Abstract
Cytosolic sulfotransferase (SULT)-mediated sulfation is generally known to involve the transfer of a sulfonate group from the active sulfate, 3’-phosphoadenosine 5’-phosphosulfate (PAPS), to a hydroxyl group or an amino group of a substrate compound. We report here that human SULT2A1, in addition to being able to sulfate dehydroepiandrosterone (DHEA) and other hydroxysteroids, could also catalyze the sulfation of Δ4-3-ketosteroids, which carry no hydroxyl groups in their chemical structure. Among a panel of Δ4-3-ketosteroids tested as substrates, 4-androstene-3,17-dione and progesterone were found to be sulfated by SULT2A1. Mass spectrometry analysis and structural modeling supported a reaction mechanism which involves the isomerization of Δ4-3-ketosteroids from the keto form to an enol form, prior to being subjected to sulfation. Results derived from this study suggested a potential role of SULT2A1 as a Δ4-3-ketosteroid sulfotransferase in steroid metabolism.
Keywords: sulfotransferase, ketosteroid, 4-androstene-3, 17-dione, ketosteroid isomerase
1. Introduction
In humans and other vertebrates, sulfate conjugation is critically involved in the metabolism of key endogenous compounds such as steroid/thyroid hormones, catecholamine neurotransmitters, cholesterol, and bile acids, as well as in the detoxification and excretion of drugs and other dietary/environmental xenobiotics [1]. The responsible enzymes have been shown to be the cytosolic sulfotransferases (SULTs) that catalyze the transfer of the sulfonate group from the sulfate donor, 3-phsphoadenosine-5’-phosphosulfate (PAPS), to substrate compounds [2].
All SULTs identified in vertebrates have been proposed to constitute a gene superfamily, with SULT members categorized into several SULT families [3–4]. In humans, SULT1 and SULT2 families represent two major SULT families whose constituent members have been rather well characterized [5–6]. In general, members of the SULT1 family showed substrate preference for phenolic compounds and used to be described as the phenol sulfotransferases [7–8]. Members belonging to the SULT2 family, on the other hand, exhibited substrate preference for steroid compounds and were called the hydroxysteroid sulfotransferases [1]. Of the three SULT2 family members, SULT2A1 showed strong sulfating activity toward dehydroepiandrosterone (DHEA), whereas SULT2B1a and SULT2B1b, displayed sulfating activity preferentially toward cholesterol and pregnenolone, respectively [9–10]. These latter enzymes play an important role in the metabolism of steroids in the body [11]. For example, DHEA sulfate (DHEAS), produced primarily under the action of SULT2A1, is the most abundant adrenal steroids in circulation, and when transported to peripheral tissues it may be converted to DHEA by steroid sulfatase (STS) [12–13]. DHEA serves as a principal precursor for the synthesis of downstream sex steroids such as 17β-estradiol and testosterone [14]. In neuronal tissues, DHEAS and pregnenolone sulfate (members of the so-called neurosteroids), in addition to their unsulfated counterparts, function as key modulators of neurotransmitter release and act on an array of neurotransmitter receptors and voltage-dependent ion channels [15]. The presence of large amount of steroid sulfates in neuronal tissues and the contributions of sulfated steroids to neuronal activities suggest that sulfation as catalyzed by the SULTs is a direct regulator of the activity of neurosteroids. A consensus feature of steroids that have been shown to be sulfated is the presence of the hydroxyl group in their chemical structures (and thus are called hydroxysteroids). It is an interesting question whether other steroids that do not carry hydroxyl group may also be subjected to SULT-mediated sulfation.
In this communication, we report for the first time the sulfation of two Δ4-3-ketosteroids, 4-androstene-3,17-dione and progesterone, by human SULT2A1. Kinetic parameters of the sulfation of 4-androstene-3,17-dione and DHEA by SULT2A1 were determined. The chemical structure of sulfated 4-androstene-3,17-dione generated by SULT2A1 was determined by mass spectrometry. A hypothetical reaction mechanism for SULT2A1-mediated Δ4-3-ketosteroid sulfation is proposed based on molecular modeling in conjunction with mutational analysis of potential catalytic amino acid residues of SULT2A1 in mediating the sulfation of 4-androstene-3,17-dione.
2. Materials and Methods
2.1. Materials
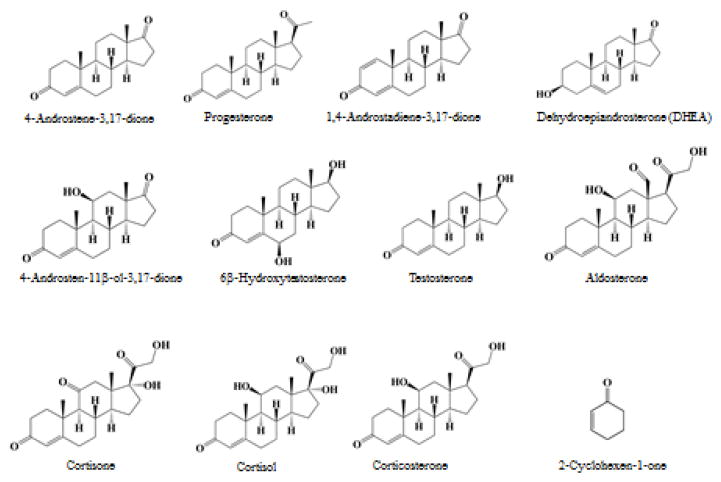
4-Androstene-3,17-dione and 1,4-androstadiene-3,17-dione were purchased from Tokyo Chemical Industry Company. Cortisol, progesterone, and 2-cyclohexen-1-one were products of Wako Pure Chemical Industries. 4-Androsten-11β-ol-3,17-dione, cortisone, corticosterone, aldosterone, testosterone, 6β-hydroxytestosterone, and dehydroepiandrosterone (DHEA) were from Sigma Chemical Company. Isopropyl β-D-thiogalactopyranoside (IPTG) was from TAKARA. BL21 Escherichia coli host strain was obtained from Stratagene. pGEX-2TK prokaryotic GST fusion vectors and glutathione Sepharose 4B were from GE Healthcare Biosciences. KOD-Plus-Mutagenesis Kit was a product of TOYOBO. Cellulose and silica gel thin-layer chromatography (TLC) plates were from Merck. All other chemicals were of the highest grade commercially available.
2.2. Bacterial expression and purification of recombinant human SULT2A1
E. coli BL21 cells transformed with pGEX-2TK harboring human SULT2A1 cDNA (ORF) was prepared as described in our previous study [16]. Transformed BL21 cells were grown to OD600nm = ~ 0.3 in 100 mL LB medium supplemented with 100 μg/mL ampicillin, and induced with 0.1 mM IPTG. After a 12-hour induction at 24°C, the cells were collected by centrifugation and homogenized in 15 mL of ice-cold lysis buffer (50 mM Tris-HCl, pH 8.0, 150 mM NaCl, and 1 mM EDTA) using an Ohtake French Press. The crude homogenate was subjected to centrifugation at 20,400×g for 15 min at 4°C. The supernatant collected was fractionated using 0.5 mL of glutathione Sepharose 4B, and the bound GST fusion protein was treated with 0.5 mL of a thrombin digestion buffer (50 mM Tris-HCl, pH 8.0, 150 mM NaCl, and 2.5 mM CaCl2) containing 5 units/mL bovine thrombin. Following a 2-hour incubation at 4°C with constant agitation, the preparation was subjected to centrifugation, and the supernatant containing purified recombinant SULT2A1 was collected and used in the enzymatic assay.
2.3. Enzymatic assay
Sulfating activity of human SULT2A1 was assayed using 35S-PAPS as the sulfate donor. The standard assay mixture, with a final volume of 25 μL, contained 50 mM sodium phosphate buffer, pH 7.5, 0.2 μM 35S-PAPS (45 Ci/mmol), and 100 μM substrate. The reaction was started by the addition of the enzyme, allowed to proceed for 30 min at 37°C, and terminated by heating at 100°C for 3 min. The precipitates formed were removed by centrifugation, and the supernatant was subjected to analysis of 35S-sulfated product by using a previously developed TLC separation procedure [17], with ethyl acetate/n-butanol (2:1; by volume) or n-butanol/isopropanol/formic acid/water (3:1:1:1; by volume) as the solvent system. Afterwards, the plate was air-dried and analyzed using a Fluoro Image Analyzer (FLA-3000).
2.4. Kinetic data analysis
For the kinetic studies, the kinetic parameters of the sulfation of 4-androstene-3,17-dione by SULT2A1 were determined using varying substrate concentrations ranging from 2 μM to 500 μM. For determining the kinetic parameters of the sulfation of DHEA by SULT2A1, varying substrate concentrations ranging from 0.02 μM to 500 μM were used. Kinetic parameters were determined by nonlinear regression analysis, fitting the data to Michaelis-Menten or substrate inhibition model using the software GraphPad Prism 6 (GraphPad Software Inc.). Eadie-Hofstee plots were concomitantly analyzed.
2.5. Mass spectrometry analysis of sulfated 4-androstene-3,17-dione. Sulfated
4-androstene-3,17-dione generated under the action of SULT2A1 was analyzed in order to determine its chemical structure. The enzymatic sulfation of 4-androstene-3,17-dione with SULT2A1 was performed using non-radioactive PAPS as the sulfonate donor based on the above-mentioned procedure. The final reaction mixture was applied to a Waters Sep-Pak C18 cartridge, followed by washing with water and elution with 80% methanol. The eluate was dried using a SpeedVac concentrator, reconstituted using 2% formic acid/10% methanol, and loaded to an Oasis WAX cartridge. The cartridge was sequentially washed by 2% formic acid solution and 80% methanol and finally eluted by 1% NH4OH/80% methanol. The eluate was dried, reconstituted with 10% methanol, and analyzed by liquid chromatography coupled to Q-Exactive hybrid quadrupole-orbitrap mass spectrometry (LC-MS) (Thermo Fisher Scientific). An Accucore XL C18 column (150 mm x 2.1 mm, 4 μm), coupled with a guard column, was used for the separation of the sample at 0.2 ml/min at 40 °C. The mobile phase was water (A) and methanol (B). The sample was eluted with a linear gradient: 0–2 min, 0–40% B; 2–22 min, 40–80% B; 22–24 min, 80–100% B; 24–27 min, 100% B; 27–30 min, 100-0% B. In the mass spectrometric analysis, a heated electrospray ionization source was used in negative mode with the following conditions; spray voltage, 2.0 kV; sheath gas flow rate, 40; AUX gas flow rate, 10; capillary temperature, 350°C; heater temperature, 300°C.
2.6. Modeling of the binding of hSULT2A1 with 4-androstene-3,17-dione
4-Androstene-3,17-dione molecular structure was obtained from the crystal structure of 3-ketosteroid-delta4-(5alpha)-dehydrogenase, from Rhodococcus jostii RHA1, in complex with 4-androstene-3,17-dione (PDBid:4AT2) deposited in the Protein Data Bank (PDB). Two crystal structures of human SULT2A1 were obtained from those in complex with, respectively, DHEA and lithocholic acid (LCA) (hSULT2A1-DHEA; 1J99 and hSULT2A1-LCA; 3F3Y). The structure of SULT2A1-LCA complex was used for superimposing the 3-oxygen atom of 4-androstene-3,17-dione molecule on 3-OH of LCA molecule, and 4-androstene-3,17-dione molecule was adequately oriented to suit the LCA orientation (LCA-up-type model). The structure of SULT2A1-DHEA complex was used for superimposing the 4-androstene-3,17-dione molecule on the DHEA orientation (DHEA-down-type model).
2.7. Preparation of hSULT2A1 mutant enzymes
Site-directed mutagenesis was performed in order to prepare the hSULT2A1 mutant enzymes, W77A, H99A, H99S, Y160F, and Y231F. PCR reactions for the generation of mutated hSULT2A1 cDNAs were carried out using pGEX-2TK-hSULT2A1 plasmid as the template in conjunction with mutagenic primer sets (Table 1), based on the manufacture’s procedure of KOD-Plus-Mutagenesis Kit. Amplification conditions were 2 min at 94°C and 7 cycles of 98°C for 10 sec, 55°C for 30 sec, and 68°C for 7 min. The PCR products thus generated were treated with DpnI and individually transformed into MRF’ E. coli competent cells for the amplification and purification of pGEX-2TK harboring mutated SULT2A1 cDNAs. Upon verification of the authenticity of mutated SULT2A1 cDNA sequences by nucleotide sequencing, pGEX-2TK harboring individual mutated SULT2A1 cDNA was transformed into BL21 cells, and SULT2A1 mutant enzymes were expressed and purified based on the above-mentioned procedure.
Table 1.
Oligonucleotides used for the site-directed mutagenesis of hSULT2A1 cDNA.
| Oligo | Sequence |
|---|---|
| W77F-sense | 5’-CCCTTCGTAGAGAGTGAGATTGGGTATACAGC-3’ |
| W77-antisense | 5’-TGATCGCTCCCAGATGGGCACAGATTGGATCC-3’ |
| H99A-sense | 5’-TCCGCCCTCCCCATCCAGTTATTCCCCAAGTC-3’ |
| H99S-sense | 5’-TCCAGCCTCCCCATCCAGTTATTCCCCAAGTC-3’ |
| H99-antisense | 5’-GGAGAATAAACGTGGACTCTCCGTTTCACTGA -3’ |
| Y160F-sense | 5’-CTATTTGGGTCATGGTTTGACCACATTCATGG-3’ |
| Y160-antisense | 5’-CACAGTTCCTTGACAAAACCATTCAAAATATT-3’ |
| Y231F-sense | 5’-AATTTTTCCCTCCTGAGTGTTGATTATGTAGT-3’ |
| Y231-antisense | 5’-GGACATCTTGTTTTCTTTCATGCTCTGAAAGG-3’ |
2.8. Miscellaneous methods
35S-PAPS (45 Ci/mmol) was synthesized from ATP and 35S-sulfate using recombinant human bifunctional ATP sulfurylase/adenosine 5’-phosphosulfate kinase, as previously described [18]. Protein determination was performed based on Lowry’s method with bovine serum albumin as the standard [19].
3. Results and Discussion
Over the past several decades, it has been generally accepted that SULT-mediated sulfation occurs only for hydroxyl group- or amino group-containing compounds. For example, SULT2A1, which used to be referred to as the DHEA SULT, has been shown to sulfate 3α- or 3β-hydroxyl group of hydroxysteroids, as well as 17-hydroxyl group of testosterone [20–21]. Recent studies indicated that SULT2A1 could also sulfate 24-hydroxyl group of hydroxycholesterol [22], and could mediate the N-sulfoconjugation of quinolones and other amine drugs in human liver [23]. As described below, we recently made an unexpected finding that Δ4-3-ketosteroids, which contain no hydroxyl or amino groups in their chemical structure (cf. Figure 1), could also be sulfated by SULT2A1.
Figure 1. Chemical structures of substrates tested in the present study.
Dehydroepiandrosterone (DHEA) was used as positive control for sulfation.
3.1. Sulfation of Δ4-3-ketosteroids and related compounds by human SULT2A1
In a preliminary experiment, two ketosteroids, 4-androstene-3,17-dione and 1,4-androstadiene-3,17-dione, were tested as substrates for SULT2A1 under assays conditions described in Materials and Methods. The final reaction mixtures were subjected to TLC separation. Figure 2 shows the autoradiograph taken upon completion of TLC. Interestingly, in spite of the absence of the hydroxyl group, 4-androstene-3,17-dione was apparently sulfated by SULT2A1. In contrast to 4-androstene-3,17-dione, 1,4-androstadiene-3,17-dione which contains an additional 1,2-double bond in the A ring was not sulfated. A panel of Δ4-3-keto steroids was subsequently tested as substrates for SULT2A1. Activity data obtained are compiled in Table 2. It was noted that progesterone, which also lacks a hydroxyl group, could also be sulfated by SULT2A1. Of the other ketosteroids tested, cortisone, cortisol, aldosterone, testosterone, and 6β-hydroxytestosterone contain both a hydroxyl group as well as a 3-carbonyl group. Whether the 3-carbonyl group in these compounds, in addition to the 3β-hydroxyl group, can also be sulfated by SULT2A1 remains to be clarified. Moreover, SULT2A1 showed no sulfating activity toward 2-cyclohexen-1-one, a non-steroidal α,β-unsaturated carbonyl compound. It was noted that the specific activity determined with progesterone (5.5 pmol/min/mg the enzyme) was much lower than that determined with 4-androstene-3,17-dione (170.5 pmol/min/mg enzyme). These results may imply that the intact steroid nucleus is important for the binding of Δ4-3-keto steroids by SULT2A1. Another interesting issue is whether human SULT1E1 and SULT2B1b, which are also capable of sulfating steroids, may also utilize 4-androstene-3,17-dione and progesterone as substrates. Similar enzymatic assays were performed and results showed that SULT1E1 could not catalyze the sulfation of either of these two Δ4-3-ketosteroids, whereas SULT2B1b exhibited weak activity toward only 4-androstene-3,17-dione (7.1 pmol/min/mg enzyme), but not progesterone. It therefore appears that SULT2A1 may be the only major human SULT that is capable of sulfating Δ4-3-ketosteroids, particularly 4-androstene-3,17-dione.
Figure 2. Separation of sulfated 4-androstene-3,17-dione and related steroids by thin layer chromatography (TLC).
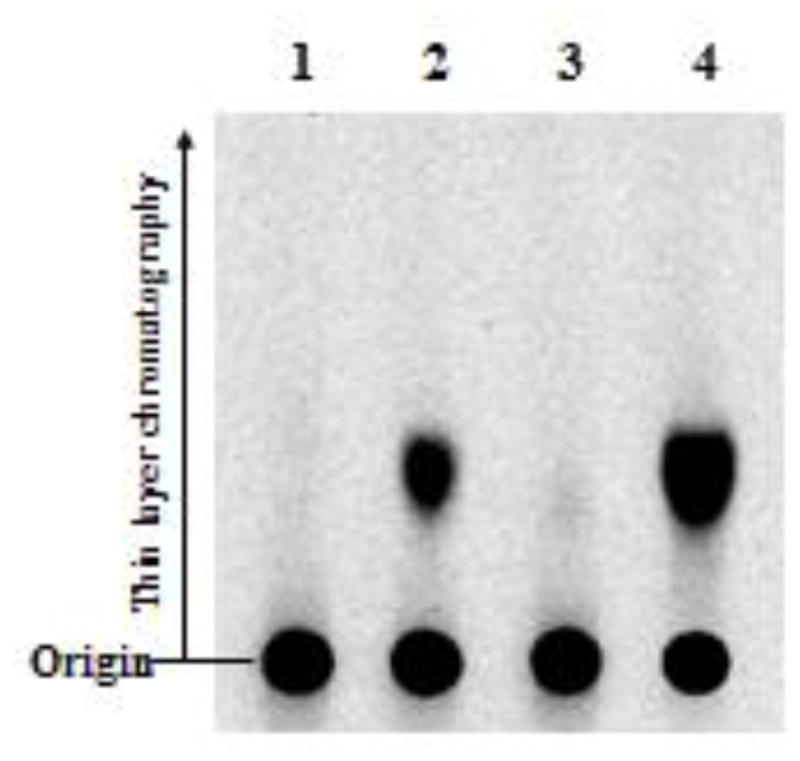
The reaction mixtures following the sulfotransferase assay were analyzed by TLC procedures as described in Materials and Methods. The signals were detected by autoradiography using the Fluoro Image Analyzer (FLA-3000). Compounds analyzed were: lane 1, DMSO (as a control); lane 2, sulfated 4-androstene-3,17-dione; lane 3, sulfated 1,4-androstadiene-3,17-dione; lane 4, sulfated DHEA.
Table 2.
Specific activities of human SULT2A1 toward Δ4-3-ketosteroids and related-compounds as substrates
| Substrate | Specific Activity (pmol/min/mg enzyme) |
|---|---|
|
|
|
| hSULT2A1 | |
| 4-Androstene-3,17-dione | 170.5 ± 6.6 |
| Progesterone | 5.5 ± 0.4 |
| 1,4-Androstadiene-3,17-dione | N.D. |
| 4-Androsten-11β-ol-3,17-dione | N.D. |
| Cortisone | 33.1 ± 2.6 |
| Cortisol | 8.5 ± 0.7 |
| Corticosterone | 228.8 ± 10.5 |
| Aldosterone | 93.2 ± 4.0 |
| Testosterone | 163.9 ± 5.6 |
| 6β-Hydroxytestosterone | 106.1 ± 4.0 |
| Dehydroepiandrosterone | 543.1 ± 16.2 |
| 2-Cyclohexen-1-one | N.D. |
Specific activity refers to pmol of sulfated product formed/min/mg enzyme. The data shown represent means ±S.D. from three determinations. N.D. refers to activity not detected (< 5.0 pmol/min/mg).
3.2. Kinetic parameters of the sulfation of 4-androstene-3,17-dione and DHEA by SULT2A1
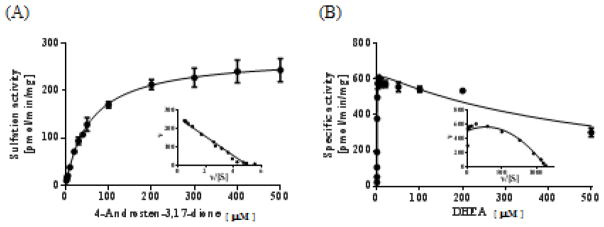
The above-mentioned results showed clearly that SULT2A1 can utilize conventional hydroxyl group-containing substrates, i.e., hydroxysteroids, as well as unconventional substrates, i.e., ketosteroids. An interesting question that ensues is with regard to the catalytic efficiency of SULT2A1 with these two groups of compounds as substrates. Kinetics of SULT2A1 in catalyzing the sulfation of DHEA vs. 4-androstene-3,17-dione was investigated. Michaelis-Menten and substrate inhibition kinetics with Eadie-Hofstee plots are shown in Figure 3. The apparent Km, Vmax, and Vmax/Km values obtained are compiled in Table 3. Plots of the sulfating activity versus the concentration of 4-androstene-3,17-dione showed hyperbolic curve and Eadie-Hofstee plots indicated monophasic kinetics [24], indicating that the sulfation of 4-androstene-3,17-dione followed a typical Michaelis-Menten kinetics. In contrast, a previous study [25] showed that the sulfation of DHEA followed the substrate inhibition kinetics, which was confirmed by a hook in the upper quadrant of the Eadie-Hofstee plot (Figure 3). As shown in Table 3, the apparent Km with 4-androstene-3,17-dione (57.6 μM) was two orders of magnitude higher than that with DHEA (0.38 μM), and, as a result, the Vmax/Km, which reflects catalytic efficiency, was much lower with 4-androstene-3,17-dione than with DHEA.
Figure 3. Kinetic analysis for the sulfation of 4-androstene-3,17-dione and DHEA by human SULT2A1.
The fitting curves were generated based on (A) Michaelis-Menten and (B) substrate inhibition kinetics. Eadie-Hofstee plots are inserted under each fitting curve. Data shown represent calculated mean±S.D from three determination.
Table 3.
Kinetic data of hSULT2A1 toward 4-androstene-3,17-dione, and DHEA as substrates.
| Substrate | Km (μM) | Vmax (pmol/min/mg) | Vmax/Km |
|---|---|---|---|
| 4-Androstene-3,17-dione | 57.6 ± 3.5 | 272.8 ± 5.0 | 4.7 |
| DHEA | 0.38 ± 0.03 | 642.0 ± 12.1 | 1689.5 |
Data shown represent means ± S.D from three determinations.
3.3. Chemical structure of sulfated 4-androstene-3,17-dione
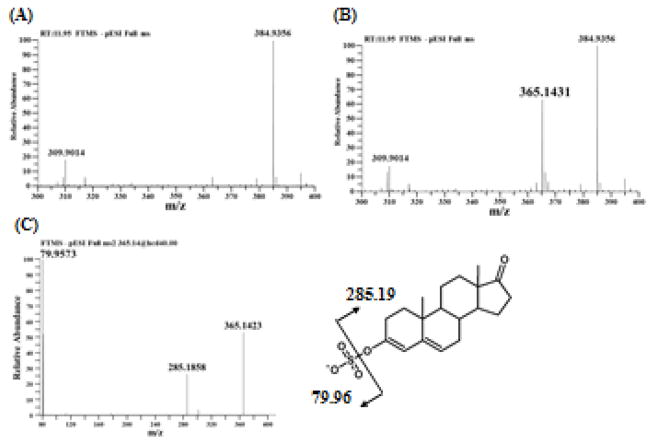
To clarify the chemical structure of sulfated 4-androstene-3,17-dione, produced enzymatically with SULT2A1, was analyzed by mass spectrometry. As shown in Panels A and B of Figure 4, a sufficient abundant ion with m/z of 365.1431, corresponding to a deprotonated sulfated 4-androstene-3,17-dione (C19H25O5S), was specifically detected at 11.95 min retention time. MS/MS analysis of the ion with m/z of 365.1431 showed two major fragment ions with m/z of 79.9573 and 285.1858 (Figure 4C), demonstrating clearly that the parent ion carries a sulfonate moiety (SO3) (theoretical mono isotopic mass, 79.9568) and 4-androstene-3,17-dione moiety (C19H25O2) (285.1855). It should be pointed out that, while 4-androstene-3,17-dione itself was only detected in positive mode but not in negative mode (data not shown), the fragmentation of sulfated 4-androstene-3,17-dione generated a 4-androstene-3,17-dione moiety detected in negative mode. Androgens including androstenedione and DHEA are usually analyzed in positive mode due to the less ion effectiveness or ion suppression in negative mode, whereas estrogens are analyzed in both positive and negative modes [26-28]. MS/MS analysis of sulfated estrone and sulfated DHEA was performed. The fragmentation of estrone sulfate generated the estrone and sulfonate moiety (79.96 m/z), while the fragmentation of DHEA sulfate generated the sulfate moiety (96.96 m/z) but not the DHEA moiety in negative mode (data not shown). These findings indicated that sulfated 4-androstene-3,17-dione may carry an enolic sulfate group and the fragmentation of m/z of 365.1431 may produce the 4-androstene-3,17-dione moiety (C19H25O2:285.1858 m/z) with enolic hydroxyl group. Molecular mass of sulfated 4-androstene-3,17-dione and MS/MS fragment of 4-androstene-3,17-dione moiety also suggested that deprotonation occurred in the structure of 4-androstene-3,17-dione moiety. Since the enol form is not chemically stable so as to be detected by MS/MS analysis, C-C double bond of the enol may be conjugated with another C-C double bond. Therefore, sulfated 4-androstene-3,17-dione may form a 3,4-double bond in the A-ring and a 5,6-double bond in the B-ring as shown in Figure 4C. The chemical structure determined suggests that sulfation of 4-androstene-3,17-dione may occur though the deprotonation of C-6.
Figure 4. Mass spectrometry analysis of sulfated 4-androstene-3,17-dione produced by SULT2A1.
(A, B) Full mass spectrum (300-400 m/z window) of the product extracted from the assay mixture (A) without or (B) with SULT2A1. (C) MS/MS spectrum of 365.14 m/z.
3.4. Binding models of hSULT2A1 with 4-androstene-3,17-dione
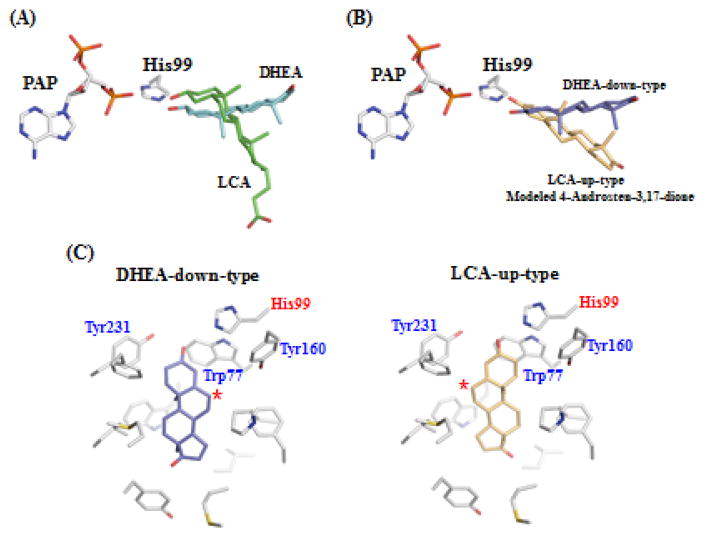
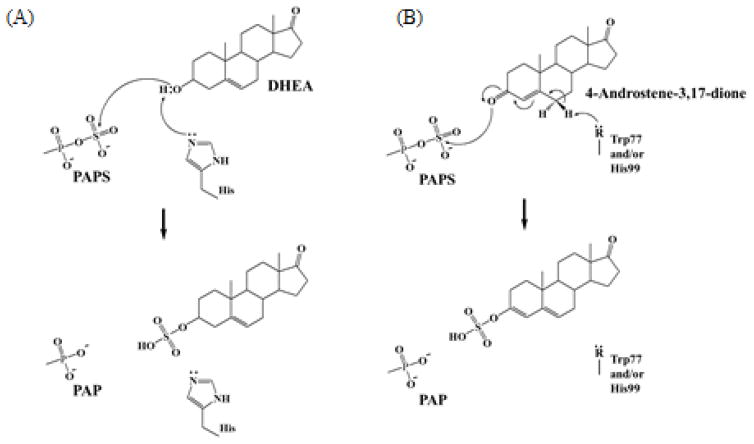
To probe further the mechanism of the sulfation of 4-androstene-3,17-dione, molecular modeling was performed. Several crystal structures of human SULT2A1 in complex with different steroid substrates have been resolved (e.g., hSULT2A1-DHEA; (PDBid) 1J99, hSULT2A1-androsterone; 1OV4, and hSULT2A1-LCA; 3F3Y). These hSULT2A1-steroid substrates complex structures showed that DHEA and lithocholic acid (LCA) are present in different catalytic competent manners (Figure 5A). The orientation of the LCA steroid A-ring is similar to that of the DHEA orientation. In contrast, the B, C and D-ring of the LCA molecule rotated by approximately 80° to form an orientation different from that of DHEA. It is important to point out that both the two methyl groups, C-18 and C-19, of DHEA and LCA, point perpendicularly, downward and upward, respectively, to the plane of the rings and point (cf. Figure 5A). We therefore performed two kinds of molecular modeling of 4-androstene-3,17-dione in the acceptor substrate binding site of SULT2A1. One binding mode was almost identical to that of DHEA (herein named DHEA-down-type) and the other one was similar to the binding mode of LCA (herein named LCA-up-type) (Figure 5B). These binding models indicated that Trp77, His99, and Tyr160 may be located relatively close to the C-6 position, and ready to be deprotonated, in LCA-up-type and Tyr231 may be located relatively close to the C-6 position in DHEA-down-type (Figure 5C). It is possible that these residues may be important in the catalytic process of the sulfation of ketosteroids by SULT2A1.
Figure 5. Binding models of hSULT2A1 with 4-androstene-3,17-dione.
(A) Superposition of DHEA (down-type) and LCA (up-type) with PAP and His99 in the active site of hSULT2A1. The position of DHEA and LCA were superimposed from the hSULT2A1 structure with DHEA (1J99) and LCA (3F3Y). (B) Superposition of modeled DHEA-down-type and LCA-up-type with 4-androstene-3,17-dione (C) Two types of the modeling of 4-androstene-3,17-dione with the active site of hSULT2A1. Left panel shows the position of DHEA-down-type and right panel shows the position of LCA-up-type of 4-androstene-3,17-dione. C6 position of 4-androstene-3,17-dione is marked with an asterisk.
3.5. Proposed mechanism of Δ4-3-ketosteroid sulfation by SULT2A1
That Δ4-3-ketosteroids representing a new class of substrates lacking hydroxyl or amino groups raises an interesting issue regarding the mechanism of their sulfation by SULT2A1. Previous studies have shown that sulfation as mediated by the SULTs proceeds via an SN2 in-line displacement mechanism [29] in which the occurring phenoxide ion of the substrate attacks the sulfate of PAPS. The question then is whether SULT2A1, which also sulfates conventional substrates, particularly DHEA that carry a hydroxyl group, may utilize the same or a different reaction mechanism for the sulfation of Δ4-3-ketosteroids. Chemical structure of sulfated 4-androstene-3,17-dione (Figure 4) and molecular binding modeling of SULT2A1 with 4-androstene-3,17-dione (Figure 5) suggested that H99, W77, Y160, and/or Y231 residues may be involved in the deprotonation at the C6 position. To verify their contributions to the catalysis, we performed a mutational analysis using W77F, H99A, H99S, Y160F, and T231F. W77F and H99A mutant showed a 50% reduction in the sulfating activity, while H99S showed the complete loss of the sulfating activity toward 4-androstene-3,17-dione (Table 4). In contrast, Y160F and Y231F showed no reduction in the sulfating activity. These results indicated that W77 and H99 are likely involved in the catalysis, possibly in the deprotonation of C-6. Further studies are needed in order to define unequivocally the catalytic residue(s). Based on the chemical structure, molecular modeling, and mutational analysis, a hypothetical mechanism for the sulfation of Δ4-3-ketosteroids by SULT2A1 is proposed as shown in Figure 6. Assuming that the sulfation of 4-androstene-3,17-dione might progress through the formation of a hydroxyl group which is a typical target for sulfation, it would require the isomerization of 4-androstene-3,17-dione from a keto form to an enol form. In mammals including humans, a bifunctional 3β-hydroxysteroid dehydrogenase/Δ5-Δ4-isomerase (3β-HSD) has been identified. Its involvement in the conversion of DHEA to 4-androstene-3,17-dione, as well as pregnenolone to progesterone, represents an important step in the steroid biosynthesis in the body [30]. It is noted that the reaction mechanism of a ketosteroid isomerase (KSI) from the bacterium Comamonas testosteroni, which catalyzes the isomerization of Δ5-3-ketosteroid to Δ4-3-ketosteroid, has been thoroughly investigated [31]. In the active site of C. testosteroni ketosteroid isomerase, an aspartate base, Asp38, is in position to attract the C4β proton of the steroid substrate, thereby generating an enolate intermediate. The subsequent reketonization may result in the protonation at the C6β position by Asp38 [31]. In almost all SULTs, a highly conserved catalytic histidine residue located at around 100th amino acid position has been reported to deprotonate directly the hydroxyl group of substrate compounds including phenolic compounds and hydroxysteroids [32]. However, binding models of SULT2A1 with 4-androstene-3,17-dione indicated that His99 alone may not be sufficient to attract the C-6 proton. We therefore hypothesize that, in catalyzing 4-androstene-3,17-dione sulfation, the His99 and W77 of SULT2A1 might cooperatively attract the proton at C6-position in a way similar to that of Asp38 in mediating ketosteroid isomerization in C. testosteroni KSI. The resultant electron lone pair then forms a 5,6-double bond in the B-ring, which in turn drives the 3,4-double bond-formation of A-ring, followed by the formation of a nucleophilic enolate intermediate. The latter can then execute a SN2 nucleophilic attack on the sulfur atom of PAPS, resulting in generation of 4-androstene-3,17-dione sulfate. Since 4-androstene-3,17-dione needs to undergo isomerization forming the 3-hydroxylgroup which then serve as the target for sulfation, it may not be surprising that Δ4-3-ketosteroids are less effective substrates than hydroxysteroids (Table 3). As mentioned above (cf. Table 2), 1,4-androstadiene-3,17-dione could not be sulfated by SULT2A1. This might be due to its electron-rich 1,2-double bond sustaining the C3-carbonyl group thereby preventing keto-enol isomerization.
Table 4.
Specific activity of hSULT2A1 mutant enzymes with 4-androstene-3,17-dione.
| Specific Activity (pmol/min/mg) | |
|---|---|
|
| |
| SULT2A1 mutants | 4-Androstene-3,17-dione |
| WT | 17.8 ± 0.4 |
| W77F | 9.0 ± 1.2 |
| H99A | 9.3 ± 0.2 |
| H99S | N.D. |
| Y160F | 15.4 ± 1.1 |
| Y231F | 17.8 ± 0.6 |
Data shown represent means ± S.D from three determinations. N.D. refers to activity not detected (< 1.0 pmol/min/mg). Substrate concentration used in the enzyme assay was 50 μM.
Figure 6. Proposed sulfation mechanism for steroids in the active site of hSULT2A1.
(A) Conventional enzymatic mechanism of SULT-mediated sulfation of DHEA [29]. (B) Proposed mechanism for the sulfation of ketosteroid by SULT2A1. In this model, the highly conserved His99 and W77 may collaboratively act to deprotonate the C-6 proton of 4-androstene-3,17-dione to form a enolate intermediate. The resulting nucleophilic oxyanion then attacks the sulfur of PAPS via a SN2 reaction, followed by the production and release of sulfated 4-androstene-3,17-dione from the active site.
4. Conclusions
To our knowledge, this is the first report on the identification of a new class of steroid substrates, which lack the hydroxyl or amino group in their chemical structure, for SULTs. Human SULT2A1, as elaborated above, may play a dual role as a hydroxysteroid (DHEA) sulfotransferase and as a Δ4-3-ketosteroid (4-androstene-3,17-dione and progesterone) sulfotransferase. While the present study provided unequivocal evidence for the Δ4-3-ketosteroid-sulfating activity of SULT2A1, it will be important to show that sulfation of 4-androstene-3,17-dione and progesterone may indeed occur in vivo and how SULT2A1-mediated sulfation may influence the biological effects of these steroids. To this end, SULT2A1 has been shown to be highly expressed in the adrenal glands and the liver [33]. Studies have shown that 4-androstene-3,17-dione is produced mainly in the adrenal glands and gonads, and serves as the biosynthetic precursor for androgens such as testosterone as well as estrogens such as 17β-estradiol [34]. Progesterone, on the other hand, is the principal hormone released from the corpus luteum after ovulation during the menstrual cycle [35], and also plays a role during pregnancy and embryogenesis [36]. Its sulfation would likely have an impact on these physiological processes. Other than SULT2A1, SULT2B1b, previously called the cholesterol sulfotransferase [10], was also shown to be capable of sulfating 4-androstene-3,17-dione. That SULT2B1b is expressed in the brain may imply a role of this enzyme in the sulfation and regulation of 4-androstene-3,17-dione as a neurosteroid. More work is warranted in order to clarify these important issues.
Supplementary Material
Highlights.
This is first discovery of carbonyl group as a target structure for sulfotransferase.
This study revealed that sulfation is directly related to the metabolic pathway of ketosteroid.
Results derived from this study suggested a potential role of SULT2A1 as a Δ4-3-ketosteroid sulfotransferase in steroid metabolism.
Acknowledgments
This work was supported by JSPS KAKENHI Grant Numbers 23580138 (M.S.), 21580114 (Y.S.), 15H04502 (Y.S.), 17H05028 (K.K.) and for JSPS Research Fellows Grant Number 24·1404 (T.H.) and National Institutes of Health grant R03HD071146 (M.C.L.).
Abbreviations
- SULT
cytosolic sulfotransferase
- PAPS
3’-phosphoadenosine 5’-phosphosulfate
- DHEA
dehydroepiandrosterone
- KSI
Ketosteroid isomerase
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Gamage N, Barnett A, Hempel N, Duggleby RG, Windmill KF, Martin JL, McManus ME. Human Sulfotransferases and Their Role in Chemical Metabolism. Toxicol Sci. 2006;90:5–22. doi: 10.1093/toxsci/kfj061. [DOI] [PubMed] [Google Scholar]
- 2.LIPMANN F. Biological sulfate activation and transfer. Science. 1958;128:575–580. doi: 10.1126/science.128.3324.575. [DOI] [PubMed] [Google Scholar]
- 3.Yamazoe Y, Nagata K, Ozawa S, Kato R. Structural similarity and diversity of sulfotransferases. Chem Biol Interact. 1994;92:107–117. doi: 10.1016/0009-2797(94)90057-4. [DOI] [PubMed] [Google Scholar]
- 4.Blanchard RL, Freimuth RR, Buck J, Weinshilboum RM, Coughtrie MW. A proposed nomenclature system for the cytosolic sulfotransferase (SULT) superfamily. Pharmacogenetics. 2004;14:199–211. doi: 10.1097/00008571-200403000-00009. [DOI] [PubMed] [Google Scholar]
- 5.Kurogi K, Chepak A, Hanrahan MT, Liu MY, Sakakibara Y, Suiko M, Liu MC. Sulfation of opioid drugs by human cytosolic sulfotransferases: Metabolic labeling study and enzymatic analysis. Eur J Pharm Sci. 2014;62:40–48. doi: 10.1016/j.ejps.2014.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yamamoto A, Liu MY, Kurogi K, Sakakibara Y, Saeki Y, Suiko M, Liu MC. Sulphation of acetaminophen by the human cytosolic sulfotransferases: a systematic analysis. J Biochem. 2015 doi: 10.1093/jb/mvv062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wilborn TW, Comer KA, Dooley TP, Reardon IM, Heinrikson RL, Falany CN. Sequence analysis and expression of the cDNA for the phenol-sulfating form of human liver phenol sulfotransferase. Mol Pharmacol. 1993;43:70–77. [PubMed] [Google Scholar]
- 8.Ozawa S, Nagata K, Shimada M, Ueda M, Tsuzuki T, Yamazoe Y, Kato R. Primary structures and properties of two related forms of aryl sulfotransferases in human liver. Pharmacogenetics. 1995;5:135–140. doi: 10.1097/00008571-199512001-00015. [DOI] [PubMed] [Google Scholar]
- 9.Otterness DM, Wieben ED, Wood TC, Watson WG, Madden BJ, McCormick DJ, Weinshilboum RM. Human liver dehydroepiandrosterone sulfotransferase: molecular cloning and expression of cDNA. Mol Pharmacol. 1992;41:865–872. [PubMed] [Google Scholar]
- 10.Her C, Wood TC, Eichler EE, Mohrenweiser HW, Ramagli LS, Siciliano MJ, Weinshilboum RM. Human hydroxysteroid sulfotransferase SULT2B1: two enzymes encoded by a single chromosome 19 gene. Genomics. 1998;53:284–295. doi: 10.1006/geno.1998.5518. [DOI] [PubMed] [Google Scholar]
- 11.Barker EV, Hume R, Hallas A, Coughtrie WH. Dehydroepiandrosterone sulfotransferase in the developing human fetus: quantitative biochemical and immunological characterization of the hepatic, renal, and adrenal enzymes. Endocrinology. 1994;134:982–989. doi: 10.1210/endo.134.2.8299591. [DOI] [PubMed] [Google Scholar]
- 12.Hobkirk R. Steroid sulfation Current concepts. Trends Endocrinol Metab. 1993;4:69–74. doi: 10.1016/s1043-2760(05)80018-9. [DOI] [PubMed] [Google Scholar]
- 13.Purohit A, Dauvois S, Parker MG, Potter BV, Williams GJ, Reed MJ. The hydrolysis of oestrone sulphate and dehydroepiandrosterone sulphate by human steroid sulphatase expressed in transfected COS-1 cells. J Steroid Biochem Mol Biol. 1994;50:101–104. doi: 10.1016/0960-0760(94)90177-5. [DOI] [PubMed] [Google Scholar]
- 14.Hanukoglu I. Steroidogenic enzymes: structure, function, and role in regulation of steroid hormone biosynthesis. J Steroid Biochem Mol Biol. 1992;43:779–804. doi: 10.1016/0960-0760(92)90307-5. [DOI] [PubMed] [Google Scholar]
- 15.Zheng P. Neuroactive steroid regulation of neurotransmitter release in the CNS: action, mechanism and possible significance. Prog Neurobiol. 2009;89:134–152. doi: 10.1016/j.pneurobio.2009.07.001. [DOI] [PubMed] [Google Scholar]
- 16.Hashiguchi T, Kurogi K, Sakakibara Y, Yamasaki M, Nishiyama K, Yasuda S, Liu MC, Suiko M. Enzymatic Sulfation of Tocopherols and Tocopherol Metabolites by Human Cytosolic Sulfotransferases. Biosci Biotechnol Biochem. 2011;75:1951–1956. doi: 10.1271/bbb.110352. [DOI] [PubMed] [Google Scholar]
- 17.Liu MC, Lipmann F. Decrease of tyrosine-O-sulfate-containing proteins found in rat fibroblasts infected with Rous sarcoma virus or Fujinami sarcoma virus. Proc Natl Acad Sci U S A. 1984;81:3695–3698. doi: 10.1073/pnas.81.12.3695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yanagisawa K, Sakakibara Y, Suiko M, Takami Y, Nakayama T, Nakajima H, Takayanagi K, Natori Y, Liu MC. cDNA cloning, expression, and characterization of the human bifunctional ATP sulfurylase/adenosine 5'-phosphosulfate kinase enzyme. Biosci Biotechnol Biochem. 1998;62:1037–1040. doi: 10.1271/bbb.62.1037. [DOI] [PubMed] [Google Scholar]
- 19.Lowry OH, Rosebrough NJ, Farr AL, Randall RJ. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951;193:265–275. [PubMed] [Google Scholar]
- 20.Radominska A, Comer KA, Zimniak P, Falany J, Iscan M, Falany CN. Human liver steroid sulphotransferase sulphates bile acids. Biochem J. 1990;272:597–604. doi: 10.1042/bj2720597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Falany CN, Wheeler J, Oh TS, Falany JL. Steroid sulfation by expressed human cytosolic sulfotransferases. J Steroid Biochem Mol Biol. 1994;48:369–375. doi: 10.1016/0960-0760(94)90077-9. [DOI] [PubMed] [Google Scholar]
- 22.Cook IT, Duniec-Dmuchowski Z, Kocarek TA, Runge-Morris M, Falany CN. 24-Hydroxycholesterol Sulfation by Human Cytosolic Sulfotransferases: Formation of Monosulfates and Disulfates, Molecular Modeling, Sulfatase Sensitivity, and Inhibition of Liver X Receptor Activation. Drug Metab Dispos. 2009;37:2069–2078. doi: 10.1124/dmd.108.025759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Senggunprai L, Yoshinari K, Yamazoe Y. Selective Role of Sulfotransferase 2A1 (SULT2A1) in the N-Sulfoconjugation of Quinolone Drugs in Humans. Drug Metab Dispos. 2009;37:1711–1717. doi: 10.1124/dmd.109.027441. [DOI] [PubMed] [Google Scholar]
- 24.Hutzler JM, Tracy TS. Atypical Kinetic Profiles in Drug Metabolism Reactions. Drug Metab Dispos. 2002;30:355–362. doi: 10.1124/dmd.30.4.355. [DOI] [PubMed] [Google Scholar]
- 25.Gulcan HO, Duffel MW. Substrate inhibition in human hydroxysteroid sulfotransferase SULT2A1: studies on the formation of catalytically non-productive enzyme complexes. Arch Biochem Biophys. 2010;507:232–240. doi: 10.1016/j.abb.2010.12.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Díaz-Cruz MS, López de Alda MJ, López R, Barceló D. Determination of estrogens and progestogens by mass spectrometric techniques (GC/MS, LC/MS and LC/MS/MS) J Mass Spectrom. 2003;38:917–923. doi: 10.1002/jms.529. [DOI] [PubMed] [Google Scholar]
- 27.Gao W, Stalder T, Foley P, Rauh M, Deng H, Kirschbaum C. Quantitative analysis of steroid hormones in human hair using a column-switching LC-APCI-MS/MS assay. J Chromatogr B Analyt Technol Biomed Life Sci. 2013;928:1–8. doi: 10.1016/j.jchromb.2013.03.008. [DOI] [PubMed] [Google Scholar]
- 28.Büttler RM, Martens F, Kushnir MM, Ackermans MT, Blankenstein MA, Heijboer AC. Simultaneous measurement of testosterone, androstenedione and dehydroepiandrosterone (DHEA) in serum and plasma using Isotope-Dilution 2-Dimension Ultra High Performance Liquid-Chromatography Tandem Mass Spectrometry (ID-LC-MS/MS) Clin Chim Acta. 2014;438:157–159. doi: 10.1016/j.cca.2014.08.023. [DOI] [PubMed] [Google Scholar]
- 29.Kakuta Y, Pedersen LG, Carter CW, Negishi M, Pedersen LC. Crystal structure of estrogen sulphotransferase. Nat Struct Biol. 1997;4:904–908. doi: 10.1038/nsb1197-904. [DOI] [PubMed] [Google Scholar]
- 30.Thomas JL, Duax WL, Addlagatta A, Brandt S, Fuller RR, Norris W. Structure/Function Relationships Responsible for Coenzyme Specificity and the Isomerase Activity of Human Type 1–3β-Hydroxysteroid Dehydrogenase/Isomerase. J Biol Chem. 2003;278:35483–35490. doi: 10.1074/jbc.M304752200. [DOI] [PubMed] [Google Scholar]
- 31.Ha NC, Choi G, Choi KY, Oh BH. Structure and enzymology of Delta5-3-ketosteroid isomerase. Curr Opin Struct Biol. 2001;11:674–678. doi: 10.1016/s0959-440x(01)00268-8. [DOI] [PubMed] [Google Scholar]
- 32.Negishi M, Pedersen LG, Petrotchenko E, Shevtsov S, Gorokhov A, Kakuta Y, Pedersen LC. Structure and function of sulfotransferases. Arch Biochem Biophys. 2001;390:149–157. doi: 10.1006/abbi.2001.2368. [DOI] [PubMed] [Google Scholar]
- 33.Parker CR, Falany CN, Stockard CR, Stankovic AK, Grizzle WE. Immunohistochemical localization of dehydroepiandrosterone sulfotransferase in human fetal tissues. J Clin Endocrinol Metab. 1994;78:234–236. doi: 10.1210/jcem.78.1.8288708. [DOI] [PubMed] [Google Scholar]
- 34.Do Rego JL, Seong JY, Burel D, Leprince J, Luu-The V, Tsutsui K, Tonon MC, Pelletier G, Vaudry H. Neurosteroid biosynthesis: enzymatic pathways and neuroendocrine regulation by neurotransmitters and neuropeptides. Front Neuroendocrinol. 2009;30:259–301. doi: 10.1016/j.yfrne.2009.05.006. [DOI] [PubMed] [Google Scholar]
- 35.Maybin JA, Critchley HO. Progesterone: a pivotal hormone at menstruation. Ann N Y Acad Sci. 2011;1221:88–97. doi: 10.1111/j.1749-6632.2011.05953.x. [DOI] [PubMed] [Google Scholar]
- 36.Wetendorf M, DeMayo FJ. Progesterone receptor signaling in the initiation of pregnancy and preservation of a healthy uterus. Int J Dev Biol. 2014;58:95–106. doi: 10.1387/ijdb.140069mw. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.