Abstract
Oral squamous cell carcinoma is one of the most malignant tumors with high mortality rate worldwide. Biomarker discovery is critical for early diagnosis and precision treatment of this disease. MicroRNAs are small noncoding RNA molecules which often regulate essential biological processes and are good candidates for biomarkers. By integrative analysis of both the cancer-associated gene expression data and microRNA-mRNA network, miR-148b-3p, miR-629-3p, miR-27a-3p, and miR-142-3p were screened as novel diagnostic biomarkers for oral squamous cell carcinoma based on their unique regulatory abilities in the network structure of the conditional microRNA-mRNA network and their important functions. These findings were confirmed by literature verification and functional enrichment analysis. Future experimental validation is expected for the further investigation of their molecular mechanisms.
1. Introduction
Oral squamous cell carcinoma (OSCC) is the sixth most common cancer with more than 300,000 cases worldwide each year [1]. It is the most malignant tumor in the oral and maxillofacial regions and accounts for 90% of oral cancers [2, 3]. The risk factors for OSCC could be tobacco, alcohol consumption, betel quid (BQ) chewing, Bidi smoking, and genetic predisposition [4, 5]. OSCC can metastasize to lymph-node, even to remote organs with high mortality rate. The present diagnosis of OSCC often happened at late stage and the treatment can be unsuccessful due to its local recurrence. The precise early diagnosis is critical and essential to the future prevention and personalized treatment of this disease.
MicroRNA is a family of functional noncoding RNA molecules containing about 22 nucleotides, which play roles in the posttranscriptional gene regulation. Since many key biological processes including the development, differentiation, and cell cycles are regulated by microRNAs, the abnormal expression of microRNAs is often associated with the initialization and progression of many diseases [6, 7]. Thus miRNAs usually could serve as suitable biomarkers for many diseases, such as neurodevelopmental disorders [8], cancer, and cardiovascular disease [9–11].
Previous studies have demonstrated that microRNAs played important roles in OSCC. For example, microRNA-23b/27b cluster is reported as tumor suppressive and regulates the MET oncogene in OSCC [12]. MicroRNA-27a-3p can regulate transition from epithelial to mesenchymal in OSCC by targeting YAP1 [13]. The apoptosis-related protein expression and radiosensitivity in BQ-associated OSCC are regulated by microRNA-17-5p [14]. Metabolic shift in OSCC is mediated by microRNA-340 targeting glucose transporter-1 [15]. Tumor growth and activation of NF-κB signaling were promoted via the regulation of NLK by microRNA-92b in OSCC [16]. In addition, microRNA-17/20a was suggested as a prognostic marker since it can inhibit cell migration in OSCC [2]. Circulating microRNA-21 and PTEN (phosphatase and tensin homolog) are reported as promising biomarkers for detection of OSCC [17].
From the above introduction, we believe that microRNAs are good candidates to act as diagnostic and prognostic biomarkers of OSCC. As we know, OSCC is a complex and heterogeneous disease. For this reason, more precise and personalized biomarkers are needed for the diagnosis, prognosis, and treatment of OSCC. Until now, very few studies have focused on the expression data of OSCC to integrate it with the microRNA-mRNA network structural analysis for biomarker discovery in OSCC, especially the application of bioinformatics and network analysis to the study of the functions of microRNAs in the OSCC initialization and progression.
The experimental methods for biomarker discovery are time-consuming and costly. Bioinformatics screening will be helpful to the efficient biomarker screening. Previously, several models have been developed to infer key and biomarker microRNAs in complex diseases from conditional gene expression data. Differential expression genes (DEGs) are often used to screen biomarker genes, but only few DEGs are validated as biomarkers; therefore integrative analysis of DEGs with other information is very necessary for efficient biomarker discovery. As described in previous work [18–21], these models screen the potential biomarker based on the scrutinizing of the structure of the conditional microRNA-mRNA network. By statistical analysis of the network structure and functions of the biomarker microRNA's targets, the model can very effectively identify novel putative microRNA biomarkers for the diagnosis of complex diseases. So we here apply the model to the biomarker microRNA discovery for diagnosis of OSCC.
2. Materials and Methods
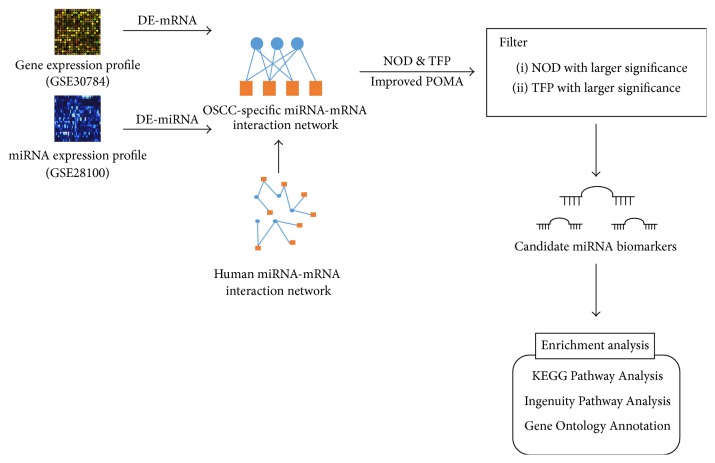
The schematic pipeline of the present work for the data collection, model construction, biomarker microRNAs prediction, and validation and enrichment analysis of the targets of the predicted microRNAs is presented in Figure 1. The details of the step-by-step procedures for the screening of OSCC diagnostic biomarker microRNAs are described as follows.
Figure 1.
Schematic pipeline for the identification of oral squamous cell carcinoma (OSCC) microRNA biomarkers.
2.1. Gene Expression Data Collection
The data for the OSCC gene expression and microRNA expression were extracted from the GEO database [22]. The OSCC associated expression data in the GEO database are listed in Table 1. Eight OSCC associated data sets measured from different microarray platforms are available in the GEO database. After the condition filtering, the final data used for the construction of OSCC-specific microRNA-mRNA network are GSE30784 and GSE28100. The former is the mRNA expression data including 167 OSCC samples and 45 samples as control [23] and the latter is the microRNA expression data with 17 OSCC samples and 3 control samples [24]. The data were normalized and the differentially expressed mRNAs were identified based on linear models in Limma R package [25, 26]. The p value and other parameters were calculated with the empirical Bayes (eBayes) method. The Benjamini-Hochberg correction was applied to adjust the p values. The adjusted p value less than 0.05 was chosen as the cut-off criteria.
Table 1.
OSCC gene expression data collected from GEO database.
| Accession/ID | PMID | Platform | OSCC | Control | Gene/microRNA |
|---|---|---|---|---|---|
| GSE3524 | 15381369 | GPL96 | n = 16 | n = 4 | Gene |
| GSE70604 | 26700817 | GPL2986 | n = 7 | n = 6 | Gene |
| 23624915 | |||||
| GSE37991 | 25204733 | GPL6883 | n = 40 | n = 40 | Gene |
| 23362108 | |||||
| GSE28100 | 22761427 | GPL10850 | n = 17 | n = 3 | MicroRNA |
| GSE23558 | 22072328 | GPL6480 | n = 27 | n = 5 | Gene |
| GSE25099 | 21853135 | GPL5175 | n = 57 | n = 22 | Gene |
| GSE30784 | 18669583 | GPL570 | n = 167 | n = 45 | Gene |
| GSE10121 | 18472963 | GPL6353 | n = 35 | n = 6 | Gene |
The reported OSCC associated microRNAs were also collected from PubMed citations by the search criteria “(Oral squamous cell carcinoma OR OSCC) AND (miRNA OR microRNA) AND (biomarker∗ OR marker∗)”. They were checked manually and listed in Table 2.
Table 2.
Previously reported microRNA biomarkers for OSCC from PubMed.
| MicroRNA | PMID | Biomarker Type | Samples | Expression in OSCC | NOD (Ref.)a |
TFP (Ref.)a |
|---|---|---|---|---|---|---|
| miR-155-5p | 26307116 24692283 |
Prognostic biomarker | Tissue | Upregulated | 71 | 0.211 |
|
| ||||||
| miR-483-5p | 26224475 | Diagnostic/prognostic biomarker | Serum | Upregulated | 4 | 0.135 |
|
| ||||||
| miR-216a | 25955794 | Prognostic biomarker | Tissue | Downregulated | 6 | 0.087 |
|
| ||||||
| miR-21-3p miR-96-5p miR-141-3p miR-130b-3p |
25532855 | Prognostic biomarker | Tissue | Upregulated | N/A 8 22 2 |
N/A 0.159 0.167 0.119 |
|
| ||||||
| miR-21-5p | 24755828 | Prognostic biomarker | Tissue | Upregulated | 38 | 0.135 |
|
| ||||||
| miR-31-5p | 22083872 | Diagnostic biomarker | Saliva | Upregulated | 10 | 0.142 |
aThe NOD and TFP are calculated based on the reference human microRNA-mRNA network, while the measurements in Section 3.2 are calculated based on the OSCC-specific network.
2.2. Prediction of microRNA Biomarkers for Diagnosis of OSCC
As reported in the previous researches in Shen's group [18–21], two measurements are important for candidate biomarker microRNA. The first one is the novel of degree measurement (NOD). It measures the number of genes solely targeted by certain microRNA [19, 20]. This character is reasonable since the abnormal alteration of this type of interaction cannot be compensated by another microRNA-mRNA interaction pair as most of the microRNA-mRNA interactions are synergic. The other measurement is the transcription factor percentage (TFP), which was defined as the percentage of transcription factor (TF) genes of all targets of the microRNA [18].
According to the above hypothesis, the OSCC-specific microRNA-mRNA network was constructed by mapping the detected differentially expressed microRNAs in OSCC onto the reference human microRNA-mRNA network. The reference network was constructed with the integration of the experimentally validated and computational predicted microRNA-mRNA pairs. The experimentally validated data included information from miRecords [27], TarBase [28], miR2Disease [29], and miRTarBase [30], while the computational predicted microRNA-mRNA pairs are extracted from no fewer than 2 databases among HOCTAR [31], ExprTargetDB [32], and starBase [33]. With the reconstructed conditional network, the above-mentioned measurements, that is, the NOD and TFP, were calculated for each microRNA in the OSCC-specific network. MicroRNAs with significantly large NOD and TFP values (Wilcoxon signed-rank test, p value < 0.05) were screened as putative biomarkers.
2.3. Functional Enrichment Investigation of the Targets of Predicted OSCC Diagnostic MicroRNA Biomarkers
Functional enrichment analysis of the genes targeted by the identified candidate biomarker microRNAs from the OSCC-specific microRNA-mRNA networks was performed through three different tools: Gene Ontology Annotation, KEGG Pathway Analysis, and Ingenuity Pathway Analysis (IPA). Here, the first two analyses were conducted on the DAVID (Database for Annotation, Visualization, and Integrated Discovery) online analysis webpage [34]. The significantly enriched pathways and ontologies for OSCC with p value less than 0.05 were ranked. The p value was calculated based on the hypergeometric test and FDR adjustment was used for multiple test correction.
3. Result and Discussion
3.1. The Characterization of the Previous Reported OSCC Diagnostic Biomarker MicroRNAs
We checked the PubMed citations and the previously reported biomarker microRNAs for OSCC were listed in Table 2. From Table 2, it is clear that all the reported microRNAs have high NOD and TFP values except miR-21-3p, which cannot be extracted from the reconstructed microRNA-mRNA network. This observation confirmed that the model using the NOD and TFP as two measurements for the evaluation of the potential biomarkers is applicable for OSCC biomarker discovery.
3.2. Predicted Diagnostic Biomarker MicroRNAs for OSCC
We first identified 56 dysregulated microRNAs and 3375 differentially expressed genes in OSCC by using the pipeline presented in Figure 1. Five microRNAs were identified through Wilcoxon signed-rank test with p value less than 0.05. These microRNAs were predicted to be candidate biomarkers for the diagnosis of OSCC. Their network structural characteristics in the microRNA-mRNA network, including the number of targets and NOD and TFP values calculated based on the conditional OSCC-specific microRNA-mRNA network, are listed in Table 3. Among the five microRNAs, miR-155-5p was reported in previous work as biomarker [35, 36]. The other four microRNAs, that is, miR-148b-3p, miR-629-3p, miR-27a-3p, and miR-142-3p, are the novel putative biomarkers identified for OSCC.
Table 3.
Candidate OSCC microRNA biomarkers identified by our model.
| MicroRNA | NOD | p value | TFP | p value | Number of targets |
|---|---|---|---|---|---|
| miR-148b-3p | 15 | 5.06E-06 | 0.168 | 0.00912 | 95 |
| miR-155-5p | 27 | 1.25E − 12 | 0.259 | 1.53E − 05 | 58 |
| miR-629-3p | 10 | 0.00845 | 0.214 | 4.58E − 05 | 14 |
| miR-27a-3p | 10 | 0.00845 | 0.194 | 3.81E − 04 | 31 |
| miR-142-3p | 10 | 0.00845 | 0.164 | 0.0168 | 55 |
3.3. Literature-Based Validation of Identified MicroRNA Biomarkers
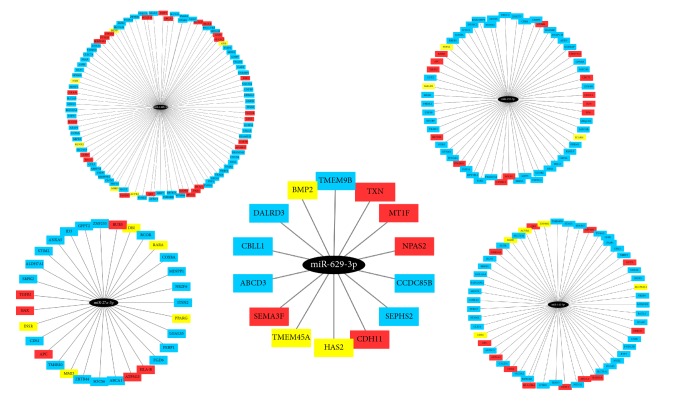
The targets of the five putative microRNAs are presented in Figure 2. From the figure, we can see that some of the targets of these microRNAs have been reported to be associated with OSCC (genes colored red) or other oral diseases (genes colored yellow) according to the PubMed citations.
Figure 2.
Candidate oral squamous cell carcinoma (OSCC) microRNA biomarkers with their target genes. Here, ellipses in black represent 5 microRNA biomarkers. Genes in red are associated with OSCC according to literature reports while genes in yellow are reported to be associated with other oral diseases.
3.4. Functional Enrichment Analysis of Target Genes of Candidate MicroRNA Biomarkers
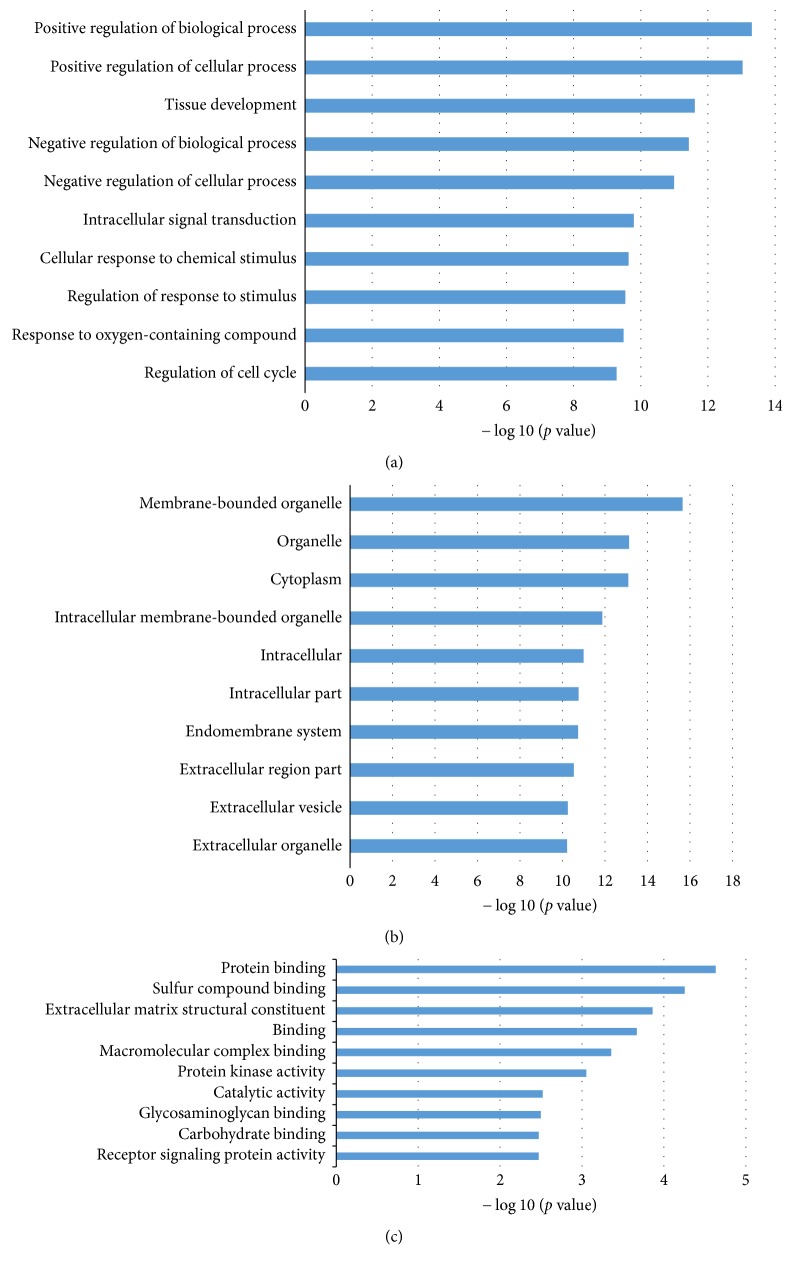
The functional enrichment analysis was further performed to explore the roles of the uniquely regulated genes of the identified microRNAs in OSCC with DAVID and IPA tools. Figure 3 presented the Gene Ontology (GO) Annotation for targets of the identified microRNA biomarkers in OSCC. The three levels of GO analysis are presented in Figures 3(a)–3(c), respectively, for biological process, cellular component, and molecular function. The top 10 significantly enriched items are listed for each level. Most of the dysregulated biological processes are the positive/negative biological or cellular processes, the regulation of cell cycle, and the response to oxygen-containing compound. The former are well-known popular cancer-associated processes, while the latter is associated specifically with OSCC [37–39]. The most enriched molecular functions are the general cancer-associated items, such as protein binding, protein kinase activity, and receptor signaling protein activity. The functions for carbohydrate binding [40–42] and glycosaminoglycan binding [43, 44] were also discovered in the OSCC studies.
Figure 3.
Gene Ontology (GO) Annotation for genes targeted by identified microRNA biomarkers. (a), (b), and (c), respectively, represent three levels of GO: biological process, cellular component, and molecular function. The statistical significance value (p value) has been negative 10-based log-transformed. The top 10 significantly enriched items are listed for each level.
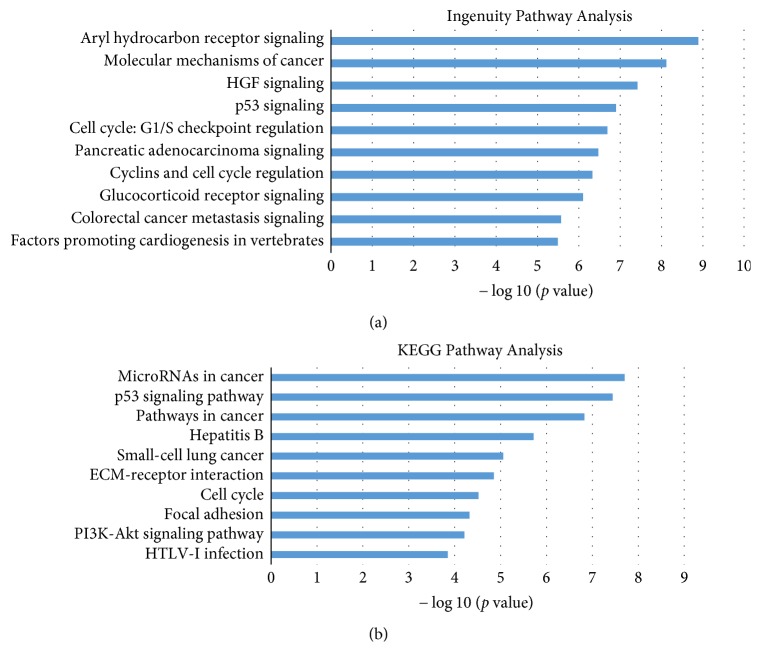
The result of the pathway enrichment analysis of the targeted genes of the putative microRNA biomarkers is displayed in Figure 4. The most common cancer-associated pathways like p53 signaling pathway and cell cycle pathway are enriched in both the DAVID and IPA methods. There are still other pathways such as PI3K-Akt signaling pathways and colorectal cancer metastasis were screened by these two enrichment analyses. The Aryl hydrocarbon receptor [45–47], the HGF [48, 49], ECM receptor interaction [50, 51], Hepatitis B [52, 53], and glucocorticoid receptor signaling [54] are all supported by the PubMed citations.
Figure 4.
KEGG Pathway Enrichment Analysis and Ingenuity Pathway Analysis for genes targeted by identified microRNA biomarkers. The statistical significance value (p value) has been negative 10-based log-transformed. The top 10 significantly enriched pathways are listed, respectively, in (a) and (b).
4. Conclusions
In this research, we applied the concepts of NOD and TPF to the integrative analysis of OSCC gene expression and the microRNA-mRNA network. We identified five microRNAs that could be putative biomarkers for OSCC. Among them, one has been reported as biomarker and two are reported as associated microRNAs. The other two are the novel finding microRNA biomarkers. As a result, four novel biomarker microRNAs, that is, miR-148b-3p, miR-629-3p, miR-27a-3p, and miR-142-3p, are discovered in our work. The literature checking and the functional enrichment analysis confirmed our finding. Therefore, further experimental verification and clinical testing were suggested for these putative OSCC microRNA biomarkers.
Abbreviations
- OSCC:
Oral squamous cell carcinoma
- TF:
Transcription factor
- NOD:
Novel out degree
- TFP:
Transcription factor gene percentage
- KEGG:
Kyoto Encyclopedia of Genes and Genomes
- DAVID:
Database for Annotation, Visualization, and Integrated Discovery
- IPA:
Ingenuity Pathway Analysis.
Conflicts of Interest
The authors declare no conflicts of interest.
Authors' Contributions
Xiangya Huang and Linqing Zheng conceived and designed the experiments; Hui Zhang performed the experiments; Hui Zhang and Tangxin Li analyzed the data; Xiangya Huang wrote the paper. Hui Zhang and Tangxin Li contributed equally to this work.
References
- 1.Wang Q., Gao P., Wang X., Duan Y. Investigation and identification of potential biomarkers in human saliva for the early diagnosis of oral squamous cell carcinoma. Clinica Chimica Acta. 2014;427:79–85. doi: 10.1016/j.cca.2013.10.004. [DOI] [PubMed] [Google Scholar]
- 2.Chang C.-C., Yang Y.-J., Li Y.-J., et al. MicroRNA-17/20a functions to inhibit cell migration and can be used a prognostic marker in oral squamous cell carcinoma. Oral Oncology. 2013;49(9):923–931. doi: 10.1016/j.oraloncology.2013.03.430. [DOI] [PubMed] [Google Scholar]
- 3.Li Y., Li B., Xu B., et al. Expression of p53, p21(CIP1/WAF1) and eIF4E in the adjacent tissues of oral squamous cell carcinoma: establishing the molecular boundary and a cancer progression model. International Journal of Oral Science. 2015;7(3):161–168. doi: 10.1038/ijos.2015.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Subapriya R., Thangavelu A., Mathavan B., Ramachandran C. R., Nagini S. Assessment of risk factors for oral squamous cell carcinoma in Chidambaram, southern India: a case-control study. European Journal of Cancer Prevention. 2007;16(3):251–256. doi: 10.1097/01.cej.0000228402.53106.9e. [DOI] [PubMed] [Google Scholar]
- 5.Santos H.-B., dos Santos T.-K., Paz A.-R., et al. Clinical findings and risk factors to oral squamous cell carcinoma in young patients: A 12-year retrospective analysis. Medicina Oral, Patologia Oral y Cirugia Bucal. 2016;21(2):e151–e156. doi: 10.4317/medoral.20770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tang Y., Yan W., Chen J., Luo C., Kaipia A., Shen B. Identification of novel microRNA regulatory pathways associated with heterogeneous prostate cancer. BMC Systems Biology. 2013;7(supplement 3, article S6) doi: 10.1186/1752-0509-7-s3-s6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kong Y. W., Ferland-McCollough D., Jackson T. J., Bushell M. microRNAs in cancer management. The Lancet Oncology. 2012;13(6):e249–e258. doi: 10.1016/S1470-2045(12)70073-6. [DOI] [PubMed] [Google Scholar]
- 8.Shen L., Lin Y., Sun Z., Yuan X., Chen L., Shen B. Knowledge-guided bioinformatics model for identifying autism spectrum disorder diagnostic microrna biomarkers. Scientific Reports. 2016;6, article 39663 doi: 10.1038/srep39663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhao X.-M., Liu K.-Q., Zhu G., et al. Identifying cancer-related microRNAs based on gene expression data. Bioinformatics. 2015;31(8):1226–1234. doi: 10.1093/bioinformatics/btu811. [DOI] [PubMed] [Google Scholar]
- 10.Qin G.-M., Li R.-Y., Zhao X.-M. Identifying Disease Associated miRNAs Based on Protein Domains. IEEE/ACM Transactions on Computational Biology and Bioinformatics. 2016;13(6):1027–1035. doi: 10.1109/TCBB.2016.2515608. [DOI] [PubMed] [Google Scholar]
- 11.Becker S., Florian A., Patrascu A., et al. Identification of cardiomyopathy associated circulating miRNA biomarkers in patients with muscular dystrophy using a complementary cardiovascular magnetic resonance and plasma profiling approach. Journal of Cardiovascular Magnetic Resonance. 2016;18(1, article no. 244) doi: 10.1186/s12968-016-0244-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fukumoto I., Koshizuka K., Hana Zawa T., et al. The tumor-suppressive microRNA-23b/27b cluster regulates the MET oncogene in oral squamous cell carcinoma. International Journal of Oncology. 2016;49(3):1119–1129. doi: 10.3892/ijo.2016.3602. [DOI] [PubMed] [Google Scholar]
- 13.Zeng G., Xun W., Wei K., Yang Y., Shen H. MicroRNA-27a-3p regulates epithelial to mesenchymal transition via targeting YAP1 in oral squamous cell carcinoma cells. Oncology Reports. 2016;36(3):1475–1482. doi: 10.3892/or.2016.4916. [DOI] [PubMed] [Google Scholar]
- 14.Wu S.-Y., Wu A. T. H., Liu S.-H. Microrna-17-5p regulated apoptosis-related protein expression and radiosensitivity in oral squamous cell carcinoma caused by betel nut chewing. Oncotarget. 2016;7(32):51482–51493. doi: 10.18632/oncotarget.9856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xu P., Li Y., Zhang H., Li M., Zhu H. MicroRNA-340 Mediates Metabolic Shift in Oral Squamous Cell Carcinoma by Targeting Glucose Transporter-1. Journal of Oral and Maxillofacial Surgery. 2016;74(4):844–850. doi: 10.1016/j.joms.2015.09.038. [DOI] [PubMed] [Google Scholar]
- 16.Liu Z., Diep C., Mao T., et al. MicroRNA-92b promotes tumor growth and activation of NF-κB signaling via regulation of NLK in oral squamous cell carcinoma. Oncology Reports. 2015;34(6):2961–2968. doi: 10.3892/or.2015.4323. [DOI] [PubMed] [Google Scholar]
- 17.Ren W., Qiang C., Gao L., et al. Circulating microRNA-21 (MIR-21) and phosphatase and tensin homolog (PTEN) are promising novel biomarkers for detection of oral squamous cell carcinoma. Biomarkers. 2014;19(7):590–596. doi: 10.3109/1354750X.2014.955059. [DOI] [PubMed] [Google Scholar]
- 18.Yan W., Xu L., Sun Z., et al. MicroRNA biomarker identification for pediatric acute myeloid leukemia based on a novel bioinformatics model. Oncotarget. 2015;6(28):26424–26436. doi: 10.18632/oncotarget.4459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang W., Zang J., Jing X., et al. Identification of candidate miRNA biomarkers from miRNA regulatory network with application to prostate cancer. Journal of Translational Medicine. 2014;12, article 66 doi: 10.1186/1479-5876-12-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhu J., Wang S., Zhang W., et al. Screening key microRNAs for castration-resistant prostate cancer based on miRNA/mRNA functional synergistic network. Oncotarget. 2015;6(41):43819–43830. doi: 10.18632/oncotarget.6102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen J., Zhang D., Zhang W., et al. Clear cell renal cell carcinoma associated microRNA expression signatures identified by an integrated bioinformatics analysis. Journal of Translational Medicine. 2013;11(1, article 169) doi: 10.1186/1479-5876-11-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Barrett T., Suzek T. O., Troup D. B., et al. NCBI GEO: mining millions of expression profiles—database and tools. Nucleic Acids Research. 2005;33:D562–D566. doi: 10.1093/nar/gki022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen C., Méndez E., Houck J., et al. Gene expression profiling identifies genes predictive of oral squamous cell carcinoma. Cancer Epidemiology Biomarkers and Prevention. 2008;17(8):2152–2162. doi: 10.1158/1055-9965.EPI-07-2893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jung H. M., Phillips B. L., Patel R. S., et al. Keratinization-associated miR-7 and miR-21 regulate tumor suppressor reversion-inducing cysteine-rich protein with kazal motifs (RECK) in oral cancer. The Journal of Biological Chemistry. 2012;287(35):29261–29272. doi: 10.1074/jbc.m112.366518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Smyth G. K. Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Statistical Applications in Genetics and Molecular Biology. 2004;3:Art. 3, 29. doi: 10.2202/1544-6115.1027. [DOI] [PubMed] [Google Scholar]
- 26.Ritchie M. E., Phipson B., Wu D., et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Research. 2015 doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xiao F., Zuo Z., Cai G., Kang S., Gao X., Li T. miRecords: an integrated resource for microRNA-target interactions. Nucleic Acids Research. 2009;37(1):D105–D110. doi: 10.1093/nar/gkn851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sethupathy P., Corda B., Hatzigeorgiou A. G. TarBase: a comprehensive database of experimentally supported animal microRNA targets. RNA. 2006;12(2):192–197. doi: 10.1261/rna.2239606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jiang Q., Wang Y., Hao Y., et al. miR2Disease: a manually curated database for microRNA deregulation in human disease. Nucleic Acids Research. 2009;37(1):D98–D104. doi: 10.1093/nar/gkn714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hsu S.-D., Lin F.-M., Wu W.-Y., et al. MiRTarBase: a database curates experimentally validated microRNA-target interactions. Nucleic Acids Research. 2011;39(supplement 1):D163–D169. doi: 10.1093/nar/gkq1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gennarino V. A., Sardiello M., Avellino R., et al. MicroRNA target prediction by expression analysis of host genes. Genome Research. 2009;19(3):481–490. doi: 10.1101/gr.084129.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gamazon E. R., Im H.-K., Duan S., et al. ExprTarget: an integrative approach to predicting human microRNA targets. PLoS ONE. 2010;5(10) doi: 10.1371/journal.pone.0013534.e13534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang J.-H., Li J.-H., Shao P., Zhou H., Chen Y.-Q., Qu L.-H. StarBase: a database for exploring microRNA-mRNA interaction maps from Argonaute CLIP-Seq and Degradome-Seq data. Nucleic Acids Research. 2011;39(1):D202–D209. doi: 10.1093/nar/gkq1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dennis Jr. G., et al. DAVID: database for annotation, visualization, and integrated discovery. Genome Biology. 2003;4(5) [PubMed] [Google Scholar]
- 35.Rather M. I., Nagashri M. N., Swamy S. S., Gopinath K. S., Kumar A. Oncogenic microRNA-155 down-regulates tumor suppressor CDC73 and promotes oral squamous cell carcinoma cell proliferation: implications for cancer therapeutics. Journal of Biological Chemistry. 2013;288(1):608–618. doi: 10.1074/jbc.m112.425736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Baba O., Hasegawa S., Nagai H., et al. MicroRNA-155-5p is associated with oral squamous cell carcinoma metastasis and poor prognosis. Journal of Oral Pathology and Medicine. 2016;45(4):248–255. doi: 10.1111/jop.12351. [DOI] [PubMed] [Google Scholar]
- 37.Sasabe E., Yang Z., Ohno S., Yamamoto T. Reactive oxygen species produced by the knockdown of manganese-superoxide dismutase up-regulate hypoxia-inducible factor-1α expression in oral squamous cell carcinoma cells. Free Radical Biology and Medicine. 2010;48(10):1321–1329. doi: 10.1016/j.freeradbiomed.2010.02.013. [DOI] [PubMed] [Google Scholar]
- 38.Hua K.-F., Liao P.-C., Fang Z., et al. Generation of Reactive Oxygen Species by Polyenylpyrroles Derivatives Causes DNA Damage Leading to G2/M Arrest and Apoptosis in Human Oral Squamous Cell Carcinoma Cells. PLoS ONE. 2013;8(6) doi: 10.1371/journal.pone.0067603.e67603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Czesnikiewicz-Guzik M., Lorkowska B., Zapala J., et al. NADPH oxidase and uncoupled nitric oxide synthase are major sources of reactive oxygen species in oral squamous cell carcinoma. Potential implications for immune regulation in high oxidative stress conditions. Journal of Physiology and Pharmacology. 2008;59(1):139–152. [PubMed] [Google Scholar]
- 40.Li P., Xiao L. Y., Tan H. Muc-1 promotes migration and invasion of oral squamous cell carcinoma cells via PI3K-Akt signaling. International Journal of Clinical and Experimental Pathology. 2015;8(9):10365–10374. [PMC free article] [PubMed] [Google Scholar]
- 41.Ding Y.-M., Dong J.-H., Chen L.-L., Zhang H.-D. Increased expression of galectin-1 is associated with human oral squamous cell carcinoma development. Oncology Reports. 2009;21(4):983–987. doi: 10.3892/or_00000312. [DOI] [PubMed] [Google Scholar]
- 42.Aggarwal S., Sharma S. C., Das S. N. Galectin-1 and galectin-3: Plausible tumour markers for oral squamous cell carcinoma and suitable targets for screening high-risk population. Clinica Chimica Acta. 2015;442:13–21. doi: 10.1016/j.cca.2014.12.038. [DOI] [PubMed] [Google Scholar]
- 43.Ueda K. Heparin induces apoptosis through suppression of AKt in oral squamous cell carcinoma cells. Anticancer Research. 2009;29(4):1079–1088. [PubMed] [Google Scholar]
- 44.Mori S., Nose M., Morikawa H., et al. A novel evaluation system of metastatic potential of oral squamous cell carcinoma according to the histopathological and histochemical grading. Oral Oncology. 1998;34(6):549–557. doi: 10.1016/S1368-8375(98)00048-7. [DOI] [PubMed] [Google Scholar]
- 45.Kimura Y., Kasamatsu A., Nakashima D., et al. ARNT2 regulates tumoral growth in oral squamous cell carcinoma. Journal of Cancer. 2016;7(6):702–710. doi: 10.7150/jca.14208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stanford E. A., Ramirez-Cardenas A., Wang Z., et al. Role for the aryl hydrocarbon receptor and diverse ligands in oral squamous cell carcinoma migration and tumorigenesis. Molecular Cancer Research. 2016;14(8):696–706. doi: 10.1158/1541-7786.MCR-16-0069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Xie X., Jiang Y., Yuan Y., et al. MALDI imaging reveals NCOA7 as a potential biomarker in oral squamous cell carcinoma arising from oral submucous fibrosis. Oncotarget. 2016;7(37):59987–60004. doi: 10.18632/oncotarget.11046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Klosek S., Nakashiro K., Hara S., Li C., Shintani S., Hamakawa H. Constitutive activation of Stat3 correlates with increased expression of the c-Met/HGF receptor in oral squamous cell carcinoma. Oncology Reports. 2004 doi: 10.3892/or.12.2.293. [DOI] [PubMed] [Google Scholar]
- 49.Borgiel-Marek H., Marek B., Niedzielska I., Kajdaniuk D. Serum concentration of fibroblastic growth factor in oral squamous cell carcinoma before and after surgery. International Journal of Oral and Maxillofacial Surgery. 2009;38(5):p. 537. doi: 10.1016/j.ijom.2009.03.493. [DOI] [Google Scholar]
- 50.Liu Z., Niu Y., Li C., Yang Y., Gao C. Integrating multiple microarray datasets on oral squamous cell carcinoma to reveal dysregulated networks. Head and Neck. 2012;34(12):1789–1797. doi: 10.1002/hed.22013. [DOI] [PubMed] [Google Scholar]
- 51.He Y., Shao F., Pi W., et al. Largescale transcriptomics analysis suggests over-expression of BGH3, MMP9 and PDIA3 in oral squamous cell carcinoma. PLoS ONE. 2016;11(1) doi: 10.1371/journal.pone.0146530.e0146530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bakri M. M., Hussaini H. M., Holmes A., Cannon R. D., Rich A. M. Revisiting the association between candidal infection and carcinoma, particularly oral squamous cell carcinoma. Journal of Oral Microbiology. 2010;2(2010, article no. 5780) doi: 10.3402/jom.v2i0.5780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fang F.-M., Li C.-F., Huang H.-Y., et al. Overexpression of a chromatin remodeling factor, RSF-1/HBXAP, correlates with aggressive oral squamous cell carcinoma. American Journal of Pathology. 2011;178(5):2407–2415. doi: 10.1016/j.ajpath.2011.01.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Li B., et al. [Assessment of mental health status in oral squamous cell carcinoma patients and its correlation with catecholamines level] Shanghai Kou Qiang Yi Xue. 2013;22(6):671–675. [PubMed] [Google Scholar]