Abstract
The well‐known link between longevity and the Sir2 histone deacetylase family suggests that histone deacetylation, a modification associated with repressed chromatin, is beneficial to longevity. However, the molecular links between histone acetylation and longevity remain unclear. Here, we report an unexpected finding that the MYST family histone acetyltransferase complex (MYS‐1/TRR‐1 complex) promotes rather than inhibits stress resistance and longevity in Caenorhabditis elegans. Our results show that these beneficial effects are largely mediated through transcriptional up‐regulation of the FOXO transcription factor DAF‐16. MYS‐1 and TRR‐1 are recruited to the promoter regions of the daf‐16 gene, where they play a role in histone acetylation, including H4K16 acetylation. Remarkably, we also find that the human MYST family Tip60/TRRAP complex promotes oxidative stress resistance by up‐regulating the expression of FOXO transcription factors in human cells. Tip60 is recruited to the promoter regions of the foxo1 gene, where it increases H4K16 acetylation levels. Our results thus identify the evolutionarily conserved role of the MYST family acetyltransferase as a key epigenetic regulator of DAF‐16/FOXO transcription factors.
Keywords: DAF‐16, FOXO transcription factor, longevity, MYST family histone acetyltransferase, stress resistance
Subject Categories: Ageing; Chromatin, Epigenetics, Genomics & Functional Genomics
Introduction
Aging is associated with a loss of chromatin repression, and chromatin status is controlled by alterations in epigenetic marks, including histone acetylation. The well‐known link between longevity and the Sir2 histone deacetylase gene family suggests that histone deacetylation, a modification associated with repressed chromatin, is beneficial to longevity 1, 2, 3, 4, 5. However, the role of Sir2 in regulating invertebrate lifespan is controversial 6, and a positive link between histone acetylation and longevity has also been reported 7.
In the nematode Caenorhabditis elegans, reduced insulin/IGF‐like signaling (IIS) greatly extends the organism's lifespan 8. This effect depends on the activity of the FOXO transcription factor DAF‐16, which is a key pro‐longevity protein. The insulin–FOXO pathway is evolutionarily conserved, and posttranslational control mechanisms of DAF‐16/FOXO, such as the control of DAF‐16 nuclear translocation by AKT‐mediated phosphorylation, have been well established 8. However, the regulation of DAF‐16/FOXO at the transcriptional level has remained unclear.
Dietary restriction is an effective and reproducible intervention for extending lifespans in multiple diverse species. In C. elegans, an intermittent fasting (IF) regimen effectively extends the organism's lifespan, and can be used as a model system to identify lifespan regulators. Our previous work has shown that target of rapamycin (TOR) and Rheb are involved in IF‐induced longevity 9. TOR is a member of the highly conserved family of Ser/Thr‐protein kinases, termed phosphatidylinositol 3‐kinase‐related kinase (PIKK). Because the down‐regulation of let‐363 (C. elegans TOR) results in partial inhibition of IF‐induced longevity 9, we hypothesized that other PIKK family proteins might also be involved in IF‐induced longevity.
In this study, we show that the MYST family acetyltransferase complex, the MYS‐1/TRR‐1 complex, up‐regulates the expression of daf‐16 and its target genes. Our results demonstrate that both MYS‐1 and TRR‐1 are recruited to the promoter regions of the daf‐16 gene, where they play a key role in histone acetylation, including histone H4 lysine 16 acetylation. Our results also show that the human MYST family Tip60/TRRAP complex promotes oxidative stress resistance by up‐regulating the expression of FOXO transcription factors in human cells. Tip60 is recruited to the promoter regions of foxo1 gene, where it increases H4K16 acetylation levels. These results identify the evolutionarily conserved role of the MYST family HAT complex in the control of FOXO transcription factors, which are key regulators of stress resistance and longevity in diverse organisms.
Results and Discussion
To examine the possibility that other PIKK family proteins might be involved in IF‐induced longevity, we measured the lifespan of worms fed RNAi bacteria expressing dsRNA targeting PIKK family members under both ad libitum (AL) and IF conditions. There are six PIKKs in mammals: ATM, ATR, DNA‐PKcs, SMG‐1, mTOR, and TRRAP. Five out of the six are conserved in C. elegans, and ATM‐1, ATL‐1, SMG‐1, LET‐363, and TRR‐1 are the C. elegans homologues of ATM, ATR, SMG‐1, mTOR, and TRRAP, respectively. trr‐1 RNAi as well as let‐363 RNAi suppressed IF‐induced longevity, whereas the RNAi of other PIKK family members did not (Figs 1A and EV1A).
Figure 1. The MYS‐1 HAT complex regulates longevity and oxidative stress resistance.
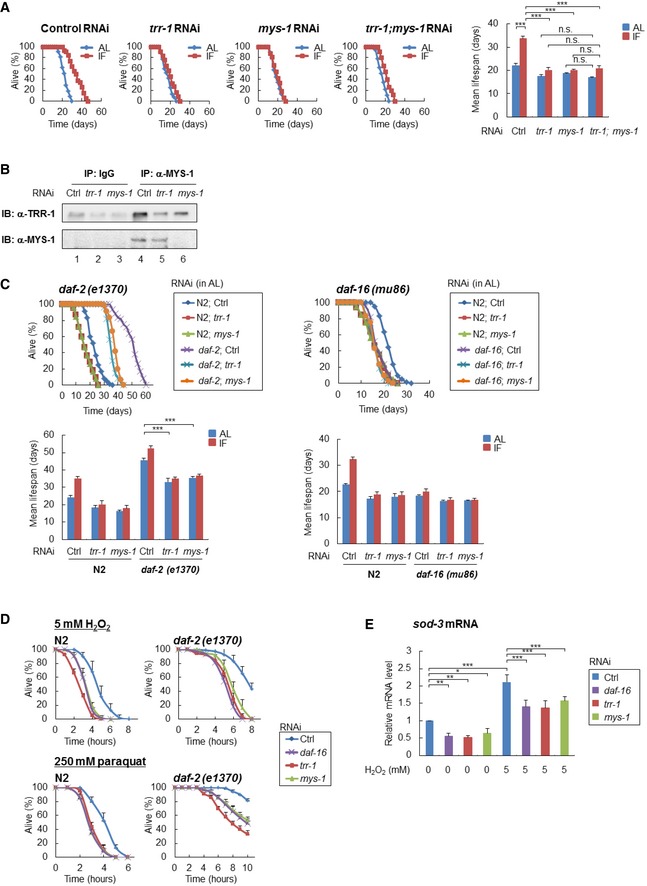
- Survival curves of the indicated RNAi‐treated N2 worms under AL and IF are shown [control RNAi, n = 117 (AL), 98 (IF); trr‐1 RNAi, n = 117 (AL), 107 (IF); mys‐1 RNAi, n = 118 (AL), 104 (IF); trr‐1;mys‐1 RNAi, n = 111 (AL), 106 (IF)]. The bars represent the mean lifespan of three independent experiments. n, total number of worms in three independent experiments. Error bars, SD. ***P < 0.001, n.s., not significant; one‐way ANOVA followed by Tukey's test.
- Immunoprecipitates with anti‐Tip60/MYS‐1 antibody from extracts of the indicated RNAi‐treated N2 worms at day 2 adulthood were subjected to immunoblot analysis using anti‐TRR‐1 antibody. Representative images of two independent experiments are shown.
- Survival curves of the indicated RNAi‐treated daf‐2(e1370) mutants (left) or daf‐16(mu86) mutants (right) under AL are shown [daf‐2(e1370) mutants n = 180 (control RNAi), 180 (trr‐1RNAi), 180 (mys‐1RNAi); daf‐16(mu86) mutants n = 178 (control RNAi), 180 (trr‐1RNAi), 180 (mys‐1RNAi)]. The bars represent the mean lifespan from three independent experiments. n, total number of worms in three independent experiments. Error bars, SD ***P < 0.001, one‐way ANOVA followed by Tukey's test.
- Survival curves of the indicated RNAi‐treated N2 worms or daf‐2(e1370) mutants, which were exposed to 5 mM hydrogen peroxide (H2O2) [N2 worms n = 180 (control RNAi), 180 (trr‐1RNAi), 180 (mys‐1RNAi); daf‐2(e1370) mutants n = 180 (control RNAi), 180 (trr‐1RNAi), 180 (mys‐1RNAi)] or 250 mM paraquat [N2 worms n = 128 (control RNAi), 128 (trr‐1RNAi), 128 (mys‐1RNAi); daf‐2(e1370) mutants n = 90 (control RNAi), 90 (trr‐1RNAi), 90 (mys‐1RNAi)] at the young adult stage, are shown. The number of surviving worms was counted every hour. Error bars represent the SD derived from three independent experiments. n, total number of worms in three independent experiments.
- Indicated RNAi‐treated N2 worms at the young adult stage were exposed to 5 mM H2O2 for 30 min, and sod‐3 mRNA expression levels were determined by qRT–PCR. The value of the control RNAi‐treated N2 worms (0 mM H2O2) was set to 1. Error bars represent the SD derived from three independent experiments. *P < 0.05, **P < 0.01, ***P < 0.001, one‐way ANOVA followed by Tukey's test. See also Fig EV1.
Source data are available online for this figure.
Figure EV1. The MYS‐1 HAT complex regulates longevity in Caenorhabditis elegans .
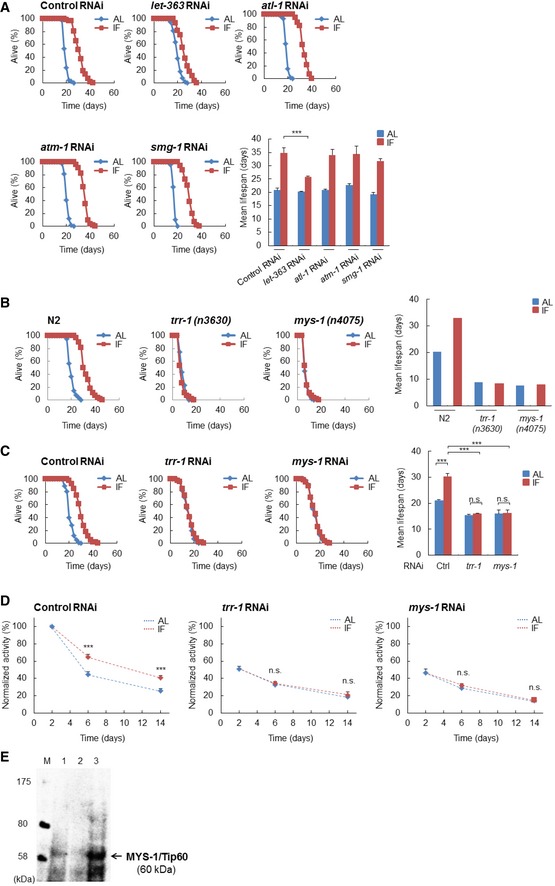
- Survival curves of the indicated RNAi‐treated N2 worms under AL and IF are shown [control RNAi, n = 117 (AL), 167 (IF); let‐363 RNAi, n = 98 (AL), 101 (IF); atl‐1 RNAi, n = 125 (AL), 162 (IF); atm‐1 RNAi, n = 117 (AL), 136 (IF); smg‐1 RNAi, n = 103 (AL), 138 (IF)]. The bars represent the mean lifespan from three independent experiments. n, total number of worms in three independent experiments. Error bars, SD ***P < 0.001, one‐way ANOVA followed by Tukey's test.
- Survival curves of the N2, trr‐1(n3630) mutants and mys‐1(n4075) mutants under AL and IF are shown [N2, n = 60 (AL), 54 (IF); trr‐1(n3630) mutants, n = 59 (AL), 50 (IF); mys‐1(n4075) mutants, n = 60 (AL), 58 (IF)]. The bars represent the mean lifespan from two independent experiments. n, total number of worms in two independent experiments.
- Survival curves of the indicated RNAi‐treated N2 worms under AL and IF are shown [control RNAi, n = 88 (AL), 83 (IF); trr‐1 RNAi, n = 87 (AL), 77 (IF); mys‐1 RNAi, n = 84 (AL), 81 (IF)]. To avoid potential developmental abnormalities, RNAi feeding was started at the young adult stage to day 2 adulthood. The bars represent the mean lifespan from three independent experiments. n, total number of worms in three independent experiments. Error bars, SD ***P < 0.001, n.s., not significant; one‐way ANOVA followed by Tukey's test.
- The value of the control RNAi‐treated worms at day 2 was set to 100. The bars represent the mean body movement from three independent experiments (n = 90 in every condition at each stage). n is a total of three independent experiments. Error bars, SD ***P < 0.001, n.s., not significant; one‐way ANOVA followed by Tukey's test.
- Whole‐cell lysates isolated from HeLa cells and whole‐worm lysates isolated from N2 and mys‐1(n4075) mutants were subjected to immunoblot analysis using an antibody against human Tip60. This antibody recognizes both human Tip60 and C. elegans MYS‐1 (lanes 1 and 3, respectively); however, the band at 60 kDa was not found in the mys‐1(n4075) deletion mutant (lane 2). See also Table EV1.
TRRAP protein is a member of the PIKK family; however, it does not possess kinase activity, because it lacks the conserved amino acids required for ATP binding and catalytic activity for phosphate transfer. In mammals, TRRAP serves as a common component of the Tip60 (also called KAT5) histone acetyltransferase (HAT) complex, which regulates transcription by modifying local chromatin through histone acetylation. In worms, a loss‐of‐function mutation in trr‐1 causes a multivulva phenotype, which is strikingly similar to that of the mys‐1 mutant 10 (a C. elegans orthologue of Tip60), thus suggesting that TRR‐1 and MYS‐1 act cooperatively as a HAT complex in vivo. To investigate the role of mys‐1 in lifespan regulation, we performed mys‐1 knockdown. Similar to trr‐1 RNAi, mys‐1 RNAi almost completely suppressed IF‐induced longevity (Fig 1A). In addition, IF‐induced longevity was completely suppressed in trr‐1(n3630) and mys‐1(n4075) mutants (Fig EV1B). These results demonstrate the involvement of trr‐1 and mys‐1 in IF‐induced longevity. However, the suppression of IF‐induced longevity in the trr‐1(n3630) and mys‐1(n4075) mutants may also have been related to developmental abnormalities in these mutants because their lifespan under AL conditions was reduced compared with that of the wild‐type N2 animals. To avoid potential developmental abnormalities, we then suppressed trr‐1 or mys‐1 expression, after the completion of development, by feeding RNAi. Down‐regulating trr‐1 or mys‐1 after the completion of development also resulted in near‐complete suppression of IF‐induced longevity (Fig EV1C). Therefore, we concluded that the suppression of IF‐induced longevity via trr‐1 or mys‐1 depletion is unlikely to be caused by developmental abnormalities.
An IF regimen delays the age‐dependent decline in locomotor activity; hence, we next investigated the locomotor activity of worms subjected to trr‐1 or mys‐1 RNAi. The results showed that trr‐1 RNAi and mys‐1 RNAi suppressed the IF‐induced delay in the age‐dependent decline in locomotor activity (Fig EV1D), thus indicating that trr‐1 and mys‐1 are necessary for IF to slow aging.
To examine whether TRR‐1 and MYS‐1 form a complex, similar to the mammalian Tip60 HAT complex, we performed immunoprecipitation experiments using an anti‐Tip60 antibody that specifically recognized the MYS‐1 protein in C. elegans extracts (Fig EV1E). Protein immunoprecipitated with this anti‐Tip60/MYS‐1 antibody from extracts of control RNAi‐treated worms contained the TRR‐1 protein in addition to the MYS‐1 protein (Fig 1B), indicating that TRR‐1 and MYS‐1 are in the same complex. The amounts of TRR‐1 and MYS‐1 protein in the complex were greatly reduced when the worms were treated with trr‐1 RNAi or mys‐1 RNAi (Fig 1B), thus confirming their complex formation in vivo. The lifespan measurements showed that double knockdown of trr‐1 and mys‐1 did not show an additive effect on lifespan compared to the single knockdown of trr‐1 or mys‐1 (Fig 1A). This result is also consistent with the idea that TRR‐1 and MYS‐1 exist in the same complex.
To analyze the role of the TRR‐1‐containing MYS‐1 HAT complex in lifespan regulation, we focused on the IIS effector DAF‐16. As shown in Fig 1A, the RNAi of trr‐1 or mys‐1 led to a slight reduction in the mean lifespan, even under AL conditions, suggesting that this HAT complex is required to maintain a normal lifespan, in which DAF‐16 plays a role. Moreover, the longevity effect of IF is mediated at least in part by DAF‐16 9. Therefore, we speculated that the MYS‐1 HAT complex might regulate longevity by controlling DAF‐16 activity. To test this idea, we examined whether trr‐1 and mys‐1 were required for the extended lifespan of daf‐2 mutants, which depends on DAF‐16 activity. The lifespan of daf‐2(e1370) mutants was markedly reduced by trr‐1 or mys‐1 RNAi (Fig 1C, left), whereas the lifespan of daf‐16(mu86) null mutants was only slightly decreased by trr‐1 or mys‐1 RNAi (Fig 1C, right). These results indicate that the MYS‐1 HAT complex regulates lifespans at least partly through DAF‐16.
Because previous studies have demonstrated a correlation between increased stress resistance and extended lifespan 11, 12, we examined the role of the MYS‐1 HAT complex in oxidative stress resistance. The trr‐1 RNAi or mys‐1 RNAi treatment markedly reduced the resistance to oxidative stress, such as exposure to H2O2 or paraquat, in both the wild‐type N2 worms and daf‐2(e1370) mutants (Fig 1D). The extent of the reduced oxidative stress resistance caused by trr‐1 or mys‐1 RNAi was similar to that caused by daf‐16 RNAi (Fig 1D). Moreover, the expression of superoxide dismutase (SOD‐3), which plays an important role in protection against oxidative stress, was markedly reduced by trr‐1 or mys‐1 RNAi in both the H2O2‐treated and untreated worms (Fig 1E), in which the extent of reduction was similar to that caused by daf‐16 RNAi (Fig 1E). Thus, these results demonstrate a requirement for the MYS‐1 HAT complex in oxidative stress resistance.
We next examined the mRNA expression of hsp‐12.6, a well‐known DAF‐16 transcriptional target, and found that hsp‐12.6 mRNA expression levels were markedly reduced in trr‐1 RNAi‐ and mys‐1 RNAi‐treated worms, as well as in daf‐16 RNAi‐treated worms in both the N2 worms and daf‐2(e1370) mutants (Fig 2A, left). Additionally, the increased fluorescence of HSP‐12.6::GFP in the fasting worms was markedly decreased by the treatments with trr‐1 or mys‐1 RNAi, as well as with daf‐16 RNAi (Fig 2A, right).
Figure 2. The MYS‐1 HAT complex regulates the expression of DAF‐16 and its transcriptional targets.
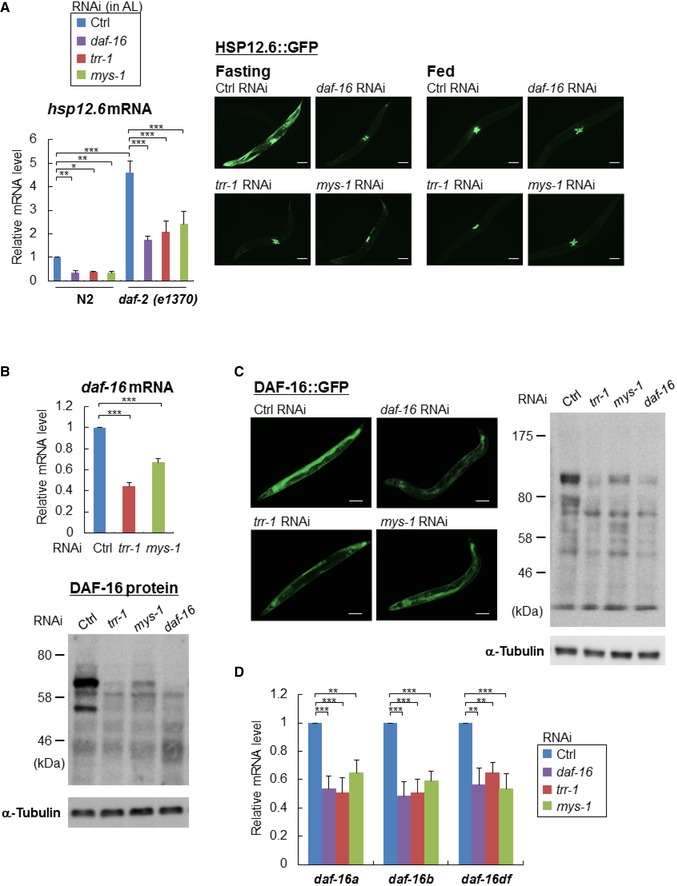
- Total RNA was extracted from the indicated RNAi‐treated N2 worms or daf‐2(e1370) mutants at day 2 adulthood, and hsp‐12.6 mRNA expression level was determined by qRT–PCR (left). Error bars represent the SD derived from three independent experiments. *P < 0.05, **P < 0.01, ***P < 0.001, one‐way ANOVA followed by Tukey's test. The value of the control RNAi‐treated N2 worms was set to 1. Representative images of HSP‐12.6::GFP expression in the indicated RNAi‐treated HSP‐12.6::GFP worms under fasting and feeding conditions are shown (right). Scale bar, 100 μm.
- Total RNA was extracted from the indicated RNAi‐treated N2 worms at day 2 adulthood, and the daf‐16 mRNA expression level was determined by qRT–PCR (upper). Error bars represent the SD derived from three independent experiments. ***P < 0.001, one‐way ANOVA followed by Tukey's test. The value of the control RNAi‐treated N2 worms was set to 1. Worm extracts from the indicated RNAi‐treated N2 worms or DAF‐16::GFP worms at day 2 adulthood were subjected to immunoblot analysis using an anti‐DAF‐16 antibody (bottom). Representative images of three independent experiments are shown.
- Representative images of DAF‐16::GFP expression in the indicated RNAi‐treated DAF‐16::GFP worms at day 2 adulthood are shown (left). Scale bar, 100 μm. Worm extracts from the indicated RNAi‐treated DAF‐16::GFP worms at day 2 adulthood were subjected to immunoblot analysis using an anti‐DAF‐16 antibody (right).
- Total RNA was extracted from the indicated RNAi‐treated N2 worms at day 2 adulthood, and the mRNA expression levels of daf‐16 isoforms were determined by qRT–PCR. Error bars represent the SD derived from three independent experiments. **P < 0.01, ***P < 0.001, one‐way ANOVA followed by Tukey's test. The value of the control RNAi‐treated N2 worms was set to 1. See also Fig EV2.
All of the above results led us to hypothesize that the MYS‐1 HAT complex regulates the expression of daf‐16 itself. Therefore, we performed quantitative PCR analyses and immunoblotting experiments, and the results clearly showed that trr‐1 RNAi or mys‐1 RNAi markedly reduced daf‐16 expression at both the mRNA (Fig 2B, upper) and protein (Fig 2B, lower) levels. Moreover, the expression level of DAF‐16::GFP in the DAF‐16::GFP worms was markedly reduced by trr‐1 RNAi or mys‐1 RNAi (Fig 2C). The reduced expression of DAF‐16::GFP was most clearly observed in the intestine (Fig 2C, left). Furthermore, to examine whether MYS‐1‐ overexpression affects the mRNA levels of daf‐16 and its transcriptional target, sod‐3, we produced transgenic worms which overexpressed MYS‐1. Expression levels of both daf‐16 and sod‐3 were markedly increased in MYS‐1‐overexpressing worms (Fig EV2A). We then examined whether the MYS‐1 HAT complex might regulate other genes that are known to be involved in the regulation of longevity and stress resistance. Our analysis showed that none of these genes, including eat‐2, daf‐2, pha‐4, hsf‐1, and skn‐1, were significantly affected by trr‐1 RNAi or mys‐1 RNAi (Fig EV2B). Therefore, transcriptional regulation by the MYS‐1 HAT complex appears to be specific to daf‐16 among longevity/stress resistance‐regulating genes.
Figure EV2. The MYS‐1 HAT complex regulates the expression of DAF‐16 but not other well‐known genes involved in lifespan regulation.
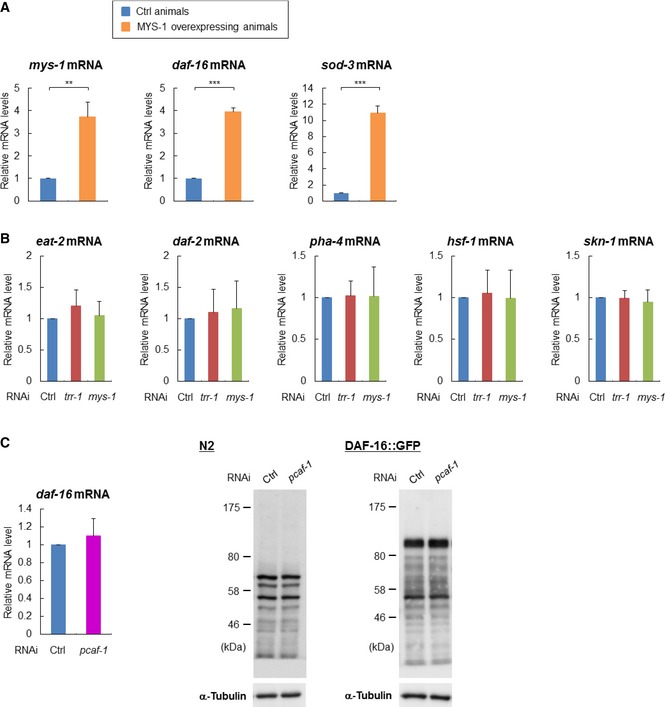
- Total RNA was extracted from the control or MYS‐1‐overexpressing worms, and the expression levels of daf‐16 mRNA or sod‐3 mRNA were determined by qRT–PCR. Error bars represent the SD derived from two independent experiments. **P < 0.01, ***P < 0.001, one‐way ANOVA followed by Tukey's test. The value of the control worms was set to 1.
- N2 worms were treated with the indicated RNAi, and total RNA was extracted at day 2 adulthood. The level of each mRNA was determined by qRT–PCR. The value of the control RNAi‐treated worms was set to 1. The bars represent the relative mRNA level from three independent experiments. Error bars, SD.
- Total RNA was extracted from the indicated RNAi‐treated N2 worms at day 2 adulthood, and the expression level of daf‐16 mRNA was determined by qRT–PCR. Error bars represent the SD derived from three independent experiments. The value of the control RNAi‐treated worms was set to 1 (left). Worm extracts from the indicated RNAi‐treated N2 worms or DAF‐16::GFP worms at day 2 adulthood were subjected to immunoblot analysis using an anti‐DAF‐16 antibody, respectively (right). Representative images of two independent experiments are shown.
In mammals, TRRAP is also a component of a Gcn5/PCAF HAT complex, and thus, TRR‐1 may also function in a Gcn5/PCAF HAT complex. To address this issue, we knocked down the pcaf‐1 gene that encodes the sole C. elegans Gcn5/PCAF‐like histone acetyltransferase. The result showed that pcaf‐1 RNAi did not alter the expression of daf‐16 at both the mRNA and protein levels (Fig EV2C), and thus suggests that the MYS‐1 HAT complex, but not the PCAF‐1 HAT complex, appears to be a critical regulator for daf‐16 expression.
Recently, multiple daf‐16 isoforms have been shown to function cooperatively, with two particular daf‐16 isoforms, daf‐16a and daf‐16d/f, playing a major role in the regulation of longevity and stress resistance 13. Therefore, we examined the effect of knocking down trr‐1 or mys‐1 on the expression of each daf‐16 isoform at day 2 adulthood and found that trr‐1 or mys‐1 RNAi markedly reduced the expression levels of all daf‐16 isoforms (Fig 2D). Together, these results indicate that the MYS‐1 HAT complex up‐regulates the expression of all daf‐16 isoforms and promotes longevity and stress resistance.
The mammalian Tip60 (MYST) HAT complex co‐activates gene expression through histone H4 acetylation 14, and Tip60‐mediated acetylation of histone H4 at lysine 16 (H4K16) is well known to facilitate gene expression 15. Thus, we performed an immunoblotting analysis using whole‐worm lysates to determine the acetylation state of the histones. RNAi knockdown of trr‐1 or mys‐1 dramatically decreased histone H4K16 acetylation and modestly decreased histone H4K12 acetylation (Fig 3A). Changes in the overall acetylation levels of histone H4 and H3 by trr‐1 or mys‐1 RNAi were not evident (Fig 3A). Moreover, the acetylation levels of histone H3K9 and H3K14 were not affected by trr‐1 or mys‐1 RNAi (Fig 3A).
Figure 3. The MYS‐1 HAT complex is recruited to the daf‐16 promoter region and contributes to histone acetylation.
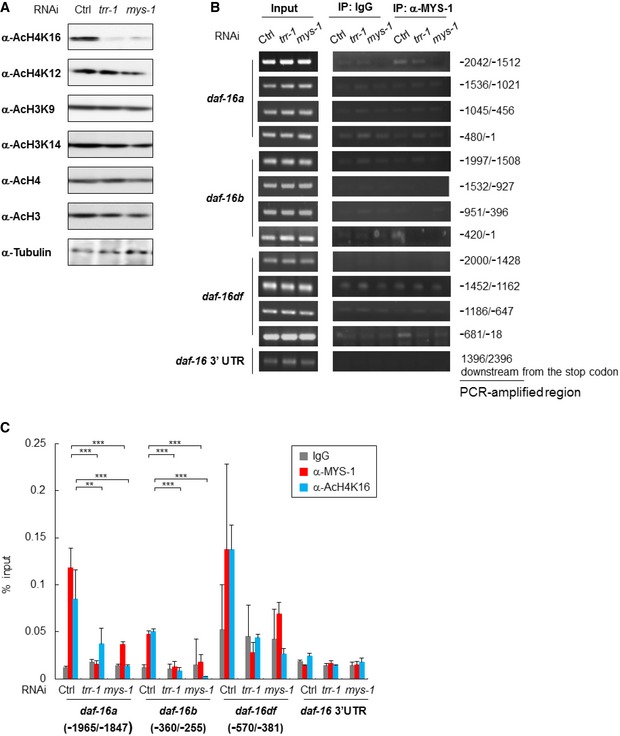
- Whole‐worm lysates isolated from the indicated RNAi‐treated N2 worms at day 2 adulthood were subjected to immunoblot analysis using the indicated antibodies. Representative images of three independent experiments are shown.
- MYS‐1 binding was examined by ChIP–PCR using crosslinked DNA–protein complexes isolated from the indicated RNAi‐treated N2 worms at day 2 adulthood with anti‐Tip60/MYS‐1 and control IgG antibody. PCR amplification was done with specific primers for the daf‐16 promoter region and the daf‐16 3′UTR. Representative images of two independent experiments are shown. Genomic DNA in the input samples was used as a positive control.
- MYS‐1 binding and histone H4K16 acetylation status were examined by ChIP–qPCR using crosslinked DNA–protein complexes isolated from the indicated RNAi‐treated N2 worms at day 2 adulthood with anti‐Tip60/MYS‐1, anti‐AcH4K16 and control IgG antibody. The bars represent the percentage of total input DNA for each ChIP sample, and error bars represent the SD derived from three independent experiments. **P < 0.01, ***P < 0.001, one‐way ANOVA followed by Tukey's test. See also Fig EV3.
Source data are available online for this figure.
We then performed chromatin immunoprecipitation (ChIP) assays on the daf‐16 genes. The analyses using anti‐MYS‐1 antibody (anti‐Tip60 antibody) revealed the physical interaction between MYS‐1 and the promoter regions of the daf‐16a, daf‐16b, and daf‐16d/f genes and demonstrated that the interaction with the daf‐16a and daf‐16d/f promoter regions was strong and the interaction with the daf‐16b promoter region was relatively weak (Fig 3B and C). Notably, this assay also revealed that TRR‐1 was recruited to the daf‐16 promoter region, because the interactions between MYS‐1 and the daf‐16 promoter regions were inhibited by trr‐1 RNAi (Fig 3B and C). Remarkably, the assays using anti‐AcH4K16 antibody showed that both MYS‐1 and TRR‐1 were required for histone H4K16 acetylation in these promoter regions (Fig 3C). Therefore, these results strongly suggest that the MYS‐1 HAT complex is recruited to daf‐16 promoter regions and involved in the transcriptional regulation of daf‐16 genes through histone acetylation.
We then examined whether overexpression of DAF‐16, which has been shown to be up‐regulated by the MYS‐1 HAT complex, could rescue the defects in trr‐1/mys‐1 RNAi worms. We used DAF‐16::GFP worms as worms overexpressing DAF‐16. The results clearly showed that DAF‐16 overexpression promoted lifespan and restored the response to IF (Fig EV3).
Figure EV3. DAF‐16 overexpression rescues the defects in trr‐1/mys‐1 RNAi worms.
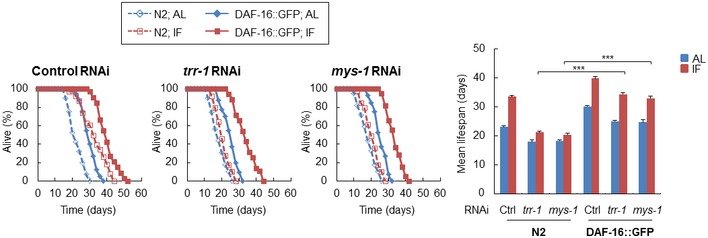
Survival curves of the indicated RNAi‐treated N2 (dashed line) or DAF‐16::GFP (solid line) worms under AL and IF are shown (left) [control RNAi, n = 80 (N2 in AL), 74 (N2 in IF), 117 (DAF‐16::GFP in AL), 99 (DAF‐16::GFP in IF); trr‐1 RNAi, n = 79 (N2 in AL), 77 (N2 in IF), 120 (DAF‐16::GFP in AL), 109 (DAF‐16::GFP in IF); mys‐1 RNAi, n = 75 (N2 in AL), 77 (N2 in IF), 118 (DAF‐16::GFP in AL), 108 (DAF‐16::GFP in IF)]. The bars represent the mean lifespan of three independent experiments (right). n, total number of worms in three independent experiments. Error bars, SD, ***P < 0.001, one‐way ANOVA followed by Tukey's test.
DAF‐16/FOXO, TRR‐1/TRRAP, and MYS‐1/Tip60 are all highly conserved proteins that are expressed in diverse eukaryotic organisms. Hence, the regulation of foxo genes by the MYST family HAT complex is likely to be conserved in mammals. Caenorhabditis elegans has one foxo gene (daf‐16), whereas mammals have four foxo family members: foxo1, foxo3, foxo4, and foxo6. We sought to determine whether the expression levels of foxo family members might be regulated by the Tip60 HAT complex, by using siRNAs against trrap and tip60 in HeLa cells. trrap siRNA and tip60 siRNA led to a 39% and a 55% decrease in their respective target gene's expression (Fig 4A). Our results showed that the histone H4K16 and H4K12 acetylation levels were markedly and modestly decreased, respectively, by trrap siRNA or tip60 siRNA in HeLa cells (Fig 4A). The results were consistent with those in trr‐1 RNAi‐treated or mys‐1 RNAi‐treated worms (see Fig 3A). Both siRNAs significantly reduced FOXO1 expression at both the mRNA and protein levels (Fig 4A and B) and slightly reduced foxo6 mRNA levels (Fig 4A). In addition, the mRNA levels of the FOXO1 transcriptional targets Mnsod, p21, and p27 16 were also reduced by trrap siRNA or tip60 siRNA (Fig 4C), whereas the mRNA levels of foxo4 and foxa1, another forkhead family member, were not reduced by trrap siRNA or tip60 siRNA (Fig 4A). These results show that the Tip60 HAT complex regulates the FOXO1 expression in HeLa cells.
Figure 4. The Tip60 HAT complex regulates FOXO1 expression in mammalian cells.
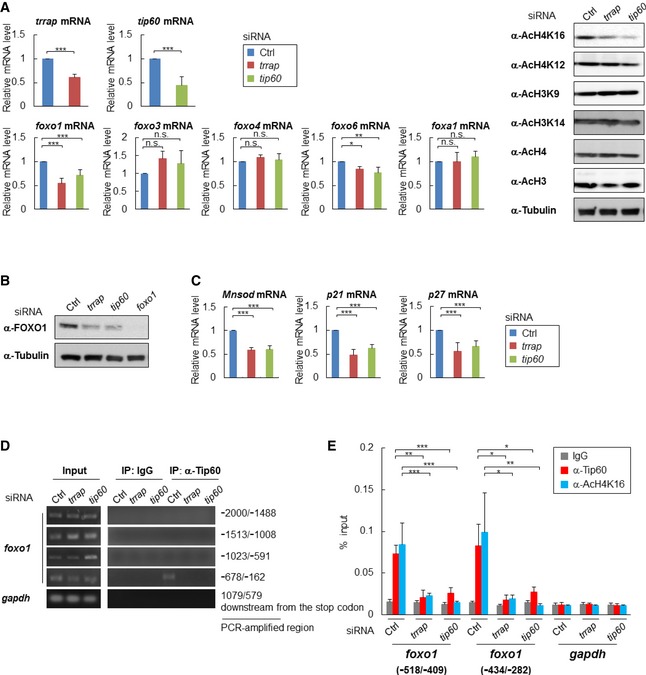
- HeLa cells were transfected with the indicated siRNA, and each sample was subjected to qRT–PCR (left) or immunoblot analysis (right). Error bars represent the SD derived from three independent experiments. *P < 0.05, **P < 0.01, ***P < 0.001, n.s., not significant; one‐way ANOVA followed by Tukey's test. The value of the control siRNA‐transfected cells was set to 1.
- Total cell lysates extracted from the indicated siRNA‐transfected HeLa cells were subjected to immunoblot analysis using anti‐FOXO1 antibodies. Representative images of three independent experiments are shown.
- Total RNA was extracted from the indicated siRNA‐transfected HeLa cells, and the mRNA expression levels of Mnsod, p21, and p27 were determined by qRT–PCR. Error bars represent the SD derived from three independent experiments. ***P < 0.001, one‐way ANOVA followed by Tukey's test. The value of the control siRNA‐transfected cells was set to 1.
- Tip60 binding was examined by ChIP–PCR using crosslinked DNA–protein complexes isolated from the indicated siRNA‐transfected HeLa cells with anti‐Tip60 and control IgG antibody. PCR amplification was done with specific primers for the foxo1 and gapdh promoter regions. Representative images of two independent experiments are shown. Genomic DNA in the input samples was used as a positive control.
- Tip60 binding and histone H4K16 acetylation were examined by ChIP–qPCR using crosslinked DNA–protein complexes isolated from the indicated siRNA‐transfected HeLa cells with anti‐Tip60, anti‐AcH4K16, and control IgG antibody. The bars represent the percentage of total input DNA for each ChIP sample, and error bars represent the SD derived from three independent experiments. *P < 0.05, **P < 0.01, ***P < 0.001, one‐way ANOVA followed by Tukey's test.
Source data are available online for this figure.
Because FOXO1 expression was shown to be markedly down‐regulated by trrap siRNA or tip60 siRNA, we then performed ChIP assays on the foxo1 gene. Assays using anti‐Tip60 antibody showed that Tip60, along with TRRAP, was recruited to the promoter region of foxo1 (Fig 4D and E). In addition, ChIP assays using anti‐AcH4K16 antibody showed that both Tip60 and TRRAP were required for histone H4K16 acetylation in the foxo1 promoter region (Fig 4E). These results strongly suggest that the Tip60 HAT complex is recruited to the foxo1 promoter region and plays an important role in the transcriptional regulation of the foxo1 gene through histone acetylation in human cells.
FOXO transcription factors have multiple functions, including the regulation of stress resistance 17. Because we identified the Tip60 HAT complex as an important epigenetic regulator of foxo1, we then examined the possibility that the Tip60 HAT complex might regulate cellular defense against oxidative stress in HeLa cells. The results showed that tip60 siRNA and trrap siRNA markedly reduced cellular resistance to oxidative stress at rates similar to those induced by foxo1 siRNA (Fig 5A). We next investigated the role of the Tip60 HAT complex and FOXO1 in the oxidative stress‐induced up‐regulation of the antioxidant genes Mnsod and catalase. After treatment with H2O2, Mnsod and catalase mRNA expression increased and reached a maximum level after 12 and 6 h, respectively, and these increases were suppressed by tip60 siRNA and trrap siRNA, as well as by foxo1 siRNA (Fig 5B). Finally, we examined whether overexpression of FOXO1 could induce the expressions of its transcriptional targets and rescue the response to oxidative stress in the absence of trrap or tip60. HeLa cells were transfected with a FOXO1 expression vector (Fig EV4). FOXO1 overexpression increased the mRNA levels of its transcriptional targets and rescued the oxidative stress resistance in the absence of tip60 or trrap (Fig 5C and D). Collectively, these results show that the Tip60 (MYST) HAT complex regulates cellular stress resistance, at least partly through up‐regulating FOXO1 expression.
Figure 5. The Tip60 HAT complex regulates the cellular defense against oxidative stress in mammalian cells.
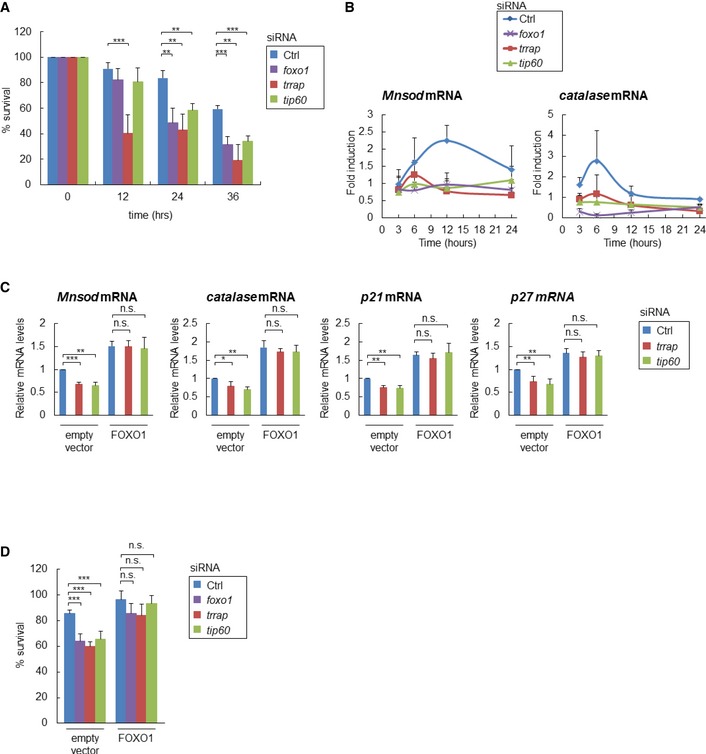
- The indicated siRNA‐transfected HeLa cells were exposed to 100 μM H2O2, and the percentage of surviving cells was measured by crystal violet staining at the indicated times. Error bars represent the SD derived from three independent experiments. **P < 0.01, ***P < 0.001, one‐way ANOVA followed by Tukey's test.
- The indicated siRNA‐transfected HeLa cells were exposed to 100 μM H2O2, and the mRNA expression level of Mnsod and catalase was determined by qRT–PCR at the indicated times. Error bars represent the SD derived from three independent experiments. The value of the control siRNA‐transfected cells (0 μM H2O2) was set to 1.
- Total RNA was extracted from the indicated RNAi‐ or plasmid‐transfected HeLa cells, and the mRNA expression levels of Mnsod, catalase, p21, and p27 were determined by qRT–PCR. Error bars represent the SD derived from three independent experiments. *P < 0.05, **P < 0.01, ***P < 0.001, n.s., not significant; one‐way ANOVA followed by Tukey's test. The value of the control siRNA‐/empty vector‐transfected cells was set to 1.
- The indicated RNAi‐ or plasmid‐transfected HeLa cells were exposed to 100 μM H2O2, and the percentage of surviving cells was measured by crystal violet staining after 24 h. Error bars represent the SD derived from three independent experiments. ***P < 0.001, n.s., not significant; one‐way ANOVA followed by Tukey's test. See also Fig EV4.
Figure EV4. Overexpression of FOXO1.
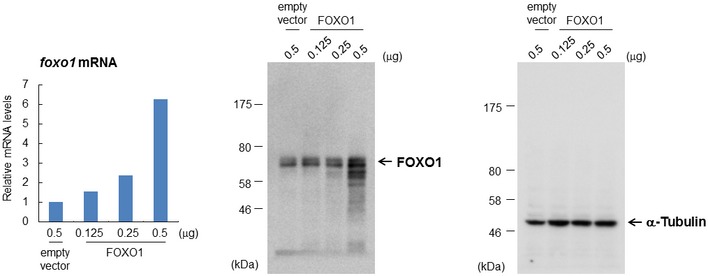
HeLa cells were transfected with the indicated plasmid, and each sample was subjected to qRT–PCR (left) or immunoblot analysis (middle and right). The value of the control vector‐transfected cells was set to 1. Data from one experiment are shown.
This study demonstrates that the MYST family HAT complex (MYS‐1/TRR‐1‐containing complex in C. elegans or Tip60/TRRAP‐containing complex in human) up‐regulates FOXO transcription factors (daf‐16 in C. elegans and foxo1 in human) and thus regulates stress resistance in both species, as well as longevity in C. elegans. Our analyses showed that the MYS‐1/TRR‐1 complex up‐regulates the expression of daf‐16 and its target genes and demonstrated that both MYS‐1 and TRR‐1 are recruited to the promoter regions of the daf‐16 gene, where they play a key role in histone acetylation, including histone H4 lysine 16 acetylation. Our results also show that the human MYST family Tip60/TRRAP complex promotes oxidative stress resistance by up‐regulating the expression of FOXO transcription factors in human cells. Tip60 is recruited to the promoter regions of the foxo1 gene, where it increases H4K16 acetylation levels.
Thus, our study illustrates the evolutionarily conserved role of the MYST family HAT complex in the control of FOXO transcription factors, which are key regulators of stress resistance and longevity in diverse organisms. Although a large number of previous studies have elucidated posttranslational control mechanisms of DAF‐16/FOXO 18, its transcriptional control mechanisms, especially its epigenetic control mechanisms, have remained unclear. In fact, the present study is the first to identify an epigenetic regulator of daf‐16/foxo gene expression. In conclusion, our findings provide novel insights into the regulatory mechanisms of aging and stress resistance by uncovering a new player, the MYST family HAT complex.
Materials and Methods
Caenorhabditis elegans strains
All of the nematodes were cultured using standard C. elegans methods 19. The following C. elegans strains were used for this study: wild‐type N2; MT12352, trr‐1(n3630)/mIn1[dpy‐10(e128) mIs14]; MT13172, mys‐1(n4075) V/nT1[qIs51]; CF1038, daf‐16(mu86); CB1370, daf‐2(e1370); TJ356, zls356[daf‐16::gfp; rol‐6]; and NIS1013, kytEx1013[Phsp‐12.6::hsp12.6::gfp; rol‐6]. To generate MYS‐1‐overexpressing worms, the fragment of the 5′ upstream sequence and the fragment of the coding region were amplified using PCR and the obtained fragments were cloned into the pPD95.75 vector. Transgenic worms were generated by microinjecting the plasmid into wild‐type N2 worms at 25 ng/μl, with the transformation marker.
Cell culture
HeLa cells were maintained in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal calf serum, penicillin, and streptomycin at 37°C in 5% CO2.
RNA interference
RNAi was performed by the feeding method as previously described 20. The first 500 nucleotides from the coding regions of trr‐1, mys‐1, let‐363, atl‐1, atm‐1, and smg‐1 complementary DNA were used for RNAi. The primers used were as follows: trr‐1 left, 5′‐CCGCGGATGGATCCGGCTATGGCTT‐3′, and trr‐1 right, 5′‐GGTACCCACTCGCCGTCAGATCAAT‐3′; mys‐1 left, 5′‐CCGCGGATGACCGAGCCGAAGAAGG‐3′, and mys‐1 right, 5′‐GGTACCATTGCATCTTCACTATGGC‐3′; let‐363 left, 5′‐CCGCG GATGCTCCAACAACACGGAA‐3′, and let‐363 right, 5′‐GGTACCGCTTTTGAAGCCATCTTGA‐3′; atl‐1 left, 5′‐CCGCGGATCGATAAGGATGTTTCAG‐3′, and atl‐1 right, 5′‐GGTACCATTCCACTTCAAATGATAT‐3′; atm‐1 left, 5′‐CCGCGGGAACCTGATAGAACAGTTT‐3′, and atm‐1 right, 5′‐GGTACCAACATTGCAGCATAAACAT‐3′; smg‐1 left, 5′‐CCGCGGATGATAACATCGAGAAACA‐3′, and smg‐1 right, 5′‐GGTACCTTCAAACAGTGGTTATAAA‐3′. To generate the RNAi construct for double knockdown of trr‐1 and mys‐1, the first 500 nucleotides from the coding region of mys‐1 cDNA were amplified by PCR and inserted into the trr‐1 RNAi construct. The primers used were described above. The pcaf‐1 gene silencing was done using RNAi library 21. For RNAi experiments, HT115 bacteria with RNAi plasmids were seeded on RNAi plates, and worms were fed with RNAi bacteria from egg to day 2 adulthood, unless otherwise mentioned. Animals fed HT115 bacteria carrying an empty pPD129.36 vector were used as controls.
Small interfering RNAs (siRNAs) were transfected into HeLa cells with Lipofectamine 2000 (Invitrogen) according to the manufacturer's protocol and harvested 48 h later. In all experiments, each siRNA was transfected using a final concentration of 20 nM. The following siRNAs were used: human trrap siRNA 22, 5′‐AAUGGCUCGAAAGGAAUGG‐3′; human tip60 siRNA 23, 5′‐GACGUAAGAACAAGAGUUAUU‐3′; and human foxo1 siRNA 24, 5′‐GAGCGUGCCCUACUUCAAGTT‐3′ (all RNA oligonucleotides were purchased from Sigma). MISSION siRNA Universal Negative Control #1 (Sigma) was used as a control.
Transfection
Complementary DNA (cDNA) of foxo1 was isolated by PCR and ligated into pcDNA3 vector. HeLa cells were transfected by using Lipofectamine 2000 (Invitrogen) according to the manufacturer's protocol and harvested 48 h later.
Lifespan analysis
The lifespan analysis and intermittent fasting were performed as previously described 9. All of the experiments were conducted at 20°C. P‐values were calculated using one‐way ANOVA followed by Tukey's test with the mean lifespan.
Locomotion activity
The worms were picked into M9 buffer in individual wells of 35‐mm culture dishes and allowed to equilibrate for 1 min. A single bend was defined as a complete movement through the midpoint and back, and the number of body bends in 1 min was counted. IF was started at day 2 adulthood. This assay was performed at day 2, day 6, and day 14 of adulthood at 20°C.
Fluorescence microscopy
The worms expressing DAF‐16::GFP or HSP‐12.6::GFP were synchronized and grown on RNAi bacteria from hatching until the L4/young adult stage and then transferred to FUdR‐containing plates. For the DAF‐16::GFP strains, the worms were collected at day 2 adulthood. For the HSP‐12.6::GFP strains, the worms were transferred to new plates with or without food at day 2 adulthood and collected after 2 days of fasting. Both strains were fixed with 2% formaldehyde for 2 min at room temperature and observed with a Zeiss Axioplan2.
Assays for oxidative stress resistance
To assay H2O2 or paraquat sensitivity, worms were transferred to M9 buffer containing 5 mM H2O2 or 250 mM paraquat and incubated at 20°C until death. The worms that did not move after a gentle mechanical touch were scored as dead, and the number of surviving worms was counted every hour.
Crystal violet assay
To assess the sensitivity to H2O2, HeLa cells transfected with siRNAs were exposed to 100 μM H2O2 for 0–36 h and fixed with 4% paraformaldehyde and then stained with 0.1% crystal violet. The dye was dissolved in 10% acetic acid, and absorbance was measured at OD590 nm.
Quantitative RT–PCR
Total RNA was isolated using TRIzol (Invitrogen). Reverse transcription was performed with a QuantiTect® Reverse Transcription kit (Qiagen) according to the manufacturer's instructions. cDNA was subjected to quantitative PCR analysis using an ABI 7300 Real‐Time PCR System (Applied Biosystems) with a SYBR Green PCR kit (Roche). Each value was normalized to act‐1 or gapdh. The primer sequences are available on request.
Immunoprecipitation and immunoblot analyses
For the immunoprecipitation analysis, the worms were lysed in IP buffer (0.1% NP‐40, 150 mM NaCl, 1 mM EDTA, 5% glycerol, 20 mM HEPES (pH 7.6), 1 mM PMSF, and 1 μg/ml aprotinin and 10 μM MG132) and the supernatant was incubated with anti‐Tip60 antibody (Abcam) or control rabbit IgG (Santa Cruz Biotechnology) for 4 h at 4°C. Protein G‐Sepharose beads (GE Healthcare) were then added to the sample and rotated overnight at 4°C. After several washes with wash buffer (0.1% NP‐40, 150 mM NaCl, and 20 mM HEPES (pH 7.6)), the pellet was lysed with SDS‐sample buffer and subjected to immunoblot analysis using anti‐Tip60 or anti‐TRR‐1 antibodies (a kind gift from Dr. Horvitz, H.R.). The immunoblot analysis was performed according to standard protocols using anti‐DAF‐16 (Santa Cruz Biotechnology), anti‐AcH4K16 (Millipore), anti‐AcH4K12 (Millipore), anti‐AcH3K9 (Millipore), anti‐AcH3K14 (Millipore), anti‐AcH4 (Millipore), anti‐AcH3 (Millipore), anti‐FOXO1 (Cell Signaling), and anti‐tubulin antibodies (Sigma). α‐Tubulin was used as a loading control.
Chromatin immunoprecipitation assay
The chromatin immunoprecipitation assays were performed as previously described 25. Briefly, the worms were grown on RNAi bacteria from hatching until the L4/young adult stage and harvested at day 2 adulthood. HeLa cells were transfected with scrambled or target gene‐specific siRNA and harvested 48 h later. After fixation with 1% formaldehyde, the worms and cells were lysed and sonicated (Diagenode, Bioruptor). Then, 10% of the total lysate was removed for input control, and the remaining lysates were diluted in IP dilution buffer. Immunoprecipitation was performed overnight at 4°C with 2 μg of specific antibodies conjugated to Protein G‐Sepharose beads. The antibodies used for immunoprecipitation are as follows: anti‐Tip60 antibody, anti‐AcH4K16 antibody, and normal rabbit IgG. The beads were washed, and the DNA–protein complexes were eluted. After reverse crosslinking and purification, the DNA was amplified and separated on 1% agarose gels. The relative enrichment levels were measured by quantitative PCR. Amplification of the daf‐16 3′UTR or gapdh promoter regions served as negative controls.
Author contributions
TI, MU, and SH designed the study. TI performed experiments with the help of MU and SH. TI, MU, and EN wrote the manuscript. EN supervised the project. All of the authors discussed the results and commented on the manuscript.
Conflict of interest
The authors declare that they have no conflict of interest.
Supporting information
Expanded View Figures PDF
Table EV1
Review Process File
Source Data for Figure 1
Source Data for Figure 3
Source Data for Figure 4
Acknowledgements
We thank Dr. H.R. Horvitz (Massachusetts Institute of Technology, Cambridge) for providing anti‐TRR‐1 antibody and the members of our laboratory for providing technical comments and helpful discussions. This work was supported by grants from the Japan Society for the Promotion of Science (to E.N.) and by a Research Fellow of the Japan Society for the Promotion of Science (to T.I.). The nematode strains used in this work were provided by the Caenorhabditis Genetics Center.
EMBO Reports (2017) 18: 1716–1726
References
- 1. Imai S, Armstrong CM, Kaeberlein M, Guarente L (2000) Transcriptional silencing and longevity protein Sir2 is an NAD‐dependent histone deacetylase. Nature 403: 795–800 [DOI] [PubMed] [Google Scholar]
- 2. Hekimi S, Guarente L (2003) Genetics and the specificity of the aging process. Science 299: 1351–1354 [DOI] [PubMed] [Google Scholar]
- 3. Kenyon C (2005) The plasticity of aging: insights from long‐lived mutants. Cell 120: 449–460 [DOI] [PubMed] [Google Scholar]
- 4. Chang KT, Min KT (2002) Regulation of lifespan by histone deacetylase. Ageing Res Rev 1: 313–326 [DOI] [PubMed] [Google Scholar]
- 5. Berdichevsky A, Viswanathan M, Horvitz HR, Guarente L (2006) Caenorhabditis elegans SIR‐2.1 interacts with 14‐3‐3 proteins to activate DAF‐16 and extend life span. Cell 125: 1165–1177 [DOI] [PubMed] [Google Scholar]
- 6. Burnett C, Valentini S, Cabreiro F, Goss M, Somogyvari M, Piper MD, Hoddinott M, Sutphin GL, Leko V, McElwee JJ et al (2011) Absence of effects of Sir2 overexpression on lifespan in C. elegans and Drosophila . Nature 477: 482–485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Zhang M, Poplawski M, Yen K, Cheng H, Bloss E, Zhu X, Patel H, Mobbs CV (2009) Role of CBP and SATB‐1 in aging, dietary restriction, and insulin‐like signaling. PLoS Biol 7: e1000245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kenyon CJ (2010) The genetics of ageing. Nature 464: 504–512 [DOI] [PubMed] [Google Scholar]
- 9. Honjoh S, Yamamoto T, Uno M, Nishida E (2009) Signalling through RHEB‐1 mediates intermittent fasting‐induced longevity in C. elegans . Nature 457: 726–730 [DOI] [PubMed] [Google Scholar]
- 10. Ceol CJ, Horvitz HR (2004) A new class of C. elegans synMuv genes implicates a Tip60/NuA4‐like HAT complex as a negative regulator of Ras signaling. Dev Cell 6: 563–576 [DOI] [PubMed] [Google Scholar]
- 11. Murphy CT, McCarroll SA, Bargmann CI, Fraser A, Kamath RS, Ahringer J, Li H, Kenyon C (2003) Genes that act downstream of DAF‐16 to influence the lifespan of Caenorhabditis elegans . Nature 424: 277–283 [DOI] [PubMed] [Google Scholar]
- 12. Hsu AL, Murphy CT, Kenyon C (2003) Regulation of aging and age‐related disease by DAF‐16 and heat‐shock factor. Science 300: 1142–1145 [DOI] [PubMed] [Google Scholar]
- 13. Kwon ES, Narasimhan SD, Yen K, Tissenbaum HA (2010) A new DAF‐16 isoform regulates longevity. Nature 466: 498–502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Frank SR, Parisi T, Taubert S, Fernandez P, Fuchs M, Chan HM, Livingston DM, Amati B (2003) MYC recruits the TIP60 histone acetyltransferase complex to chromatin. EMBO Rep 4: 575–580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Sapountzi V, Logan IR, Robson CN (2006) Cellular functions of TIP60. Int J Biochem Cell Biol 38: 1496–1509 [DOI] [PubMed] [Google Scholar]
- 16. Storz P (2011) Forkhead homeobox type O transcription factors in the responses to oxidative stress. Antioxid Redox Signal 14: 593–605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Martins R, Lithgow GJ, Link W (2016) Long live FOXO: unraveling the role of FOXO proteins in aging and longevity. Aging Cell 15: 196–207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lee SS, Kennedy S, Tolonen AC, Ruvkun G (2003) DAF‐16 target genes that control C. elegans life‐span and metabolism. Science 300: 644–647 [DOI] [PubMed] [Google Scholar]
- 19. Brenner S (1974) The genetics of Caenorhabditis elegans . Genetics 77: 71–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kamath RS, Martinez‐Campos M, Zipperlen P, Fraser AG, Ahringer J (2001) Effectiveness of specific RNA‐mediated interference through ingested double‐stranded RNA in Caenorhabditis elegans . Genome Biol 2: RESEARCH0002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Fraser AG, Kamath RS, Zipperlen P, Martinez‐Campos M, Sohrmann M, Ahringer J (2000) Functional genomic analysis of C. elegans chromosome I by systematic RNA interference. Nature 408: 325–330 [DOI] [PubMed] [Google Scholar]
- 22. Murr R, Loizou JI, Yang YG, Cuenin C, Li H, Wang ZQ, Herceg Z (2006) Histone acetylation by Trrap‐Tip60 modulates loading of repair proteins and repair of DNA double‐strand breaks. Nat Cell Biol 8: 91–99 [DOI] [PubMed] [Google Scholar]
- 23. Sun Y, Jiang X, Chen S, Fernandes N, Price BD (2005) A role for the Tip60 histone acetyltransferase in the acetylation and activation of ATM. Proc Natl Acad Sci USA 102: 13182–13187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Potente M, Urbich C, Sasaki K, Hofmann WK, Heeschen C, Aicher A, Kollipara R, DePinho RA, Zeiher AM, Dimmeler S (2005) Involvement of Foxo transcription factors in angiogenesis and postnatal neovascularization. J Clin Invest 115: 2382–2392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Oh SW, Mukhopadhyay A, Dixit BL, Raha T, Green MR, Tissenbaum HA (2005) Identification of direct DAF‐16 targets controlling longevity, metabolism and diapause by chromatin immunoprecipitation. Nat Genet 38: 251–257 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Expanded View Figures PDF
Table EV1
Review Process File
Source Data for Figure 1
Source Data for Figure 3
Source Data for Figure 4


