Abstract
H4Nx viruses were reported in swine in Canada and China, but had not been recognized in swine in the USA. In late 2015, an avian-origin H4N6 influenza A virus was isolated from pigs in the United States during a routine diagnostic investigation of clinical respiratory disease in the herd. Serological analysis from additional pigs at the farm and other pigs within the swine production system indicated that the virus did not efficiently transmit from pig to pig and the mode of transmission to swine could not be determined. The isolate was characterized at the molecular level and the pathogenesis and transmission was experimentally evaluated in pigs. Although the virus replicated in the lungs of pigs and caused mild pulmonary lesions, there was no evidence of replication in the upper respiratory tract or transmission to indirect contacts, supporting the findings on the farm.
Keywords: avian influenza, H4N6, swine influenza, pathogenesis
1. Introduction
Influenza A virus (IAV) is an enveloped virus with a negative-sense RNA genome comprised of 8 segments. Wild aquatic bird populations are the natural reservoir for most IAV strains, including those of the subtypes H1-16/N1-9 (Joseph et al., 2016; Yoon et al., 2014), while bats have recently been described as a reservoir for H17-18/N10-11 (Tong et al., 2012; Tong et al., 2013). Despite the large amount of diversity of HA and NA subtypes circulating in birds, only H1N1, H1N2 and H3N2 subtypes are endemic in swine worldwide (Nelson et al., 2015b). Even though there are only few HA and NA subtypes co-circulating in swine, the ecology of IAV in swine is complex and is characterized by large genetic and antigenic diversity due primarily to human-to-swine spillover events. The diversity of contemporary strains co-circulating in the United States has been relatively well characterized in large part due to the US Department of Agriculture (USDA) surveillance efforts operated through the National Animal Health Laboratory Network and the National Veterinary Services Laboratories. The increased reporting and accessibility to publically available sequence data resulted in a more comprehensive genetic and antigenic understanding of circulating strains (Abente et al., 2016; Anderson et al., 2015; Anderson et al., 2013; Lewis et al., 2014). Furthermore, it facilitated the early detection of novel HA introductions into the swine population, such as a 2010–11 human H3 that has sustained onward transmission in the pig population (Rajao et al., 2015). Early detection of novel incursions such as these, particularly of antigenically distinct HA’s for which there is little to no population immunity through vaccination or exposure to circulating strains, are critical to prevent and intervene in such events and to identify potential vaccine strains for preventing spread of new introductions.
Avian lineage IAV have also played an important role in the ecology of IAV in swine through incursion of surface and internal genes. Historically, the first report of IAV in swine was concomitant with the spread of the 1918 “Spanish flu” (Koen, 1919), and an H1N1 was later isolated (Shope, 1931) and identified as belonging to what is now described as the classical swine H1N1 lineage. While the details of how the pandemic 1918 strain emerged have not been fully determined (Smith et al., 2009; Taubenberger, 2006; Taubenberger et al., 2005; Worobey et al., 2014a, b), genes of avian-origin were associated with the genesis of the classical swine H1N1 and have been associated with the emergence of other pandemic strains (Smith et al., 2009). Another major IAV swine lineage endemic in pigs on multiple continents is a direct incursion of an avian lineage IAV, the Eurasian avian-like H1N1 (Castrucci et al., 1993; Guan et al., 1996; Vincent et al., 2014), but spillover and onward transmission of non-reassortant avian viruses in swine is not common. Additionally, avian-origin PB2 and PA genes comprise the well-established triple reassortant internal gene cassette that continues to circulate in North America (Zhou et al., 1999), migrated to parts of Asia, and contributed to the 2009 H1N1 pandemic genome constellation.
Avian H4 IAV are known to have been circulating at least since 1956 when an H4N6 virus was isolated from a duck, with two well supported lineages: North American and Eurasian (Donis et al., 1989). Among the H4 HA sequences detected worldwide, publicly available in the Influenza Research Database, the majority were identified in avian species (2140/2235), a small number were found in mammals (5 in swine, 3 in sea mammals, and 2 in muskrats), 84 were detected in environmental samples and one was of unknown origin (Zhang et al., 2017). Pigs were experimentally challenged with 5 avian H4 strains from ducks in a comprehensive study that assessed the susceptibility of swine to 38 avian IAV strains (Kida et al., 1994). Virus was isolated from nasal swabs in 4 out of 5 H4-challenged pigs and duration of shedding varied from 4–7 days post infection. A recent study characterized 32 contemporary H4 viruses that were isolated from live poultry markets in China, and consistent with previous findings, all viruses could replicate in mice without adaptation (Liang et al., 2016). Additionally, they determined from a subset of 10 H4 viruses that 6 efficiently transmitted in guinea pigs, an animal model used to assess the potential risk of IAV to infect and circulate in mammals.
To date, three reports have documented the isolation of avian H4 viruses from naturally infected pigs exhibiting influenza-like symptoms (Hu et al., 2012; Karasin et al., 2000; Su et al., 2012). All three cases were self-limiting without sustained onward transmission or spread from the index herds. In two cases, H4 viruses were isolated from commercial farms during an outbreak that displayed respiratory disease (Karasin et al., 2000; Su et al., 2012). Pre- and post-outbreak serologic data were reported only from the outbreak that occurred in Ontario, Canada (Karasin et al., 2000). In that report, a small subset of sera (10) tested post-outbreak were all HI-positive against the H4N6 virus associated with the outbreak suggesting potential pig-to-pig transmission, although this was not experimentally confirmed.
Molecular determinants that allow adaptation of avian IAV to a new host species, particularly humans, have been described. Most studies examined the HA and its interaction with the host receptor, but the requirements for host-switch are complex. Interactions between the HA and sialic acids are important for initiating infection and can define species tropisms, and while this interaction has been studied extensively (de Graaf and Fouchier, 2014), other viral genes are also involved in host-switch events. The polymerase and non-structural genes are associated with the ability of viruses to replicate efficiently in the host cell, and specific interactions with host cellular proteins and modifications of the virus genome have been described (Long et al., 2016; Xu et al., 2016). Reassortment of gene segments is a hallmark of genetic evolution of IAV and can play a role in cross-species spread of IAV (Steel and Lowen, 2014).
Here, we report an H4N6 that was detected and isolated in December 2015 from pigs on a farm in Missouri that exhibited influenza-like symptoms through the USDA IAV surveillance system in swine. We describe the epidemiology associated with this avian H4N6 detection, the genetic characterization of the virus genome, and the results of a pathogenesis and transmission challenge study in weaned pigs.
2. Materials and methods
2.1. Collection and processing of clinical samples
On December 16, 2015, five nasal swab and five serum samples were submitted to the Iowa State University Veterinary Diagnostic Laboratory (ISU VDL) from a group of female pigs that had not yet given birth to a litter of piglets (gilts). The five gilts (approximately 7–8 months of age), pregnant at the time, demonstrated respiratory disease at a breeding and gestation farm located in Northeast Missouri. Clinical signs were observed only in the replacement gilt population and included coughing, lethargy, and anorexia. RNA was extracted from the five nasal swab samples and evaluated for the presence of porcine reproductive and respiratory syndrome virus (PRRSV) by RT-rPCR (VetMAX™ NA and EU PRRSV, ThermoFisher Scientific, Waltham, MA) and serum was used to test for PRRSV-specific antibodies by ELISA (IDEXX PRRS X3 Ab Test, IDEXX Laboratories, Inc., Westbrook, ME). Samples tested negative for PRRS virus and antibody. The samples were also tested by real-time RT-PCR for the presence of influenza A virus (VetMAX™-Gold SIV detection Kit, ThermoFisher Scientific, Waltham, MA), then tested for HA and NA subtyping specific for H1/H3 and N1/N2 (VetMAX™-Gold SIV Subtyping Kit, ThermoFisher Scientific, Waltham, MA). Samples were then tested for IAV isolation on Madin Darby Canine Kidney (MDCK) cells, and whole-genome sequencing and sequence analysis were performed on the successfully recovered virus isolate (described below).
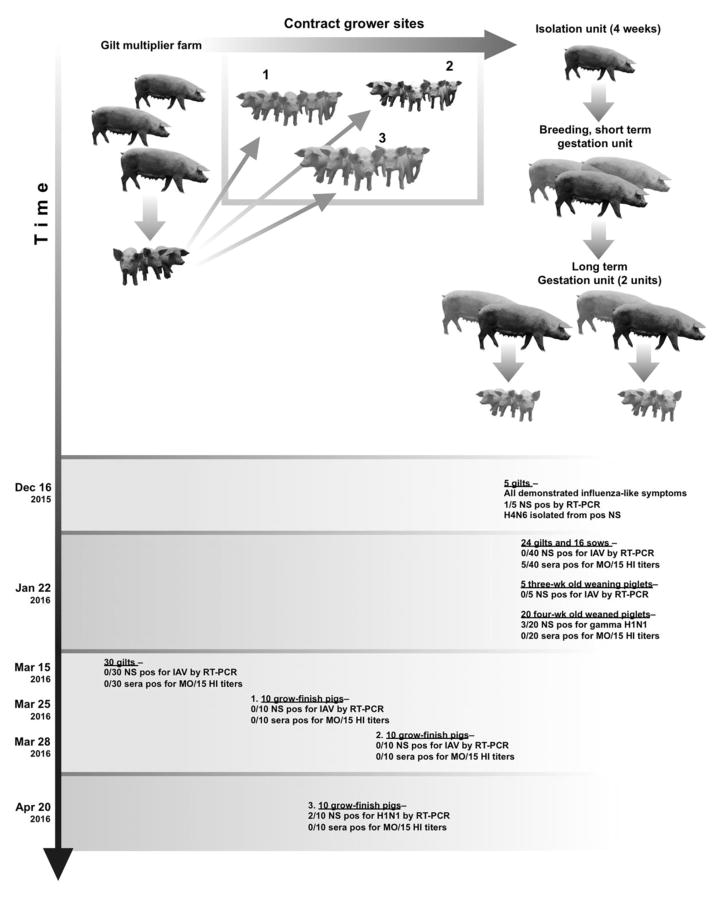
As a follow-up investigation, additional nasal swab and sera diagnostic samples were collected from the commercial breeding and gestation farm (index farm) to investigate the origin and presence of the H4N6. Diagnostic samples were also collected from additional production sites linked to the flow of pigs associated with the index farm, including an upstream breeding and gestation farm (gilt multiplier) that supplied new breeding-age females to the index herd and its associated nursery (~3–10 weeks of age) and grow-finish sites (~11–30 weeks of age), designated 1, 2 and 3 and known as contract facilities (Fig. 1). On January 22, 2016, thirty-seven days after the initial H4N6 detection, the submitting veterinarian collected sixty-five nasal swab samples and sixty sera from the H4-positive index farm. Samples were collected from twenty-four gilts approximately eight-months-old that included four of the five original gilts tested in December 2015 (one died prior to re-sampling), from sixteen sows (approximately 1–2 years of age and older) that had farrowed at least one litter, from five three-week-old piglets at weaning, and from twenty four-week-old piglets that were already weaned and moved to a downstream commercial nursery. On March 15, 2016, thirty nasal swabs and sera were collected from randomly selected gilts and sows approximately 1–4 years of age from the upstream gilt multiplier breeding and gestation farm. On March 25, March 28, and April 20, 2016, ten nasal swabs and sera were randomly collected from nursery pigs and grow-finish pigs located in the contract facilities 1, 2 and 3, respectively. Samples were processed and nasal swabs tested individually or pooled in groups of 5 for RT-PCR and sera tested individually for antibody to investigate if the H4N6 virus continued to circulate.
Figure 1.
Schematic diagram depicting the flow of pigs through the breeding and gestation farm. Replacement gilts were born on the upstream gilt multiplier breeding farm, and were transferred to three different contract grower facilities. When gilts reached the appropriate age, they were then moved to the downstream isolation unit where they were vaccinated against influenza (FluSure XP® Zoetis) and kept a minimum of 4 weeks. Gilts are then transferred to a breeding and early gestation unit where pigs are inseminated, and this is where the H4N6 index cases were detected. Once pregnancy is confirmed, pigs are transferred to one of two long-term gestation units. Piglets are sold and sows return to the breeding and gestation unit. Dates of clinical sample collection are noted and virologic and serologic results are described. Gilt, female pig that has not produced a litter. Sow, female pig that has already produced a litter.
2.2 Viruses and cell lines
A virus was isolated on Madin-Darby canine kidney (MDCK) cells from one RT-PCR positive nasal swab specimen collected on December 16, 2015 and submitted to the Swine IAV Surveillance System repository held at the USDA National Veterinary Service Laboratories (NVSL) in conjunction with the USDA National Animal Health Laboratory Network. Whole genome sequencing was performed at NVSL (in accordance with guidelines stated in the National Swine Influenza Virus Surveillance Plan (Anderson et al., 2013; APHIS, 2010) and submitted to GenBank (Benson et al., 2017; Clark et al., 2016). All eight segments of the virus were amplified using universal primers for each segment. cDNA was purified and cDNA libraries were prepared using the Nextera XT DNA Library Preparation Kit with Nextera XT Index Kit barcode adapters (Illumina, San Diego, CA). Prepared libraries were pooled and loaded onto a MiSeq Reagent Cartridge and a 500–cycle (2 × 250) PE kit v2 was used for sequencing. The isolated virus, A/swine/Missouri/A01727926/2015 (H4N6) (MO/15; GenBank Accession Numbers KU641618-KU641625), was passaged once in MDCK cells, and stored in the virus repository at NVSL. The virus isolate was passaged once more in MDCK cells to prepare inoculum for the animal study.
The following endemic swine viruses were used in addition to MO/15 H4N6 to test for hemagglutination inhibition (HI) titers against sera collected from pigs from the index herd as well as upstream and downstream epidemiologically-linked farms: A/swine/Ohio/511445/2007 (H1N1, gamma genetic lineage), A/swine/Illinois/003200/2010 (H1N2, delta1 genetic lineage), A/swine/Illinois/00685/2005 (H1N1, delta2 genetic lineage), A/swine/Missouri/A01410819/2014 (H3N1, human-like genetic lineage), A/swine/New York/A01104005/2011 (H3N2, IV-A genetic lineage), A/swine/Iowa/A01480656/2014 (H3N2, IV-A genetic lineage) (Abente et al., 2016; Anderson et al., 2015; Rajao et al., 2015).
2.3. Animal study
Twenty 4-week old crossbred pigs were obtained from a healthy herd for the challenge study. Pigs were seronegative to IAV antibodies against the nucleoprotein (NP) as measured by ELISA (Swine Influenza Virus antibody test, IDEXX, Westbrook, ME) and were negative (<10 reciprocal titer) by hemagglutination inhibition (HI) assay against MO/15 prior to challenge. Pigs were divided into three groups: non-challenged (n=5), challenged with MO/15 (n=10), and contact pigs (n=5). Pigs challenged with MO/15 were inoculated simultaneously via intranasal (1 ml via slowly dripping 0.5 ml of virus into each nostril) and intratracheal (2 ml via laryngoscope and tracheal tube) routes with 105 50% tissue culture infective dose (TCID50) per ml of virus. Inoculation was performed while the pigs were under anesthesia by an intramuscular injection of a cocktail of ketamine (8 mg/kg of body weight; Zoetis Animal Health, Florham Park, NJ), xylazine (4 mg/kg), and tiletamine-zolazepam (Telazol; 6 mg/kg) (Zoetis Animal Health, Florham Park, NJ). On 2 dpi, five naïve contact pigs were placed in a separate raised deck in the same room but with separate food and water and approximately 2 feet (0.6 meters) away from the inoculated group to evaluate indirect contact transmission.
Nasal swab samples were collected daily 1–5 days post infection (dpi) for negative control and challenged pigs, and 1–5, 7, and 9 days post contact (dpc) for contact pigs. Five challenged pigs and 5 naïve negative control pigs were humanely euthanized and necropsied at 5 dpi, at which time bronchoalveolar lavage fluid (BALF) and tissue samples from the distal trachea and right cardiac or affected lung lobe were collected. The remaining 5 challenged pigs and the 5 contact pigs were euthanized at 14 dpi and 12 dpc, respectively, to assess seroconversion.
2.4. Pathological examination of lungs
Tissue samples collected at necropsy were evaluated for the percentage of the lung affected by purple-red consolidation typical of IAV infection. The percentage of the surface of the entire lung affected by pneumonia was calculated on the basis of the weighted proportions of each lobe to the total lung volume as described previously (Gauger et al., 2012; Halbur et al., 1995). Microscopic lesions were evaluated and scored by a veterinary pathologist blinded to the treatment groups. The presence of IAV-specific antigen was examined in the trachea and lung tissues using immunohistochemistry (IHC) with a mouse monoclonal antibody (clone HB65) to detect NP. Microscopic lesions and IHC results were scored as described previously (Khurana et al., 2013).
2.5. Virus isolation and titers in nasal swab and BALF samples
For virus isolation, nasal swab samples were filtered (0.45 μm), and plated onto confluent MDCK cells in 24-well plates. Virus titration was performed in triplicate on confluent MDCK cells in 96-well plates. Virus isolation and viral titer assays were performed with 1 μg/ml tosylsulfonyl phenylalanyl chloromethyl ketone (TPCK)-trypsin. At 48 h, plates were fixed with 4% phosphate-buffered formalin and stained using a mouse monoclonal antibody (clone HB65) to detect NP.
2.6. Serology
Sera collected from the swine farm were treated with receptor-destroying enzyme (Denka Seiken, Japan), heat inactivated at 56°C for 30 min, and adsorbed with 50% turkey red blood cells (RBCs) to remove nonspecific hemagglutinin inhibitors and natural serum agglutinins. HI assays were performed with MO/15 and other reference swine H1 and H3 viruses as antigens and 0.5% turkey RBCs using standard techniques.
Homologous anti-sera against MO/15 were produced as described previously for use as controls to test for cross-reactivity to endemic swine reference antigens (Lewis et al., 2014). Briefly, two primary challenged pigs were immunized with 128 to 256 hemagglutinin units (HAU) of UV-inactivated wild type MO/15 combined with 20% commercial adjuvant (Emulsigen D; MVP Laboratories, NE, USA) by the intramuscular route and tested 2 weeks later. HI titers to homologous virus were 160, so the pigs were humanely euthanized with pentobarbital sodium (Fatal Plus; Vortech Pharmaceuticals, MI, USA) for blood collection. Sera were collected and stored at −20°C.
2.7. Phylogenetic analysis
The phylogenetic relationships of A/swine/Missouri/A01727926/2015 (H4N6; GenBank Accession Numbers KU641618-KU641625) to selected North American and Eurasian lineage reference strains were estimated from the nucleotide sequences of each viral gene. Blastn was used to identify gene sequences that were genetically similar to the gene segments of A/swine/Missouri/A01727926/2016. Sequences from 2012 to present were downloaded. We then manually subsampled to include different spatial and temporal representative sequences. Eurasian lineage reference strains were selected from a publication examining H4 avian influenza viruses isolated from live poultry markets in China (Liang et al., 2016). Nucleotide sequences were aligned using MAFFT v7.222 (Katoh and Standley, 2013) and then manually corrected in MEGA7 (Kumar, Stecher, and Tamura 2015). Maximum likelihood trees were inferred using IQ-TREE v1.4.3 (Nguyen et al., 2015) under a GTR+G+I evolutionary model. To assess branch support, all IQ-TREE analysis used the ultrafast bootstrap approximation (Minh et al., 2013) with 1000 replicates. The consensus trees were visualized and annotated using MEGA7.
3. Results
3.1. H4N6 detections in the index farm
Nasal swabs and sera were collected from 5 replacement gilts exhibiting influenza-like symptoms on December 16, 2015. One nasal swab from the diagnostic submission was RT-PCR positive in the IAV screening assay, so it was examined further to determine the HA and NA subtype using commercially available swine subtyping reagents. Subtyping results were unexpectedly negative, so a second test of the original sample using the screening real-time RT-PCR kit was conducted and remained positive with a similar cycle threshold value compared to the original positive result (33.8 and 31.1, respectively). Virus isolation was initiated only on the PCR-positive sample using MDCK cells per the USDA IAV surveillance protocol, and confirmed positive after cytopathic effect was observed. After two freeze-thaw cycles, 128 hemagglutination units were detected in the supernatant. RNA extracted from the virus isolation cell culture supernatant was tested with the real-time RT-PCR assay and was again confirmed IAV-positive. However, the H1/H3 and N1/N2 subtyping assays on the virus isolate remained negative. The virus was submitted to the IAV Swine Isolate Repository held at the USDA NVSL where whole-genome sequencing was carried out. The virus was characterized as a H4N6 IAV.
3.2. Pig-to-pig transmission of H4N6 was limited on index farm
Subsequent nasal swabs and sera were collected from the index farm and pig-flow related farms at different time points to examine if the H4N6 transmitted from pig-to-pig or if the source farm of its emergence could be determined (Fig. 1). Nasal swabs randomly collected at the index farm on January 22, 2016 from the same cohort of replacement gilts as the index case, adult-breeding sows, and five three-week-old piglets were negative for IAV by real-time RT-PCR. However, 15% (3/20) of the nasal swabs collected from four-week-old piglets that originated at the index farm and held at a separate off-site commercial nursery were positive for H1N1 based on RT-PCR. Subsequent sequencing of the HA gene, and submission to a publicly available subtyping tool (Anderson et al., 2016) identified the HA as a gamma H1. Sera were tested for HI activity against H4N6 antigen (Table 1). The control antisera raised against MO/15 were not cross-reactive with reference H1 and H3 antigens used with the farm sera samples (Table S2). Among the 24 gilts sampled on January 22, 4 were previously tested with the virus positive index case submitted to the ISU VDL in December. Three out of these 4 demonstrated positive HI titers against the H4 antigen (reciprocal titers 80, 80, 160). Another gilt from the same cohort of replacement gilts also had a positive HI titer against the H4 antigen (reciprocal titer 80). Additionally, 1/16 sows had an HI titer against the H4 antigen (reciprocal titer 160), for a total of 5 seropositive adult females at the index farm. This set of sera collected on January 22 was also evaluated for HI titers to six genetically and antigenically representative endemic IAV strains. Swine with reciprocal HI antibody titers ≥40 are reported in Table 1.
Table 1.
Number of pigs positive for hemagglutination inhibition titers ≥40 against selected antigens (HI titers shown in Table S1).
| Antigen tested by HI (subtype, genetic lineage) | *# Gilts (4) | * Gilts (20) | * Sows (16) | *4 wk (20) |
|---|---|---|---|---|
| A/Swine/Missouri/A01727926/2015 (H4N6) | 3 | 1 | 1 | 0 |
| A/Swine/Ohio/511445/2007 (H1N1, gamma) | 2 | 20 | 11 | 9 |
| A/Swine/Illinois/003200/2010 (H1N2, delta 1) | 0 | 4 | 4 | 5 |
| A/Swine/Illinois/00685/2005 (H1N1, delta 2) | 2 | 9 | 7 | 6 |
| A/Swine/Missouri/A01410819/2014 (H3N1, human-like swine virus) | 0 | 7 | 3 | 0 |
| A/Swine/New York/A01104005/2011 (H3N2, IV-A) | 4 | 4 | 7 | 4 |
| A/Swine/Iowa/A01480656/2014 (H3N2, IV-A) | 4 | 5 | 10 | 4 |
Sera collected from pigs at the breeding and gestation index farm on January 22 2016 with number of samples in parentheses.
Replacement gilts that were experiencing influenza-like symptoms at the index farm on December 16 2015, including the gilt from which MO/15 was isolated.
Thirty adult breeding sows were randomly sampled from the gilt multiplier farm for sera and nasal swabs and submitted on March 15, 2016. All serum samples tested negative for H4N6 HI antibody and RNA from pooled nasal swab samples tested by real-time RT-PCR were also negative. Ten nasal swab samples and sera were randomly collected from nursery-grow-finish pigs currently located in the contract facilities 1, 2 and 3 on March 25, March 28 and April 20, 2016, respectively, and were processed to investigate evidence of circulation of the H4N6 virus. All of the tested sera were negative for HI antibodies against H4N6. RNA extracted from pooled nasal swab samples from contract grower 1 and 3 were negative for IAV by real-time RT-PCR. RNA from three different pooled nasal swab samples from contract grower 2 were positive for H1N1 by sub-typing real-time RT-PCR assays. Sequencing attempts were negative, most likely due to low levels of virus in the samples (Ct values 35.0 – 36.5).
3.3. All gene segments of MO/15 were of avian-origin
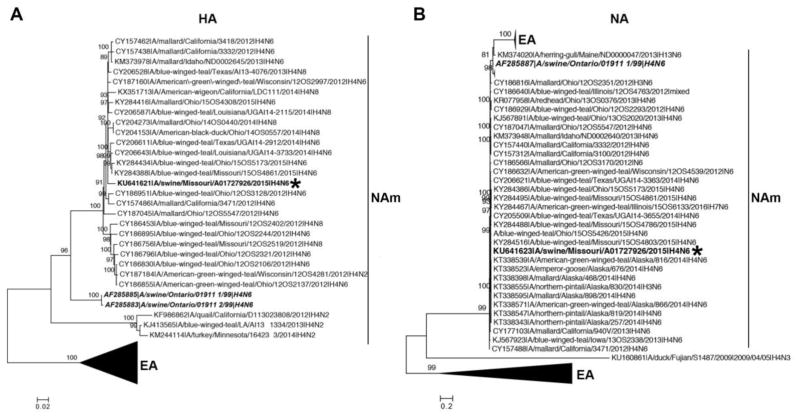
Nucleotide sequence data from all eight gene segments of MO/15 H4N6 were used to perform phylogenetic analyses. All surface and internal genes of MO/15 grouped within well-supported monophyletic clades comprised of North American avian gene segments (Fig. 2–3). The protein sequences of MO/15 were aligned with A/swine/Ontario/01911_1/1999(H4N6) (ONT/99_1), an avian-like virus also isolated from swine that was characterized in vitro, to identify putative markers associated with infectivity in swine cells (Table S3). MO/15 encodes L226 in the HA1, previously shown to be a major determinant of infectivity of primary swine and human respiratory epithelial cells in ONT/99_1 H4N6 (Bateman et al., 2008). MO/15, like ONT/99_1, also contained an asparagine at position 193 in the HA1, which was recently shown to be critical for a Eurasian lineage avian H4N2 strain to bind to α-2,6-linked sialic acids and is a possible molecular marker for predicting α-2,6-linked sialic acids binding for avian H4 viruses (Liang et al., 2016). Both MO/15 and ONT/99_1 contained PB1 gene segments that encode full-length 90 amino acid PB1-F2 proteins, although they vary at position 66. ONT/99_1 encodes S66, and in contrast MO/15 encodes N66 in the PB1-F2 and N66 has been previously associated with increased pathogenicity in a mouse IAV infection model (Conenello et al., 2007). An analysis of sequence derived phenotype markers (Squires et al., 2012) between the two strains found no other significant differences in viral protein sequences.
Figure 2.
Phylogenetic analysis of the surface genes of A/swine/Missouri/A01727926/2015 (MO/15). Maximum likelihood phylogeny of the HA (A) and the NA gene (B) of MO/15. Numbers on the branches represent measures of robustness based on an ultrafast bootstrap approach implemented in IQ-TREE. Only bootstrap values greater than 80 are shown. Well-supported monophyletic clades that consist of sequences from North American (NAm) and Eurasian (EA) regions are indicated. MO/15 is shown in bold font and labeled with an asterisk, and A/swine/Ontario/01911_1/1999 (ONT/99_1) and A/swine/Ontario/01911_2/1999 (ONT/99_2) are shown in bold italic font.
Figure 3.
Phylogenetic analysis of the internal genes of A/swine/Missouri/A01727926/2015 (MO/15). Maximum likelihood phylogeny of the PB2 (A), PB1 (B), PA (C), NP (D), M (E), and NS gene (F) of MO/15. Numbers on the branches represent measures of robustness based on an ultrafast bootstrap approach implemented in IQ-TREE. Only bootstrap values greater than 80 are shown. Well-supported monophyletic clades that consist of sequences from North American (NAm) and Eurasian (EA) regions are indicated. MO/15 genes are shown in bold font and labeled with an asterisk, and A/swine/Ontario/01911_1/1999 (ONT/99_1) and A/swine/Ontario/01911_2/1999 (ONT/99_2) are shown in bold italic font.
3.4. Limited replication and transmissibility of MO/15 in experimentally infected pigs
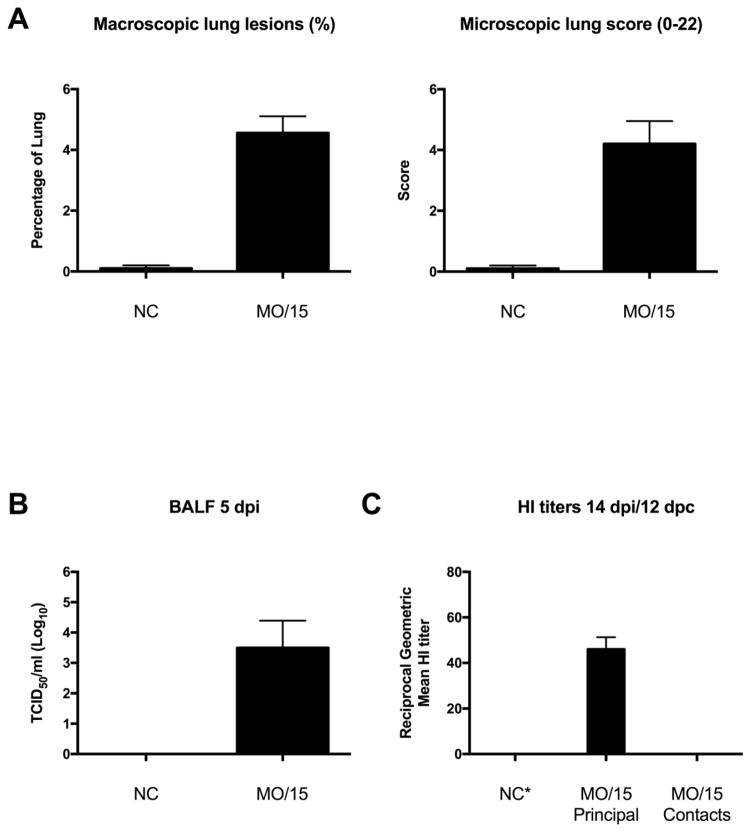
Virus was not isolated from nasal swab samples collected from principal pigs on any dpi (Table 2), suggesting little to no replication in the upper respiratory tract. At 5 dpi, mild macroscopic and microscopic lesions were observed in challenged pigs (Fig. 4A). Consistent with the pathology observed in the lungs, virus was detected in 4/5 lungs with a group mean titer of 3.5 log10 TCID50/ml (Fig. 4B). The 5 principal pigs tested at 14 dpi were sero-positive against the MO/15 H4N6 by HI assay (Fig. 4C). There was no evidence of transmission to indirect contact pigs as assessed by absence of virus isolation from all nasal swab samples and lack of seroconversion by nucleoprotein ELISA (Table 2) and HI assay against homologous antigen (Fig. 4C).
Table 2.
Virus isolation from nasal swab samples and nucleoprotein serological analysis
| Nasal swab samplesa | NP serology | ||
|---|---|---|---|
|
| |||
| Virus isolation | NP ELISA positive at 5 dpi | NP ELISA positive at 14 dpi/12 dpc | |
| Principal (10) | 0/10 | 1/5 | 5/5 |
| Indirect contact (5) | 0/5 | - | 0/5 |
| Non-challenged (5) | 0/5 | 0/5 | - |
Nasal swab samples were collected on days 1–5 post infection from principal pigs, days 1–5 and 7 and 9 days post-contact from indirect contact pigs, and day 1 and 5 of the study from non-challenged pigs.
Figure 4.
Analysis of swine susceptibility to A/swine/Missouri/A01727926/2015 (MO/15). Mean percentage of macroscopic lung lesions and microscopic pneumonia score (A) in negative control pigs (NC) and MO/15-challenged pigs. Mean virus titers in bronchoalveolar lavage fluid (BALF) at 5 dpi (B). Reciprocal geometric mean hemagglutination inhibition (HI) titer (C). Data presented as mean with standard error of the mean error bars.
4. Discussion
Avian H4 influenza viruses are detected frequently in wild waterfowl (Liang et al., 2016; Shelton et al., 2011). Although infrequently detected in mammals in nature, several H4 strains tested in the guinea pig model were able to replicate and transmit to direct contacts and by respiratory droplet without further adaptation (Liang et al., 2016). Including this report, there are now four documented cases of H4 viruses detected in pigs (Hu et al., 2012; Karasin et al., 2000; Su et al., 2012). Understanding the risk of H4 IAVs infecting and being sustained in a mammalian species such as swine is important for risk assessment because of the financial impact influenza can have on the commercial aspect of pork products as well as due to the zoonotic potential of viruses adapted to swine. The H4N6 MO/15 strain used in this study was isolated from a young gilt in a breeding herd with signs of respiratory disease. In an experimental setting, we found that MO/15 replicated in the lungs of inoculated pigs, caused mild pathological lesions in the lung, but there was no evidence of replication in the upper respiratory tract or transmission to indirect contacts.
The breeding and gestation index farm under diagnostic investigation housed approximately 1,200 adult females of various ages and received replacement gilts every six weeks from a single gilt multiplier farm. The gilt multiplier farm was located approximately thirty miles southwest of the H4N6-positive farm. Weaned gilts were transferred from the multiplier to one of three contract nursery-grow-finish facilities to mature until transport to the breeding and gestation index farm located approximately 10–15 miles away. Replacement gilts, approximately 120 per group, entered an on-farm isolation facility for a minimum of four weeks. Groups of approximately 30 gilts were selected for breeding through artificial insemination and moved into a gilt gestation facility located approximately 100–300 yards from the isolation barn. The breeding and gestation index farm was supplied with drinking water from wells (not ponds or recycled lagoon water) and pig manure was transported into lagoon storage located on-site. Weaned piglets were transferred to new owners for the commercial nursery/grow/finish phase of production.
The upstream gilt multiplier farm and breeding and gestation index farm vaccinated all sows for IAV twice annually with a commercially available, inactivated, adjuvanted whole virus product (FluSure XP® Zoetis). The contract growers did not use IAV vaccines in the replacement gilts prior to transport to the breeding index farm, but replacement gilts received two doses of the commercial inactivated IAV vaccine: one dose upon arrival to the on-farm isolation facility and a booster dose three weeks later prior to entry into the gilt gestation facility.
Clinical influenza-like illness had not been observed at the breeding/gestation index farm in the time period immediately prior to detection of the H4N6 and no endemic strain of swine IAV had been detected in this production system for approximately two years. Affected gilts were six weeks pregnant and housed in the center of the gilt gestation facility. In this group of gilts selected for breeding, morbidity was observed in 60% (18/30), abortions were observed in 40% (12/30), and one gilt from the affected group with respiratory illness died during that time (1/5). No additional gilts or sows located on the index farm were affected with clinical influenza-like illness. No new swine had entered the farm between the time the current group of replacement gilts had been introduced into the herd and the detection of the H4N6, and no additional clinical signs were observed in other animals located on the site. Despite HI titers in gilts against several swine IAV antigens that were likely vaccine-induced (Table 1), the presence of antibodies against a recently emerged, and antigenically distinct, human-like H3 swine virus (Rajao et al., 2015) that is not included in commercial swine vaccines suggests a non-detected circulation of IAV on the farm. Additionally, there was low detection rate of IAV RNA in the contract facility pigs and commercial weaned pigs from the multiplier and index breeding/gestation farms, respectively (Fig. 1).
Currently, it is unknown how or when the replacement gilts were exposed to an avian-lineage H4N6. A different breeding and gestation farm site (same owner as the H4N6-positive farm) received the same source of gilts, but did not experience clinical signs suggestive of IAV infection and diagnostic tests were IAV negative during the same time frame (data not shown). The breeding gestation index farm did not have prior history or continued detection of the H4N6 avian-lineage IAV, nor did the contract facilities and the gilt multiplier farm. The sow that also had a positive MO/15 HI titer was in a distinct older cohort of pigs compared to the replacement gilts that included the index case, although different cohorts of pigs comingled in the breeding, short term gestation unit. This is suggestive of a common source of exposure, although this could not be verified. The owner and manager of the H4N6-positive farm did not notice waterfowl populations in the production area in December 2015 or within proximity to the farm. A lagoon located on-site may be visited by migrating or resident avian populations that went unnoticed by farm personnel, but recycled lagoon water was not used as a source of drinking water or for flushing manure from the facility. No other surface water was used for cleaning facilities or as a source of drinking water. Bird barriers used in the isolation or breeding/gestation facilities were not damaged or impaired; however, small birds may enter through fan shutters used for ventilation, although this was unlikely in December in Missouri.
H4N6 is one of the common subtypes detected in wild waterfowl in North America. H4 HA avian IAV represented ~17% (1206/6971) of all avian viruses detected between 2000–2017 in North America (Fig. S1A). Among those H4 HA viruses detected, the majority (878/1206, 72%) were detected with an N6 NA (Fig. S1B). The frequency of detection of H4 HA avian viruses in the state of Missouri (22/96, 23%) is comparable to what is observed in North America (Fig. S1C). While the abundance of H4N6 viruses in North America does not alone suggest a higher probability of an avian-to-mammalian species spillover event, it is more likely that sporadic cases of detection of H4N6 viruses in swine are due to the propensity of the H4 HA to bind α-2,6-linked sialic acids in combination with its prevalence and other unknown ecological factors (Liang et al., 2016).
Swine located at the various sites involved in this case (the gilt multiplication farm, contract facilities 1, 2 and 3 and breeding and gestation index farm) were not housed outdoors at any time during production and sources of uncovered feed that may be exposed to avian feces were not observed. Transportation equipment such as livestock trailers were cleaned and disinfected and required to follow downtime protocols between each use. However, transport trailers were stored outside with potential exposure to birds and avian feces. Prior to the H4N6 infection, gilts in isolation or gilts/sows in breeding and gestation were not exposed to unusual environmental stress or management procedures (vaccination or treatment) that may have increased their susceptibility to an avian IAV. In addition, influenza-like illness was not reported by any farm personnel, including a new employee working with swine at a different location that followed a two-week downtime protocol before entering the current breeding and gestation index farm.
Serologic evidence of H4N6 transmission from pig to pig was demonstrated in a previously characterized H4N6 spillover event in 1999 that occurred in Canada (Karasin et al., 2000). A subsequent study with a strain isolated from that outbreak, ONT/99_1, identified leucine at position 226 in the HA1 as critical for high-binding affinity to α-2,6-linked sialic acids and for efficient replication in swine and human primary cells (Bateman et al., 2008), although the significance of position 226 was not tested in vivo. Among 1788 full-length H4 HA sequences from viruses isolated from avian species available on Influenza Research Database, all 1788 HA’s encoded Q226. The only H4 HA accessions that encode L226 are the strains isolated from pigs: ONT/99_1 and ONT/99_2 (Karasin et al., 2000), and the MO/15 described in the present study. While this suggests that L226 may play an important role for infection in a mammalian species, its presence alone was not sufficient for the virus to transmit and sustain infection in pigs as evidenced by the experimental challenge and epidemiological results presented in this study.
Studies characterizing the sialic acid distribution in the pig respiratory tract have varied in their conclusions of the presence of α-2,3-linked sialic acids in the upper respiratory tract (nose, trachea), but they have consistently observed α-2,6-linked sialic acids in both the upper and lower respiratory tract (bronchi, bronchioles and alveoli) and α-2,3-linked sialic acids in the lower respiratory tract (Ito et al., 1998; Nelli et al., 2010; Van Poucke et al., 2010). In pigs challenged with an avian H4N6, detection of virus antigen by immunohistochemistry was concomitant with localization of α-2,3-linked sialic acids in the bronchioles and alveoli (Trebbien et al., 2011), although virus titer was not measured and serological analysis was not performed. This is consistent with our finding that MO/15 caused lesions in the lung and replicated in the lower respiratory tract (Fig. 4A–B), but there was no detection of replication in the upper respiratory tract. It remains to be determined what the minimum requirements are for an avian H4N6 virus to replicate and transmit efficiently in swine.
Although avian virus spillover rarely leads to establishment in pigs (Nelson et al., 2015a; Nelson and Vincent, 2015), cross-species transmission and reassortment in a mammalian host, particularly swine, is a concern for public health and commercial livestock producers. The distribution of migratory birds, density of swine in some major migration flyways, transcontinental pig movement, and global live-swine trade (Nelson et al., 2015b; Ren et al., 2016) make these rare events in swine important to detect and investigate before the novel virus can be widely disseminated or contribute internal genes to endemic swine strains. Fortunately, in this case, the virus quickly disappeared from the source herd and no other detections were found in any other swine herds submitting samples to the ISU Veterinary Diagnostic Lab or in herds participating in the USDA IAV swine surveillance program. However, where, when, and how the replacement gilts became infected with the H4N6 remains undetermined. Continued surveillance efforts are important to monitor and better understand the dynamics of cross-species spread of IAV.
Supplementary Material
Highlights.
An avian H4N6 influenza A virus was detected and isolated from a pig in Midwestern United States
Analysis of clinical samples suggests the H4N6 virus was not widespread in swine
The H4N6 replicated efficiently in the lungs of challenged pigs and caused lesions
Virus was not detected in the upper respiratory tract of challenged pigs
There was no evidence of transmission in contact pigs exposed to challenged pigs
Acknowledgments
Funding
This study was supported by USDA-ARS, USDA-APHIS, and by an NIH-National Institute of Allergy and Infectious Diseases (NIAID) interagency agreement (R21AI098079) associated with Center of Research in Influenza Pathogenesis, an NIAID funded Center of Excellence in Influenza Research and Surveillance (HHSN272201400008C). EJA and RRW were supported in part by an appointment to the ARS-USDA Research Participation Program administered by the Oak Ridge Institute for Science and Education through an interagency agreement between the U.S. Department of Energy and USDA under contract number DE-AC05-06OR23100. Mention of trade names or commercial products in this article is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the U.S. Government. USDA is an equal opportunity provider and employer.
The authors thank Michelle Harland and Gwen Nordholm for technical assistance and Jason Huegel, Justin Miller, Aaron Hebeisen and Keiko Sampson for assistance with animal studies. We thank Michael Marti for providing graphic design assistance and Kerrie Franzen for sequencing support.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abente EJ, Santos J, Lewis NS, Gauger PC, Stratton J, Skepner E, Anderson TK, Rajao DS, Perez DR, Vincent AL. The Molecular Determinants of Antibody Recognition and Antigenic Drift in the H3 Hemagglutinin of Swine Influenza A Virus. Journal of virology. 2016;90:8266–8280. doi: 10.1128/JVI.01002-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson TK, Campbell BA, Nelson MI, Lewis NS, Janas-Martindale A, Killian ML, Vincent AL. Characterization of co-circulating swine influenza A viruses in North America and the identification of a novel H1 genetic clade with antigenic significance. Virus research. 2015;201:24–31. doi: 10.1016/j.virusres.2015.02.009. [DOI] [PubMed] [Google Scholar]
- Anderson TK, Macken CA, Lewis NS, Scheuermann RH, Van Reeth K, Brown IH, Swenson SL, Simon G, Saito T, Berhane Y, Ciacci-Zanella J, Pereda A, Davis CT, Donis RO, Webby RJ, Vincent AL. A Phylogeny-Based Global Nomenclature System and Automated Annotation Tool for H1 Hemagglutinin Genes from Swine Influenza A Viruses. mSphere. 2016:1. doi: 10.1128/mSphere.00275-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson TK, Nelson MI, Kitikoon P, Swenson SL, Korslund JA, Vincent AL. Population dynamics of cocirculating swine influenza A viruses in the United States from 2009 to 2012. Influenza and other respiratory viruses. 2013;7(Suppl 4):42–51. doi: 10.1111/irv.12193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- APHIS; Agriculture, U.S.D.o, editor. National Surveillance Plan for Swine Influenza Virus in Pigs. 2010. Version 3.2 ed. [Google Scholar]
- Bateman AC, Busch MG, Karasin AI, Bovin N, Olsen CW. Amino acid 226 in the hemagglutinin of H4N6 influenza virus determines binding affinity for alpha2,6-linked sialic acid and infectivity levels in primary swine and human respiratory epithelial cells. Journal of virology. 2008;82:8204–8209. doi: 10.1128/JVI.00718-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benson DA, Cavanaugh M, Clark K, Karsch-Mizrachi I, Lipman DJ, Ostell J, Sayers EW. GenBank. Nucleic acids research. 2017;45:D37–D42. doi: 10.1093/nar/gkw1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castrucci MR, Donatelli I, Sidoli L, Barigazzi G, Kawaoka Y, Webster RG. Genetic reassortment between avian and human influenza A viruses in Italian pigs. Virology. 1993;193:503–506. doi: 10.1006/viro.1993.1155. [DOI] [PubMed] [Google Scholar]
- Clark K, Karsch-Mizrachi I, Lipman DJ, Ostell J, Sayers EW. GenBank. Nucleic acids research. 2016;44:D67–72. doi: 10.1093/nar/gkv1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conenello GM, Zamarin D, Perrone LA, Tumpey T, Palese P. A single mutation in the PB1-F2 of H5N1 (HK/97) and 1918 influenza A viruses contributes to increased virulence. PLoS pathogens. 2007;3:1414–1421. doi: 10.1371/journal.ppat.0030141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Graaf M, Fouchier RA. Role of receptor binding specificity in influenza A virus transmission and pathogenesis. EMBO J. 2014;33:823–841. doi: 10.1002/embj.201387442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donis RO, Bean WJ, Kawaoka Y, Webster RG. Distinct lineages of influenza virus H4 hemagglutinin genes in different regions of the world. Virology. 1989;169:408–417. doi: 10.1016/0042-6822(89)90166-9. [DOI] [PubMed] [Google Scholar]
- Gauger PC, Vincent AL, Loving CL, Henningson JN, Lager KM, Janke BH, Kehrli ME, Jr, Roth JA. Kinetics of lung lesion development and pro-inflammatory cytokine response in pigs with vaccine-associated enhanced respiratory disease induced by challenge with pandemic (2009) A/H1N1 influenza virus. Veterinary pathology. 2012;49:900–912. doi: 10.1177/0300985812439724. [DOI] [PubMed] [Google Scholar]
- Guan Y, Shortridge KF, Krauss S, Li PH, Kawaoka Y, Webster RG. Emergence of avian H1N1 influenza viruses in pigs in China. Journal of virology. 1996;70:8041–8046. doi: 10.1128/jvi.70.11.8041-8046.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halbur PG, Paul PS, Frey ML, Landgraf J, Eernisse K, Meng XJ, Lum MA, Andrews JJ, Rathje JA. Comparison of the pathogenicity of two US porcine reproductive and respiratory syndrome virus isolates with that of the Lelystad virus. Veterinary pathology. 1995;32:648–660. doi: 10.1177/030098589503200606. [DOI] [PubMed] [Google Scholar]
- Hu Y, Liu X, Li S, Guo X, Yang Y, Jin M. Complete genome sequence of a novel H4N1 influenza virus isolated from a pig in central China. Journal of virology. 2012;86:13879. doi: 10.1128/JVI.02726-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito T, Couceiro JN, Kelm S, Baum LG, Krauss S, Castrucci MR, Donatelli I, Kida H, Paulson JC, Webster RG, Kawaoka Y. Molecular basis for the generation in pigs of influenza A viruses with pandemic potential. Journal of virology. 1998;72:7367–7373. doi: 10.1128/jvi.72.9.7367-7373.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joseph U, Su YC, Vijaykrishna D, Smith GJ. The ecology and adaptive evolution of influenza A interspecies transmission. Influenza and other respiratory viruses. 2016 doi: 10.1111/irv.12412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karasin AI, Brown IH, Carman S, Olsen CW. Isolation and characterization of H4N6 avian influenza viruses from pigs with pneumonia in Canada. Journal of virology. 2000;74:9322–9327. doi: 10.1128/jvi.74.19.9322-9327.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khurana S, Loving CL, Manischewitz J, King LR, Gauger PC, Henningson J, Vincent AL, Golding H. Vaccine-induced anti-HA2 antibodies promote virus fusion and enhance influenza virus respiratory disease. Science translational medicine. 2013;5:200ra114. doi: 10.1126/scitranslmed.3006366. [DOI] [PubMed] [Google Scholar]
- Kida H, Ito T, Yasuda J, Shimizu Y, Itakura C, Shortridge KF, Kawaoka Y, Webster RG. Potential for transmission of avian influenza viruses to pigs. The Journal of general virology. 1994;75(Pt 9):2183–2188. doi: 10.1099/0022-1317-75-9-2183. [DOI] [PubMed] [Google Scholar]
- Koen JS. A practical method for field diagnosis of swine diseases. American Journal of Veterinary Medicine. 1919;14:468–470. [Google Scholar]
- Lewis NS, Anderson TK, Kitikoon P, Skepner E, Burke DF, Vincent AL. Substitutions near the hemagglutinin receptor-binding site determine the antigenic evolution of influenza A H3N2 viruses in U.S. swine. Journal of virology. 2014;88:4752–4763. doi: 10.1128/JVI.03805-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang L, Deng G, Shi J, Wang S, Zhang Q, Kong H, Gu C, Guan Y, Suzuki Y, Li Y, Jiang Y, Tian G, Liu L, Li C, Chen H. Genetics, Receptor Binding, Replication, and Mammalian Transmission of H4 Avian Influenza Viruses Isolated from Live Poultry Markets in China. Journal of virology. 2016;90:1455–1469. doi: 10.1128/JVI.02692-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long JS, Giotis ES, Moncorge O, Frise R, Mistry B, James J, Morisson M, Iqbal M, Vignal A, Skinner MA, Barclay WS. Species difference in ANP32A underlies influenza A virus polymerase host restriction. Nature. 2016;529:101–104. doi: 10.1038/nature16474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minh BQ, Nguyen MA, von Haeseler A. Ultrafast approximation for phylogenetic bootstrap. Mol Biol Evol. 2013;30:1188–1195. doi: 10.1093/molbev/mst024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelli RK, Kuchipudi SV, White GA, Perez BB, Dunham SP, Chang KC. Comparative distribution of human and avian type sialic acid influenza receptors in the pig. BMC Vet Res. 2010;6:4. doi: 10.1186/1746-6148-6-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson MI, Stratton J, Killian ML, Janas-Martindale A, Vincent AL. Continual Reintroduction of Human Pandemic H1N1 Influenza A Viruses into Swine in the United States, 2009 to 2014. Journal of virology. 2015a;89:6218–6226. doi: 10.1128/JVI.00459-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson MI, Viboud C, Vincent AL, Culhane MR, Detmer SE, Wentworth DE, Rambaut A, Suchard MA, Holmes EC, Lemey P. Global migration of influenza A viruses in swine. Nat Commun. 2015b;6:6696. doi: 10.1038/ncomms7696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson MI, Vincent AL. Reverse zoonosis of influenza to swine: new perspectives on the human-animal interface. Trends in microbiology. 2015;23:142–153. doi: 10.1016/j.tim.2014.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 2015;32:268–274. doi: 10.1093/molbev/msu300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajao DS, Gauger PC, Anderson TK, Lewis NS, Abente EJ, Killian ML, Perez DR, Sutton TC, Zhang J, Vincent AL. Novel Reassortant Human-Like H3N2 and H3N1 Influenza A Viruses Detected in Pigs Are Virulent and Antigenically Distinct from Swine Viruses Endemic to the United States. Journal of virology. 2015;89:11213–11222. doi: 10.1128/JVI.01675-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren H, Jin Y, Hu M, Zhou J, Song T, Huang Z, Li B, Li K, Zhou W, Dai H, Shi W, Yue J, Liang L. Ecological dynamics of influenza A viruses: cross-species transmission and global migration. Scientific reports. 2016;6:36839. doi: 10.1038/srep36839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shelton H, Ayora-Talavera G, Ren J, Loureiro S, Pickles RJ, Barclay WS, Jones IM. Receptor binding profiles of avian influenza virus hemagglutinin subtypes on human cells as a predictor of pandemic potential. Journal of virology. 2011;85:1875–1880. doi: 10.1128/JVI.01822-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shope RE. Swine Influenza : Iii. Filtration Experiments and Etiology. The Journal of experimental medicine. 1931;54:373–385. doi: 10.1084/jem.54.3.373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith GJ, Bahl J, Vijaykrishna D, Zhang J, Poon LL, Chen H, Webster RG, Peiris JS, Guan Y. Dating the emergence of pandemic influenza viruses. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:11709–11712. doi: 10.1073/pnas.0904991106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Squires RB, Noronha J, Hunt V, Garcia-Sastre A, Macken C, Baumgarth N, Suarez D, Pickett BE, Zhang Y, Larsen CN, Ramsey A, Zhou L, Zaremba S, Kumar S, Deitrich J, Klem E, Scheuermann RH. Influenza research database: an integrated bioinformatics resource for influenza research and surveillance. Influenza and other respiratory viruses. 2012;6:404–416. doi: 10.1111/j.1750-2659.2011.00331.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steel J, Lowen AC. Influenza A virus reassortment. Current topics in microbiology and immunology. 2014;385:377–401. doi: 10.1007/82_2014_395. [DOI] [PubMed] [Google Scholar]
- Su S, Qi WB, Chen JD, Cao N, Zhu WJ, Yuan LG, Wang H, Zhang GH. Complete genome sequence of an avian-like H4N8 swine influenza virus discovered in southern China. Journal of virology. 2012;86:9542. doi: 10.1128/JVI.01475-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taubenberger JK. The origin and virulence of the 1918 “Spanish” influenza virus. Proc Am Philos Soc. 2006;150:86–112. [PMC free article] [PubMed] [Google Scholar]
- Taubenberger JK, Reid AH, Lourens RM, Wang R, Jin G, Fanning TG. Characterization of the 1918 influenza virus polymerase genes. Nature. 2005;437:889–893. doi: 10.1038/nature04230. [DOI] [PubMed] [Google Scholar]
- Tong S, Li Y, Rivailler P, Conrardy C, Castillo DA, Chen LM, Recuenco S, Ellison JA, Davis CT, York IA, Turmelle AS, Moran D, Rogers S, Shi M, Tao Y, Weil MR, Tang K, Rowe LA, Sammons S, Xu X, Frace M, Lindblade KA, Cox NJ, Anderson LJ, Rupprecht CE, Donis RO. A distinct lineage of influenza A virus from bats. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:4269–4274. doi: 10.1073/pnas.1116200109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong S, Zhu X, Li Y, Shi M, Zhang J, Bourgeois M, Yang H, Chen X, Recuenco S, Gomez J, Chen LM, Johnson A, Tao Y, Dreyfus C, Yu W, McBride R, Carney PJ, Gilbert AT, Chang J, Guo Z, Davis CT, Paulson JC, Stevens J, Rupprecht CE, Holmes EC, Wilson IA, Donis RO. New world bats harbor diverse influenza A viruses. PLoS pathogens. 2013;9:e1003657. doi: 10.1371/journal.ppat.1003657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trebbien R, Larsen LE, Viuff BM. Distribution of sialic acid receptors and influenza A virus of avian and swine origin in experimentally infected pigs. Virology journal. 2011;8:434. doi: 10.1186/1743-422X-8-434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Poucke SG, Nicholls JM, Nauwynck HJ, Van Reeth K. Replication of avian, human and swine influenza viruses in porcine respiratory explants and association with sialic acid distribution. Virology journal. 2010;7:38. doi: 10.1186/1743-422X-7-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincent A, Awada L, Brown I, Chen H, Claes F, Dauphin G, Donis R, Culhane M, Hamilton K, Lewis N, Mumford E, Nguyen T, Parchariyanon S, Pasick J, Pavade G, Pereda A, Peiris M, Saito T, Swenson S, Van Reeth K, Webby R, Wong F, Ciacci-Zanella J. Review of influenza A virus in swine worldwide: a call for increased surveillance and research. Zoonoses and public health. 2014;61:4–17. doi: 10.1111/zph.12049. [DOI] [PubMed] [Google Scholar]
- Worobey M, Han GZ, Rambaut A. Genesis and pathogenesis of the 1918 pandemic H1N1 influenza A virus. Proceedings of the National Academy of Sciences of the United States of America. 2014a;111:8107–8112. doi: 10.1073/pnas.1324197111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worobey M, Han GZ, Rambaut A. A synchronized global sweep of the internal genes of modern avian influenza virus. Nature. 2014b;508:254–257. doi: 10.1038/nature13016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu G, Zhang X, Sun Y, Liu Q, Sun H, Xiong X, Jiang M, He Q, Wang Y, Pu J, Guo X, Yang H, Liu J. Truncation of C-terminal 20 amino acids in PA-X contributes to adaptation of swine influenza virus in pigs. Scientific reports. 2016;6:21845. doi: 10.1038/srep21845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon SW, Webby RJ, Webster RG. Evolution and ecology of influenza A viruses. Current topics in microbiology and immunology. 2014;385:359–375. doi: 10.1007/82_2014_396. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Aevermann BD, Anderson TK, Burke DF, Dauphin G, Gu Z, He S, Kumar S, Larsen CN, Lee AJ, Li X, Macken C, Mahaffey C, Pickett BE, Reardon B, Smith T, Stewart L, Suloway C, Sun G, Tong L, Vincent AL, Walters B, Zaremba S, Zhao H, Zhou L, Zmasek C, Klem EB, Scheuermann RH. Influenza Research Database: An integrated bioinformatics resource for influenza virus research. Nucleic acids research. 2017;45:D466–D474. doi: 10.1093/nar/gkw857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou NN, Senne DA, Landgraf JS, Swenson SL, Erickson G, Rossow K, Liu L, Yoon K, Krauss S, Webster RG. Genetic reassortment of avian, swine, and human influenza A viruses in American pigs. Journal of virology. 1999;73:8851–8856. doi: 10.1128/jvi.73.10.8851-8856.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.