Abstract
A central goal of evolutionary biology is to understand the genetic origin of morphological novelties – i.e. anatomical structures unique to a taxonomic group. Elaboration of morphology during development depends on networks of regulatory genes that activate patterned gene expression through transcriptional enhancer regions. We summarize recent case studies and genome-wide investigations that have uncovered diverse mechanisms though which new enhancers arise. We also discuss how these enhancer-originating mechanisms have clarified the history of genetic networks underlying diversification of genital structures in flies, limbs and neural crest in chordates, and plant leaves. These studies have identified enhancers that were pivotal for morphological divergence and highlighted how novel genetic networks shaping form emerged from pre-existing ones.
Keywords: enhancer origination, morphological novelty, gene regulatory network co-option
Introduction
A key problem in biology is to discern how the distinct features of different organisms arose at the genetic level. Of particular importance to morphological traits are the networks of transcription factors that control the expression of hundreds to thousands of downstream genes that confer upon each cell its distinctive physical properties [1]. Transcriptional control is mediated by cis-regulatory sequences, often called enhancers in cases of transcriptional activation and silencers in cases of repression, that recruit combinations of transcription factors to short binding sites that collectively determine when, where, and how much each gene is transcribed during development [2]. Thus, determining the evolutionary history of a morphological feature’s regulatory network at the level of its participating enhancers provides key information on the origins of morphological novelty. We will review progress in studying the origins of enhancers and morphological novelties in the last two years.
How to build a new enhancer
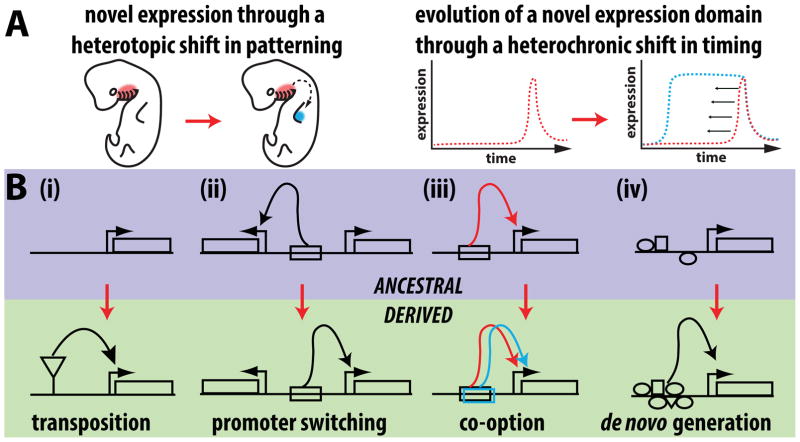
New gene expression patterns may evolve through spatial or temporal changes in transcription (Figure 1A). Recent studies have highlighted a surprisingly wide range of molecular mechanisms that modify regulatory DNA (Figure 1B). Early papers by Britten and Davidson proposed that repetitive sequences such as those provided by transposons could contribute to gene regulation [3,4]. Many reports have since implicated transposable elements in the evolution of gene regulation [5–7]. Recently, genomic studies have investigated the significance of transposon-related enhancer birth genome wide. For example, transcriptome comparisons of uterine cells showed that thousands of genes gained expression during the evolution of mammalian pregnancy [8]. Transposons are enriched in the presumed regulatory regions of these genes, suggesting that they may have contributed to this major evolutionary transition. On the other hand, a comparative survey of enhancer-associated histone marks in the mammalian neocortex revealed that transposons were underrepresented in newly evolved neocortex enhancers [9]. These studies highlight that lineage specific trends may constrain the contribution of transposons to novel expression patterns, and raise the question of what direct impact they have on expression.
Figure 1. The genetic underpinnings of novel gene expression patterns and enhancer activities.
(A) Novel expression patterns can arise through heterotopic shifts that cause a gene to be expressed in a spatially distinct pattern (left) or by temporal shifts that cause a gene to be expressed much earlier or later during development (right). (B) Genetic models for the origins of new enhancer activities. The ancestral state for each model depicts the status of a locus before a new expression domain evolved by changes in its cis-regulatory sequences. (i) A gene may gain a novel expression pattern through the introduction of a transposon that can carry regulatory information resulting in a new enhancer activity. (ii) Changes in mechanisms targeting enhancers to specific promoters (e.g. point mutations or large-scale chromosomal rearrangements) can cause a pre-existing enhancer to target a different promoter. (iii) A pre-existing enhancer active in an ancestral tissue may gain or lose inputs that allow it to be expressed in a novel domain. (iv) A stretch of DNA that ancestrally lacked regulatory function may evolve a de novo enhancer activity.
One recent study presented evidence that multiple families of transposons in mammalian genomes carry an interferon response element [10], and used CRISPR/Cas9 genome editing to demonstrate that these sequences are necessary for immune responses in cell culture. A striking example of a transposon insertion causing altered enhancer activity and concomitant morphological evolution was found in stickleback fish. Specifically, a change in body armor size that accompanied the transition from marine to freshwater environments was caused by a transposon insertion in the BMP-like GDF6 gene which was associated with its increased expression [11]. While the insertion was necessary for this increase, it was itself insufficient to recapitulate the novel expression of GDF6 in a transgenic assay, indicating that other changes were involved in this morphological shift. The above findings underline how genome-wide and single gene approaches can provide complimentary insights into how molecular patterns shape differences in gene expression.
While transposition has made considerable contributions to the evolution of novel gene expression features, many additional mechanisms have been identified in mammals and other systems (Figure 1B). Although the evolution of enhancers de novo from mutations in non-functional sequences represents an obvious null hypothesis (Figure 1B), this mechanism has been difficult to study, and it more often turns out that new regulatory sequences have evolved from pre-existing ancestral ones. Studies of the domesticated chicken have shown how large-scale chromosome rearrangements and structural variations have contributed to diverse phenotypes by rearranging or duplicating regulatory elements and placing them in association with new genes (“promoter switching” in Figure 1B) [12–14]. Multiple studies have shown how novel expression domains can evolve from pre-existing enhancers that derive additional tissue specificities (“co-option” in Figure 1B) [15–18]. For example, novel domains of Wingless expression associated with unique spots of pigmentation in the Drosophila guttifera wing evolved by modifying a pre-existing enhancer that was ancestrally active in a distinct wing location [17]. A study of the fly Zaprionus capensis uncovered an expression pattern in the larval wing disc that evolved through an extreme heterochronic shift (Figure 1A) that drove a pupal pattern much earlier into the larval life stage [18]. There is a growing appreciation that the introduction of mutations to an enhancer can often elicit its ectopic activation in additional locations [19,20]. Two recent papers by Farley and Levine illustrate how enhancers may either employ suboptimal binding affinity or suboptimal spacing between binding sites, which prevents ectopic activity [21,22].
Considering how frequently enhancers evolve by co-opting pre-existing activities, the role of pleiotropy in constraining their subsequent evolution has become increasingly appreciated. The loss of trichomes in Drosophila sechellia compared to other fly species, including Drosophila melanogaster, is one of the most well understood examples of morphological differences that have been dissected to the level of participating enhancers, mutations, and the binding sites they affect [23–25]. In an elegant set of experiments, Preger-Ben Noon et al. [26] performed a high-resolution identification of the binding sites that were gained and lost as an enhancer of the shavenbaby (svb) gene lost its trichome-patterning activity in the dorsal surface of D. sechellia larvae. Combining transcriptomic data on sorted epithelial cells with a computational analyses, they found that binding sites for the transcriptional activator Arrowhead were lost in D. sechellia [26]. Additionally, through a functional assay involving RNA interference for all detected transcription factors, they also found that the complete inactivation of this enhancer required evolution of a binding site for the spatially restricted repressor Abrupt. This transition was speculated to involve the gain of repressive inputs that allow maintenance of this enhancer’s pleiotropic function in other tissues. In a similar case, the inactivation of a limb enhancer of the Tbx4 transcription factor in snakes occurred while preserving its function in the genitalia, likely contributing to evolution of the characteristic limbless body plan of these animals [27]. These findings illustrate how enhancers can disable linkages in gene regulatory networks, while maintaining pleiotropic functions in other tissues.
Network origins as a window into morphological novelties
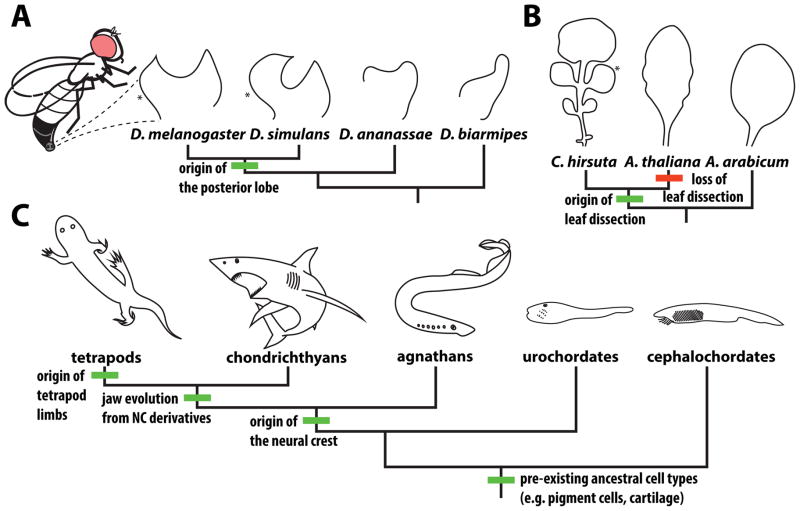
To understand how new structures (i.e. “morphological novelties” [28–30]) evolve, one promising avenue of investigation is to trace the evolutionary history of their developmental networks. Several recent examples have leveraged the principles of enhancer evolution discussed above to study how novelties arose at a network level (Figure 2).
Figure 2. Morphological structures whose origins have been illuminated through the study of gene regulatory networks and their constituent enhancers.
(A) The posterior lobe (*) is a genital outgrowth unique to males of Drosophila (D.) melanogaster and its close relatives that is required for mating. (B) Leaf dissection and the consequent presence of distinct leaflets (*) has arisen multiple times in seed plants. A convenient model for studying this trait is the complex leaf of the Cardamine (C.) hirsuta, which likely evolved from an ancestral simple leaf exemplified by Aethionema arabicum, a basally branching species in the Brassicaceae family. The well-studied model organism Arabidopsis thaliana has lost leaf dissection and has simple leaves with only slight serrations at their margins. (C) Chordate novelties. The neural crest (NC) is a novel migratory cell population that invades multiple tissues along the body axis, differentiating into several different cell types. NC derivatives play key roles in the development of the vertebrate jaw. Several of the cell types that neural crest cells ultimately adopt pre-date the origins of this dynamic cell population, which arose in the ancestor of vertebrates. The tetrapod limb evolved from fins that first appeared in jawless fish.
The posterior lobe of Drosophila male genitalia
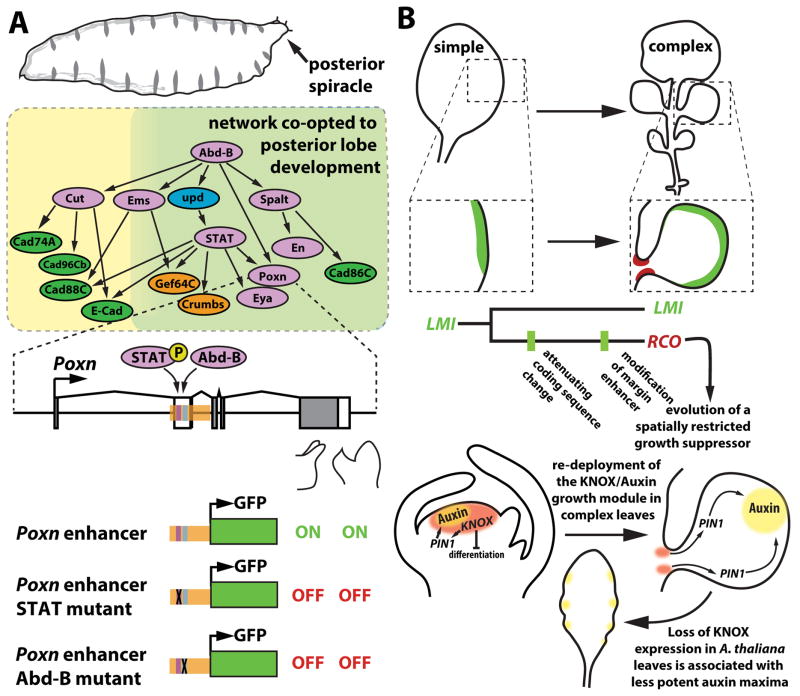
Genital traits represent some of the most rapidly evolving morphologies in the animal kingdom, and among insects, these characters are key to species identification [31,32]. A recent study investigated the origins of a genital appendage, the posterior lobe (Figure 2A), present in the model organism D. melanogaster [33]. Development of this structure requires the transcription factor Pox neuro (Poxn) [34], and the authors used this pivotal gene to trace the network’s evolutionary history. By examining an enhancer of Poxn that drives expression in the posterior lobe during pupal development, they found that its function had been co-opted from another network deployed in the posterior spiracle, a structure that forms during embryonic development (Figure 3A). Interestingly, both the spiracle and lobe form in posterior regions of the Drosophila body plan, in a zone specified by the Hox gene Abdominal-B (Abd-B), a known regulator of several genes in the spiracle network [35]. The authors found that several genes of this ancestral network are active in the posterior lobe, and showed that at least seven enhancers active in this structure can be traced to activities in the posterior spiracle. In two enhancers, individual transcription factor binding sites were required for activity in both the spiracle and lobe contexts (Figure 3A). This demonstrates how tracing the origin of a network’s enhancers can illuminate ancestral functions that would have been impossible to predict a priori.
Figure 3. Tracing the origins of networks that participate in novelties.
(A) Co-option of genes to the posterior lobe network. (top) Multiple genes of the posterior lobe network participate in the development of a larval breathing structure termed the posterior spiracle. Green shading delineates genes shared between the two networks. (bottom) The posterior lobe enhancer of the Pox neuro (Poxn) gene (orange shading) contains binding sites for STAT and Abdominal-B that are required for gene activity in both the novel posterior lobe and the ancestral posterior spiracle contexts. (B) The genetic basis for diversification of leaf shape in the Brassicaceae family. (top) In simple leaves, LMI is expressed at the leaf margin (green shading) and in floral tissues (not shown). The duplication of LMI resulted in a second copy of this transcription factor, named RCO, which evolved an enhancer specific to the base of leaflets (red shading), as well as an amino-acid substitution that reduced its pleiotropic effects on growth. The relative order of these regulatory and coding changes is unknown. (bottom) KNOX transcription factors have an ancestral role in the apical meristem, which involves suppressing differentiation and enabling organization of activity maxima of auxin that supports leaf initiation through the PIN1 transporter. This module was re-deployed in complex leaves to generate distal foci of auxin that promote growth. The secondary loss of complex leaves in A. thaliana is partly due to loss of KNOX expression in developing leaf primordia. This loss likely reduced the morphogenetic potency of marginal auxin foci, contributing to shallower outgrowths. Note that leaves likely first evolved from ancestral branched shoots expressing meristem genes [65] which may account for the predisposition to reactivate meristem genes that contributes to repeated independent origins of leaf dissection in seed plants.
Rewiring networks through cis and trans co-evolution during leaf shape evolution
Land plants show striking morphological variation, presenting attractive opportunities to study the contribution of regulatory evolution to morphological diversity in parallel to animals. One trait that has been extensively studied is leaf shape, particularly complex leaves with multiple leaflets, which have evolved repeatedly in seed plant lineages (Figure 2B). In the Brassicaceae family, complex leaves evolved from simpler forms, and work on two families of homeodomain transcription factors, KNOX (KNOTTED1-like homeobox) and REDUCED COMPLEXITY/LATE-MERISTEM IDENTITY1 (RCO/LMI1), has illuminated the molecular basis of this transition. Through a genetic screen in C. hirsuta, a complex-leaved A. thaliana relative [36] (Figure 2B), Vlad et al. identified the RCO gene [37]. RCO encodes an HD-ZIP class I transcription factor that promotes leaflet formation by repressing growth at focal points along leaf margins (Figure 3B). RCO arose in Brassicaceae through gene duplication of the floral regulator LMI1, and was secondarily lost in A. thaliana. Transgenic re-introduction of RCO from C. hirsuta into the genome of A. thaliana increased leaf complexity, indicating that its loss was a critical change for causing the simple leaf phenotype of this species. Diversification of RCO from LMI1 arose through cis-regulatory evolution, which generated a novel and specific RCO expression domain at the base of developing leaflets in a region pivotal for shape determination. To investigate how this occurred, Vuolo et al. [38] discovered that a leaf-margin enhancer of LMI1 which drives gene expression distally in leaf primordia was repurposed in the RCO paralog to drive expression proximally, flanking the emerging leaflets (Figure 3B). They also showed that a single amino acid substitution reduced RCO protein stability, which suppressed the potential pleiotropic effects of its altered expression. Both the regulatory and coding sequence changes in RCO show hallmarks of positive selection. Thus in this case, a potentially adaptive path for morphological evolution involved the neo-functionalization of an enhancer coupled with changes to its associated coding sequence, steps that limited pleiotropy while exploiting a novel expression domain. Interestingly, an RCO-like gene was also shown to underlie variation in leaf complexity between sister species in the related Capsella genus [39]. Hence, the LMI1/RCO genes likely define key nodes in an often-utilized network to modulate leaf shape.
While RCO restricts growth locally along the leaf margin, KNOX proteins actively promote outgrowth and patterning of leaflets. The expression of KNOX genes is associated with complex leaf forms, while simple leaves lack expression [40]. They are also active in the pluripotent Shoot Apical Meristem (SAM) from which leaves initiate. Their role in leaf complexity involves the partial redeployment of their SAM functions (for a review see [41]) of repressing differentiation and influencing cell polarity [42]. Previous work showed that cis-regulatory divergence at two KNOX genes, SHOOTMERISTMLESS (STM) and BREVIPEDICELLUS (BP), correlated with differences in leaf shape between A. thaliana and C. hirsuta [43]. However the functional significance of these regulatory differences were unknown. Rast-Somsich et al. showed that regulatory changes in the C. hirsuta BP gene were more potent than those at STM in terms of restoring complexity to A. thaliana leaves [44]. This result contrasted with the relative pleiotropy of these two genes, as mutations in BP had less widespread effects than STM on plant development in both C. hirsuta and A. thaliana. The resulting changes in BP expression introduced a new node in a small GRN that shapes leaf growth and promotes activity maxima of the indolic hormone auxin, which supports both leaf and leaflet initiation [44–46]. These findings indicate that regulatory divergence of weakly pleiotropic regulators like BP might offer favorable paths for morphological divergence to occur. Ichihashi et al. took a complementary genomics approach to study evolution of leaf complexity in the tomato lineage where this trait arose independently [47]. By conducting comparative transcriptome analyses between three species differing in leaflet number, they detected evolutionary changes in KNOX-related gene co-expression networks and identified a BOP transcription factor as an upstream modulator of KNOX activity and leaf shape [47].
The neural crest
The cartilage and skeletal elements of the vertebrate head embody an exceptionally complex novelty that allowed this group to transition to a predatory life style (Figure 2C). Of the cell types that contribute extensively to these structures, the neural crest stands out as a new tissue type whose origination was crucial to the evolution of this novelty [48]. Neural crest cells comprise a multipotent migratory population that invades multiple tissues along the anterior-posterior axis of the embryo, and subsequently differentiate into several different cell types. The neural domain that produces neural crest cells, the neural border, implements a highly conserved network that appears to predate the neural crest’s emergence [49]. In contrast, the gene regulatory network underlying neural crest formation and migration (the NC-GRN) seems to be unique to vertebrates. Based upon comparative analyses of gene expression, it has been argued that many of the cell types derived from the neural crest (such as melanocytes, cartilage, connective tissues, and sensory neurons) already existed in basal lineages, and were redeployed in descendants of this new cell type [50]. This suggests many of the gene regulatory sub-circuits conferring neural crest-like behaviors and potencies predate final assembly of the full NC-GRN in the vertebrate lineage. One gene, SoxE plays an important role in the specification of neural crest fate. A recent study introduced a 186kb fragment encompassing the SoxE gene from amphioxus into zebrafish [51]. While this genomic fragment recapitulated the amphioxus pattern of SoxE expression, it failed to drive neural crest expression, suggesting that novel neural crest enhancers arose at this key gene. However, a large number of cell types descend from the neural crest, and recent studies have made arguments for migratory populations in outgroup species that may share ancestry with the neural crest [52,53]. Detailed studies of enhancers within these networks may unveil their underlying homology relationships.
Novelties among vertebrate appendages
Some of the most striking morphological novelties reside in the appendages of vertebrates. Specifically, the tetrapod limb has novel elements in the wrist, ankle, and digits. As such, it represents a remarkably complex elaboration of the fin from an aquatic ancestor that had fewer skeletal and muscular elements (Figure 2C). The role of Hox genes in the evolution of the tetrapod limb has long been thought to correlate with a late phase of Hox expression in distal portions that form digits [54,55]. Two recent studies elegantly demonstrated that fish indeed have a late phase of Hox gene activity that is controlled by elements conserved with tetrapods [56,57]. Previous studies reported that the zebrafish versions of these elements fail to drive gene expression in the mouse limb bud, suggesting that these regions were novel to tetrapods [58]. However, this interpretation may have been complicated by derived features of zebrafish. Using the genome of the spotted gar [59], Gehrke et al. identified late Hox enhancers that drive distal fin expression [57]. While the zebrafish version of this enhancer lacks activity in a mouse reporter assay, the gar version is able to produce a pattern similar to that driven by the endogenous mouse enhancer. This result suggests that the lack of zebrafish activity likely reflects derived differences in zebrafish that cause its enhancer to no longer function in the mouse limb bud [57]. Such findings highlight the problem that tests for novelty in gene regulatory elements implicitly depend upon negative results (lack of activity), which may result from drift in the lineage displaying the ancestral trait rather than active changes in the lineage developing the novelty. Lineage tracing of cell populations marked by Hoxd13a enhancers, coupled with CRISPR/Cas9 knockout of Hox13 paralogs in fish confirmed how this late phase of expression is required to pattern distal fin elements [56]. Collectively, these new findings suggest that known networks regulating limb development are ancient and that the changes underlying the evolution of the tetrapod limb lie in genes outside the Hox loci in this network. This work also illustrates how tracing a network’s enhancers can clarify homology relationships among highly divergent traits. Given the age of the tetrapod limb (~370 MYA) it is likely that its evolution required multiple changes of small effect scattered throughout the genome.
Concluding remarks
The above studies show how the examination of enhancer history provides an important perspective into network origins and diversification. They underscore how enhancers lie at the heart of pleiotropic connections between different networks and also direct our attention towards their most evolutionarily relevant feature: the nodes that underlie trait diversity. The morphological features discussed above arose millions, or hundreds of millions of years ago, and their evolution probably involved coordinated changes in dozens to hundreds of genes. Thus, an important future challenge is to understand and quantify how the integration of multiple genetic changes produced such complex traits. In parallel, studies of the effectors of these GRNs that mediate morphogenesis will be a critical area of research. What are the key genes causing cells to grow, collapse, mineralize or move to produce phenotypic diversity and how do they exert their effects? Answering these questions will require the combination of classical genetic approaches and genomics coupled with recently developed methods for quantitative and computational studies of development [60–62], cell biology, and precision genome engineering [63,64].
Highlights.
Genome-wide and single gene studies have revealed a variety of mechanisms by which new expression patterns arise
Studying newly evolved morphologies (novelties) at the level of their regulatory sequences has provided key insights into the history of their genetic networks
Pleiotropic connections between networks have resulted both from wholesale network co-options and expansion of regulatory sequence activity to new developmental contexts
Targeted developmental changes in regulatory sequences have been shown to underlie morphological novelty in both animals and plants
Acknowledgments
We thank members of the Tsiantis and Rebeiz lab groups for helpful discussions, and Sheila McCormick, Thomas M. Williams, and Daniel M. Medeiros for comments on the manuscript. Work on Drosophila genital evolution in the Rebeiz lab is supported by grants from the National Institutes of Health (GM107387 and GM112758). The Tsiantis lab is supported by Deutsche Forschungsgemeinschaft grants “Adaptomics” TS 229/1-1, SFB (Sonderforschungbereit) 680, and a core grant by the Max Planck Society. We thank Nicolas Gompel and Benjamin Prud’homme for organizing the 2014 Fondation de Treilles meeting on “The mechanisms of evolutionary changes and adaptation”, where discussions relating to this review started.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Davidson EH, Peter IS. Genomic Control Process [Internet] Elsevier; 2015. [Google Scholar]
- 2.Levine M. Transcriptional enhancers in animal development and evolution. Curr Biol. 2010;20:R754–R763. doi: 10.1016/j.cub.2010.06.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Britten RJ, Davidson EH. Repetitive and non-repetitive DNA sequences and a speculation on the origins of evolutionary novelty. Q Rev Biol. 1971;46:111–138. doi: 10.1086/406830. [DOI] [PubMed] [Google Scholar]
- 4.Britten RJ, Davidson EH. Gene regulation for higher cells: a theory. Science (80- ) 1969;165:349–357. doi: 10.1126/science.165.3891.349. [DOI] [PubMed] [Google Scholar]
- 5.Emera D, Wagner GP. Transformation of a transposon into a derived prolactin promoter with function during human pregnancy. Proc Natl Acad Sci U S A. 2012;109:11246–51. doi: 10.1073/pnas.1118566109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lynch VJ, Leclerc RD, May G, Wagner GP. Transposon-mediated rewiring of gene regulatory networks contributed to the evolution of pregnancy in mammals. Nat Genet. 2011;43:1154–9. doi: 10.1038/ng.917. [DOI] [PubMed] [Google Scholar]
- 7.de Souza FSJ, Franchini LF, Rubinstein M. Exaptation of transposable elements into novel cis-regulatory elements: is the evidence always strong? Mol Biol Evol. 2013;30:1239–51. doi: 10.1093/molbev/mst045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lynch VJ, Nnamani MC, Kapusta A, Brayer K, Plaza SL, Mazur EC, Emera D, Sheikh SZ, Grützner F, Bauersachs S, et al. Ancient transposable elements transformed the uterine regulatory landscape and transcriptome during the evolution of mammalian pregnancy. Cell Rep. 2015;10:551–61. doi: 10.1016/j.celrep.2014.12.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Emera D, Yin J, Reilly SK, Gockley J, Noonan JP. Origin and evolution of developmental enhancers in the mammalian neocortex. Proc Natl Acad Sci U S A. 2016;113:E2617–26. doi: 10.1073/pnas.1603718113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ••10.Chuong EB, Elde NC, Feschotte C. Regulatory evolution of innate immunity through co-option of endogenous retroviruses. Science. 2016;351:1083–7. doi: 10.1126/science.aad5497. This paper integrates genomic approaches with genetic manipulations to demonstrate the impact of recently inserted transposable element sequences on gene expression. Performing ChIP-seq analyses on three cell lines that respond to interferon gamma, the authors found that multiple endogenous retrovirus families associated with binding for two key regulators of the interferon response. CRISPR/Cas9 deletion of four identified transposable elements confirmed that these regions contribute to the interferon response of these genes. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ••11.Indjeian VB, Kingman GA, Jones FC, Guenther CA, Grimwood J, Schmutz J, Myers RM, Kingsley DM. Evolving New Skeletal Traits by cis-Regulatory Changes in Bone Morphogenetic Proteins. Cell. 2016;164:45–56. doi: 10.1016/j.cell.2015.12.007. This paper provides a nuanced understanding of the role of transposons in morphological evolution by examining the contribution of GDF6 to skeletal trait diversification in sticklebacks and humans. Genetic mapping and transgenic assays showed a that transposon insertion and point mutations contributed increased expression of GDF6, leading to armor plate reduction in freshwater sticklebacks. In humans, a distinct conserved enhancer element active in the foot was deleted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Imsland F, Feng C, Boije H, Bed’hom B, Fillon V, Dorshorst B, Rubin C-J, Liu R, Gao Y, Gu X, et al. The Rose-comb mutation in chickens constitutes a structural rearrangement causing both altered comb morphology and defective sperm motility. PLoS Genet. 2012;8:e1002775. doi: 10.1371/journal.pgen.1002775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dorshorst B, Harun-Or-Rashid M, Bagherpoor AJ, Rubin C-J, Ashwell C, Gourichon D, Tixier-Boichard M, Hallböök F, Andersson L. A genomic duplication is associated with ectopic eomesodermin expression in the embryonic chicken comb and two duplex-comb phenotypes. PLoS Genet. 2015;11:e1004947. doi: 10.1371/journal.pgen.1004947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guo Y, Gu X, Sheng Z, Wang Y, Luo C, Liu R, Qu H, Shu D, Wen J, Crooijmans RPMA, et al. A Complex Structural Variation on Chromosome 27 Leads to the Ectopic Expression of HOXB8 and the Muffs and Beard Phenotype in Chickens. PLoS Genet. 2016;12:e1006071. doi: 10.1371/journal.pgen.1006071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rebeiz M, Jikomes N, Kassner VA, Carroll SB. The evolutionary origin of a novel gene expression pattern through co-option of the latent activities of existing regulatory sequences. Proc Natl Acad Sci U S A. 2011;108:10036. doi: 10.1073/pnas.1105937108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gompel N, Prud’homme B, Wittkopp PJ, Kassner VA, Carroll SB. Chance caught on the wing: cis-regulatory evolution and the origin of pigment patterns in Drosophila. Nature. 2005;433:481–487. doi: 10.1038/nature03235. [DOI] [PubMed] [Google Scholar]
- 17.Koshikawa S, Giorgianni MW, Vaccaro K, Kassner VA, Yoder JH, Werner T, Carroll SB. Gain of cis -regulatory activities underlies novel domains of wingless gene expression in Drosophila. Proc Natl Acad Sci. 2015;112:201509022. doi: 10.1073/pnas.1509022112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pham T, Day SM, Glassford WJ, Williams TM, Rebeiz M. The evolutionary origination of a novel expression pattern through an extreme heterochronic shift. Evol Dev. 2017;19:43–55. doi: 10.1111/ede.12215. [DOI] [PubMed] [Google Scholar]
- 19.Glassford WJ, Rebeiz M. Assessing constraints on the path of regulatory sequence evolution. Philos Trans R Soc Lond B Biol Sci. 2013;368:20130026. doi: 10.1098/rstb.2013.0026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu F, Posakony JW. Role of architecture in the function and specificity of two Notch-regulated transcriptional enhancer modules. PLoS Genet. 2012;8:e1002796. doi: 10.1371/journal.pgen.1002796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Farley EK, Olson KM, Zhang W, Rokhsar DS, Levine MS. Syntax compensates for poor binding sites to encode tissue specificity of developmental enhancers. Proc Natl Acad Sci U S A. 2016;113:6508–13. doi: 10.1073/pnas.1605085113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ••22.Farley EK, Olson KM, Zhang W, Brandt AJ, Rokhsar DS, Levine MS. Suboptimization of developmental enhancers. Science. 2015;350:325–8. doi: 10.1126/science.aac6948. The authors show that activity of a key developmental enhancer of the Otx gene in Ciona (sea squirt) is constrained by trade-offs between the specificity of gene activation and the level of transcriptional activity. Their findings highlight how biochemical level functions were sub-optimized to facilitate the development of tissue- and organism-level traits. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sucena E, Delon I, Jones I, Payre F, Stern DL. Regulatory evolution of shavenbaby/ovo underlies multiple cases of morphological parallelism. Nature. 2003;424:935–938. doi: 10.1038/nature01768. [DOI] [PubMed] [Google Scholar]
- 24.Frankel N, Erezyilmaz DF, McGregor AP, Wang S, Payre F, Stern DL. Morphological evolution caused by many subtle-effect substitutions in regulatory DNA. Nature. 2011;474:598–603. doi: 10.1038/nature10200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McGregor AP, Orgogozo V, Delon I, Zanet J, Srinivasan DG, Payre F, Stern DL. Morphological evolution through multiple cis-regulatory mutations at a single gene. Nature. 2007;448:587–590. doi: 10.1038/nature05988. [DOI] [PubMed] [Google Scholar]
- ••26.Preger-Ben Noon E, Davis FP, Stern DL. Evolved Repression Overcomes Enhancer Robustness. Dev Cell. 2016 doi: 10.1016/j.devcel.2016.10.010. A textbook example of how evolution of transcriptional repression can contribute to morphological shifts by disabling robust gene regulatory modules. The authors show that a two-pronged process underpinned loss of robust activity of an enhancer in the Drosophila shavenbaby gene. First, activator sites were lost, weakening enhancer activity. Second, the evolution of a binding site for a body wall-specific repressor eliminated expression. [DOI] [PubMed] [Google Scholar]
- 27.Infante CR, Mihala AG, Park S, Wang JS, Johnson KK, Lauderdale JD, Menke DB. Shared Enhancer Activity in the Limbs and Phallus and Functional Divergence of a Limb-Genital cis-Regulatory Element in Snakes. Dev Cell. 2015;35:107–119. doi: 10.1016/j.devcel.2015.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wagner GP, Lynch VJ. Evolutionary novelties. Curr Biol. 2010;20:R48–52. doi: 10.1016/j.cub.2009.11.010. [DOI] [PubMed] [Google Scholar]
- 29.Shubin N, Tabin C, Carroll S. Deep homology and the origins of evolutionary novelty. Nature. 2009;457:818–823. doi: 10.1038/nature07891. [DOI] [PubMed] [Google Scholar]
- 30.Moczek AP. On the origins of novelty in development and evolution. BioEssays. 2008;30:432–47. doi: 10.1002/bies.20754. [DOI] [PubMed] [Google Scholar]
- 31.Eberhard WG. Sexual selection and animal genitalia. Harvard University Press; 1985. [Google Scholar]
- 32.Masly JP. 170 Years of “Lock-and-Key”: Genital Morphology and Reproductive Isolation. Int J Evol Biol. 2012;2012:247352. doi: 10.1155/2012/247352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ••33.Glassford WJ, Johnson WC, Dall NR, Smith SJ, Liu Y, Boll W, Noll M, Rebeiz M. Co-option of an Ancestral Hox-Regulated Network Underlies a Recently Evolved Morphological Novelty. Dev Cell. 2015 doi: 10.1016/j.devcel.2015.08.005. This study provided evidence that gene regulatory networks are co-opted during the evolution of novelties by redeploying pre-existing enhancers from one context to another. The authors found that multiple genes activated during the development of a Drosophila genital structure do so through the action of enhancers that are used to activate these genes in an unrelated structure that is patterned during embryonic development. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Boll W, Noll M. The Drosophila Pox neuro gene: control of male courtship behavior and fertility as revealed by a complete dissection of all enhancers. Development. 2002;129:5667–5681. doi: 10.1242/dev.00157. [DOI] [PubMed] [Google Scholar]
- 35.Lovegrove B, Simões S, Rivas ML, Sotillos S, Johnson K, Knust E, Jacinto A, Hombría JC-G. Coordinated control of cell adhesion, polarity, and cytoskeleton underlies Hox-induced organogenesis in Drosophila. Curr Biol. 2006;16:2206–2216. doi: 10.1016/j.cub.2006.09.029. [DOI] [PubMed] [Google Scholar]
- 36.Gan X, Hay A, Kwantes M, Haberer G, Hallab A, Dello Ioio R, Hofhuis H, Pieper B, Cartolano M, Neumann U, et al. The Cardamine hirsuta genome offers insight into the evolution of morphological diversity. Nat Plants. 2016;2:16167. doi: 10.1038/nplants.2016.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vlad D, Kierzkowski D, Rast MI, Vuolo F, Dello Ioio R, Galinha C, Gan X, Hajheidari M, Hay A, Smith RS, et al. Leaf Shape Evolution Through Duplication, Regulatory Diversification, and Loss of a Homeobox Gene. Science (80- ) 2014;343:780–783. doi: 10.1126/science.1248384. [DOI] [PubMed] [Google Scholar]
- ••38.Vuolo F, Mentink RA, Hajheidari M, Bailey CD, Filatov DA, Tsiantis M. Coupled enhancer and coding sequence evolution of a homeobox gene shaped leaf diversity. Genes Dev. 2016;30:2370–2375. doi: 10.1101/gad.290684.116. This study provides a pioneering example of how the interplay of regulatory and coding sequence diversification can drive morphological evolution. The novel expression domain conferred by diversification of a specific enhancer in the Reduced Complexity homeobox gene allowed changes in leaf form, while coding changes that reduced RCO protein stability mitigated the pleiotropic effects this change likely entailed. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sicard A, Thamm A, Marona C, Lee YW, Wahl V, Stinchcombe JR, Wright SI, Kappel C, Lenhard M. Repeated Evolutionary Changes of Leaf Morphology Caused by Mutations to a Homeobox Gene. Curr Biol. 2014;24:1880–1886. doi: 10.1016/j.cub.2014.06.061. [DOI] [PubMed] [Google Scholar]
- 40.Bharathan G. Homologies in Leaf Form Inferred from KNOXI Gene Expression During Development. Science (80- ) 2002;296:1858–1860. doi: 10.1126/science.1070343. [DOI] [PubMed] [Google Scholar]
- 41.Bar M, Ori N. Leaf development and morphogenesis. Development. 2014;141:4219–4230. doi: 10.1242/dev.106195. [DOI] [PubMed] [Google Scholar]
- 42.Barkoulas M, Hay A, Kougioumoutzi E, Tsiantis M. A developmental framework for dissected leaf formation in the Arabidopsis relative Cardamine hirsuta. Nat Genet. 2008;40:1136–1141. doi: 10.1038/ng.189. [DOI] [PubMed] [Google Scholar]
- 43.Hay A, Tsiantis M. The genetic basis for differences in leaf form between Arabidopsis thaliana and its wild relative Cardamine hirsuta. Nat Genet. 2006;38:942–947. doi: 10.1038/ng1835. [DOI] [PubMed] [Google Scholar]
- 44.Rast-Somssich MI, Broholm S, Jenkins H, Canales C, Vlad D, Kwantes M, Bilsborough G, Dello Ioio R, Ewing RM, Laufs P, et al. Alternate wiring of a KNOXI genetic network underlies differences in leaf development of A. thaliana and C. hirsuta. Genes Dev. 2015;29:2391–2404. doi: 10.1101/gad.269050.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bilsborough GD, Runions A, Barkoulas M, Jenkins HW, Hasson A, Galinha C, Laufs P, Hay A, Prusinkiewicz P, Tsiantis M. Model for the regulation of Arabidopsis thaliana leaf margin development. Proc Natl Acad Sci. 2011;108:3424–3429. doi: 10.1073/pnas.1015162108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Heisler MG, Ohno C, Das P, Sieber P, Reddy GV, Long JA, Meyerowitz EM. Patterns of Auxin Transport and Gene Expression during Primordium Development Revealed by Live Imaging of the Arabidopsis Inflorescence Meristem. Curr Biol. 2005;15:1899–1911. doi: 10.1016/j.cub.2005.09.052. [DOI] [PubMed] [Google Scholar]
- 47.Ichihashi Y, Aguilar-Martinez JA, Farhi M, Chitwood DH, Kumar R, Millon LV, Peng J, Maloof JN, Sinha NR. Evolutionary developmental transcriptomics reveals a gene network module regulating interspecific diversity in plant leaf shape. Proc Natl Acad Sci. 2014;111:E2616–E2621. doi: 10.1073/pnas.1402835111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gans C, Northcutt RG. Neural Crest and the Origin of Vertebrates: A New Head. Science (80- ) 1983;220:268–273. doi: 10.1126/science.220.4594.268. [DOI] [PubMed] [Google Scholar]
- 49.Green SA, Simoes-Costa M, Bronner ME. Evolution of vertebrates as viewed from the crest. Nature. 2015;520:474–482. doi: 10.1038/nature14436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Medeiros DM. The evolution of the neural crest: new perspectives from lamprey and invertebrate neural crest-like cells. Wiley Interdiscip Rev Dev Biol. 2013;2:1–15. doi: 10.1002/wdev.85. [DOI] [PubMed] [Google Scholar]
- ••51.Jandzik D, Garnett AT, Square TA, Cattell MV, Kai Yu, Medeiros DM. Evolution of the new vertebrate head by co-option of an ancient chordate skeletal tissue. Nature. 2015:518. doi: 10.1038/nature14000. This paper took important steps to pinpoint a key change in the neural crest network. The authors identified a cartilage-like cell type in the cephalochordate amphioxus and showed that the SOXE gene is expressed in this tissue. Through an elegant assay that tested 186kb of regulatory sequence surrounding SOXE, the authors show that although aspects of amphioxus SOXE expression are recapitulated in zebrafish, that the capability to drive neural crest expression must be a vertebrate novelty. [DOI] [PubMed] [Google Scholar]
- 52.Stolfi A, Ryan K, Meinertzhagen IA, Christiaen L. Migratory neuronal progenitors arise from the neural plate borders in tunicates. Nature. 2015;527:371–374. doi: 10.1038/nature15758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Abitua PB, Wagner E, Navarrete IA, Levine M. Identification of a rudimentary neural crest in a non-vertebrate chordate. Nature. 2012;492:104–107. doi: 10.1038/nature11589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sordino P, van der Hoeven F, Duboule D. Hox gene expression in teleost fins and the origin of vertebrate digits. Nature. 1995;375:678–681. doi: 10.1038/375678a0. [DOI] [PubMed] [Google Scholar]
- 55.Shubin N, Tabin C, Carroll S. Fossils, genes and the evolution of animal limbs. Nature. 1997;388:639–48. doi: 10.1038/41710. [DOI] [PubMed] [Google Scholar]
- 56.Nakamura T, Gehrke AR, Lemberg J, Szymaszek J, Shubin NH. Digits and fin rays share common developmental histories. Nature. 2016;537:225–228. doi: 10.1038/nature19322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ••57.Gehrke AR, Schneider I, de la Calle-Mustienes E, Tena JJ, Gomez-Marin C, Chandran M, Nakamura T, Braasch I, Postlethwait JH, Gómez-Skarmeta JL, et al. Deep conservation of wrist and digit enhancers in fish. Proc Natl Acad Sci U S A. 2015;112:803–8. doi: 10.1073/pnas.1420208112. This study showed how the autopod (wrist/digit) function of Hox loci, previously posited to be a tetrapod novelty, is conserved to fish. By examining sequence conservation and chromatin accessibility, the authors identified autopod enhancers in zebrafish and gar genomes, and showed that they drive patterns very similar to those found in tetrapods. Strikingly, while the zebrafish enhancer fails to drive mouse limb bud expression, the gar sequence remains active, suggesting drift in zebrafish limb patterning. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Woltering JM, Noordermeer D, Leleu M, Duboule D. Conservation and divergence of regulatory strategies at Hox Loci and the origin of tetrapod digits. PLoS Biol. 2014;12:e1001773. doi: 10.1371/journal.pbio.1001773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Braasch I, Gehrke AR, Smith JJ, Kawasaki K, Manousaki T, Pasquier J, Amores A, Desvignes T, Batzel P, Catchen J, et al. The spotted gar genome illuminates vertebrate evolution and facilitates human-teleost comparisons. Nat Genet. 2016;48:427–437. doi: 10.1038/ng.3526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Barbier de Reuille P, Routier-Kierzkowska A-L, Kierzkowski D, Bassel GW, Schüpbach T, Tauriello G, Bajpai N, Strauss S, Weber A, Kiss A, et al. MorphoGraphX: A platform for quantifying morphogenesis in 4D. Elife. 2015;4:5864. doi: 10.7554/eLife.05864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Barkoulas M, Vargas Velazquez AM, Peluffo AE, Félix M-A. Evolution of New cis-Regulatory Motifs Required for Cell-Specific Gene Expression in Caenorhabditis. PLOS Genet. 2016;12:e1006278. doi: 10.1371/journal.pgen.1006278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kennaway R, Coen E, Green A, Bangham A. Generation of Diverse Biological Forms through Combinatorial Interactions between Tissue Polarity and Growth. PLoS Comput Biol. 2011;7:e1002071. doi: 10.1371/journal.pcbi.1002071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mali P, Esvelt KM, Church GM. Cas9 as a versatile tool for engineering biology. Nat Methods. 2013;10:957–63. doi: 10.1038/nmeth.2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gilles AF, Averof M. Functional genetics for all: engineered nucleases, CRISPR and the gene editing revolution. Evodevo. 2014;5:43. doi: 10.1186/2041-9139-5-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kenrick P, Crane PR. The origin and early diversification of land plants: a cladistic study. 1997 no volume. [Google Scholar]