Abstract
Recent studies have described the important multiple roles of long non‐coding RNAs (lncRNAs) during oncogenic transformation. Because the coding genome accounts for a small amount of total DNA, and many mutations leading to cancer occur in the non‐coding genome, it is plausible that the dysregulation of such non‐coding transcribes might also affect tumor phenotypes. Indeed, to date, lncRNAs have been reported to affect diverse biological processes through the regulation of mRNA stability, RNA splicing, chromatin structure, and miRNA‐mediated gene regulation by acting as miRNA sponges. Furthermore, accumulating studies have described the roles of lncRNAs in tumorigenesis; however, the precise mechanisms of many lncRNAs are still under investigation. Here, we discuss recently reported mechanistic insights into how lncRNAs regulate gene expression and contribute to tumorigenesis through interactions with other regulatory molecules. We especially highlight the role of taurine upregulated gene 1, which was recently reported to have biological functions related to gene regulation, and discuss the future clinical implications of lncRNAs in cancer treatments.
Keywords: Chromatin structure, epigenetics, non‐coding RNA, transcription modulators, treatment
Recent comprehensive molecular profiling in many types of cancers has revealed oncogenic mutations and the aberrant expressions of protein‐coding genes. However, because the coding genome accounts for <2% of all DNA sequences and many mutations were reported in non‐coding sequences,1 the dysregulation of non‐coding RNAs might affect tumor phenotypes. For example, significant numbers of non‐coding RNAs such as microRNA (miRNA, miR), siRNA, piwi‐interacting RNA, and long non‐coding RNAs (lncRNAs), were recently discovered and identified as biologically functional non‐coding RNAs, some of which had a significant impact on tumor biology.2
Among such non‐coding RNAs, miRNAs are short non‐coding RNAs (approximately 22 nt long) that bind to short regions of complementary sequences of target mRNAs to post‐transcriptionally regulate the expression of target genes. MicroRNAs have important roles in a wide variety of pathological processes related to tumor formation. The aberrant expression of miRNA was shown to induce tumor suppression or induce oncogenic effects resulting in tumor formation.2 Extensively studied examples of tumor‐suppressive miRNAs include the let‐7, miR‐15a/16‐1, miR‐26a, or miR‐34 family members, which are associated with antiproliferative, antitumorigenic, and pro‐apoptotic activities and whose expressions are sometimes aberrantly suppressed in many types of cancers. Examples of oncogenic miRNAs include the miR‐17‐92 cluster, miR‐155, and miR‐21, the functions of which are associated with the repression of tumor‐suppressor genes such as PTEN and CDKN1A, and whose expressions are sometimes aberrantly upregulated in cancers.3
In addition to miRNAs, lncRNAs, defined as transcripts >200 nt in length, are also functional. Although the precise roles of the vast majority of ~40 000 lncRNAs are still under investigation,4 some of these transcripts were shown to be potential key regulators of cellular differentiation and proliferation, as well as having oncogenic functions in many types of cancers.5, 6 Furthermore, studies have shown that lncRNAs affect chromatin structure and RNA interactions, such as the miRNA sponge, which upregulates protein expression by inhibiting the binding of miRNAs to their targets.6, 7
With regard to RNA interactions between lncRNAs and miRNA in cancers, Hox transcript antisense intergenic RNA (HOTAIR), an lncRNA, has an oncogenic role in tumor formation by upregulating fibroblast growth factor 1 by sponging miR‐326, which activates the PI3K/AKT and MEK1/2 pathways.8 We also recently found that Notch1 activation in glioma cells specifically induced the expression of lncRNA for taurine upregulated gene 1 (TUG1), which coordinately promoted self‐renewal by sponging miR‐145.9
Thus, accumulating studies of cancer‐associated lncRNAs have reported their roles in the multiple pathological steps of tumorigenesis, including cell proliferation, cellular signaling, angiogenesis, and metastasis, which may provide a strong rationale for the targeting of lncRNAs as a specific and potent therapeutic approach to eliminate cancer cells.6 In this review, we provide a summary of the current understanding of lncRNAs, including TUG1, which were recently reported to have pathological roles in cancer, and we discuss the future perspective of targeting lncRNAs as a new approach for cancer treatment.
Roles of lncRNA in gene regulation
Recent studies reported the regulatory mechanisms of gene expression by lncRNAs in at least seven pathways (Fig. 1).10, 11, 12, 13 These functions are closely associated with the subcellular localization of lncRNAs, although the precise factors (e.g. binding proteins) and sequence elements in lncRNAs that determine their localization remain largely unknown.14 Generally, lncRNAs have a more nuclear‐biased localization pattern when compared with mRNAs, although large amounts of lncRNAs are located in the cytoplasm.14 This localization bias may support the idea that lncRNAs function as essential molecules in the scaffolding and recruitment of multiple proteins to specific genomic loci to form 3‐D nuclear structures, which affect gene expression (Fig. 1). Furthermore, nascent RNA transcripts in the nucleus are processed by different methods including splicing, polyadenylation, 5ʹ‐capping, and methylation. During these processes, lncRNAs affect RNA splicing and mRNA stability. For example, metastasis‐associated lung adenocarcinoma transcript 1 (MALAT1), which is restricted to the nucleus, especially to nuclear speckles, affects alternative splicing through interactions with serine/arginine‐rich splicing factors and pre‐mRNA.13, 15 In addition to their nuclear functions, cytoplasmic lncRNAs function as a modulator by interacting with other types of RNAs (e.g. competitive endogenous RNAs [ceRNAs]). These functions of lncRNAs are exemplified and explained in more detail below.
Figure 1.
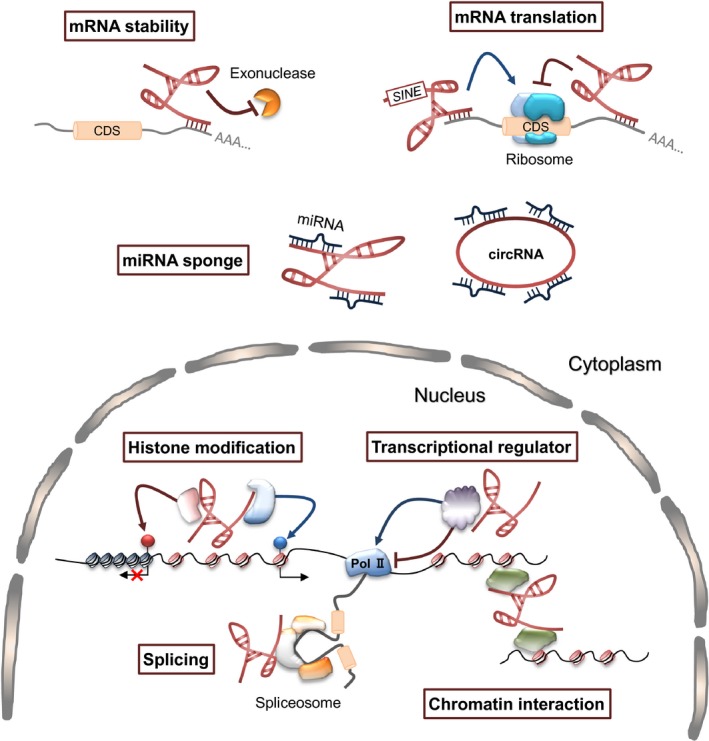
Multiple long non‐coding RNA (lncRNA) mechanisms of gene regulation, which rely on interactions with multiple molecules. In the nucleus, lncRNAs regulate gene expression by controlling the local chromatin structure or recruiting regulatory molecules to specific loci. In the cytoplasm, lncRNAs interact with other types of RNA and affect functions including mRNA stability, mRNA translation, or microRNA (miRNA) sponge. CDS, coding sequence; circRNA, circular RNA; Pol II, RNA polymerase II.
Interactions between chromatin regulatory proteins and lncRNAs
Long non‐coding RNAs regulate gene expression through their scaffolding activity for chromatin modifying proteins (e.g. methyltransferases, demethylases, acetyltransferases, and deacetylases), and recruiting these proteins to target loci through cis‐regulation (regulation of the transcription of nearby genes) or trans‐regulation (regulation of the transcription of genomically distant genes). These interactions occur by affecting the nuclear structure.16
One of the most studied lncRNAs is X‐inactive specific transcript (Xist), which is expressed from one of the two X chromosomes at the initial phase of X chromosome inactivation (XCI) in early female embryonic development. Xist binds to chromatin by scaffold attachment factor A (also known as hnRNPU) and recruits SMRT/histone deacetylase 1 (HDAC1)‐associated repressor protein (SHARP), which interacts with HDAC3, polycomb repressive complex 1 (PRC1), and PRC2.17, 18 Intriguingly, although the induction of Xist expression and the recruitment of SHARP‐HDAC3 are prerequisites for the initiation of XCI, Xist appears to be dispensable for the maintenance of transcriptional inactivation.16, 19 Furthermore, it appears that the presence of PRC2 is not required for the initiation of XCI, because the genetic deletion of PRC2 had no effect on the initiation of transcriptional silencing,20, 21 although it was required for the maintenance of transcriptional inactivation.22
Similar to Xist, a set of lncRNAs are thought to interact with PRC2, resulting in gene inactivation of certain specific genome loci. HOTAIR is transcribed in the Homeobox (HOX) C gene cluster region on chromosome 12 and is co‐expressed with the HOXC genes.23 HOTAIR regulates the expression of HOXD genes in chromosome 2 through transregulation, whereas interactions between HOTAIR, lysine‐specific demethylase 1, and PRC2 promotes coordinated H3K27 methylation and H3K4 demethylation. Many cancers, including breast cancer, pancreatic cancer, non‐small‐cell lung cancer (NSCLC), and gastrointestinal stromal tumor overexpress HOTAIR, which affects tumor behavior.24, 25, 26, 27 For example, the overexpression of HOTAIR in breast cancer cells increased their invasive and metastatic abilities and reprogramed PRC2 occupancy throughout the genome, which is similar to embryonic fibroblasts.24
A recent study showed that although HOTAIR binds to PRC2 with a high affinity to silence target loci by H3K27me3 deposition, the genetic deletion of PRC2 components did not affect the silencing activity of HOTAIR, indicating that PRC2 is dispensable for the initiation of HOTAIR‐mediated silencing machinery.28 This is similar to Xist‐mediated gene silencing, as mentioned above. These two examples of functional interactions between an lncRNA (Xist and HOTAIR) and PRC2 suggest interesting mechanistic consequences of lncRNA‐guided gene regulation, in which many chromatin regulatory proteins are involved. Thus, in addition to the dysregulation of many chromatin modifiers such as histone modification enzymes and chromatin remodelers that have been comprehensively analyzed in many cancers, the dysregulation of lncRNAs may also play an important functional role in tumorigenesis.
Interactions between lncRNAs and other types of RNA
Long non‐coding RNAs interact with other types of RNA molecules in cells, such as mRNA and miRNA, and modulate their stability, splicing, translation, and metabolism (Fig. 1). MALAT1, a highly abundant lncRNA, regulates alternative splicing through interactions with serine/arginine‐rich splicing factors and pre‐mRNA.15, 29 Details of MALAT1 functions were well documented in a recent review.30 In cancer studies, the high expression of MALAT1 in NSCLC was associated with metastatic progression.31 The genetic loss or systemic knockdown of Malat1 in a mouse cancer model resulted in slower tumor growth and a reduction in the metastasis of lung cancer and breast cancer.32, 33 Long non‐coding RNAs also stabilize mRNA. Terminal tissue differentiation‐inducing ncRNA (TINCR) is a characteristic lncRNA that binds to mRNAs with a 25‐nt TINCR box motif.34 TINCR recruits Staufen‐1 protein, a regulator of tissue differentiation, to mRNA with a TINCR box motif, and stabilizes the target mRNAs to promote its translation.
Recently, a model of ceRNA was proposed, where abundant cytoplasmic lncRNAs containing miRNA‐binding sites interacted with miRNAs through their seed sequences (i.e. sequence‐specific sequester) to reduce their regulatory effect on target mRNA, the so‐called miRNA sponge.13 Phosphatase and tensin homolog (PTEN) is a well‐known tumor‐suppressor gene. Studies have shown that PTEN pseudogene 1 (PTENP1) increased PTEN protein levels by competing for a set of PTEN‐targeting miRNAs, which downregulate PTEN independent of its protein‐coding function.35, 36 In colon cancer, the loss of focal copy number at the PTENP1 locus was associated with the downregulation of PTEN expression in colon cancer patients. A similar relationship was shown between the oncogene KRAS and its pseudogene KRAS1P in colon cancer.35
These lncRNA contributions to tumorigenesis through mechanisms including ceRNA have generated substantial interest and have been reported in many cancers.6, 37 However, the ceRNA hypothesis should consider the physiological stoichiometry of miRNAs and ceRNAs in cells because the suppressive activity of ceRNAs might be closely associated with miRNA cellular abundance.38, 39 Generally, high amounts of miRNAs are unlikely to be susceptible to ceRNA competition.40 Therefore, a similar abundance of ceRNAs is thought to be required for efficient competition with target miRNAs.41 Although further studies are required to clarify more precisely the regulatory cross‐talk between transcripts, including mRNAs, miRNAs, and lncRNAs, the ceRNA mechanism might explain the complexity of the dysregulation of mRNA through its 3ʹ‐UTR in cancers.
Mechanisms of dysregulated lncRNA expression in cancer
Increasing evidence has shown that the dysregulation of lncRNAs is associated with cancer pathogenesis and that lncRNAs function as regulators of cancer‐related genes.6, 42 Several lines of evidence showed that aberrant signal transduction induces lncRNA dysregulation (Fig. 2). The Notch signaling pathway plays a dominant role in inhibiting neural stem cell differentiation through the activities of its downstream effectors, such as Hairy and enhancer of split 1/5.43 A recent study showed that Notch triggered oncogenic activity through lncRNA activation in leukemia (Fig. 2a). In human T‐cell acute lymphoblastic leukemia, a set of lncRNAs, including leukemia‐induced non‐coding activator RNA (LUNAR1), are directly controlled by the Notch1 signaling. LUNAR1 is required for T‐cell acute lymphoblastic leukemia growth through the enhancement of insulin‐like growth factor 1 receptor expression and sustained insulin‐like growth factor 1 signaling.44 Notch1 signaling also induces another lncRNA, TUG1,9 which is a cancer‐related lncRNA that binds to PRC2 or PRC1 and also represses gene expression in glioma cells.45, 46
Figure 2.
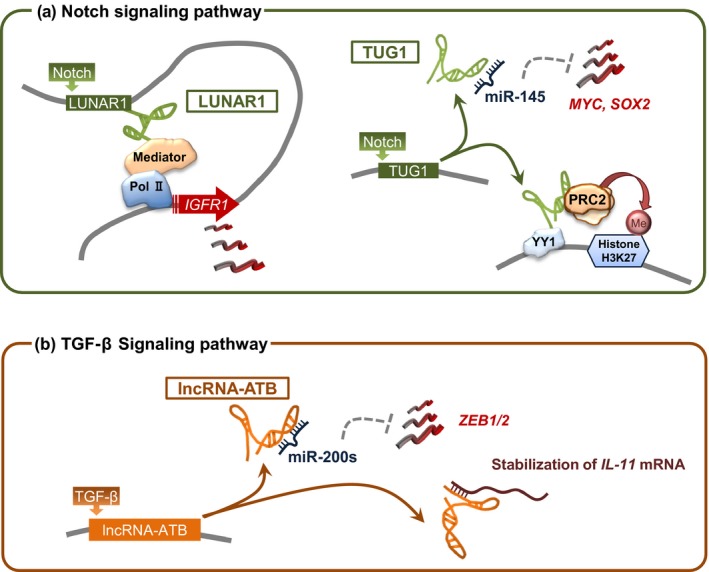
Aberrant signal transduction induces long non‐coding RNA (lncRNA) dysregulation in cancer cells. (a) Notch triggers oncogenic activity by the activation of two different lncRNAs (leukemia‐induced non‐coding activator RNA [LUNAR1] and taurine upregulated gene 1 [TUG1]) in cancers. LUNAR1 enhances insulin‐like growth factor 1 receptor (IGF1R) expression through a cis‐activation mechanism in leukemia (left). TUG1 coordinately promotes self‐renewal by sponging microRNA‐145 (miR‐145) in the cytoplasm and recruiting polycomb repressive complex 2 (PRC2) to repress differentiation genes by the locus‐specific methylation of histone H3K27 by YY1 binding activity in glioma stem cells. Me, methylation; Pol II, RNA polymerase II. (b) LncRNA activated by transforming growth factor‐β (TGF‐β) (lncRNA‐ATB) is upregulated by TGF‐β signaling in hepatocellular carcinoma. LncRNA‐ATB upregulates Zinc finger E‐box‐binding homeobox (ZEB)1 and ZEB2 by sequestering miR‐200 family members (miR‐200s) and inducing epithelial mesenchymal transition and invasion. In addition, lncRNA‐ATB promotes the organ colonization of tumor cells by binding to interleukin (IL)‐11 mRNA.
Long non‐coding RNA activated by TGF‐β (lncRNA‐ATB) was upregulated by transforming growth factor‐β signaling in hepatocellular carcinoma metastases. This lncRNA upregulates Zinc finger E‐box‐binding homeobox 1 and 2 by competitively binding to miR‐200 family members to induce epithelial–mesenchymal transition and invasion. Furthermore, lncRNA‐ATB promoted the organ colonization of tumor cells by binding to interleukin‐11 mRNA and triggering signal transducer and activator of transcription 3 signaling (Fig. 2b).10
These studies indicate that a set of lncRNAs may act as key regulators of signaling pathways. Downstream of the cancer‐promoting signals, lncRNAs may sustain cancer cell proliferation and enhance viability and motility, which are linked to the clinically relevant cancer subtypes that predict tumor behavior and prognosis.
Taurine upregulated gene 1 plays important roles in tumorigenesis
We recently identified the Notch‐regulated lncRNA, TUG1, in glioma cells by whole‐genome RNA sequencing and comprehensively characterized its function in relation to gliomagenesis.9 Taurine upregulated gene 1 is a cancer‐related lncRNA that binds to PRC2 or PRC1.45, 46 This lncRNA was originally identified as a transcript upregulated by taurine, whose function is associated with retinal development.47 It is overexpressed in bladder cancer, gastric cancer, and osteosarcoma;48, 49, 50 in contrast, it is downregulated in NSCLC,51 suggesting context‐dependent roles in different types of cancers (Table 1). Because the length of TUG1 lncRNA is long (approximately 7.1 kb), it is plausible that TUG1 has multiple functions.
Table 1.
Function of taurine upregulated gene 1 (TUG1) in human cancers
| Cancer type | Molecular function |
|---|---|
| Oncogenic function | |
| Glioma53 | miRNA sponge (miR‐144) |
| Glioma9 | Recruitment of PRC2, miRNA sponge (miR‐145) |
| Glioma54 | miRNA sponge (miR‐26a) |
| Glioma55 | miRNA sponge (miR‐299) |
| Oral squamous cell carcinoma56 | Unknown |
| Esophageal squamous cell carcinoma57, 58 | Unknown |
| Cervical cancer59 | Unknown |
| Small cell lung cancer60 | Recruitment of PRC2 |
| Gastric cancer61 | miRNA sponge (miR‐144) |
| Gastric cancer62 | Recruitment of PRC2 |
| Hepatocellular carcinoma63 | Recruitment of PRC2 |
| Hepatoblastoma64 | miRNA sponge (miR‐34a) |
| Gallbladder carcinoma65 | miRNA sponge (miR‐300) |
| Breast cancer66 | miRNA sponge (miR‐9) |
| Breast cancer67 | Unknown |
| Colorectal cancer68, 69, 70 | Unknown |
| Ovarian cancer71 | Unknown |
| Renal cell carcinoma72 | Unknown |
| Bladder cancer49 | Unknown |
| Bladder cancer48 | miRNA sponge (miR‐145) |
| Bladder cancer73 | Unknown |
| Osteosarcoma50 | Unknown |
| Osteosarcoma74 | miRNA sponge (miR‐9) |
| Osteosarcoma75 | miRNA sponge (miR‐335) |
| Tumor‐suppressive function | |
| Cervical cancer46 | Recruitment of PRC1 and PRC2 |
| Non‐small‐cell lung carcinoma51, 76 | Recruitment of PRC2 |
| Prostate cancer77 | miRNA sponge (unknown) |
Text in parentheses shows target microRNA (miRNA, miR) of TUG1 in each cancer type. PRC, polycomb repressive complex.
Expression of TUG1 is regulated by the Notch signaling pathway, and TUG1 was highly expressed in glioma stem cell populations and downregulated during its differentiation. Intriguingly, in cell nuclei, TUG1 physically interacts with PRC2, which might direct it to modify histone H3K27me3 levels in neuronal differentiation‐associated genes. In the cytoplasm, TUG1 shares miR‐145‐response elements with the mRNAs of several stemness markers (MYC and SOX2) and prevents them from miR‐145‐mediated degradation (Fig. 2). Importantly, the inhibition of TUG1 expression impaired stemness and tumorigenesis in gliomas both in vitro and in vivo, indicating that targeting TUG1 is a potent therapeutic approach to eliminate glioma stem cell populations.9 Indeed, antisense oligonucleotide (ASO) targeting TUG1, especially coupled with a potent drug delivery system, is an effective novel strategy for glioblastoma (GBM) treatment9 (Fig. 3). Cyclic Arg‐Gly‐Asp (cRGD) peptides are promising ligand molecules for targeting αvβ3 and αvβ5 integrins, which are frequently overexpressed in GBM cells. We used cRGD ligand‐conjugated polymeric micelles for delivery. These targetable polymeric micelles retained ASO accumulation within tumors. Although further investigations are required, cRGD‐mediated drug delivery is a powerful strategy for targeting GBMs through facilitated ASO delivery beyond the blood–brain tumor barrier.52
Figure 3.
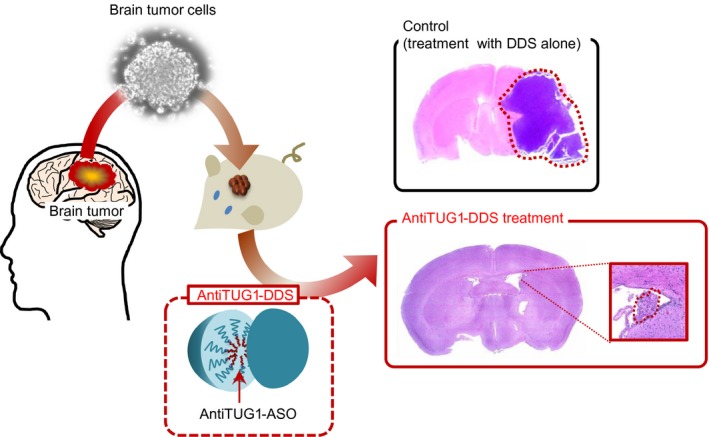
Inhibition of taurine upregulated gene 1 (TUG1) by an antiTUG1 drug delivery system (DDS) in a mouse xenograft model. Mice bearing brain tumors were given i.v. antisense oligonucleotide (ASO) targeting TUG1 coupled with a potent DDS using cyclic Arg‐Gly‐Asp peptide‐conjugated polymeric micelle (antiTUG1‐DDS). AntiTUG1‐DDS was specifically accumulated and retained in the tumors and markedly reduced tumor growth.9 Tumor areas are surrounded by red dotted line.
Similar to miR‐145, TUG1 interacts with other miRNAs such as miR‐144, miR‐26a, miR‐299, miR‐34a, miR‐300, miR‐9, and miR‐335, in different types of cancers (Table 1). Although further experimental validation is required to clarify the impact of ceRNA mechanisms on tumorigenesis, TUG1 functions through a ceRNA mechanism that can dynamically change the transcriptome. Therefore, it will be particularly interesting to understand the pathologies of plastic cancer cells.
Concluding remarks
In this review, we exemplified and explained the functional roles of lncRNAs (Table 2) and discussed the future clinical implications of lncRNAs in cancers. Recent comprehensive studies have shown that, in addition to genetic alterations, the spatial and temporal epigenetic regulation of gene functions in pre‐cancer and cancer cells is particularly important in tumorigenesis. In particular, given that cancer cells are dynamic in response to extracellular signals, the plastic epigenetic control of gene expression plays a central role in cancer cell adaptation to new microenvironments. A better understanding of lncRNA pathways and other epigenetic mechanisms in cancer cells will hopefully provide multiple novel therapeutic strategies for devastating cancers in the near future.
Table 2.
Summary of key functions of the exemplified long non‐coding RNAs (lncRNA)
| LncRNA | Biological roles | Molecular functions |
|---|---|---|
| HOTAIR | Promotion of cell invasion and metastasis | Recruitment of PRC2, miRNA sponge |
| TUG1 | Promotion of cell proliferation | Recruitment of PRC2, miRNA sponge |
| MALAT1 | Regulation of alternative splicing | Splicing |
| XIST | Inactivation of X chromosome | Recruitment of PRC2 |
| TINCR | Regulation of epidermal differentiation | Stabilization of mRNA |
| PTENP1 | Inhibition of cell proliferation | miRNA sponge |
| KRAS1P | Promotion of cell proliferation | miRNA sponge |
| LUNAR1 | Promotion of cell proliferation | Chromosome looping |
| lncRNA‐ATB | Promotion of cell invasion and metastasis | Stabilization of mRNA, miRNA sponge |
HOTAIR, Hox transcript antisense intergenic RNA; lncRNA‐ATB, lncRNA activated by transforming growth factor‐β; LUNAR1, leukemia‐induced non‐coding activator RNA; MALAT1, metastasis‐associated lung adenocarcinoma transcript 1; miRNA, microRNA; PRC2, polycomb repressive complex 2; PTENP1, phosphatase and tensin homolog pseudogene 1; TINCR, tissue differentiation‐inducing ncRNA; TUG1, taurine upregulated gene 1; XIST, X‐inactive specific transcript.
Disclosure Statement
The authors have no conflict of interest.
Acknowledgments
This study was funded as a research program of the Project for Development of Innovative Research on Cancer Therapeutics (P‐Direct) and Project for Cancer Research and Therapeutic Evolution (P‐Create), Japan Agency for Medical Research and Development (Y. Kondo, K. Shinjo, and K. Katsushima), and by a Grant‐in‐Aid for Scientific Research, the Japan Society for the Promotion of Science (2617H03582, Y. Kondo).
Cancer Sci 108 (2017) 1927–1933
References
- 1. Fujimoto A, Furuta M, Totoki Y et al Whole‐genome mutational landscape and characterization of noncoding and structural mutations in liver cancer. Nat Genet 2016; 48: 500–9. [DOI] [PubMed] [Google Scholar]
- 2. Ha M, Kim VN. Regulation of microRNA biogenesis. Nat Rev 2014; 15: 509–24. [DOI] [PubMed] [Google Scholar]
- 3. Lujambio A, Lowe SW. The microcosmos of cancer. Nature 2012; 482: 347–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Schlosser K, Hanson J, Villeneuve PJ et al Assessment of Circulating LncRNAs Under Physiologic and Pathologic Conditions in Humans Reveals Potential Limitations as Biomarkers. Sci Rep 2016; 6: 36596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Wahlestedt C. Targeting long non‐coding RNA to therapeutically upregulate gene expression. Nat Rev Drug Discov 2013; 12: 433–46. [DOI] [PubMed] [Google Scholar]
- 6. Schmitt AM, Chang HY. Long Noncoding RNAs in Cancer Pathways. Cancer Cell 2016; 29: 452–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Guttman M, Donaghey J, Carey BW et al lincRNAs act in the circuitry controlling pluripotency and differentiation. Nature 2011; 477: 295–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ke J, Yao YL, Zheng J et al Knockdown of long non‐coding RNA HOTAIR inhibits malignant biological behaviors of human glioma cells via modulation of miR‐326. Oncotarget 2015; 6: 21934–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Katsushima K, Natsume A, Ohka F et al Targeting the Notch‐regulated non‐coding RNA TUG1 for glioma treatment. Nat Commun 2016; 7: 13616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Yuan JH, Yang F, Wang F et al A long noncoding RNA activated by TGF‐beta promotes the invasion‐metastasis cascade in hepatocellular carcinoma. Cancer Cell 2014; 25: 666–81. [DOI] [PubMed] [Google Scholar]
- 11. Rybak‐Wolf A, Stottmeister C, Glazar P et al Circular RNAs in the Mammalian Brain Are Highly Abundant, Conserved, and Dynamically Expressed. Mol Cell 2015; 58: 870–85. [DOI] [PubMed] [Google Scholar]
- 12. Carrieri C, Cimatti L, Biagioli M et al Long non‐coding antisense RNA controls Uchl1 translation through an embedded SINEB2 repeat. Nature 2012; 491: 454–7. [DOI] [PubMed] [Google Scholar]
- 13. Quinn JJ, Chang HY. Unique features of long non‐coding RNA biogenesis and function. Nat Rev Genet 2016; 17: 47–62. [DOI] [PubMed] [Google Scholar]
- 14. Ulitsky I, Bartel DP. lincRNAs: genomics, evolution, and mechanisms. Cell 2013; 154: 26–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Tripathi V, Ellis JD, Shen Z et al The nuclear‐retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. Mol Cell 2010; 39: 925–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Engreitz JM, Ollikainen N, Guttman M. Long non‐coding RNAs: spatial amplifiers that control nuclear structure and gene expression. Nat Rev 2016; 17: 756–70. [DOI] [PubMed] [Google Scholar]
- 17. McHugh CA, Chen CK, Chow A et al The Xist lncRNA interacts directly with SHARP to silence transcription through HDAC3. Nature 2015; 521: 232–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Chu C, Zhang QC, da Rocha ST et al Systematic discovery of Xist RNA binding proteins. Cell 2015; 161: 404–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Splinter E, de Wit E, Nora EP et al The inactive X chromosome adopts a unique three‐dimensional conformation that is dependent on Xist RNA. Genes Dev 2011; 25: 1371–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Schoeftner S, Sengupta AK, Kubicek S et al Recruitment of PRC1 function at the initiation of X inactivation independent of PRC2 and silencing. EMBO J 2006; 25: 3110–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kalantry S, Magnuson T. The Polycomb group protein EED is dispensable for the initiation of random X‐chromosome inactivation. PLoS Genet 2006; 2: e66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kalantry S, Mills KC, Yee D, Otte AP, Panning B, Magnuson T. The Polycomb group protein Eed protects the inactive X‐chromosome from differentiation‐induced reactivation. Nat Cell Biol 2006; 8: 195–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Tsai MC, Manor O, Wan Y et al Long noncoding RNA as modular scaffold of histone modification complexes. Science 2010; 329: 689–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Gupta RA, Shah N, Wang KC et al Long non‐coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature 2010; 464: 1071–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kim K, Jutooru I, Chadalapaka G et al HOTAIR is a negative prognostic factor and exhibits pro‐oncogenic activity in pancreatic cancer. Oncogene 2013; 32: 1616–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Nakagawa T, Endo H, Yokoyama M et al Large noncoding RNA HOTAIR enhances aggressive biological behavior and is associated with short disease‐free survival in human non‐small cell lung cancer. Biochem Biophys Res Comm 2013; 436: 319–24. [DOI] [PubMed] [Google Scholar]
- 27. Niinuma T, Suzuki H, Nojima M et al Upregulation of miR‐196a and HOTAIR drive malignant character in gastrointestinal stromal tumors. Cancer Res 2012; 72: 1126–36. [DOI] [PubMed] [Google Scholar]
- 28. Portoso M, Ragazzini R, Brencic Z et al PRC2 is dispensable for HOTAIR‐mediated transcriptional repression. EMBO J 2017; 36: 981–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Quinn JJ, Zhang QC, Georgiev P, Ilik IA, Akhtar A, Chang HY. Rapid evolutionary turnover underlies conserved lncRNA‐genome interactions. Genes Dev 2016; 30: 191–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Yoshimoto R, Mayeda A, Yoshida M, Nakagawa S. MALAT1 long non‐coding RNA in cancer. Biochem Biophys Acta 2016; 1859: 192–9. [DOI] [PubMed] [Google Scholar]
- 31. Ji P, Diederichs S, Wang W et al MALAT‐1, a novel noncoding RNA, and thymosin beta4 predict metastasis and survival in early‐stage non‐small cell lung cancer. Oncogene 2003; 22: 8031–41. [DOI] [PubMed] [Google Scholar]
- 32. Gutschner T, Hammerle M, Eissmann M et al The noncoding RNA MALAT1 is a critical regulator of the metastasis phenotype of lung cancer cells. Cancer Res 2013; 73: 1180–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Arun G, Diermeier S, Akerman M et al Differentiation of mammary tumors and reduction in metastasis upon Malat1 lncRNA loss. Genes Dev 2016; 30: 34–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kretz M, Siprashvili Z, Chu C et al Control of somatic tissue differentiation by the long non‐coding RNA TINCR. Nature 2013; 493: 231–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Poliseno L, Salmena L, Zhang J, Carver B, Haveman WJ, Pandolfi PP. A coding‐independent function of gene and pseudogene mRNAs regulates tumour biology. Nature 2010; 465: 1033–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Tay Y, Kats L, Salmena L et al Coding‐independent regulation of the tumor suppressor PTEN by competing endogenous mRNAs. Cell 2011; 147: 344–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Thomson DW, Dinger ME. Endogenous microRNA sponges: evidence and controversy. Nat Rev Genet 2016; 17: 272–83. [DOI] [PubMed] [Google Scholar]
- 38. Mullokandov G, Baccarini A, Ruzo A et al High‐throughput assessment of microRNA activity and function using microRNA sensor and decoy libraries. Nat Methods 2012; 9: 840–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Thomson DW, Pillman KA, Anderson ML et al Assessing the gene regulatory properties of Argonaute‐bound small RNAs of diverse genomic origin. Nucleic Acids Res 2015; 43: 470–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Bosson AD, Zamudio JR, Sharp PA. Endogenous miRNA and target concentrations determine susceptibility to potential ceRNA competition. Mol Cell 2014; 56: 347–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Ala U, Karreth FA, Bosia C et al Integrated transcriptional and competitive endogenous RNA networks are cross‐regulated in permissive molecular environments. Proc Natl Acad Sci USA 2013; 110: 7154–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Prensner JR, Chinnaiyan AM. The emergence of lncRNAs in cancer biology. Cancer Discov 2011; 1: 391–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Louvi A, Artavanis‐Tsakonas S. Notch signalling in vertebrate neural development. Nat Rev Neurosci 2006; 7: 93–102. [DOI] [PubMed] [Google Scholar]
- 44. Trimarchi T, Bilal E, Ntziachristos P et al Genome‐wide mapping and characterization of Notch‐regulated long noncoding RNAs in acute leukemia. Cell 2014; 158: 593–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Khalil AM, Guttman M, Huarte M et al Many human large intergenic noncoding RNAs associate with chromatin‐modifying complexes and affect gene expression. Proc Natl Acad Sci USA 2009; 106: 11667–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Yang L, Lin C, Liu W et al ncRNA‐ and Pc2 methylation‐dependent gene relocation between nuclear structures mediates gene activation programs. Cell 2011; 147: 773–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Young TL, Matsuda T, Cepko CL. The noncoding RNA taurine upregulated gene 1 is required for differentiation of the murine retina. Curr Biol 2005; 15: 501–12. [DOI] [PubMed] [Google Scholar]
- 48. Tan J, Qiu K, Li M, Liang Y. Double‐negative feedback loop between long non‐coding RNA TUG1 and miR‐145 promotes epithelial to mesenchymal transition and radioresistance in human bladder cancer cells. FEBS Lett 2015; 589: 3175–81. [DOI] [PubMed] [Google Scholar]
- 49. Han Y, Liu Y, Gui Y, Cai Z. Long intergenic non‐coding RNA TUG1 is overexpressed in urothelial carcinoma of the bladder. J Surg Oncol 2013; 107: 555–9. [DOI] [PubMed] [Google Scholar]
- 50. Zhang Q, Geng PL, Yin P, Wang XL, Jia JP, Yao J. Down‐regulation of long non‐coding RNA TUG1 inhibits osteosarcoma cell proliferation and promotes apoptosis. Asian Pac J Cancer Prev 2013; 14: 2311–15. [DOI] [PubMed] [Google Scholar]
- 51. Zhang EB, Yin DD, Sun M et al P53‐regulated long non‐coding RNA TUG1 affects cell proliferation in human non‐small cell lung cancer, partly through epigenetically regulating HOXB7 expression. Cell Death Dis 2014; 5: e1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Miura Y, Takenaka T, Toh K et al Cyclic RGD‐linked polymeric micelles for targeted delivery of platinum anticancer drugs to glioblastoma through the blood‐brain tumor barrier. ACS Nano 2013; 7: 8583–92. [DOI] [PubMed] [Google Scholar]
- 53. Cai H, Xue Y, Wang P et al The long noncoding RNA TUG1 regulates blood‐tumor barrier permeability by targeting miR‐144. Oncotarget 2015; 6: 19759–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Li J, An G, Zhang M, Ma Q. Long non‐coding RNA TUG1 acts as a miR‐26a sponge in human glioma cells. Biochem Biophys Res Comm 2016; 477: 743–8. [DOI] [PubMed] [Google Scholar]
- 55. Cai H, Liu X, Zheng J et al Long non‐coding RNA taurine upregulated 1 enhances tumor‐induced angiogenesis through inhibiting microRNA‐299 in human glioblastoma. Oncogene 2017; 36: 318–31. [DOI] [PubMed] [Google Scholar]
- 56. Liang S, Zhang S, Wang P et al LncRNA, TUG1 regulates the oral squamous cell carcinoma progression possibly via interacting with Wnt/beta‐catenin signaling. Gene 2017; 608: 49–57. [DOI] [PubMed] [Google Scholar]
- 57. Xu Y, Wang J, Qiu M et al Upregulation of the long noncoding RNA TUG1 promotes proliferation and migration of esophageal squamous cell carcinoma. Tumour Biol 2015; 36: 1643–51. [DOI] [PubMed] [Google Scholar]
- 58. Jiang L, Wang W, Li G et al High TUG1 expression is associated with chemotherapy resistance and poor prognosis in esophageal squamous cell carcinoma. Cancer Chemother Pharmacol 2016; 78: 333–9. [DOI] [PubMed] [Google Scholar]
- 59. Hu Y, Sun X, Mao C et al Upregulation of long noncoding RNA TUG1 promotes cervical cancer cell proliferation and migration. Cancer Med 2017; 6: 471–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Niu Y, Ma F, Huang W et al Long non‐coding RNA TUG1 is involved in cell growth and chemoresistance of small cell lung cancer by regulating LIMK2b via EZH2. Mol Cancer 2017; 16: 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Ji TT, Huang X, Jin J, Pan SH, Zhuge XJ. Inhibition of long non‐coding RNA TUG1 on gastric cancer cell transference and invasion through regulating and controlling the expression of miR‐144/c‐Met axis. Asian Pac J Trop Med 2016; 9: 508–12. [DOI] [PubMed] [Google Scholar]
- 62. Zhang E, He X, Yin D et al Increased expression of long noncoding RNA TUG1 predicts a poor prognosis of gastric cancer and regulates cell proliferation by epigenetically silencing of p57. Cell Death Dis 2016; 7: e2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Huang MD, Chen WM, Qi FZ et al Long non‐coding RNA TUG1 is up‐regulated in hepatocellular carcinoma and promotes cell growth and apoptosis by epigenetically silencing of KLF2. Mol Cancer 2015; 14: 165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Dong R, Liu GB, Liu BH et al Targeting long non‐coding RNA‐TUG1 inhibits tumor growth and angiogenesis in hepatoblastoma. Cell Death Dis 2016; 7: e2278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Ma F, Wang SH, Cai Q et al Long non‐coding RNA TUG1 promotes cell proliferation and metastasis by negatively regulating miR‐300 in gallbladder carcinoma. Biomed Pharmacother 2017; 88: 863–9. [DOI] [PubMed] [Google Scholar]
- 66. Zhao XB, Ren GS. LncRNA Taurine‐Upregulated Gene 1 Promotes Cell Proliferation by Inhibiting MicroRNA‐9 in MCF‐7 Cells. J Breast Cancer 2016; 19: 349–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Li T, Liu Y, Xiao H, Xu G. Long non‐coding RNA TUG1 promotes cell proliferation and metastasis in human breast cancer. Breast Cancer (Tokyo, Japan) 2017; 24: 535–43. [DOI] [PubMed] [Google Scholar]
- 68. Sun J, Ding C, Yang Z et al The long non‐coding RNA TUG1 indicates a poor prognosis for colorectal cancer and promotes metastasis by affecting epithelial‐mesenchymal transition. J Transl Med 2016; 14: 42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Zhai HY, Sui MH, Yu X et al Overexpression of Long Non‐Coding RNA TUG1 Promotes Colon Cancer Progression. Med Sci Monit 2016; 22: 3281–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Wang L, Zhao Z, Feng W et al Long non‐coding RNA TUG1 promotes colorectal cancer metastasis via EMT pathway. Oncotarget 2016; 7: 51713–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Kuang D, Zhang X, Hua S, Dong W, Li Z. Long non‐coding RNA TUG1 regulates ovarian cancer proliferation and metastasis via affecting epithelial‐mesenchymal transition. Exp Mol Pathol 2016; 101: 267–73. [DOI] [PubMed] [Google Scholar]
- 72. Zhang M, Lu W, Huang Y et al Downregulation of the long noncoding RNA TUG1 inhibits the proliferation, migration, invasion and promotes apoptosis of renal cell carcinoma. J Mol Histol 2016; 47: 421–8. [DOI] [PubMed] [Google Scholar]
- 73. Iliev R, Kleinova R, Juracek J et al Overexpression of long non‐coding RNA TUG1 predicts poor prognosis and promotes cancer cell proliferation and migration in high‐grade muscle‐invasive bladder cancer. Tumour Biol 2016; 37: 13385–90. [DOI] [PubMed] [Google Scholar]
- 74. Xie CH, Cao YM, Huang Y et al Long non‐coding RNA TUG1 contributes to tumorigenesis of human osteosarcoma by sponging miR‐9‐5p and regulating POU2F1 expression. Tumour Biol 2016; 37: 15031–41. [DOI] [PubMed] [Google Scholar]
- 75. Wang Y, Yang T, Zhang Z et al Long non‐coding RNA TUG1 promotes migration and invasion by acting as a ceRNA of miR‐335‐5p in osteosarcoma cells. Cancer Sci 2017; 108: 859–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Lin PC, Huang HD, Chang CC et al Long noncoding RNA TUG1 is downregulated in non‐small cell lung cancer and can regulate CELF1 on binding to PRC2. BMC Cancer 2016; 16: 583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Du Z, Sun T, Hacisuleyman E et al Integrative analyses reveal a long noncoding RNA‐mediated sponge regulatory network in prostate cancer. Nat Commun 2016; 7: 10982. [DOI] [PMC free article] [PubMed] [Google Scholar]


