Abstract
Germ layer formation and axial patterning are biological processes that are tightly linked during embryonic development of most metazoans. In addition to canonical WNT, it has been proposed that ERK-MAPK signaling is involved in specifying oral as well as aboral territories in cnidarians. However, the effector and the molecular mechanism underlying latter phenomenon is unknown. By screening for potential effectors of ERK-MAPK signaling in both domains, we identified a member of the ETS family of transcription factors, Nverg that is bi-polarily expressed prior to gastrulation. We further describe the crucial role of NvERG for gastrulation, endomesoderm as well as apical domain formation. The molecular characterization of the obtained NvERG knock-down phenotype using previously described as well as novel potential downstream targets, provides evidence that a single transcription factor, NvERG, simultaneously controls expression of two different sets of downstream targets, leading to two different embryonic gene regulatory networks (GRNs) in opposite poles of the developing embryo. We also highlight the molecular interaction of cWNT and MEK/ERK/ERG signaling that provides novel insight into the embryonic axial organization of Nematostella, and show a cWNT repressive role of MEK/ERK/ERG signaling in segregating the endomesoderm in two sub-domains, while a common input of both pathways is required for proper apical domain formation. Taking together, we build the first blueprint for a global cnidarian embryonic GRN that is the foundation for additional gene specific studies addressing the evolution of embryonic and larval development.
Keywords: ERK signaling, embryonic development, gastrulation, endomesoderm, apical organ, ERG, ETS transcription factor, gene regulatory network, Nematostella vectensis, sea anemone, cnidarian, gene expression, evolution
Introduction
Fibroblast Growth Factor (FGF) induced ERK signaling plays a crucial role in various aspects of mesoderm formation and coordinating cell movements in bilaterian animals [1–11]. FGFs bind to FGF Receptors (FGFRs) that are part of the RTK (Receptor Tyrosine Kinase) family, in order to activate an intracellular MAP Kinase (RAS/MEK/ERK) signaling cascade leading to the phosphorylation of transcription factors and thus the repression or activation of downstream targets [12]. Well known transcriptional regulators whose activity can be controlled by MEK/ERK signaling, belong to the ETS domain containing family of transcription factors [13].
Cnidarians are the extant sister group to all bilaterians and their phylogenetic position makes them very interesting for understanding the evolution of biological novelties [14–16]. One intensely used cnidarian model is the anthozoan sea anemone Nematostella vectensis that can easily be cultured and manipulated under laboratory conditions and for which functional genomic tools are well established [16–21]. Based on the sequenced Nematostella genome [19], 15 putative FGF ligands and three potential receptors have been identified [22,23] with the spatial expression patterns reported for three ligands and two receptors [22–24]. Two ligands and one receptor (NvfgfA1, NvfgfA2, NvfgfrA) are expressed in the vegetal hemisphere/apical domain, the ligand Nvfgf8 is expressed in the animal hemisphere and its descendants, and the receptor NvfgfrB in derivatives of both poles. Interestingly, Nvsprouty, a well described downstream target and modulator of FGF signaling [25–27], is also expressed in both extremities of the developing embryo/larvae [22]. Only based on these expression patterns, FGF signaling has been suggested to play a role in gastrulation and neural development [22]. However, the importance of the FGF pathway has so far only been analyzed at late stages of development and shown to be crucial for apical organ formation and metamorphosis [23,28,29].
The vegetal pole of cnidarian embryos gives rise to the apical organ, characterized by an apical tuft, a group of long cilia, at the aboral most part of the planula larvae [17,23,30]. Recent studies have shown that a gene regulatory module involving the transcription factor Six3/6, FGF signaling as well as Frizzled 5/8, that potentially signals through ß-catenin, is required to specify and pattern the apical domain, form the apical tuft and subsequently allow the process of metamorphosis into a sessile juvenile [23,28,29,31]. Unfortunately, little is known about the role of MEK/ERK signaling in the specification of the apical domain prior to the onset of gastrulation.
Cnidarians are so-called diploblastic animals that, although they possess the genetic toolkit involved in bilaterian mesoderm formation, lack a true mesodermal germ layer [24,30,32]. A precise embryonic cell lineage analysis has yet to be performed in cnidarians due to the lack of a stereotyped cleavage program but existing labeling experiments clearly indicate that derivatives of cells from the animal hemisphere in Nematostella gives rise to the epitheliomuscular gut, the pharynx and the mouth of the planula larva [33,34]. In a previous study, we have defined three gene expression domains within the animal hemisphere of the blastula prior to the onset of gastrulation; the central domain, the central ring and the external ring that appears to give rise to the gut (bodywall endomesoderm), pharynx and mouth respectively [24]. This work also showed that canonical WNT signaling (cWnt) is required for proper gene expression within all three domains, in particular for genes expressed within the central ring domain and normal pharynx formation [24]. Interestingly, cWnt/TCF represses expression of the potential FGF ligand, fgf8A, in the central ring (animal hemisphere) restricting its expression to the central domain, suggesting a role of FGF induced ERK/MAPK signaling in endomesoderm formation [24]. However, a recent study that focuses on the role of ERK/MAPK signaling in the initiation of the neurogenic program in Nematostella development, suggest that FGFR might not be the (sole) activator of this pathway in the presumptive endomesoderm [35]. The same authors have also shown that pharmacologically inhibition of ERK/MAPK signaling using U0126, a potent inhibitor of the ERK activating kinase MEK [36,37], after fertilization blocks gastrulation and endomesoderm formation [35]. As this treatment perturbs gene expression within the animal hemisphere as well as the apical domain, this further suggests a dual role of this pathway in germ layer formation and axial patterning [35]. In addition, by using a genome wide expression array approach, the authors have identified a large set of putative downstream targets of this pathway of which only the genes potentially involved in neurogenesis have been reported [35].
In this study, we present the spatio-temporal expression of NvERG, a member of the ETS family of transcription factors that is expressed in both, the central domain of the animal hemisphere as well as in the apical domain of the vegetal hemisphere. Inhibition of NvERG phenocopies the effects of disrupting MEK/ERK signaling, causing the failure of gastrulation and endomesoderm formation as well as the perturbation of apical tuft development. Fine scale temporal and spatial gene expression analysis of genes identified in a differential genome wide expression array comparing DMSO (control) and U0126 treated embryos [35] enabled us to describe 39 potential downstream targets of this pathway that are expressed in the presumptive endomesoderm as well as in the apical domain. Finally, molecular analysis of the resulting phenotype in NvERG morphants, highlights its crucial role for setting up the gene regulatory networks (GRNs) underlying endomesoderm forming within the central domain as well as apical domain patterning. Interestingly, we functionally confirmed a computational prediction [38] that NvERG negatively regulates Nvbra expression in the central domain, in order to restrict its expression in the central ring. This work enables us today to draw a global blueprint of genetic interactions governing specification, patterning and morphogenic events underlying embryonic development of Nematostella.
Materials and Methods
Culture and spawning of Nematostella vectensis
Adult Nematostella were cultivated either at the Kewalo Marine Laboratory/PBRC of the University of Hawaii (USA), the Whitney Laboratory for Marine Bioscience of the University of Florida (USA) or the Institute for Research on Cancer and Aging of the University of Nice-Sophia-Antipolis (FRA). Culture and spawning/fertilization was performed according to the protocol described in [24]. Fertilized eggs were kept in dark in filtered 1/3 seawater at 16°C until the desired stage.
RNA Extraction and quantitative PCR (qPCR)
RNA Extraction and quantitative PCR (qPCR) was performed following protocols described in [24]: For the fine scale temporal analysis total RNA was extracted from the following stages (in hours post fertilization, hpf): 0, 2, 4, 6, 8, 10, 12, 14, 16, 18, 20, 24, 28, 32, 40, 48. For the molecular phenotype analysis, total RNA was extracted 24hpf. Samples were obtained from three biological replicates and performed in three technical replicates. qPCR analysis using a LightCycler 480 (Roche) utilizing LightCycler 480 SYBR Green 1 Master mix (Roche, #04887352001) was carried out as described previously [39]. The full list of qPCR primer pairs and their efficiency used in this study can be found in Table S2 or [35]. The houskeeping genes Nvactin and/or Nvgadph were used to normalize relative fold changes between control and manipulated embryos and each qPCR analysis was repeated on independent biological replicates.
In situ hybridization, actin and nuclear staining
Previously described gene sequences were used to sub-clone into pGemT (Promega, #A3600) from mixed stage cDNA. All other sequences used in this study were isolated in the course of a microarray analysis [35]. Genome predictions as well as EST sequence information were combined to design primer pairs (Table S3, [35]) that allow the amplification and cloning of genes between 05.kb and 2kb. PCR amplified cDNA fragments, corresponding to partial or full-length sequences of the gene of interest, have been cloned into pGEMT vectors (Promega, #1360). T7 and SP6 primers have been used to amplify the insert and subsequently used for anti-sense probe synthesis using either the T7 or SP6 promoters (Ambion, #AM1330, #AM1333). Probe integrity was validated by RNA electrophoresis and presented expression patterns observed in at least three independent experiments. Accession numbers for all analyzed genes in this study can be found in [35] as well as in Table S4, S5). Embryo fixation, probe synthesis and in situ hybridization were performed as previously described [24,30]. To analyze embryonic and larval morphology, we used Biodipy FL Phallacidin (Molecular Probes/Invitrogen, #B607), propidium iodide (Sigma, #81845) and an anti-acetylated tubulin antibody (Sigma, #T6793), to stain f-actin, the cell nuclei and the apical tuft respectively following the protocols described previously [40].
Nomenclature
Nomenclature of the newly identified genes follows the approach used in (24). To distinguish between previously published genes, and newly identified putative TFs and signaling molecules, we used the best Blast Hit identification, followed by “- like” to designate the newly identified gene sequences. While “Blast hit” approaches can be used to provide a general idea of the protein family, a detailed phylogenetic analysis is required to better resolve these gene orthologies, especially when paralogy issues or when multiple gene predictions are present for one gene family.
cDNA construction, MO-NvERG design and microinjection
cDNA constructs encoding the wild type ORF (NvERG) and a dominant negative form (NvERG-DB1) of NvERG, were generated by PCR using the following primers:
NvERG_FWD 5′ATGTATGGTTTAAGTTCAGAATC-3′
NvERG-DB1_FWD 5′-ATGTTCAATGCCAGCCCGATG-3′
NvERG_REV 5′GGCGTAGTAGGTCATACTGGC-3′
The reverse primer used in combination with both forward primers was lacking the stop codon for fusion with a C-terminal Venus fluorescent tag. All cDNA constructs were cloned, linearized and transcribed according to [20,24]. mRNAs were injected in zygotes at final concentrations of 0.2–0.5 mg/ml. A splice blocking morpholino antisense oligonucleotide (Gene Tools) was designed (MO-NvERG 5′-CTTACTTTTTCTCAAGACGCACAGA-3′) to target the exon3-intron3 boundary of NvERG and used from 0.3 – 0.9mM without noticeable toxicity. A control MO (MO-CTRL 5′-AGAGGAAGAATAACATACCCTGTCC-3′ (35)) was also injected at a concentration of 0,9 mM. Animals were sorted after injection to eliminate the un-injected animals as indicated by the lack of fluorescence. Microinjections were carried out as described in [20,24] and the following primers used to test the splice-blocking efficiency of the MO (Figure 1):
NvERG_Mosplice_FWD 5′-ACCAAAGAACACGTTCGCCAGTGGA-3′,
NvEtsA_Mosplice_REV 5′ATCGCAAACCCCCAGGCTCTCC-3′
Figure 1. NvErg is required for endomesoderm formation, gastrulation and participates in apical tuft development.
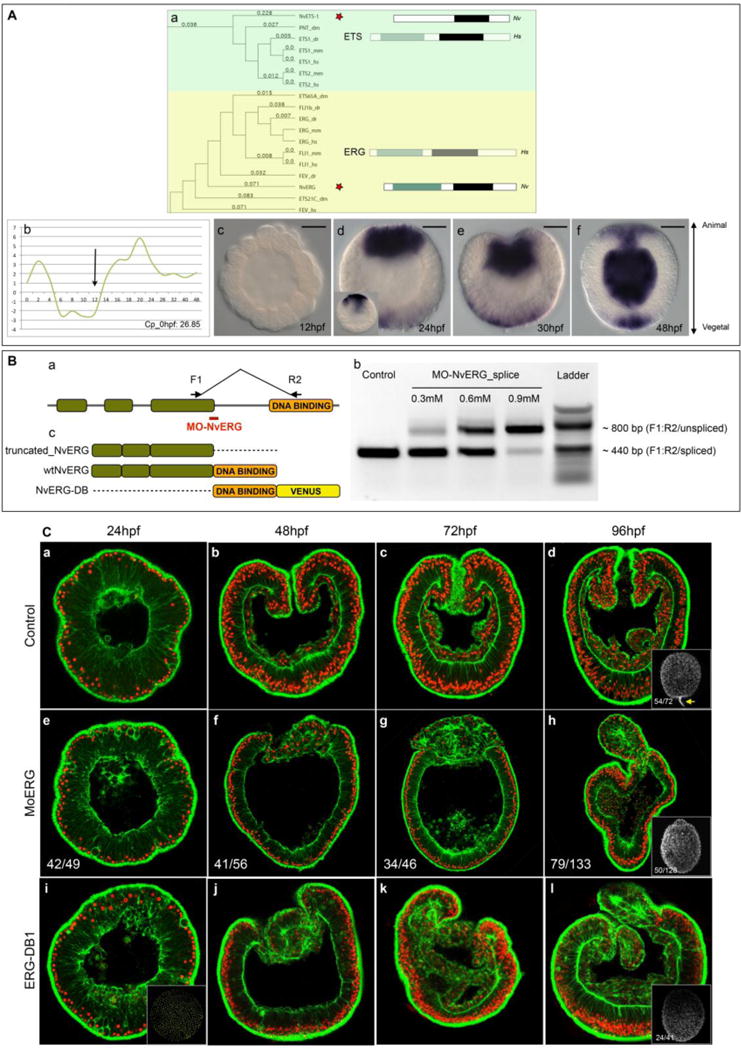
(A) Identification of NvErg and analysis of its spatiotemporal expression. (Aa) Excerpt of the phylogenetic analysis of the ETS transcription factor complement in Nematostella, indicating the existence of NvERG and NvETS1 orthologs in cnidarians. The full analysis can be found in Fig. S1. To the right of the tree, the bars indicated the protein domain organization of either the human representative (Hs, greyed out rectangles) of the subfamily as well as the protein domain organization of the Nematostella ortholog (Nv). Green rectangles indicate the Pointed domain, black rectangles the ETS domain. (Ab) Temporal expression of Nverg analyzed by qPCR during embryonic development of the first 48 hours post fertilization. The y-axis indicates relative fold changes compared to fertilized eggs. (Ac–Af) Spatial (in situ hybridization) expression of Nverg at early (Ac) and late (Ad) blastula, mid (Ae) and late (Af) gastrula stages. Orientation of blastula stages was determined by the thickening of the animal pole prior to its invagination that was observable in certain embryos of a given batch at the analyzed time point. Animal pole to the top and vegetal pole to the bottom. The black bar in the upper right corner of Ac–Af indicates the scale bars: 50μm (Ba) Schematic representation of the genomic organization of NvERG, and the recognition site of MO-NvERG and the position of the PCR primers to verify the efficiency of the splice blocking morpholino. (Bb) Splice blocking efficiency of MO-NvERG analyzed by RT-PCR at increasing concentrations of MO-NvERG injected embryos. (Bc) Schematic representation of the protein structure of the various tools used in Fig. 1C and Fig. S2. (C) Morphological effects of inhibiting NvERG function during early Nematostella development. (Ca–Cd) Control embryos injected with MO-CTRL, (Ce–Ch) embryos injected with MO-NvERG or (Ci–Cl) mRNA encoding a dominant negative version of NvERG (NvERG-DB1) at 24 (Ca,Ce,Ci, late blastula), 48 (Cb,Cf,Cj, late gastrula), 72 (Cc,Cg,Ck, early planula) and 96 (Cd,Ch,Cl, late planula) hpf. All images are lateral views with the animal/oral pole to the top and confocal z-sections using phalloidin (green) to show f-actin filaments and propidium iodide (red) to visualize the nuclei. The insets in (Cd, Ch, Ci) correspond to lateral views of embryos stained with acetylated tubulin to visualize the presence or absence of the apical tuft (yellow arrow in Cd). The numbers in the insets indicate the number or embryos with the represented phenotype/total amount of analyzed animals.
Imaging
in situ hybridization images were taken on either a Zeiss AxioScop 2 or a Zeiss Axio Imager A2 mounted with an Axiocam camera triggered by Axiovision software (Carl Zeiss). All expression patterns described here have been submitted to Kahi Kai, a comparative invertebrate gene expression database [41] hosted at http://www.kahikai.org/index.php?content=genes. Scoring of treatment phenotypes was performed on either a Zeiss Z-1 Axio imager or a Zeiss Axio Imager A2 microscope and confocal imaging was conducted on either a Zeiss LSM710 or Zeiss LSM Exciter microscope running the LSM ZEN software (Carl Zeiss). Fluorescent images were false-colored, the fluorescent channels merged using ImageJ (http://rsbweb.nih.gov/ij/) and cropped to final size in Photoshop Cs6 (Adobe Inc.).
Results
Nverg is expressed in the central and the apical domains
In recent reports from Nematostella, genes belonging to the family of ETS transcription factors have been identified and reported [24,35]. However, the entire complement of this transcription factor family in cnidarians is currently unknown. We have identified 12 genes that encode proteins predicted to contain an ETS DNA binding domain defining this family of transcription factors. A phylogenetic analysis of those factors has revealed that Nematostella possess members of 8 out of 11 ETS sub-familes (Fig. 1Aa, Fig. S1) [42,43]. We have further analyzed their spatio-temporal expression at late blastula stages and observed clear expression patterns for 3 genes (Nvets-likeA (previously called NvelkA-like [24]), Nvpea3 (previously called Nvpea3-like [35]) and Nverg. Nvets-likeA was identified to respond to over-activated canonical Wnt signaling (1-azakenpaullone treatments) and is expressed in the central domain [24]. Nvpea3, identified to be downstream of ERK/MAPK signaling (U0126 treatments) is expressed in individual cells of a circumferential territory within the ectodermal body wall [35]. One gene that retained our particular attention was Nverg, whose transcripts were detected in a bi-polar manner within the central as well as the apical domains of the late blastula/very early gastrula (Fig. 1Ad,Ae). While it is only detected in the central domain at blastula stages (Fig. 1Ad, inset), the apical expression appears progressively when the animal regions flattens prior to its invagination (Fig. 1Ad, Ae). At later stages this gene was expressed within the forming mouth opening, the gastrodermis as well as the apical pole (Fig. 1Af). Interestingly, this gene has not been previously identified in any of the microarray studies analyzing the effects of blocking FGF signaling in the apical domain [29] or general ERK/MAPK inhibition [35], suggesting that it may have been missed or that its activity might be regulated by post-transcriptional modifications. Fine-scale qPCR analysis of Nverg temporal gene expression revealed that it is maternally expressed and that the onset of zygotic expression occurs between 12–14 hours post fertilization (hpf, at 17C, Fig1Ab).
Inhibition of NvERG prevents formation of the gut and perturbs the genesis of the apical tuft
In order to block activity of NvERG during Nematostella development, we used a splice blocking morpholino (MO-NvERG) targeting exon three of this gene and thus creating a truncated version of the protein that is lacking the DNA binding domain (Fig. 1Ba). Microinjecting increasing concentrations of MO-NvERG into oocytes followed up by PCR revealed that MO-NvERG injection at 0.9mM causes drastic splice defects (Fig. 1Bb). We further analyzed the morphological phenotypes induced by the disruption of NvERG function and observed that MO-NvERG morphants entirely failed to gastrulate and form a gut (Fig. 1Cf–h) compared to control embryos (Fig. 1Cb–d). This resulting phenotype can be the direct consequence of the absence of morphogenetic movements required for gastrulation or indirectly, caused by the lack of bodywall endomesoderm that becomes the future gut. In addition, MO-NvERG also perturbs the formation of the apical tuft (Fig. 1Ch) as revealed by acetylated tubulin staining [23]. In order to confirm the specificity of MO-NvERG, we performed a rescue experiment by overexpressing mRNA encoding a wild-type, Venus tagged version of NvERG after MO-NvERG injection, and observed that gastrulation movements are restored in the majority of analyzed embryos (Fig. S2A–G). A control rescue experiment using TomatoNLS mRNA instead of NvERG:Venus, failed to rescue the phenotype (Fig. S2D,H), showing that the rescued phenotype is NvERG specific. We also microinjected a dominant negative form of NvERG (NvERG-DB) that is lacking the transactivation domain in the N-terminus of the protein [44]. While not as efficient as MO-NvERG, injection of NvERG-DB caused i) severe perturbations of gut formation and gastrulation movements (that appear to recover partially during later embryonic development) and ii) the perturbation of apical tuft development (Fig. 1i–l). All together these experiments show that NvERG is required for gut formation and participates in apical tuft genesis. Interestingly, this loss of NvERG function in Nematostella causes a phenotype that is strikingly similar to the one described from blocking ERK/MAPK signaling using U0126 [35], supporting the idea that NvERG might be one of the effectors of ERK/MAPK signaling in the animal as well as the vegetal hemispheres.
Differential expression of ERK/MAPK targets along the animal-vegetal axis
A previous study has reported the differentially expressed genes identified in a genome-wide expression array comparing DMSO treated controls with U0126 treated blastula stages [35]. The spatial expression pattern presented in this study were focused on genes that were expressed in individual cells and that might be involved in early neurogenesis in Nematostella [35]. However, our in situ hybridization screen also revealed genes that were specifically expressed in the animal or vegetal hemispheres at blastula stages (24hpf) and at the end of gastrulation (48hpf) (Fig. 2, 3). While we focused on newly identified genes, we included previously published genes in our analysis to either obtain additional spatial information (e.g. Nvgata [30]) or because their expression domains had not been characterized during the initial period of our analysis (e.g. Nvsix3/6, Nvfoxq2a [28], Nvsfrp1 [29] and Nvfz5/8 [31]). All original publications corresponding to a given gene (sequence identification and/or gene expression pattern) can be found in [35], Tables S4 and S5.
Figure 2. MEK/ERK signaling targets expressed in the presumptive endomesoderm.
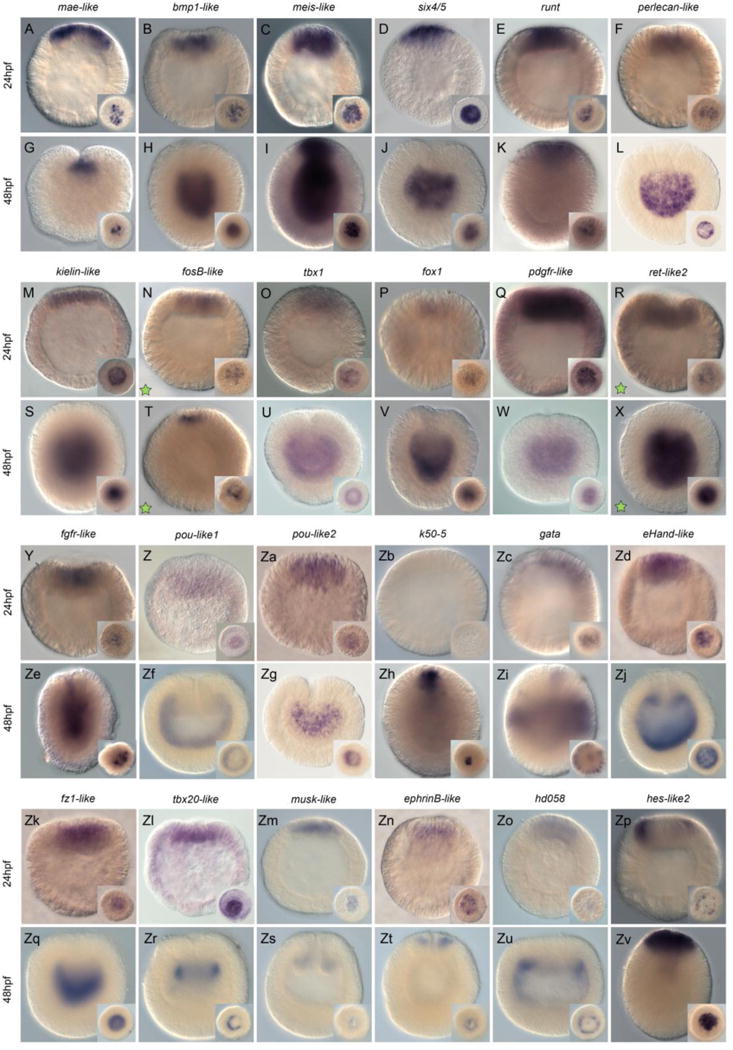
Wild-type endomesodermal gene expression analysis by in situ hybridization of genes differentially regulated by U0126 treatments. All animals are either blastula (24hpf - A–F, M–R, Y–Zd, Zk–Zp) or gastrula (48hpf – G–L, S–X, Ze–Zj, Zq–Zv) stages. All images are lateral views with the animal pole (presumptive endomesoderm) to the top. The insets correspond to animal pole views (A–G, I, K, M–R, T, Y–Zd, Zh, Zv, Zk–Zp) or optical cross-sections. Antisense probes used as indicated at the top of each pair of embryos. Green stars in N, T and R, X indicate that these genes were upregulated under U0126 conditions. All other genes were downregulated.
Figure 3. MEK/ERK signaling targets expressed in broad ectodermal domains.
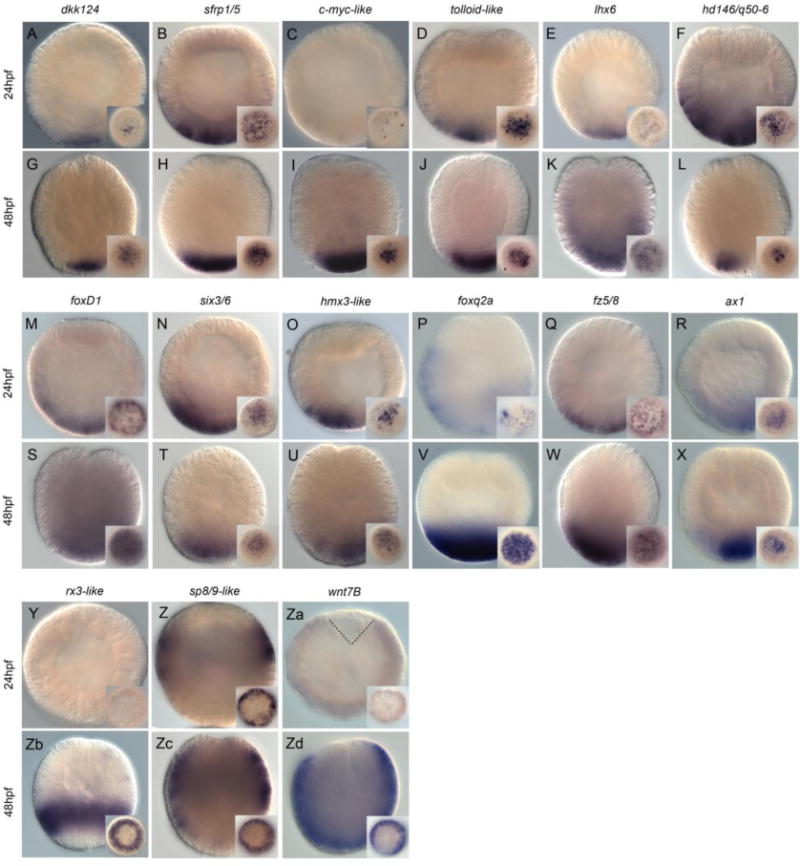
Wild type ectoderm gene expression analysis by in situ hybridization of genes downregulated by U0126 treatments. All animals are either blastula (24hpf - A–F, M–R, Y–Za) or gastrula (48hpf – G–L, S–X, Zb–Zd) stages. All images are lateral views with the animal pole to the top. All insets correspond to vegetal pole/aboral views. Antisense probes used as indicated at the top of each pair of embryos.
Genes expressed within the animal hemisphere (endomesoderm)
We observed 24 localized expression patterns within the animal hemisphere (Nvmae-like, Nvbmp1-like, Nvmeis-like, Nvsix4/5, Nvrunt, Nvperlecan-like, Nvkielin-like, NvfosB-like, Nvtbx1, Nvfox1, NvpdgfR-like, Nvret-like2, Nvfgfr-like, Nvpou-like1, Nvpou-like2, Nvk50-5, Nvgata, NveHand-like, Nvfz1-like, Nvtbx20-like, Nvmusk-like, NvephrinB-like, Nvhd058, Nvhes-like2, Fig. 2). While some genes were only faintly detected (e.g. Nvtbx1, Nvfox1 (Fig. 2O,P), only one gene, Nvk50-5 was not detected at the blastula stage (but was by the gastrula stage)(Fig. 2Zb). However, among the 23 genes that displayed localized gene expression within the animal hemisphere at the blastula stage (24hpf @17°C), only Nvhes-like2 was detected in the central ring (Fig. 2Zp). All other analyzed genes were expressed in the central domain prior to the onset of gastrulation (Nvmae-like, Nvbmp1-like, Nvmeis-like, Nvsix4/5, Nvrunt, Nvperlecan-like, Nvkielin-like, NvfosB-like, Nvtbx1, Nvfox1, Nvpdgfr-like, Nvret-like2, Nvfgfr-like, Nvpou-like1, Nvpou-like2, Nvgata, NveHand-like, Nvfz1-like, Nvtbx20-like2, Nvmusk-like, NvephrinB-like, Nvhd058, Fig. 2A–F,M,O,Q,R,Y,Za,Zc,Zd,Zk–Zo).
However, at the end of gastrulation (48hpf at 17°C), the expression domains of the same set of genes are not only restricted to a single domain as seen at the blastula stage, but are expressed in at least five distinct territories (Fig. 2G–L,S–X,Ze,Zj,Zq–Zv). Only Nvgata is expressed in individual cells within the ectoderm (Fig. 2Zi) confirming a previous description [30]. Six genes, NvfosB-like, Nvhes-like2, Nvmae-like, runt, Nvk50-5 and NvephrinB-like are expressed only within the oral ectoderm (Fig. 2T,Zh,K,Zh,Zt). Nvmeis is expressed in the oral ectoderm, pharyngeal ectoderm, as well as pharyngeal and body wall endomesoderm (Fig. 2I). Transcripts of Nvmusk-like are only detected in the pharyngeal ectoderm (Fig. 2Zs) and expression of Nvtbx20-like2 only in the pharyngeal endomesoderm (Fig. 2Zr). Nvbmp1-like, Nvsix4/5, Nvperlecan-like, Nvkielin-like, Nvtbx1-like, Nvfox1, Nvpdgfr-like, Nvret-like2, Nvfgfr-like, Nvpou-like1, Nvpou-like2, NveHand-like, Nvfz1-like, Nvhd058, transcripts are clearly detected in body wall endomesoderm and potentially also in the pharyngeal endomesoderm (Fig. 2H,J,L,S,U,V,W,X,Ze,Zf,Zg,Zj,Zq,Zu).
Genes expressed within the ectoderm/apical domain
Our in situ hybridization screen also revealed expression patterns of 15 genes in continuous territories within the ectoderm/apical domain (Nvdkk124, Nvsfrp1/5, Nvc-myc-like, Nvtolloid-like, Nvlhx6, Nvhd146, NvfoxD1, Nvsix3/6, Nvhmx3-like, Nvfoxq2a, Nvfz5/8, Nvax1, Nvrx3-like, Nvsp8/9-like, Nvwnt7B, Fig. 3). At the blastula stage, we observed very restricted expression of Nvdkk124, Nvc-myc-like and Nvlhx6 towards the vegetal most part of the embryo (Fig. 3A,C,E) and broader expression within the apical domain of Nvsfrp1/5, Nvtolloid-like, Nvhd146. NvfoxD1, Nvsix3/6, Nvhmx3, Nvfoxq2, Nvfz5/8 and Nvax1 (Fig. 3B,D,F,M–R). While we did not observe localized expression prior to gastrulation for Nvrx3-like (Fig. 3Y) transcripts of Nvsp8/9-like were detected in a circumferential ring (Fig. 3Z) and those of NvWnt7B faintly throughout the entire blastula but lacking a territory that appears to correspond to the central domain (Fig. 3Za).
Of the twelve genes that were exclusively expressed within the apical domain of the blastula stage, one can distinguish two groups of genes based on their expression domains at the end of gastrulation (48hpf). While transcripts of Nvlhx6, NvfoxD1, Nvfoxq2a and Nvfz5/8 are detected in a broader domain that appears to correspond to the sub-apical as well as the apical pole domains (Fig. 3K,S,V,W) expression of Nvdkk124, Nvsfrp1/5, Nvcmyc-like, Nvtolloid-like, Nvhd146, Nvsix3/6, Nvhmx3-lik3 and Nvax1 seem more restricted to only the apical pole (Fig. 3G,H,I,J,L,T,U,X). Nvrx3-like is restricted to a region of the gastrula stage that corresponds to the sub-apical pole domain (Fig. 3Zb), while Nvsp8/9-like expression is localized in a broader territory that spans the sub-apical pole and the body wall ectoderm (Fig. 3Zc) and NvWnt7B within the bodywall ectoderm, the sub- as well as the apical pole domains (Fig. 3Zd).
Of all gene expression patterns described here only NvfosB-like (Fig. 2N,T) and Nvret-like2 (Fig. 2R,X) have been identified from the set of genes that were up regulated after U0126 treatments [35]. Thus, taken together these data strongly suggest that functional ERK/MAPK signaling is crucial for specification and patterning events throughout the entire embryo by the blastula stage. Additional double in situ hybridization experiments are required to fine-tune the precise boundaries of expression domains and the relationships that may exist with neighboring domains.
Temporal gene expression of endomesodermal and ectodermal genes
Spatial expression data, providing information about the presence of maternal transcripts or zygotic upregulation of a given gene, is crucial for the design of functional studies, to predict potential genetic interactions, and build gene regulatory networks [45]. We thus performed a fine scale RT-qPCR analysis (0–48hpf, every two hours) of 23 endomesodermal and 18 apical domain genes (Fig. 4C,D, Fig. S3). This set of analyzed genes contains most of the genes characterized above (Fig. 2, 3) but for the sake of enhancing the current view of the endomesodermal and ectodermal GRNs we also included genes whose ectodermal expression patterns were previously reported (see Fig. 4C,D and Tables S4 and S5 in [35] for references).
Figure 4. High density gene expression profiling.
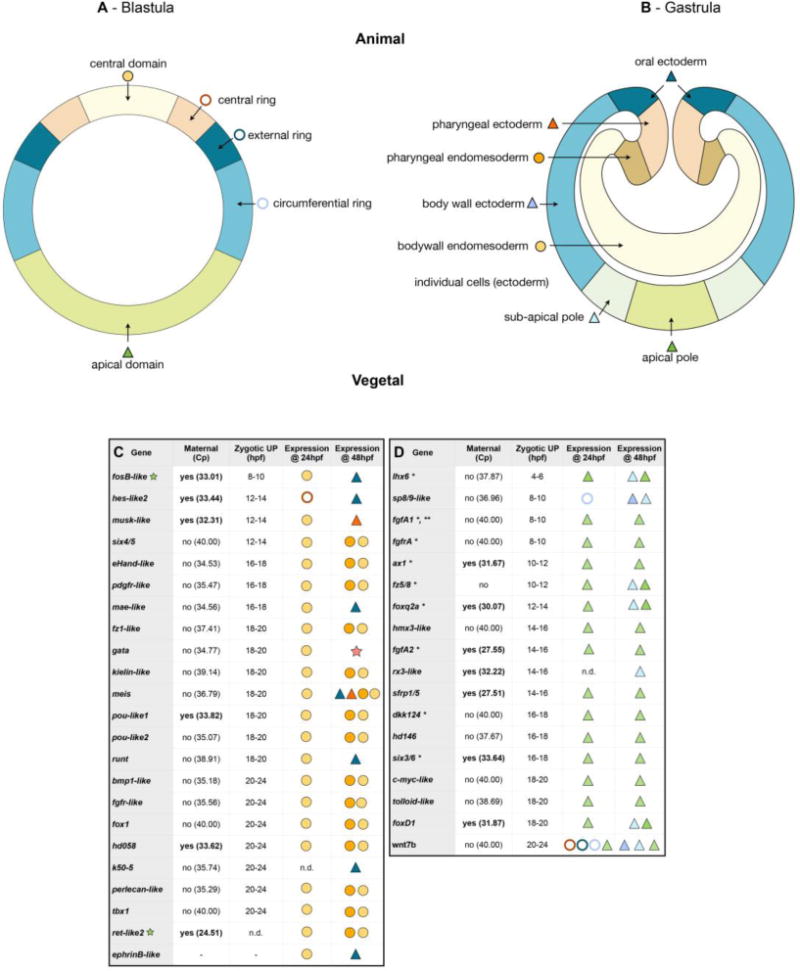
(A, B) Schematic representation of stage-specific expression domains prior to, and after gastrulation movements. (C, D) Summarized results of the temporal high density profiling (qPCR) used to determine the presence of maternal transcripts and significant zygotic up-regulation of a given gene expressed in (C) endomesodermal, or (D) broad ectodermal domains (see Fig. S3 for details). The Cp value corresponds to the cycle number at detection threshold (crossing point). (hpf) hours post fertilization. Visual keys used to describe the spatial expression domain determined by in situ hybridization at 24hpf or 48hpf same as in A,B. (n.d.) Not determined. (*) Indicate genes that have been identified and their spatial blastula and gastrula expression patterns characterized elsewhere (see Tables S4 and S5 in [35] for references). However, to include them into our GRNs we performed qPCRs also for these genes (i.e. fgfA1, fgfA2, ax1 etc…). (**) qPCR value from [24].
From the 41 analyzed genes, the vast majority (68%; n=28/41) were not expressed maternally (Cp value, corresponding to the cycle number at detection threshold > 34.00), only one gene did not show significant zygotic upregulation, while all remaining analyzed genes were zygotically expressed before the onset of gastrulation (20–24hpf, Fig. 4C,D). Within the endomesodermal genes (Fig. 4C), the first gene zygotically upregulated was NvfosB-like (8–10hpf), followed a few hours later first by Nvhes-like2, Nvmusk-like, Nvsix4/5 (12–14hpf), then by NveHand-like, Nvpdgfr-like, Nvmae-like and Nvtbx20-like2 (16–18hpf). The vast majority (14/23) within this group of genes, are zygotically upregulated either 18–20hpf (Nvfz1-like, Nvgata, Nvkielin-like, Nvmeis, Nvpou-like1, Nvpou-like2, Nvrunt) or 20–24hpf (Nvbmp1-like, Nvfgfr-like, Nvfox1, Nvhd058, Nvk50-5, Nvperlecan-like, Nvtbx1-like). Interestingly, the general zygotic activation pattern of the 18 genes expressed in the ectoderm (Fig. 4D) was earlier than the other group of genes. In fact, Nvlhx6 was zygotically upregulated 4–6hpf, followed by Nvsp8/9-like, NvfgfA1, NvfgfrA (8–10hpf), Nvax1, Nvfz5/8 (10–12hpf), Nvfoxq2a (12–14hpf), Nvhmx3-like, NvfgfA2, Nvrx3-like and Nvsfrp1/5 (14–16hpf). While Nvdkk124, Nvhd146 and Nvsix3/6 are upregulated at 16–18hpf the last set of genes, Nvc-myc-like, Nvtolloid-like, NvfoxD1 and NvWnt7b are zygotically regulated between 18–20hpf and 20–24hpf respectively.
NvErg is required to specify the central domain within the animal hemisphere
In order to identify downstream targets of NvERG required for gastrulation, endomesoderm as well as apical domain formation, we injected MO-NvERG into the fertilized egg and performed qPCR analysis on genes expressed in distinct domains along the animal-vegetal axis of the blastula stage (see Fig. 4A). Of the 47 genes expressed in the central domain, 18 genes (Nvadmp-related, Nvfox1, Nvrunt, Nvmae-like, Nvpou-like2, NvotxA, Nvret-like2, Nvgli, Nvhd050, Nvtolloid, NvelkA-like, Nvkielin-like, Nvbmp1-like, Nvgsc, NvotxB, Nvtbx1, Nvperlecan-like and NvotxC) were significantly down-regulated (Fig. 5A). The only gene that is up-regulated in this context is Nverg which might be caused by a stabilizing effect of the morpholino targeting Nverg [28]. In order to confirm that NvERG-DB causes a similar phenotype than Mo-NvERG even at the molecular levels, we performed qPCR analysis on NvERG-DB injected embryos at 24hpf. In line with the milder phenotype observed in latter animals at later developmental stages, the downregulation of gene expression shows a similar trend but not the same amplitude (Fig. S4). Genes that are expressed in both, the central domain as well as the central ring, or only restricted to the central, external or circumferential rings were not significantly affected by NvERG down-regulation (Fig. 5A).
Figure 5. Molecular phenotype analysis of Mo-NvERG injected embryos on genes expressed in the endomesoderm.
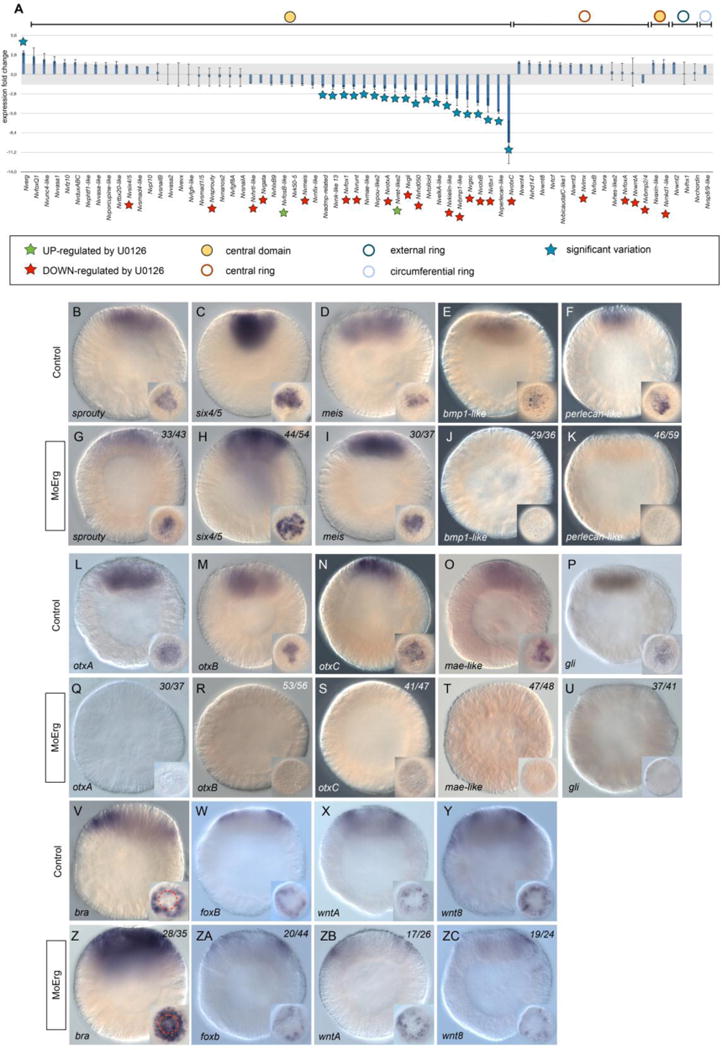
(A) Changes in gene expression of 66 potential components of the cnidarian endomesoderm GRN within the animal hemisphere after NvERG knock-down compared to control embryos analyzed by qPCR. Changes in gene expression are indicated as relative fold changes compared to MO-CTRL injected embryos (x ± sem, n = 3 per gene). The grey bar indicates no significant change in gene expression (−1.5,1.5). Information on the iconography (stars and circles) are indicated below the graph. Gene expression domains at the blastula stage are the same as Figure 4A. (B) Analysis of the molecular effects of NvERG inhibition (G–K, Q–U, ZA–ZE) compared to control injections (B–F, L–P, V–Z) on endomesodermal gene expression analyzed by in situ hybridization. Antisense probes used as indicated in the bottom left corner of each image (also valid for the corresponding inset). The red dashed circle in (V,Z) indicates the central ring expression of Nvbra showing extension of its expression domain into the central domain. The numbers in the upper right corner indicates the ratio of embryos with perturbed gene expression to the total number of analyzed embryos. All images are lateral views with the presumptive endomesoderm (animal pole) to the top. Insets are animal pole views.
We have further analyzed the molecular effect of perturbing NvERG function by in situ hybridization in order to confirm the qPCR data and to gain additional spatial insight. Analyzing the spatial expression patterns of central domain genes are in line with the qPCR data and showed that nvsprouty, nvsix4/5 and nvmeis were unaffected in NvERG morphants (Fig. 5B–D,G–I). Further in agreement with the quantitative expression information, Nvbmp1-like, Nvperlecan-like, NvotxA, NvotxB, NvotxC, Nvmae-like and Nvgli are no longer/faintly detected in the majority of MO-NvERG injected embryos (Fig. 5E,F,J,K, L–P, Q–U). A report using computational approaches to predict gene interactions in Nematostella has suggested that NvERG in the central domain might repress Nvbra in order to restrict its expression to the central ring [38]. We therefore analyzed central ring gene expression (Fig. 5V–Y,Z–ZC) in MO-NvERG injected embryos, even though their expression levels didn’t vary significantly in our qPCR assays. The in situ information obtained for NvfoxB, NvwntA, Nvwnt8 (Fig. 5W‐Y, ZA‐ZC) were in line with the qPCR data and revealed no variations in response to NvERG inhibition. Interestingly though, Nvbra transcripts in NvERG morphants were now also detected in the central domain as well as the central ring (Fig. 5V,Z). The expanded expression domain of this gene highlights the importance of NvERG not only to induce expression of central domain genes, but also to repress specific gene expression in that domain. Thus, NvERG is crucial for the segregation of the central domain and central ring territories during early Nematostella development.
NvERG is a key player of the apical domain gene regulatory network
In addition to its expression in the central domain, Nverg transcripts are also detected in the apical domain (Fig. 1Ad–Af) raising the question about its role in patterning this territory. Quantitative molecular analysis of 15 genes expressed in the apical domain revealed that only five genes (Nvsix3/6, Nvtolloid-like, Nvhd146 and NvfoxD1, NvfgfA2) are significantly downregulated after MO-NvERG injection prior to the onset of gastrulation (24hpf, Fig. 6A). Consistent with this result, spatial expression analysis revealed that no transcripts were detected in the apical domain for Nvsix3/6, NvfoxD1, Nvhd146 and NvfgfA2 for the majority of NvERG morphants (Fig. 6A, G–I,K). Surprisingly, we also observed a visible downregulation of Nvsfrp1/5 by in situ hybridization, although there was no striking effect observed by qPCR (Fig. 6A, J). These effects of NvERG down-regulation on genes expressed in the vegetal most domains are in line with the bipolar gene expression of this gene. In addition, these data suggest that the phenotype of MO-NvERG on apical tuft formation (Fig. 2h) is a direct consequence of this knockdown, rather than an indirect effect caused by the failure of gastrulation.
Figure 6. Molecular phenotype analysis of MO-NvERG, MO-NvFGFRA and NvSix3/6 injected embryos on genes expressed in the apical domain.
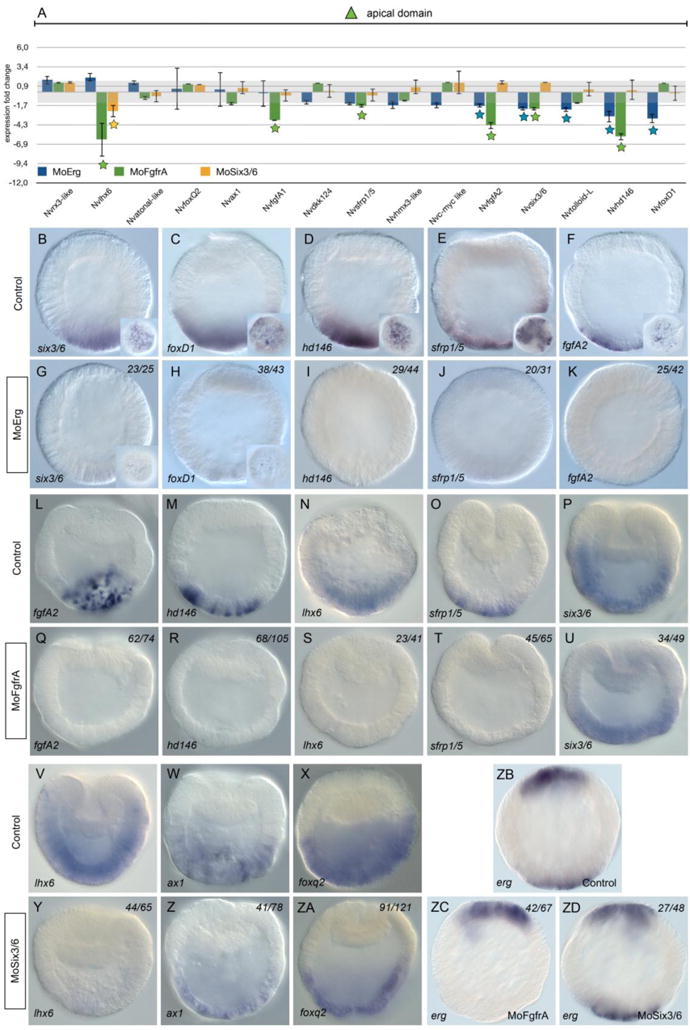
(A) Changes in gene expression of 15 potential components of the cnidarian apical domain GRN within the vegetal hemisphere after NvERG (blue), NvFGFRA (green) or NvSix3/6 (yellow) knock-downs compared to control embryos analyzed by qPCR. Changes in gene expression are indicated as relative fold changes compared to MO-CTRL injected control embryos (x ± sem, n = 3 per gene). The grey bar indicates no significant change in gene expression (−1.5,1.5). Stars below the bars indicate significant variation. Analysis of the molecular effects of NvERG (G–K), NvFGFRA (Q–U) and NvSix3/6 (Y–ZA) inhibition compared to control injections (B–F, L–P, V–X) on apical domain gene expression analyzed by in situ hybridization. Antisense probes used as indicated in the bottom left corner of each image (also valid for the corresponding inset). The numbers in the upper right corner indicates the ratio of embryos with perturbed gene expression to the total number of analyzed embryos. All images are lateral views with the presumptive endomesoderm (animal pole) to the top.
FGF/FGFR signaling and the transcription factor NvSix3/6 have been shown to play a crucial role for apical domain patterning at the end of gastrulation [23,28]. However, no information is available concerning the roles of those genes prior to the onset of gastrulation. In order to gain a better understanding about the relationship between NvFGFRA, NvSix3/6 and NvERG, we inhibited NvFGFRA and NvSix3/6 using previously described morpholinos [23,28] and analyzed gene expression of the same set of apical domain genes (Fig. 6A, Q–U, Y–ZA) as for NvERG morphants. In NvSix3/6 morphant early gastrula, only Nvlhx6 expression is affected (Fig. 6A, Y). However, inhibition of NvFGFRA shows that expression of Nvlhx6, NvfgfA1, NvfgfA2, Nvsix3/6, Nvhd146 and Nvsfrp1/5 is downregulated (Fig. 6A). While in situ expression analysis clearly confirmed the loss of NvfgfA2, Nvhd146, Nvlhx6 and Nvsfrp1/5 expression, the reduction of Nvsix3/6 expression after blocking NvERG function seems subtle (Fig. 6Q–U). The clear overlap of NvFGFRA and NvERG downstream targets, strongly suggest that NvERG activity is partly mediated by NvFGFRA/MAPK signaling in the apical domain. In addition, we have assessed whether apical domain expression of Nverg is NvSix3/6 or FgfrA dependent. While erg expression is not affected in Six3/6 morphants, the apical domain expression of erg is not detected anymore in MO-FgfrA injected embryos, suggesting that apical erg expression requires functional FgfrA signaling.
Discussion
The ETS gene family in Nematostella
The ETS family of genes is evolutionarily conserved [42,43,46,47] and formed by transcriptional regulators involved in various aspects of development, differentiation, hormone responses and tumorgenesis [4,48–55]. Our genome wide survey of ETS transcription factors present in Nematostella revealed the presence of 12 members of this gene family (Fig S1). Nine of them belong to eight out of the eleven described subfamilies (SPI, ESE, TEL, TCF, ETS, ERG, ELG, PEA3). While we were not able to identify members of the PDEF, ELF and ERF subfamilies, we identified three additional genes (Nvets-like-A, Nvets-like-B and Nvets-like-C) that do not group in neither of the subfamilies but are predicted to contain an ETS domain (Fig S1). Expression information for Nvpea3, Nvets-likeA and Nverg have been reported ([24,35], this study), however, additional work is needed to gain insight into the spatial and temporal expression dynamics of those transcription factors during cnidarian development.
The PDEF, ESE, TEL, ETS, ERG and ELG subfamilies in bilaterian animals are characterized, in addition to the ETS domain, by the presence of a Pointed domain that is involved in a series of complex interactions with co-factors to modulate gene expression of downstream targets [47,56,57]. To our surprise, the only ETS gene product in Nematostella predicted to possess a Pointed domain is NvERG (Fig S1), suggesting a potential modulation of its transcriptional activity by other Pointed domain containing proteins. Interestingly, one of the downstream targets of MEK/ERK signaling and NvERG we identified is NvMae-like, a Pointed domain containing protein described to regulate transcriptional activity of YAN (Drosophila ortholog of TEL) and Pointed (Drosophila ortholog of ETS-2) [58]. Nvmae-like is zygotically expressed in the central domain under the control of NvERG suggesting that both proteins interact to potentiate transcriptional activity in this territory and further enhance segregation of specific domains within the animal domain.
A global gene regulatory network orchestrating specification and patterning events of the early embryo
Our data show that MEK/ERK signaling upstream of the transcription factor ERG is required during early embryogenesis in both hemispheres of the blastula: in the animal pole for specifying endomesoderm that probably lead indirectly to the observed failure of gastrulation and gut formation; in the vegetal pole, for specifying the apical domain and participating in apical tuft development. Thus, the present spatial and temporal expression data combined with the molecular characterization of NvERG specific knock-down experiments enabled us i) to add new genes to the existing endomesoderm GRN [24] in particular within the central domain of the blastula stage (Fig. 7), ii) extend this GRN to genes involved in early apical domain (ectoderm) specification at the same stage (Fig. 7), and iii) draft a global GRN framework for body wall endomesoderm, pharynx (endomesoderm and ectoderm), mouth, body wall ectoderm, sub-apical and apical domains, including components of the Nematostella nervous system (Fig. S5). In order to provide an up to date view of the genetic interactions during Nematostella development, the present networks (Fig. 7, Fig. S5) also include previously published functional data [24,39], [23,28,35,39,59]. In the current version, no claim about direct genetic interactions is made, and additional experiments such as cis-regulatory or CHIP-seq analysis are required.
Figure 7. Updated gene regulatory network orchestrating embryonic development in the cnidarian N. vectensis.
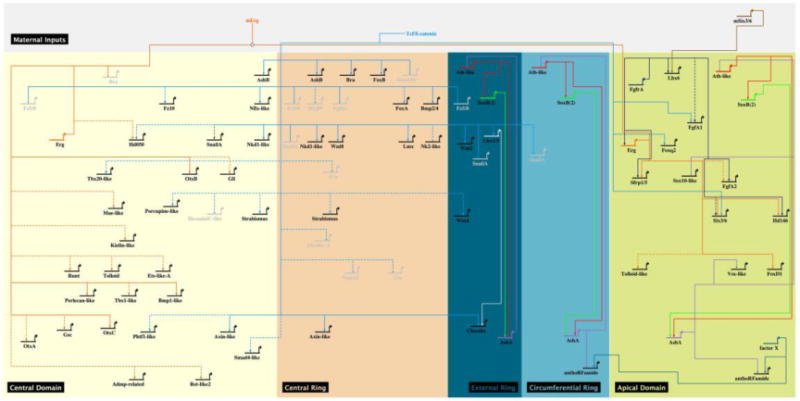
Enhanced Biotapestry diagram [86] of the gene regulatory network describing the gene deployment at 24hpf and regulatory interactions of endomesodermal, ectodermal and neuronal genes identified in previous studies [24,31,35,39,74]. No assumption on whether these interactions are direct or indirect is made. Solid lines indicate functional evidence obtained by qPCR as well as in situ hybridization, dashed lines indicate evidence obtained only by qPCR. The colored boxes represent the spatial domains as described in Figure 4A. Genes inactivated by repression in a given territory are represented in light grey. Controversial results [24,76] about the role of cWnt/TCF signaling on NvsnailA expression is indicated by a red dashed arrow. The same GRN, including non-connected genes that are expressed within the specific territories is provided in Figure S5. A first draft of the global GRN framework for body wall endomesoderm, pharynx (endomesoderm and ectoderm), mouth, body wall ectoderm, sub-apical and apical domain including components of the Nematostella nervous system at the end of gastrulation (48 hpf) is provided in Figure S6.
The present results clearly show that NvERG is one of the main transcription factors involved in this bi-polar activity and might be one of the effectors of MEK/ERK signaling. In fact, MEK/ERK signaling is known to control transcriptional activity (enhancer or repression) of ETS transcription factors by phosphorylating specific residues [4] [60] [13] [61]. Nverg appears not to be transcriptionally controlled by MEK/ERK signaling as it was not identified as being one of the MEK/ERK downstream targets [35]. Nonetheless, we identified certain downstream targets of MEK/ERK [35] that are also controlled by NvERG (e.g. NvotxA, Fig. 5), suggesting a functional control of NvERG by ERK/MEK signaling. Using a phosphorylation motif prediction software (PhosphoMotif Finder, www.hprd.org), we identified 196 potential Serine/Threonine Kinase/phosphatase motifs in NvERG, 15 of which might be more prone to be sensitive to regulation by ERK [61]. However, a precise and systematic approach is required to identify the residue(s) responsible for the activation of ERG in Nematostella. Interestingly, we also observed that a few genes (e.g. Nvsprouty) downstream of MEK/ERK signaling [35] are not sensitive to NvERG knock-down (Fig. 5), suggesting that other MEK/ERK transcriptional effectors are also involved in initiating the embryonic GRN in Nematostella.
Relation between cWnt and ERK pathway in endomesoderm specification
We have previously shown that cWNT controls expression of genes in different domains within the animal hemisphere, with a predominant role for proper gene expression within the central ring domain [24]. In the present study, focusing only at the animal hemisphere, we have identified and characterized genes downstream of MEK/ERK signaling that are primarily expressed in the central domain at the blastula stage (Fig. 2). Among the genes down-regulated by U0126 treatments for which previous expression patterns have been reported, we can find genes expressed in the central domain (Nvgli, Nvgsc, Nvhd50, Nvfix-like, NvotxA, NvotxB, NvotxC, Nvsprouty, [22,24,62–67]), in the central ring (Nvbmp2/4, NvfoxA, Nvlmx, NvwntA, Nvwnt3, [24,30,66,68–73]), in both central ring and central domain (Nvnkd1-like, [24]) as well as those just in the external ring (Nvlhx1, [24,74]). NvERG specific knockdown experiments show that a large part of MEK/ERK downstream targets within the central domain are also NvERG targets suggesting that within the central domain, MEK/ERK signaling might be mediated via NvERG at the transcriptional level. This in turn also suggests that in the other domains of the animal hemisphere, MEK/ERK signaling is mediated by other transcription factors that are yet to be identified.
Temporal qPCR data show that massive zygotic up-regulation of genes within the animal hemisphere identified from the UO126 array [35] begins at about 16hpf (Fig. 4). Interestingly, a temporal qPCR analysis obtained from genes downstream of cWNT signaling show a similar massive zygotic up-regulation of genes but at an earlier stage of development (prior to 14hpf) [24]. Thus, it appears that cWNT specifies a broad “endomesodermal” domain within the animal hemisphere early during embryonic development, and subsequently MEK/ERK signaling is activated to specify a sub-domain (the central domain) to restrict cWNT activity to the central ring domain. The fact that MEK/ERK and NvERG inhibition expand expression of the cWNT target and central ring gene Nvbra towards the central domain ([35], Fig. 5) support the idea that MEK/ERK/ERG signaling has a major impact on specifying the central domain (the future gut) and preventing cWNT activity in this domain.
Within the animal hemisphere at blastula stages, perturbing cWNT signaling has a major impact on central ring gene expression [24], while inhibiting MEK/ERK/ERG activity blocks expression of mainly central domain genes (Fig. 5). The role of cWNT signaling on gastrulation movements has been addressed using different approaches obtaining various degrees of phenotypes [24,31,33,75,76]. Nonetheless, the latest study showed that by performing a morpholino-mediated inhibition of Nvßcatenin, gastrulation was blocked [31]. The present study shows that inhibition of ERG (Fig. 1) phenocopies inhibition of MEK/ERK signaling [35] by also blocking (directly or indirectly) the invagination of the endomesoderm. It would be of importance to decipher the precise mechanisms and the molecular interplay between cWNT and MEK/ERK/ERG signaling in governing morphogenetic movements of gastrulation in Nematostella.
Endomesoderm GRN evolution
Comparative gene regulatory analyses in echinoderm embryos, has suggested the presence of an evolutionarily conserved network “kernel” required for endomesoderm formation [77]. This Kernel is composed of five transcription factors (bra, foxA, otx, blimp1 and gataE) that tightly interact via feedback loops and that severely affect endomesoderm formation when individually knocked-down [77]. Based on the observation that Nvblimp orthologs appear not to be expressed prior to the end of gastrulation (Martindale, unpublished) and that Nvgata transcripts were only described in individual cells of the ectoderm [30], we have previously proposed that the cnidarians endomesodermal kernel is only composed of NvfoxA, Nvbra and NvOtx (A,B and C) [24]. The present study, clearly shows that in addition to its ectodermal expression during gastrulation [30] Nvgata transcripts are detected in the animal hemisphere at the blastula stage, strongly suggesting that this gene could be part of the cnidarian endomesoderm kernel. At the blastula stage Nvbra and NvfoxA (both downstream of cWNT signaling [24]) are expressed in the central ring while Nvotx (A,B and C) and Nvgata are expressed in the central domain. As NvERG is required to repress Nvbra expression in the central domain ([38], this study), these observations foster the idea that rather than being a general endomesoderm kernel connected by feedback loops, those genes are required for the segregation of a central domain and a central ring prior to the onset of gastrulation. A careful analysis of their spatial expression during earlier developmental stages and importantly, functional studies to decipher the precise genetic interactions between those genes is required to better understand a evolutionary conservation of the endomesoderm kernel.
In sea urchins, MEK/ERK signaling, activated during later stages by FGF/FGFR and VEGF/VEGFR [4,7,78,79] is required for mesoderm formation. Interestingly, ETS1 is activated in the primary mesenchyme cells (PMC, mesoderm) by MEK/ERK and crucial for ERG expression (http://sugp.caltech.edu/endomes/#Veg-6-18-NetworkDiagram), PMC ingression and differentiation into the specific mesodermal lineage [4]. On the other hand echinoderm cWNT signaling is required to initiate the general endomesoderm GRN and drive endoderm specification once the mesoderm specification program has been launched [80,81]. Our observations about i) cWNT initiating a broad endomesoderm GRN with a particular emphasis on central ring domain expression [24] and ii) MEK/ERK/ERG signaling being required to repress central ring fate (Nvbra expression) and specify the central domain illustrate the strong evolutionary conservation of this mechanism between cnidarians and echinoderms. Cnidarians are described not to form a true mesodermal germ layer. However, the expression of the classical mesodermal marker brachyury [82] in the central ring, as well as the activation of a MEK/ERK/ERG pathway (required for mesoderm formation in bilaterians [83]) in the central domain, provide additional compelling evidence that both domains together form sub-regionalized territories of the cnidarian endomesoderm. Tissue tracking experiments using photo-convertible fluorescent proteins [84] are required to determine the fate of the central domain and ring. It might also be important to carry out a precise physiological analysis of the differentiated tissues that originated from the endomesodermal territory, to gain more insight in the evolutionary origin of mesoderm in bilaterians.
Relationship between cWnt and MEK/ERK/ERG signaling in patterning the apical domain
Several studies suggested a role of cWNT signaling in ectoderm patterning, as ectopically activating or inhibiting cWNT [24,33,75,76,85], perturbs gene expression within the body wall ectoderm and apical domain. In particular, inhibiting ßcat function causes the loss of Nvfgfa1, NvfoxQ2 and Nvsix3/6 expression in the apical domain [31]. In a previous study, we predominantly reported genes that were up-regulated following ectopic cWnt activation [24]. However, about 30 genes were also downregulated under those conditions (Table S1). Interestingly, 14 out of the 30 identified genes are also down regulated after MEK/ERK inhibition [35]. Among these 14 genes downregulated following MEK/ERK inhibition, seven are expressed in the apical domain (Nvtolloid-like, Nvsfrp1/5, Nvlhx6, Nvhmx3, Nvhd146, NvfoxD1, Nvax1 (Fig. 3)), four in individual cells throughout the ectoderm (Nvhd145, Nvgfi-like, Nvgcm, Nvcoup-like2 [35]) and two in derivatives of the animal pole (Nvhes-like2, Nvgsc (Fig 2, [24]). Inhibition of NvERG has no detectable effect on genes expressed in the individual cells of the ectoderm (data not shown) or Nvhes-like2 of the central ring (Fig. 5). However, perturbing NvERG function clearly blocks Nvgsc expression in the central domain (Fig. 5) and expression of Nvtolloid-like, Nvsfrp1/5, Nvhmx3, Nvhd146 and NvfoxD1 (Fig. 6) within the apical domain. These observations strongly support the idea of an antagonistic effect of cWnt and MEK/ERK/ERG signaling in a specific set of genes and that this signaling pathway interplay is crucial for patterning the apical domain. Importantly though, Nvsix3/6 expression (a key regulator of apical domain formation (28)) requires the combined activity of cWnt (31) as well as MEK/ERK/ERG signaling further underlining the complex interplay of these two pathways to pattern the cnidarian embryonic body axis. Additional gene specific experiments as well as cis-regulatory analysis of the above mentioned genes would shed further light on the direct or indirect antagonistic effects of these two major developmental signaling pathways for cnidarian embryogenesis.
Supplementary Material
Highlights.
Characterization of NvERG, a bi-polarily expressed transcription factor in cnidarians
NvERG is required for endomesoderm and apical domain formation
Identification of ERG downstream targets, distinctly expressed in both poles
NvERG simultaneously controls two different GRNs in opposite poles
Acknowledgments
The authors thank individuals in the MQM, ER and ML labs and Dominic van Essen for discussions and careful reading of the manuscript. The authors also acknowledge Valérie Carlin for animal care and the IRCAN’s Molecular and Cellular Core Imaging (PICMI) Facility. PICMI was supported financially by Cancéropole PACA, Région Provence Alpes-Côte d’Azur, Conseil Départemental 06 and INSERM.
Funding
This project was funded by an NIH grant GM093116 to MQM, and supported by an ATIP-Avenir award (CNRS/INSERM - ITMO Plan Cancer C13992AS), a Marie-Curie Career Integration Grant (CIG – FP7 European Commission #631665) as well as by the Fondation pour la recherche sur le cancer (ARC PJA2014120186) to ER. Individual fellowships were provided by the FRM (Fondation pour la Recherche Médicale SPF20130526781) and the MESR (French Ministry of higher education and science) to ARA and HJ respectively.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author contributions
ER, MJL and MQM – Conceived, designed and performed experiments. ARA, HJ, TC, PD, MI – generated, collected and analyzed data. ER, MQM - Contributed reagents/materials/analysis tools. ER, ARA, HJ, MQM - Drafted the manuscript. All authors read and approved the final manuscript
Competing interests
The author(s) declare(s) that they have no competing interests.
Bibliography
- 1.Schulte-Merker S, Smith JC. Mesoderm formation in response to Brachyury requires FGF signalling. Curr Biol. 1995;5:62–67. doi: 10.1016/s0960-9822(95)00017-0. [DOI] [PubMed] [Google Scholar]
- 2.Burdine RD, Chen EB, Kwok SF, Stern MJ. egl-17 encodes an invertebrate fibroblast growth factor family member required specifically for sex myoblast migration in Caenorhabditis elegans. Proceedings of the National Academy of Sciences. 1997;94:2433–2437. doi: 10.1073/pnas.94.6.2433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Draper BW, Stock DW, Kimmel CB. Zebrafish fgf24 functions with fgf8 to promote posterior mesodermal development. Development. 2003;130:4639–4654. doi: 10.1242/dev.00671. [DOI] [PubMed] [Google Scholar]
- 4.Röttinger E, Besnardeau L, Lepage T. Development. Vol. 131. The Company of Biologists Limited; 2004. A Raf/MEK/ERK signaling pathway is required for development of the sea urchin embryo micromere lineage through phosphorylation of the transcription factor Ets; pp. 1075–1087. [DOI] [PubMed] [Google Scholar]
- 5.Stathopoulos A, Tam B, Ronshaugen M, Frasch M, Levine M. pyramus and thisbe: FGF genes that pattern the mesoderm of Drosophila embryos. Genes Dev. 2004;18:687–699. doi: 10.1101/gad.1166404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yasuo H, Hudson C. FGF8/17/18 functions together with FGF9/16/20 during formation of the notochord in Ciona embryos. Dev Biol. 2007;302:92–103. doi: 10.1016/j.ydbio.2006.08.075. [DOI] [PubMed] [Google Scholar]
- 7.Röttinger E, Saudemont A, Duboc V, Besnardeau L, McClay D, Lepage T. FGF signals guide migration of mesenchymal cells, control skeletal morphogenesis [corrected] and regulate gastrulation during sea urchin development. Development. 2008;135:353–365. doi: 10.1242/dev.014282. [DOI] [PubMed] [Google Scholar]
- 8.Ota S, Tonou-Fujimori N, Yamasu K. The roles of the FGF signal in zebrafish embryos analyzed using constitutive activation and dominant-negative suppression of different FGF receptors. Mech Dev. 2009;126:1–17. doi: 10.1016/j.mod.2008.10.008. [DOI] [PubMed] [Google Scholar]
- 9.Bertrand S, Camasses A, Somorjai I, Belgacem MR, Chabrol O, Escande M-L, et al. Amphioxus FGF signaling predicts the acquisition of vertebrate morphological traits. Proceedings of the National Academy of Sciences. 2011;108:9160–9165. doi: 10.1073/pnas.1014235108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Green SA, Norris RP, Terasaki M, Lowe CJ. FGF signaling induces mesoderm in the hemichordate Saccoglossus kowalevskii. Development (Cambridge, England) 2013 doi: 10.1242/dev.083790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Röttinger E, DuBuc TQ, Amiel AR, Martindale MQ. Nodal signaling is required for mesodermal and ventral but not for dorsal fates in the indirect developing hemichordate, Ptychodera flava. Biology Open. 2015 doi: 10.1242/bio.011809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bertrand S, Iwema T, Escriva H. FGF Signaling Emerged Concomitantly with the Origin of Eumetazoans. Mol Biol Evol. 2014;31:310–318. doi: 10.1093/molbev/mst222. [DOI] [PubMed] [Google Scholar]
- 13.Selvaraj N, Kedage V, Hollenhorst PC. Comparison of MAPK specificity across the ETS transcription factor family identifies a high-affinity ERK interaction required for ERG function in prostate cells. Cell Commun Signal BioMed Central. 2015;13:12. doi: 10.1186/s12964-015-0089-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Martindale MQ, Hejnol A. A developmental perspective: changes in the position of the blastopore during bilaterian evolution. Dev Cell. 2009;17:162–174. doi: 10.1016/j.devcel.2009.07.024. [DOI] [PubMed] [Google Scholar]
- 15.Technau U, Steele RE. Development. Vol. 138. Oxford University Press for The Company of Biologists Limited; 2011. Evolutionary crossroads in developmental biology: Cnidaria; pp. 1447–1458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Layden MJ, Rentzsch F, Röttinger E. WIREs Dev Biol. John Wiley & Sons, Inc; 2016. The rise of the starlet sea anemone Nematostella vectensis as a model system to investigate development and regeneration. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hand C, Uhlinger KR. The culture, sexual and asexual reproduction, and growth of the sea anemone Nematostella vectensis. Biol Bull MBL. 1992;182:169–176. doi: 10.2307/1542110. [DOI] [PubMed] [Google Scholar]
- 18.Darling JA, Reitzel AR, Burton PM, Mazza ME, Ryan JF, Sullivan JC, et al. Rising starlet: the starlet sea anemone, Nematostella vectensis. Bioessays. 2005;27:211–221. doi: 10.1002/bies.20181. [DOI] [PubMed] [Google Scholar]
- 19.Putnam NH, Srivastava M, Hellsten U, Dirks B, Chapman J, Salamov A, et al. Sea anemone genome reveals ancestral eumetazoan gene repertoire and genomic organization. Science. 2007;317:86–94. doi: 10.1126/science.1139158. [DOI] [PubMed] [Google Scholar]
- 20.Layden MJ, Röttinger E, Wolenski FS, Gilmore TD, Martindale MQ. Microinjection of mRNA or morpholinos for reverse genetic analysis in the starlet sea anemone, Nematostella vectensis. Nat Protoc. 2013;8:924–934. doi: 10.1038/nprot.2013.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ikmi A, McKinney SA, Delventhal KM, Gibson MC. TALEN and CRISPR/Cas9-mediated genome editing in the early-branching metazoan Nematostella vectensis. Nature Communications. 2014;5:5486. doi: 10.1038/ncomms6486. [DOI] [PubMed] [Google Scholar]
- 22.Matus DQ, Thomsen GH, Martindale MQ. FGF signaling in gastrulation and neural development in Nematostella vectensis, an anthozoan cnidarian. Dev Genes Evol. 2007;217:137–148. doi: 10.1007/s00427-006-0122-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rentzsch F, Fritzenwanker JH, Scholz CB, Technau U. FGF signalling controls formation of the apical sensory organ in the cnidarian Nematostella vectensis. Development. 2008;135:1761–1769. doi: 10.1242/dev.020784. [DOI] [PubMed] [Google Scholar]
- 24.Röttinger E, Dahlin P, Martindale MQ. A Framework for the Establishment of a Cnidarian Gene Regulatory Network for “Endomesoderm” Specification: The Inputs of ß-Catenin/TCF Signaling. In: Mullins MC, editor. PLoS Genet. Vol. 8. 2012. p. e1003164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hacohen NN, Kramer SS, Sutherland DD, Hiromi YY, Krasnow MAM. sprouty encodes a novel antagonist of FGF signaling that patterns apical branching of the Drosophila airways. Cell. 1998;92:253–263. doi: 10.1016/S0092-8674(00)80919-8. [DOI] [PubMed] [Google Scholar]
- 26.Casci T, Vinós J, Freeman M. Sprouty, an intracellular inhibitor of Ras signaling. Cell. 1999;96:655–665. doi: 10.1016/s0092-8674(00)80576-0. [DOI] [PubMed] [Google Scholar]
- 27.Kramer S, Okabe M, Hacohen N, Krasnow MA, Hiromi Y. Sprouty: a common antagonist of FGF and EGF signaling pathways in Drosophila. Development. 1999;126:2515–2525. doi: 10.1242/dev.126.11.2515. [DOI] [PubMed] [Google Scholar]
- 28.Sinigaglia C, Busengdal H, Leclère L, Technau U, Rentzsch F. The Bilaterian Head Patterning Gene six3/6 Controls Aboral Domain Development in a Cnidarian. In: Levine M, editor. PLoS Biol. Vol. 11. 2013. p. e1001488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sinigaglia C, Busengdal H, Lerner A, Oliveri P, Rentzsch F. Molecular characterization of the apical organ of the anthozoan Nematostella vectensis. Dev Biol. 2014 doi: 10.1016/j.ydbio.2014.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Martindale MQ, Pang K, Finnerty JR. Investigating the origins of triploblasty: “mesodermal” gene expression in a diploblastic animal, the sea anemone Nematostella vectensis (phylum, Cnidaria; class, Anthozoa) Development (Cambridge, England) 2004;131:2463–2474. doi: 10.1242/dev.01119. [DOI] [PubMed] [Google Scholar]
- 31.Leclère L, Bause M, Sinigaglia C, Steger J, Rentzsch F. Development of the aboral domain in Nematostella requires β-catenin and the opposing activities of six3/6 and frizzled5/8. Development (Cambridge, England) 2016;143:1766–1777. doi: 10.1242/dev.120931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Technau U, Scholz CB. Origin and evolution of endoderm and mesoderm. Int J Dev Biol. 2003;47:531–539. [PubMed] [Google Scholar]
- 33.Lee PN, Kumburegama S, Marlow HQ, Martindale MQ, Wikramanayake AH. Asymmetric developmental potential along the animal-vegetal axis in the anthozoan cnidarian, Nematostella vectensis, is mediated by Dishevelled. Dev Biol. 2007;310:169–186. doi: 10.1016/j.ydbio.2007.05.040. [DOI] [PubMed] [Google Scholar]
- 34.Fritzenwanker JH, Genikhovich G, Kraus Y, Technau U. Early development and axis specification in the sea anemone Nematostella vectensis. Dev Biol. 2007;310:264–279. doi: 10.1016/j.ydbio.2007.07.029. [DOI] [PubMed] [Google Scholar]
- 35.Layden MJ, Johnston H, Amiel AR, Havrilak J, Steinworth B, Chock T, et al. MAPK signaling is necessary for neurogenesis in Nematostella vectensis. BMC Biol. 2016;14:61. doi: 10.1186/s12915-016-0282-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Davies SP, Reddy H, Caivano M, Cohen P. Specificity and mechanism of action of some commonly used protein kinase inhibitors. Biochem J. 2000;351:95–105. doi: 10.1042/0264-6021:3510095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.DeSilva DR, Jones EA, Favata MF, Jaffee BD, Magolda RL, Trzaskos JM, et al. Inhibition of mitogen-activated protein kinase kinase blocks T cell proliferation but does not induce or prevent anergy. The Journal of Immunology. Am Assoc Immnol. 1998;160:4175–4181. [PubMed] [Google Scholar]
- 38.A novel technique to combine and analyse spatial and temporal expression datasets: A case study with the sea anemone Nematostella vectensis to identify potential gene interactions. Dev Biol. 2017 doi: 10.1016/j.ydbio.2017.06.004. [DOI] [PubMed] [Google Scholar]
- 39.Layden MJ, Boekhout M, Martindale MQ. Nematostella vectensis achaete-scute homolog NvashA regulates embryonic ectodermal neurogenesis and represents an ancient component of the metazoan neural specification pathway. Development (Cambridge, England) 2012;139:1013–1022. doi: 10.1242/dev.073221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Magie CR, Daly M, Martindale MQ. Gastrulation in the cnidarian Nematostella vectensis occurs via invagination not ingression. Dev Biol. 2007;305:483–497. doi: 10.1016/j.ydbio.2007.02.044. [DOI] [PubMed] [Google Scholar]
- 41.Ormestad M, Martindale M, Rottinger E. A comparative gene expression database for invertebrates. EvoDevo. 2011 doi: 10.1186/2041-9139-2-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Laudet V, Niel C, Duterquecoquillaud M. Evolution of the ets gene family. Biochemical and …. 1993 doi: 10.1006/bbrc.1993.1002. [DOI] [PubMed] [Google Scholar]
- 43.Laudet V, Hänni C, Stéhelin D, Duterque-Coquillaud M. Molecular phylogeny of the ETS gene family. Oncogene. 1999;18:1351–1359. doi: 10.1038/sj.onc.1202444. [DOI] [PubMed] [Google Scholar]
- 44.Pourtier-Manzanedo A, Vercamer C, Van Belle E, Mattot V, Mouquet F, Vandenbunder B. Expression of an Ets-1 dominant-negative mutant perturbs normal and tumor angiogenesis in a mouse ear model. Oncogene Nature Publishing Group. 2003;22:1795–1806. doi: 10.1038/sj.onc.1206215. [DOI] [PubMed] [Google Scholar]
- 45.Oliveri P, Davidson E. Gene regulatory network controlling embryonic specification in the sea urchin. Curr Opin Genet Dev. 2004 doi: 10.1016/j.gde.2004.06.004. [DOI] [PubMed] [Google Scholar]
- 46.Degnan BM, Degnan SM, Naganuma T, Morse DE. The ets multigene family is conserved throughout the Metazoa. Nucleic Acids Res. 1993;21:3479–3484. doi: 10.1093/nar/21.15.3479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rizzo F, Fernandez-Serra M, Squarzoni P, Archimandritis A, Arnone MI. Identification and developmental expression of the ets gene family in the sea urchin (Strongylocentrotus purpuratus) Dev Biol. 2006;300:35–48. doi: 10.1016/j.ydbio.2006.08.012. [DOI] [PubMed] [Google Scholar]
- 48.Kiyota T, Kato A, Kato Y. Ets-1 regulates radial glia formation during vertebrate embryogenesis. Organogenesis. 2007;3:93–101. doi: 10.4161/org.3.2.5171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kataoka H, Hayashi M, Nakagawa R, Tanaka Y, Izumi N, Nishikawa S, et al. Blood. Vol. 118. American Society of Hematology; 2011. Etv2/ER71 induces vascular mesoderm from Flk1+PDGFRα+ primitive mesoderm; pp. 6975–6986. [DOI] [PubMed] [Google Scholar]
- 50.Kar A, Gutierrez-Hartmann A. Critical Reviews in Biochemistry and Molecular Biology. Vol. 48. Taylor & Francis; 2013. Molecular mechanisms of ETS transcription factor-mediated tumorigenesis; pp. 522–543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chen Y, Chi P, Rockowitz S, Iaquinta PJ, Shamu T, Shukla S, et al. ETS factors reprogram the androgen receptor cistrome and prime prostate tumorigenesis in response to PTEN loss. Nat Med Nature Research. 2013;19:1023–1029. doi: 10.1038/nm.3216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kawahara T, Shareef HK, Aljarah AK, Ide H, Li Y, Kashiwagi E, et al. ELK1 is up-regulated by androgen in bladder cancer cells and promotes tumor progression. Oncotarget Impact Journals. 2015;6:29860–29876. doi: 10.18632/oncotarget.5007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Koh B, Hufford MM, Pham D, Olson MR, Wu T, Jabeen R, et al. J Immunol. Vol. 197. American Association of Immunologists; 2016. The ETS Family Transcription Factors Etv5 and PU.1 Function in Parallel To Promote Th9 Cell Development; pp. 2465–2472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Peng C, Zeng W, Su J, Kuang Y, He Y, Zhao S, et al. Cyclin-dependent kinase 2 (CDK2) is a key mediator for EGF-induced cell transformation mediated through the ELK4/c-Fos signaling pathway. Oncogene. 2016;35:1170–1179. doi: 10.1038/onc.2015.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rizzo F, Coffman JA, Arnone MI. An Elk transcription factor is required for Runx-dependent survival signaling in the sea urchin embryo. Dev Biol. 2016;416:173–186. doi: 10.1016/j.ydbio.2016.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sharrocks AD. The ETS-domain transcription factor family. Nat Rev Mol Cell Biol. 2001;2:827–837. doi: 10.1038/35099076. [DOI] [PubMed] [Google Scholar]
- 57.Gutierrez-Hartmann A, Duval DL, Bradford AP. ETS transcription factors in endocrine systems. Trends Endocrinol Metab. 2007;18:150–158. doi: 10.1016/j.tem.2007.03.002. [DOI] [PubMed] [Google Scholar]
- 58.Baker DA, Mille-Baker B, Wainwright SM, Ish-Horowicz D, Dibb NJ. Mae mediates MAP kinase phosphorylation of Ets transcription factors in Drosophila. Nature. 2001;411:330–334. doi: 10.1038/35077122. [DOI] [PubMed] [Google Scholar]
- 59.Kumburegama S, Wijesena N, Xu R, Wikramanayake AH. Strabismus-mediated primary archenteron invagination is uncoupled from Wnt/β-catenin-dependent endoderm cell fate specification in Nematostella vectensis (Anthozoa, Cnidaria): Implications for the evolution of gastrulation. EvoDevo. 2011;2:2. doi: 10.1186/2041-9139-2-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hollenhorst P. RAS/ERK pathway transcriptional regulation through ETS/AP-1 binding sites. smallgtpases. 2012;3:154–158. doi: 10.4161/sgtp.19630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Huang Y, Thoms JAI, Tursky ML, Knezevic K, Beck D, Chandrakanthan V, et al. MAPK/ERK2 phosphorylates ERG at serine 283 in leukemic cells and promotes stem cell signatures and cell proliferation. Leukemia. 2016;30:1552–1561. doi: 10.1038/leu.2016.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Matus DQ, Magie CR, Pang K, Martindale MQ, Thomsen GH. The Hedgehog gene family of the cnidarian, Nematostella vectensis, and implications for understanding metazoan Hedgehog pathway evolution. Dev Biol. 2008;313:501–518. doi: 10.1016/j.ydbio.2007.09.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Matus DQ, Pang K, Marlow H, Dunn CW, Thomsen GH, Martindale MQ. Molecular evidence for deep evolutionary roots of bilaterality in animal development. Proc Natl Acad Sci USA. 2006;103:11195–11200. doi: 10.1073/pnas.0601257103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ryan JF, Burton PM, Mazza ME, Kwong GK, Mullikin JC, Finnerty JR. The cnidarian-bilaterian ancestor possessed at least 56 homeoboxes: evidence from the starlet sea anemone, Nematostella vectensis. Genome Biol. 2006;7:R64. doi: 10.1186/gb-2006-7-7-R64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ryan JF, Mazza ME, Pang K, Matus DQ, Baxevanis AD, Martindale MQ, et al. Pre-bilaterian origins of the Hox cluster and the Hox code: evidence from the sea anemone, Nematostella vectensis. PLoS ONE. 2007;2:e153. doi: 10.1371/journal.pone.0000153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chourrout D, Delsuc F, Chourrout P, Edvardsen RB, Rentzsch F, Renfer E, et al. Minimal ProtoHox cluster inferred from bilaterian and cnidarian Hox complements. Nature. 2006;442:684–687. doi: 10.1038/nature04863. [DOI] [PubMed] [Google Scholar]
- 67.Mazza ME, Pang K, Martindale MQ, Finnerty JR. Genomic organization, gene structure, and developmental expression of three clustered otx genes in the sea anemone Nematostella vectensis. J Exp Zool. 2007;308:494–506. doi: 10.1002/jez.b.21158. [DOI] [PubMed] [Google Scholar]
- 68.Matus DQ, Thomsen GH, Martindale MQ. Dorso/ventral genes are asymmetrically expressed and involved in germ-layer demarcation during cnidarian gastrulation. Curr Biol. 2006;16:499–505. doi: 10.1016/j.cub.2006.01.052. [DOI] [PubMed] [Google Scholar]
- 69.Rentzsch F, Anton R, Saina M, Hammerschmidt M, Holstein TW, Technau U. Asymmetric expression of the BMP antagonists chordin and gremlin in the sea anemone Nematostella vectensis: implications for the evolution of axial patterning. Dev Biol. 2006;296:375–387. doi: 10.1016/j.ydbio.2006.06.003. [DOI] [PubMed] [Google Scholar]
- 70.Fritzenwanker JHJ, Saina MM, Technau UU. Analysis of forkhead and snail expression reveals epithelial-mesenchymal transitions during embryonic and larval development of Nematostella vectensis. Dev Biol. 2004;275:389–402. doi: 10.1016/j.ydbio.2004.08.014. [DOI] [PubMed] [Google Scholar]
- 71.Srivastava M, Larroux C, Lu DR, Mohanty K, Chapman J, Degnan BM, et al. Early evolution of the LIM homeobox gene family. BMC Biol. 2010;8:4. doi: 10.1186/1741-7007-8-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lee PN, Pang K, Matus DQ, Martindale MQ. A WNT of things to come: evolution of Wnt signaling and polarity in cnidarians. Semin Cell Dev Biol. 2006;17:157–167. doi: 10.1016/j.semcdb.2006.05.002. [DOI] [PubMed] [Google Scholar]
- 73.Kusserow A, Pang K, Sturm C, Lentfer J, Schmidt HA, Technau U, et al. Unexpected complexity of the Wnt gene family in a sea anemone. Nature. 2005;433:156–160. doi: 10.1038/nature03158. [DOI] [PubMed] [Google Scholar]
- 74.Yasuoka Y, Kobayashi M, Kurokawa D, Akasaka K, Saiga H, Taira M. Evolutionary origins of blastoporal expression and organizer activity of the vertebrate gastrula organizer gene lhx1 and its ancient metazoan paralog lhx3. Development. 2009;136:2005–2014. doi: 10.1242/dev.028530. [DOI] [PubMed] [Google Scholar]
- 75.Wikramanayake AH, Hong M, Lee PN, Pang K, Byrum CA, Bince JM, et al. An ancient role for nuclear beta-catenin in the evolution of axial polarity and germ layer segregation. Nature. 2003;426:446–450. doi: 10.1038/nature02113. [DOI] [PubMed] [Google Scholar]
- 76.Kumburegama S, Wijesena N, Xu R. Strabismus-mediated primary archenteron invagination is uncoupled from Wnt/β-catenin-dependent endoderm cell fate specification in Nematostella …. EvoDevo. 2011 doi: 10.1186/2041-9139-2-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Hinman VF, Nguyen A, Davidson EH. Caught in the evolutionary act: precise cis-regulatory basis of difference in the organization of gene networks of sea stars and sea urchins. Dev Biol. 2007;312:584–595. doi: 10.1016/j.ydbio.2007.09.006. [DOI] [PubMed] [Google Scholar]
- 78.Fernandez-Serra M, Consales C, Livigni A, Arnone MI. Role of the ERK-mediated signaling pathway in mesenchyme formation and differentiation in the sea urchin embryo. Dev Biol. 2004;268:384–402. doi: 10.1016/j.ydbio.2003.12.029. [DOI] [PubMed] [Google Scholar]
- 79.Duloquin L, Lhomond G, Gache C. Localized VEGF signaling from ectoderm to mesenchyme cells controls morphogenesis of the sea urchin embryo skeleton. Development (Cambridge, England) 2007;134:2293–2302. doi: 10.1242/dev.005108. [DOI] [PubMed] [Google Scholar]
- 80.Davidson EH, Rast JP, Oliveri P, Ransick A, Calestani C, Yuh C-H, et al. A provisional regulatory gene network for specification of endomesoderm in the sea urchin embryo. Dev Biol. 2002;246:162–190. doi: 10.1006/dbio.2002.0635. [DOI] [PubMed] [Google Scholar]
- 81.Davidson EH, Rast JP, Oliveri P, Ransick A, Calestani C, Yuh C-H, et al. A genomic regulatory network for development. Science. 2002;295:1669–1678. doi: 10.1126/science.1069883. [DOI] [PubMed] [Google Scholar]
- 82.Herrmann BG, Labeit S, Poustka A, King TR, Lehrach H. Cloning of the T gene required in mesoderm formation in the mouse. Nature. 1990;343:617–622. doi: 10.1038/343617a0. [DOI] [PubMed] [Google Scholar]
- 83.Dorey K, Amaya E. Development. Vol. 137. Cambridge, England: Oxford University Press for The Company of Biologists Limited; 2010. FGF signalling: diverse roles during early vertebrate embryogenesis; pp. 3731–3742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Amiel AR, Johnston HT, Nedoncelle K, Warner JF, Ferreira S, Röttinger E. Characterization of Morphological and Cellular Events Underlying Oral Regeneration in the Sea Anemone, Nematostella vectensis. Int J Mol Sci. 2015;16:28449–28471. doi: 10.3390/ijms161226100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Marlow H, Matus DQ, Martindale MQ. Ectopic activation of the canonical wnt signaling pathway affects ectodermal patterning along the primary axis during larval development in the anthozoan Nematostella vectensis. Dev Biol. 2013;380:324–334. doi: 10.1016/j.ydbio.2013.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Longabaugh W, Bolouri H. Understanding the dynamic behavior of genetic regulatory networks by functional decomposition. 2006;7:333–341. doi: 10.2174/138920206778948718. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


