Abstract
Long non-coding RNAs (lncRNAs) are rapidly emerging as important regulators of a diverse array of cellular functions. Here, we describe a meta-analysis of two independent RNA-seq studies to identify lncRNAs that are differentially expressed upon HIV-1 infection. Only three lncRNA genes exhibited altered expression of ≥ 2-fold in HIV-1-infected cells. Of these, the uncharacterized lncRNA LINC00173 was chosen for further study. Both transcript variants of LINC00173 (lnc173 TSV1 and 2) could be detected by qPCR, localized predominantly to the nucleus and were reproducibly up-regulated during infection. Knock-out of the LINC00173 locus did not have detectable effects on HIV-1 replication. Interestingly, however, stimulation of Jurkat T cells with PMA/ionomycin resulted in a decrease of lnc173 expression, and Jurkat cells deficient for lnc173 on average expressed higher levels of specific cytokines than control cells. These data suggest that lnc173 may have a role in the regulation of cytokines in T cells.
Keywords: Long non-coding RNA, lncRNA, lincRNA, Human immunodeficiency virus, HIV, RNA-seq, Gene expression, Cytokine
Highlights
-
•
Meta-analysis of RNA-seq studies reveals 3 lncRNAs increased by HIV-1 infection.
-
•
Lnc173 is up-regulated in a time- and dose-dependent manner during HIV-1 infection.
-
•
Loss of lnc173 does not affect any stage of the viral replication cycle.
-
•
Lnc173 is down-regulated in stimulated T cells.
-
•
Lnc173 may be involved in cytokine regulation.
1. Introduction
Long non-coding RNAs (lncRNAs) are defined as RNA transcripts that are not translated and are at least 200 nucleotides long. The current GENCODE release (version 26) has identified 15,787 lncRNA genes in the human genome, encoding 27,720 different transcripts, excluding transcribed pseudogenes (https://www.gencodegenes.org/stats/current.html) (Harrow et al., 2012). Only a few hundred of these lncRNAs have been functionally characterized to date, but it has become evident that this class of biomolecules includes central regulators of varied biological processes, such as the regulation of cellular proliferation, immunity, development and even nuclear organization (Atianand et al., 2017, Chaudhary and Lal, 2016, Goff and Rinn, 2015, Perry and Ulitsky, 2016, Pircher et al., 2014). Based on their genomic context, lncRNAs can be grouped into four simple classes: antisense RNAs, which are transcribed in the opposite direction of overlapping protein-coding genes; large intergenic non-coding RNAs (lincRNAs), which are not flanked by other genes in close proximity; intronic lncRNAs, which are encoded within an intron of a protein-coding gene; and overlapping lncRNAs, which contain a protein-coding gene within a lncRNA intron (Atianand et al., 2017, Spurlock et al., 2016). More complex classification schemes have also been proposed (Chen, 2016). Most lncRNAs are thought to be capped, polyadenylated, and spliced using the same cellular machinery as protein-coding transcripts, although distinct differences have been noted (Mukherjee et al., 2017, Schlackow et al., 2016, Ulitsky and Bartel, 2013). Depending on their mechanism of action, lncRNAs are located in the cytoplasm or in the nucleus. While cytoplasmic lncRNAs tend to function as scaffolds for protein complexes or as microRNA “sponges”, nuclear lncRNAs often aid or interfere with transcription in cis or in trans . For instance, nuclear lncRNAs have been reported to bind protein regulators of transcription and either guide them to their intended target site or act as a decoy to sequester them (Atianand et al., 2017, Goff and Rinn, 2015).
Recent studies have only begun to elucidate the role of lncRNAs during viral infections. Several viruses have been shown to encode lncRNAs in their genome, including both DNA and RNA viruses (Tycowski et al., 2015). Multiple groups have reported that the genome of HIV-1 is transcribed in the antisense direction to yield various lncRNA species (Kobayashi-Ishihara et al., 2012, Landry et al., 2007, Ludwig et al., 2006, Saayman et al., 2014). While the length of the identified transcripts differed between studies, there is evidence to suggest that at least some of these antisense RNAs may interfere with viral transcription by establishing repressive histone methylation at the 5′ LTR (Kobayashi-Ishihara et al., 2012, Saayman et al., 2014, Zapata et al., 2017).
Beyond virally encoded lncRNAs, the expression levels of lncRNAs encoded by the host have been shown to be altered upon infection with a variety of viral pathogens (Fortes and Morris, 2015). Specific examples include influenza A virus (IAV), hepatitis C virus, severe acute respiratory syndrome-related coronavirus, adenovirus, herpes simplex virus 1 (HSV-1), human cytomegalovirus, and HIV-1 (Carnero et al., 2016, Chang et al., 2011, Hu et al., 2016, Ouyang et al., 2015, Peng et al., 2010, Peng et al., 2014, Trypsteen et al., 2016, Winterling et al., 2014; Zhang et al., 2016; Zhao et al., 2016). Of the host-encoded lncRNAs reported to be differentially expressed upon HIV-1 infection, only nuclear enriched abundant transcript 1 (NEAT1) and noncoding repressor of nuclear factor of activated T cells (NRON) have been functionally characterized (Lazar et al., 2016). NEAT1 is a nuclear lncRNA which stabilizes paraspeckles and is believed thereby to sequester unspliced and singly spliced HIV-1 transcripts (Clemson et al., 2009, Sasaki et al., 2009, West et al., 2014; Zhang et al., 2013; Zolotukhin et al., 2003). Of note, NEAT1 expression has also been shown to enhance the expression of antiviral genes during infection with IAV and HSV-1, as well as after poly(I:C) treatment (Imamura et al., 2014). NRON, by contrast, is a cytoplasmic lncRNA that forms part of a high-molecular-weight complex including nuclear factor of activated T cells (NFAT), a transcription factor of central importance to T-cell activation. NRON stabilizes the inactive form of NFAT, and correspondingly loss of NRON results in the hyperactivation of T cells (Macian, 2005, Sharma et al., 2011, Willingham et al., 2005). A complex interplay of HIV-1 and NRON has been described, where NRON levels are decreased early during infection by the protein Nef and increased by the late gene product Vpu (Imam et al., 2015). Recently, Li et al. (2016) demonstrated that NRON specifically induces the degradation of Tat and thus contributes to HIV-1 latency. Other lncRNAs that have been documented to be up-regulated in HIV-1-infected cells, such as MALAT1 and MIAT, have not been functionally analyzed in the context of infection.
Most prior reports specifically investigating the effect of HIV-1 on lncRNA expression have focused on a small panel of previously characterized lncRNAs rather than employing a genome-wide screen. Furthermore, there was only limited overlap between the results of these studies, likely reflecting different experimental conditions (Imam et al., 2015; Zhang et al., 2013). In this report, we describe a meta-analysis of two independent RNA-seq studies of HIV-1-infected cells and show that, unexpectedly, only three lncRNAs are differentially expressed in both of these datasets (Chang et al., 2011, Mohammadi et al., 2013). Of those three candidates, LINC00173 exhibits the highest degree of evolutionary conservation. We demonstrate that both transcript variants of LINC00173 are indeed detectable in a variety of human cell lines, are located predominantly in the nucleus, and are up-regulated upon infection with HIV-1. While, somewhat surprisingly, loss of LINC00173 did not affect the viral replication cycle directly in cell culture, Jurkat T cells deficient for LINC00173 on average expressed higher levels of a subset of cytokines than control cells, while other cytokines remained unaffected. These data indicate a possible involvement of LINC00173 in the regulation of cytokine expression.
2. Results
2.1. Meta-analysis of two independent RNA-seq studies of HIV-1-infected T cells
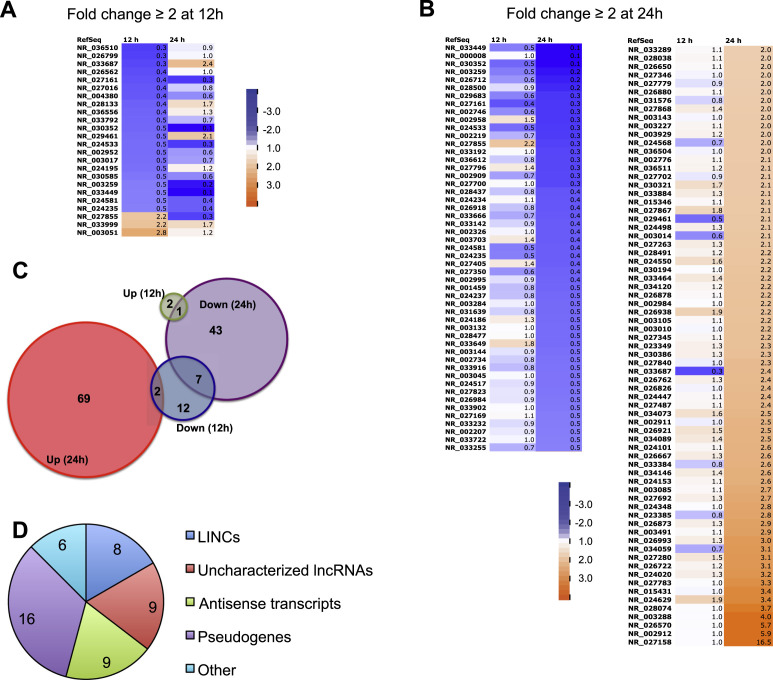
To obtain an unbiased understanding of any changes in lncRNA expression during HIV-1 infection, we performed a meta-analysis of two independent RNA-seq studies conducted by two different groups using two different virus strains (Supplementary Fig. S1) (Chang et al., 2011, Mohammadi et al., 2013). Chang et al. (2011) infected SUP-T1 cells with the fully replication-competent HIV-1 isolate LAI and performed RNA-seq with samples harvested at 12 and 24 h p.i.. Using this dataset, we identified non-coding transcripts based on their Genbank classification. Transcripts with a fold change of ≥ 2 were considered differentially expressed. Twelve hours after infection, 21 non-coding transcripts were down-regulated, 3 were up-regulated ( Fig. 1A). At 24 h, 51 transcripts were down-regulated, while 71 had increased expression levels (Fig. 1B). Unexpectedly, only 10 transcripts were differentially expressed at both time points (Fig. 1C). Next, we filtered out snoRNAs, miRNAs, and non-coding transcript variants of protein-coding genes. The remaining transcripts comprised 8 large intergenic non-coding RNAs (LINCs), 9 uncharacterized lncRNAs, 9 antisense transcripts, 16 pseudogenes and 6 other transcripts, such as a miRNA polycistron (Fig. 1D). We further filtered these results using the findings of Mohammadi et al. (2013), who infected SUP-T1 cells with VSV-G-pseudotyped HIV-1 NL4-3 Δenv/EGFP and performed RNA-seq with samples harvested every 2 h for 24 h. The processed RNA-seq data from that study have been made available via the dedicated online platform Patterns of Expression and Analysis of Clusters of HIV/Host interactions (PEACHi, http://www.peachi.labtelenti.org). After excluding pseudogenes and the transcripts in the “other” category, we queried the remaining 26 lncRNA transcripts from our analysis of the study by Chang et al. in PEACHi. Of these, only four were also identified as differentially expressed in PEACHi (11 were not differentially expressed and 11 were not catalogued). Two of those 4 transcripts were variants of one gene, LINC00173, identified by Genbank as a large intergenic non-coding RNA. The remaining 2 transcripts were both antisense RNAs, CCDC18-AS1 and MCM3AP-AS1 ( Table 1).
Fig. 1.
RNA-seq data reveal multiple non-coding RNA transcripts differentially regulated during HIV-1 infection of SupT1 cells. RNA-seq data from a study by Chang et al. (2011) was used to identify RNA transcripts with changes in expression ≥ 2-fold at 12 and 24 h after infection with replication-competent HIV-1. (A) Non-coding RNA transcripts differentially expressed 12 h after infection. (B) Non-coding RNA transcripts differentially expressed 24 h after infection. Colors correspond to the level of linear fold change, with blue indicating a relative decrease and orange a relative increase in transcript levels in HIV-1-infected samples compared to controls. (C) Venn diagram of the transcripts listed in (A) and (B), grouped by time point and direction of expression change. Numbers represent total transcript count in each group. (D) After filtering out transcripts representing snoRNAs, miRNAs and non-coding variants of protein-coding genes, the remaining transcripts were categorized as large intergenic non-coding RNAs (LINCs), uncharacterized long non-coding RNAs (lncRNAs), antisense transcripts, pseudogenes, and others (transcripts that do not fall in any of the above categories). The pie chart represents total numbers of transcripts in each category.
Table 1.
lncRNA transcripts with differential expression (fold change ≥ 2) in the RNA-seq data sets of both Chang et al. (2011) and Mohammadi et al. (2013).
| Name | RefSeq ID | Transcript length (nt) | Genomic length (bp) | Exon count | Type | PhyloCSF score | Bazzini small ORFs | Conservation across vertebrates |
|---|---|---|---|---|---|---|---|---|
| LINC00173 (transcript variant 1) | NR_027345 | 1597 | 2100 | 2 | intergenic | 0.2084(non-coding) | 0 | Partial |
| LINC00173 (transcript variant 2) | NR_027346 | 435 | 3079 | 3 | intergenic | − 91.861(non-coding) | 0 | Low |
| CCDC18-AS1 | NR_034089 | 4033 | 35,703 | 9 | antisense | 11.4184 | 0 | Low |
| MCM3AP-AS1 | NR_002776 | 2338 | 22,471 | 3 | antisense | N/Aa | N/Aa | Low |
| (transcript variant 2 only) |
Transcript variant 2 of MCM3AP-AS1 is not listed in LNCipedia. Most transcripts overlapping this genomic region are rated non-coding.
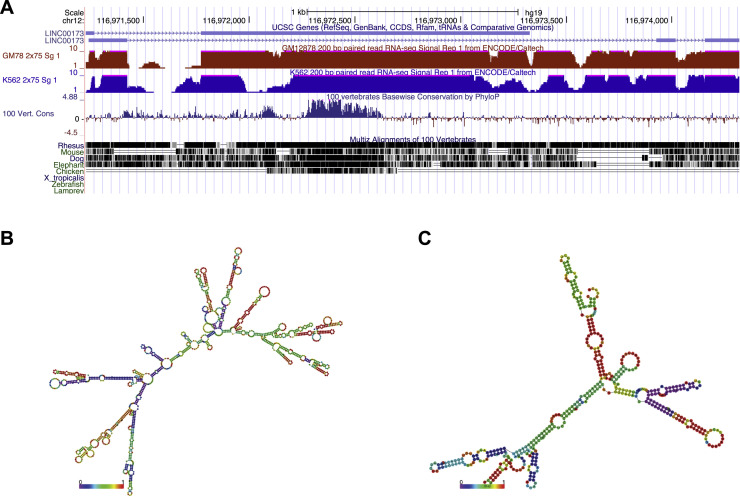
2.2. In silico analysis of the LINC00173 locus indicates active transcription and evolutionary conservation
We chose to further investigate LINC00173 as both of its transcript variants were differentially expressed after HIV-1 infection in the RNA-seq studies and it showed the highest degree of conservation across vertebrate species among the 3 candidate genes (Table 1). Transcript variant 1 (lnc173 TSV1) is encoded by 2 exons and 1597 nt long, transcript variant 2 (lnc173 TSV2) is encoded by 3 exons and reaches a length of 435 nt. The central section of TSV1 is particularly well conserved across species, with a substantially similar region present even in the chicken genome ( Fig. 2A). RNA-seq data from GM78 and K562 cells available in the UCSC Genome Browser indicate robust transcription and splicing (Fig. 2A) (Kent et al., 2002). Recorded histone modifications in multiple cell types are also consistent with expression, including histone H3K27 acetylation and H3K4 methylation marks near the transcriptional start site (Supplementary Fig. S2). DNase I hypersensitivity levels suggest an open chromatin conformation at this locus (Supplementary Fig. S2). All of these data point to active transcription of LINC00173. The predicted secondary structures of lnc173 TSV1 and TSV2 are drastically different, reflecting the minimal sequence overlap restricted to the first exon (Fig. 2B, C).
Fig. 2.
In silicoanalysis ofLINC00173is consistent with active transcription. (A) RNA-seq expression data and conservation at the LINC00173 locus. Data obtained from and visualized with UCSC Genome Browser (http://genome.ucsc.edu) (Kent et al., 2002). (B–C) Prediction for secondary structure of (B) lnc173 transcript variant 1 (TSV1) and (C) lnc173 TSV2. Data obtained from LNCipedia (www.lncipedia.org) (Volders et al., 2015, Volders et al., 2013).
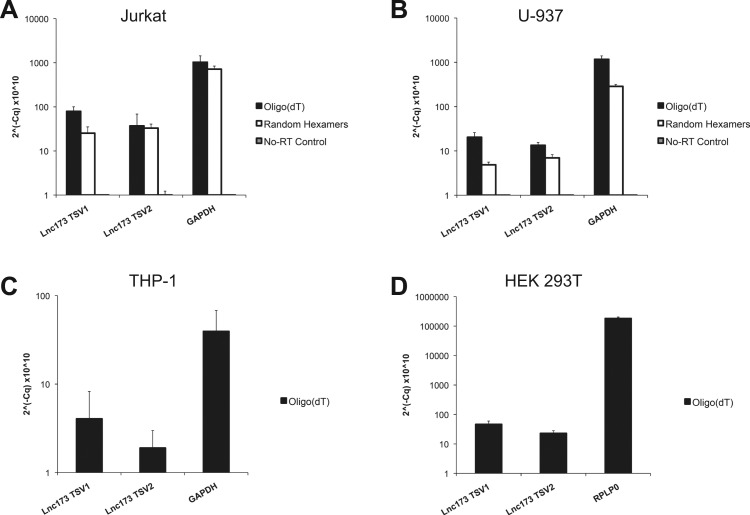
2.3. Both transcript variants of lnc173 are detectable in several human cell lines, are polyadenylated and localize predominantly to the nucleus
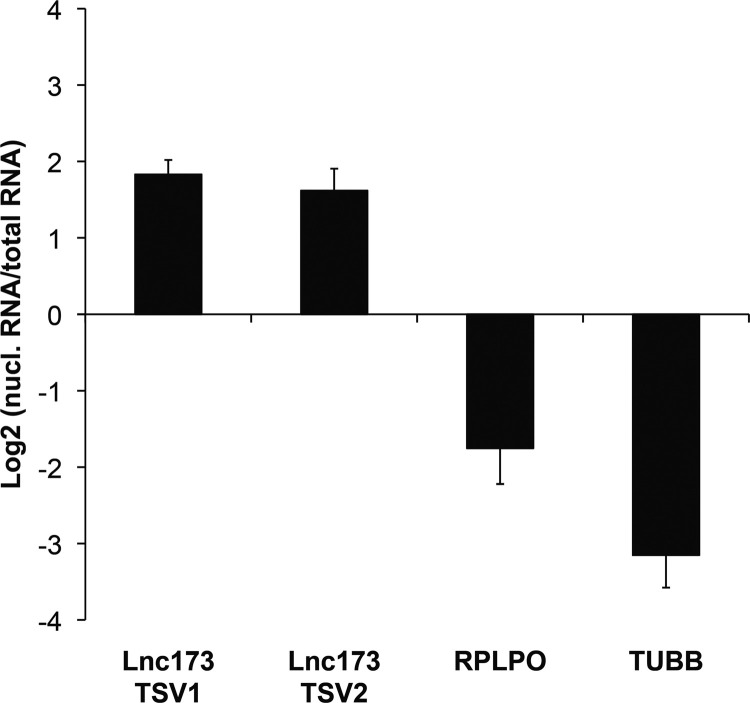
To validate the results of the in silico analysis of lnc173 expression, we isolated RNA from 4 human cell lines and probed for the presence of both transcript variants by quantitative RT-PCR (qRT-PCR; Fig. 3A–D). TSV1 and TSV2 were readily detectable in Jurkat T cells after reverse transcription with random-hexamer or oligo(dT) primers, demonstrating that both transcript variants are expressed and polyadenylated (Fig. 3A). Control samples without reverse transcriptase yielded no signal, confirming specificity of the assay for RNA rather than genomic DNA. Similarly, lnc173 TSV1 and TSV2 were present in the monocyte-derived cell lines U-937 and THP-1, as well as in the renal epithelial cell line HEK 293T (Fig. 3B–D). Both transcript variants were highly enriched in the nuclear fraction of unstimulated Jurkat cells, identifying lnc173 as a nuclear lncRNA ( Fig. 4).
Fig. 3.
Lnc173 is polyadenylated and expressed in varied human cell lines. qRT-PCR with cDNA derived from unstimulated (A) Jurkat, (B) U-937, (C) THP-1 and (D) HEK 293T cells. Oligo(dT) or random-hexamer primers were used for reverse transcription, as indicated. TSV, transcript variant. Cq, threshold cycle. Data are the average of 3 (Jurkat, U-937) or 4 (THP-1, HEK 293T) independent experiments. Error bars indicate standard deviation. Note logarithmic scale.
Fig. 4.
Both transcript variants of lnc173 are located predominantly in the nucleus. RNA was isolated from whole-cell lysates or nuclear fractions of unstimulated Jurkat cells. Equivalent amounts of RNA were reverse-transcribed and cDNA used for qRT-PCR. Shown is the relative amount of RNA in the nuclear fraction compared to whole-cell RNA. Data are the average of 3 independent experiments. Error bars indicate standard deviation.
2.4. Both transcript variants are up-regulated during infection with HIV-1 in a dose- and time-dependent manner
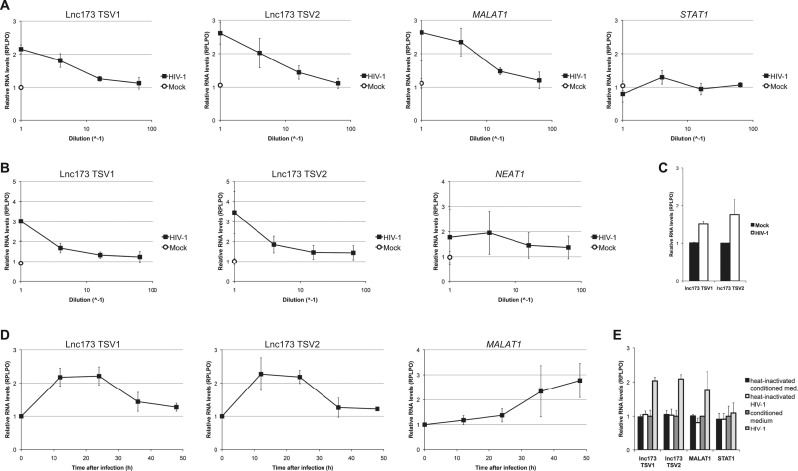
To confirm the RNA-seq data by Chang et al. and Mohammadi et al., we infected HEK 293T cells with serial 4-fold dilutions of VSV-G-pseudotyped HIV-1 NL4-3 Δenv/EGFP and performed qRT-PCR with samples harvested 24 h p.i. (Zhang et al., 2004). Both lnc173 transcript variants were indeed up-regulated in a dose-dependent manner compared to uninfected control cells. The lncRNA MALAT1, which has also been reported to be induced upon HIV-1 infection, showed a dose-dependent increase of similar magnitude (Zhang et al., 2013). STAT1 mRNA levels remained unchanged ( Fig. 5A). Infection of the monocytic THP-1 cell line mirrored the results of HEK 293T infection in magnitude and dose dependence (Fig. 5B). Infection of Jurkat cells resulted in a reproducible but less pronounced increase in lnc173 expression (Fig. 5C). This was expected, as the efficiency of infection for this cell type ranged only from 50% to 70%, whereas the qRT-PCR analysis reflected the average mRNA levels of both uninfected and infected cells (cf. Supplementary Fig. S3C). Infection efficiency for HEK 293T cells, by contrast, reached nearly 100% (cf. Supplementary Fig. S4B). To determine the temporal kinetics of lnc173 expression during viral replication, we infected HEK 293T cells in a 48-h time course and harvested samples every 12 h. Levels of lnc173 TSV1 and TSV2 were increased compared to time-matched controls at 12 h and 24 h p.i., followed by a decline towards control levels. Expression levels of MALAT1, by contrast, continued to increase throughout the entire time course (Fig. 5D). Exposure to heat-inactivated viral particles did not result in an increase of lnc173 expression (Fig. 5E). Taken together, these data establish that HIV-1 infection does induce an increase in the cellular levels of lnc173 TSV1 and TSV2 in a time- and dose-dependent manner and in multiple cell lines representing several cell types.
Fig. 5.
Infection with HIV-1 induces up-regulation of lnc173 expression in a manner dependent on dose and time. qRT-PCR with RNA from different cell lines after infection with VSV-G-pseudotyped HIV-1 NL4-3 Δenv/EGFP. Cells were lysed and RNA was harvested 24 h after infection, unless stated otherwise. (A) Infection of HEK 293T cells with serial 4-fold dilutions. Results are the average of 4 independent experiments. (B) Infection of THP-1 cells with serial 4-fold dilutions. Results are the average of 4 independent experiments. (C) Infection of Jurkat cells with undiluted HIV-1. Results are the average of 3 independent experiments. (D) Infection of HEK 293T cells in a time course with harvest after 12, 24, 36 and 48 h. Expression values are relative to time-matched control samples. Results are the average of 3 independent experiments. (E) Exposure of HEK 293T cells to heat-inactivated HIV-1. Virus was heat-inactivated for 30 min at 65 °C prior to infection where indicated. Results are the average of 3 independent experiments. Error bars indicate standard deviation. RPLP0 was used as reference gene.
2.5. Loss of lnc173 does not affect any part of the HIV-1 life cycle
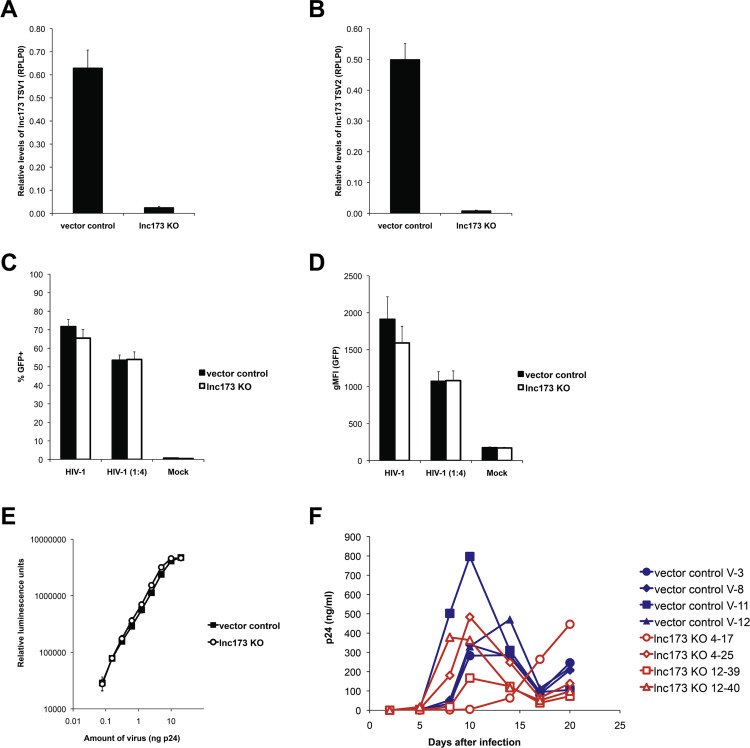
To determine whether either transcript variant of lnc173 is involved in an important part of the HIV-1 life cycle, we used CRISPR-Cas9-mediated deletion to remove the entire LINC00173 gene in Jurkat cells. Specifically, we targeted both ends of the gene with separate guide RNAs simultaneously, derived monoclonal lines from the population and identified clones with no remaining copies of LINC00173 by PCR. Two separate combinations of guide RNAs were used, set 4 and set 12 (Supplementary Table S1). A total of 6 lnc173 knock-out (KO) clones was obtained, 2 with set 4 and 4 with set 12. Additionally, 4 control clones were derived from Jurkat cells transfected with an sgRNA-free Cas9 vector. The absence of lnc173 transcripts was confirmed by qRT-PCR ( Fig. 6A, B and Supplementary Fig. S3A, B). The residual signal detected with lnc173 KO clone 12–17 was close to the limit of detection but may reflect incomplete deletion of the gene. We then proceeded to systematically probe all parts of the HIV-1 replication cycle. To assess the post-entry steps of replication up to viral gene expression, we infected lnc173 KO cells or controls with VSV-G-pseudotyped HIV-1 NL4-3 Δenv/EGFP. This construct carries the gene encoding EGFP in the open reading frame of the env gene and thus serves as a reliable reporter for the efficiency of reverse transcription, nuclear import, integration and expression of viral gene products (Zhang et al., 2004). A defect at any of these steps would result in a lower number of GFP+ cells and/or a lower level of GFP expression overall, and enhancement would result in the opposite phenotype. As expected with a collection of monoclonal lines derived from a heterogeneous parent population, the percentage of GFP+ cells and the intensity of GFP expression varied considerably between the clones (Supplementary Fig. S3C, D). However, the average values obtained for the 6 lnc173 KO clones and for the 4 control clones were remarkably similar and gave no indication of a defect or enhancement (Fig. 6C, D). Analogously, we created two monoclonal HEK 293T-derived cell lines with complete deletion of LINC00173 (Supplementary Fig. S4A). When these lines were infected with VSV-G-pseudotyped HIV-1 NL4-3 Δenv/EGFP, the percentage of GFP+ cells and GFP levels were also indistinguishable from two control lines (Supplementary Fig. S4B, C). Next, we tested whether viral infectivity or particle release might be affected by the absence of lnc173. We produced fully infectious HIV-1 NL4-3 particles in the HEK 293T lnc173 KO clone 11-2B3 and compared yield and infectivity to virus produced in HEK 293T control clone 1B5. Stocks produced from both cell lines yielded similar concentrations of viral particles, demonstrating that virion production and release were unaffected by deletion of LINC00173 (Supplementary Fig. S5A). When virus from these stocks, normalized for p24 content, was used to infect the TZM-bl reporter cell line to determine infectivity, no difference was detectable (Fig. 6E) (Derdeyn et al., 2000, Platt et al., 1998). To understand whether the presence of lnc173 influences viral growth over multiple rounds of replication, we infected 4 Jurkat lnc173 KO clones and 4 control clones with fully replication-competent HIV-1 NL4-3 and tracked viral replication over a period of three weeks. While results varied between the clones of each group, no clear defect or enhancement was associated with the deletion of LINC00173 (Fig. 6F). Infection of the same clones with virus produced in lnc173-deficient HEK 293T cells yielded a highly similar growth curve (Supplementary Fig. S5B). These results demonstrate that both transcript variants of lnc173 are dispensable for replication of HIV-1 in cell culture.
Fig. 6.
Deletion of LINC00173 does not affect integration, viral gene expression, infectivity or growth of HIV-1. (A and B) Levels of lnc173 (A) TSV1 and (B) TSV2 after deletion with CRISPR-Cas9 in Jurkat cells, as determined by qRT-PCR. Shown is the average of 6 monoclonal knock-out lines (lnc173 KO) and 4 monoclonal vector control lines, including every data point from 3 independent experiments. Error bars indicate SEM. RPLP0 was used as reference gene. (C and D) Monoclonal lnc173 knock-out and vector control Jurkat lines were infected with VSV-G-pseudotyped HIV-1 NL4-3 Δenv/EGFP (undiluted or diluted 1:4). Cells were harvested 48 h later and GFP levels were determined by flow cytometry. Shown is the average of 6 monoclonal knock-out lines (lnc173 KO) and 4 monoclonal vector control lines, including every data point from 3 independent experiments. Error bars indicate SEM. (C) Percentage of GFP+ cells in live gate. (D) Geometric mean fluorescence intensity (gMFI) of cells in live gate. (E) Infectivity of fully replication-competent HIV-1 NL4-3 produced in monoclonal lnc173-deficient HEK 293T cells (lnc173 KO) or a vector control line. TZM-bl cells were infected in triplicate with equivalent virus amounts normalized by p24 content, as indicated. Luciferase was measured 48 h after infection. Error bars indicate standard deviation. (F) Growth curve of HIV-1 NL4-3 in 4 monoclonal Jurkat lines deficient for lnc173 and in 4 monoclonal control lines. p24 levels in the supernatant of infected cells was quantified on the indicated days post infection.
2.6. PMA/ionomycin activation of T cells decreases cellular levels of lnc173
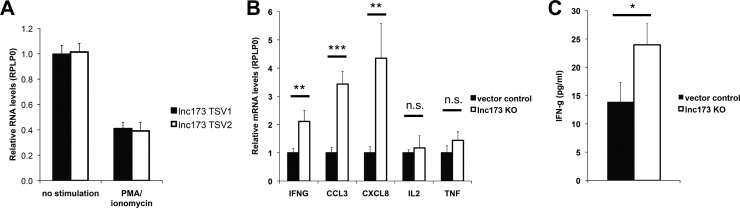
HIV-1 replicates efficiently only in activated cells, and consequently has evolved multiple redundant mechanisms to increase the state of cellular activation in its host cell (Alexander et al., 1997, Fenard et al., 2005, McDougal et al., 1985, Postler and Desrosiers, 2012, Simmons et al., 2001, Stevenson et al., 1990, Zack et al., 1990, Zagury et al., 1986). We therefore investigated whether the increase in lnc173 levels was associated with general T-cell activation. Surprisingly, stimulation of Jurkat T cells with PMA and ionomycin resulted in a marked decrease in the levels of lnc173 TSV1 and TSV2 ( Fig. 7A). Similarly, RNA levels of a previously unknown, likely lnc173 ortholog in the mouse genome decreased upon stimulation with PMA and ionomycin in EL4 cells, indicating at least some degree of evolutionary conservation of this mechanism (Supplementary Fig. S6). Interestingly, the lnc173 KO Jurkat clones on average expressed higher levels of mRNA encoding a subset of cytokines than the cognate control lines. Specifically, average mRNA levels of IFNG, CCL3 and CXCL8 upon stimulation with PMA and ionomycin were significantly higher in lnc173 KO clones than in control clones, whereas mRNA levels of IL2 and TNF did not differ (Fig. 7B and Supplementary Fig. S7A-E). Due to the central role of IFN-γ in the differentiation of naive T cells into TH1 cells during the immune response to viral and other intracellular infections, we tested whether the observed increase in IFNG mRNA translated into an increase in IFN-γ protein levels secreted by lnc173 KO clones (Schoenborn and Wilson, 2007). Indeed, the average concentration of IFN-γ in the supernatant of lnc173 KO Jurkat clones was modestly but reproducibly elevated compared to the average of control clones, consistent with the mRNA results (Fig. 7C and Supplementary Fig. S7F). Although there was considerable variability between the clones of each group, these results raise the intriguing possibility that lnc173 may be involved in the transcriptional regulation of a subset of cytokines. Further studies are under way to address this question in detail.
Fig. 7.
Jurkat cells deficient for lnc173 tend to express higher levels of specific cytokines upon stimulation. (A) Levels of lnc173 TSV1 and TSV2 in Jurkat cells after stimulation with 25 ng/ml PMA and 500 ng/ml ionomycin for 4 h, as quantified by qRT-PCR. RPLP0 was used as reference gene. Results are the average of 3 independent experiments. Error bars indicate standard deviation. (B) mRNA levels of the cytokine-encoding genes IFNG, CCL3, CXCL8, IL2 and TNF after stimulation with 25 ng/ml PMA and 500 ng/ml ionomycin for 4 h in lnc173 knock-out and control Jurkat cells, as quantified by qRT-PCR. RPLP0 was used as reference gene. Shown is the average of 6 monoclonal knock-out lines (lnc173 KO) and 4 monoclonal vector control lines, including every data point from 5 to 8 independent experiments. Error bars indicate SEM. (C) Concentration of IFN-γ in the supernatant of lnc173 knock-out and control Jurkat cells after stimulation with 25 ng/ml PMA and 500 ng/ml ionomycin for 24 h, as determined by ELISA. Shown is the average of 6 monoclonal knock-out lines (lnc173 KO) and 4 monoclonal vector control lines, including every data point from 5 independent experiments. Error bars indicate SEM. p-values were determined with Welch's t-test. ***, p ≤ 0.001; **, p ≤ 0.01; *, p ≤ 0.05; n.s., not significant.
3. Discussion
Of the thousands of lncRNAs expressed in the human body, the vast majority remains uncharacterized, in particular in the context of viral infections. Until the recently published microarray study by Trypsteen et al., all reports investigating the effect of HIV-1 infection on lncRNA expression scrutinized only a small panel of previously characterized lncRNAs and thus, while yielding important insights, were limited in scope (Imam et al., 2015, Trypsteen et al., 2016; Zhang et al., 2013). To our knowledge, this is the first study using RNA-seq data to address specifically this aspect of the HIV-1 life cycle, although others have touched on the subject (Peng et al., 2014). To minimize the effect of interexperimental variability, we utilized two separate data sets created by two independent groups and employing two different strains of HIV-1 (Chang et al., 2011, Mohammadi et al., 2013). The finding that only three lncRNAs exhibited differential expression in both studies is somewhat surprising, especially considering that Trypsteen et al. (2016) found 328 lncRNA-coding genes to be altered (including those identified in this study, namely LINC00173, CCDC18-AS1 and MCM3AP-AS1). The reason for this discrepancy is not clear, but it may stem from the inherent differences between RNA-seq and microarray technology. Individual lncRNA expression levels tend to be lower than mRNA levels, and while microarrays afford a lower dynamic range than RNA-seq, they provide higher accuracy of quantification for low-abundance transcripts (Derrien et al., 2012, Jiang et al., 2011; Łabaj et al., 2011; Ravasi et al., 2006; Toung et al., 2011; Xu et al., 2011). Additionally, using two independent data sets is likely to reduce the number of false positives but increase the number of false negatives.
LINC00173, a previously uncharacterized lncRNA, is conserved amongst mammals and at least a partial homolog appears to be present in chickens. As cross-species conservation is often an indicator of functional importance, we chose to further investigate the role of LINC00173 during HIV-1 infection. The experimental results presented here clearly demonstrate that its two transcript variants are robustly expressed in various human cell types, in accordance with the available in silico data. Consistent with the data mined from the studies by Chang et al. (2011), Mohammadi et al. (2013), and Trypsteen et al. (2016), lnc173 TSV1 and TSV2 are up-regulated during the course of HIV-1 infection. This effect is dose- and, more importantly, time-dependent. The observation that lnc173 TSV1 and TSV2 levels increase within the first 12 h after infection, remain increased at 24 h, but then decrease progressively towards baseline at 36 and 48 h, indicates targeted control of expression. Nonetheless, loss of the LINC00173 locus does not affect any aspect of the HIV-1 replication cycle in cell culture, from entry to particle production. Based on their nuclear localization, lnc173 TSV1 and/or TSV2 are likely involved in transcriptional regulation, but their precise role in this process, if any, remains unclear. It is conceivable that lnc173 may contribute to the regulation of viral transcription only under specific conditions, such as entering or exiting viral latency – an intriguing possibility that remains to be explored.
It is also possible that lnc173 affects the expression or function of proteins that are not directly involved in viral replication on a cellular level but are important on an organismal level, such as cytokines that coordinate the immune response. Activation of T cells by exposure to PMA and ionomycin, which increases the expression of several cytokines, decreases the levels of both lnc173 transcript variants, opening up the possibility that lnc173 TSV1 and/or TSV2 may be negative regulators of cytokine expression. Indeed, upon activation the monoclonal Jurkat cell lines deficient for the LINC00173 locus on average produced higher mRNA levels of several cytokines than cognate controls, lending preliminary support to this hypothesis. Ongoing experiments outside the scope of this study will further explore the role of LINC00173 in cytokine regulation.
In this context, it is also interesting to note that the expression levels of both lnc173 transcript variants are increased during HIV-1 infection but decreased during T-cell activation, even though HIV-1 has evolved several independent mechanisms to induce the activation of host cells (Alexander et al., 1997, Fenard et al., 2005, Postler and Desrosiers, 2012, Simmons et al., 2001). This unexpected antagonistic phenotype may imply a specific function of lnc173 to be exploited or blunted by HIV-1. Of note, the cytokines exhibiting increased expression in the lnc173 KO Jurkat clones include IFN-γ, a cytokine of central importance to the development and maintenance of the adaptive immune response to intracellular pathogens, including viruses (Schoenborn and Wilson, 2007). It is therefore tempting to speculate that HIV-1 has evolved the ability to increase levels of lnc173 to impede the immune response.
4. Materials and methods
4.1. In silico analysis
The normalized FPKM values of the RNA-seq experiments carried out by Chang et al. were obtained through the NCBI Gene Expression Omnibus portal (https://www.ncbi.nlm.nih.gov/gds, dataset GSE38006) (Barrett et al., 2013, Chang et al., 2011, Edgar et al., 2002). Gene information was analyzed and filtered by individual evaluation of associated RefSeq identifiers and records. All lncRNA candidates identified in this data set were queried in the online platform created for the RNA-seq data by Mohammadi et al. (2013), Patterns of Expression and Analysis of Clusters of HIV/Host interactions (PEACHi, http://www.peachi.labtelenti.org). To that end, RefSeq identifiers of candidate genes were matched to corresponding Ensembl identifiers (Ensembl release 75, www.ensembl.org), which could be entered into PEACHi (Yates et al., 2016). Phylogenetic conservation, expression data, ChIP-seq data and DNase hypersensitivity data were obtained from publicly accessible tracks on the UCSC Genome Browser (GRCh38, hg38; http://genome.ucsc.edu) (Kent et al., 2002). PhyloCSF score, number of Bazzini small ORFs and secondary structure prediction were obtained from LNCipedia (www.lncipedia.org) (Volders et al., 2013, Volders et al., 2015).
4.2. Cell culture and production of virus stocks
Jurkat cells (a kind gift from Dr. Stephen Goff, Columbia University), EL4 cells, U937 cells and THP-1 cells (both ATCC, Manassas, VA, USA) were maintained in RPMI-1640 medium (Gibco/Thermo Fisher Scientific, Waltham, MA, USA) supplemented with 10% fetal bovine serum (FBS; Atlanta Biologicals, Flowery Branch, GA, USA). HEK293T/17 (ATCC) and TZM-bl cells (obtained through the NIH AIDS Reagent Program, Division of AIDS, NIAID, NIH) were maintained in DMEM (Gibco/Thermo Fisher Scientific) with 10% FBS (Derdeyn et al., 2000, Platt et al., 1998). The plasmids encoding HIV-1 NL4-3 Δenv/EGFP (pNL4-3-deltaE-EGFP) and VSV-G (pVSV-G) also were generous gifts from Dr. Stephen Goff (Haili Zhang et al., 2004). To produce pseudotyped virions, HEK 293T/17 cells were co-transfected with pNL4-3-deltaE-EGFP and pVSV-G at a mass ratio of 3:1 using FuGENE 6 (Promega, Madison, WI, USA). Transfection reagent was washed out after 6 h, and virion-containing supernatant was harvested 48 h after transfection. Fully replication-competent HIV-1 NL4-3 was produced from plasmid pNL4-3, also obtained through the NIH AIDS Reagent Program (GenBank ID AF324493) (Adachi et al., 1986). NL4-3 stocks were produced from two monoclonal derivates of HEK293T/17, LINC00173 knock-out clone 11-2B3 and wild-type control clone 1B5. These cells were transfected using jetPRIME (Polyplus Transfection, Illkirch, France) and supernatant was harvested after 72 h. The concentration of p24 in fully replication-competent virus stocks was determined by p24 Antigen Capture Assay (Advanced Bioscience Laboratories, Rockville, MD, USA).
4.3. Isolation of nuclear RNA
To isolate nuclear RNA, 107 Jurkat cells were resuspended in 1 ml phosphate-buffered saline (PBS), 1 ml of lysis buffer C1 (1.28 M sucrose, 40 mM Tris-HCl pH 7.5, 20 mM MgCl2, 4% Triton X-100) and 3 ml ddH2O. After incubation on ice for 15 min, cells were centrifuged at 4 °C and 580×g for 15 min. Supernatant containing the cytoplasmic fraction was discarded. The pellet containing the nuclear fraction was resuspended in 350 µl Buffer RLT + β-mercaptoethanol (QIAGEN, Germantown, MD, USA). In parallel, 107 Jurkat cells were resuspended directly in 350 µl Buffer RLT + β-mercaptoethanol to obtain whole-cell RNA. RNA was isolated from both sample sets using the RNeasy Mini Kit (QIAGEN), following the manufacturer's instructions. This included an on-column DNase I digest during purification.
4.4. Quantitative reverse-transcription polymerase chain reaction (qRT-PCR)
Cells were lysed directly in Buffer RLT + β-mercaptoethanol and RNA was isolated using the RNeasy Mini Kit (QIAGEN), according to the manufacturer's instructions, with on-column DNase I digest. Reverse transcription of usually 1 µg of purified RNA into cDNA was performed with SuperScript III Reverse Transcriptase and oligo(dT)12–18 primers, unless noted otherwise. VeriQuest Fast SYBR Green qPCR Master Mix was used for qRT-PCR (all Thermo Fisher Scientific). Relative cDNA levels were determined using the ΔΔCt method (Livak and Schmittgen, 2001). See Supplementary Table S2 for primer sequences.
4.5. CRISPR-Cas9 knock-out
In HEK 293T cells, the LINC00173 locus was deleted using plasmid pSpCas9(BB)-2A-Puro (PX459), provided by Dr. Feng Zhang (Massachusetts Institute of Technology) through Addgene (plasmid #48139) (Ran et al., 2013). Specifically, cells were transfected with pairs of plasmids targeting each end of the transcribed sequence, using Lipofectamine 2000 (Thermo Fisher Scientific). Supplementary Table S1 lists the sgRNA sequences and combinations used. Cells transfected with PX459 without an sgRNA sequence served as control. Successfully transfected cells were selected with 4 µg/ml puromycin for three days. Surviving cells were separated into monoclonal cultures by limiting dilution, which were then screened for successful deletion of the LINC00173 locus with the primers listed in Supplementary Table S3. For deletion of LINC00173 in Jurkat cells, the same sgRNA sequences were introduced into plasmid pSpCas9(BB)-2A-GFP (PX458), also provided by Dr. Feng Zhang through Addgene (plasmid #48138) (Ran et al., 2013). Jurkat cells were transfected with the same pairs of plasmids detailed in Supplementary Table S1 using an Amaxa Nucleofector II device and the Cell Line Nucleofector Kit V (both from Lonza, Basel, Switzerland). After 48 h, cells with high levels of GFP expression were identified by FACS and sorted into monoclonal cultures. Screening for successful deletion of LINC00173 was performed as described above.
4.6. Infection with VSV-G-pseudotyped virus particles
HEK 293T cells were infected by incubation with VSV-G-pseudotyped HIV-1 NL4-3 Δenv/EGFP for 4 h at 37 °C, while THP-1 and Jurkat cells were usually infected by spin inoculation for 2 h at 34 °C and 1000×g. Polybrene was added at a concentration of 6 µg/ml to all pseudotype infections. After incubation or spin inoculation, supernatant was replaced with fresh medium. Unless otherwise noted, cells were harvested 24 h after infection for RNA isolation as described above, or 48 h after infection for flow cytometry.
4.7. Infectivity assay
The infectivity of virus produced in lnc173-deficient cells or control cells was quantified by infection of TZM-bl reporter cells, essentially as previously described (Bixby et al., 2010). Cells were infected in 2-fold serial dilutions with virus amounts normalized for p24 content, each in triplicate. Luciferase activity was quantified 48 h after infection using the Britelite Plus Reporter Gene Assay System (PerkinElmer, Waltham, MA, USA).
4.8. Virus growth curve
Monoclonal Jurkat cells deficient for lnc173 or controls were infected with virus amounts equivalent to 25 ng p24. Supernatant from the infected cells was harvested every 2–4 days and the concentration of p24 was determined by p24 Antigen Capture Assay (Advanced Bioscience Laboratories).
4.9. Enzyme-linked immunosorbent assay (ELISA)
To determine the concentration of IFN-γ secreted by lnc173 KO and control Jurkat clones, cells were stimulated with 25 ng/ml PMA and 500 ng/ml ionomycin for 24 h. Clarified supernatant was harvested and IFN-γ concentration was quantified using the IFN gamma Human Uncoated ELISA Kit with Plates (Thermo Fisher Scientific).
Acknowledgements
The authors wish to thank Drs. Yosef Sabo and Stephen Goff (both Columbia University) for the generous gifts of their reagents, time and advice. This work was supported by NIH grants R01AI068977 (to SG) and R01AI104523 (to RCD), as well as institutional support from Columbia University.
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.virol.2017.08.006.
Appendix A. Supplementary material
Supplementary material Fig. S1. Work flow for the meta-analysis of the RNA-seq data byChang et al. (2011)andMohammadi et al. (2013). See text for detail.
Supplementary material Fig. S2. The chromatin signature of theLINC00173locus is consistent with active transcription. Tracks from top to bottom: H3K27Ac signature; digital DNase I hypersensitivity clusters; transcription factor ChIP-seq; DNase I hypersensitivity uniform peaks; DNase I/ FAIRE/ ChIP synthesis; chromatin state segmentation; histone modifications by ChIP-seq; nuclease-accessible sites; DNase I hypersensitivity; ChIA-PET; DNA/CpG methylation; histone modifications by ChIP-seq; 5C; FAIRE. All data obtained from and visualized with UCSC Genome Browser (http://genome.ucsc.edu).
Supplementary material Fig. S3. Deletion of lnc173 does not affect integration or viral gene expression of HIV-1 in Jurkat cells. (A and B) Levels of lnc173 (A) TSV1 and (B) TSV2 after deletion with CRISPR-Cas9 in Jurkat cells, as determined by qRT-PCR. Results are the average of 3 independent experiments. RPLP0 was used as reference gene. Error bars indicate standard deviation. (C and D) Monoclonal lnc173 knock-out and vector control Jurkat lines were infected with VSV-G-pseudotyped HIV-1 NL4-3 Δenv/EGFP (undiluted or diluted 1:4). Cells were harvested 48 h after infection and GFP levels were determined by flow cytometry. (C) Percentage of GFP+ cells in live gate. (D) Geometric mean fluorescence intensity (gMFI) of cells in live gate. Results are the average of 3 independent experiments. Error bars indicate standard deviation.
Supplementary material Fig. S4. Deletion of lnc173 does not affect integration or viral gene expression of HIV-1 in HEK 293T cells. (A) PCR with genomic DNA from monoclonal HEK293T lines. Reactions on the left side detect the wild-type sequence (blue), whereas primers on the right side will only give a product if lnc173 has been deleted (red). Two lnc173 knock-out lines (4-2FB and 11-2B3) and two corresponding control lines (1B5 and 1D6) were used in further experiments. (B and C) Monoclonal lnc173 knock-out and vector control HEK 293T lines were infected with VSV-G-pseudotyped HIV-1 NL4-3 Δenv/EGFP in 4-fold serial dilutions. Cells were lysed 48 h after infection and GFP levels were determined by flow cytometry. (B) Percentage of GFP+ cells in live gate. (D) Geometric mean fluorescence intensity (gMFI) of cells in live gate. Results are representative of 2 independent experiments.
Supplementary material Fig. S5. Deletion of lnc173 does not affect particle production or growth of HIV-1. (A) Concentration of p24 in the supernatant of monoclonal HEK 293T cells with (vector control, clone 1B5) or without lnc173 (lnc173 KO, clone 11-2B3) producing HIV-1 NL4-3. (B) Growth curve of HIV-1 NL4-3 produced in lnc173-deficient HEK 293T cells (clone 11-2B3) in four monoclonal Jurkat lines deficient for lnc173 and in four monoclonal control lines. p24 levels in the supernatant of infected cells was quantified on the indicated days post infection.
Supplementary material Fig. S6. RNA levels of a likely murine ortholog ofLINC00173are decreased after stimulation. Levels of “murine lnc173” in EL4 cells after stimulation with 25 ng/ml PMA and 500 ng/ml ionomycin for 4 h, as quantified by qRT-PCR. Hprt was used as reference gene. Results are the average of 3 independent experiments. Error bars indicate standard deviation.
Supplementary material Fig. S7. Jurkat cells deficient for lnc173 tend to express lower levels of specific cytokines upon stimulation. (A-E) mRNA levels of the cytokine-encoding genes (A) IFNG, (B) CCL3, (C) CXCL8, (D) IL2 and (E) TNF, as quantified by qRT-PCR. lnc173 knock-out and control Jurkat clones were stimulated with 25 ng/ml PMA and 500 ng/ml ionomycin for 4 h. RPLP0 was used as reference gene. Results are the average of 5–8 independent experiments. Error bars indicate standard deviation. Note logarithmic scale. (F) Concentration of IFN-γ in the supernatant of individual lnc173 knock-out and control Jurkat clones after stimulation with 25 ng/ml PMA and 500 ng/ml ionomycin for 24 h, as determined by ELISA. Results are the average of 5 independent experiments. Error bars indicate standard deviation.
Supplementary material
References
- Adachi A., Gendelman H.E., Koenig S., Folks T., Willey R., Rabson A., Martin M.A. Production of acquired immunodeficiency syndrome-associated retrovirus in human and nonhuman cells transfected with an infectious molecular clone. J. Virol. 1986;59:284–291. doi: 10.1128/jvi.59.2.284-291.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander L., Du Z., Rosenzweig M., Jung J.U., Desrosiers R.C. A role for natural simian immunodeficiency virus and human immunodeficiency virus type 1 nef alleles in lymphocyte activation. J. Virol. 1997;71:6094–6099. doi: 10.1128/jvi.71.8.6094-6099.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atianand M.K., Cafferey D.R., Fitzgerald K.A. Immunobiology of long noncoding RNAs. Annu. Rev. Immunol. 2017 doi: 10.1146/annurev-immunol-041015-055459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett T., Wilhite S.E., Ledoux P., Evangelista C., Kim I.F., Tomashevsky M., Marshall K.A., Phillippy K.H., Sherman P.M., Holko M., Yefanov A., Lee H., Zhang N., Robertson C.L., Serova N., Davis S., Soboleva A. NCBI GEO: archive for functional genomics data sets–update. Nucleic Acids Res. 2013;41:D991–D995. doi: 10.1093/nar/gks1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bixby J.G., Laur O., Johnson W.E., Desrosiers R.C. Diversity of envelope genes from an uncloned stock of SIVmac251. AIDS Res. Hum. Retrovir. 2010;26:1115–1131. doi: 10.1089/aid.2010.0029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carnero E., Barriocanal M., Prior C., Pablo Unfried J., Segura V., Guruceaga E., Enguita M., Smerdou C., Gastaminza P., Fortes P. Long noncoding RNA EGOT negatively affects the antiviral response and favors HCV replication. EMBO Rep. 2016 doi: 10.15252/embr.201541763. (e201541763) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang S.T., Sova P., Peng X., Weiss J., Law G.L., Palermo R.E., Katze M.G. Next-generation sequencing reveals HIV-1-mediated suppression of T cell activation and RNA processing and regulation of noncoding RNA expression in a CD4+ T cell line. mBio. 2011:2. doi: 10.1128/mBio.00134-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhary, R., Lal, A., 2016. Long noncoding RNAs in the p53 network. Wiley Interdisciplinary Reviews. RNA. 〈 10.1002/wrna.1410〉. [DOI] [PMC free article] [PubMed]
- Chen L.-L. Linking long noncoding RNA localization and function. Trends Biochem. Sci. 2016 doi: 10.1016/j.tibs.2016.07.003. [DOI] [PubMed] [Google Scholar]
- Clemson C.M., Hutchinson J.N., Sara S.A., Ensminger A.W., Fox A.H., Chess A., Lawrence J.B. An architectural role for a nuclear noncoding RNA: NEAT1 RNA is essential for the structure of paraspeckles. Mol. Cell. 2009;33:717–726. doi: 10.1016/j.molcel.2009.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derdeyn C.A., Decker J.M., Sfakianos J.N., Wu X., O'Brien W.A., Ratner L., Kappes J.C., Shaw G.M., Hunter E. Sensitivity of human immunodeficiency virus type 1 to the fusion inhibitor T-20 is modulated by coreceptor specificity defined by the V3 loop of gp120. J. Virol. 2000;74:8358–8367. doi: 10.1128/jvi.74.18.8358-8367.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derrien T., Johnson R., Bussotti G., Tanzer A., Djebali S., Tilgner H., Guernec G., Martin D., Merkel A., Knowles D.G., Lagarde J., Veeravalli L., Ruan X., Ruan Y., Lassmann T., Carninci P., Brown J.B., Lipovich L., Gonzalez J.M., Thomas M., Davis C.A., Shiekhattar R., Gingeras T.R., Hubbard T.J., Notredame C., Harrow J., Guigó R. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res. 2012;22:1775–1789. doi: 10.1101/gr.132159.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R., Domrachev M., Lash A.E. Gene expression omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30:207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenard D., Yonemoto W., de Noronha C., Cavrois M., Williams S.A., Greene W.C. Nef is physically recruited into the immunological synapse and potentiates T cell activation early after TCR engagement. J. Immunol. 2005;175:6050–6057. doi: 10.4049/jimmunol.175.9.6050. [DOI] [PubMed] [Google Scholar]
- Fortes P., Morris K. Long noncoding RNAs in viral infections. Virus Res. 2015;212:1–11. doi: 10.1016/j.virusres.2015.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goff L.A., Rinn J.L. Linking RNA biology to lncRNAs. Genome Res. 2015;25:1456–1465. doi: 10.1101/gr.191122.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrow J., Frankish A., Gonzalez J.M., Tapanari E., Diekhans M., Kokocinski F., Aken B.L., Barrell D., Zadissa A., Searle S., Barnes I., Bignell A., Boychenko V., Hunt T., Kay M., Mukherjee G., Rajan J., Despacio-Reyes G., Saunders G., Steward C., Harte R., Lin M., Howald C., Tanzer A., Derrien T., Chrast J., Walters N., Balasubramanian S., Pei B., Tress M., Rodriguez J.M., Ezkurdia I., van Baren J., Brent M., Haussler D., Kellis M., Valencia A., Reymond A., Gerstein M., Guigó R., Hubbard T.J. GENCODE: the reference human genome annotation for The ENCODE Project. Genome Res. 2012;22:1760–1774. doi: 10.1101/gr.135350.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu B., Huo Y., Chen G., Yang L., Wu D., Zhou J. Functional prediction of differentially expressed lncRNAs in HSV-1 infected human foreskin fibroblasts. Virol. J. 2016;13:137. doi: 10.1186/s12985-016-0592-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imam H., Shahr Bano A., Patel P., Holla P., Jameel S. The lncRNA NRON modulates HIV-1 replication in a NFAT-dependent manner and is differentially regulated by early and late viral proteins. Sci. Rep. 2015;5:8639. doi: 10.1038/srep08639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imamura K., Imamachi N., Akizuki G., Kumakura M., Kawaguchi A., Nagata K., Kato A., Kawaguchi Y., Sato H., Yoneda M., Kai C., Yada T., Suzuki Y., Yamada T., Ozawa T., Kaneki K., Inoue T., Kobayashi M., Kodama T., Wada Y., Sekimizu K., Akimitsu N. Long noncoding RNA NEAT1-dependent SFPQ relocation from promoter region to paraspeckle mediates IL8 expression upon immune stimuli. Mol. Cell. 2014;53:393–406. doi: 10.1016/j.molcel.2014.01.009. [DOI] [PubMed] [Google Scholar]
- Jiang L., Schlesinger F., Davis C.A., Zhang Y., Li R., Salit M., Gingeras T.R., Oliver B. Synthetic spike-in standards for RNA-seq experiments. Genome Res. 2011;21:1543–1551. doi: 10.1101/gr.121095.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent W.J., Sugnet C.W., Furey T.S., Roskin K.M., Pringle T.H., Zahler A.M., Haussler D. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi-Ishihara M., Yamagishi M., Hara T., Matsuda Y., Takahashi R., Miyake A., Nakano K., Yamochi T., Ishida T., Watanabe T. HIV-1-encoded antisense RNA suppresses viral replication for a prolonged period. Retrovirology. 2012;9:38. doi: 10.1186/1742-4690-9-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Łabaj P.P., Leparc G.G., Linggi B.E., Markillie L.M., Wiley H.S., Kreil D.P. Characterization and improvement of RNA-Seq precision in quantitative transcript expression profiling. Bioinformatics. 2011;27:i383–i391. doi: 10.1093/bioinformatics/btr247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landry S., Halin M., Lefort S., Audet B., Vaquero C., Mesnard J.-M., Barbeau B. Detection, characterization and regulation of antisense transcripts in HIV-1. Retrovirology. 2007;4:71. doi: 10.1186/1742-4690-4-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazar D.C., Morris K.V., Saayman S.M. The emerging role of long non-coding RNAs in HIV infection. Virus Res. 2016;212:114–126. doi: 10.1016/j.virusres.2015.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Chen C., Ma X., Geng G., Liu B., Zhang Y., Zhang S., Zhong F., Liu C., Yin Y., Cai W., Zhang H. Long noncoding RNA NRON contributes to HIV-1 latency by specifically inducing tat protein degradation. Nat. Commun. 2016;7:11730. doi: 10.1038/ncomms11730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Ludwig L.B., Ambrus J.L., Krawczyk K.A., Sharma S., Brooks S., Hsiao C.-B., Schwartz S.A. Human Immunodeficiency Virus-Type 1 LTR DNA contains an intrinsic gene producing antisense RNA and protein products. Retrovirology. 2006;3:80. doi: 10.1186/1742-4690-3-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macian F. NFAT proteins: key regulators of T-cell development and function. Nat. Rev. Immunol. 2005;5:472–484. doi: 10.1038/nri1632. [DOI] [PubMed] [Google Scholar]
- McDougal J.S., Mawle A., Cort S.P., Nicholson J.K., Cross G.D., Scheppler-Campbell J.A., Hicks D., Sligh J. Cellular tropism of the human retrovirus HTLV-III/LAV. I. role of T cell activation and expression of the T4 antigen. J. Immunol. 1985;135:3151–3162. [PubMed] [Google Scholar]
- Mohammadi P., Desfarges S., Bartha I., Joos B., Zangger N., Muñoz M., Günthard H.F., Beerenwinkel N., Telenti A., Ciuffi A. 24 h in the life of HIV-1 in a T cell line. PLoS Pathog. 2013;9:e1003161. doi: 10.1371/journal.ppat.1003161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee N., Calviello L., Hirsekorn A., de Pretis S., Pelizzola M., Ohler U. Integrative classification of human coding and noncoding genes through RNA metabolism profiles. Nat. Struct. Mol. Biol. 2017;24:86–96. doi: 10.1038/nsmb.3325. [DOI] [PubMed] [Google Scholar]
- Ouyang, J., Hu, J., Chen, J.-L., 2015. lncRNAs regulate the innate immune response to viral infection. Wiley Interdisciplinary Reviews. RNA. n/a–n/a. 〈 10.1002/wrna.1321〉. [DOI] [PMC free article] [PubMed]
- Peng X., Gralinski L., Armour C.D., Ferris M.T., Thomas M.J., Proll S., Bradel-Tretheway B.G., Korth M.J., Castle J.C., Biery M.C., Bouzek H.K., Haynor D.R., Frieman M.B., Heise M., Raymond C.K., Baric R.S., Katze M.G. Unique signatures of long noncoding RNA expression in response to virus infection and altered innate immune signaling. mBio. 2010:1. doi: 10.1128/mBio.00206-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng X., Sova P., Green R.R., Thomas M.J., Korth M.J., Proll S., Xu J., Cheng Y., Yi K., Chen L., Peng Z., Wang J., Palermo R.E., Katze M.G. Deep sequencing of HIV infected cells: insights into nascent transcription and host-directed therapy. J. Virol. 2014 doi: 10.1128/JVI.00768-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry R.B.-T., Ulitsky I. The functions of long noncoding RNAs in development and stem cells. Development. 2016;143:3882–3894. doi: 10.1242/dev.140962. [DOI] [PubMed] [Google Scholar]
- Pircher A., Gebetsberger J., Polacek N. Ribosome-associated ncRNAs: an emerging class of translation regulators. RNA Biol. 2014;11:1335–1339. doi: 10.1080/15476286.2014.996459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platt E.J., Wehrly K., Kuhmann S.E., Chesebro B., Kabat D. Effects of CCR5 and CD4 cell surface concentrations on infections by macrophagetropic isolates of human immunodeficiency virus type 1. J. Virol. 1998;72:2855–2864. doi: 10.1128/jvi.72.4.2855-2864.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Postler T.S., Desrosiers R.C. The cytoplasmic domain of the HIV-1 glycoprotein gp41 induces NF-κB activation through TGF-β-activated kinase 1. Cell Host Microbe. 2012;11:181–193. doi: 10.1016/j.chom.2011.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran F.A., Hsu P.D., Wright J., Agarwala V., Scott D.A., Zhang F. Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 2013;8:2281–2308. doi: 10.1038/nprot.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravasi T., Suzuki H., Pang K.C., Katayama S., Furuno M., Okunishi R., Fukuda S., Ru K., Frith M.C., Gongora M.M., Grimmond S.M., Hume D.A., Hayashizaki Y., Mattick J.S. Experimental validation of the regulated expression of large numbers of non-coding RNAs from the mouse genome. Genome Res. 2006;16:11–19. doi: 10.1101/gr.4200206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saayman S., Ackley A., Turner A.-M.W., Famiglietti M., Bosque A., Clemson M., Planelles V., Morris K.V. An HIV-encoded antisense long noncoding RNA epigenetically regulates viral transcription. Mol. Ther. 2014 doi: 10.1038/mt.2014.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki Y.T.F., Ideue T., Sano M., Mituyama T., Hirose T. MENepsilon/beta noncoding RNAs are essential for structural integrity of nuclear paraspeckles. Proc. Natl. Acad. Sci. USA. 2009;106:2525–2530. doi: 10.1073/pnas.0807899106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlackow M., Nojima T., Gomes T., Dhir A., Carmo-Fonseca M., Proudfoot N.J. Distinctive patterns of transcription and RNA processing for human lincRNAs. Mol. Cell. 2016 doi: 10.1016/j.molcel.2016.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoenborn J.R., Wilson C.B. Regulation of interferon-gamma during innate and adaptive immune responses. Adv. Immunol. 2007;96:41–101. doi: 10.1016/S0065-2776(07)96002-2. [DOI] [PubMed] [Google Scholar]
- Sharma S., Findlay G.M., Bandukwala H.S., Oberdoerffer S., Baust B., Li Z., Schmidt V., Hogan P.G., Sacks D.B., Rao A. Dephosphorylation of the nuclear factor of activated T cells (NFAT) transcription factor is regulated by an RNA-protein scaffold complex. Proc. Natl. Acad. Sci. USA. 2011;108:11381–11386. doi: 10.1073/pnas.1019711108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmons A., Aluvihare V., McMichael A. Nef triggers a transcriptional program in T cells imitating single-signal T cell activation and inducing HIV virulence mediators. Immunity. 2001;14:763–777. doi: 10.1016/s1074-7613(01)00158-3. [DOI] [PubMed] [Google Scholar]
- Spurlock C.F., Crooke P.S., Aune T.M. Biogenesis and transcriptional regulation of long noncoding RNAs in the human immune system. J. Immunol. 2016;197:4509–4517. doi: 10.4049/jimmunol.1600970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevenson M., Stanwick T.L., Dempsey M.P., Lamonica C.A. HIV-1 replication is controlled at the level of T cell activation and proviral integration. EMBO J. 1990;9:1551–1560. doi: 10.1002/j.1460-2075.1990.tb08274.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toung J.M., Morley M., Li M., Cheung V.G. RNA-sequence analysis of human B-cells. Genome Res. 2011;21:991–998. doi: 10.1101/gr.116335.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trypsteen W., Mohammadi P., Van Hecke C., Mestdagh P., Lefever S., Saeys Y., De Bleser P., Vandesompele J., Ciuffi A., Vandekerckhove L., De Spiegelaere W. Differential expression of lncRNAs during the HIV replication cycle: an underestimated layer in the HIV-host interplay. Sci. Rep. 2016;6:36111. doi: 10.1038/srep36111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tycowski K.T., Guo Y.E., Lee N., Moss W.N., Vallery T.K., Xie M., Steitz J.A. Viral noncoding RNAs: more surprises. Genes Dev. 2015;29:567–584. doi: 10.1101/gad.259077.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulitsky I., Bartel D.P. lincRNAs: genomics, evolution, and mechanisms. Cell. 2013;154:26–46. doi: 10.1016/j.cell.2013.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volders P.-J., Helsens K., Wang X., Menten B., Martens L., Gevaert K., Vandesompele J., Mestdagh P. LNCipedia: a database for annotated human lncRNA transcript sequences and structures. Nucleic Acids Res. 2013;41:D246–D251. doi: 10.1093/nar/gks915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volders P.-J., Verheggen K., Menschaert G., Vandepoele K., Martens L., Vandesompele J., Mestdagh P. An update on LNCipedia: a database for annotated human lncRNA sequences. Nucleic Acids Res. 2015;43:D174–D180. doi: 10.1093/nar/gku1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West J.A., Davis C.P., Sunwoo H., Simon M.D., Sadreyev R.I., Wang P.I., Tolstorukov M.Y., Kingston R.E. The long noncoding RNAs NEAT1 and MALAT1 bind active chromatin sites. Mol. Cell. 2014 doi: 10.1016/j.molcel.2014.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willingham A.T., Orth A.P., Batalov S., Peters E.C., Wen B.G., Aza-Blanc P., Hogenesch J.B., Schultz P.G. A strategy for probing the function of noncoding RNAs finds a repressor of NFAT. Science. 2005;309:1570–1573. doi: 10.1126/science.1115901. [DOI] [PubMed] [Google Scholar]
- Winterling C., Koch M., Koeppel M., Garcia-Alcalde F., Karlas A., Meyer T.F. Evidence for a crucial role of a host non-coding RNA in influenza A virus replication. RNA Biol. 2014;11:66–75. doi: 10.4161/rna.27504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, W., Seok, J., Mindrinos, M.N., Schweitzer, A.C., Jiang, H., Wilhelmy, J., Clark, T.A., Kapur, K., Xing, Y., Faham, M., Storey, J.D., Moldawer, L.L., Maier, R.V., Tompkins, R.G., Wong, W.H., Davis, R.W., Xiao, W., Inflammation and Host Response to Injury Large-Scale Collaborative Research Program, 2011. Human Transcriptome Array for High-throughput Clinical Studies, vol. 108, pp. 3707–3712. 〈 10.1073/pnas.1019753108〉. [DOI] [PMC free article] [PubMed]
- Yates A., Akanni W., Amode M.R., Barrell D., Billis K., Carvalho-Silva D., Cummins C., Clapham P., Fitzgerald S., Gil L., Girón C.G., Gordon L., Hourlier T., Hunt S.E., Janacek S.H., Johnson N., Juettemann T., Keenan S., Lavidas I., Martin F.J., Maurel T., McLaren W., Murphy D.N., Nag R., Nuhn M., Parker A., Patricio M., Pignatelli M., Rahtz M., Riat H.S., Sheppard D., Taylor K., Thormann A., Vullo A., Wilder S.P., Zadissa A., Birney E., Harrow J., Muffato M., Perry E., Ruffier M., Spudich G., Trevanion S.J., Cunningham F., Aken B.L., Zerbino D.R., Flicek P. Ensembl 2016. Nucleic Acids Res. 2016;44:D710–D716. doi: 10.1093/nar/gkv1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zack J.A., Arrigo S.J., Weitsman S.R., Go A.S., Haislip A., Chen I.S. HIV-1 entry into quiescent primary lymphocytes: molecular analysis reveals a labile, latent viral structure. Cell. 1990;61:213–222. doi: 10.1016/0092-8674(90)90802-l. [DOI] [PubMed] [Google Scholar]
- Zagury D., Bernard J., Leonard R., Cheynier R., Feldman M., Sarin P.S., Gallo R.C. Long-term cultures of HTLV-III – infected T cells: a model of cytopathology of T-cell depletion in AIDS. Science. 1986;231:850–853. doi: 10.1126/science.2418502. [DOI] [PubMed] [Google Scholar]
- Zapata J.C., Campilongo F., Barclay R.A., DeMarino C., Iglesias-Ussel M.D., Kashanchi F., Romerio F. The human immunodeficiency virus 1 ASP RNA promotes viral latency by recruiting the Polycomb Repressor Complex 2 and promoting nucleosome assembly. Virology. 2017;506:34–44. doi: 10.1016/j.virol.2017.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Haili, Zhou Y., Alcock C., Kiefer T., Monie D., Siliciano J., Li Q., Pham P., Cofrancesco J., Persaud D., Siliciano R.F. Novel single-cell-level phenotypic assay for residual drug susceptibility and reduced replication capacity of drug-resistant human immunodeficiency virus type 1. J. Virol. 2004;78:1718–1729. doi: 10.1128/JVI.78.4.1718-1729.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Qi, Lai M.-M., Lou Y.-Y., Guo B.-H., Wang H.-Y., Zheng X.-Q. Transcriptome altered by latent human cytomegalovirus infection on THP-1 cells using RNA-seq. Gene. 2016;594:144–150. doi: 10.1016/j.gene.2016.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Quan, Chen C.-Y., Yedavalli V.S.R.K., Jeang K.-T. NEAT1 long noncoding RNA and paraspeckle bodies modulate HIV-1 posttranscriptional expression. mBio. 2013;4 doi: 10.1128/mBio.00596-12. (e00596-12) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H., Chen M., Lind S.B., Pettersson U. Distinct temporal changes in host cell lncRNA expression during the course of an adenovirus infection. Virology. 2016;492:242–250. doi: 10.1016/j.virol.2016.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zolotukhin A.S., Michalowski D., Bear J., Smulevitch S.V., Traish A.M., Peng R., Patton J., Shatsky I.N., Felber B.K. PSF acts through the human immunodeficiency virus type 1 mRNA instability elements to regulate virus expression. Mol. Cell. Biol. 2003;23:6618–6630. doi: 10.1128/MCB.23.18.6618-6630.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material Fig. S1. Work flow for the meta-analysis of the RNA-seq data byChang et al. (2011)andMohammadi et al. (2013). See text for detail.
Supplementary material Fig. S2. The chromatin signature of theLINC00173locus is consistent with active transcription. Tracks from top to bottom: H3K27Ac signature; digital DNase I hypersensitivity clusters; transcription factor ChIP-seq; DNase I hypersensitivity uniform peaks; DNase I/ FAIRE/ ChIP synthesis; chromatin state segmentation; histone modifications by ChIP-seq; nuclease-accessible sites; DNase I hypersensitivity; ChIA-PET; DNA/CpG methylation; histone modifications by ChIP-seq; 5C; FAIRE. All data obtained from and visualized with UCSC Genome Browser (http://genome.ucsc.edu).
Supplementary material Fig. S3. Deletion of lnc173 does not affect integration or viral gene expression of HIV-1 in Jurkat cells. (A and B) Levels of lnc173 (A) TSV1 and (B) TSV2 after deletion with CRISPR-Cas9 in Jurkat cells, as determined by qRT-PCR. Results are the average of 3 independent experiments. RPLP0 was used as reference gene. Error bars indicate standard deviation. (C and D) Monoclonal lnc173 knock-out and vector control Jurkat lines were infected with VSV-G-pseudotyped HIV-1 NL4-3 Δenv/EGFP (undiluted or diluted 1:4). Cells were harvested 48 h after infection and GFP levels were determined by flow cytometry. (C) Percentage of GFP+ cells in live gate. (D) Geometric mean fluorescence intensity (gMFI) of cells in live gate. Results are the average of 3 independent experiments. Error bars indicate standard deviation.
Supplementary material Fig. S4. Deletion of lnc173 does not affect integration or viral gene expression of HIV-1 in HEK 293T cells. (A) PCR with genomic DNA from monoclonal HEK293T lines. Reactions on the left side detect the wild-type sequence (blue), whereas primers on the right side will only give a product if lnc173 has been deleted (red). Two lnc173 knock-out lines (4-2FB and 11-2B3) and two corresponding control lines (1B5 and 1D6) were used in further experiments. (B and C) Monoclonal lnc173 knock-out and vector control HEK 293T lines were infected with VSV-G-pseudotyped HIV-1 NL4-3 Δenv/EGFP in 4-fold serial dilutions. Cells were lysed 48 h after infection and GFP levels were determined by flow cytometry. (B) Percentage of GFP+ cells in live gate. (D) Geometric mean fluorescence intensity (gMFI) of cells in live gate. Results are representative of 2 independent experiments.
Supplementary material Fig. S5. Deletion of lnc173 does not affect particle production or growth of HIV-1. (A) Concentration of p24 in the supernatant of monoclonal HEK 293T cells with (vector control, clone 1B5) or without lnc173 (lnc173 KO, clone 11-2B3) producing HIV-1 NL4-3. (B) Growth curve of HIV-1 NL4-3 produced in lnc173-deficient HEK 293T cells (clone 11-2B3) in four monoclonal Jurkat lines deficient for lnc173 and in four monoclonal control lines. p24 levels in the supernatant of infected cells was quantified on the indicated days post infection.
Supplementary material Fig. S6. RNA levels of a likely murine ortholog ofLINC00173are decreased after stimulation. Levels of “murine lnc173” in EL4 cells after stimulation with 25 ng/ml PMA and 500 ng/ml ionomycin for 4 h, as quantified by qRT-PCR. Hprt was used as reference gene. Results are the average of 3 independent experiments. Error bars indicate standard deviation.
Supplementary material Fig. S7. Jurkat cells deficient for lnc173 tend to express lower levels of specific cytokines upon stimulation. (A-E) mRNA levels of the cytokine-encoding genes (A) IFNG, (B) CCL3, (C) CXCL8, (D) IL2 and (E) TNF, as quantified by qRT-PCR. lnc173 knock-out and control Jurkat clones were stimulated with 25 ng/ml PMA and 500 ng/ml ionomycin for 4 h. RPLP0 was used as reference gene. Results are the average of 5–8 independent experiments. Error bars indicate standard deviation. Note logarithmic scale. (F) Concentration of IFN-γ in the supernatant of individual lnc173 knock-out and control Jurkat clones after stimulation with 25 ng/ml PMA and 500 ng/ml ionomycin for 24 h, as determined by ELISA. Results are the average of 5 independent experiments. Error bars indicate standard deviation.
Supplementary material