in the Xenopus A6 kidney cell line pretreated with a X. laevis recombinant type IFN in vitro and by partially protecting pre-treated tadpoles subsequently infected with FV3 (Grayfer et al., 2014a). The interferon system in Xenopus as in mammals and birds also includes type III or IFN-λ, which is more prominently involved in tadpoles than adult frogs during FV3 infection (Grayfer et al., 2015). Pre-injection of recombinant IFN-λ, can also confer some protection against FV3 infection in tadpoles, but not as efficiently as type I recombinant IFN (Grayfer et al., 2015). This is possibly because FV3 infection readily impairs IFN-λ, receptor gene expression in tadpoles and in X. laevis A6 kidney cell lines. Interestingly, recent evidence using water exposure to FV3 as a natural route of infection of skin mucosa has revealed a distinctive reliance on IFN between a X. laevis adult predominant type I-based and a tadpole mainly type III-based antiviral IFN systems (Wendel et al., 2017). In addition, the rapid decrease of viral loads in tadpole skin mucosa over 72 hr post-water infection indicate an active and efficient antiviral response (Wendel et al., 2017). Therefore, as in the case of adaptive immunity, it is likely that in response to developmental and evolutionary pressures, the regulation and activation of the innate components of antiviral immunity in tadpoles is distinct from that of adult frogs.
2.2. Complex role of macrophages
A key immune effector cell type in host response to FV3 in both tadpoles and adult X. laevis are monocytic phagocytes of the myeloid lineage or macrophages. Compared to mammals where many types of tissue resident and circulating macrophages have been identified, the understanding of distinct macrophage subsets in cold blooded vertebrates, including Xenopus, is limited. However, monopoiesis is fundamentally conserved between amphibians and mammals (reviewed in (Grayfer and Robert, 2016; Huber and Zon, 1998; Robert and Ohta, 2009)). Macrophages reside in most organs and tissues throughout the amphibian body, where they provide a core foundation of the first line of host immune defenses against infectious agents such as ranavirus. Indeed, macrophages play a key role in orchestrating antiviral immunity against FV3 (Grayfer and Robert, 2016). Notably, the macrophage-colony stimulating factor (CSF) receptor 1 (CSF-R1) that drives the differentiation and function of macrophages was shown to interact in Xenopus as in mammals, with two distinct, evolutionarily unrelated ligands, CSF-1 and Interleukin 34 (IL-34; (Grayfer et al., 2014b)). Using recombinant X. laevis proteins, we showed that, both in adults and tadpoles, macrophages stimulated in vitro or in vivo by IL-34 exhibited stronger antiviral activity characterized by the production of antimicrobial factors (iNOS) and by preventing viral replication when infected with FV3 (Grayfer and Robert, 2014, 2015). In contrast, CSF-1 derived macrophages were more phagocytic and, as a possible consequence, more susceptible to FV3 infection.
This functional disparity is of particular relevance since macrophages are not only crucial antiviral effector cells but appear to be specially targeted by ranavirus pathogens in Xenopus and other species including mammals (Gendrault et al., 1981; Grayfer and Robert, 2016; Gut et al., 1981). Considering the indiscriminate infection mode and apparent absence of specific cellular receptor(s) requirement, we have proposed that macrophages with their ability to acquire exogenous particles and antigens through various pathways such as micropinocytosis and phagocytosis represent an advantageous cellular target for viral dissemination and persistence into the host. Consistent with this postulate, dissemination of FV3 into the X. laevis tadpole brain is associated with and can be promoted by macrophages infiltration into brain tissue (De Jesus Andino et al., 2016). Furthermore, accumulating evidence indicates that macrophages are critical for viral persistence in asymptomatic adult X. laevis (Robert et al., 2007). FV3 infecting peritoneal macrophages in vivo is found to become rapidly transcriptionally silent or quiescent (Morales et al., 2010). Similarly, little to no replication of FV3 occurs following in vitro infection of peritoneal macrophages despite FV3 efficient penetration into the cells as indicated by the viral load recovered at early stage of infection (Robert et al., 2014). This non-permissiveness of viral replication is enhanced in IL-34-derived (Grayfer and Robert, 2014, 2015). The possible role of macrophages as reservoirs for viral persistence in resistant hosts is of relevance given that ranavirus is increasingly detected in asymptomatic amphibian populations in the wild (Brenes et al., 2014; Forzan and Wood, 2013) as well as in captive turtle species (Hausmann et al., 2015). The threat of resistant animal carriers in propagating ranavirus infection is underscored by the fact that X. laevis adults can harbor quiescent virus prone to reactivation. This was demonstrated by provoking inflammation with heat-killed bacteria in adult X. laevis one month past the time of viral clearance of a primary infection, as a way to reactivate FV3 (Robert et al., 2014). This treatment of apparently healthy animals not only induces the reappearance of active FV3 infection in peritoneal macrophages, but also results in significant increase in mortality from systemic viral infection. The increased susceptibility of previously infected, but apparently healthy, adults frogs to reactivated FV3 infection suggests that conditions altering the immune status such as stress or pollution can contribute to the dissemination of emerging ranavirus infections. In this regard, we have reported that exposure during early development of tadpoles to water contaminants including the herbicide atrazine and the insecticide carbaryl, even at very low subtoxic doses, induces long lasting defect of antiviral immunity that persists into adults (De Jesus Andino et al., 2017; Sifkarovski et al., 2014). For example, exposure of tadpoles for only 3 weeks to low (0.1 and 1.0 ppb) subtoxic concentrations of carbaryl induced long lasting defects of expression response of pro-inflammatory (IL-1β) and antiviral type I interferon (IFN) genes persisting after metamorphosis (De Jesus Andino et al., 2017). This was accompanied by a significant increase in viral loads in infected tissues of young adult frogs several months after being exposed to the contaminant.
3. Ranavirus genes involved in immune evasion
The other side of the host-pathogen equation is represented by the ranavirus pathogens themselves, and more specifically by their putative virulence and immune evasion genes. As large double stranded DNA viruses, ranavirus genomes encode as many as 100 genes, the majority of which are of unknown functions and do not share any sequence similarity outside iridoviruses (Chinchar et al., 2009). We and others have developed a convenient and reliable method for generating ranavirus recombinant by site-specific integration of a fluorescent gene (GFP) reporter fused to a drug resistance gene (puromycin or neomycin) under the control of the immediate early 18K FV3 promoter (Chen et al., 2011; Jancovich and Jacobs, 2011). To date, the putative immune evasion function of the viral homolog of the cellular translation factor eIF-2α (vIF-2α) that can antagonize protein kinase R (PKR) is one of the best documented examples (Jancovich and Jacobs, 2011; Rothenburg et al., 2011; Rothenburg et al., 2008).
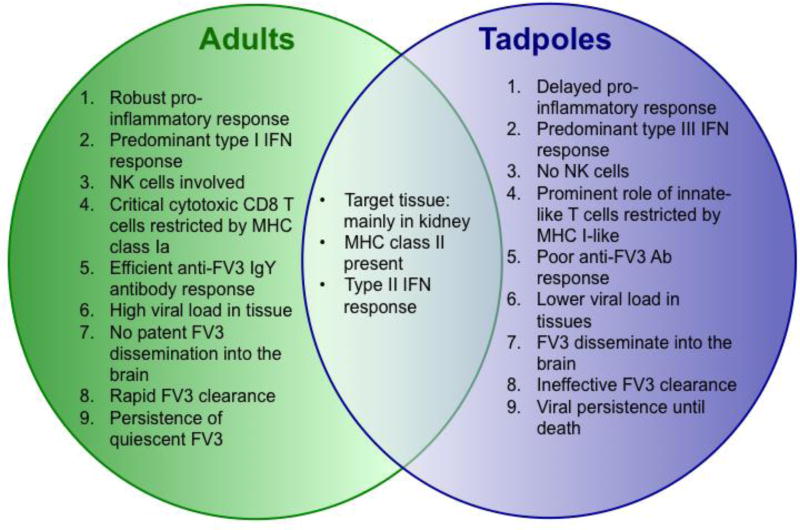
Among the potential other FV3 immune evasion gene candidates for which homology to eukaryotic gene could be inferred, two have retained our attention: 64R encoding a Caspase-like Activation and Recruitment Domain decoy (vCARD)-like molecule; and 52L encoding a β-hydroxysteroid dehydrogenase (βHSD)-like molecule. Using the technique described above we generated FV3 knock-out (KO) recombinants for 64R and 52L (Andino Fde et al., 2015). Because of its robust immediate early expression pattern and dispensable role for productive infection in cell culture (Cheng et al., 2014; Sample et al., 2007), the FV3 ICP18 or 18K (82R) gene was also selected as a virulence gene candidate and knocked out (Chen et al., 2011). Evidence from initial characterization of two FV3 KO viruses (Δ64R-, and Δ52L-FV3) is consistent with their involvement in immune evasion, whereas results obtained with Δ18-FV3 suggest a more complex role of the 18K gene. While deficiency of each of these viral genes results in different degrees of attenuated viral replication in X. laevis hosts, closer examination in vitro indicates distinct, albeit not fully defined, functions. Infection studies of A6 kidney cells suggest that the CARD-like molecule encoded by 64R interferes with IFN-induced apoptosis, whereas 18K may indirectly alter IFN responses by regulating other viral genes (Andino Fde et al., 2015). The role of the βHSD-like molecule encoded by 52L has remained elusive to date. Figure 1 summarizes what has been learned to date about FV3-Xenopus host pathogen interactions contrasting the adult and tadpole stages.
Fig. 1. Overview of the different interactions of FV3 pathogens with the distinct immune systems of X. laevis adult and tadpole hosts.
To further elucidate the role of these 3 putative immune evasion genes, we have assessed in detail replication, dissemination and persistence of each FV3 KO in tadpole and adult X. laevis.
Results
Effects of vCARD (ORF64R) and β–HSD (ORF52L) gene deletions on FV3 infectivity in X. laevis tadpoles
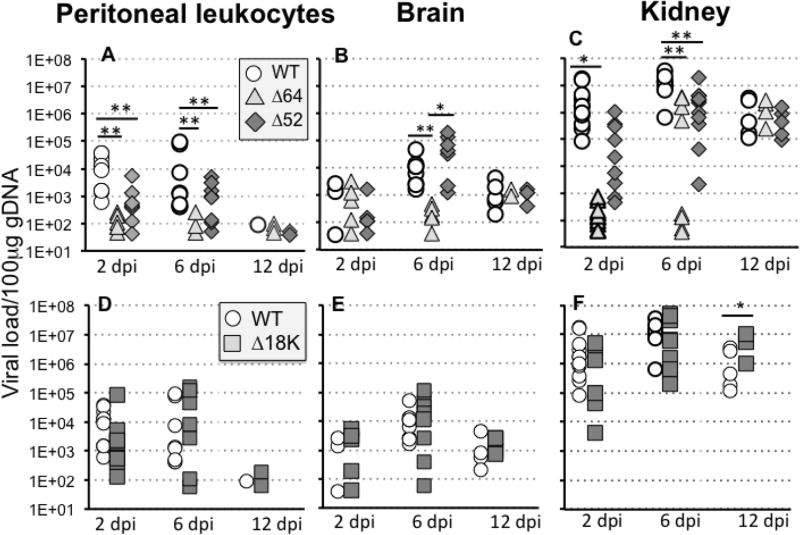
We previously reported that deletion of either 64R or 52L genes reduced FV3 replication in tadpole kidneys, which is the main site of viral replication in X. laevis (Gantress et al., 2003). Since larval peritoneal macrophages are particularly susceptible to FV3 infection (De Jesus Andino et al., 2012) and that FV3 is able to disseminate in tadpole brain (De Jesus Andino et al., 2016), we thought to first examine the ability of knock-out (KO) recombinant FV3 Δ64R- or Δ52L-FV3 compared to FV3-WT to actively infect these sites and induce host antiviral responses. Accordingly, we determined the viral loads and expression of several key host antiviral genes in peritoneal leukocytes, brain and kidneys of tadpoles at different times postinfection. Defects were more dramatic for Δ64R-FV3, which was barely detected in PLs and brain at all time points tested, suggesting a weak ability to infect and persist in infected peritoneal cells as well as a poor dissemination (Fig. 2). In kidney, the increase in viral load was markedly delayed and became detectable only for a fraction (3 individuals from each experiment) of animals at 6 dpi, whereas it stayed close to base line levels the remaining tadpoles. In all animals tested, the Δ64R-FV3 genome copy number reached similar level to WT-FV3 at 12 dpi. Unlike Δ64R-FV3, Δ52L-FV3 disseminated more efficiently in the brain, but was significantly impaired in persisting in PLs and in replicating in kidneys compared to FV3-WT. It is noteworthy that viral loads in each group exhibited substantial individual variability especially at early time points (2 and 6 dpi). This has been observed before (De Jesus Andino et al., 2012) and may reflect both experimental errors (small variations in the infectious doses injected in each animals by the investigator) and genetic variations of the hosts since outbred animals were used. Importantly, however, comparable variability in each group was observed in the two experiments that were combined to achieve statistical significance.
Fig. 2. Viral loads in peritoneal leukocytes (PLs), brain and kidneys of tadpoles at different time following infection with WT-, Δ52L-, Δ64R- or Δ18-FV3.
Outbred pre-metamorphic tadpoles were infected by i.p. injection of 1 × 104 PFU of each virus type and FV3 genome copy numbers in PLs, brains and kidneys at 2, 6 and 12 dpi were determined by absolute qPCR using primers specific for FV3 vDNA Pol II. (A–C) Viral loads for Δ52L- Δ64R- and WT-FV3. (D–F) Viral loads for Δ18K- and the same WT-FV3 control as in A. Results are means ± SE of genome copy number/100µ of genomic DNA of 6 to 10 animals per group and are representative from two different experiments. **; P <0.001 and *; P <0.05 significant differences between WT and KO-FV3 using one-way ANOVA test and Tukey’s post hoc test.
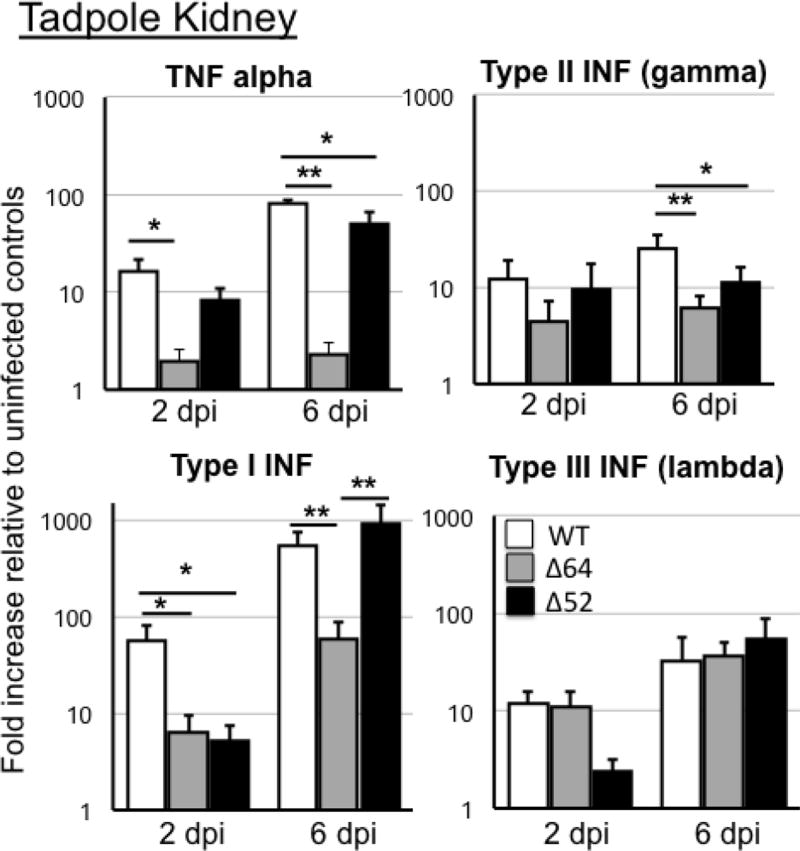
The distinctive defects associated with 64R and 52L deletions were further revealed by the changes in expression of critical innate immune genes in tadpole kidneys (Fig. 3). Consistent with the low viral loads, gene expression responses for TNFα was ablated in kidneys of tadpoles infected with Δ64R-FV3, and significantly reduced for type I IFN at 2 and 6 dpi. In addition, IFN-γ (type II) gene expression was significantly decreased at 6 dpi in Δ64R-FV3 compared to FV3-WT. However, the 64R deletion did not significantly affected IFN-λ, gene response in kidney, which is a critical antiviral response in tadpoles. For Δ52L-FV3, there was only a significant diminished type I IFN gene expression at 2 dpi compared to FV3-WT.
Fig. 3. Changes in expression by RT-qPCR of TNFα, type I, II and III IFN genes in tadpole kidneys during infection with Δ52L- or Δ64R-FV3 compared to WT-FV3.
Outbred pre-metamorphic tadpoles were infected by i.p. injection of 1 ×104 PFU of each virus type for 2 and 6 days (dpi). Results are average ± SEM fold increase relative to uninfected controls of 6 to 10 animals per group and are representative from two different experiments. **; P <0.001 and *; P <0.05 significant differences between WT- and KO-FV3 using one-way ANOVA test and Tukey’s post hoc test.
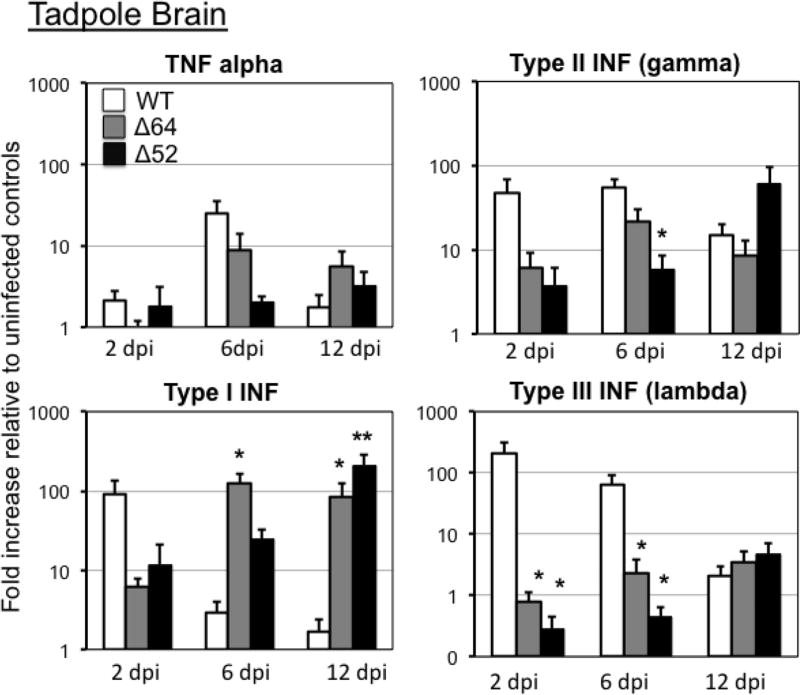
In tadpole brains, while 64R or 52L deficiency did not result in statistically significant alteration the inflammatory gene TNFα expression response compared to WT-FV3, type I IFN gene expression was exacerbated at 6 dpi and remained high at 12 dpi in tadpole infected with Δ64R-FV3 (Fig. 4). Enhanced type I IFN gene expression was also triggered by Δ52L-FV3 at late stage of infection (12 dpi). In contrast, the two KO FV3 recombinants induced significantly less increase of IFN-λ, gene expression than WT-FV3 at 2 and 6 dpi, whereas only Δ52L-FV3 infection had a significant defect in the IFN-γ gene response at 6 dpi (Fig. 4). Thus, 64R and 52L deletions result in different alteration of IFN gene response in kidneys and brain. No significant changes in the gene expression patterns was observed in PLs at 2 and 6 dpi for the different KO FV3, although there was a high individual variability (Suppl. Fig 1).
Fig. 4. Changes in expression by RT-qPCR of TNFα, type I, II and III IFN genes in tadpole brains during infection with Δ52L- or Δ64R-FV3 compared to WT-FV3.
Outbred pre-metamorphic tadpoles were infected by i.p. injection of 1 ×104 PFU of each virus type for 2, 6 and 12 days (dpi). Results are average ± SEM fold increase relative to uninfected controls of 6 to 10 animals per group and are representative from two different experiments. **; P <0.001 and *; P <0.05 significant differences between WT- and KO-FV3 using one-way ANOVA test and Tukey’s post hoc test.
Effects of 18K gene deletion on FV3 infectivity and IFN response in X. laevis tadpoles
To date, the potential function of the immediate-early18K gene remains unclear. However, the absence of sequence similarity with any eukaryotic or prokaryotic genes compared to the high degree of conservation of this gene within the ranavirus genus suggests a biologically important species-specific role of 18K molecules. Based on our previous studies with a Δ18K-FV3 KO mutant, we postulate that 18K play a role in immune evasion by interfering with the IFN response (Andino Fde et al., 2015). We further examined the ability of Δ18K-FV3 to infect tadpole PLs, replicate in kidney and disseminate into the brain. In contrast to Δ64R-FV3 and Δ52L-FV3, replication of Δ18K-FV3 in kidneys, as determined by genome copy number using absolute qPCR was minimally affected with a slight but no statistically significant decrease at 2 dpi compared to WT-FV3 in this group of animals (Fig. 2). Interestingly, higher Δ18K-FV3 than WT-FV3 genome copy number was detected at late stage (12 dpi) of infection. Infection in PLs and brain was similar between Δ18K-FV3 and WT-FV3.
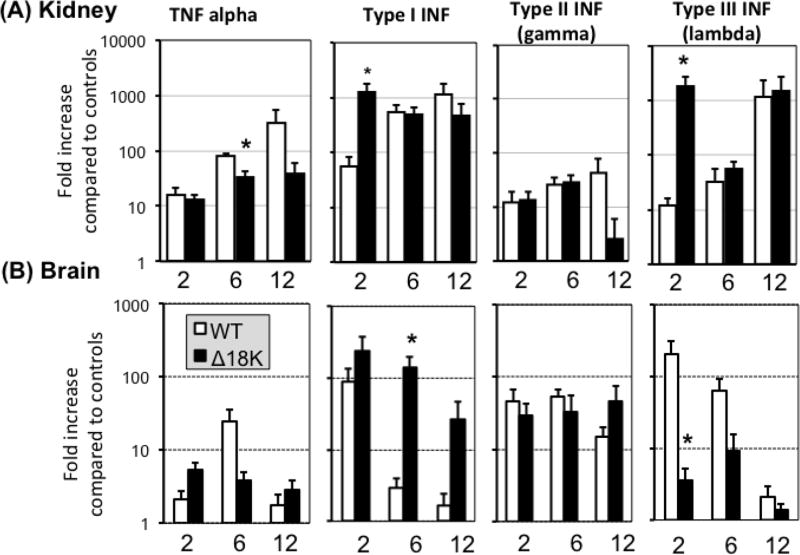
Reminiscent to what was observed in vitro using the A6 kidney cell line (Andino Fde et al., 2015), Δ18K-FV3 had the tendency to exacerbate tadpole IFN responses. In PLs, although high individual variation prevented to reach statistical significance, there was a trend toward higher expression of type I and type II IFN genes at 2 dpi in Δ18K-compared to WT-FV3 infected tadpoles (Suppl. Fig. 2). In kidneys, however, both type I and type III INF gene expression response was significantly increased at early stage of infection (2 dpi) with Δ18K-compared to WT-FV3 (Fig. 5A). Similarly, significantly higher type I IFN gene expression was triggered by Δ18K-FV3 than WT-FV3 the brain at 6 but not 2 dpi (Fig. 5B). In addition and in contrast to what was observed in PLs and kidneys, gene expression of type III IFN in the tadpole brain was significantly reduced in Δ18K-FV3 compared to WT-FV3 at 2 dpi.
Fig. 5. Changes in expression by RT-qPCR of TNFα, type I, II and III IFN genes in tadpole kidneys and brains during infection with Δ18K-FV3 compared to WT-FV3.
Outbred pre-metamorphic tadpoles were infected by i.p. injection of 1 ×104 PFU of Δ18K- or WT-FV3 for 2, 6 and 12 days. Results are average ± SEM fold increase relative to uninfected controls of 6 to 10 animals per group and are representative from two different experiments. **; P <0.001 and *; P <0.05 significant differences between WT- and Δ18K-FV3 using one-way ANOVA test and Tukey’s post hoc test.
Comparably, TNFα gene expression induced by Δ18K-FV3 was significantly reduced compared to WT-FV3 at 6 dpi in kidneys while no statistical difference was observed in the TNFα gene expression in the brain (Fig. 5 and Suppl. Fig. 2).
Infectivity KO-FV3 in X. laevis adults
To date the consequence of specifically deleting 64R, 52L and 18K genes on FV3 infectivity has been mainly characterized in X. laevis tadpoles, which are more susceptible than adult frogs and, as detailed in the introduction, rely on distinct antiviral immunity. Therefore, we were interested to determine in more detail the infection and replication efficiency of Δ64R-, Δ52- and Δ18K-FV3 in adult frogs.
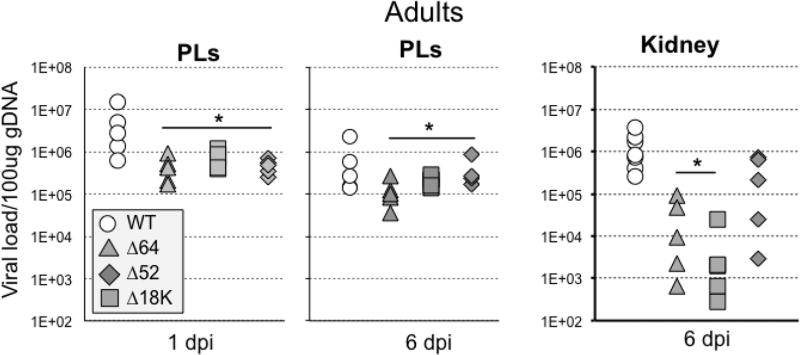
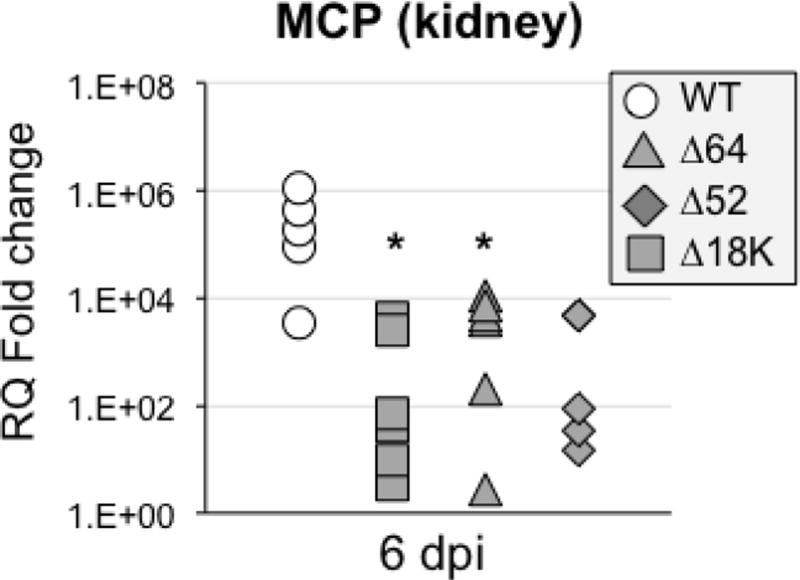
For Δ64R-, Δ52-FV3, while as previously reported the replication of these 2 KO mutant FV3 was impaired in adult kidneys (Andino Fde et al., 2015), the capacity to infect PLs at early (1 dpi) and later (6 dpi) stage of infection was also significantly reduced (Fig. 6). For Δ18K-FV3, the genome copy number was also significantly reduced in PLs at 1 and 6 dpi. Although Δ18K-FV3 viral load was lower at 6 dpi in kidney, it did not reach statistical significance in this experiment (Fig. 6). This may indicate some variability since in an independent smaller experiment, Δ18K-FV3 infection result in significantly lower genome copy numbers compared to WT-FV3 (Suppl. Fig 3). The drastic replicative impairment of Δ64R-FV3 and more moderate defect of Δ52L-FV3 are reflected in the number of infectious particles recovered from kidneys of infected tadpoles at 6 dpi (Table 1). Intriguingly, however, despite a relatively high genome copy number in experiment of Fig. 6, we barely detected any infectious Δ18K-FV3 by plaques assay at 6 dpi in the kidney of 6 animals in two independent experiments (Table 1). Moreover, consistent with a possible defect in the final generation of infectious viral particles, expression of the late viral gene MCP, an essential gene, was significantly reduced in Δ18K-FV3 infected kidney at 6 dpi (Fig. 7).
Fig. 6. Viral loads in PLs and kidneys of adult frogs at 1 and 6 days post-infection with WT-, Δ52L-, Δ64R- or Δ18-FV3.
Outbred adult frogs were infected by i.p. injection of 1 ×106 PFU of each virus type and FV3 genome copy numbers of PLs and kidneys at 1 and 6 dpi. were determined by absolute qPCR using primers specific for FV3 vDNA Pol II. Results are means ± SE of genome copy number/100µ of genomic DNA of 6 to 10 animals per group and are representative from two different experiments. **; P <0.001 and *; P <0.05 significant differences between WT and KO-FV3 using one-way ANOVA test and Tukey as post hoc test.
Table 1.
Number of FV3 infectious particles recovered from adult kidney at 6 dpi determined by plaques assay (2 independent experiments using 6 and 3 individual per group, respectively)
| Virus | Nb. animals | PFU/ml |
|---|---|---|
| FV3-WT | 6 | 1948±90 |
| FV3-Δ64 | 6 | 88±5 |
| FV-3Δ52 | 6 | 144±5 |
| FV3-Δ18K | 6 | ND |
| FV3-WT | 3 | 2440±890 |
| FV3-Δ64 | 3 | 305±91 |
| FV3-Δ18K | 3 | 70±35 |
Fig. 7. Changes in expression by RT-qPCR of the viral late gene MCP in infected adult frog kidneys.
Outbred adult frogs (6 individuals per group) were infected by i.p. injection of 1 ×106 PFU of each virus type for 6 days (dpi). Results are average ± SEM of RQ values. **; P <0.001 and *; P <0.05 significant differences between WT- and Δ18K-FV3 using one-way ANOVA test and Tukey’s post hoc test
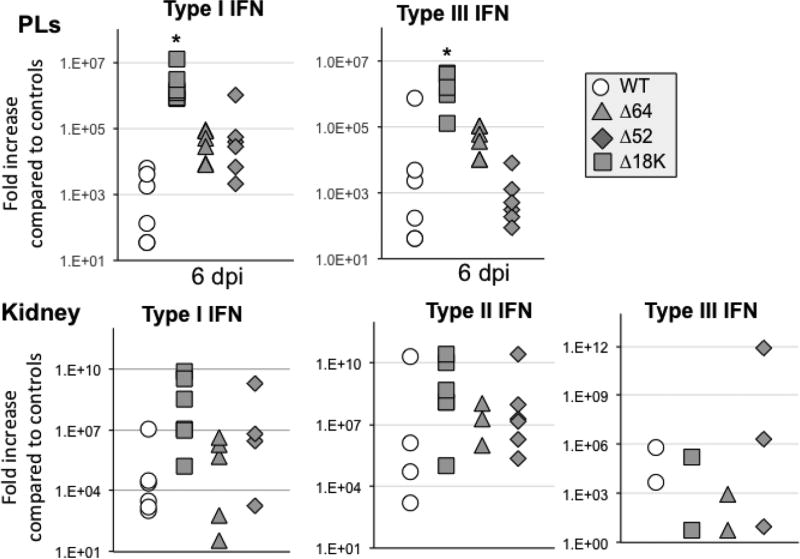
The induction of antiviral gene expression responses was not significantly altered at 6 dpi in PLs and kidneys of adult infected with either Δ64R- or Δ52-FV3 when compared to WT-FV3. However, the examination of host antiviral gene expression revealed that infection with Δ18K-FV3 in adults as in tadpoles resulted in an exacerbated expression of type I as well as type III INF genes at 6 dpi both in kidneys and PLs (Fig. 8).
Fig. 8. Changes in expression by RT-qPCR of type I and III IFN genes in adult frog PLs and kidneys during infection with Δ52L- or Δ64R- and Δ18K-FV3 compared to WT-FV3.
Outbred adult frogs (6 individuals per group) were infected by i.p. injection of 1 ×106 PFU of each virus type for 6 days (dpi). Results are average ± SEM fold increase relative to uninfected controls. **; P <0.001 and *; P <0.05 significant differences between WT- and KO-FV3 using one-way ANOVA test and Tukey’s post hoc test.
Discussion
Identifying and deciphering the function of viral genes that enable ranavirus pathogens to overcome host immune responses is in its infancy. The establishment of reliable methodologies to generate ranavirus recombinant deficient for specific ORFs constitutes a first critical step that not only permits the identification of non-essential genes candidates contributing to virulence or immune evasion but also provide stable deficient virus recombinant invaluable for uncovering mechanisms by which ranavirus interfere with host defenses. Although, investigation in vitro with host cell lines cultures can provide useful information, the complexity of the immune system ultimately requires in vivo studies using whole animals such as the amphibian Xenopus. Building upon our previous characterization of KO FV3 recombinants, here we have further investigated the infection patterns and host immune responses to 3 KO FV3 recombinants Δ64R, Δ52- and 18KΔ-FV3, both in tadpoles and adult frogs. Our data substantiate our initial, mainly in vitro, characterization, suggesting that these viral genes play distinct non-overlapping roles in promoting successful infection of X. laevis hosts.
64R vCARD-like encoding gene
Typically in eukaryotic cells CARD motifs are involved in interactions among various CARD-containing cellular proteins (Kawai and Akira, 2009, 2010). Cellular signaling molecules containing CARD domains of particular interest in antiviral responses include pro-apoptotic proteins, pro-inflammatory molecules and proteins participating in the cellular interferon responses (Besch et al., 2009; Meylan et al., 2005). Based on sequence similarity, the ranavirus vCARD-like protein could interact with one or several of these signaling molecules to circumvent the cellular antiviral responses. Our study using the X. laevis A6 kidney cells lines showed that in absence of the 64R gene encoding vCARD, FV3 was more susceptible to both type I and III IFN response and induced more apoptosis, suggesting that vCARD contributes in subverting the antiviral IFN response within infected cells (Andino Fde et al., 2015; Grayfer et al., 2015). But because as much IFN is induced by the vCARD KO mutant as WT virus, it was concluded that vCARD may not directly interfere with type I IFN synthesis (Andino Fde et al., 2015). In the present study, considering that IFN-λ gene expression in tadpoles at the site of infection in PLs and at the primary site of infection in kidneys is mostly unaltered, it is tempting to conclude that vCARD is critical for FV3 to overcome the tadpole IFN-λ antiviral system, which is prominent over type I IFN in tadpoles. The weaker type I IFN gene expression response induced by Δ64R-FV3 is likely related to the low viral loads. 64R-FV3 replication and persistence is also impaired in adult. However, it is interesting to note the higher viral load in adult PLs compared to tadpoles. The significant decrease in genome copy numbers in adult kidneys at 6dpi compared to adult PLs suggests a faster clearance of 64R-FV3. The higher type IFN gene response resulting from Δ64R-FV3 in the tadpole brain is more puzzling given the barely detectable viral load and IFN-λ gene expression. Further study will be needed to determine whether this could be due to some secondary effect deregulated viral gene expression and/or tissue specific effect.
52L vβHSD-like encoding gene
The ranavirus homolog of β-hydroxysteroid dehydrogenase (vβHSD) has been identified as another possible immune evasion protein. The eukaryotic gene β-hydroxysteroid dehydrogenase is required for the synthesis of progesterone, mineralocorticoids, and glucocorticoids (GCs; (Rhen and Cidlowski, 2005)). βHSD homologs are present within poxviruses and have been shown to play a role in dampening host immune responses (Moore and Smith, 1992; Sroller et al., 1998; Sun et al., 2006). Whether the putative ranavirus homolog of βHSD functions in the same way remains to be fully determined. Constitutive expression of the Rana grylio virus (RGV) β-HSD homolog slightly suppressed cytopathic effect and prolonged the viability of infected cells (Sun et al., 2006). Although, similar to vCARD, deletion of vβHSD encoded by 52L gene reduces the ability of FV3 to infect X. laevis A6 kidney cells in vitro and tadpole in vivo, the loss of vβHSD did not markedly increased apoptosis of infect A6 cells (Andino Fde et al., 2015). The present study provides further evidence of the distinct roles between vCARD and vβHSD in promoting FV3 infection. FV3 lacking vβHSD is impaired in viral replication, dissemination and persistence, albeit not as dramatically as vCARD deficient FV3.
The last putative virulence or immune evasion FV3 gene examined in this study is the enigmatic 18K or ICP18 gene. In FV3 as in other virus, this is an immediate early gene (Cheng et al., 2014). While the function of 18K remains unknown, its abundance and temporal class suggests that it is likely an important viral regulatory protein. Phylogenetic analysis of the amino acid sequence of the FV3 18K open reading frame indicates that it is conserved among members of the genus Ranavirus, but is not found among other genera (Lymphocystivirus, Megalocytivirus, or Iridovirus) within the family. Collectively, these data indicate that the 18K gene encodes a protein unique to the genus Ranavirus. The 18 kDa immediate-early protein is thought to be non-essential for replication in BHK and FHM cells (Sample et al., 2007). However, in vitro study with the X. laevis A6 kidney cell line has shown that Δ18K-FV3 is as resistant to type I and type III IFN inhibition as WT-FV3, but also markedly induces type IFN gene expression and apoptosis (Andino Fde et al., 2015). In the present in vivo study, we also found that infection of both tadpole and adults X. laevis with Δ18K-FV3 exacerbate the expression of the type I IFN gene. Notably, replication of Δ18K-FV3 is not as impaired as the two other KO FV3 as indicated by the substantial genome copy number found in different tissues of tadpoles and adults frogs. However, in adult we barely detected the production of infectious viral particles in kidneys by plaque assay, suggesting that the final step of viral assembly was not effective. Defect in release of infectious virus is also supported by the significant down expression of the FV3 MCP gene that is essential for the synthesis of the formation of the viral capside. These data are consistent with the idea that 18K defective FV3 exhibit a deregulated and overabundant production of viral genomes that may lead to a greater intracellular PKR detection/signaling and thus trigger increases in cellular apoptosis as well as an increase of IFN-I response. As such, 18K is a viral regulatory gene rather than a typical immune evasion gene.
Materials and Methods
Animals
Outbred (OB) pre-metamorphic X. laevis tadpoles (stage 54–56 / 3 weeks-old) and two-year adult frogs were obtained from our X. laevis research resource for immunology at the University of Rochester (https://www.urmc.rochester.edu/microbiology-immunology/xenopus-laevis.aspx). All animals were handled under strict laboratory and University Committee on Animal Resources regulations (100577/2003–151), and discomfort was minimized at all times.
Animal Infections
Three weeks-old tadpoles (developmental stage 54–56; (Nieuwkoop and Faber, 1994)) were infected by intraperitoneal (i.p.) injection with 1 × 104 PFU of FV3 in 10 µL volume of amphibian phosphate-buffered saline (APBS) using a glass Pasteur pipette with the extremity elongated by flame and attached to rubber tubing (Gantress et al., 2003). Adult frogs were infected by i.p. injection with 1 × 106 PFU of FV3 in 100 µL volume APBS using a 1 ml sterile syringe with a 22 gauge, 1½ inch needle. Mock-infected controls (0 days post-infection, d.p.i.) were ip injected with the same amount of amphibian phosphate-buffered saline (APBS). Animals were euthanized by immersion in 1% tricaine methane sulfonate (TMS-222) buffered with bicarbonate. At the indicated times, peritoneal leukocytes were collected by peritoneal lavage alternatively, animals were euthanized by immersion in 1% tricane methane sulfonate (MS-222), and tissues were removed and processed for RNA and DNA isolation.
Cell lines and FV3 Stocks
High titer of WT-FV3 (Granoff et al., 1965); ATCCVR-569), Δ64R-FV3, Δ52-FV3 (Andino Fde et al., 2015) stocks were produced using Baby hamster kidney-21 cells (BHK-21; ATCC no. CCL-10) that were cultured in Dulbecco’s modified Eagle’s medium (DMEM; Invitrogen) supplemented with 10% fetal bovine serum (FBS; Invitrogen), penicillin (100 U/mL) and streptomycin (100 µg/mL) with 5% CO2 at 37°C. FV3 was grown by a single passage on BHK-21 cells, purified by ultracentrifugation on a 30% sucrose gradient. Virus was quantified by plaque assay on BHK-21 monolayers in 6-well plates under an overlay of 1% methylcellulose (Morales et al., 2010). Infected cells were cultured 7 days at 30°C in 5% CO2. Overlay media was aspirated and the cells stained for 10 min with 1% crystal violet in 20% ethanol.
Genome copy number and gene expression analysis
Genomic DNA and total RNA were isolated from tissues or cells using Trizol reagent (Invitrogen) following the manufacturer's protocol. 1 µg total RNA was transcribed into cDNA with iScript reverse transcriptase using oligo-dT primers (Bio-Rad). Quantitative PCR parameters were as follows: 2 min at 95°C followed by 40 cycles of 95°C for 15 s and 60°C for 1 min. Relative quantitative PCR (qPCR) gene expression analysis (type I IFN, type II IFN, type III IFN and TNF-α) was performed using the ΔΔCT method. Expression of the different genes was examined relative to the endogenous GAPDH control and normalized against the expression of each gene compared to APBS injected control groups. Absolute qPCR was performed to measure FV3 viral loads in isolated genomic DNA, using a serially diluted standard curve, as previously described (Grayfer et al., 2014a). All primers were validated prior to use. All primer sequences are listed in Table 1.
Statistical analysis
All quantitative data were analyzed using by a one-way test of variance (ANOVA) followed by pos-hoc analysis using the Vassar Stat software (http://faculty.vassar.edu/lowry//anovalu.html). A p value < 0.05 was considered significant.
Supplementary Material
Table 2.
List of primer sequences
| PRIMER | SEQUENCE (5’– 3’) |
|---|---|
| DNA Pol II | F: ACGAGCCCGACGAAGACTACA |
| R: TGGTGGTCCTCAGCATCC T | |
| GAPDH | F: GACATCAAGGCCGCCATTAAGACT |
| R: AGATGGAGGAGTGAGTGTCACCAT | |
| Type I IFN | F: GCTGCTCCTGCTCAGTCTCA |
| R: GAAAGCCTTCAGGATCTGTGTGT | |
| Type II (gamma) | F:CTGAGGAAATACTTTAACTCCATTGACC |
| R:TTGTAACATCTCCCACCTGTATTGTC | |
| Type II IFN (lambda) | F: TCCCTCCCAACAGCTCATG |
| R: CCGACACACTGAGCGGAAA |
F: Forward; R: Reverse. Sequence and more information also available online at https://www.urmc.rochester.edu/microbiology-immunology/xenopus-laevis/primers.aspx
Highlights.
The X. laevis-FV3 model permits basic understanding of immunity to ranavirus
64R and 52L counteract type III IFN in tadpoles and type I IFN in adult frogs
64R and 52L are critical immune evasion genes in Xenopus adults and tadpoles
18K deletion exacerbates IFN responses and impairs release of infectious FV3
18K is a viral regulatory gene rather than immune evasion gene
Acknowledgments
This research was supported by National Institutes of Health Grant R25-GM064133 (to O. T.-L. and J.S.), R24-AI-059830 and National Sciences Foundation IOB-074271 (to E.-S.E., F.D.A. and J.R.). We would like to thank Tina Martin for animal husbandry and Let Yi for her input in the design of Fig. 1.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Andino Fde J, Grayfer L, Chen G, Chinchar VG, Edholm ES, Robert J. Characterization of Frog Virus 3 knockout mutants lacking putative virulence genes. Virology. 2015;485:162–170. doi: 10.1016/j.virol.2015.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayley AE, Hill BJ, Feist SW. Susceptibility of the European common frog Rana temporaria to a panel of ranavirus isolates from fish and amphibian hosts. Diseases of aquatic organisms. 2013;103:171–183. doi: 10.3354/dao02574. [DOI] [PubMed] [Google Scholar]
- Besch R, Poeck H, Hohenauer T, Senft D, Hacker G, Berking C, Hornung V, Endres S, Ruzicka T, Rothenfusser S, Hartmann G. Proapoptotic signaling induced by RIG-I and MDA-5 results in type I interferon-independent apoptosis in human melanoma cells. The Journal of clinical investigation. 2009;119:2399–2411. doi: 10.1172/JCI37155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blomberg B, Bernard CC, Du Pasquier L. In vitro evidence for T-B lymphocyte collaboration in the clawed toad, Xenopus. European journal of immunology. 1980;10:869–876. doi: 10.1002/eji.1830101112. [DOI] [PubMed] [Google Scholar]
- Brenes R, Gray MJ, Waltzek TB, Wilkes RP, Miller DL. Transmission of ranavirus between ectothermic vertebrate hosts. PloS one. 2014;9:e92476. doi: 10.1371/journal.pone.0092476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen G, Robert J. Antiviral immunity in amphibians. Viruses. 2011;3:2065–2086. doi: 10.3390/v3112065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen G, Robert J. Antiviral immunity in amphibians. Viruses. 2012;3:2065–2086. doi: 10.3390/v3112065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen G, Ward BM, Yu KH, Chinchar VG, Robert J. Improved knockout methodology reveals that frog virus 3 mutants lacking either the 18K immediate-early gene or the truncated vIF-2alpha gene are defective for replication and growth in vivo. Journal of virology. 2011;85:11131–11138. doi: 10.1128/JVI.05589-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng K, Escalon BL, Robert J, Chinchar VG, Garcia-Reyero N. Differential transcription of fathead minnow immune-related genes following infection with frog virus 3, an emerging pathogen of ectothermic vertebrates. Virology. 2014;456–457:77–86. doi: 10.1016/j.virol.2014.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinchar VG, Hyatt A, Miyazaki T, Williams T. Family Iridoviridae: poor viral relations no longer. Curr Top Microbiol Immunol. 2009;328:123–170. doi: 10.1007/978-3-540-68618-7_4. [DOI] [PubMed] [Google Scholar]
- Chinchar VG, Waltzek TB. Ranaviruses: not just for frogs. PLoS pathogens. 2014;10:e1003850. doi: 10.1371/journal.ppat.1003850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins JP. Amphibian decline and extinction: what we know and what we need to learn. Dis Aquat Organ. 92:93–99. doi: 10.3354/dao02307. [DOI] [PubMed] [Google Scholar]
- Cuesta A, Tafalla C. Transcription of immune genes upon challenge with viral hemorrhagic septicemia virus (VHSV) in DNA vaccinated rainbow trout (Oncorhynchus mykiss) Vaccine. 2009;27:280–289. doi: 10.1016/j.vaccine.2008.10.029. [DOI] [PubMed] [Google Scholar]
- Daszak P, Berger L, Cunningham AA, Hyatt AD, Green DE, Speare R. Emerging infectious diseases and amphibian population declines. Emerg Infect Dis. 1999;5:735–748. doi: 10.3201/eid0506.990601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Jesus Andino F, Chen G, Li Z, Grayfer L, Robert J. Susceptibility of Xenopus laevis tadpoles to infection by the ranavirus Frog-Virus 3 correlates with a reduced and delayed innate immune response in comparison with adult frogs. Virology. 2012;432:435–443. doi: 10.1016/j.virol.2012.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Jesus Andino F, Jones L, Maggirwar SB, Robert J. Frog Virus 3 dissemination in the brain of tadpoles, but not in adult Xenopus, involves blood brain barrier dysfunction. Scientific reports. 2016;6:22508. doi: 10.1038/srep22508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Jesus Andino F, Lawrence BP, Robert J. Long term effects of carbaryl exposure on antiviral immune responses in Xenopus laevis. Chemosphere. 2017;170:169–175. doi: 10.1016/j.chemosphere.2016.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duffus A, Waltzek T, Stöhr A, Allender M, Gotesman M, Whittington R, Hick P, Hines M, Marschang R. Distribution and Host Range of Ranaviruses. In: Gray MJ, Chinchar VG, editors. Ranaviruses: Lethal Pathogens of Ectothermic Vertebrates. Springer: 2015. pp. 9–59. [Google Scholar]
- Edholm ES, Albertorio Saez LM, Gill AL, Gill SR, Grayfer L, Haynes N, Myers JR, Robert J. Nonclassical MHC class I-dependent invariant T cells are evolutionarily conserved and prominent from early development in amphibians. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:14342–14347. doi: 10.1073/pnas.1309840110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edholm ES, Grayfer L, De Jesus Andino F, Robert J. Nonclassical MHC-Restricted Invariant Valpha6 T Cells Are Critical for Efficient Early Innate Antiviral Immunity in the Amphibian Xenopus laevis. Journal of immunology (Baltimore, Md: 1950) 2015;195:576–586. doi: 10.4049/jimmunol.1500458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forzan MJ, Wood J. Low detection of ranavirus DNA in wild postmetamorphic green frogs, Rana (Lithobates) clamitans, despite previous or concurrent tadpole mortality. Journal of wildlife diseases. 2013;49:879–886. doi: 10.7589/2013-03-051. [DOI] [PubMed] [Google Scholar]
- Gantress J, Maniero GD, Cohen N, Robert J. Development and characterization of a model system to study amphibian immune responses to iridoviruses. Virology. 2003;311:254–262. doi: 10.1016/s0042-6822(03)00151-x. [DOI] [PubMed] [Google Scholar]
- Gendrault JL, Steffan AM, Bingen A, Kirn A. Penetration and uncoating of frog virus 3 (FV3) in cultured rat Kupffer cells. Virology. 1981;112:375–384. doi: 10.1016/0042-6822(81)90284-1. [DOI] [PubMed] [Google Scholar]
- Granoff A, Came PE, Rafferty KA., Jr The isolation and properties of viruses from Rana pipiens: their possible relationship to the renal adenocarcinoma of the leopard frog. Ann N Y Acad Sci. 1965;126:237–255. doi: 10.1111/j.1749-6632.1965.tb14278.x. [DOI] [PubMed] [Google Scholar]
- Gray MJ, Miller DL, Hoverman JT. Ecology and pathology of amphibian ranaviruses. Dis Aquat Organ. 2009;87:243–266. doi: 10.3354/dao02138. [DOI] [PubMed] [Google Scholar]
- Grayfer L, De Jesus Andino F, Robert J. The amphibian (Xenopus laevis) type I interferon response to frog virus 3: new insight into ranavirus pathogenicity. Journal of virology. 2014a;88:5766–5777. doi: 10.1128/JVI.00223-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grayfer L, De Jesus Andino F, Robert J. Prominent amphibian (Xenopus laevis) tadpole type III interferon response to the frog virus 3 ranavirus. Journal of virology. 2015;89:5072–5082. doi: 10.1128/JVI.00051-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grayfer L, Edholm ES, Robert J. Mechanisms of amphibian macrophage development: characterization of the Xenopus laevis colony-stimulating factor-1 receptor. The International journal of developmental biology. 2014b;58:757–766. doi: 10.1387/ijdb.140271jr. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grayfer L, Robert J. Divergent antiviral roles of amphibian (Xenopus laevis) macrophages elicited by colony-stimulating factor-1 and interleukin-34. Journal of leukocyte biology. 2014;96:1143–1153. doi: 10.1189/jlb.4A0614-295R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grayfer L, Robert J. Distinct functional roles of amphibian (Xenopus laevis) colony-stimulating factor-1- and interleukin-34-derived macrophages. Journal of leukocyte biology. 2015;98:641–649. doi: 10.1189/jlb.4AB0315-117RR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grayfer L, Robert J. Amphibian macrophage development and antiviral defenses. Developmental and comparative immunology. 2016;58:60–67. doi: 10.1016/j.dci.2015.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gut JP, Anton M, Bingen A, Vetter JM, Kirn A. Frog virus 3 induces a fatal hepatitis in rats. Laboratory investigation; a journal of technical methods and pathology. 1981;45:218–228. [PubMed] [Google Scholar]
- Hausmann JC, Wack AN, Allender MC, Cranfield MR, Murphy KJ, Barrett K, Romero JL, Wellehan JF, Blum SA, Zink MC, Bronson E. EXPERIMENTAL CHALLENGE STUDY OF FV3-LIKE RANAVIRUS INFECTION IN PREVIOUSLY FV3-LIKE RANAVIRUS INFECTED EASTERN BOX TURTLES (TERRAPENE CAROLINA CAROLINA) TO ASSESS INFECTION AND SURVIVAL. Journal of zoo and wildlife medicine : official publication of the American Association of Zoo Veterinarians. 2015;46:732–746. doi: 10.1638/2015-0022.1. [DOI] [PubMed] [Google Scholar]
- Hellsten Uffe ea. The genome of the western clawed frog Xenopus tropicalis. 2009 doi: 10.1126/science.1183670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoverman JT, Gray MJ, Haislip NA, Miller DL. Phylogeny, life history, and ecology contribute to differences in amphibian susceptibility to ranaviruses. Ecohealth. 8:301–319. doi: 10.1007/s10393-011-0717-7. [DOI] [PubMed] [Google Scholar]
- Huber TL, Zon LI. Transcriptional regulation of blood formation during Xenopus development. Seminars in immunology. 1998;10:103–109. doi: 10.1006/smim.1998.0111. [DOI] [PubMed] [Google Scholar]
- Jancovich JK, Jacobs BL. Innate immune evasion mediated by the Ambystoma tigrinum virus eukaryotic translation initiation factor 2alpha homologue. Journal of virology. 2011;85:5061–5069. doi: 10.1128/JVI.01488-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson AJ, Pessier AP, Wellehan JF, Childress A, Norton TM, Stedman NL, Bloom DC, Belzer W, Titus VR, Wagner R, Brooks JW, Spratt J, Jacobson ER. Ranavirus infection of free-ranging and captive box turtles and tortoises in the United States. J Wildl Dis. 2008;44:851–863. doi: 10.7589/0090-3558-44.4.851. [DOI] [PubMed] [Google Scholar]
- Kawai T, Akira S. The roles of TLRs, RLRs and NLRs in pathogen recognition. International immunology. 2009;21:317–337. doi: 10.1093/intimm/dxp017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawai T, Akira S. The role of pattern-recognition receptors in innate immunity: update on Toll-like receptors. Nature immunology. 2010;11:373–384. doi: 10.1038/ni.1863. [DOI] [PubMed] [Google Scholar]
- Kik M, Martel A, Sluijs AS, Pasmans F, Wohlsein P, Grone A, Rijks JM. Ranavirus-associated mass mortality in wild amphibians, the Netherlands, 2010: a first report. Veterinary journal (London, England : 1997) 2011;190:284–286. doi: 10.1016/j.tvjl.2011.08.031. [DOI] [PubMed] [Google Scholar]
- Kolby JE, Smith KM, Berger L, Karesh WB, Preston A, Pessier AP, Skerratt LF. First evidence of amphibian chytrid fungus (Batrachochytrium dendrobatidis) and ranavirus in Hong Kong amphibian trade. PloS one. 2014;9:e90750. doi: 10.1371/journal.pone.0090750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolby JE, Smith KM, Ramirez SD, Rabemananjara F, Pessier AP, Brunner JL, Goldberg CS, Berger L, Skerratt LF. Rapid Response to Evaluate the Presence of Amphibian Chytrid Fungus (Batrachochytrium dendrobatidis) and Ranavirus in Wild Amphibian Populations in Madagascar. PloS one. 2015;10:e0125330. doi: 10.1371/journal.pone.0125330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koubourli DV, Wendel ES, Yaparla A, Ghaul JR, Grayfer L. Immune roles of amphibian (Xenopus laevis) tadpole granulocytes during Frog Virus 3 ranavirus infections. Developmental and comparative immunology. 2017 doi: 10.1016/j.dci.2017.02.016. [DOI] [PubMed] [Google Scholar]
- Landsberg JH, Kiryu Y, Tabuchi M, Waltzek TB, Enge KM, Reintjes-Tolen S, Preston A, Pessier AP. Co-infection by alveolate parasites and frog virus 3-like ranavirus during an amphibian larval mortality event in Florida, USA. Diseases of aquatic organisms. 2013;105:89–99. doi: 10.3354/dao02625. [DOI] [PubMed] [Google Scholar]
- Mahalingam S, Foster PS, Lobigs M, Farber JM, Karupiah G. Interferon-inducible chemokines and immunity to poxvirus infections. Immunological reviews. 2000;177:127–133. doi: 10.1034/j.1600-065x.2000.17720.x. [DOI] [PubMed] [Google Scholar]
- Maniero GD, Morales H, Gantress J, Robert J. Generation of a long-lasting, protective, and neutralizing antibody response to the ranavirus FV3 by the frog Xenopus. Developmental and comparative immunology. 2006;30:649–657. doi: 10.1016/j.dci.2005.09.007. [DOI] [PubMed] [Google Scholar]
- Mao J, Green DE, Fellers G, Chinchar VG. Molecular characterization of iridoviruses isolated from sympatric amphibians and fish. Virus Res. 1999;63:45–52. doi: 10.1016/s0168-1702(99)00057-x. [DOI] [PubMed] [Google Scholar]
- Marr S, Morales H, Bottaro A, Cooper M, Flajnik M, Robert J. Localization and differential expression of activation-induced cytidine deaminase in the amphibian Xenopus upon antigen stimulation and during early development. Journal of immunology (Baltimore, Md.: 1950) 2007;179:6783–6789. doi: 10.4049/jimmunol.179.10.6783. [DOI] [PubMed] [Google Scholar]
- Meylan E, Curran J, Hofmann K, Moradpour D, Binder M, Bartenschlager R, Tschopp J. Cardif is an adaptor protein in the RIG-I antiviral pathway and is targeted by hepatitis C virus. Nature. 2005;437:1167–1172. doi: 10.1038/nature04193. [DOI] [PubMed] [Google Scholar]
- Moore JB, Smith GL. Steroid hormone synthesis by a vaccinia enzyme: a new type of virus virulence factor. The EMBO journal. 1992;11:1973–1980. doi: 10.1002/j.1460-2075.1992.tb05251.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morales HD, Abramowitz L, Gertz J, Sowa J, Vogel A, Robert J. Innate immune responses and permissiveness to ranavirus infection of peritoneal leukocytes in the frog Xenopus laevis. Journal of virology. 2010;84:4912–4922. doi: 10.1128/JVI.02486-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morales HD, Robert J. Characterization of primary and memory CD8 T-cell responses against ranavirus (FV3) in Xenopus laevis. Journal of virology. 2007;81:2240–2248. doi: 10.1128/JVI.01104-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieuwkoop PD, Faber J. Normal tables of Xenopus laevis (Daudin) Garland Publishing; New York, London: 1994. [Google Scholar]
- Panchanathan V, Chaudhri G, Karupiah G. Protective immunity against secondary poxvirus infection is dependent on antibody but not on CD4 or CD8 T-cell function. Journal of virology. 2006;80:6333–6338. doi: 10.1128/JVI.00115-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panchanathan V, Chaudhri G, Karupiah G. Correlates of protective immunity in poxvirus infection: where does antibody stand? Immunology and cell biology. 2008;86:80–86. doi: 10.1038/sj.icb.7100118. [DOI] [PubMed] [Google Scholar]
- Price SJ, Garner TW, Balloux F, Ruis C, Paszkiewicz KH, Moore K, Griffiths AG. A de novo Assembly of the Common Frog (Rana temporaria) Transcriptome and Comparison of Transcription Following Exposure to Ranavirus and Batrachochytrium dendrobatidis. PloS one. 2015;10:e0130500. doi: 10.1371/journal.pone.0130500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price SJ, Garner TW, Nichols RA, Balloux F, Ayres C, Mora-Cabello de Alba A, Bosch J. Collapse of amphibian communities due to an introduced Ranavirus. Current biology : CB. 2014;24:2586–2591. doi: 10.1016/j.cub.2014.09.028. [DOI] [PubMed] [Google Scholar]
- Reeve BC, Crespi EJ, Whipps CM, Brunner JL. Natural stressors and ranavirus susceptibility in larval wood frogs (Rana sylvatica) EcoHealth. 2013;10:190–200. doi: 10.1007/s10393-013-0834-6. [DOI] [PubMed] [Google Scholar]
- Rhen T, Cidlowski JA. Antiinflammatory action of glucocorticoids--new mechanisms for old drugs. The New England journal of medicine. 2005;353:1711–1723. doi: 10.1056/NEJMra050541. [DOI] [PubMed] [Google Scholar]
- Robert J, Abramowitz L, Gantress J, Morales HD. Xenopus laevis: a possible vector of Ranavirus infection? Journal of wildlife diseases. 2007;43:645–652. doi: 10.7589/0090-3558-43.4.645. [DOI] [PubMed] [Google Scholar]
- Robert J, Edholm ES. A prominent role for invariant T cells in the amphibian Xenopus laevis tadpoles. Immunogenetics. 2014;66:513–523. doi: 10.1007/s00251-014-0781-6. [DOI] [PubMed] [Google Scholar]
- Robert J, Grayfer L, Edholm ES, Ward B, De Jesus Andino F. Inflammation-induced reactivation of the ranavirus Frog Virus 3 in asymptomatic Xenopus laevis. PloS one. 2014;9:e112904. doi: 10.1371/journal.pone.0112904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert J, Gregory Chinchar V. “Ranaviruses: an emerging threat to ectothermic vertebrates” report of the First International Symposium on Ranaviruses, Minneapolis MN July 8, 2011. Dev Comp Immunol. 2012;36:259–261. doi: 10.1016/j.dci.2011.08.008. [DOI] [PubMed] [Google Scholar]
- Robert J, Jancovich JK. Recombinant Ranaviruses for Studying Evolution of Host-Pathogen Interactions in Ectothermic Vertebrates. Viruses. 2016:8. doi: 10.3390/v8070187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert J, Morales H, Buck W, Cohen N, Marr S, Gantress J. Adaptive immunity and histopathology in frog virus 3-infected Xenopus. Virology. 2005;332:667–675. doi: 10.1016/j.virol.2004.12.012. [DOI] [PubMed] [Google Scholar]
- Robert J, Ohta Y. Comparative and developmental study of the immune system in Xenopus. Dev Dyn. 2009;238:1249–1270. doi: 10.1002/dvdy.21891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothenburg S, Chinchar VG, Dever TE. Characterization of a ranavirus inhibitor of the antiviral protein kinase PKR. BMC microbiology. 2011;11:56. doi: 10.1186/1471-2180-11-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothenburg S, Deigendesch N, Dey M, Dever TE, Tazi L. Double-stranded RNA-activated protein kinase PKR of fishes and amphibians: varying the number of double-stranded RNA binding domains and lineage-specific duplications. BMC biology. 2008;6:12. doi: 10.1186/1741-7007-6-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sample R, Bryan L, Long S, Majji S, Hoskins G, Sinning A, Olivier J, Chinchar VG. Inhibition of iridovirus protein synthesis and virus replication by antisense morpholino oligonucleotides targeted to the major capsid protein, the 18 kDa immediate-early protein, and a viral homolog of RNA polymerase II. Virology. 2007;358:311–320. doi: 10.1016/j.virol.2006.07.009. [DOI] [PubMed] [Google Scholar]
- Session AM, Uno Y, Kwon T, Chapman JA, Toyoda A, Takahashi S, Fukui A, Hikosaka A, Suzuki A, Kondo M, van Heeringen SJ, Quigley I, Heinz S, Ogino H, Ochi H, Hellsten U, Lyons JB, Simakov O, Putnam N, Stites J, Kuroki Y, Tanaka T, Michiue T, Watanabe M, Bogdanovic O, Lister R, Georgiou G, Paranjpe SS, van Kruijsbergen I, Shu S, Carlson J, Kinoshita T, Ohta Y, Mawaribuchi S, Jenkins J, Grimwood J, Schmutz J, Mitros T, Mozaffari SV, Suzuki Y, Haramoto Y, Yamamoto TS, Takagi C, Heald R, Miller K, Haudenschild C, Kitzman J, Nakayama T, Izutsu Y, Robert J, Fortriede J, Burns K, Lotay V, Karimi K, Yasuoka Y, Dichmann DS, Flajnik MF, Houston DW, Shendure J, DuPasquier L, Vize PD, Zorn AM, Ito M, Marcotte EM, Wallingford JB, Ito Y, Asashima M, Ueno N, Matsuda Y, Veenstra GJ, Fujiyama A, Harland RM, Taira M, Rokhsar DS. Genome evolution in the allotetraploid frog Xenopus laevis. Nature. 2016;538:336–343. doi: 10.1038/nature19840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sifkarovski J, Grayfer L, De Jesus Andino F, Lawrence BP, Robert J. Negative effects of low dose atrazine exposure on the development of effective immunity to FV3 in Xenopus laevis. Developmental and comparative immunology. 2014;47:52–58. doi: 10.1016/j.dci.2014.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somamoto T, Koppang EO, Fischer U. Antiviral functions of CD8(+) cytotoxic T cells in teleost fish. Developmental and comparative immunology. 2014;43:197–204. doi: 10.1016/j.dci.2013.07.014. [DOI] [PubMed] [Google Scholar]
- Sroller V, Kutinova L, Nemeckova S, Simonova V, Vonka V. Effect of 3-beta-hydroxysteroid dehydrogenase gene deletion on virulence and immunogenicity of different vaccinia viruses and their recombinants. Archives of virology. 1998;143:1311–1320. doi: 10.1007/s007050050377. [DOI] [PubMed] [Google Scholar]
- Sun W, Huang Y, Zhao Z, Gui J, Zhang Q. Characterization of the Rana grylio virus 3beta-hydroxysteroid dehydrogenase and its novel role in suppressing virus-induced cytopathic effect. Biochemical and biophysical research communications. 2006;351:44–50. doi: 10.1016/j.bbrc.2006.09.169. [DOI] [PubMed] [Google Scholar]
- Teacher AG, Garner TW, Nichols RA. Evidence for directional selection at a novel major histocompatibility class I marker in wild common frogs (Rana temporaria) exposed to a viral pathogen (Ranavirus) PloS one. 2009;4:e4616. doi: 10.1371/journal.pone.0004616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendel ES, Yaparla A, Koubourli DV, Grayfer L. Amphibian (Xenopus laevis) tadpoles and adult frogs mount distinct interferon responses to the Frog Virus 3 ranavirus. Virology. 2017;503:12–20. doi: 10.1016/j.virol.2017.01.001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.