Abstract
The Solfatara volcano near Naples (Italy), the origin of the recently discovered verrucomicrobial methanotroph Methylacidiphilum fumariolicum SolV was shown to contain ammonium () at concentrations ranging from 1 to 28 mM. Ammonia (NH3) can be converted to toxic hydroxylamine (NH2OH) by the particulate methane monooxygenase (pMMO), the first enzyme of the methane (CH4) oxidation pathway. Methanotrophs rapidly detoxify the intermediate NH2OH. Here, we show that strain SolV performs ammonium oxidation to nitrite at a rate of 48.2 nmol .h−1.mg DW−1 under O2 limitation in a continuous culture grown on hydrogen (H2) as an electron donor. In addition, strain SolV carries out nitrite reduction at a rate of 74.4 nmol .h−1.mg DW−1 under anoxic condition at pH 5–6. This range of pH was selected to minimize the chemical conversion of nitrite () potentially occurring at more acidic pH values. Furthermore, at pH 6, we showed that the affinity constants (Ks) of the cells for NH3 vary from 5 to 270 μM in the batch incubations with 0.5–8% (v/v) CH4, respectively. Detailed kinetic analysis showed competitive substrate inhibition between CH4 and NH3. Using transcriptome analysis, we showed up-regulation of the gene encoding hydroxylamine dehydrogenase (haoA) cells grown on H2/ compared to the cells grown on CH4/ which do not have to cope with reactive N-compounds. The denitrifying genes nirk and norC showed high expression in H2/ and CH4/ grown cells compared to cells growing at μmax (with no limitation) while the norB gene showed downregulation in CH4/ grown cells. These cells showed a strong upregulation of the genes in nitrate/nitrite assimilation. Our results demonstrate that strain SolV can perform ammonium oxidation producing nitrite. At high concentrations of ammonium this may results in toxic effects. However, at low oxygen concentrations strain SolV is able to reduce nitrite to N2O to cope with this toxicity.
Keywords: Methylacidiphilum, methanotroph, ammonia, methane, nitrite, reactive N compounds
Introduction
Methane (CH4) is a powerful greenhouse gas, which is released in to the atmosphere both from natural and anthropogenic sources (Conrad, 2009). Understanding sources and sinks of CH4 is important for future models of climate change on our planet. Methane oxidizing microorganisms are one of the most important biological sinks of CH4 (Murrell and Jetten, 2009).
Aerobic methanotrophic bacteria belong to a physiological group of bacteria recognized as methylotrophs. The proteobacterial methanotrophs are distinctive in their ability to exploit CH4 as the only carbon and energy source (Hanson and Hanson, 1996). Recently, three independent research groups discovered extreme acidophilic methanotrophic Verrucomicrobia in geothermal regions (Dunfield et al., 2007; Pol et al., 2007; Islam et al., 2008). Prior to this finding, obligate aerobic methanotrophs were speculated to be exclusively represented in the Alpha and Gamma subclasses of the Proteobacteria. Analysis of the 16S ribosomal RNA and pmoA genes demonstrated that the new Verrucomicrobia species do not form a monophyletic group with this subclasses (Heyer et al., 2005), and the new genus name Methylacidiphilum was suggested (Op den Camp et al., 2009). Furthermore, it has been shown that growth of the new acidophilic methanotrophic bacterium Methylacidiphilum fumariolicum SolV is strictly dependent on the presence of lanthanides acting as a cofactor of the methanol dehydrogenase (Keltjens et al., 2014; Pol et al., 2014). Recently, new species of mesophilic acidophilic verrucomicrobial methanotrophs were isolated and characterized from a volcanic region in Italy and the new genus Methylacidimicrobium was proposed (Sharp et al., 2014; van Teeseling et al., 2014). This finding expands the diversity of verrucomicrobial methanotrophs and demonstrates that they could be present in more ecosystems than formerly supposed (Chistoserdova et al., 2009). The new verrucomicrobial strains from both genera were shown to be autotrophs that use CH4 as the sole energy source and fix CO2 using the Calvin-Benson-Bassham Cycle (Khadem et al., 2011; Sharp et al., 2012, 2013, 2014; van Teeseling et al., 2014), and strain SolV was shown to be able to fix N2 (Khadem et al., 2010).
Methanotrophic and nitrifying microorganisms share many similarities. They grow obligately on the specific substrates, CH4 for methanotrophs and NH3 for nitrifiers. These molecules are structurally comparable and both are highly reduced. Many of these types of microorganisms have intracellular membrane structures where the membrane bound ammonia monooxygenase (AMO) or CH4 monooxygenase (pMMO) are localized. In the first step of aerobic CH4 or NH3 oxidation, the monooxygenase enzymes introduce a single oxygen atom from O2 into CH4 or NH3, producing methanol from CH4 and hydroxylamine from NH3 (Stein et al., 2012). Both microorganisms are able to co-oxidize a range of other substrates and are inhibited by similar compounds (Bédard and Knowles, 1989; Stein et al., 2012). Nitrifiers are able to oxidize CH4, and methanotrophs are capable of nitrification. It has been shown that in nutrient limited situations, methanotrophs do participate in soil nitrification, mainly in the production of N2O. Nitrification by aerobic methanotrophs relies on CH4, because they cannot grow on NH3 (Stein et al., 2012). Recent studies of CH4 oxidation and N2O production in soils using stable isotopes and particular inhibitors offered more evidence for a role of methanotrophic bacteria in nitrification (Mandernack et al., 2000; Lee et al., 2009; Acton and Baggs, 2011; Im et al., 2011).
is a nitrogen source for methanotrophic bacteria but was also shown to inhibit CH4 oxidation in the model organism Methylosinus sporium, especially due to accumulation of (He et al., 2017). The pMMO enzyme catalyzing the first step of CH4 oxidation in methanotrophs, also oxidizes NH3 () to hydroxylamine (NH2OH; Hanson and Hanson, 1996; Nyerges and Stein, 2009; Stein and Klotz, 2011; Stein et al., 2012). Ammonia-oxidizers can convey electrons from hydroxylamine oxidation to the quinone pool to conserve energy and support cellular growth (Klotz and Stein, 2008), but methanotrophs lack this system and cannot conserve energy from this oxidation. Since the intermediate NH2OH is highly toxic, methanotrophs use mechanisms to quickly detoxify it. In the natural environment strain SolV cells are faced with 1–28 mM concentrations (Khadem et al., 2010) meaning that the cells have to balance assimilation and tolerance in response to reactive-N molecules. Detoxification can be achieved by conversion of NH2OH back to or to using a hydroxylamine dehydrogenase enzyme. Nitrite, which is also toxic, can be further converted to nitrous oxide (N2O) via toxic nitric oxide (NO) by denitrification enzymes under anoxic conditions (Campbell et al., 2011). Recently, Kits et al. (2015) reported the reduction of nitrate coupled with aerobic methane oxidation under extreme oxygen limited conditions in which N2O production was directly supported by CH4 oxidation in Methylomonas denitrificans strain FJG1T.
In the genome of strain SolV, genes encoding enzymes responsible for reduction (nirK) and NO reduction (norB encoding the catalytic subunit, norC encoding the electron-accepting subunit), were identified but the gene encoding N2O reductase was absent. A haoAB gene cluster encoding hydroxylamine dehydrogenase was also identified, suggesting the ability of nitrification and handling of reactive N-compounds (Khadem et al., 2012c; Anvar et al., 2014). Previously a pH of 2–3 has been used for physiological studies of strain SolV (Khadem et al., 2010, 2011, 2012a,b,c). However, since strain SolV has a rather broad pH range for growth (Pol et al., 2007) and can be easily adapted to grow at higher pH values, we used the pH range of 5–6 in the present study. This minimized the chemical conversion of occurring at acidic pH (Matthew et al., 2005; Ryabenko et al., 2009).
Recently, using growth experiments (batch and continuous cultures) together with transcriptome and kinetics analyses, M. fumariolicum SolV was shown to be able to grow as a real “Knallgas” bacterium on hydrogen/carbon dioxide, without addition of CH4 (Mohammadi et al., 2017). Cells grown on H2 still express active pMMO similar to the CH4 culture (Mohammadi et al., 2017). Since we hypothesized that the oxidation is limited by the presence of CH4, we tested oxidation to using a continuous culture grown on hydrogen in the absence of CH4 (Mohammadi et al., 2017). Furthermore, we examined the affinity of cells for using batch cultures with different concentrations of CH4 in a range of 0.5–8% (v/v). The aim of this study was first to investigate whether strain SolV can perform oxidation, and secondly, how it could detoxify the reactive N-compounds resulting from this oxidation using physiological experiments and transcriptome analysis.
Materials and methods
Microorganism and medium composition
M. fumariolicum strain SolV used in this study was initially isolated from the volcanic region Campi Flegrei, near Naples, Italy (Pol et al., 2007). In this study the medium to obtain an OD600 of 1.0 was composed of 0.2 mM MgCl2.6H2O; 0.2 mM CaCl2.2H2O; 1 mM Na2SO4; 2 mM K2SO4; 2 mM (NH4)2SO4 (or 5 mM KNO3) and 1 mM NaH2PO4.H2O. A trace element solution containing 1 μM NiCl2, CoCl2, MoO4Na2, ZnSO4 and CeCl3; 5 μM MnCl2 and FeSO4; 10 μM CuSO4 and 40–50 μM nitrilotriacetic acid (NTA). The pH of medium was adjusted to 2.7 using 1 M H2SO4 (1 ml H2SO4 per 1 L medium). To avoid precipitation, CaCl2.2H2O and the rest of medium were autoclaved separately and mixed after cooling. This medium composition was used in batch and continuous cultures, unless otherwise stated.
Chemostat cultivation
The continuous culture with CH4 as an electron donor and nitrate () as N-source (CH4/), liquid volume 500 ml, was operated at 55°C with stirring at 900 rpm with a stirrer bar. The chemostat was supplied with medium at a flow rate of 14.5 ml.h−1 (D = 0.026 h−1), using a peristaltic pump. The cell-containing medium was removed automatically from the chemostat by a peristaltic pump when the liquid level reached the 500 ml level sensor in the reactor. A supply of 10% CH4 (v/v), 8% O2 (v/v), and 68% CO2 (v/v) took place by mass flow controllers through a sterile filter and was sparged into the medium just above the stirrer bar (total gas flow rate ≈20 ml.min−1). The initial pH was 3.4 and was regulated with 1 M carbonate connected to the vessel by a peristaltic pump. The pH was gradually increased to 6 and after obtaining a steady state, all experiments were performed at this pH. In the continuous culture with H2 as an electron donor and as N-source (H2/), liquid volume was 1.2 L and this culture was operated at 55°C with stirring at 1,000 rpm. The chemostat was supplied with medium at a flow rate of 29.9 ml.h−1 (D = 0.023 h−1). A gas supply of 12% H2 (v/v), 10% air (v/v), and 5% CO2 (v/v) was provided by mass flow controllers through a sterile filter and sparged into the medium (total gas flow rate ≈16.5 ml.min−1). The initial pH was 2.9 and the pH was regulated by 1 M NaOH. A pH range from 3 to 5.5 was investigated in the steady state. In the continuous culture with CH4 as an electron donor and as N-source (CH4/), the liquid volume was 0.3 L and the culture was operated at 55°C with stirring at 700 rpm at pH 2.7. The chemostat was supplied with medium at a flow rate of 0.35 ml.h−1 (D = 0.0012 h−1). A gas supply of 0.16% CH4 (v/v), 0.6% O2 (v/v), and 5% CO2 (v/v) was directed by mass flow controllers through a sterile filter and sparged into the medium (total gas flow rate ≈10 ml.min−1). An O2 sensor in the liquid was coupled to a Biocontroller (Applikon) regulating the O2 mass controller in each reactor.
Batch cultivation
In order to obtain cells growing at maximum growth rate (μmax), cells were grown without any limitation in 250-ml serum bottles containing 40 ml medium (4 mM ; pH 2.7), and sealed with red butyl rubber stoppers. The headspace contained air with (v/v) 10% CH4, 5% CO2 at 55°C with shaking at 250 rpm. Incubations were performed in duplicate.
Gas analysis
Nitric oxide and nitrous oxide (NO and N2O) were analyzed on an Agilent series 6890 gas chromatograph (Agilent, USA) equipped with a Porapak Q and a Molecular sieve column, coupled to a thermal conductivity detector and a mass spectrometer (MS; Agilent 5975 Cinert MSD; Agilent, USA) as described before (Ettwig et al., 2008). For all gas analyses, 100 μl gas samples were injected into the gas chromatograph. Furthermore, nitric oxide production was monitored directly from the gas outlet of the reactors using a nitric oxide analyzer (NOA 280i, GE) with a suction rate of 11.6 ml.min−1.
Dry-weight determination and elemental analysis
To determine the dry weight, samples of 8–10 ml from the culture suspension were filtered through pre-weighed 0.45 μm filters and dried to constant weight in a vacuum oven at 70°C (n = 3). In order to determine the total content of carbon and nitrogen, 10 ml of the culture suspension (duplicate) was centrifuged at 4,500 g for 30 min and the clear supernatant was used for the analysis. The nitrogen and carbon content in the supernatant was compared with the corresponding values in the whole cell suspension. The total carbon and nitrogen contents were measured using TOC-L and TNM-1 analyzers (Shimadzu).
Nitrite, ammonium, and hydroxylamine analysis
To determine nitrite () concentrations, 50 μl of sample, and 450 μl of MilliQ water were added to a cuvette. Then, 500 μl of reagent A [1% (w/v) sulfanilic acid in 1M HCl; kept in the dark] and 500 μl of reagent B [0.1% (w/v) naphtylethylene diaminedihydrochloride (NED) in water; kept at 4°C in the dark] were added to the same cuvette and mixed well. After incubation for 10 min at room temperature, the absorbance at 540 nm was measured and the values were compared with a calibration curve using known concentrations of nitrite in a range of 0–0.5 mM. If necessary, the sensitivity of this assay could be increased 10-fold using 500 μl samples without addition of water. concentrations were measured using the ortho phthaldialdehyde (OPA) method (Taylor et al., 1974). In order to determine hydroxylamine concentrations, 200 μl reagent A (50 mM potassium phosphate buffer pH 7), 160 μl demineralized water, 200 μl sample, 40 μl reagent B [12% (w/v) trichloroacetic acid in water, kept in the dark], 200 μl reagent C (1% w/v 8-hydroxyquinoline (quinolinol) in 100% ethanol, kept in the dark) and 200 μl reagent D (1 M Na2CO3) were mixed and incubated at 100°C for 1 min. The absorption was measured at 705 nm and the values were compared to a calibration curve using hydroxylamine concentrations 0.02–0.1 mM.
Activity assays
To determine the affinity constant of pMMO for NH3 of each sample, a volume of 5 ml of cells from the CH4/ continuous culture were washed and resuspended in the same medium at pH 6 (The pH of the medium was adjusted to 6 using MES buffer at a final concentration of 25 mM), transferred to a 60-ml serum bottle and capped. After a pre-incubation for 30 min, CH4 was added to each bottle at final concentrations of 0.5, 1, 2, 3, 4, and 8% (v/v). To each incubation, with a certain concentration of CH4, was added in a range of 0.5–16 mM. The initial production of was measured, and the values were normalized to the total protein content of the cells. Incubations were performed at 55°C and shaking at 380 rpm. Each condition was performed in duplicate and values did not deviate more than 5%.
RNA isolation and transcriptome analysis
The complete genome sequence of strain SolV (Anvar et al., 2014), which is also available at the MicroScope annotation platform (https://www.genoscope.cns.fr/agc/microscope/home/), was used as the template for the transcriptome analysis (RNA-seq). A 4-ml volume of cells (OD600 = 1) was sampled from the continuous cultures (H2 and CH4 grown cells under O2 limitation) and from a batch culture (cells at μmax grown on CH4 without limitation) and harvested by centrifugation. The pellet was further used for mRNA isolation using the RiboPure™-Bacteria Kit according to the manufacturer's protocol (ThermoFisher, Waltham, USA). Briefly, cells were disrupted by cold Zirconia beads and after centrifugation, 0.2 volumes of chloroform was added to the supernatant for initial RNA purification. Next, 0.5 volumes of 100% ethanol was added to the aqueous phase obtained after chloroform addition and the whole sample was transferred to a filter cartridge. After washing, the RNA was eluted from the filter cartridge. Afterwards, using MICROBexpress™ kit (ThermoFisher, Waltham, USA) the ribosomal RNAs were removed from the total RNA. The rRNA removal efficiency was checked using the Agilent 2100 Bioanalyzer (Agilent, Santa Clara, USA). Next, Ion Total RNA-Seq Kit v2 (ThermoFisher, Waltham, USA) was used to construct the cDNA libraries from rRNA-depleted total RNA. Briefly, the rRNA-depleted total RNA was fragmented using RNase III and then, reverse transcription was performed on the fragmented RNAs. The obtained cDNAs were amplified and further purified to prepare barcoded libraries. To prepare the template for the Ion Personal Genome Machine® (PGM™) System, a volume of 15 μl from two sample libraries with a concentration of 14 pM were mixed. This mixture of two libraries was used to prepare the template-positive Ion Sphere™ particles (ISPs) using the Ion OneTouch™ 2 instrument. Afterwards, the template-positive ISPs were enriched using the Ion OneTouch™ ES instrument. Both template preparation and enrichment were performed using the Ion PGM™ Template OT2 200 Kit (Ion Torrent, Life technologies). Enriched templates were sequenced on an Ion 318™ Chip v2 using the Ion PGM™ sequencing 200 Kit v2. Expression analysis was performed with the RNA-seq Analysis tool from the CLC Genomic Work bench software (version 7.0.4, CLC-Bio, Aarhus, Denmark). The sequencing reads were first mapped to the ribosomal RNA operon and all tRNA and ncRNA genes, and mapped reads were discarded. The remaining reads were mapped to the CDS sequences extracted from the genome sequence of strain SolV (Anvar et al., 2014). Expression values are given as RPKM (Reads per Kilo base of exon model per Million mapped reads; Mortazavi et al., 2008). The total number of reads obtained and mapped on the coding sequences of the genome for each sample together with the calculated expression levels (RPKM) is provided in the Supplementary Material (Table S1).
Results
Physiological tests regarding ammonium oxidation to nitrite and nitrite reduction to N2O
To study the effect and conversion of nitrogenous compounds, three different continuous cultures were used which are referred to as CH4/, H2/, and CH4/. In the second and third cultures, oxygen was limiting. Using a NOx analyzer and GC-MS, we demonstrated that in the CH4/ culture with low actual CH4 concentrations in the liquid (0.3 μM) and with (4 mM), was not detected, and N2O production rate was only 0.015 nmol N2O.h−1.mg DW−1 (Table 1) which was 12,000-fold less than the CH4 conversion rate (180 nmol.h−1.mg DW−1). To increase concentrations and study potential toxic effects of this compound, we used the H2/ continuous culture applying different conditions. Initially, the production of , NO, and N2O were measured under steady state conditions at a pH range of 3–5.5 under O2 limitation (Figure 1). We showed that the , NO and N2O concentrations were elevated by increasing the pH from 3 to 5.5 in the presence of 4 mM . Changing pH from 3 to 5.5 introduces more NH3 in the medium. The NH3 concentration in a range of 12 nM to 5 μM was calculated using the Henderson–Hasselbalch equation (Hütter, 1992), considering the temperature of 55°C at pH 3 to 5.5, respectively. At pH 5.5, we measured a concentration at steady state of about 420 μM in the reactor (Figure 2) resulting from a production rate of ≈ 48 nmol .h−1.mg DW−1, while nitrite production at pH 3 was very limited. Based on the clear effect of increasing pH on the production of , one could speculate that the real substrate for pMMO to produce is NH3 (not ). Furthermore, the reduction activities (NO and N2O production) were measured at 0.81 nmol .h−1.mg DW−1 (1.7% of oxidation rate) which is 53-fold higher than that in the CH4/ culture (Table 1). A rapid consumption (≈83 nmol .h−1.mg DW−1) was observed when O2 supply was switched off completely (Figure 2), and the reduction rate (as NO and N2O) increased about 100-fold (74.4 nmol .h−1.mg DW−1). A rapid initial increase of NO suggests that conversion to N2O is the rate limiting step. The decrease of N2O levels was due to the continuous dilution of the gas present in the reactor headspace (total gas flow rate in the outlet ≈ 15 ml.min−1). Concentrations of 1–5 μM NH2OH were measured in data points before and after switching off O2 supply.
Table 1.
Overview of oxidation and reduction rates calculated in each continuous culture at two different pH values.
| Continuous cultures | ||||
|---|---|---|---|---|
| CH4/ | H2/ | |||
| pH 3 | pH 5.5 | pH 3 | pH 5.5 | |
| (NH3)a | 4 (0.02) | 4 (5) | 4 (0.02) | 4 (5) |
| oxidationb | BDLd | NDe | 0.12f | 48.2 |
| reductionb | ND | ND | BDL | 0.8 |
| reductionbc | 0.015 | ND | 0.011 | 74.4 |
and NH3 concentrations are in mM and μM, respectively.
production and N2O production values are in nmol.h−1.mg DW−1.
reduction rates under anoxic conditions.
BDL, below detection limit.
ND, not determined.
All values are the average of two replicates of the same continuous culture with <5% difference between duplicates.
Figure 1.
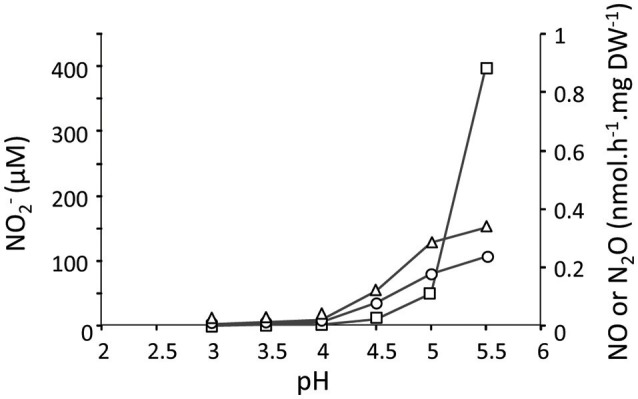
The concentration of (open rectangles) and production rates of NO (open triangles) and N2O (open circles) at pH values from 3 to 5.5 in the H2/ continuous culture (1.2 L; D = 0.023 h−1; OD600 = 0.85; O2 limited; 4 mM ). The amounts of , NO and N2O were determined when cells in the reactor reached the steady state. Each data point represents the average of two replicates with deviation of individual values below 5%.
Figure 2.
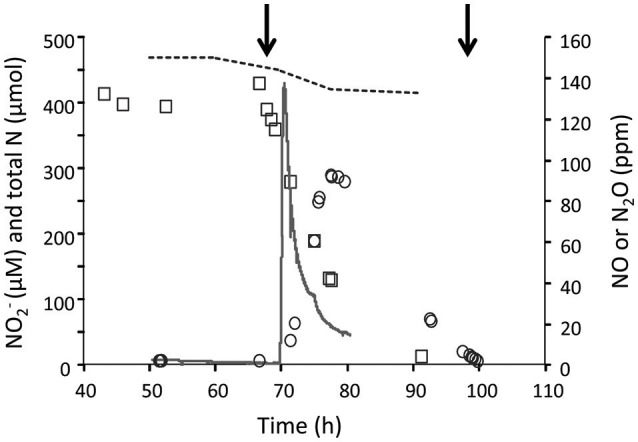
The concentrations of , NO and N2O in the H2/ continuous culture at pH 5.5 (1.2 L; D = 0.023 h−1; OD600 = 0.85; O2 limited; 4 mM ). Nitrite (open rectangles), NO (solid line), N2O (open circles) were determined under O2 limitation and anoxic conditions. The first arrow indicates the oxic to anoxic, the second arrow indicates the anoxic to oxic condition and the dashed line shows the total N during the experimental phase. The decrease of N2O levels was because of the continuous dilution of the gas present in the reactor headspace (total gas flow rate in the outlet ≈15 ml.min−1).
We further tested the effect of different concentrations of (4–20 mM) on the , NO and N2O production at pH 4 under oxygen limitation in the H2/ continuous culture (Figure 3). We showed that the concentrations of , NO, and N2O slightly increased once the concentration was gradually elevated. This observation indicates that at pH 4, even a 4-fold increase in the concentration did not result in a high production of similar to what we observed at pH 5.5 supporting our assumption that pH plays an important role regarding the availability of NH3 molecules. Furthermore, we showed that the cells in the CH4/ continuous culture were able to perform reduction at a rate of 120 nmol .h−1.mg DW−1 by converting the added (50 μM) to NO and further to N2O in the absence of oxygen (Figure 4). Table 1 shows an overview of rates of ammonium oxidation to nitrite and nitrite reduction to NO/N2O) in the different continuous culture.
Figure 3.
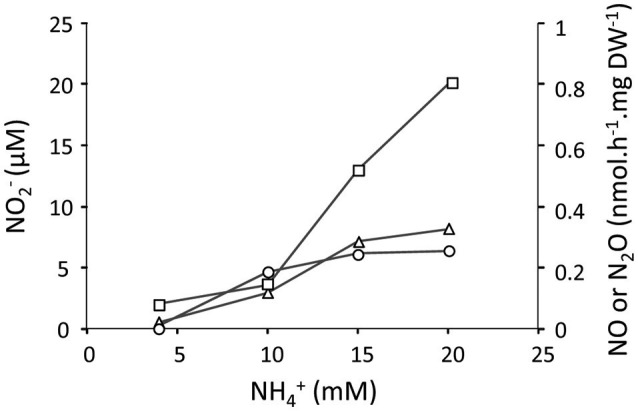
The concentration of (open rectangles) and production rates of NO (open triangles) and N2O (open circles) in the H2/ chemostat culture (1.2 L; D = 0.023 h−1; OD600 = 0.85; O2 limited) at concentrations ranging 4–20 mM at pH 4. The amounts of , NO, and N2O were determined when cells in the reactor reached the steady state. Each data point represents the average of two replicates with deviation of individual values below 5%.
Figure 4.
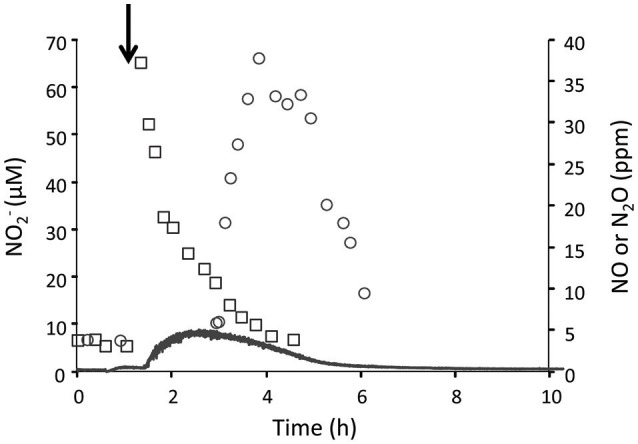
Cells from the CH4/ continuous culture (0.6 L; D = 0.026 h−1; OD600 = 1.3; O2 limited) perform denitrification when nitrite was added to the reactor vessel. The concentrations of (open rectangles), NO (solid line) and N2O (open circles) were measured before and after addition of 50 μM (arrow). Duplicates of individual values do not deviate more than 5 and 10% for and N2O, respectively.
Kinetics of ammonia oxidation
The affinity constants (Ks) for and NH3 were determined using SolV cells from the CH4/ continuous culture. From the initial production rates of nitrite the best fitting curves to Michaelis–Menten kinetics were predicted (Figure S1). Since part of the is present as NH3 at pH 6 (1 M is about 3 mM NH3 at pH 6), the Michaelis–Menten curves were also produced based on the NH3 concentrations (Figure S1). Therefore, we calculated apparent affinity constants (Ks) for both and NH3 in strain SolV (Table 2).
Table 2.
Kinetics of oxidation with variable CH4 supply at pH 6.
| CH4 | Affinity constantc [Ks(app)] | Vmaxd | |
|---|---|---|---|
| (mM) | NH3 (μM) | ||
| 0.5a(0.005)b | 1.25 | 4.9 | 1.61 |
| 1 (0.01) | 1.50 | 5.8 | 1.61 |
| 2 (0.02) | 6 | 23.3 | 1.43 |
| 3 (0.03) | 9 | 35.0 | 1.43 |
| 4 (0.04) | 30 | 116.7 | 1.43 |
| 8 (0.08) | 70 | 272.3 | 1.43 |
CH4 concentrations in % (v/v).
CH4 concentrations in the liquid in mM.
Affinity constants were calculated based on two independent experiments
Vmax values are in μmol .h−1.mg protein−1.
To identify the type of inhibition, the Michaelis–Menten curves were transformed to Lineweaver-Burk plots. Figure S2 shows a set of double reciprocal plots, obtained with different concentrations in the presence of CH4 at a range of 2, 4, and 8% (v/v). Increasing the CH4 concentration resulted in a group of lines with a common intercept on the 1/V0 axis but with different slopes. The intercept is 1/Vmax and Vmax is constant regardless of increasing CH4 concentration (Vmax = 1.61 ± 0.05 μmol.h−1.mg protein−1). The constant intercept of all lines suggests a competitive inhibition between CH4 and NH3. The production rate in the absence of CH4 was about 3- to 4-fold lower compared to the rate in the presence of 0.5% (v/v) CH4 suggesting that traces of CH4 are essential for the pMMO activation. Table 2 shows an overview of affinity constants calculated for NH3 () obtained in the incubations with different CH4 concentrations. Affinity constants for NH3 were calculated based on the Henderson–Hasselbalch equation considering a temperature of 55°C (Hütter, 1992). These results showed that increasing CH4 concentration limits the affinity of pMMO for NH3 significantly, which correlates with the observed competitive inhibition between CH4 and NH3.
Whole genome transcriptome analysis of strain SolV
Expression levels of housekeeping genes and genes involved in metabolism of nitrogenous compounds were determined for H2- and CH4-grown cells (both under O2 limited conditions). These values were compared to the expression values in cells growing at μmax on CH4 (without limitation). To compare baseline expression levels, we selected a group of 384 housekeeping genes (in total 437.9 kbp) involved in energy generation, ribosome assembly, carbon fixation (CBB cycle), C1 metabolism (except for pmo), amino acid synthesis, cell wall synthesis, translation, transcription, DNA replication, and tRNA synthesis (Khadem et al., 2012a,b). All ratios of expression levels of the housekeeping genes under these conditions were between 0.5 and 2 (Table S1). The robustness of the transcriptome data were tested using the method of Chaudhuri et al. (2011). In this method, the logarithmic value of RPKM + 1 of each condition (in duplicates) was calculated and the values were plotted against each other. This resulted in correlation coefficients of 0.80, 0.82, and 0.87 (Figure S3), showing the high robustness of the transcriptome data.
The transcriptome data showed that genes encoding the enzymes involved in assimilation in strain SolV including glutamine synthase (GlnA)/glutamate synthase (GltB) and the alanine and glutamate dehydrogenases (Ald, Gdh) were equally expressed under all conditions (Table 3). Among these genes, only glnA was about 2.5-fold less expressed in the continuous cultures compared to the cells grown at μmax (Table 3). We also found that the carAB operons (encoding the glutamine hydrolyzing carbamoyl-phosphate synthase) were constitutively expressed. The conversion of glutamine and carbon dioxide into glutamate and carbamoyl phosphate is performed by this enzyme (Khadem et al., 2012a). Similarly, the argDHFG operons (encoding enzymes from the urea cycle) were expressed under all conditions. Interestingly, we detected the ammonium/ammonia transporter (amtB) was at least 3-fold up-regulated in the CH4/ continuous culture compared to the other conditions reflecting that cells may have a preference for as N-source. In addition, the genes encoding the / transport (nasA) and the assimilatory nitrite and nitrate reductases were 9- to 45-fold up-regulated in the CH4/ continuous culture compared to the cells at μmax (Table 3). Both latter observations correlate with the fact that nitrate was used as N-source under this condition. Interestingly, the transcriptome analysis showed that the nirK and norC genes were up-regulated in the chemostat continuous culture compared to those at μmax, while results for norB (encoding the catalytic subunit) were less clear. This may imply that other NO reductases were active. We also found that the haoA gene was about 2-fold down-regulated in the CH4/ continuous culture compared to the H2/ and μmax cultures, likely due to the absence of in this condition. The haoA gene showed comparable high expression levels in the H2/ continuous and batch μmax culture (Table 3).
Table 3.
The transcriptome analysis of the genes involved in nitrogen metabolism in Methylacidiphilum fumariolicum SolV.
| Enzyme | Gene name | GenBank identifier | Expression level (RPKM)a | ||
|---|---|---|---|---|---|
| H2/ | CH4/ | Cells at μmaxb | |||
| Glutamine synthetase type I (EC 6.3.1.2) | glnA | Mfumv2_1420 | 764 | 893 | 2,065 |
| Glutamine synthetase regulatory protein PII | glnB | Mfumv2_1419 | 943 | 719 | 883 |
| [Protein-PII] uridylyltransferase (EC 2.7.7.59) | glnD | Mfumv2_1837 | 124 | 136 | 156 |
| Nitrogen regulatory protein PII | glnK | Mfumv2_1285 | 371 | 125 | 193 |
| Alanine dehydrogenase (EC 1.4.1.1) | ald | Mfumv2_2049 | 107 | 106 | 171 |
| Glutamate dehydrogenase (EC 1.4.1.2; EC 1.4.1.4) | gdhA | Mfumv2_0663 | 227 | 231 | 421 |
| Glutamate synthase [NADPH] large chain (EC 1.4.1.13) | gltB | Mfumv2_2397 | 906 | 696 | 1,300 |
| Glutamate synthase beta chain | gltD | Mfumv2_1978 | 192 | 328 | 198 |
| Ornithine-acetylornithine aminotransferase (EC 2.6.1.11) | argD1 | Mfumv2_1148 | 279 | 271 | 627 |
| Ornithine-acetylornithine aminotransferase (EC 2.6.1.11) | argD2 | Mfumv2_0135 | 145 | 273 | 357 |
| Argininosuccinate lyase (EC 4.3.2.1) | argH | Mfumv2_2465 | 78 | 68 | 203 |
| Ornithine carbamoyltransferase (EC 2.1.3.3) | argF | Mfumv2_0136 | 161 | 239 | 278 |
| Argininosuccinate synthase (EC 6.3.4.5) | argG | Mfumv2_1907 | 666 | 654 | 645 |
| Carbamoyl-phosphate synthase small chain (EC 6.3.5.5) | carA | Mfumv2_1926 | 318 | 350 | 453 |
| Carbamoyl-phosphate synthase large chain (EC 6.3.5.5) | carB | Mfumv2_0408 | 347 | 674 | 514 |
| Ammonium-Ammonia transporter | amtB | Mfumv2_1275 | 294 | 1,082 | 391 |
| Nitrate ABC transporter, nitrate-binding protein | tauA | Mfumv2_1299 | 28 | 41 | 34 |
| Assimilatory nitrate reductase catalytic subunit (EC 1.7.99.4) | nasC | Mfumv2_1297 | 20 | 105 | 13 |
| Nitrate-nitrite transporter | nasA | Mfumv2_1294 | 67 | 321 | 23 |
| Nitrite reductase [NAD(P)H] large subunit (EC 1.7.1.4) | nirB | Mfumv2_1296 | 140 | 854 | 19 |
| Nitrite reductase [NAD(P)H], small subunit (EC 1.7.1.4) | nirD | Mfumv2_1295 | 63 | 308 | 33 |
| Signal transduction histidine kinase with PAS domain | ntrB | Mfumv2_0271 | 275 | 180 | 291 |
| Signal transduction response regulator, NtrC family | ntrC1 | Mfumv2_1349 | 98 | 84 | 103 |
| Sigma-54 dependent transcriptional regulator-response regulator | ntrC2 | Mfumv2_1221 | 65 | 59 | 100 |
| Transcriptional regulator, NifA subfamily, Fis Family | ntrC3 | Mfumv2_2103 | 581 | 400 | 533 |
| Sigma-54 dependent transcriptional regulator-response regulator | ntrC4 | Mfumv2_0272 | 264 | 387 | 293 |
| Hydroxylamine dehydrogenase (EC 1.7.2.6) | haoA | Mfumv2_2472 | 402 | 109 | 351 |
| Hydroxylamine dehydrogenase associated protein | haoB | Mfumv2_2471 | 179 | 163 | 302 |
| Nitric-oxide reductase subunit B (EC 1.7.99.7) | norB | Mfumv2_0037 | 125 | 84 | 178 |
| Nitric-oxide reductase subunit C (EC 1.7.99.7) | norC | Mfumv2_0036 | 429 | 372 | 197 |
| Copper-containing nitrite reductase (EC 1.7.2.1) | nirK | Mfumv2_1973 | 379 | 520 | 136 |
| DNA-binding response regulator, NarL family | mxaB | Mfumv2_1738 | 163 | 291 | 288 |
| DNA-binding response regulator, LuxR family | citB1 | Mfumv2_1799 | 7,016 | 4,126 | 1,063 |
| DNA-binding response regulator, LuxR family | citB2 | Mfumv2_0457 | 137 | 133 | 307 |
The mRNA expression is shown as RPKM according to Mortazavi et al. (2008). Changes in expression in the continuous cultures (H2/ and CH4/) compared to batch culture cells growing at μmax are demonstrated by shading [up-regulation >2-fold dark gray; down-regulation <0.5 (light gray)].
Cells grown on CH4 with as N-source.
The transcriptome data showed different expression levels of two of the three different pmo operons in strain SolV (Table 4). We found that the pmoCAB2 operon including the mfumv2_1793, mfumv2_1792 and mfumv2_1791 subunits was significantly expressed (RPKM values 14,899–37,218) in the cells growing at μmax with no limitation and the pmoCAB1 operon showed very low expression. In contrast, cells in the continuous cultures on H2/ and CH4/ under O2 limitation showed a significantly different expression pattern of the pmoCAB operons. We found that the pmoCAB1 operon including mfumv2_1796, mfumv2_1795 and mfumv2_1794 subunits was very highly expressed under these conditions (RPKM values 5,003–47,785), whereas the expression levels of the pmoCAB2 operon was found to be 2- to 19-fold lower in comparison to the cells growing at μmax. The pmoCAB3 operon including the mfumv2_1606, mfumv2_1605 and mfumv2_1604 subunits showed low expressed under all conditions although expression in H2/ grown cells seems to be slightly up-regulated. The conversion of methanol to formaldehyde is the second step in CH4 oxidation pathway. Interestingly, it has been shown that strain SolV contains a XoxF-type methanol dehydrogenase (MDH) that can convert methanol directly to formate (Pol et al., 2014). We found that the xoxFGJ operon encoding the methanol dehydrogenase and pqqABCDEF operon encoding the proteins involved in biosynthesis of the methanol dehydrogenase cofactor pyrroloquinoline quinone were expressed more or less similar under all conditions tested. The last step of the CH4 oxidation pathway is conversion of formate to CO2 catalyzed by NAD-dependent formate dehydrogenase and a membrane-bound formate dehydrogenase. The genes encoding these enzymes were expressed under all conditions, although the expression levels of these enzymes (except for fdsD and fdh) in continuous cultures under O2 limitation was 2- to 2.5-fold lower compared to cells grown at μmax (Table 4).
Table 4.
The transcriptome analysis of the genes involved in the methane oxidation pathway of Methylacidiphilum fumariolicum SolV.
| Enzyme | Gene name | GenBank identifier | Expression level (RPKM)a | ||
|---|---|---|---|---|---|
| H2/ | CH4/ | Cells at μmaxb | |||
| Particulate CH4 monooxygenase_1 (EC 1.14.13.25) | pmoC1 | Mfumv2_1796 | 47,785 | 34,734 | 207 |
| pmoA1 | Mfumv2_1795 | 9,772 | 3,775 | 41 | |
| pmoB1 | Mfumv2_1794 | 9,550 | 5,003 | 164 | |
| Particulate CH4 monooxygenase_2 (EC 1.14.13.25) | pmoC2 | Mfumv2_1793 | 18,136 | 5,462 | 37,218 |
| pmoA2 | Mfumv2_1792 | 2,383 | 1,119 | 21,207 | |
| pmoB2 | Mfumv2_1791 | 2,139 | 1,265 | 14,899 | |
| Particulate CH4 monooxygenase_3 (EC 1.14.13.25) | pmoC3 | Mfumv2_1606 | 539 | 209 | 181 |
| pmoA3 | Mfumv2_1605 | 143 | 17 | 57 | |
| pmoB3 | Mfumv2_1604 | 58 | 13 | 28 | |
| Methanol dehydrogenase XoxF-type (EC 1.1.99.8) | xoxF | Mfumv2_1183 | 6,220 | 5,291 | 6,041 |
| Extracellular solute-binding protein family 3 | xoxJ | Mfumv2_1184 | 714 | 1,057 | 1,478 |
| Cytochrome c1 protein fused with XoxJ | xoxGJ | Mfumv2_1185 | 611 | 829 | 1,042 |
| Coenzyme PQQ precursor peptide | ppqA | Mfumv2_1461a | 2,920 | 1,919 | 2,133 |
| Coenzyme PQQ synthesis proteins | pqqB | Mfumv2_1461 | 1,308 | 588 | 620 |
| pqqC | Mfumv2_1462 | 1,165 | 560 | 622 | |
| pqqD | Mfumv2_0766 | 144 | 242 | 60 | |
| pqqD | Mfumv2_1463 | 451 | 153 | 249 | |
| pqqE | Mfumv2_1464 | 747 | 514 | 509 | |
| pqqF | Mfumv2_0519 | 408 | 680 | 718 | |
| NADPH:quinone oxidoreductase (EC 1.6.5.5) | qor1 | Mfumv2_1937 | 253 | 287 | 315 |
| qor2 | Mfumv2_2088 | 338 | 300 | 432 | |
| qor3 | Mfumv2_0618 | 60 | 12 | 130 | |
| Zn-dependent alcohol dehydrogenase (EC 1.1.1.1) | adh1 | Mfumv2_2176 | 160 | 154 | 208 |
| adh2 | Mfumv2_0724 | 252 | 218 | 288 | |
| Aldehyde dehydrogenase (EC 1.2.1.3) | dhaS1 | Mfumv2_2408 | 130 | 317 | 108 |
| dhaS2 | Mfumv2_0597 | 1,310 | 1,503 | 1,125 | |
| Dihydropteroate synthase (EC 2.5.1.15) | folP1 | Mfumv2_0503 | 161 | 233 | 167 |
| folP2 | Mfumv2_2400 | 126 | 95 | 208 | |
| Formate–tetrahydrofolate ligase (EC 6.3.4.3) | fhs | Mfumv2_2082 | 396 | 457 | 282 |
| Methylenetetrahydrofolate dehydrogenase (NADP+) (EC 1.5.1.5) - methenyltetrahydrofolate cyclohydrolase (EC 3.5.4.9) | folD | Mfumv2_1033 | 257 | 173 | 261 |
| GTP cyclohydrolase I (EC 3.5.4.16) type 2 | folE | Mfumv2_0074 | 1,485 | 1,477 | 795 |
| NAD-dependent formate dehydrogenase alpha subunit | fdsA | Mfumv2_1457 | 568 | 665 | 1,342 |
| NAD-dependent formate dehydrogenase beta subunit | fdsB | Mfumv2_1458 | 569 | 435 | 1,149 |
| NAD-dependent formate dehydrogenase gamma subunit | fdsC | Mfumv2_1459 | 475 | 240 | 672 |
| NAD-dependent formate dehydrogenase delta subunit | fdsD | Mfumv2_1456 | 593 | 979 | 588 |
| NAD-dependent formate dehydrogenase (EC 1.2.1.2) | fdh | Mfumv2_1567 | 738 | 863 | 1,110 |
| Methylamine dehydrogenase light chain (EC 1.4.99.3) | mauA | Mfumv2_0350 | 119 | 450 | 108 |
| Methylamine dehydrogenase heavy chain (EC 1.4.99.3) | mauB | Mfumv2_0347 | 99 | 135 | 235 |
The mRNA expression is shown as RPKM according to Mortazavi et al. (2008). Changes in expression in the continuous cultures (H2/ and CH4/) compared to batch culture cells growing at μmax are demonstrated by shading [up-regulation >2-fold (dark gray), down-regulation <0.5 (light gray)].
Cells grown on CH4 with as N-source.
Discussion
In the present study, the physiological data of the H2/ continuous culture showed that strain SolV is able to oxidize to at a rate of 48.2 nmol .h−1.mg DW−1 at pH 5.5. At pH 3, with less NH3 available this rate was about 400-fold lower (Table 1). We also detected a very limited oxidation rate in the cells of the CH4/ chemostat in comparison to the H2/ cells. These observations indicate that the higher oxidation activity occurs when CH4 is replaced by H2 as the electron donor. Nitrification was previously reported in methanotrophs. CH4-dependent nitrification was detected in a humisol that was enriched with CH4 (Megraw and Knowles, 1987). It has been shown that methanotrophs are efficient nitrifiers and produce NH2OH as a product of NH3 monooxygenation (Bédard and Knowles, 1989; Nyerges and Stein, 2009).
We observed a similar pattern in the batch experiments using cells from the CH4/ continuous culture. In these batch tests, we found higher production rates when the CH4 concentration was limited, although traces of CH4 seemed to be essential for activation of pMMO. In these batch tests, the calculated apparent affinity constants [Ks(app)] for were approximately between 1.25 and 70 mM. At increasing pH values the equilibrium shifts toward higher NH3 concentrations and the calculated Ks values for NH3 in the same tests were 4–273 μM. Comparable values have been reported in literature (Table 5). Our data showed that increasing the pH from 3 to 5.5 significantly affects the rates of oxidation to . This reflects the fact that the pMMO of strain SolV might use NH3 as a substrate (and not ). This assumption could explain why at low pH, when is present, we observed very limited nitrification. In a study from O'Neill and Wilkinson (1977), they also showed that by increasing pH the rate of oxidation by M. trichosporium OB3B increased, and they also suggested the active species to be NH3.
Table 5.
Comparison of apparent Ks values for .
| Organism | Ks () mM | CH4 % (v/v) | pH | Calculated Ks (NH3) μM | References |
|---|---|---|---|---|---|
| M. fumariolicum | 1.25–70 | 0.5–8 | 6 | 4–273 | This studya |
| Mm. album | 2 and 3.9 | 0.5 and 5 | – | – | Nyerges and Stein, 2009 |
| Methylocystis sp. | 0.5 and 1.1 | 0.5 and 5 | – | – | Nyerges and Stein, 2009 |
| Ms. trichosporium | 4.1 | – | 6.5 | – | O'Neill and Wilkinson, 1977 |
| 0.6 | – | 7.5 | – | ||
| Mb. capsulatus | 87b | – | 7 | – | Dalton, 1977 |
See also Table 1.
At concentrations between 20 and 200 mM, −, not reported.
In the present study, we showed that strain SolV performs reduction to N2O usingcells from CH4/ and H2/ continuous cultures (Table 1). Under anoxic condition, higher reduction rates were observed in cells from the CH4/ and H2/ cultures (Table 1). The reduction of to N2O may provide a way to remove potentially toxic . The lower reduction rate in H2/ compared to the CH4/ continuous cultures in the absence of oxygen could be explained by the fact that cells in the H2 reactor were confronted with NH2OH and over a relatively long term. Cells might suffer under these conditions and show a decrease in reduction rate. Many methanotrophs possess partial denitrification pathways and they are able to reduce to N2O via NO (Nyerges et al., 2010; Campbell et al., 2011). Recently, two methanotrophic strains were cultured together (Methylomicrobium album ATCC 33003 and Methylocystis sp. strain ATCC 49242), one with high tolerance to and one with high tolerance to , and the nitrite-tolerant strain was shown to be more competitive and produced more N2O compared to the other strain (Nyerges et al., 2010). The highest N2O production rate was reported at about 0.4 nmol.h−1 per 106 cells in M. album ATCC 33003 (Nyerges et al., 2010). Campbell et al. (2011) reported a headspace production of 26.3 μM N2O after 48 h (≈0.24 ppb.h−1 per 106 cells) in Methylococcus capsulatus Bath. Recently, Kits et al. (2015) reported the reduction of nitrate coupled to aerobic CH4 oxidation under extreme oxygen limited conditions in which N2O production (0.414 μmol.h−1.L−1) was directly supported by CH4 oxidation in M. denitrificans strain FJG1T. The latter N2O production rate is about 60-fold lower compared to our results obtained under anoxic condition in the absence of CH4.
In this study, the transcriptome data showed that the pmoCAB1 and pmoCAB2 operons were tightly regulated by oxygen as observed previously (Khadem et al., 2012a). Recently, the down-regulation of pmoCAB gene was detected in response to 30 mM concentration in the medium compared to 10 mM in Methylocystis sp. strain SC2 (Dam et al., 2014). It has been shown that CH4 oxidation in Methylocystis sp. strain SC2 cells supplied with 30 mM was inhibited at CH4 concentrations <400 ppm (v/v; Dam et al., 2014). Our results in all cases showed no expression of the pmoCAB3 operon, suggesting other growth conditions could be examined to elucidate the regulation and role of this pmo operon. Recently, the concurrent growth of the methanotroph Methylocella silvestris was described on CH4 and propane (Crombie and Murrell, 2014). Two soluble di-iron center monooxygenase gene clusters (sMMO) were identified with different expression during bacterial growth on these alkanes, although both gene sets were essential for efficient propane utilization (Crombie and Murrell, 2014).
In our study, the haoAB genes encoding hydroxylamine dehydrogenase (HAO) and an associated protein were constitutively expressed in cells grown in the H2/ continuous and batch cultures (Table 3). In M. capsulatus Bath the haoAB genes were shown to respond to addition of 5 mM of (Poret-Peterson et al., 2008). The currently accepted model for oxidation of NH3 to proceeds via the intermediate NH2OH which in a follow up reaction catalyzed by HAO is oxidized to . Recently, evidence was provided that HAO oxidizes NH2OH by only three electrons to NO under both aerobic and anaerobic conditions using purified Nitrosomonas europaea HAO (Caranto and Lancaster, 2017). This also implies the need for an enzyme converting NO to . For future research we aim at purifying the HAO from strain SolV to test its properties.
The assimilatory nitrite and nitrate reductase genes were found 9- to 45-fold up-regulated in the CH4/ continuous culture compared to the cells at μmax. These observations are similar to the down-regulation of assimilatory nitrite and nitrate reductase genes in Methylocystis sp. strain SC2 under 30 mM compared to 10 mM nitrate or (Dam et al., 2014). It has been proposed that methanotrophs with denitrifying capacity might surpass other methanotrophs in ecosystems with high concentrations of nitrogen, because they have the ability to deal with reactive N-compounds (Nyerges et al., 2010). The reducing capacity of strain SolV helps this microorganism to balance assimilation and tolerance in response to reactive-N molecules in the extreme conditions of its habitat. Our experiments show that strain SolV is well adapted to cope with the fluctuating conditions (presence of H2, differences in and O2 concentrations and pH) that may occur in its natural environment.
Author contributions
SM, AP, MJ, and HO designed the project and experiments. Experimental work was performed by SM, TvA, and AP. SM and AP maintained the chemostat cultures. SM, TvA, AP, MJ, and HO performed data analysis and data interpretation. SM and HO wrote the manuscript with input from AP, TvA, and MJ. HO and MJ supervised the research.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
Funding. SM was supported by the Spinoza grant of MJ (Netherlands Organization for Scientific Research) and the European Research Council (ERC Advanced Grant project VOLCANO 669371), MJ by the European Research Council (ERC Advanced Grant Eco_MoM 339880) and HO by the European Research Council (ERC Advanced Grant project VOLCANO 669371).
Supplementary material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2017.01901/full#supplementary-material
References
- Acton S. D., Baggs E. M. (2011). Interactions between N application rate, CH4 oxidation and N2O production in soil. Biogeochemistry 103, 15–26. 10.1007/s10533-010-9442-5 [DOI] [Google Scholar]
- Anvar S. Y., Frank J., Pol A., Schmitz A., Kraaijeveld K., den Dunnen J. T., et al. (2014). The genomic landscape of the verrucomicrobial methanotroph Methylacidiphilum fumariolicum SolV. BMC Genomics 15:914. 10.1186/1471-2164-15-914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bédard C., Knowles R. (1989). Physiology, biochemistry, and specific inhibitors of CH4, and CO oxidation by methanotrophs and nitrifiers. Microbiol. Rev. 53, 68–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell M. A., Nyerges G., Kozlowski J. A., Poret-Peterson A. T., Stein L. Y., Klotz M. G. (2011). Model of the molecular basis for hydroxylamine oxidation and nitrous oxide production in methanotrophic bacteria. FEMS Microbiol. Lett. 322, 82–89. 10.1111/j.1574-6968.2011.02340.x [DOI] [PubMed] [Google Scholar]
- Caranto J. D., Lancaster K. M. (2017). Nitric oxide is an obligate bacterial nitrification intermediate produced by hydroxylamine oxidoreductase. Proc. Natl. Acad. Sci. U.S.A. 114, 8217–8222. 10.1073/pnas.1704504114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhuri R. R., Yu L., Kanji A., Perkins T. T., Gardner P. P., Choudhary J., et al. (2011). Quantitative RNA- seq analysis of the Campylobacter jejuni transcriptome. Microbiology 157, 2922–2932. 10.1099/mic.0.050278-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chistoserdova L., Kalyuzhnaya M. G., Lidstrom M. E. (2009). The expanding world of methylotrophic metabolism. Annu. Rev. Microbiol. 63, 477–499. 10.1146/annurev.micro.091208.073600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conrad R. (2009). The global methane cycle: recent advances in understanding the microbial processes involved. Environ. Microbiol. Rep. 1, 285–292. 10.1111/j.1758-2229.2009.00038.x [DOI] [PubMed] [Google Scholar]
- Crombie A. T., Murrell J. C. (2014). Trace-gas metabolic versatility of the facultative methanotroph Methylocella silvestris. Nature 510, 148–151. 10.1038/nature13192 [DOI] [PubMed] [Google Scholar]
- Dalton H. (1977). Ammonia oxidation by the methane oxidising bacterium Methylococcus capsulatus strain bath. Arch. Microbiol. 114, 273–279. 10.1007/BF00446873 [DOI] [PubMed] [Google Scholar]
- Dam B., Dam S., Kim Y., Liesack W. (2014). Ammonium induces differential expression of methane and nitrogen metabolism-related genes in Methylocystis sp. strain SC2. Environ. Microbiol. 16, 3115–3127. 10.1111/1462-2920.12367 [DOI] [PubMed] [Google Scholar]
- Dunfield P. F., Yuryev A., Senin P., Smirnova A. V., Stott M. B., Hou S., et al. (2007). Methane oxidation by an extremely acidophilic bacterium of the phylum Verrucomicrobia. Nature 450, 879–883. 10.1038/nature06411 [DOI] [PubMed] [Google Scholar]
- Ettwig K. F., Shima S., van de Pas-Schoonen K. T., Kahnt J., Medema M. H., Op den Camp H. J. M., et al. (2008). Denitrifying bacteria anaerobically oxidize methane in the absence of Archaea. Environ. Microbiol. 10, 3164–3173. 10.1111/j.1462-2920.2008.01724.x [DOI] [PubMed] [Google Scholar]
- Hanson R. S., Hanson T. E. (1996). Methanotrophic bacteria. Microbiol. Rev. 60, 439–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He R., Chen M., Ma R. C., Sum Y., Zhang X. (2017). Ammonium conversion and its feedback effect on methane oxidation of Methylosinus sporium. J. Biosci. Bioeng. 123, 466–473. 10.1016/j.jbiosc.2016.11.003 [DOI] [PubMed] [Google Scholar]
- Heyer J., Berger U., Hardt M., Dunfield P. F. (2005). Methylohalobius crimeensis gen. nov., sp. nov., a moderately halophilic, methanotrophic bacterium isolated from hypersaline lakes of Crimea. Int. J. Syst. Evol. Microbiol. 55, 1817–1826. 10.1099/ijs.0.63213-0 [DOI] [PubMed] [Google Scholar]
- Hütter L. A. (1992). Wasser und Wasseruntersuchung – Methodik, Theorie und Praxis chemischer, Chemisch-Physikalischer und Bakteriologischer Untersuchungsverfahren. Frankfurt: Salle. [Google Scholar]
- Im J., Lee S. W., Bodrossy L., Barcelona M. J., Semrau J. D. (2011). Field application of nitrogen and phenylacetylene to mitigate greenhouse gas emissions from landfill cover soils: effects on microbial community structure. Appl. Microbiol. Biotechnol. 89, 189–200. 10.1007/s00253-010-2811-0 [DOI] [PubMed] [Google Scholar]
- Islam T., Jensen S., Reigstad L. J., Larsen O., Birkeland N. K. (2008). Methane oxidation at 55 degrees C and pH 2 by a thermoacidophilic bacterium belonging to the Verrucomicrobia phylum. Proc. Natl. Acad. Sci. U.S.A. 105, 300–304. 10.1073/pnas.0704162105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keltjens J. T., Pol A., Reimann J., Op den Camp H. J. M. (2014). PQQ-dependent methanol dehydrogenases: rare-earth elements make a difference. Appl. Microbiol. Biotechnol. 98, 6163–6183. 10.1007/s00253-014-5766-8 [DOI] [PubMed] [Google Scholar]
- Khadem A. F., Pol A., Jetten M. S. M., Op den Camp H. J. M. (2010). Nitrogen fixation by the verrucomicrobial methanotroph “Methylacidiphilum fumariolicum” SolV. Microbiology 156, 1052–1059. 10.1099/mic.0.036061-0 [DOI] [PubMed] [Google Scholar]
- Khadem A. F., Pol A., Wieczorek A., Mohammadi S. S., Francoijs K. J., Stunnenberg H. G., et al. (2011). Autotrophic methanotrophy in verrucomicrobia: Methylacidiphilum fumariolicum SolV uses the Calvin-Benson-Bassham cycle for carbon dioxide fixation. J. Bacteriol. 193, 4438–4446. 10.1128/JB.00407-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khadem A. F., Pol A., Wieczorek A. S., Jetten M. S. M., Op den Camp H. J. M. (2012a). Metabolic regulation of “Ca. Methylacidiphilum fumariolicum” SolV cells grown under different nitrogen and oxygen limitations. Front. Microbiol. 3:266 10.3389/fmicb.2012.00266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khadem A. F., van Teeseling M. C., van Niftrik L., Jetten M. S. M., Op den Camp H. J. M. (2012b). Genomic and physiological analysis of carbon storage in the verrucomicrobial methanotroph “Ca. Methylacidiphilum fumariolicum” SolV. Front. Microbiol. 3:345 10.3389/fmicb.2012.00345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khadem A. F., Wieczorek A. S., Pol A., Vuilleumier S., Harhangi H. R., Dunfield P. F., et al. (2012c). Draft genome sequence of the volcano-inhabiting thermoacidophilic methanotroph Methylacidiphilum fumariolicum strain SolV. J. Bacteriol. 194, 3729–3730. 10.1128/JB.00501-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kits K. D., Klotz M. G., Stein L. Y. (2015). Methane oxidation coupled to nitrate reduction under hypoxia by the Gammaproteobacterium Methylomonas denitrificans, sp. nov. type strain FJG1. Environ. Microbiol. 17, 3219–3232. 10.1111/1462-2920.12772 [DOI] [PubMed] [Google Scholar]
- Klotz M. G., Stein L. Y. (2008). Nitrifier genomics and evolution of the nitrogen cycle. FEMS Microbiol. Lett. 278, 146–156. 10.1111/j.1574-6968.2007.00970.x [DOI] [PubMed] [Google Scholar]
- Lee S.-W., Im J., Dispirito A. A., Bodrossy L., Barcelona M. J., Semrau J. D. (2009). Effect of nutrient and selective inhibitor amendments on methane oxidation, nitrous oxide production, and key gene presence and expression in landfill cover soils: characterization of the role of methanotrophs, nitrifiers, and denitrifiers. Appl. Microbiol. Biotechnol. 85, 389–403. 10.1007/s00253-009-2238-7 [DOI] [PubMed] [Google Scholar]
- Mandernack K. W., Kinney C. A., Coleman D., Huang Y. S., Freeman K. H., Bogner J. (2000). The biogeochemical controls of N2O production and emission in landfill cover soils: the role of methanotrophs in the nitrogen cycle. Environ. Microbiol. 2, 298–309. 10.1046/j.1462-2920.2000.00106.x [DOI] [PubMed] [Google Scholar]
- Matthew R., McIlvin M. R., Altabet M. A. (2005). Chemical conversion of nitrate and nitrite to nitrous oxide for nitrogen and oxygen isotopic analysis in freshwater and seawater. Anal. Chem. 77, 5589–5595. 10.1021/ac050528s [DOI] [PubMed] [Google Scholar]
- Megraw S. R., Knowles R. (1987). Methane production and consumption in a cultivated humisol. Biol. Fertil. Soils 5, 56–60. 10.1007/BF00264347 [DOI] [Google Scholar]
- Mohammadi S., Pol A., van Alen T. A., Jetten M. S. M., Op den Camp H. J. M. (2017). Methylacidiphilum fumariolicum SolV, a thermoacidophilic ‘Knallgas’ methanotroph with both an oxygen-sensitive and -insensitive hydrogenase. ISME J. 11, 945–958. 10.1038/ismej.2016.171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortazavi A., Williams B. A., McCue K., Schaeffer L., Wold B. (2008). Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods 5, 621–628. 10.1038/nmeth.1226 [DOI] [PubMed] [Google Scholar]
- Murrell J. C., Jetten M. S. M. (2009). The microbial methane cycle. Environ. Microbiol. Rep. 1, 279–284. 10.1111/j.1758-2229.2009.00089.x [DOI] [PubMed] [Google Scholar]
- Nyerges G., Han S. K., Stein L. Y. (2010). Effects of ammonium and nitrite on growth and competitive fitness of cultivated methanotrophic bacteria. Appl. Env. Microbiol. 76, 5648–5651. 10.1128/AEM.00747-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nyerges G., Stein L. Y. (2009). Ammonia cometabolism and product inhibition vary considerably among species of methanotrophic bacteria. FEMS Microbiol. Lett. 297, 131–136. 10.1111/j.1574-6968.2009.01674.x [DOI] [PubMed] [Google Scholar]
- O'Neill J. G., Wilkinson J. F. (1977). Oxidation of ammonia by methane-oxidizing bacteria and the effects of ammonia on methane oxidation. J. Gen. Microbiol. 100, 407–412. 10.1099/00221287-100-2-407 [DOI] [Google Scholar]
- Op den Camp H. J. M., Islam T., Stott M. B., Harhangi H. R., Hynes A., Schouten S., et al. (2009). Environmental, genomic and taxonomic perspectives on methanotrophic Verrucomicrobia. Environ. Microbiol. Rep. 1, 293–306. 10.1111/j.1758-2229.2009.00022.x [DOI] [PubMed] [Google Scholar]
- Pol A., Barends T. R., Dietl A., Khadem A. F., Eygensteyn J., Jetten M. S. M., et al. (2014). Rare earth metals are essential for methanotrophic life in volcanic mudpots. Environ. Microbiol. 16, 255–264. 10.1111/1462-2920.12249 [DOI] [PubMed] [Google Scholar]
- Pol A., Heijmans K., Harhangi H. R., Tedesco D., Jetten M. S. M., Op den Camp H. J. M. (2007). Methanotrophy below pH 1 by a new Verrucomicrobia species. Nature 450, 874–878. 10.1038/nature06222 [DOI] [PubMed] [Google Scholar]
- Poret-Peterson A. T., Grahamm J. E., Gulledge J., Klotz M. G. (2008). Transcription of nitrification genes by the methane-oxidizing bacterium, Methylococcus capsulatus strain Bath. ISME J. 2, 1213–1220. 10.1038/ismej.2008.71 [DOI] [PubMed] [Google Scholar]
- Ryabenko E., Altabet M. A., Wallace D. W. R. (2009). Effect of chloride on the chemical conversion of nitrate to nitrous oxide for 15N analysis. Limnol. Oceanogr. Methods 7, 545–552. 10.4319/lom.2009.7.545 [DOI] [Google Scholar]
- Sharp C. E., Op den Camp H. J. M., Tamas I., Dunfield P. F. (2013). Unusual members of the PVC superphylum: the methanotrophic Verrucomicrobia genus “Methylacidiphilum”, in Planctomycetes: Cell Structure, Origins and Biology, ed Fuerst J. A. (New York, NY: Humana Press; Springer; ), 211–227. [Google Scholar]
- Sharp C. E., Smirnova A. V., Graham J. M., Stott M. B., Khadka R., Moore T. R., et al. (2014). Distribution and diversity of Verrucomicrobia methanotrophs in geothermal and acidic environments. Environ. Microbiol. 16, 1867–1878. 10.1111/1462-2920.12454 [DOI] [PubMed] [Google Scholar]
- Sharp C. E., Stott M. B., Dunfield P. F. (2012). Detection of autotrophic verrucomicrobial methanotrophs in a geothermal environment using stable isotope probing. Front. Microbiol. 3:303. 10.3389/fmicb.2012.00303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein L. Y., Klotz M. G. (2011). Nitrifying and denitrifying pathways of methanotrophic bacteria. Biochem. Soc. Trans. 39, 1826–1831. 10.1042/BST20110712 [DOI] [PubMed] [Google Scholar]
- Stein L. Y., Roy R., Dunfield P. F. (2012). Aerobic methanotrophy and nitrification processes and connections, in eLS (Chichester: John Wiley Sons, Ltd; ). 10.1002/9780470015902.a0022213 Available online at: http://www.els.net [DOI] [Google Scholar]
- Taylor S., Ninjoor V., Dowd D. M., Tappel A. L. (1974). Cathepsin B2 measurement by sensitive fluorometric ammonia analysis. Anal. Biochem. 60, 153–162. 10.1016/0003-2697(74)90140-7 [DOI] [PubMed] [Google Scholar]
- van Teeseling M. C. F., Pol A., Harhangi H. R., van der Zwart S., Jetten M. S. M., Op den Camp H. J. M., et al. (2014). Expanding the verrucomicrobial methanotrophic world: description of three novel species of Methylacidimicrobium gen. nov. Appl. Environ. Microbiol. 80, 6782–6791. 10.1128/AEM.01838-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


