Abstract
Droplet digital polymerase chain reaction (ddPCR) was adapted for quantifying the number of orthopoxviral genomes in purified virus samples, infected cell lysates and tissues of infected animals. In contrast to the more commonly used qPCR, the newer ddPCR provides absolute numbers of DNA copies in samples without need for standard curves and has the ability to detect rare mutants in a population. The genome/infectious unit ratio for several sucrose gradient-purified orthopoxviruses varied from 5 to 10, which correlated well with values obtained using the Virocyt, a dedicated fluorescence flow cytometer. By employing a nuclease step to digest unencapsulated DNA, the genome/infectious unit ratios of virus in crude cell lysates approached that of purified virus particles. The speed, accuracy, sensitivity, and dynamic range of less than one to millions of infectious units in a sample make this semi-automated method well suited to a variety of laboratory, animal and clinical studies.
Keywords: poxvirus quantification, poxvirus particle infectivity ratio, digital droplet polymerase chain reaction, vaccinia virus quantification
1. INTRODUCTION
Quantification of virus particles is essential for research, vaccine production, therapeutics and other biotechnology applications. Four types of viral quantification methods exist based on: infectivity, particle number, protein and nucleic acid (Heider and Metzner, 2014). Infectious particles are enumerated by plaque, focus forming or end-point infectivity assays. These methods are specific and can be performed with unpurified or purified preparations and simple equipment. Limitations include the need for appropriate cell substrates and relatively long times, frequently several days. A faster adaptation, involving recombinant viruses that express reporter genes such as green fluorescent protein or that use antibodies conjugated to fluorescent dyes for detection of infected cells by flow cytometry has been described and exploited for neutralization assays (Dominguez et al., 1998; Earl et al., 2003; Lambeth et al., 2005). Enumeration of physical particles, originally carried out by electron microscopy (Overman and Tamm, 1956), has been extended by several other detection methods including epifluorescence microscopy, atomic force microscopy, laser light scattering, nanoparticle tracking and flow cytometry (Heider and Metzner, 2014). The latter methods count non-infectious as well as infectious particles and may require purified, non-aggregated preparations for accuracy. Advantages include speed and ability to dispense with cell cultures. A third method that analyzes viral proteins by ELISA or other antibody-mediated detection is relatively fast and amenable to scale up but also counts molecules that are not associated with virions and requires standard curves. The fourth method detects nucleic acids, usually by qPCR and has advantages and some of the disadvantages of specific protein determination. ddPCR, a newer technology than qPCR, provides absolute quantification without need for standard curves as well as high sensitivity, reproducibility and semiautomation (Hindson et al., 2013; Strain et al., 2013). For ddPCR, a nucleic acid sample is partitioned in 20,000 water-in-oil droplets in which PCR reactions are carried out. After PCR amplification, the individual fluorescent positive and negative droplets are quantified to determine the number of DNA copies in the starting sample.
Although there have been a number of studies using qPCR to detect or quantify poxvirus DNA particularly for diagnostic and epidemiologic purposes (Baker and Ward, 2014; Lamien et al., 2011; Nitsche et al., 2004; Olson et al., 2004; Orba et al., 2015; Usme-Ciro et al., 2017), we are unaware of reports on the use of ddPCR. Members of the poxvirus family contain a single double-stranded DNA genome of approximately 200,000 bp enclosed within the core of a complex enveloped particle (Moss, 2013). The orthopoxvirus genus includes the best studied poxviruses including variola virus, the causative agent of smallpox; vaccinia virus (VACV) used as the vaccine to eradicate smallpox, currently employed as a vector for vaccines against other pathogens and cancer and enzootic in rodents in Brazil; cowpox virus (CPXV), the putative original smallpox vaccine and enzootic in northern Europe where it infects rodents and is transmitted to felines and humans; and monkeypox virus, which primarily infects rodents in central Africa and is transmitted to humans causing a smallpox-like disease. Here we describe an adaptation of ddPCR for quantification of poxvirus genomes. The specificity for DNA enclosed within virus particles was enhanced by incorporating nucleic acid digestion in the protocol and the ratio of total to infectious particles compared well with the values we obtained with the Virocyt, a dedicated virus flow cytometer (Ferris et al., 2011), and with previous data obtained by electron microscopy (Joklik, 1962) and qPCR (Baker and Ward, 2014). ddPCR offers advantages over other methods for the accurate quantification of poxvirus genomes and determination of the ratios of particles to infectious units for laboratory- and animal-based research and standardization of recombinant vaccines.
2. MATERIALS & METHODS
2.1. Cells and viruses
African green monkey kidney cells (BS-C-1) were grown and maintained at 37°C and 5% CO2 in modified Eagle minimal essential medium supplemented with 8% heat inactivated fetal bovine serum, 10 U of penicillin/ml, 10 mg streptomycin/ml, and 2 mM L-glutamine (Quality Biologicals, Gaithersburg, MD). The Western Reserve (WR) (ATCC #VR1354), modified vaccinia Ankara (MVA) (ATCC #VR-1508), recombinant GFP strains of vaccinia virus (VACV), and the Brighton (ATCC #VR-302) and CPXV_GER2002_MKY (Carroll et al., 2011) strains of cowpox virus (CPXV) were employed. For purification, virus particles were centrifuged once through a sucrose cushion for CPXV_GER2002_MKY or once through a sucrose cushion followed by two sucrose gradient sedimentations for all other viruses as described (Cotter et al., 2017). Virus titrations were performed in triplicate and dilutions made in duplicate for plaque assay on chick embryo cells for MVA and on BS-C-1 cells for all other viruses.
2.2. Extraction of DNA from infected cell lysates and purified virus particles
DNA was extracted from infected cell lysates or sucrose gradient purified virus containing 5×10−1 to 5×106 plaque forming units (PFU) using the DNeasy Blood and Tissue Kit or the QiaAMP Minelute Virus Spin Kit (Qiagen, Valencia, CA), respectively. Nucleic acids were eluted from the binding column with 80–120 μl of nuclease-free H2O (Thermo Fisher, Waltham, MA), divided into several vials and stored at −30°C until use. Where indicated, prior to DNA extraction the lysates or purified virus particles were treated with 18 units of Benzonase nuclease (Sigma, St. Louis, MO) for 1 h at 37°C in filter-sterilized sample buffer comprised of 50 mM Tris-HCl pH 8.0, 20 mM MgCl2 and 0.1 mg/ml bovine serum albumin (BSA). After 1 h, Benzonase was deactivated by the addition of EDTA (5 mM final) for 20 min at room temperature.
2.3. Quantification of DNA using ddPCR
Primers generating a 120 bp amplicon within the conserved VACV WR E11L open reading frame were synthesized and used for each of the orthopoxviruses in this study. The ddPCR reaction components and a small aliquot of each DNA sample were removed from −30°C storage, allowed to thaw at room temperature and diluted in nuclease-free H2O. For each reaction, 11 μl of 2× QX200 ddPCR EvaGreen supermix (Bio-rad, Hercules, CA), 0.3 μl of 20 μM forward primer (5′-GAATACATTCACATTGACCAATCAGAA-3′), 0.3 μl of 20 μM reverse primer (5′-GGTTCGTCAAAGACATAAAACTCATT-3′) and 1.5 μl of nuclease-free H2O were added to reaction wells of a semi-skirted twin-tec PCR plate (Eppendorf, MA). After addition of the ddPCR reagents, 9 μl of diluted DNA sample was added and mixed by manually pipetting the mixture up and down several times. The PCR plate was heat-sealed with pierceable foil and centrifuged for 1 min at 300×g to remove bubbles from the bottom of each well. The plate was then loaded into an Automated Droplet Generator (Bio-Rad) where samples were emulsified with oil and deposited into a new semi-skirted twin-tec PCR plate. After droplet generation, plates were heat-sealed with pierceable foil and placed in a thermocycler. With a ramp rate of 2°C/sec, PCR was performed under the following conditions: 95°C for 5 min, 40 cycles of 95°C for 30 sec, 40 cycles of 60°C for 1 min, 4°C for 5 min, 90°C for 5 min, 4°C standby. Upon completion of thermal cycling, the plate was seated into a QX200 Droplet Reader (Bio-Rad) for droplet acquisition and fluorescence quantitation. Data analysis was performed with QuantaLife software that uses Poisson distribution statistics to calculate the number of genome copies within a sample well. Analyses were performed on at least two occasions with three dilutions for each sample.
2.4. Quantification of virus particles with the Virocyt virus counter
Virus particles were quantified with a Virocyt 2100 Virus Counter (Virocyt, Boulder, CO). Prior to sample preparation and device operation, Virocyt performance was ensured by running startup/shutdown rinse solution, a performance validation standard and cleanliness verification fluid. Sample staining solution was prepared by adding 5 μl acetonitrile to a vial of combo dye and 500 μl combo dye buffer. The mixture was vortexed thoroughly and used within 2 h of preparation. Because of the narrow linear range of the device, gradient purified viruses were sonicated and diluted in sample buffer to achieve a concentration between 106 and 109 virus particles/ml. A sample to dye ratio of 2:1 was established by adding 100–300 μl of diluted sample to 50–150 μl of combo dye in a glass vial. Samples were vortexed and allowed to incubate in the dark at room temperature for 30 min before analysis. To eliminate carryover, inter-sample wash solution and cleanliness verification fluid were run between each sample. Virocyt software was used to calculate virus particles/ml.
2.5. Animal infections
Sucrose cushion or gradient purified VACV-WR, CPXV-BR, and CPXV_GER2002_MKY were diluted to their respective challenge dose concentration in sterile phosphate buffered saline containing 0.05% BSA. The virus concentration of each dilution used for inoculation was verified by plaque assay prior to inoculation of mice. CAST/EiJ (M. m. castaneous) or BALB/c mice were anesthetized by inhalation of isoflurane and intranasal or intraperitoneal infections were performed by instillation of 10–20 μl of virus into one nostril or 100–200 μl of virus into the peritoneum. Animals were weighed every other day and observed daily for morbidity and mortality. Animals that lost 30% of their starting weight were euthanized in accordance with NIAID Animal Care and Use protocols.
2.6. Sample processing and titration of virus from infected organs
On the day of death or euthanasia, infected tissues (lung, spleen, liver, ovary, and kidney) were extracted from animals and placed in 2 ml of Hanks Balanced Salt Solution with HEPES containing 0.1% BSA. Organ weights were determined and samples were frozen until further use. Organs were thawed at 37°C and subsequently homogenized using a mechanical grinder equipped with a hard-tissue probe. Homogenized samples were aliquoted, frozen and stored at −80°C until further use. On the day of titration, sample aliquots were thawed at 37°C and sonicated for 45 sec × 3 in ice-water. Sonicates were further clarified by centrifugation for 10 sec at 400×g. Several 10-fold dilutions of each sample were prepared in Eagle minimal essential medium supplemented with 2% heat inactivated fetal bovine serum, 10 U of penicillin/ml, 10 mg streptomycin/ml, and 2 mM L-glutamine. BS-C-1 cells were infected in duplicate with each dilution. Virus titers were determined by crystal violet staining of cell monolayers 48–72 h after infection.
2.7. DNA extraction from animal tissue
DNA was extracted from 2 to 15 mg of tissue using the DNeasy Blood and Tissue Kit (Qiagen) as per the manufacturer instructions. Bound DNA was eluted from the column with 165 μl of nuclease-free H2O, divided into several fresh vials and stored at −80°C. DNA samples were diluted and ddPCR was performed as described above.
3. RESULTS/DISCUSSION
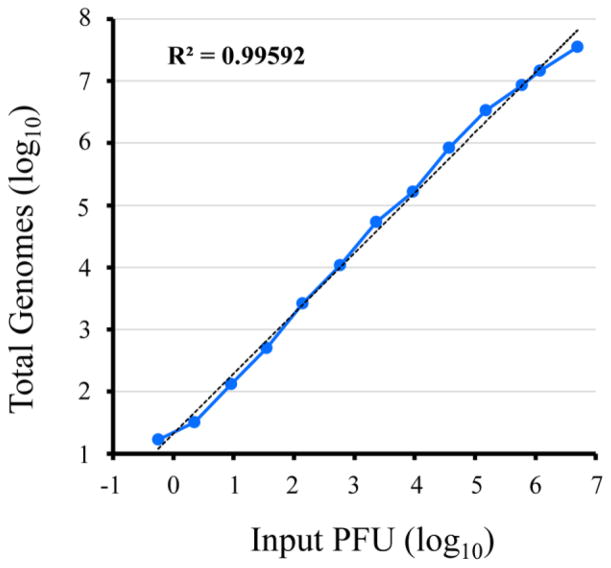
To adapt ddPCR for analysis of poxvirus genomes, we used the well characterized WR strain of VACV, which was purified from infected cells by sedimentation through a sucrose cushion followed by banding twice on sucrose gradients. PCR primer sets from several different open reading frames (A3L, E11L, I2L, D13L) gave comparable results with VACV WR DNA. The primer set derived from the E11L open reading frame, which was also tested with DNA from MVA, cowpox virus, monkeypox virus and ectromelia virus, was used for all experiments presented here. VACV was titered by plaque formation and dilutions were made to encompass a wide range of input virus prior to DNA extraction. Fig. 1 shows a plot of the number of VACV genomes versus PFU; linearity (R2=0.996) was obtained for the range of less than 1 PFU to 5 million PFU (Fig. 1). Departure from linearity occurred at higher PFU because the amount of DNA in the sample exceeded the binding capacity of the Qiagen Minelute Virus Spin Kit. The high dynamic range should enable the screening of large numbers of samples with a single dilution.
Fig. 1.
Dynamic range of ddPCR. A series of dilutions of sucrose gradient purified VACV strain WR was made to provide a range of 5.6×10−1 to 5×106 PFU. Each dilution was treated with Benzonase nuclease, which was then inactivated with EDTA. Viral DNA was extracted from each sample using the Qiagen Minelute Virus Spin Kit and then amplified with 40 cycles of PCR using primers derived from the VACV E11L gene. The ddPCR analysis was performed on reaction droplets. The plot represents the total number of extracted genomes detected from the corresponding input PFUs. Coefficient of determination (R2) from trend line analysis (dashed line) is provided in the upper left corner of plot.
For many reasons, the number of total viral particles of orthopoxviruses exceed the number of infectious particles (Lulf et al., 2016) and the determination of particle/PFU ratios are important when comparing different virus strains and mutant versus wild type virus. The genome/PFU ratios determined by ddPCR were compared to the particle/PFU ratios determined with the Virocyt, a dedicated flow cytometer that scores virions by coincidence of DNA and protein fluorescent staining. The average values for various PFU inputs were approximately 10 with both methods (Table 1) in agreement with the number determined by electron microscopy for orthopoxviruses (Joklik, 1962).
TABLE 1.
Comparison of genome and particle to PFU ratios
| PFU Input | Genomes/PFU | Particles/PFU |
|---|---|---|
|
| ||
| ddPCR (SEM) | Virocyt (SEM) | |
| 5.0 × 106 | 7 (.26) | 12 (2.2) |
| 2.5 × 106 | 10 (3.3) | 12 (2.4) |
| 1.2 × 106 | 12 (1.7) | 9 (1.8) |
| 6.0 × 105 | 14 (0.9) | 6 (1.7) |
| Average | 10.7 (1.5) | 9.7 (2.1) |
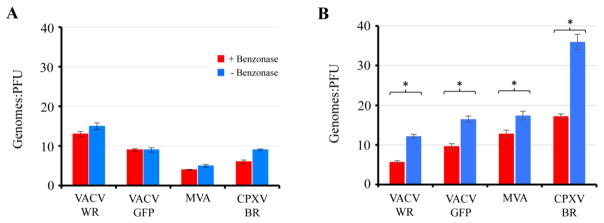
Benzonase, a nuclease that degrades all forms of DNA and RNA, does not diminish the infectivity of orthopoxviruses (Hughes et al., 2017). The effect of Benzonase treatment on genome quantification was assessed for both sucrose gradient purified virus, which may have adventitious DNA, and crude lysates in which there is unpackaged DNA-protein complexes and immature virions. Wild type and recombinant VACV WR, MVA and CPXV strain Brighton were analyzed using the same primer set. With purified virus particles, Benzonase had only a slight effect on the genome/PFU ratio (Fig. 2A). For MVA, the genome/PFU ratio was about 5, lower than that of VACV WR and similar to that obtained in another laboratory with the Virocyt (Lulf et al., 2016). Benzonase had a greater effect on crude lysates than on gradient-purified virus particles but the genome/PFU ratio remained higher in some cases perhaps because of non-infectious immature forms in the lysates (Fig. 2B).
Fig. 2.
Genome/PFU ratios of orthopoxviruses determined from purified particles and cell lysates. Approximately 6×105 to 1.2×106 PFU of VACV-WR, WR-GFP, MVA, and CPXV-BR were treated (+) or mock treated (−) with Benzonase nuclease for 1 h, after which viral DNA was isolated and amplified with 40 cycles of PCR using primers derived from the VACV E11L open reading frame. Genome/PFU ratios were determined for sucrose gradient purified virus particles (A) and crude lysate preparations (B). PFUs were determined in BS-C-1 cells for VACV and CPXV and in chick embryo cells for MVA. All data points were determined in triplicate and error bars indicate standard deviations. Asterisks represent p≦0.01.
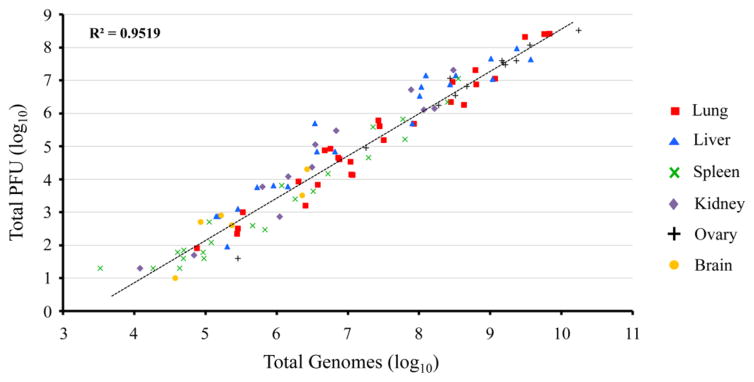
Animal experiments typically involve experimental and control virus infections, multiple animals in each group, and multiple infectious doses. For example, 300 samples would need to be assayed in a single experiment with one experimental and one control virus, three virus doses each, five animals per group and analysis of five organs at two time points. Furthermore, multiple dilutions of each sample would be needed for infectivity assays usually carried out in 6-well plates followed by manual counting of plaques. In contrast, ddPCR is carried out in 96-well plates and both analysis and data collection are semi-automated. To compare infectivity assays with genome numbers determined by ddPCR, we re-analyzed frozen tissue homogenates from a number of our previous unpublished experiments in which mice were infected with various doses of VACV-WR, CPXV-BR or CPXV_GER2002_MKY by intranasal or intraperitoneal routes. Because the homogenates were not treated with Benzonase, the genome numbers represent replicated DNA rather than mature virus particles. Nevertheless, a good correlation between the relative values for total genomes and PFU (R2=0.95) was obtained in the lung, liver, spleen, kidney, ovary, and brain (Fig. 3).
Fig. 3.
Correlation of viral genomes with total PFU in orthopoxvirus-infected organs. Mice were infected with varying doses of VACV-WR, CPXV-BR or CPXV_GER2002_MKY by intranasal or intraperitoneal routes of inoculation. Infected tissues were harvested on the day of death or euthanasia. Viral DNA from homogenized organs was isolated, amplified with 40 cycles of PCR using primers for the VACV E11L open reading frame, and ddPCR analysis performed in reaction droplets. Organ titers were determined by plaque assay on BS-C-1 cells. The relationship of total genomes and total PFU was determined and plotted for infected organs. Coefficient of determination (R2) from trend line analysis (dashed line) is provided in upper left corner of plot.
CONCLUSIONS
The high dynamic range of ddPCR, which reduces the number of dilutions needed for analysis, and the absence of tissue culture requirements make the method labor saving and cost effective compared to traditional plaque assays. Moreover, the procedure is semi-automated and can be completed in one working day with minimal hands-on time, whereas plaque titration requires advance preparation of plates containing cell monolayers, multiple virus dilutions, two to five days of incubation depending on the virus followed by the laborious step of counting and recording plaques all under appropriate biosafety conditions. This difference is amplified when large numbers of samples need to be analyzed for animal experiments and plaque titrations can easily take a week or more. Over the past two years ddPCR has also been used in our laboratory to determine particle/PFU ratios of mutant poxviruses that have diminished infectivity. In principal, ddPCR should also be useful for quantifying individual viruses in mixtures provided specific PCR primers are used. The wide dynamic range and sensitivity of ddPCR should also enable isolation of marker-free recombinant viruses by the technique of sib selection (McCormick, 1987). Although not described here, ddPCR is also advantageous for absolute quantification of viral and cellular mRNAs. Thus, ddPCR is a valuable addition to the virologist’s toolbox.
Highlights.
ddPCR provides absolute quantification of viral genomes
Semi-automated with high sensitivity and dynamic range
Applicable to infected cell lysates and animal tissues and to purified virions
Inclusion of a DNase step allows determination of particle numbers in crude extracts
Acknowledgments
We thank Catherine Cotter for providing some of the virus samples and assistance with the Virocyt. The research was supported by the Division of Intramural Research, NIAID, NIH.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Baker JL, Ward BM. Development and comparison of a quantitative TaqMan-MGB real-time PCR assay to three other methods of quantifying vaccinia virions. J Virol Meth. 2014;197:126–132. doi: 10.1016/j.jviromet.2013.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll DS, Emerson GL, Li Y, Sammons S, Olson V, Frace M, Nakazawa Y, Czerny CP, Tryland M, Kolodziejek J, Nowotny N, Olsen-Rasmussen M, Khristova M, Govil D, Karem K, Damon IK, Meyer H. Chasing Jenner’s vaccine: revisiting cowpox virus classification. Plos One. 2011;6:e23086. doi: 10.1371/journal.pone.0023086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotter CA, Earl PL, Wyatt LS, Moss B. Preparation of cell cultures and vaccinia virus stocks. Curr Protoc Mol Biol. 2017;117:16 16 11–16 16 18. doi: 10.1002/cpmb.33. [DOI] [PubMed] [Google Scholar]
- Dominguez J, Lorenzo MD, Blasco R. Green fluorescent protein expressed by a recombinant vaccinia virus permits early detection of infected cells by flow cytometry. J Immunol Meth. 1998;220:115–121. doi: 10.1016/s0022-1759(98)00156-2. [DOI] [PubMed] [Google Scholar]
- Earl PL, Americo JL, Moss B. Development and use of a vaccinia virus neutralization assay based on flow cytometric detection of green fluorescent protein. J Virol. 2003;77:10684–10688. doi: 10.1128/JVI.77.19.10684-10688.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferris MM, Stepp PC, Ranno KA, Mahmoud W, Ibbitson E, Jarvis J, Cox MM, Christensen K, Votaw H, Edwards DP, Rowlen KL. Evaluation of the Virus Counter(R) for rapid baculovirus quantitation. J Virol Meth. 2011;171:111–116. doi: 10.1016/j.jviromet.2010.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heider S, Metzner C. Quantitative real-time single particle analysis of virions. Virology. 2014;462–463C:199–206. doi: 10.1016/j.virol.2014.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hindson CM, Chevillet JR, Briggs HA, Gallichotte EN, Ruf IK, Hindson BJ, Vessella RL, Tewari M. Absolute quantification by droplet digital PCR versus analog real-time PCR. Nat Methods. 2013;10:1003–1005. doi: 10.1038/nmeth.2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes L, Wilkins K, Goldsmith CS, Smith S, Hudson P, Patel N, Karem K, Damon I, Li Y, Olson VA, Satheshkumar PS. A rapid Orthopoxvirus purification protocol suitable for high-containment laboratories. J Virol Meth. 2017;243:68–73. doi: 10.1016/j.jviromet.2017.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joklik WK. The purification of four strains of poxvirus. Virology. 1962;18:9–18. doi: 10.1016/0042-6822(62)90172-1. [DOI] [PubMed] [Google Scholar]
- Lambeth CR, White LJ, Johnston RE, de Silva AM. Flow cytometry-based assay for titrating dengue virus. J Clin Microbiol. 2005;43:3267–3272. doi: 10.1128/JCM.43.7.3267-3272.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamien CE, Lelenta M, Goger W, Silber R, Tuppurainen E, Matijevic M, Luckins AG, Diallo A. Real time PCR method for simultaneous detection, quantitation and differentiation of capripoxviruses. J Virol Meth. 2011;171:134–140. doi: 10.1016/j.jviromet.2010.10.014. [DOI] [PubMed] [Google Scholar]
- Lulf AT, Freudenstein A, Marr L, Sutter G, Volz A. Non-plaque-forming virions of Modified Vaccinia virus Ankara express viral genes. Virology. 2016;499:322–330. doi: 10.1016/j.virol.2016.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCormick M. Sib selection. Meth Enzymol. 1987;151:445–449. doi: 10.1016/s0076-6879(87)51036-9. [DOI] [PubMed] [Google Scholar]
- Moss B. Poxviridae. In: Knipe DM, Howley PM, editors. Fields Virology. Lippincott Williams & Wilkins; 2013. pp. 2129–2159. [Google Scholar]
- Nitsche A, Ellerbrok H, Pauli G. Detection of orthopoxvirus DNA by real-time PCR and identification of variola virus DNA by melting analysis. J Clin Microbiol. 2004;42:1207–1213. doi: 10.1128/JCM.42.3.1207-1213.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson VA, Laue T, Laker MT, Babkin IV, Drosten C, Shchelkunov SN, Niedrig M, Damon IK, Meyer H. Real-time PCR system for detection of orthopoxviruses and simultaneous identification of smallpox virus. J Clin Microbiol. 2004;42:1940–1946. doi: 10.1128/JCM.42.5.1940-1946.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orba Y, Sasaki M, Yamaguchi H, Ishii A, Thomas Y, Ogawa H, Hang’ombe BM, Mweene AS, Morikawa S, Saijo M, Sawa H. Orthopoxvirus infection among wildlife in Zambia. J Gen Virol. 2015;96:390–394. doi: 10.1099/vir.0.070219-0. [DOI] [PubMed] [Google Scholar]
- Overman JR, Tamm I. Equivalence between vaccinia particles counted by electron microscopy and infectious units of the virus. Proc Soc Exp Biol Med. 1956;92:806–810. doi: 10.3181/00379727-92-22621. [DOI] [PubMed] [Google Scholar]
- Strain MC, Lada SM, Luong T, Rought SE, Gianella S, Terry VH, Spina CA, Woelk CH, Richman DD. Highly precise measurement of HIV DNA by droplet digital PCR. PLoS One. 2013;8:e55943. doi: 10.1371/journal.pone.0055943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usme-Ciro JA, Paredes A, Walteros DM, Tolosa-Perez EN, Laiton-Donato K, Pinzon MD, Petersen BW, Gallardo-Romero NF, Li Y, Wilkins K, Davidson W, Gao JX, Patel N, Nakazawa Y, Reynolds MG, Satheshkumar PS, Emerson GL, Paez-Martinez A. Detection and molecular mharacterization of zoonotic poxviruses circulating in the Amazon region of Colombia, 2014. Emerg Infect Dis. 2017;23:649–653. doi: 10.3201/eid2304.161041. [DOI] [PMC free article] [PubMed] [Google Scholar]