Abstract
Gastric cancer is one of the most common malignant tumors. Although improvement in chemotherapy has been achieved, the clinical prognosis of advanced gastric cancer remains poor. Therefore, it is increasingly important to predict the prognosis and determine whether patients should or should not receive neoadjuvant or adjuvant chemotherapy. Leucine‐rich α2‐glycoprotein‐1 (LRG1) is overexpressed during inflammation and is associated with various malignancies. In this study, we assessed LRG1 expression in cancer specimens and in the sera of patients with cancer to clarify the usefulness of LRG1 as a biomarker in gastric cancer. This study enrolled 239 (for immunohistochemical staining; IHC) and 184 (for ELISA) patients with gastric cancer. Results of IHC showed that LRG1 expression was significantly associated with histological type, lymphatic and venous invasion, tumor and node factors, and disease stage. Overall survival was significantly worse in the high LRG1 expression group than in the low LRG1 group (P = 0.0003). Cox multivariate analysis of overall survival revealed that LRG1 expression was an independent prognostic factor (P = 0.0258). Serum LRG1 was significantly higher in gastric cancer patients than in healthy volunteers, and increased as the pathological stage progressed. Furthermore, a significant correlation was revealed between serum LRG1 level and LRG1 expression with IHC (P < 0.0001). Inhibition of LRG1 significantly decreased cell proliferation in vitro (migratory and invasive capacity of gastric cancer cells). These results suggest that LRG1 expression in tumors and serum may be a useful prognostic marker in gastric cancer patients.
Keywords: Biomarker, gastric cancer, leucine‐rich α2‐glycoprotein‐1, prognosis, serum
Gastric cancer is the fifth most common malignancy and the third leading cause of cancer‐related deaths worldwide.1 The clinical prognosis of advanced gastric cancer is poor; therefore, multimodal treatments before and/or after operation are being developed.2, 3, 4 The identification of new suitable biomarkers for predicting prognosis is increasingly important to determine whether patients should or should not receive neoadjuvant or adjuvant therapy.5, 6
In 1977, leucine‐rich α2‐glycoprotein‐1 (LRG1) was the first identified member of the leucine‐rich repeat family in human serum.7 It is an approximately 50‐kDa secreted glycoprotein that contains eight repeating consensus sequences with a leucine‐rich motif.8 Leucine‐rich α2‐glycoprotein‐1 is involved in the immune response, and LRG1 expression is increased in hepatocytes in response to mediators of the acute phase as an inflammatory protein.9 The expression of LRG1 is elevated in the sera of patients with autoimmune diseases such as Crohn's disease, ulcerative colitis, and rheumatoid arthritis, and it is correlated with the disease status.10, 11 Furthermore, recent studies have reported that LRG1 can be used as a biomarker in several types of cancer such as ovarian, lung, biliary tract, and hepatocellular carcinoma.12, 13, 14, 15, 16 Serum LRG1 is also detected in patients with malignancies and associated with progression of malignancies.17 However, no reports exist on the clinical significance of LRG1 expression in gastric cancer.
In the present study, we aimed to clarify the association between LRG1 expression in tissue samples and clinicopathological factors in patients with gastric cancer and to elucidate the mechanisms of tumor progression or metastasis through LRG1.
Materials and Methods
Clinical tissue samples
Between January 2009 and December 2011, 239 tissue samples were collected from patients who underwent curative gastrectomy at the Department of Gastroenterological Surgery at Osaka University Hospital (Osaka, Japan). Patients who received neoadjuvant chemotherapy before surgery, underwent R1 or R2 resection, or died within 30 days of surgery were excluded. The pathological tumor stage was evaluated using the third English edition of the Japanese classification of gastric carcinoma, which was established by the Japanese Gastric Cancer Association.18 This study was approved by the Ethics Committee of Osaka University Hospital. All patients provided written consent (No. 08226‐6).
Immunohistochemical staining
We carried out immunohistochemical staining (IHC) as previously described.19 Rabbit anti‐LRG1 antibody (1:500; HPA001888; Atlas Antibodies, Stockholm, Sweden) was used for IHC. As LRG1 expression in tumor cells was primarily localized in the cytoplasm and the pattern of staining was homogeneous and diffuse, LRG1 expression was evaluated by the intensity of stained cancer samples as previously reported.17 Liver tissues were used as the positive control. The stain intensity was scored from 0 to 3 by reference to previous report17 with a modification: score 0 = none, no stained cancer cells in the section; score 1 = weak, ≤10% of cells immunoreactive to LRG1; score 2 = moderate, >10% of cells immunoreactive to LRG1; and score 3 = strong, >10% of cells immunoreactive to LRG1 and also found both in cytoplasm and nuclei. Expression was “low” for a score of 0–1 and “high” for a score of 2–3. Evaluation was made by two double‐blinded, independent observers who were unaware of clinicopathological data and outcome.
Enzyme‐linked immunosorbent assay
One hundred and eighty‐four preoperative blood samples taken within 30 days before surgery from patients with gastric cancer who had not received neoadjuvant therapy before surgery, had undergone gastrectomy between April 2012 and September 2015, and had no history of an inflammatory disease, and 53 serum samples from healthy volunteers (HVs) were obtained at Osaka University Hospital. Samples were measured in duplicate using a sandwich ELISA for the detection of human LRG1 as previously described20, 21 and Helicobacter pylori.
Nunc‐Immuno MicroWell 96‐well solid plates (Maxisorp, Nunc #439454; Sigma‐Aldrich, St. Louis, MO, USA) were coated with 100 μL (0.1 μg/mL) anti‐huLRG1 human mAb (huLRB0091; Chugai Pharmaceutical, Tokyo, Japan) per well overnight at 4°C. Plates were washed three times with 200 μL PBS + 0.05% Tween (P3563; Sigma‐Aldrich), and blocked by blocking buffer, which included 5 g BSA + BLOCK ACE Powder (UK‐B80; DS Pharma Biomedical, Osaka, Japan) + TBS‐T (T9039; Sigma‐Aldrich) for 1 h at room temperature. Plates were washed three times with 200 μL PBS + 0.05% Tween, with 100 μL (1.0 μg/mL) anti‐huLRG1 rabbit mAb (rbLRB0048; Chugai Pharmaceutical) added per well by shaking for 1 h at room temperature. Plates were washed three times with 200 μL PBS + 0.05% Tween, with 100 μL mouse anti‐rabbit IgG monoclonal–HRP (1:5000, 4090‐05; Southern Biotech, Birmingham, AL, USA) added by shaking for 1 h at room temperature. Plates were washed three times with 200 μL PBS + 0.05% Tween, with 100 μL 3,3′,5,5′‐tetramethylbenzidine (TMBW‐1000‐01; SurModics, Eden Prairie, MN, USA) added per well by shaking for 8 min at room temperature with blocking out light. The reaction was stopped by addition of 1N‐Sulfuric acid and absorbance at 450 nm was determined using a microplate reader Model 680 (Bio‐Rad Laboratories, Hercules, CA, USA).
An ELISA kit (I‐DQ77, E‐plate Eiken H. pylori antibody; Eiken Chemical, Tokyo, Japan) was used to measure IgG antibody against H. pylori using the recommended antibody titer cut‐off point (>10 U/mL) that indicates H. pylori infection according to the manufacturer's instructions.22, 23
Cell lines and culture conditions
The human gastric cancer cell line AGS was obtained from ATCC (Manassas, VA, USA). Human gastric cancer cell lines MKN45, NUGC3, and KATOIII and a human hepatic cancer cell line (HepG2, the positive control) were obtained from the Japanese Collection of Research Bioresources (Osaka, Japan). Each cell was cultured as follows: AGS in Ham's F‐12 medium (Nacalai Tesque, Kyoto, Japan); MKN45 and NUGC3 in RPMI‐1640 medium (Nacalai Tesque), KATOIII in RPMI and Eagle's minimum essential medium (1:1; Wako Pure Chemical Industries, Osaka, Japan), and HepG2 in DMEM (Wako Pure Chemical Industries), supplemented with 10% FBS (Thermo Fisher Scientific, Waltham, MA, USA). These cell lines were incubated in 5% CO2 at 37°C.
Small interfering RNA design
Small interfering RNA against LRG1 (siLRG1; L‐015179‐01‐0010; ON‐TARGETplus SMARTpool human LRG1 siRNA, a mixture of four siRNA provided as a single reagent) and non‐targeting siRNA (negative control, D‐001810‐01‐20; ON‐TARGETplus Nontargeting siRNA#1) were purchased from Dharmacon (Lafayette, CO, USA). The siRNA oligonucleotides (20 nM) were transfected into cells using Lipofectamine RNAiMAX reagent (Life Technologies, Carlsbad, CA, USA), based on the manufacturer's protocol.
Quantitative real‐time RT‐PCR
The primer sequences were customized by Sigma‐Aldrich, as follows: LRG1 (accession no. NM_052972) and GAPDH (accession no. NM_002046, NM_001256799, NM_001289745, NM_001289746) as the internal control. Quantitative real‐time RT‐PCR (qRT‐PCR) was carried out with FastStart DNA Master SYBR Green I (Roche Diagnostics, Basel, Switzerland) and the LightCycler System (Roche Diagnostics).
Western blot analysis
Proteins were resolved with SDS‐PAGE gels (Bio‐Rad Laboratories), transferred onto PVDF membranes (Millipore, Bedford, MA, USA), and incubated with a rabbit anti‐LRG1 antibody (1:1000, HPA001888; Atlas Antibodies). After incubation with secondary antibodies, signals were detected with the ECL Prime Western Blotting Detection Reagent (GE Healthcare, Little Chalfont, UK).
Cell proliferation assay
Cells were seeded onto 96‐well plates at 3 × 103 cells/100 μL/well for 24 h, then transfected with siRNA. After transfection, cell viability was quantified every 24 h by the WST‐8 assay using the CCK‐8 (Dojindo Laboratories, Kumamoto, Japan), based on the manufacturer's protocol.
Wound‐healing assay
The wound‐healing assay was carried out based on previous reports.24, 25 After scratching, the cells were cultured in a medium supplemented with 0.5% FBS for 48 h to allow wound healing. Forty‐eight hours after wound treatment, the degree of cell migration was analyzed as a percentage of wound confluence with ImageJ 1.50i (National Institutes of Health, Bethesda, MD, USA). The mean of five fields was calculated as the sample value.
Invasion assay
The cell invasion assay was carried out using Transwell inserts with 8‐μm pores (BD Biosciences, San Jose, CA, USA), based on the manufacturer's protocol and previous reports.26, 27 Cells were seeded into inserts for 24‐well plates at 4 × 104 cells per insert in serum‐free medium and then transferred to wells filled with the culture medium containing 10% FBS. After 24 h incubation, non‐invading cells on the top of the membrane were removed by cotton swabs. The invading cells were counted using a microscope in five random visual fields (×200 magnification).
Zymography
Matrix metalloproteinase‐2, pro‐MMP‐2, and pro‐MMP‐9 activities of tumor cell were measured with a gelatin‐zymography kit (Cosmo Bio, Sapporo, Japan) according to the manufacturer's instructions and previous reports.28 In brief, 10 μL (each sample, 5 μL; sample buffer, 5 μL) was loaded for electrophoresis with DPE‐1020 (Cosmo Bio). The gels were washed and incubated for 24 h in incubation buffer at 37°C. After staining, gels were scanned with an Epson GT‐X970 (Suwa, Nagano, Japan).
Statistical analysis
Associations between LRG1 expression and clinicopathological factors were analyzed using the χ2‐test for categorical variables and the Mann–Whitney U‐test for continuous variables. Relapse‐free survival (RFS) was the period from the date of surgery to the date of detection of the first recurrence or death from any cause. Overall survival (OS) was the period from the date of surgery to the date of death from any cause. Survival was estimated using the Kaplan–Meier method, and compared using the log–rank test. Multivariate Cox regression analysis was carried out to adjust for potential confounding factors. A statistically significant difference was indicated by P < 0.05. All reported P‐values were two‐tailed. All statistical analyses were carried out with JMP Pro 11.2.1 software (SAS Institute, Cary, NC, USA).
Results
Effect of LRG1 expression on the prognosis of patients with gastric cancer
We examined LRG1 expression in gastric cancer tissues using IHC. The expression in cancer cells was primarily localized in the cytoplasm (Fig. 1a–d). Among 239 patients with gastric cancer, 134 (56%) patients had low LRG1 expression and 105 (44%) patients had high LRG1 expression. There were significant differences between patients with low and high LRG1 expression in terms of histological type, lymphatic and venous invasion, tumor (T) and node (N) factors, and disease stage (Table 1). The mean follow‐up duration for all patients in this study was 45.8 ± 16.1 months. The high LRG1 expression group had significantly worse OS, compared to the low LRG1 expression group (hazard ratio [HR], 3.71; 95% confidence interval [CI], 1.78–8.74; log–rank, P = 0.0003) (Fig. 1e). The 5‐year OS rate was 73.8% in the high expression group and 85.1% in the low expression group. The high LRG1 expression group also had significantly worse RFS, compared to the low LRG1 expression group (HR, 5.22; 95% CI, 2.82–10.39; log–rank, P < 0.0001) (Fig. 1f). The 5‐year RFS rate was 59.8% in the high LRG1 expression group and 89.0% in the low LRG1 expression group.
Figure 1.
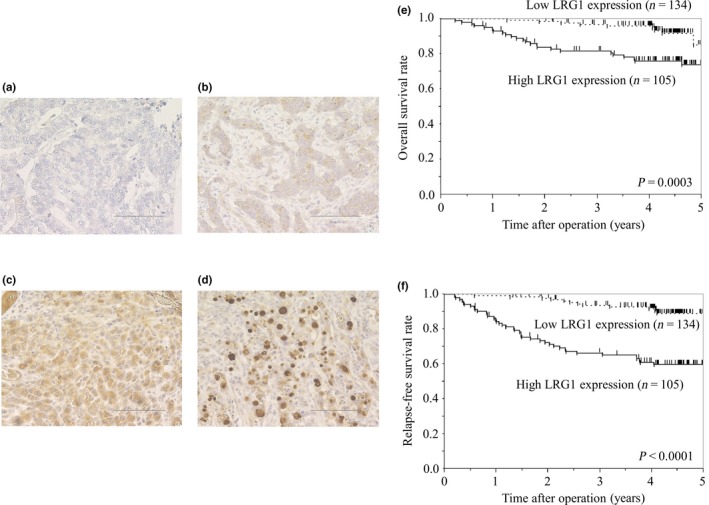
(a–d) Leucine‐rich α2‐glycoprotein‐1 (LRG1) immunohistochemical staining in gastric cancer specimens at ×200 magnification. Staining results are shown at: score 0, no stained cancer cells in the section (a); score 1, ≤10% of cells immunoreactive to LRG1 (b); score 2, >10% of cells immunoreactive to LRG1 (c); and score 3, >10% of cells immunoreactive to LRG1 and also found both in cytoplasm and nuclei (d). Bar = 100 μm. (e,f) Kaplan–Meier analyses of overall survival (e) and relapse‐free survival (f), based on LRG1 expression.
Table 1.
Correlation between Leucine‐rich α2‐glycoprotein‐1 (LRG1) expression with immunohistochemical staining (IHC) and clinicopathological features in 239 patients with gastric cancer
| LRG1 expression with IHC | |||
|---|---|---|---|
| Low (n = 134) | High (n = 105) | P‐value | |
| Age, years, median (range) | 68 (31–87) | 68 (40–89) | 0.6589 |
| Sex | 0.5106 | ||
| Male | 102 | 76 | |
| Female | 32 | 29 | |
| Location | 0.8966 | ||
| Upper | 36 | 29 | |
| Middle/lower | 98 | 76 | |
| Histological type | 0.0044 | ||
| Differentiated type | 81 | 44 | |
| Undifferentiated type | 53 | 61 | |
| Venous invasion | 0.0012 | ||
| Yes | 16 | 30 | |
| No | 118 | 75 | |
| Lymphatic invasion | <0.0001 | ||
| Yes | 42 | 68 | |
| No | 92 | 37 | |
| pT | <0.0001 | ||
| T1 | 100 | 29 | |
| T2 | 23 | 16 | |
| T3 | 10 | 48 | |
| T4 | 1 | 12 | |
| pN | <0.0001 | ||
| N0 | 113 | 64 | |
| N1–3 | 21 | 41 | |
| Pathological disease stage | <0.0001 | ||
| I | 113 | 35 | |
| II | 14 | 43 | |
| III | 7 | 27 | |
| IV | 0 | 0 | |
Multivariate Cox regression analysis revealed that high LRG1 expression was a statistically significant independent prognostic factor of poor OS, along with lymphatic invasion and disease stage III–IV (Table 2). The adjusted HR for OS in the high LRG1 expression group was 2.37 (95% CI, 1.09–5.59; P = 0.0279). Subgroup analysis by disease stage showed that, compared to the low LRG1 expression group, the high LRG1 expression group had significantly worse OS and RFS in stage II–III disease (log–rank, P = 0.0389 and P = 0.0019, respectively), but not in stage I disease (log–rank, P = 0.7622 and P = 0.9945, respectively) (Fig. 2).
Table 2.
Univariate and multivariate Cox regression analyses for overall survival and relapse‐free survival in 239 patients with gastric cancer
| Variable | Univariate analysis | P‐value | Multivariate analysis | P‐value | |||
|---|---|---|---|---|---|---|---|
| HR | 95% CI | HR | 95% CI | ||||
| Overall survival | |||||||
| Age, years | ≥65 vs <65 | 1.43 | 0.70–3.14 | 0.3371 | |||
| Sex | Male versus female | 0.89 | 0.43–2.02 | 0.7665 | |||
| Location | Upper versus middle/lower | 1.95 | 0.96–3.87 | 0.0648 | |||
| Histological type | Undifferentiated versus differentiated | 1.62 | 0.82–3.31 | 0.1656 | |||
| Venous invasion | Yes versus no | 1.61 | 0.71–3.35 | 0.2443 | |||
| Lymphatic invasion | Yes versus no | 4.19 | 1.97–9.94 | 0.0001 | 2.12 | 0.86–5.54 | 0.1014 |
| Pathological disease stage | III–IV vs I–II | 5.74 | 2.82–11.40 | <0.0001 | 3.12 | 1.42–6.95 | 0.0051 |
| LRG1 expression | High versus low | 3.71 | 1.78–8.44 | 0.0003 | 2.37 | 1.09–5.59 | 0.0279 |
| Relapse‐free survival | |||||||
| Age, years | ≥65 vs <65 | 1.38 | 0.78–2.55 | 0.2757 | |||
| Sex | Male versus female | 1.04 | 0.57–2.02 | 0.9131 | |||
| Location | Upper versus middle/low | 1.68 | 0.95–2.92 | 0.0753 | |||
| Histological type | Undifferentiated versus differentiated | 1.37 | 0.79–2.38 | 0.2574 | |||
| Venous invasion | Yes versus no | 2.35 | 1.29–4.14 | 0.0064 | 0.74 | 0.38–1.38 | 0.3409 |
| Lymphatic invasion | Yes versus no | 5.44 | 2.90–11.13 | <0.0001 | 2.98 | 1.41–6.60 | 0.0042 |
| Pathological disease stage | III–IV vs I–II | 6.00 | 3.39–10.39 | <0.0001 | 2.88 | 1.53–5.44 | 0.0012 |
| LRG1 expression | High versus low | 5.22 | 2.82–10.39 | <0.0001 | 3.28 | 1.72–6.71 | 0.0002 |
CI, confidence interval; HR, hazard ratio; LRG1, leucine‐rich α‐2‐glycoprotein‐1.
Figure 2.
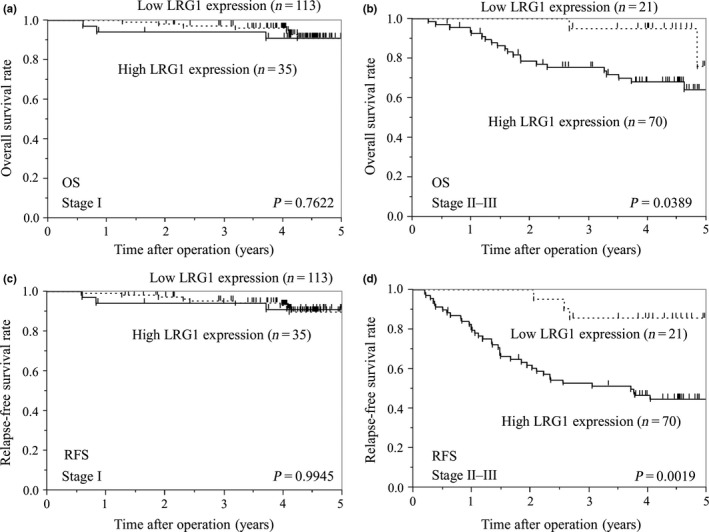
Kaplan–Meier analyses of overall survival (OS) (a,b) and relapse‐free survival (RFS) (c,d) between patients with stage I and stage II–III gastric cancer, based on leucine‐rich α2‐glycoprotein‐1 (LRG1) expression.
Serum LRG1 levels are significantly associated with LRG1 expression in tumors
Serum LRG1 was detected in the peripheral blood with ELISA. Serum LRG1 levels were significantly elevated in patients with gastric cancer (18.65 ± 9.42 μg/mL), compared to in HVs (9.86 ± 2.18 μg/mL) (P < 0.0001) (Fig. 3a). The mean LRG1 concentration increased with the progression of the pathological stage (16.08 ± 6.08, 18.13 ± 7.62, 23.16 ± 12.79, 23.28 ± 10.89 μg/mL in stage I, II, III, and IV, respectively). A significant difference existed between stage I and stage III disease and between stage I and stage IV (P < 0.01) (Fig. 3b).
Figure 3.
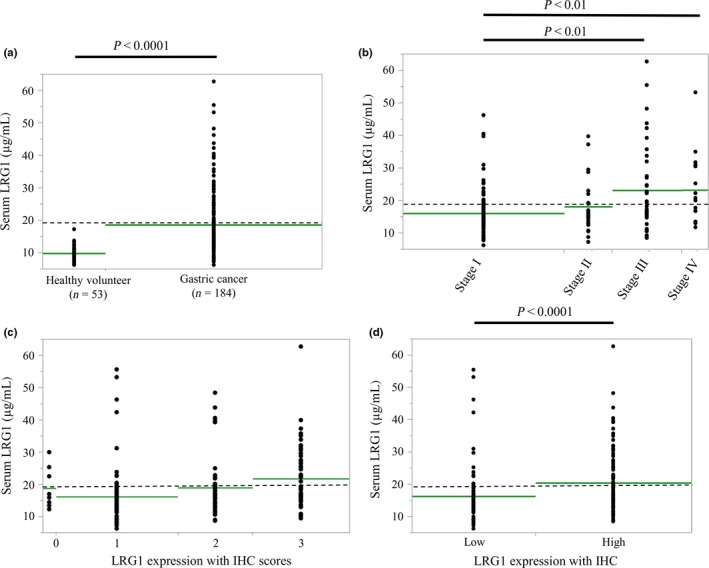
Detection of serum leucine‐rich α2‐glycoprotein‐1 (LRG1) using ELISA. Serum LRG1 concentrations in 184 patients with gastric cancer and 53 healthy volunteers. Dotted lines show the cut‐off value for LRG1 (19.1 μg/mL). (a) Mean LRG1 concentration for patients with gastric cancer was significantly higher than for healthy volunteers (P < 0.0001). (b) Mean LRG1 concentration increased with the progression of pathological stage. A significant difference was observed between stage I and stage III and between stage I and stage IV disease (P < 0.01). (c) Mean serum LRG1 concentration increased with the progression of the immunohistochemical staining (IHC) scores. (d) IHC results indicate a significant correlation between the serum LRG1 level and LRG1 expression (P < 0.0001).
Among the same patients, we examined the correlation between serum LRG1 and LRG1 expression with IHC. The mean serum LRG1 concentration increased with the progression of the IHC scores (16.29 ± 6.38, 13.55 ± 9.20, 16.61 ± 9.31, 18.84 ± 9.39 μg/mL in score 0, 1, 2, and 3, respectively) (Fig. 3c). A significant correlation was revealed between serum LRG1 level and LRG1 expression with IHC (16.35 ± 8.97 and 20.46 ± 9.41 μg/mL in low and high expression with IHC, respectively) (P < 0.0001) (Fig. 3d).
We examined the correlation between serum LRG1 and clinicopathological factors (Table 3). The cut‐off value for LRG1 was 19.1 μg/mL, based on the upper quartile. According to the cut‐off (19.1 μg/mL), the rates of high serum LRG1 was 0% in HVs and 32% in patients with gastric cancer. Moreover, in patients with gastric cancer, the rates of high serum LRG1 was 21% (stage I), 31% (stage II), 50% (stage III), and 56% (stage IV). The high serum LRG1 group included more patients with older age (P < 0.0001), venous invasion (P = 0.0093), lymphatic invasion (P = 0.0009), T factor (P = 0.0038), N factor (P < 0.0001), disease stage (P = 0.001), and C‐reactive protein (P < 0.0001). There was no significant correlation between LRG1 and H. pylori.
Table 3.
Correlation between serum leucine‐rich α2‐glycoprotein‐1 (LRG1) and clinicopathological factors in 184 patients with gastric cancer
| Serum LRG1 | P‐value | ||
|---|---|---|---|
| Negative (<19.1 μg/mL) (n = 125) | Positive (≥19.1 μg/mL) (n = 59) | ||
| Age, years, median (range) | 65 (31–89) | 72 (37–87) | <0.0001 |
| Sex | 0.3517 | ||
| Male | 91 | 39 | |
| Female | 34 | 20 | |
| Location | 0.5665 | ||
| Upper | 36 | 19 | |
| Middle/lower | 89 | 40 | |
| Histological type | 0.3209 | ||
| Differentiated type | 58 | 32 | |
| Undifferentiated type | 67 | 27 | |
| Venous invasion | 0.0093 | ||
| Yes | 37 | 30 | |
| No | 83 | 29 | |
| Lymphatic invasion | 0.0009 | ||
| Yes | 56 | 43 | |
| No | 64 | 16 | |
| pT | 0.0038 | ||
| T1 | 68 | 16 | |
| T2 | 13 | 6 | |
| T3 | 21 | 18 | |
| T4 | 23 | 19 | |
| pN | <0.0001 | ||
| N0 | 89 | 24 | |
| N1–3 | 36 | 35 | |
| Pathological disease stage | 0.0010 | ||
| I | 77 | 20 | |
| II | 20 | 9 | |
| III | 21 | 21 | |
| IV | 7 | 9 | |
| LRG1 expression with IHC | 0.0014 | ||
| Score 0 | 5 | 3 | |
| Score 1 | 61 | 12 | |
| Score 2 | 29 | 16 | |
| Score 3 | 30 | 28 | |
| Helicobacter pylori | 0.0800 | ||
| Yes | 65 | 32 | |
| No | 60 | 27 | |
| CRP, median (range) | 0.04 (0.04–1.64) | 0.12 (0.04–5.11) | <0.0001 |
CRP, C‐reactive protein; IHC, immunohistochemical staining.
Expression of LRG1 in gastric cancer and other positive control cell lines
We used qRT‐PCR analyses to compare the mRNA expression of LRG1 in four gastric cancer cell lines and HepG2 (the positive control). The expression of LRG1 mRNA was the highest in AGS and second‐highest in MKN45 (Fig. 4). We accordingly selected AGS and MKN45 cells for analysis in the LRG1 inhibition assay.
Figure 4.
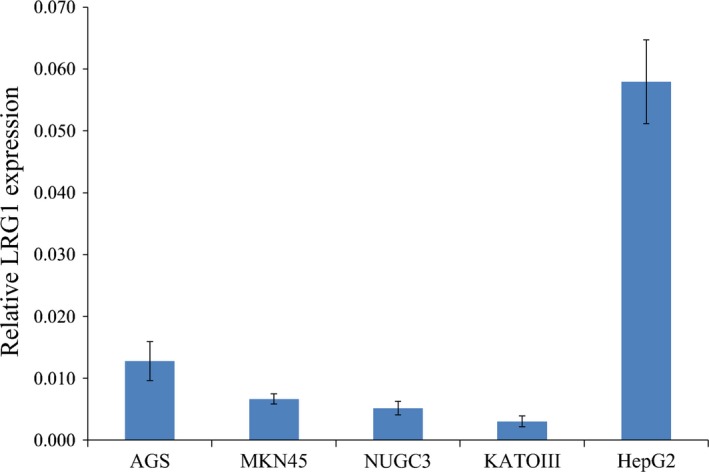
Relative leucine‐rich α2‐glycoprotein‐1 (LRG1) mRNA expression in gastric cancer cell lines AGS, MKN45, NUGC3, and KATOIII and the positive control cell line, HepG2, based on quantitative real‐time RT‐PCR.
Inhibition of LRG1 suppresses gastric cancer cell growth and invasion
The findings of qRT‐PCR and Western blot analyses showed that siLRG1 significantly reduced the expression of LRG1 mRNA and protein, compared to the negative control cells in AGS and MKN45 cell lines (Fig. 5a). We examined the effects of LRG1 inhibition on the proliferative activity of gastric cancer cell lines. Cell growth was significantly inhibited by siLRG1, compared to the negative control siRNA after transfection (Fig. 5b). Moreover, we examined the effect of LRG1 inhibition on the migratory and invasive capacity with wound‐healing and Transwell assays because cell migration and invasion are the initial steps of metastasis. Inhibition of LRG1 significantly reduced the migratory and invasive capacity of gastric cancer cell lines, compared to the negative control (Figs. 5c,S1). Zymography showed that pro‐MMP‐2 and pro‐MMP‐9 activity in AGS cells with siLRG1 decreased compared with non‐targeting siRNA (Fig. S1). However, the activity did not change in MKN45 cells with siLRG1 compared with non‐targeting siRNA.
Figure 5.
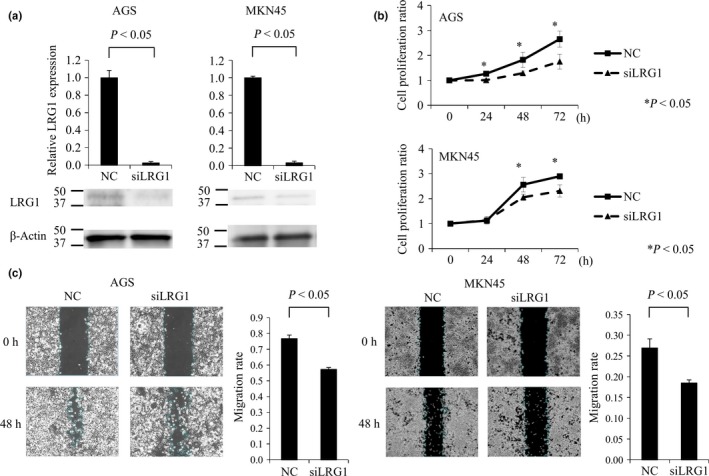
(a) Quantitative real‐time RT‐PCR and Western blot assay show that siRNA against leucine‐rich α2‐glycoprotein‐1 LRG1 (siLRG1) significantly reduced the expression of LRG1 mRNA and protein, compared to the negative control (NC), in AGS and MKN45 gastric cancer cells. (b) Cell proliferation assay of gastric cancer cells (AGS and MKN45) compared to siLRG1 and NC. Gastric cancer cells transfected with siLRG1 have significantly decreased proliferation compared to NC. Five specimens were used for each experiment, and siLRG1 was compared with NC at each time point (0, 24, 48, and 72 h). (c) Wound‐healing assay for gastric cancer cells (AGS and MKN45), compared to siLRG1 and NC. Wound‐healing assays were used to detect motility in AGS cells transfected with siLRG1.
Discussion
In this study, we used IHC analysis of gastric cancer specimens to investigate the association between the expression of LRG1 and clinicopathological characteristics. We revealed that the high LRG1 expression group had a significantly worse OS and RFS, compared to the low LRG1 expression group, and that LRG1 was a significant independent prognostic factor of OS and RFS. Moreover, subgroup analysis showed that there was no significant difference between LRG1 expression in patients with early gastric cancer (stage I) for whom surgery may be curative. However, the high LRG1 expression group had significantly worse OS and RFS for patients with advanced gastric cancer (stage II–III) (Fig. 2). Although multimodal therapy including surgery and chemotherapy is usually undertaken for these patients, there are some who have a poor prognosis.29, 30, 31 Therefore, it may be possible to select patients who should receive more intensive therapy before or after surgery, based on the evaluation of LRG1 expression by surgical samples.
Some studies have reported that the serum LRG1 levels in patients with malignancies and the LRG1 expression in tumor cells are elevated.12, 13, 14, 15, 16 We previously reported a high level of serum LRG1 in patients with pancreatic cancer, and that the serum LRG1 level was associated with the progression of tumor stage.17 However, we were not able to properly analyze the correlation between serum LRG1 and LRG1 expression in IHC with a sufficient number of samples.
With regard to gastric cancer, Uen et al. reported that LRG1 could be detected by analyzing the serum of gastric cancer patients using the proteome approach with chromatography.32 However, in that report, LRG1 was only one candidate molecule and they did not show sufficient clinical significance. We undertook the present study to compare serum LRG1 levels in patients with gastric cancer and in HVs, and to examine the association between serum LRG1 levels and clinicopathological factors in patients with gastric cancer using ELISA. First, we found that serum LRG1 levels are markedly elevated in patients with gastric cancer compared with HVs. Furthermore, the elevation of serum LRG1 is associated with the progression of the pathological stage. These findings support that LRG1 may be useful as a diagnostic marker for advanced gastric cancer.
Moreover, we examined additional immunohistochemical factors in patients with gastric cancer (these same patients' samples had been examined by ELISA) to investigate the association between serum LRG1 levels and LRG1 expression using IHC. For the first time, we successfully revealed that serum LRG1 levels were associated with LRG1 expression, based on IHC (Fig. 3d, Table 3). Therefore, serum LRG1 may be a useful biomarker in patients with gastric cancer because the expression of LRG1 with IHC was associated with prognosis, as previously indicated. Pretreatment serum LRG1 levels in patients with gastric cancer could help in decisions to undertake intensive treatment before surgery.
The clinicopathological data indicated that high LRG1 expression in tumors and in serum may be associated with lymphatic and venous invasion. Leucine‐rich α2‐glycoprotein‐1 has been reported to be correlated with inflammation9 and our results showed that serum LRG1 was associated with C‐reactive protein, an inflammatory marker. Moreover, although we examined the correlation between LRG1 and H. pylori, which has reported to induce gastritis,32, 33 we could not confirm it in this study. To clarify the relationship between LRG1 and motility in gastric cancer cells, we analyzed the inhibition of LRG1 gastric cancer cells in vitro. We found that the inhibition of LRG1 suppressed the proliferation, migration, and invasion in gastric cancer cells. We speculated that suppression of migratory and invasive activities in LRG1‐suppressed cells reflected the results indicating that patients with high LRG1 expression showed venous and lymphatic invasion. In this report, we also showed the suppression of pro‐MMP‐2 and pro‐MMP‐9 by silencing LRG1. One of the major implications of MMPs in cancer progression is their role in ECM degradation, which allows cancer cells to migrate out of the primary tumor to form metastases. It is possible that LRG1 might be related to motility in cancer cells.
The mechanism of LRG1 in tumor cells remains largely unknown, although a recent study reported that LRG1 promoted angiogenesis by modulating endothelial transforming growth factor‐β signaling.34 Therefore, we speculated that higher LRG1 expression in patients with malignancies could induce poorer prognosis because LRG1 in tumor cells stimulates the endothelial transforming growth factor‐β signaling pathway. This stimulation may promote angiogenesis and increase the activities of tumor migration and invasion, which result in local progression. Thus, LRG1 has great potential to be a new therapeutic target through suppressing angiogenesis in various cancers.
This study has some limitations. We were unable to reveal directly that serum LRG1 was associated with prognosis because the follow‐up time of these patients was not sufficient. Thus, we have continued to collect serum and will prospectively investigate the usefulness of serum LRG1 as a biomarker. In addition, as we only showed the roles of LRG1 in proliferation, inhibitory migration, and invasion in this report, further consideration will be needed to yield any findings about the detailed mechanisms of LRG1. In the future, it may be necessary to determine these mechanisms.
In conclusion, we found that patients with gastric cancer with high LRG1 expression had a poorer prognosis. Moreover, we showed that serum LRG1 could be a novel biomarker for gastric cancer.
Disclosure Statement
The authors have no conflict of interest.
Supporting information
Fig. S1. (a) Quantitative Transwell assay results of AGS gastric cancer cells transfected with siRNA against leucine‐rich α2‐glycoprotein‐1 LRG1 (siLRG1) and non‐targeting siRNA (negative control [NC]). (b) MMP‐2, pro‐MMP‐2, and pro‐MMP‐9 activities of AGS with siLRG1 and non‐targeting siRNA (NC).
Cancer Sci 108 (2017) 2052–2060
Funding information
None declared.
References
- 1. Ferlay J, Soerjomataram I, Dikshit R et al Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer 2015; 136: E359–86. [DOI] [PubMed] [Google Scholar]
- 2. Yoshikawa T, Rino Y, Yukawa N et al Neoadjuvant chemotherapy for gastric cancer in Japan: a standing position by comparing with adjuvant chemotherapy. Surg Today 2014; 44: 11–21. [DOI] [PubMed] [Google Scholar]
- 3. Honma Y, Yamada Y, Terazawa T et al Feasibility of neoadjuvant S‐1 and oxaliplatin followed by surgery for resectable advanced gastric adenocarcinoma. Surg Today 2016; 46: 1076–82. [DOI] [PubMed] [Google Scholar]
- 4. Aizawa M, Nashimoto A, Yabusaki H et al The clinical significance of potentially curative resection for gastric cancer following the clearance of free cancer cells in the peritoneal cavity by induction chemotherapy. Surg Today 2015; 45: 611–7. [DOI] [PubMed] [Google Scholar]
- 5. Shinohara S, Korenaga D, Edagawa A et al Significant prognostic factors in patients with Stage IV gastric cancer with special reference to the curability of surgery. Surg Today 2013; 43: 40–7. [DOI] [PubMed] [Google Scholar]
- 6. Suzuki T, Shimada H, Nanami T et al Hyperfibrinogenemia is associated with inflammatory mediators and poor prognosis in patients with gastric cancer. Surg Today 2016; 46: 1394–401. [DOI] [PubMed] [Google Scholar]
- 7. Haupt H, Baudner S. Isolation and characterization of an unknown, leucine‐rich 3.1‐S‐alpha2‐glycoprotein from human serum. Hoppe Seylers Z Physiol Chem 1977; 358: 639–46. [PubMed] [Google Scholar]
- 8. Takahashi N, Takahashi Y, Putnam FW. Periodicity of leucine and tandem repetition of a 24‐amino acid segment in the primary structure of leucine‐rich alpha 2‐glycoprotein of human serum. Proc Natl Acad Sci USA 1985; 82: 1906–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Shirai R, Hirano F, Ohkura N et al Upregulation of the expression of leucine‐rich alpha(2)‐glycoprotein in hepatocytes by the mediators of acute‐phase response. Biochem Biophys Res Commun 2009; 382: 776–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Serada S, Fujimoto M, Ogata A et al iTRAQ‐based proteomic identification of leucine‐rich alpha‐2 glycoprotein as a novel inflammatory biomarker in autoimmune diseases. Ann Rheum Dis 2010; 69: 770–4. [DOI] [PubMed] [Google Scholar]
- 11. Serada S, Fujimoto M, Terabe F et al Serum leucine‐rich alpha‐2 glycoprotein is a disease activity biomarker inulcerative colitis. Inflamm Bowel Dis 2012; 18: 2169–79. [DOI] [PubMed] [Google Scholar]
- 12. Andersen JD, Boylan K, Jemmerson R et al Leucine‐rich alpha‐2‐glycoprotein‐1 is upregulated in sera and tumors of ovarian cancer patients. J Ovarian Res 2010; 3: 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Okano T, Kondo T, Kakisaka T et al Plasma proteomics of lung cancer by a linkage of multi‐dimensional liquid chromatography and two‐dimensional difference gel electrophoresis. Proteomics 2006; 6: 3938–48. [DOI] [PubMed] [Google Scholar]
- 14. Sandanayake NS, Sinclair J, Andreola F et al A combination of serum leucine‐rich α‐2‐glycoprotein‐1, CA19‐9 and interleukin‐6 differentiate biliary tract cancer from benign biliary strictures. Br J Cancer 2011; 105: 1370–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Sarvari J, Mojtahedi Z, Kuramitsu Y et al Differential expression of haptoglobin isoforms in chronic active hepatitis, cirrhosis and HCC related to HBV infection. Oncol Lett 2011; 2: 871–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ladd JJ, Busald T, Johnson MM et al Increased plasma levels of the APC‐interacting protein MAPRE1, LRG1, and IGFBP2 preceding a diagnosis of colorectal cancer in women. Cancer Prev Res (Phila) 2012; 5: 655–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Furukawa K, Kawamoto K, Eguchi H et al Clinicopathological significance of leucine‐rich α2‐glycoprotein‐1 in sera of patients with pancreatic cancer. Pancreas 2015; 44: 93–8. [DOI] [PubMed] [Google Scholar]
- 18. Sano T, Aiko T. New Japanese classifications and treatment guidelines for gastric cancer: revision concepts and major revised points. Gastric Cancer 2011; 14: 97–100. [DOI] [PubMed] [Google Scholar]
- 19. Yamasaki M, Makino T, Masuzawa T et al Role of multidrug resistance protein 2 (MRP2) in chemoresistance and clinicaloutcome in oesophageal squamous cell carcinoma. Br J Cancer 2011; 104: 707–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Fujimoto M, Serada S, Suzuki K et al Leucine‐rich α2 ‐glycoprotein as a potential biomarker for joint inflammation during anti‐interleukin‐6 biologic therapy in rheumatoid arthritis. Arthritis Rheumatol 2015; 67: 2056–60. [DOI] [PubMed] [Google Scholar]
- 21. Honda H, Fujimoto M, Miyamoto S et al Sputum leucine‐rich alpha‐2 glycoprotein as a marker of airway inflammation in asthma. PLoS One 2016; 11: e0162672 https://doi.org/10.1371/journal.pone.0162672. eCollection 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Nakayama Y, Lin Y, Hongo M, Hidaka H, Kikuchi S. Helicobacter pylori infection and its related factors in junior high school students in Nagano Prefecture, Japan. Helicobacter 2017; 22 https://doi.org/10.1111/hel.12363. Epub 2016 Oct 27. [DOI] [PubMed] [Google Scholar]
- 23. Ueda J, Okuda M, Nishiyama T, Lin Y, Fukuda Y, Kikuchi S. Diagnostic accuracy of the E‐plate serum antibody test kit in detecting Helicobacter pylori infection among Japanese children. J Epidemiol 2014; 24: 47–51. Epub 2013 Nov 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Bernabé‐García Á, Armero‐Barranco D, Liarte S, Ruzafa‐Martínez M, Ramos‐Morcillo AJ, Nicolás FJ. Oleanolic acid induces migration in Mv1Lu and MDA‐MB‐231 epithelial cells involving EGF receptor and MAP kinases activation. PLoS One 2017; 12: e0172574 https://doi.org/10.1371/journal.pone.0172574. eCollection 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Pan Z, Mao W, Bao Y, Zhang M, Su X, Xu X. The long noncoding RNA CASC9 regulates migration and invasion in esophageal cancer. Cancer Med 2016; 5: 2442–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Mokutani Y, Uemura M, Munakata K et al Down‐regulation of microRNA‐132 is associated with poor prognosis of colorectal cancer. Ann Surg Oncol 2016; 23: 599–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wang Z, Lin L, Thomas DG et al The role of Dickkopf‐3 overexpression in esophageal adenocarcinoma. J Thorac Cardiovasc Surg 2015; 150: 377–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Miyazaki K, Ohta Y, Nagai M et al Disruption of neurovascular unit prior to motor neuron degeneration in amyotrophic lateral sclerosis. J Neurosci Res 2011; 89: 718–28. [DOI] [PubMed] [Google Scholar]
- 29. Sakuramoto S, Sasako M, Yamaguchi T et al Adjuvant chemotherapy for gastric cancer with S‐1, an oral fluoropyrimidine. N Engl J Med 2007; 357: 1810–20. [DOI] [PubMed] [Google Scholar]
- 30. Koizumi W, Narahara H, Hara T et al S‐1 plus cisplatin versus S‐1 alone for first‐line treatment of advanced gastric cancer (SPIRITS trial): a phase III trial. Lancet Oncol 2008; 9: 215–21. [DOI] [PubMed] [Google Scholar]
- 31. Yamada Y, Higuchi K, Nishikawa K et al Phase III study comparing oxaliplatin plus S‐1 with cisplatin plus S‐1 in chemotherapy‐naïve patients with advanced gastric cancer. Ann Oncol 2015; 26: 141–8. [DOI] [PubMed] [Google Scholar]
- 32. Uen YH, Lin KY, Sun DP et al Comparative proteomics, network analysis and post‐translational modificationidentification reveal differential profiles of plasma Con A‐bound glycoprotein biomarkers in gastric cancer. J Proteomics 2013; 83: 197–213. [DOI] [PubMed] [Google Scholar]
- 33. Warren JR, Marshall B. Unidentified curved bacilli on gastric epithelium in active chronic gastritis. Lancet 1983; 1: 1273–5. [PubMed] [Google Scholar]
- 34. Wang X, Abraham S, McKenzie JA et al LRG1 promotes angiogenesis by modulating endothelial TGF‐β signalling. Nature 2013; 499: 306–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. (a) Quantitative Transwell assay results of AGS gastric cancer cells transfected with siRNA against leucine‐rich α2‐glycoprotein‐1 LRG1 (siLRG1) and non‐targeting siRNA (negative control [NC]). (b) MMP‐2, pro‐MMP‐2, and pro‐MMP‐9 activities of AGS with siLRG1 and non‐targeting siRNA (NC).


