Abstract
Cardiac neural crest cells (CNCCs) originate at the dorsal edge of the neural tube between the otic pit and the caudal edge of the 3rd somite, and migrate into the pharyngeal arches and the heart. We have shown that fibronectin (Fn1) plays an important role in the development of the CNCC by regulating the differentiation of CNCCs into vascular smooth muscle cells around pharyngeal arch arteries (Wang and Astrof, 2016). This protocol describes the isolation of CNCCs from the neural tube and from the caudal pharyngeal arches, and the differentiation of neural crest-derived cells into smooth muscle cells. This protocol was adapted from (Newgreen and Murphy, 2000; Pfaltzgraff et al., 2012 ).
Keywords: Cardiac neural crest, Vascular smooth muscle cells, Neural tube, Pharyngeal arch, Differentiation
Background
Previous published protocols described the isolation of neural crest cells from the neural tube. However, neural crest cells in the region of the neural tube between the otic pit and the 3rd somite include neural crest cell populations that contribute to a number of different cell types; for example, vagal neural crest cells also originate from this region. In this protocol, we modified the conventional method for the isolation of cardiac neural crest cells. Instead of using the neural tube, we used the caudal pharyngeal arch region at embryonic day (E) 9.5 (22-25 somite stage). This is prior to differentiation of cardiac neural crest cells into vascular smooth muscle cells. It is common for neural crest cultures to contain contaminating mesenchymal cell types, which often express smooth muscle genes. To identify neural crest-derived cells, we isolated neural crest cells from embryos resulting from the following cross: Fn1flox/flox;ROSAmTmG/mTmG female mice x Fn1+/−;Tfap2αIRESCre/+ male mice. In 50% of the progeny from this cross, neural crest cells are lineage-labeled by the expression of GFP, so we could easily identify neural crest cells by their GFP expression without the need for cell sorting (Wang and Astrof, 2016). Additional Cre-expressing strains that can be used are Wnt1-Cre2 ( Lewis et al., 2013 ) and P3ProCre ( Li et al., 2000 ) transgenic strains, e.g., (Wang and Astrof, 2016). All experimental procedures were approved by the Institutional Animal Care and Use Committee of Thomas Jefferson University and conducted in accordance with federal guidelines for humane care of animals.
Materials and Reagents
12 mm round glass coverslips (Electron Microscopy Sciences, catalog number: 72231-01)
24-well plates (Corning, Falcon®, catalog number: 353047)
Nunc 4-well dishes untreated (Thermo Fisher Scientific, Thermo ScientificTM, catalog number: 144444)
35 cm Petri dish (Corning, Falcon®, catalog number: 353001)
1.5 ml centrifuge tube
Sterile transfer pipet (Thermo Fisher Scientific, Thermo ScientificTM, catalog number: PP89SB)
Glass pipet (Fisher Scientific, model: 13-678-6A)
Parafilm
Glass slide
0.2 μm syringe filter unit
Pregnant mice
Dulbecco’s phosphate-buffered saline (DPBS) (Thermo Fisher Scientific, GibcoTM, catalog number: 14190144), for dissection of embryos and for cell culture
Trypsin (Thermo Fisher Scientific, GibcoTM, catalog number: 25200056)
4% PFA prepared in 1x PBS
0.1% Triton X-100 in 1x PBS
Phosphate-buffered saline (PBS)
Donkey serum (Sigma-Aldrich, catalog number: D9663-10ML)
Anti-GFP antibody (Aves Labs, catalog number: GFP-1020)
Anti-αSMA antibody (Sigma-Aldrich, catalog number: SAB2500963)
Anti-calponin antibody (Abcam, catalog number: ab46794)
Donkey anti-goat IgG (H+L) secondary antibody, Alexa Fluor® 555 conjugate (Thermo Fisher Scientific, catalog number: A-21432)
Donkey anti-rabbit IgG (H+L) secondary antibody, Alexa Fluor® 647 conjugate (Thermo Fisher Scientific, catalog number: A-31573)
Alexa Fluor® 488 AffiniPure F(ab’)2 fragment donkey anti-chicken IgY (IgG) (H+L) (Jackson ImmunoResearch, catalog number: 703-546-155)
ProLong Gold Antifade Mountant with DAPI (Thermo Fisher Scientific, InvitrogeTM, catalog number: P36931)
Collagenase and dispase (Roche Diagnostics, catalog number: 10269638001)
Glacial acetic acid
Collagen I (Corning, catalog number: 354249)
DMEM-low glucose (Thermo Fisher Scientific, GibcoTM, catalog number: 11885076)
Neurobasal medium (Thermo Fisher Scientific, GibcoTM, catalog number: 21103049)
Chicken embryo extract (MP Biomedicals, catalog number: 092850145)
Penicillin/Streptomycin (Pen/Strep) (Mediatech, catalog number: 30-001-CI)
N2-supplement (Thermo Fisher Scientific, GibcoTM, catalog number: 17502048)
B27-supplement (Thermo Fisher Scientific, GibcoTM, catalog number: 17504044)
Retinoic acid (Sigma-Aldrich, catalog number: R2625)
Ethanol
2-Mercaptoethanol (Thermo Fisher Scientific, GibcoTM, catalog number: 21985023)
Fibroblast growth factor-basic (bFGF) (Sigma-Aldrich, catalog number: F0291)
Tris pH 7.6
Insulin-like growth factor 1 (IGF-1) (R&D Systems, catalog number: 291-G1)
Fetal bovine serum (FBS) (Gemini Bio-Products, catalog number: 100-125)
DMEM-high glucose (Thermo Fisher Scientific, GibcoTM, catalog number: 11965092)
Collagenase/dispase stock solution (see Recipes)
Collagen I working solution (see Recipes)
Neural crest self-renewal medium (see Recipes)
Differentiation medium (see Recipes)
Equipment
Autoclave
Biological safety hood (Thermo Fisher Scientific, Thermo ScientificTM, model: 1300 Series Class II, Type A2)
Scissors (Fisher Scientific, model: 13-804-6)
Forceps 9 cm (Fine Science Tool, model: 14060-09)
Forceps 0.1 x 0.06 mm (Fine Science Tool, model: 11251-23)
Forceps 11 cm (Fine Science Tool, model: 11254-20)
EdgeGARD® horizontal flow hood (The Baker Company, model: EdgeGARD®HF)
Flat bench
Dissection microscope and light source (Carl Zeiss, model: Stemi 2000-C)
Humidified, 37 °C tissue culture incubator (Thermo Fisher Scientific, Thermo ScientificTM, model: HeracellTM 150i)
Fluorescence microscope
Procedure
-
Coat glass coverslips and 4-well/24-well plates with collagen I working solution (150 μg/ml)
Autoclave round glass coverslips, 13 mm diameter, in advance.
Roughly estimate the desired number of round glass coverslips, and place one glass coverslip within one well of a 24-well plate in a biological safety hood. Coat each coverslip with enough collagen I (~400 μl for each well) at room temperature (RT) for 2 h.
Remove collagen I, rinse coverslips with sterile H2O twice and allow the coverslips to dry overnight.
Store coated plates and plates with coated coverslips at 4 °C for no more than 2 weeks.
-
Embryo preparation
Note: This can be done on a lab bench.
Timed pregnant mice are sacrificed by CO2. To isolate neural crest cells from the neural tube, dissect embryos at the embryonic day (E) 9.0 (12-15 somite stage). To isolate neural crest-derived cells form pharyngeal arches posterior to the 2nd arch, dissect embryos at E9.5 (24-26 somite stage).
Place the sacrificed mouse on its back, and wet the lower abdomen with 70% ethanol. Open the peritoneal cavity with fine scissors, push gut aside and expose the uterus.
Cut the uterus from the cervix with fine scissors. Hold the broken end of the uterus with forceps, pull the uterus straight, open a small hole on the uterus, insert a fine tip of scissors into the uterus, and slowly cut open the uterus on the vessel-free edge of the uterus.
Pick up the embryos and transfer to a dish with DPBS at RT.
-
Isolation of neural crest cells from neural tubes (see Movie in Pfaltzgraff et al., 2012 )
Neural tube dissection and explant culture:
Note: Dissection should be done inside a horizontal flow hood.
Prepare collagenase/dispase solution fresh for each experiment: add 50 μl of 100 mg/ml collagenase/dispase stock solution to 5 ml DPBS at RT. Pipet 500 μl into each 1.5 ml centrifuge tube. One tube will be used to digest one embryo.
Under dissection microscope, isolate E9.0 embryos by stripping of decidua and membranes with fine forceps. Trim off the tail of the embryo.
Wash each embryo once in DPBS at RT by transferring it into a new plate with fresh DPBS, and then transfer the embryo into a 1.5 ml centrifuge tube containing 500 μl of the collagenase/dispase solution by using sterile plastic transfer pipet. Incubate embryos at 37 °C water bath for 7 min. The incubation time is critical. It is difficult to separate the neural tube from the surrounding tissue if the tissue is not digested enough; however, if over-digested, the neural tube will be curled.
While waiting for the digestion, pipet 180 μl self-renewal medium into each well of a 4-well Nunc plate coated with collagen I, and label 1.5 ml centrifuge tubes, which will be used to save the rest of the embryo for genotyping.
Wash embryos with DPBS three times. And transfer embryos to a 35-mm dish in DPBS.
Under dissection microscope, hold the head of the embryo with one pair of forceps, and peel the surface ectoderm overlaying the cardiac neural tube segment (from otic vesicle to the 3rd somite) with another pair of fine forceps. Insert the tips of forceps between the lateral neural tube and the somites, and carefully separate them by moving the tip back and forth. Do the same for the other side of the neural tube. If the digestion is ideal, it should be very easy to do this step. Cut the neural tube at the level of the otic vesicle and the upper edge of the 4th somite, and free the neural tube from ventral tissues.
Rinse the neural tube with self-renewal medium ( Bixby et al., 2002 ) and transfer neural tube segments to the center of wells containing 180 μl self-renewal medium. The neural tube will dry out if too little medium is used and will float in the medium if too much is used. Place the neural tube laterally, so the neural crest cells can migrate out and attach to the bottom of the dish. Save the rest of the embryo for genotyping.
12 h later, when the neural tube is well-attached to the bottom, carefully and slowly add 350 μl of fresh self-renewal medium pre-warmed to 37 °C into each well and culture 4 more days. Replace half of the medium with fresh self-renewal medium on the 3rd day.
-
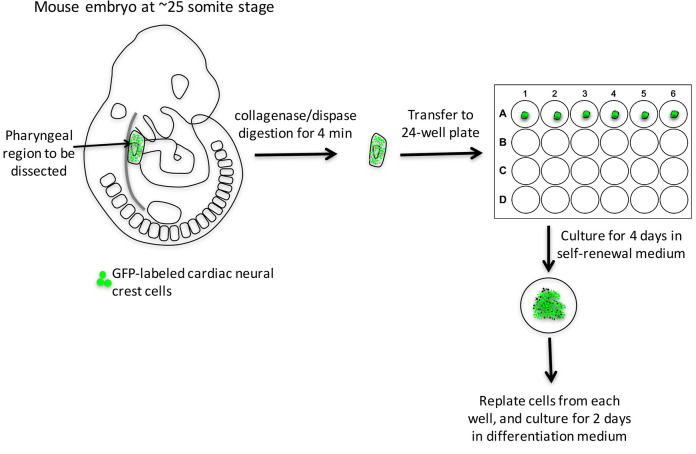
Isolation of cardiac neural crest-derived cells from pharyngeal arches (Figure 1 and Video 1)
Pharyngeal arch dissection and culture:
Note: Dissection should be done within the horizontal flow hood.
Prepare collagenase/dispase solution.
Dissect E9.5 embryos.
Digest embryos with collagenase/dispase at 37 °C in a water bath for 4 min; if multiple embryos are processed, digest one embryo at a time.
While waiting for the digestion to complete, pipet 500 μl self-renewal medium into each well of 24-well plate coated with collagen I, and label 1.5 centrifuge tubes that will be used to save a small part of each embryo for genotyping.
Wash each embryo with DPBS three times, and transfer to a 35-mm dish in DPBS for dissection.
-
Video 1 starts at this step:
Under dissection microscope, hold the head of the embryo with one pair of forceps, chop off the embryo’s tail and peel the surface ectoderm off of pharyngeal region (PA 3-6) with another pair of fine forceps.
Trim the head off from the upper edge of PA 3. Trim the heart off by cutting through the aortic sac.
Detach the pharyngeal arches from the dorsal tissue by cutting through both dorsal aortae. The pharyngeal arch tissue is composed of cardiac neural crest cells and other cell types, such as mesodermal and endodermal cells.
Transfer the dissected piece of pharyngeal tissue to a 24-well plate. The tissue will sink and attach to the bottom of the well in 12 h. Save the rest of the embryo for genotyping.
Culture pharyngeal arch tissue for 4 days. And replace half of the medium with fresh self-renewal medium on the 3rd day.
-
Smooth muscle cell differentiation
Note: This should be done within a biological safety hood.
To replate neural crest cells that have migrated from neural tube explant (Figure 2A), the neural tube should be removed by pipetting the medium up and down in the wells for about 5 times (neural crest cells will not be affected because they are tightly attached to the bottom). This step is omitted for the pharyngeal arch culture.
Wash the wells containing neural crest cells once with DPBS pre-warmed to 37 °C, and incubate with 150 μl 0.25% trypsin for 1 min at 37 °C or until all cells round up and appear to be nearly detached from the plate.
Add 350 μl differentiation medium into each well, pipet up and down about 5 times, to dissociate neural crest cells into single cells. Together with trypsin, wells now have 500 μl of medium. Cells can now be plated for differentiation, as described in the next step. There are usually enough cells to split into 4 wells of a 24-well plate.
For differentiation, plate neural crest cells onto collagen I-coated coverslips in a 24-well plate, and culture for 2 h. When cells settle down, replace the medium with fresh differentiation medium and culture for 2 days.
-
Immunostaining
To identify neural crest-derived smooth muscle cells, neural crest cultures were stained for GFP (neural crest cells are GFP+) and smooth muscle cell markers, such as αSMA and Calponin.
Rinse cells with DPBS and fix them at RT with 4% PFA pre-warmed to RT for 20 min, then rinse with DPBS 3 times, 5 min each, before proceeding for immunostaining.
Permeabilize cells with 500 μl 0.1% Triton X-100 in PBS (PBST) for 10 min at RT.
Get coverslips from wells, and place them in a flat dish covered with Parafilm. The dish should be shallow, and should be topped with a lid. Tuck a few wet Kimwipes near sides of the dish to create a humidified environment when the dish is closed. Make sure the side of the coverslip with cells faces up. Add 50 μl of blocking buffer (5% normal donkey serum in PBST) for 30 min at RT. This set up minimizes the volume of antibody solutions used for staining, only 30 μl solution is required to cover cells on the coverslip.
Remove the blocking buffer by aspiration, and add 30 μl of primary antibody solution in blocking buffer for 2 h at RT. The antibody concentrations: anti-GFP 1:500; anti-αSMA 1:300; anti-calponin 1:150.
Return coverslips to wells in a multi-well plate and wash with PBST 3 times, changing to fresh PBST every 10 min.
Get coverslips from the wells and place them on Parafilm in the staining chamber, add secondary antibodies and incubate for 1 h at RT: anti-chicken, anti-goat, anti-rabbit conjugated to Alexa 488, Alexa 555, Alexa 647, all diluted at 1:300.
Put coverslips back into the wells of a multi-well plate, and wash with PBST 3 times, changing to fresh PBST every 10 min.
Place 10 μl of anti-fade mounting reagent (with DAPI) on a glass slide, then put the coverslip upside down on the reagent.
Observe slides using a fluorescence microscope.
Figure 1. Work-flow of isolation cardiac neural crest cells from E9.5 mouse embryo and their differentiation into smooth muscle cells.
Video 1. Demonstration of dissection of pharyngeal arches 3-6 from E9.5 embryos.
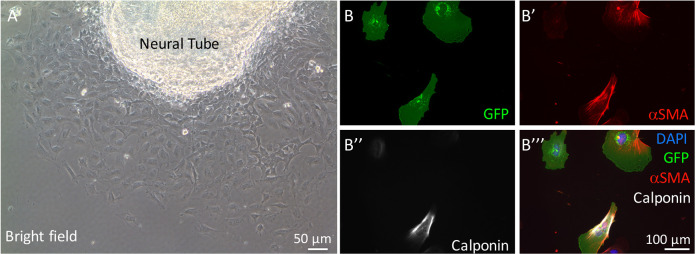
Figure 2. Neural crest cell migration from neural tube explant and staining of cultured cardiac neural crest cells for smooth muscle cell markers.
A. Neural crest cells migrate from the neural tube explant on glass coverslips. B-B’’’. Representative images of cardiac neural crest cells isolated from Fnf/+; ROSAmTmG; TFAP2αIRESCre/+ embryos. Cells were stained with antibodies to GFP (to identify neural crest-derived cells), αSMA, and calponin. DAPI was used to stain nuclei.
Data analysis
The percentage of neural crest-derived smooth muscle cells is determined by the number of αSMA+ calponin+ cells expressing GFP divided by the total number of GFP positive cells (Figures 2B-2B’’’). Detailed results can be found in our previous publication (Wang and Astrof, 2016).
Recipes
-
Collagenase/dispase stock solution (100 mg/ml)
500 mg collagenase/dispase powder
Dissolve to 5 ml Ca2+/Mg2+-free PBS
Filter through a 0.2 μm syringe filter unit
-
Collagen I working solution (150 μg/ml)
58 μl glacial acetic acid
50 ml ddH2O
Filter through a 0.2 μm filter unit
Add 0.817 ml collagen I concentrate (9.18 mg/ml)
-
Neural crest self-renewal medium (100 ml)
50 ml DMEM-low glucose
30 ml neurobasal medium
15 ml chick embryo extract
1 ml Pen/Strep (P/S)
1 ml N2
2 ml B27
100 μl retinoic acid (3.5 mg/ml, dissolve in 100% ethanol)
100 μl 2-mercaptoethanol
80 μl bFGF (25 μg/ml, dissolve in 5 mM Tris pH 7.6)
40 μl IGF-1 (50 μg/ml, dissolve in PBS)
680 μl ddH2O
-
Differentiation medium
500 μl FBS and 50 μl Pen/Strep
Add to 4,450 μl DMEM-high glucose
Get a final concentration of 10% FBS and 1% P/S
Acknowledgments
This work was supported by the funding from the National Institutes of Health [NHLBI RO1 HL103920 to S.A.], X.W. was supported by an American Heart Association Postdoctoral Fellowship [12POST11750033 to X.W.].
Citation
Readers should cite both the Bio-protocol article and the original research article where this protocol was used.
References
- 1.Bixby S., Kruger G. M., Mosher J. T., Joseph N. M. and Morrison S. J.(2002). Cell-intrinsic differences between stem cells from different regions of the peripheral nervous system regulate the generation of neural diversity. Neuron 35(4): 643-656. [DOI] [PubMed] [Google Scholar]
- 2.Lewis A. E., Vasudevan H. N., O'Neill A. K., Soriano P. and Bush J. O.(2013). The widely used Wnt1-Cre transgene causes developmental phenotypes by ectopic activation of Wnt signaling . Dev Biol 379(2): 229-234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li J., Chen F. and Epstein J. A.(2000). Neural crest expression of Cre recombinase directed by the proximal Pax3 promoter in transgenic mice. Genesis 26(2): 162-164. [DOI] [PubMed] [Google Scholar]
- 4.Newgreen D. F. and Murphy M.(2000). Neural crest cell outgrowth cultures and the analysis of cell migration. Methods Mol Biol 137: 201-211. [DOI] [PubMed] [Google Scholar]
- 5.Pfaltzgraff E. R., Mundell N. A. and Labosky P. A.(2012). Isolation and culture of neural crest cells from embryonic murine neural tube. J Vis Exp(64): e4134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang X. and Astrof S.(2016). Neural crest cell-autonomous roles of fibronectin in cardiovascular development. Development 143(1): 88-100. [DOI] [PMC free article] [PubMed] [Google Scholar]