Abstract
Background:
Cystic echinococcosis (CE) is one of the most important zoonotic diseases; caused by different genotypes of Echinococcus spp. Camels have an important role in transmission cycle of E. granulosus especially, in desert areas. This study aimed to investigate molecular characterization of hydatid cysts isolates from one-humped camel (Camelus dromedarius) and to show its molecular and phylogenic status in this important CE host in the central part of Iran.
Methods:
Twenty hydatid cyst samples (14 fertile and 6 calcified) were collected from 56 slaughtered camels in Central part of Iran. Extraction of DNA from 14 fertile samples was achieved followed by molecular studies on two mitochondrial genes (nad1 and cox1).
Results:
Blast and phylogenetic analysis on sequenced genes showed the presence of G1 (28.6%), G3 (28.6%) and G6 (35.7%) genotypes in the samples. However, one sample was detected as E. ortleppi (G5) with 99% homology with G5 isolated from camel in Egypt (AB921055) and Sudan (JX912709).
Conclusion:
Presence of E. ortleppi, originally the cattle genotype, is reported for the first time in Iran. Due to the potential of infecting human by E. ortleppi; more attention should be paid to this zoonotic genotype in this region.
Keywords: Echinococcus ortleppi, Mitochondrial DNA genes, Phylogenetic analysis, Camel, Iran
Introduction
Hydatidosis/cystic echinococcosis (CE), an important zoonotic helminthic disease, remains as a health problem with a large socioeconomic burden in many parts of the world including the Middle East (1–3). As an endemic area for CE, the disease is responsible for about 1% of surgical admissions in Iran (4).
Molecular studies based on nuclear and mitochondrial genomes have showed the Echinococcus granulosus as G1 to G10 strains (Sensu lato), E. granulosus sensu stricto (strain G1–G3), E. equinus (G4), E. ortleppi (G5) and E. canadensis (G6–G10) that has recently been named as E. intermedius (5–8). Hydatid cysts are usually found in sheep, camel, cattle and goat throughout Iran. The G1, G2, G3, G6 and recently G7 genotypes have been reported from Iran, so far (9–18). Camels have an important role in transmission cycle of the parasite and usually are infected with G6 genotype of E. granulosus (19). Three genotypes of E. granulosus including G6 (camel strain) with higher prevalence, G1 (sheep strain) and G3 (buffalo strain) have been reported from camels in Iran (20–25). To date, only camel strain (G6 genotype), sheep strain (G1–G2 genotypes), and buffalo strain (G3) have been detected from human in Iran (26–28). The proper environmental and ecological condition, emigrant population, none industrial abattoirs, home slaughtering, and large number of stray dogs are the major factors of distributing the disease in endemic countries (29–31).
E. ortleppi (G5 strain) as a common cattle strain that is geographically distributed in Europe, Africa, Southern Asia and the Americas (7, 32); has been reported from camel in Sudan and Egypt (33, 34). Human infection by this genotype has also been reported from several countries including Argentina, Brazil, Mexico, Netherlands, South Africa, France and India (35–37).
This study aimed to investigate molecular and phylogenetic data on hydatid cysts isolated from one-humped camel (Camelus dromedarius) in Iran.; where, G1, G3, and G6 genotypes have been reported from camel, earlier (20, 24).
Materials and Methods
Twenty hydatid cysts samples including 18 (90%) cysts from lung and 2 (10%) cysts from liver of 56 slaughtered camels were collected from February to March 2015 in Najaf Abad district abattoir, Isfahan Province, Central part of Iran. The samples were immediately transferred to the laboratory in cool condition. Hydatid cyst fluids (HCF) including protoscoleces (PSCs), were collected by sterile syringes and transferred into suitable and clean falcon tubes. The tubes were centrifuged at 3000 × g. The PSCs were collected and transferred into alcohol 70% for molecular studies.
Micro-tubes containing PSCs were centrifuged at 3000 × g followed by removing their supernatants. The packed sediment (30–100 μl) was transferred into a new 1.5 micro-tube and washed three times by distilled water for removing of excess alcohol. A total of 300 μl of lysis buffer was added to each sample. Freeze and thaw procedure was applied to each tube for five times-each for 3 min-using liquid nitrogen and boiling water for surface cracking of PSCs. A total of 25 μl of proteinase K was added to each sample and incubated at 37° C overnight. The DNA was extracted by phenol-chloroform protocol. In brief, 300 μl of phenol-chloroform-isoamyl alcohol was added to each sample and centrifuged at 2000 × g for 5 minutes. The supernatant was transferred into a new micro-tube and previous step was repeated. The same volume of absolute ethanol was added to the supernatant. Sodium acetate 3M, was added as much as 0.1 of the mentioned volume and was incubated at −20° C for 30 minutes. The sample was then centrifuged at 5000 × g for 12 minutes and its supernatant was poured off. The pellet was added a total of 300 μl of ethanol 70 % and centrifuged at 2000 × g for 5 minutes. The supernatant was discarded followed by waiting for drying the remaining alcohol from the samples and finally each sample was added 50 μl of deionized water and transferred into −20° C, until use.
The Cytochrome c oxidase subunit 1 (cox1) and NADH dehydrogenase subunit 1 (nad1) genes were amplified by two primers as follows: JB3 (5′-TTTTTTGGGCATCCTGAGGTTTAT-3′) and JB4.5 (5′-TAAAGAAAGAACATAATGAAAATG-3′) for cox1 gene and JB11 (5′-AGATTCGTAAGGGGCCTAATA-3′) and JB12 (5′-ACCACTAACTAATTCACTTTC-3′) for nad1 gene as forward and reverse primers, respectively (38, 39).
PCR reagents and thermal cycler program were similar in both cox1 and nad1 genes amplification. PCR reactions were applied in a final volume of 50 μl, including 2.5 μl genomic DNA, 3.5 mM MgCl2, 250 μM of dNTPs, 25 p mol. of each primer and 2 U of Taq polymerase. The following temperature profile was used for DNA amplification: 40 cycles of 94° C for 45 s, 51° C for 35 s, 72° C for 45 s, followed by a final extension at 72° C for 10 min. Positive (confirmed DNA samples) and negative (no added DNA) controls were used for each PCR program for accuracy. PCR products were visualized using electrophoresis with 1.5 % agarose gel in TAE buffer and stained with GelRed (Biotium®). A 100-bp molecular ladder was used as DNA size marker in each gel for estimating the size of the bands. Gels were observed and photographed using a UV-trans illuminator (Uvitec®).
All PCR primary products of both cox1 and nad1 genes were purified by purification kit (Vivantis®) and sequenced in two directions using the similar forward and reverse primers applied in the PCR. Sequence results were edited and aligned by Genius (40) and BioEdit (41) softwares.
To confirm the identity of the obtained sequences in comparison with the GenBank nucleotide database, all samples were blasted using NCBI (National Center for Biotechnology Information, Bethesda, MD, USA). Phylogenetic trees were constructed using Maximum Likelihood Tree implemented in MEGA software version 7 (42). Bootstrap analysis was used to evaluate the reliability of inferred trees from MEGA 7 software. Nucleotide sequences of cox 1 and nad1 genes belonged to Taenia saginata with GenBank accession numbers AB494480 and AM503345, were used as out groups in the phylogenetic trees, respectively.
Results
Out of 20 hydatid cyst samples, 18 (90%) cysts belonged to lung and two (10%) to the liver. A total of 70% (14/20) of the cysts were fertile while, 30% (6/20) were calcified. DNA isolation from calcified cysts was negative so, they were ruled out from molecular studies. PCR-based assay with specific primers yielded two different bands of 450-bp and 470-bp in PCR of cox1 (12 samples) and nad1 (3 samples) genes, respectively. DNA isolation from fourteen fertile cysts was successful which their PCR products were sequenced. Blast analysis of the sequenced data using GenBank database, indicated the presence of G1 in 28.6% (4/14), G3 in 28.6% (4/14), G6 in 35.7% (5/14) and G5 in 7.1% (1/14) isolates in the current study. The amplified genes and the accession numbers for the detected strains are shown in Table 1. It should be considered that detected G5 genotype, using cox1 gene, subsequently, was also confirmed by amplification of nad1 gene. The partial sequences generated from cox1 and nad1 genes for G5 strain were deposited in the GenBank under the accession numbers KT988115 and KT988119, respectively.
Table 1:
Information about sequences that used for phylogenetic analysis of cox1 and nad1 genes
| Accession number (cox1) | Genotype of Echinococcus | Reference | Accession number (nad1) | Genotype of Echinococcus | Reference |
|---|---|---|---|---|---|
| KU756222 | G1 | This study | JN579164 | G1 | Sadjjadi et al. (2013) |
| KU756223 | G1 | This study | JN579165 | G1 | Sadjjadi et al. (2013) |
| KU756224 | G1 | This study | KF731955 | G1 | Nikmanesh et al. (2014) |
| KU756225 | G1 | This study | AB921092 | G5 | Amer et al. (2015) |
| KF731903 | G1 | Nikmanesh et al. (2014) | JN637177 | G5 | Ahmed et al. (2013) |
| KT074949 | G3 | Tanzifi et al. (2015) | AB979274 | G5 | Morishima et al. (2014) |
| KT988111 | G3 | This study | KT988119 | G5 | This study |
| KT988112 | G3 | This study | HM749616 | G6 | Rostaminejad et al. (2010) |
| KT988113 | G3 | This study | KT988120 | G6 | This study |
| KT988114 | G3 | This study | KT988121 | G6 | This study |
| JX912709 | G5 | Ahmed et al. (2013) | AM503345 | Taenia saginata | Zhang et al. (2007) |
| KT988115 | G5 | This study | |||
| AB921055 | G5 | Amer et al (2015) | |||
| KP751426 | G6 | Karamian et al. (2015) | |||
| KT988116 | G6 | This study | |||
| KT988117 | G6 | This study | |||
| KT988118 | G6 | This study | |||
| AB494480 | Taenia saginata | Abe et al. (2009) |
In our G3 and G1 genotypes (obtained by cox1) there was only one alteration in nucleotide comparing to the used reference sequences: (KT074949) for G3 and (KF731903) for G1 genotypes, respectively. In five G6 samples detected by cox1 and nad1 primers in the present study, 99% homology was observed with reference sequences used in the phylogenetic trees. The G5 genotype isolated from camel in Egypt (AB921055) and Sudan (JX912709) were 99% identical to the G5 detected in the current study with only one-alteration nucleotide sequences. However, G5 of the current study (cox1) had 100% homology with G5 obtained from lemur (KU378107) and spotted deer (JX068638) in the United Kingdom.
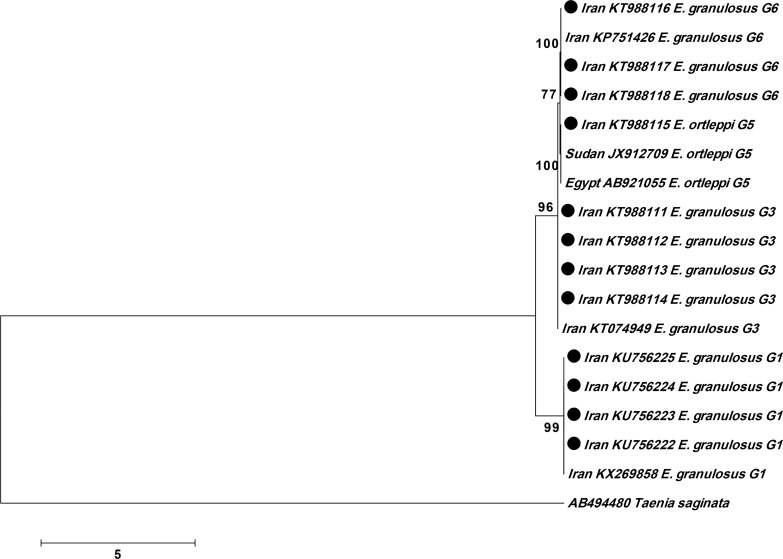
The relationship between these isolates and other similar genotypes identified worldwide are shown by phylogenetic trees for cox1 (Fig. 1) and nad1 (Fig. 2) genes. Using cox1 and nad1, 18 and 12 isolates were analyzed, respectively and related phylogenetic trees were constructed (Fig. 1 and Fig. 2).
Fig. 1:
Genetic relationships of obtained genotypes from camel in the present study and reference sequences related genotypes of E. granulosus as well as Taenia saginata as the out-group. The relationships were inferred based on phylogenetic tree (cox1 gene). The phylogenetic tree was constructed using Maximum Likelihood Tree implemented in MEGA software version 7
Fig. 2:
Genetic relationships of obtained genotypes from camel in the present study and reference sequences related genotypes of E. granulosus as well as Taenia saginata as the outgroup. The relationships were inferred based on phylogenetic tree (nad1 gene). The phylogenetic tree was constructed using Maximum Likelihood Tree implemented in MEGA software version 7
Discussion
Cystic echinococcosis (CE), affects many people throughout the world, although, advances in diagnosis and treatment of CE had been achieved in the recent years. There is still a limit to the disease control. As an endemic region with high incidence of CE, the disease is considered as a public health and socioeconomic problem in Iran, yet (2–4). To control this disease, strategies including surveying on different aspects of parasite should be more considered in the endemic regions (43). However, investigations on the epidemiology and different genotypes of parasites in the intermediate and final hosts should be considered in any endemic area to achieve the evidence-based control and management programs (36).
Different genotypes of E. granulosus including G1, G2, G3, G6 and G7 have been reported from different hosts in Iran (9–18). Camels as important intermediate hosts for CE, specially, in desert areas, have been studied in Iran and molecular studies on nuclear and mitochondrial genes have indicated the presence of G6 genotype as the dominant genotype of E. granulosus in camels (3, 23, 44, 45); however, the presence of G1 and G3 genotypes in camel hydatid cysts have also been reported from Iran (21, 24, 25).
The G6 genotype has been known as common camel strain but, in some areas, G3 genotype of E. granulosus has been shown the dominant genotype in camels (24, 44); however, G1 has also been considered as a noticeable genotype in camels (21, 22, 25). In the current study, G6 was the dominant genotype and detected in 35.7% of samples. The highest infection rate in camel has been reported from Isfahan and Khorasan Razavi and the lowest rate in Kerman and Semnan Provinces (22, 46). In the present study, the lungs were the most infected organ, which is similar to previous studies (9, 46, 47).
In the African countries, the G6 has been reported as the dominant genotype (3, 48). The genotype of all isolates from camel in Mauritania, Algeria and Sudan has been reported to be G6 (49–51). However, other studies in Kenya and Libya have shown a noticeable prevalence of G1 strains in camel isolates (51, 52). The G5 genotype, its host and distribution is different in the world. Investigation of 638 fertile cysts of cattle has shown the presence of G1 (56.6%) and G5 (43.4%) genotypes in Brazil, while the G5 was mostly isolated from lungs (53). The G5 genotype has been isolated from cattle cysts in Argentina and Italy (54, 55) and in spotted deer from UK, too (56).
Existence of G5 strain has already been reported from camel in Sudan and Egypt (33, 34). This genotype has also been reported from human in Argentina, Brazil, Mexico, Netherlands, South Africa, India and France (7, 35, 37), which makes it as an important genotype in the view of public health. The G5 genotype isolated from camel in Egypt (AB921055) and Sudan (JX912709) were 99% identical to the G5 detected in the current study with only one alteration in nucleotide sequences and similar to other studies in G5 genotypes in camel. However, G5 of the current study (cox1) had 100% homology with G5 obtained from lemur (KU378107) and spotted deer (JX068638) in the United Kingdom (56).
The camel as a natural intermediate host for E. granulosus plays an important role in the maintaining of the parasite in the nature especially in desert areas (43). On the other hand scattered camels in desert and semi desert areas, where other ruminants and carnivorous animals may live in Iran’ could be infected with this important genotype.
Conclusion
As far as our knowledge, the present study genetically showed the presence E. ortleppi (G5) in camel for the first time in Iran. However, other zoonotic genotypes including the G1, G3 and G6, which have been reported from camel, were also detected in the present study. Due to the transmission potential of G5 strain to human, the finding of E. ortleppi, in camel should be more noticed in Iran. The distribution of G5 genotype in Iran is not known. However, more studies are needed to find the distribution of G5 genotype in Iran. More molecular studies on cattle and camel hydatid cysts are needed to find the main reservoir of E. ortleppi in Iran. Moreover, molecular and parasitological studies on different final hosts will evaluate the probable existence and its circulation in this region.
Acknowledgements
This study was granted by the office of Vice Chancellor for Research at Shiraz University of Medical Sciences, Grant no: 93-7147. The present work is part of PhD. theses of MEP. The authors acknowledge Mrs S. Kazemian for her help in preparation of materials, Dr. Kabiri from Najaf Abad abattoir and the personnel of Department of Parasitology, Isfahan University of Medical Sciences.
Footnotes
Conflict of Interests
Authors declare that there is no conflict of interest.
References
- 1.Budke CM, Deplazes P, Torgerson PR. Global socioeconomic impact of cystic echinococcosis. Emerg Infect Dis. 2006; 12(2):296–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fasihi Harandi M, Budke CM, Rostami S. The monetary burden of cystic echinococcosis in Iran. PLoS Negl Trop Dis. 2012; 6(11):e1915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sadjjadi SM. Present situation of echinococcosis in the middle east and arabic north africa. Parasitol Int. 2006; 55 Suppl:S197–202. [DOI] [PubMed] [Google Scholar]
- 4.Rokni MB. Echinococcosis/hydatidosis in Iran. Iran J Parasitol. 2009;4:1–16. [Google Scholar]
- 5.Nakao M, Lavikainen A, Yanagida T, et al. Phylogenetic systematics of the genus Echinococcus (cestoda: Taeniidae). Int J Parasitol. 2013; 43(12–13):1017–29. [DOI] [PubMed] [Google Scholar]
- 6.Nakao M, McManus DP, Schantz PM, et al. A molecular phylogeny of the genus Echinococcus inferred from complete mitochondrial genomes. Parasitology. 2007; 134(Pt 5):713–22. [DOI] [PubMed] [Google Scholar]
- 7.Romig T, Ebi D, Wassermann M. Taxonomy and molecular epidemiology of Echinococcus granulosus sensu lato. Vet Parasitol. 2015; 213(3–4):76–84. [DOI] [PubMed] [Google Scholar]
- 8.Yanagida T, Mohammadzadeh T, Kamhawi S, et al. Genetic polymorphisms of Echinococcus granulosus sensu stricto in the middle east. Parasitol Int. 2012; 61(4):599–603. [DOI] [PubMed] [Google Scholar]
- 9.Ahmadi NA. Hydatidosis in camels (Camelus dromedarius) and their potential role in the epidemiology of Echinococcus granulosus in Iran. J Helminthol. 2005; 79(2):119–25. [DOI] [PubMed] [Google Scholar]
- 10.Ahmadi N, Dalimi A. Characterization of Echinococcus granulosus isolates from human, sheep and camel in Iran. Infect Genet Evol. 2006; 6(2):85–90. [DOI] [PubMed] [Google Scholar]
- 11.Farhadi M, Fazaeli A, Haniloo A. Genetic characterization of livestock and human hydatid cyst isolates from northwest Iran, using the mitochondrial cox1 gene sequence. Parasitol Res. 2015; 114(12):4363–70. [DOI] [PubMed] [Google Scholar]
- 12.Fadakar B, Tabatabaei N, 1, Borji H, et al. Genotyping of Echinococcus granulosus from goats and sheep indicating G7 genotype in goats in the northeast of Iran. Vet Parasitol. 2015; 214(1–2):204–7. [DOI] [PubMed] [Google Scholar]
- 13.Parsa F, Fasihi Harandi M, Rostami S, et al. Genotyping Echinococcus granulosus from dogs from western Iran. Exp Parasitol. 2012; 132(2):308–12. [DOI] [PubMed] [Google Scholar]
- 14.Pezeshki A, Akhlaghi L, Sharbatkhori M, et al. Genotyping of Echinococcus granulosus from domestic animals and humans from Ardabil province, northwest Iran. J Helminthol. 87(4:387–91. [DOI] [PubMed] [Google Scholar]
- 15.Rajabloo M, Hosseini SH, Jalousian F. Morphological and molecular characterisation of Echinococcus granulosus from goat isolates in Iran. Acta Trop. 2012; 123(2):67–71. [DOI] [PubMed] [Google Scholar]
- 16.Ranjbar-Bahadori S, Lotfollahzadeh S, Vaezi G, et al. Epidemiological study of the human cystic echinococcosis in Iran. Res J Parasitol. 2008;3:130–136. [Google Scholar]
- 17.Rostami S, Shariat Torbaghan S, Dabiri S, et al. Genetic characterization of Echinococcus granulosus from a large number of formalin-fixed, paraffin-embedded tissue samples of human isolates in Iran. Am J Trop Med Hyg. 2015; 92(3):588–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sharafi SM, Rostami-Nejad M, Moazeni M, et al. Echinococcus granulosus genotypes in Iran. Gastroenterol Hepatol Bed Bench. 2014; 7(2):82–8. [PMC free article] [PubMed] [Google Scholar]
- 19.Thompson RA, Lymbery AJ. Echinococcus and hydatid disease. Cab International; 1995. [Google Scholar]
- 20.Harandi MF, Hobbs RP, Adams PJ, et al. Molecular and morphological characterization of Echinococcus granulosus of human and animal origin in Iran. Parasitology. 2002; 125(Pt 4):367–73. [DOI] [PubMed] [Google Scholar]
- 21.Hajialilo E, Harandi MF, Sharbatkhori M, et al. Genetic characterization of Echinococcus granulosus in camels, cattle and sheep from the south-east of Iran indicates the presence of the g3 genotype. J Helminthol. 2012; 86(3):263–70. [DOI] [PubMed] [Google Scholar]
- 22.Moghaddas E, Borji H, Naghibi A, et al. Molecular genotyping of Echinococcus granulosus from dromedaries (Camelus dromedarius) in eastern Iran. J Helminthol. 2015; 89(1):100–4. [DOI] [PubMed] [Google Scholar]
- 23.Pestechian N, Hosseini Safa A, Tajedini M, et al. Genetic diversity of Echinococcus granulosus in center of Iran. Korean J Parasitol. 2014; 52(4):413–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sharbatkhori M, Fasihi Harandi M, Mirhendi H, et al. Sequence analysis of cox1 and nad1 genes in Echinococcus granulosus G3 genotype in camels (Camelus dromedarius) from central Iran. Parasitol Res. 2011; 108(3):521–7. [DOI] [PubMed] [Google Scholar]
- 25.Sharbatkhori M, Mirhendi H, Harandi MF, et al. Echinococcus granulosus genotypes in livestock of Iran indicating high frequency of G1 genotype in camels. Exp Parasitol. 2010; 124(4):373–9. [DOI] [PubMed] [Google Scholar]
- 26.Kia EB, Rahimi H, Sharbatkhori M, Talebi A, et al. Genotype identification of human cystic echinococcosis in Isfahan, central Iran. Parasitol Res. 2010; 107(3):757–60. [DOI] [PubMed] [Google Scholar]
- 27.Sadjjadi SM, Mikaeili F, Karamian M, et al. Evidence that the Echinococcus granulosus G6 genotype has an affinity for the brain in humans. Int J Parasitol. 2013; 43(11):875–7. [DOI] [PubMed] [Google Scholar]
- 28.Shahnazi M, Hejazi H, Salehi M, et al. Molecular characterization of human and animal Echinococcus granulosus isolates in Isfahan, Iran. Acta Trop. 2011; 117(1):47–50. [DOI] [PubMed] [Google Scholar]
- 29.Mehrabani D, Oryan A, Sadjjadi SM. Prevalence of Echinococcus granulosus infection in stray dogs and herbivores in Shiraz, Iran. Vet Parasitol. 1999; 86(3):217–20. [DOI] [PubMed] [Google Scholar]
- 30.Seimenis A. Overview of the epidemiological situation on ech-inococcosis in the Mediterranean region. Acta Trop. 2003; 85(2):191–5. [DOI] [PubMed] [Google Scholar]
- 31.Fallah M, Taherkhani H, Sadjjadi M. Echinococcosis in the stray dogs in Hamadan, west of Iran. Iran J Med Sci. 1995;20:170–172. [Google Scholar]
- 32.Mbaya H, Magambo J, Njenga S, et al. Echinococcus spp. In central Kenya: A different story. Parasitol Res. 2014; 113(10):3789–94. [DOI] [PubMed] [Google Scholar]
- 33.Ahmed ME, Eltom KH, Musa NO, et al. First report on circulation of Echinococcus ortleppi in the one humped camel (Camelus dromedaries), Sudan. BMC Vet Res. 2013; 9:127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Amer S, Helal IB, Kamau E, et al. Molecular characterization of Echinococcus granulosus sensu lato from farm animals in Egypt. PLoS One. 2015; 10(3):e0118509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Grenouillet F, Umhang G, Arbez-Gindre F, et al. Echinococcus ortleppi infections in humans and cattle, France. Emerg Infect Dis. 2014; 20(12):2100–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Alvarez Rojas CA, Romig T, Lightowlers MW. Echinococcus granulosus sensu lato genotypes infecting humans--review of current knowledge. Int J Parasitol. 2014; 44(1):9–18. [DOI] [PubMed] [Google Scholar]
- 37.Sharma M, Sehgal R, Fomda BA, et al. Molecular characterization of Echinococcus granulosus cysts in north indian patients: Identification of g1, g3, g5 and g6 genotypes. PLoS Negl Trop Dis. 2013; 7(6):e2262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bowles J, Blair D, McManus DP. Genetic variants within the genus Echinococcus identified by mitochondrial DNA sequencing. Mol Biochem Parasitol. 1992; 54(2):165–73. [DOI] [PubMed] [Google Scholar]
- 39.Bowles J, McManus DP. Nadh dehydrogenase 1 gene sequences compared for species and strains of the genus Echinococcus. Int J Parasitol. 1993; 23(7):969–72. [DOI] [PubMed] [Google Scholar]
- 40.Kearse M, Moir R, Wilson A, et al. Geneious basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 2012; 28(12):1647–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hall TA. Bioedit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/nt. Nucleic Acids Symposium Series 1999:95. [Google Scholar]
- 42.Kumar S, Stecher G, Tamura K. Mega7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 2016; 33(7):1870–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Thompson RC. The taxonomy, phylogeny and transmission of Echinococcus. Exp Parasitol. 2008; 119(4):439–46. [DOI] [PubMed] [Google Scholar]
- 44.Sharifiyazdi H, Oryan A, Ahmadnia S, et al. Genotypic characterization of Iranian camel (Camelus dromedarius) isolates of Echinoccocus granulosus. J Parasitol. 2011; 97(2):251–5. [DOI] [PubMed] [Google Scholar]
- 45.Zhang L, Eslami A, Hosseini SH, et al. Indication of the presence of two distinct strains of Echinococcus granulosus in Iran by mitochondrial DNA markers. Am J Trop Med Hyg. 1998; 59(1):171–4. [DOI] [PubMed] [Google Scholar]
- 46.Elham M, Hassan B, Ghasem NA, et al. Epidemiological study of hydatidosis in the dromedaries (Camelus dromedarius) of different regions of Iran. Asian Pac J Trop Biomed. 2014; 4(Suppl 1):S148–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mirzaei M, Rezaei H, Nematollahi A, et al. Survey of hydatidosis infection in slaughtered camel (Camelus dromedarius) in Tabriz area, northwest Iran. J Parasit Dis. 2016; 40(2):444–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Aaty HE, Abdel-Hameed DM, Alam-Eldin YH, et al. Molecular genotyping of Echinococcus granulosus in animal and human isolates from Egypt. Acta Trop. 2012; 121(2):125–8. [DOI] [PubMed] [Google Scholar]
- 49.Bardonnet K, Piarroux R, Dia L, et al. Combined eco-epidemiological and molecular biology approaches to assess Echinococcus granulosus transmission to humans in mauritania: Occurrence of the ‘camel’ strain and human cystic echinococcosis. Trans R Soc Trop Med Hyg. 2002; 96(4):383–6. [DOI] [PubMed] [Google Scholar]
- 50.Bardonnet K, Benchikh-Elfegoun MC, Bart JM, et al. Cystic echinococcosis in Algeria: Cattle act as reservoirs of a sheep strain and may contribute to human contamination. Vet Parasitol. 2003; 116(1):35–44. [DOI] [PubMed] [Google Scholar]
- 51.Dinkel A, Njoroge EM, Zimmermann A, et al. A PCR system for detection of species and genotypes of the Echinococcus granulosus-complex, with reference to the epidemiological situation in Eastern Africa. Int J Parasitol. 2004; 34(5):645–53. [DOI] [PubMed] [Google Scholar]
- 52.Tashani OA, Zhang LH, Boufana B, et al. Epidemiology and strain characteristics of Echinococcus granulosus in the Benghazi area of eastern Libya. Ann Trop Med Parasitol. 2002; 96(4):369–81. [DOI] [PubMed] [Google Scholar]
- 53.Balbinotti H, Santos GB, Badaraco J, et al. Echinococcus ortleppi (G5) and Echinococcus granulosus sensu stricto (G1) loads in cattle from southern Brazil. Vet Parasitol. 2012; 188(3–4):255–60. [DOI] [PubMed] [Google Scholar]
- 54.Andresiuk MV, Gordo FP, Saarma M, et al. Echinococcus granulosus genotype g1 dominated in cattle and sheep during 2003–2006 in Buenos Aires Province, an endemic area for cystic echinococcosis in Argentina. Acta Trop. 2013; 127(2):136–42. [DOI] [PubMed] [Google Scholar]
- 55.Casulli A, Manfredi MT, La Rosa G, et al. Echinococcus ortleppi and e. Granulosus g1, g2 and g3 genotypes in Italian Bovines. Vet Parasitol. 2008; 155(1–2):168–72. [DOI] [PubMed] [Google Scholar]
- 56.Boufana B, Stidworthy MF, Bell S, et al. Craig PS. Echinococcus and Taenia spp. from captive mammals in the United Kingdom. Vet Parasitol. 2012; 190(1–2):95–103. [DOI] [PubMed] [Google Scholar]