Abstract
Meiosis is crucial in reproduction of plants and ensuring genetic diversity. Although several genes involved in homologous recombination and DNA repair have been reported, their functions in rice (Oryza sativa) male meiosis remain poorly understood. Here, we isolated and characterized the rice OsFIGNL1 (OsFidgetin-like 1) gene, encoding a conserved AAA-ATPase, and explored its function and importance in male meiosis and pollen formation. The rice Osfignl1 mutant exhibited normal vegetative growth, but failed to produce seeds and displayed pollen abortion phenotype. Phenotypic comparisons between the wild-type and Osfignl1 mutant demonstrated that OsFIGNL1 is required for anther development, and that the recessive mutation of this gene causes male sterility in rice. Complementation and CRISPR/Cas9 experiments demonstrated that wild-type OsFIGNL1 is responsible for the male sterility phenotype. Subcellular localization showed that OsFIGNL1-green fluorescent protein was exclusively localized in the nucleus of rice protoplasts. Male meiosis in the Osfignl1 mutant exhibited abnormal chromosome behavior, including chromosome bridges and multivalent chromosomes at diakinesis, lagging chromosomes, and chromosome fragments during meiosis. Yeast two-hybrid assays demonstrated OsFIGNL1 could interact with RAD51A1, RAD51A2, DMC1A, DMC1B, and these physical interactions were further confirmed by BiFC assay. Taken together, our results suggest that OsFIGNL1 plays an important role in regulation of male meiosis and anther development.
Keywords: Oryza sativa, meiosis, male sterility, OsFIGNL1, chromosomes
Introduction
Male reproductive development is a complex biological process, an understanding of which can lend insight for breeders into traits that increase yield and ensure rice reproductive ability. Pollen fertility also is a key factor in rice grain yield (Zhang et al., 2010; Zhou et al., 2011). During male reproductive development, the anther wall consists of the epidermis, the endothecium, the middle layer and the tapetum (Zhang et al., 2011). The morphology of four somatic layers of the anther wall undergoes dynamic changes, along with the haploid male gametophyte (pollen) generated from diploid pollen mother cells (PMCs) (Tao et al., 2007; Zhang et al., 2011). The formation of pollen grains in plants includes PMC formation, male meiosis, two mitotic processes of microspore and starch filling, and maturation of pollen grains (McCormick, 2004). Meiosis is a critical step in gametogenesis, during which two rounds of cell division follow a single round of DNA replication, ending with formation of four haploid cells. It is known that meiosis plays a central role in stabilization of genomic information and establishment of genetic diversity (Nonomura et al., 2004). During meiosis, several key events, including synaptonemal complex assembly, homologous chromosome pairing, and chiasma formation, could coordinately assure accurate segregation of meiotic chromosomes (Li and Ma, 2006; Shen et al., 2012; Luo et al., 2013). Disturbing any step of this progress leads to severe dysfunction of meiotic cells, including aberrant chromosomes, abnormal pairing of homologs, reduction of chiasma frequency, and infertility.
In eukaryotes, meiotic recombination starts from meiotic prophase I, during which a conserved protein SPO11 generates double-strand breaks (DSBs) and covalently links to the 5′ends of DNA at the incision sites of the DSBs (Keeney et al., 1997). In yeast, the covalently linked SPO11 are subsequently removed by the Mre11/Rad50/Xrs2-Nbs1 (MRX/N) complex and Sae2/Com to form 3′-single-stranded tails (Neale et al., 2005; Filippo et al., 2008; Milman et al., 2009; Edlinger and Schlogelhofer, 2011). In the following step, RAD51, together with the meiosis-specific DMC1, bind to single-stranded DNA generated from a DSB, then search and attach to a homologous template to form the double Holliday junction (dHJ) (Cloud et al., 2012). And then, meiotic DSBs are finally repaired to produce crossovers (COs) and non-crossovers (NCOs). In Arabidopsis, AtRAD51, AtDMC1, AtRAD51C, and AtXRCC3 have been shown to be important for homologous recombination and DNA repair in plant meiosis (Pradillo et al., 2014). In Arabidopsis thaliana, mnd1 and hop2 mutants have been characterized that exhibit chromosome fragmentation and sterility associated with DNA repair during meiosis (Uanschou et al., 2013). In eukaryotes, Hop2-Mnd1 heterodimerizes and stimulates the DNA strand exchange activity of RAD51 and DMC1 (Pezza et al., 2010; Bugreev et al., 2014). In another study, the Arabidopsis AtBRCA2 was reported to be essential for DNA repair (Seeliger et al., 2012). AtBRCA2 is another key recombination co-factor that interacts with AtRAD51 and AtDMC1, and mediates the recruitment of AtRAD51 and AtDMC1 during meiotic recombination (Dray et al., 2006; Seeliger et al., 2012). The orthologs of these genes play conserved roles in rice. For example, two RecA homologs, the RAD51 and DMC1 proteins, also have been shown to catalyze strand exchange between homologous chromosomes (Rajanikant et al., 2008; Sakane et al., 2008; Morozumi et al., 2013). In rice, a recent study revealed that OsMRE11 is essential for homologous synapsis and DSB ends processing (Ji et al., 2013). Additionally, OsSDS are known to function in DNA DSB formation during rice meiosis (Wu et al., 2015). Mutations in these genes cause abnormalities in meiosis, including chromosome fragmentation, defective homologous pairing and synapsis, and eventually lead to severe defects in male and female fertility in rice (Couteau et al., 1999; Bleuyard and White, 2004; Li et al., 2004, 2005; Pradillo et al., 2014). However, many aspects of the process of meiosis remain poorly understood in plants.
FIDGETIN is a member of the AAA (ATPase Associated with diverse cellular Activities) protein superfamily, the mutation of which causes multiple developmental defects in mice. Fidget mice exhibit cell cycle delay, insufficient growth of the retinal neural epithelium, side-to-side head shaking and circling, and small eyes (Cox, 2000). FIDGETIN, as well as the closely related FIDGETIN-LIKE 1 (FIGNL1) and FIDGETIN-LIKE 2 (FIGNL2) proteins also were identified in mouse (Cox, 2000; Yang et al., 2005). FIGNL1 is a homolog protein of FIDGETIN (Cox, 2000), and reportedly plays a critical roles in meiosis (Girard et al., 2015). Previous studies have demonstrated that FIGNL1 plays an important role in animal meiotic cell division. In male mice, FIGNL1 is most highly expressed during the spermatocyte stage (L’Hote et al., 2011). Recently, it was reported that FIGNL1 specifically interacts with SMO-1 in Caenorhabditis elegans, with mutation of FIGNL1 resulting in defective gonad formation and the sterile phenotype (Luke-Glaser et al., 2007; Onitake et al., 2012). In Arabidopsis, the FIGNL1 was found to suppress formation of genome-wide meiotic COs, possibly by hindering the strand exchange activity of two conserved recombinases AtDMC1 and AtRAD51 (Girard et al., 2015). Despite advances in understanding the meiotic process, the potential molecular mechanism and function of FIGNL1 is still far from clear.
Considerable research has been conducted to elucidate the mechanisms underlying meiosis, particularly during male gametogenesis in plants. However, the functions of many genes involved in the meiotic process remain unknown. In this study, using a map-based cloning strategy, we isolated and functionally characterized a rice AAA-ATPase gene OsFIGNL1. Osfignl1 male meiocytes exhibited abnormal chromosomes during meiosis, which finally resulted in aborted pollen grains and entire male sterility of the plants. This work sheds new light on OsFIGNL1 regulation of meiotic chromosome behavior during male gametogenesis in monocot plants.
Materials and Methods
Plant Materials, Growth Conditions, and Mutant Phenotyping
The wild-type used in this study was Oryza sativa spp. indica cv. Zhonghui 8015 (Zh8015). Mutagenesis of Zh8015 was performed by treatment with 60Co-γ ray radiation, resulting in identification of the complete male sterility mutant Osfignl1. The Osfignl1 mutant, as the pollen acceptor, was crossed with wild-type and Zhonghua11 (japonica), respectively. The heterozygous F1 plants were then self-pollinated to generate a BC1F2, and an F2 population for genetic analysis and mapping of the target locus. All plants were grown in paddy fields in Lingshui, Hainan Province, and Hangzhou, Zhejiang Province, China. Planting density and crop management followed commercial rice production practices. The mutant exhibited genetic stability in both Hainan and Zhejiang.
To assess the viability of mature pollen grains, anthers were collected from the wild-type and Osfignl1 mutant at anthesis stage and stained with 1.2% iodine-potassium iodide solution (I2-KI) to observe starch accumulation in pollen grains by light microscopy (Li et al., 2010). The wild-type and mutant florets at different developmental stages were fixed in FAA solution (formalin:acetic acid:50% ethanol were 5:5:90, by volume) to observe developmental progression of embryo sacs as described previously (Wang C.L. et al., 2016). The ovaries were finally captured using a laser confocal scanning microscope (Zeiss, Germany).
Map-Based Cloning of the OsFIGNL1 Gene and Complementation of the Osfignl1 Mutant
The F2 mapping population was generated from a cross between the Osfignl1 mutant and O. sativa spp. japonica cv. Zhonghua11. Two thousand and three hundred plants of the F2 population showing the male sterility phenotype were selected for gene mapping. DNA was extracted from fresh leaves using the modified CTAB method (Murray and Thompson, 1980). The InDel markers were designed based on polymorphisms between japonica Nipponbare and indica 9311 (Shen et al., 2004). The InDel marker sequences are listed in Supplementary Table S1.
For functional complementation, an 11.9 kb genomic DNA amplicon containing the entire OsFIGNL1 coding region, 2.2-kb upstream and 1.0-kb downstream sequence was obtained by PCR amplification using primer pairs COM-OsFIGNL1. The PCR product was inserted into the binary vector pCAMBIA13001 to generate pCAMBIA1300-OsFIGNL1 construct using an EcoRI enzyme site. The pCAMBIA1300-OsFIGNL binary plasmids were introduced into the Agrobacterium tumefaciens strain EHA105 and transformed into calli induced from young panicles of Osfignl1. The transgenic plants were further identified by PCR amplification using the primer COM-JD (Supplementary Table S1). To confirm whether the fertility of Osfignl1 mutant was restored by OsFIGNL1 gene, the pollen grains of T0 transgenic plants were stained with 1.2% I2-KI and observed under a light microscope.
To generate the Cas9 targeting vector, we used the pCAS9-AarI vector containing a codon-optimized Cas9 driven by a maize ubiquitin promoter, the OsU3 promoter and sgRNA scaffolds, as well as the Cas9 expression backbone vector (Li et al., 2016), with AarI restriction enzyme (Fermentas). The targeting sequence primer pairs Cas-OsFIGNL1 are listed in Supplementary Table S1. The OsU3 promoter was used to drive expression of the sgRNA. The annealed gRNA oligonucleotide pair was cloned into pCAS9-AarI binary vectors and introduced into the Agrobacterium strain EHA105. Transformed calli were induced from Zh8015 seeds for Agrobacterium-mediated transformation as previously described (Lin et al., 2009). The T0 transgenic mutant plants were confirmed by PCR using primer Cr-JD (see Supplementary Table S1).
RNA Isolation and qRT-PCR Analysis
Total RNA was isolated from roots, stems, leaves, and anthers at different developmental stages using the RNAeasy Mini Kit (Tiangen, China), as per manufacturer’s instructions. Total RNA was used for reverse transcription (RT) with the ReverTra Ace quantitative PCR RT Master Mix Kit with gDNA Remover (Toyobo, Japan). qRT-PCR was performed with a LightCycler 480 (Roche, Germany) using SYBR Premix Ex Taq II (Takara, Japan) according to manufacturer’s instructions. The reaction program was run as follows: 94°C for 4 min initial denaturation, then 94°C for 15 s, 58°C for 30 s, and 72°C for 15 s for 45 cycles. The primer pairs S12 were designed to examine the expression of OsFIGNL1 in the wild-type and Osfignl1 mutant. The rice OsActin1 gene was chose as a control to normalize expression data. Each reaction was performed with three replicates. All primers are listed in Supplementary Table S1.
Resin Sections of Wild-Type and Mutant Anthers
Thin sections of developing anthers were performed on standard plastic sections according to a previously described method (Li et al., 2006). Spikelets at various development stages were collected and fixed in FAA solution overnight at room temperature. After dehydration in a graded ethanol series from 50 to 100%, the samples were embedded in Technovit 7100 resin (Heraeus, Kulzer, Germany), and polymerized at 45°C. Transverse sections of 2 μm were cut using a Leica RM2265 Fully Automated Rotary Microtome, stained with 0.25% toluidine blue (Chroma Gesellshaft Shaud), and photographed using a Leica DM2000 light microscope.
Transmission Electron Microscopy (TEM) and Scanning Electron Microscopy (SEM)
For TEM observation, spikelets at different developmental stages were fixed with 3% (w/v) paraformaldehyde and 0.25% glutaraldehyde in 0.2 M sodium phosphate buffer (pH 7.2) overnight at 4°C, then rinsed twice using 0.1 M phosphate buffer (pH 7.0). Samples were then post-fixed in 2% osmium tetroxide in PBS, pH 7.2. The samples were dehydrated using an ethanol series from 30 to 100%, then embedded in acrylic resin. Ultra-thin sections were cut and collected on uncoated nickel grids, and double stained with 2% uranyl acetate and 2.6% lead citrate aqueous solution. The pictures were examined with a Hitachi H-7650 transmission electron microscope at 80 kV.
For SEM observation, mature anthers of the wild-type and the mutant were pre-fixed in 0.1 M sodium phosphate buffer containing 2.5% glutaraldehyde (pH 7.0), overnight at 4°C, then rinsed twice using 0.1 M phosphate buffer (pH 7.0). The samples were rinsed with the same buffer and post-fixed for 1.5 h in 2% OsO4 in PBS, pH 7.2. Following ethanol dehydration and then exchanged three times with isoamyl acetate. The fixed samples were processed for critical point drying using liquid CO2, and gold coated. The samples were observed using a scanning electron microscope (TM-1000 Hitachi) with an acceleration voltage of 10 or 15 kV.
Meiotic Chromosome Preparation
Young panicles (80–90 mm), including both wild-type and Osfignl1 spikelets in meiosis were fixed with 3:1 ethanol:acetic acid (EAA), and stored at 4°C until observation. Microsporocytes undergoing meiotic division were squashed on glass slides and stained with an acetocarmine solution. Slides with chromosomes were frozen in liquid nitrogen. After removing the coverslips, the slides were dehydrated through an ethanol series (70, 90, and 100%). Male meiotic chromosomes were counterstained with 4′,6-diamidino-phenylindole (DAPI). Chromosome images were captured using Nikon Eclipse 90i microscope with Nikon Digital Camera DXM1200C and Nikon C-HGFI.
Phylogenetic Analysis
The full-length amino acid sequence of OsFIGNL1 and protein sequence of its homologs from different species were retrieved through BLASTP search using the amino acid sequence of OsFIGNL1 from the NCBI database2. The multiple sequence alignment was created using Clustalw with the default parameters3. A phylogenetic tree was constructed with the neighbor-joining algorithm using MEGA 6.0 software (Tamura et al., 2013). Bootstrap values were calculated from 1000 bootstrap replicates.
Subcellular Localization
To construct the subcellular localization plasmids, the full-length coding region of OsFIGNL1 was amplified with the primer pair GFP-OsFIGNL1 (Supplementary Table S1), with BamHI and SpeI restriction sites added to the 5′ and 3′ ends, respectively, then ligated into the pCAMBIA1305-GFP expression vector (pCAMBIA4). Mesophyll protoplasts were isolated from 2-week-old Zh8015 plants. The empty vector control and recombinant construct plasmids (pCAMBIA1305-OsFIGNL1-GFP) were transformed into rice protoplasts and incubated for 48 h in the dark (Yoo et al., 2007). Fluorescence signal in the transformed protoplasts was imaged using a confocal laser scanning microscope (Zeiss, Germany). The OsMADS3-mCherry construct was used as nuclear marker (Wu et al., 2013).
Yeast Two-Hybrid (Y2H) Assay and BiFC Assay
Full-length cDNA of OsFIGNL1 transcripts were amplified and cloned into the Y2H bait vector pGBDT7, while cDNA of RAD51A1, RAD51A2, DMC1A, and DMC1B were amplified from young panicles and cloned into the Y2H prey vector, pGADT7 (Clontech). Plasmid inserts were confirmed by DNA sequencing. The two plasmid types were then co-transformed into the Saccharomyces cerevisiae Y2HGold strain according to the manufacturer’s instruction (Clontech). Transformants were grown on tryptophan-negative and leucine-negative synthetic dropout medium (SD/–Trp–Leu) then incubated on test plates lacking Leu, Trp, His, and Ade at 30°C for 4 days. For generation of BiFC vectors, cDNA of the rice RAD51A1, RAD51A2, DMC1A, and DMC1B were amplified from young panicles of indica Zh8015 and cloned at BamHI-XhoI sites in pSPYNE to generate pSPYNE-RAD51A1, pSPYNE-RAD51A2, pSPYNE-DMC1A, and pSPYNE-DMC1B plasmids. The CDS of OsFIGNL1 was cloned into BamHI-XhoI sites of pSPYCE, resulting in pSPYCE-OsFIGNL1 vector. The different combinations of constructs were transformed into the Agrobacterium train GV3101. All constructs were expressed under the control of the 35S promoter. The procedure of BiFC was performed as described previously (Waadt et al., 2008). All primer pairs are listed in Supplementary Table S1.
Results
Morphological Characterization of the Osfignl1 Mutant
We obtained a complete male sterility mutant Osfignl1 from an indica rice cultivar, Zhonghui 8015, induced by 60Co-γ ray radiation. Compared with the wild-type plants, Osfignl1 displayed a normal phenotype during vegetative development, however, the mutant failed to set seeds (Figures 1A,B). Compared to wild-type anthers, the mutant anthers were relatively small, thin, and slightly yellow (Figures 1C,D). During the reproductive stage, mature pollen grains of wild-type anthers were darkly stained by 1.2% I2-KI, whereas the pollen grains of Osfignl1 lacked starch and could not be stained (Figures 1E,F). To estimate female fertility of the Osfignl1 mutant, we conducted reciprocal crosses between the wild-type and Osfignl1 mutant. The mutant successfully set hybrid seeds when it was pollinated with wild-type pollen grains, indicating a normal megagametogenesis in the Osfignl1 mutant (Supplementary Figure S1). To ascertain whether female organ development was influenced in the Osfignl1 mutant, a total of 300 embryo sacs of the Osfignl1 plants were examined for female fertility. Observation of normal female reproduction in Osfignl1 was further supported by our cytological analysis of the Osfignl1 embryo sac, which did not show any obviously abnormal morphology during embryo sac development (Supplementary Figure S2). We thus concluded that the sterility of spikelets in Osfignl1 plants is caused by the disruption of male reproduction in rice. Genetic analyses showed that fertile plants and sterile plants followed an approximate 3:1 ratio for phenotypic segregation in F2 progenies, indicating that the sterility phenotype was caused by a single recessive mutation (fertile: sterile = 146: 42; χ2 = 0.57 < = 3.84).
FIGURE 1.
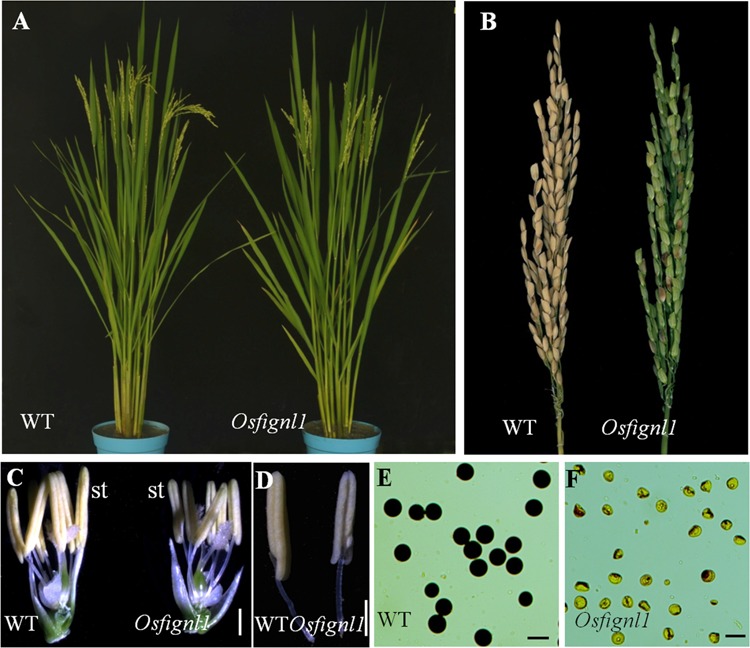
Phenotype comparisons between the wild-type (WT) and the Osfignl1 mutant. (A) Comparison between a WT (left) and a Osfignl1 mutant (right) plant after heading. (B) Comparison of a WT panicle (left) and a Osfignl1 mutant panicle (right) at the harvest stage. (C) Comparison of a WT (left) and the Osfignl1 mutant spikelet (right) after removal of palea and lemma. (D) Comparison of a WT (left) and a Osfignl1 mutant anther (right). (E) I2-KI staining of WT pollen grains. (F) I2-KI staining of Osfignl1 mutant pollen grains. st, stamen. Bars = 1 mm in (C,D), 50 μm in (E,F).
Abnormal Anther Development and Pollen Maturation of the Osfignl1 Mutant
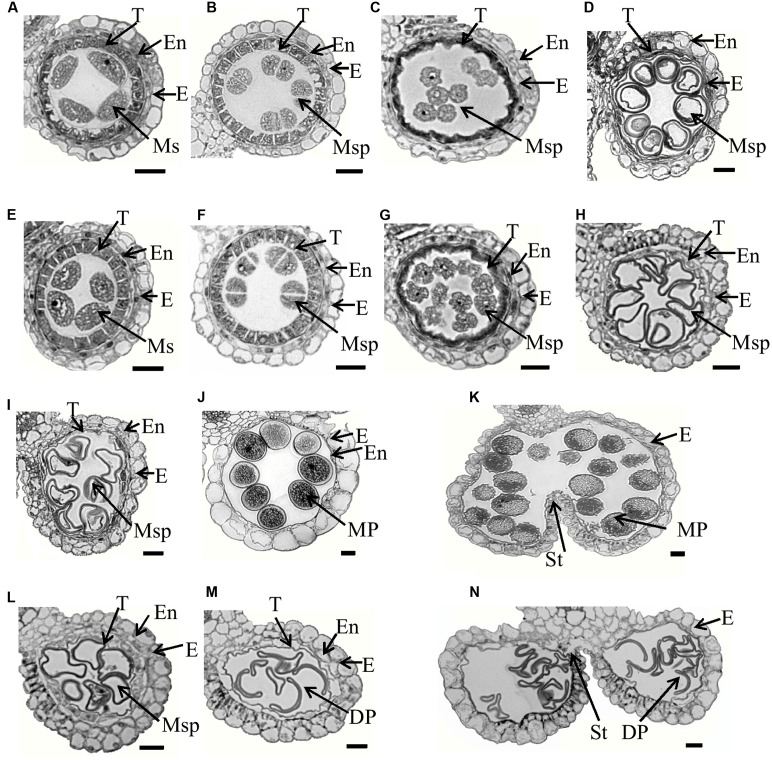
To characterize the defects in Osfignl1, we examined cross sections of anthers sampled from the wild-type and Osfignl1 mutant spikelets of different developmental stages. No obvious differences in four somatic layers of the anther wall and microsporocytes were detected between the wild-type and Osfignl1 mutant until the early microspore uninucleate stage (Figures 2A–C,E–G). PMCs of both the wild-type and the Osfignl1 mutant appeared to undergo meiosis and form tetrads. The tapetal cells became vacuolated and deeply stained, and the middle layer was hardly visible and degenerated at the tetrad stage (Figures 2B,F). At the young microspore stage, microspores were released from the tetrads, while the tapetum became condensed, less vacuolated, and degenerated normally in the Osfignl1 mutant, just like in wild-type anthers (Figures 2C,G). At the vacuolated pollen and bicellular pollen stages, the wild-type tapetum became more degenerated, the endothecium developed normally, and round-shaped, vacuolated microspores were formed in the anther locule. In contrast, the Osfignl1 tapetum was slightly expanded, and microspores formed with an abnormal shape and no vacuoles (Figures 2D,H,I,L). At the mature pollen stage, mature wild-type pollen grains filled with starch granules were densely stained, and the tapetum completely disappeared. However, at this stage in the Osfignl1 anther, the cells of the endothecium appeared expanded, the tapetum remnants abnormally persisted with no cell contents, and microspores degenerated with an irregular shrunken shape (Figures 2J,M). At the anther dehiscence stage, when the filament elongated, the endothecium in wild-type anthers was almost completely degraded and invisible. At the same stage, the Osfignl1 tapetum had not yet disappeared completely and the endothecium degenerated further (Figures 2K,N). Two adjacent pollen sacs of the wild-type were able to merge into a single locule. In contrast, pollen sacs in Osfignl1 were not linked together and aborted pollen grains were released from the anther through individual open stomium, showing that anther dehiscence also was affected in Osfignl1 (Figures 2K,N). These observations together indicate that defects in microspore formation and anther development occur in the Osfignl1 mutant.
FIGURE 2.
Semi-thin section comparison of anther development in the WT and the Osfignl1 mutant. (A–D) and (I–K) show the WT; (E–H) and (L–N) show the Osfignl1 mutant. (A,E) Meiosis stage. (B,F) Tetrads stage. (C,G) Young microspore stage. (D,H) Vacuolated pollen stage. (I,L) Bicellular pollen stage. (J,M) Mature pollen stage. (K,N) Anther dehiscence stage. E, epidermis; En, endothecium; DP, defective pollen; MP, mature pollen; T, tapetum; Ms, microsporocyte; Msp, microspore; St, stomium. Bars = 20 μm.
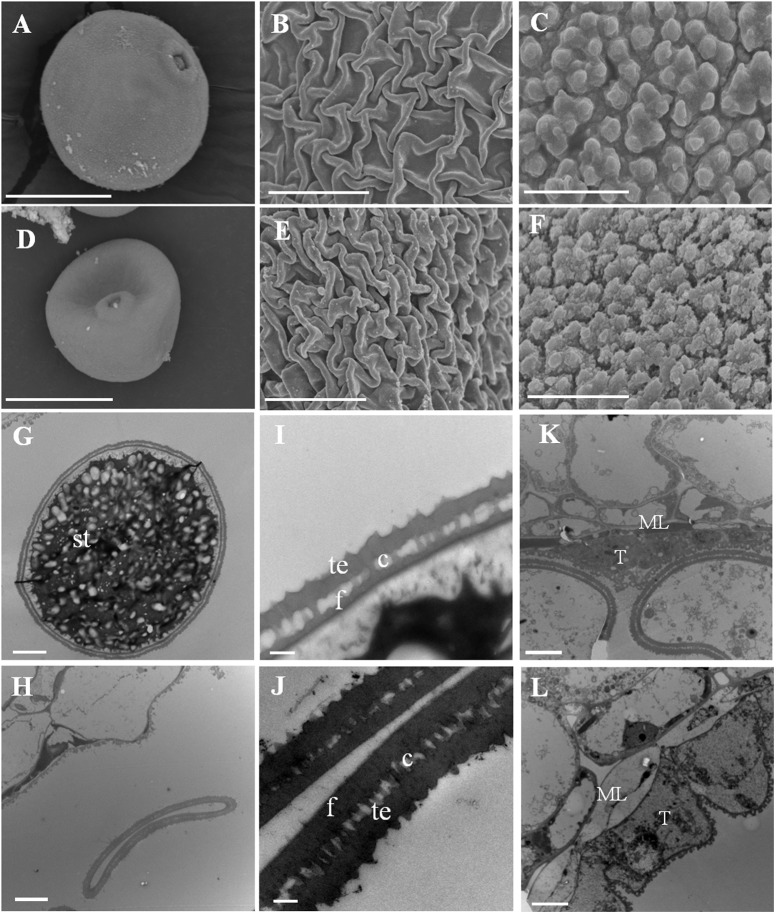
The anther and microspore abnormalities between the wild-type and Osfignl1 mutant were further analyzed by SEM and TEM observation. Mature pollen grains of the wild-type were spherical, larger, and filled with abundant starch granules, whereas Osfignl1 pollen grains were irregular, shrunken, and failed to produce internal contents and nuclei (Figures 3A,D,G,H). The wild-type anther epidermis was less compact compared to the Osfignl1 mutant (Figures 3B,E). In addition, the pollen exine of the wild-type was covered with sporopollenin which appeared typically granular, while in contrast, the Osfignl1 pollen exine had irregular or “messy” sporopollenin deposits (Figures 3C,F). The typical pollen wall is composed of exine and intine, with exine divided into the tectum and foot layer. In the wild-type, mature pollen grains developed a normal tectum and foot layer, which were linked by the columella (Figure 3I), whereas Osfignl1 pollen showed a thicker tectum and foot layer (Figure 3J). The thicker tectum and foot layer had an abnormal morphology and degenerated more than wild-type (Figure 3J), suggesting that mutation in the Osfignl1 mutant also affects the pollen wall formation. TEM micrographs also showed that the tapetum and middle layer were swollen, and thicker than those of the wild-type at bicellular pollen stage. These observations suggest that the abnormal degradation of the tapetum occurs in Osfignl1 during late anther development, which is probably an important reason for the abnormal anther maturation and sterility of pollen grains (Figures 3K,L).
FIGURE 3.
Scanning electron microscopy (SEM) and transmission electron microscopy (TEM) observation for WT and Osfignl1 mutant anther. (A) Scanning electron microscopy image of the mature WT pollen grain. (B) Scanning electron microscopy image of the WT anther epidermis. (C) Scanning electron microscopy image of the WT pollen exine. (D) Scanning electron microscopy image of the Osfignl1 pollen. (E) Scanning electron microscopy image of the Osfignl1 anther epidermis. (F) Scanning electron microscopy image of the Osfignl1 pollen exine. (G) Transmission electron microscopy showing the WT pollen. (H) Transmission electron micrograph showing the Osfignl1 pollen. (I) A higher magnification image of the WT pollen wall from (G). (J) A higher magnification image of the Osfignl1 mutant pollen grain from (H). Transmission electron micrographs of the WT (K) and Osfignl1 mutant anther (L) at bicellular pollen stage. c, columella; f, foot layer; te, tectum; st, starch granules; T, tapetum; ML, middle layer; Bars = 20 μm (A,D), 1 μm in (C,F), 5 μm in (B,E,G,H), 500 nm in (I,J), and 5 μm in (K,L).
Meiosis Is Disrupted in the Osfignl1 Mutant
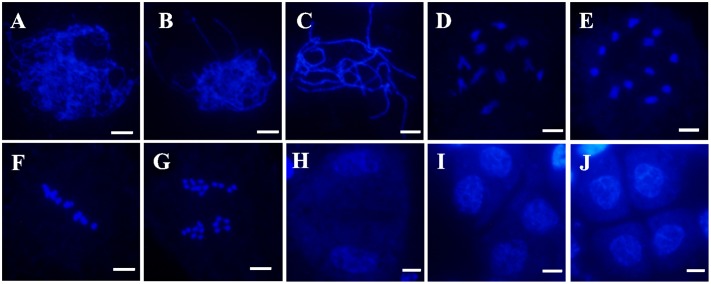
To determine whether pollen abortion resulted from defects in male meiosis, meiotic chromosomal behavior in male meiocytes of both wild-type and the Osfignl1 mutant were examined by DAPI staining. In the wild-type, meiotic chromosomes began to condense and appeared as thin thread-like structures at leptotene (Figure 4A). At the zygotene stage, homologous chromosomes underwent synapsis and concentrated around the nucleolus on one side (Figure 4B). In wild-type plants, generation of thick, thread-like chromosomes were observed at pachytene, indicating that chromosomes were paired, and fully synapsed chromosomes were completed (Figure 4C). During diplotene, the synaptonemal complex disassembled and chiasmata, which linked the paired homologous chromosomes together, were visible (Figure 4D). In the wild-type, paired chromosomes condensed into 12 bivalents at diakinesis (Figure 4E). During metaphase I, the 12 highly condensed bivalents were aligned on the equatorial plate (Figure 4F). At anaphase I, homologous chromosomes separated and migrated in opposite directions of the cell (Figure 4G), finally leading to the formation of dyads at the end of meiosis I (Figure 4H). During the second meiotic division, the sister chromatids of each chromosome separated, similar to that in mitosis. After meiosis II, the tetrads of microspores were formed (Figures 4I,J).
FIGURE 4.
4′,6-diamidino-phenylindole (DAPI)-stained chromosome spreads of male meiosis in WT plants. (A) Leptotene. (B) Zygotene. (C) Pachytene. (D) Diplotene. (E) Diakinesis. (F) Metaphase I. (G) Anaphase I. (H) Dyad. (I) Telophase II. (J) Tetrad. Bars = 5 μm.
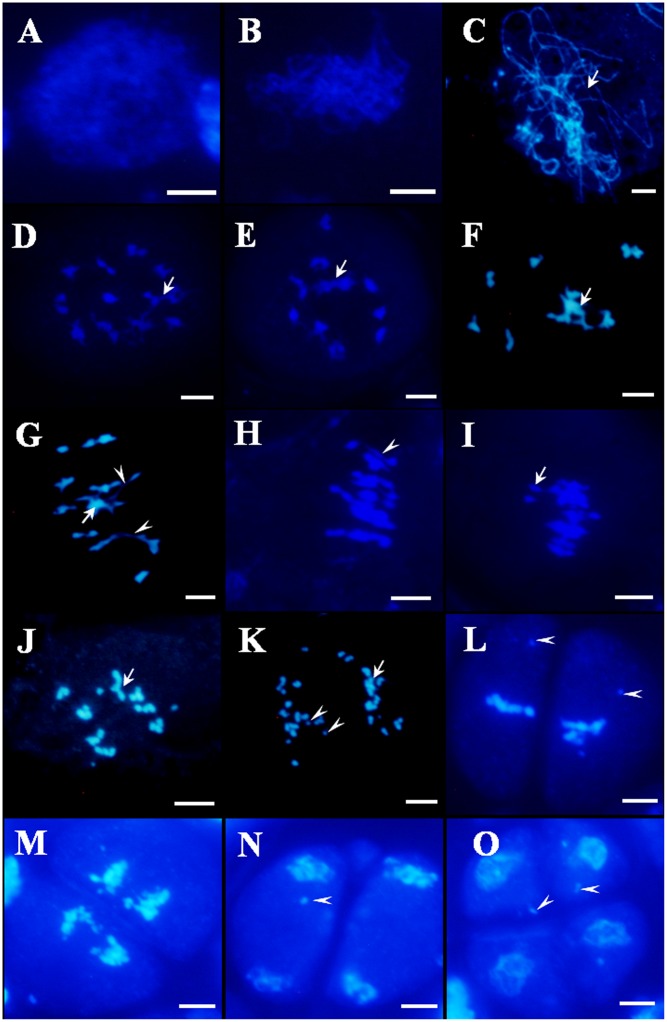
In Osfignl1 male meiocytes, meiotic chromosome behavior showed no obvious differences, compared with that of the wild-type from leptotene to zygotene (Figures 4A,B, 5A,B). However, chromosomal abnormalities appeared from the pachytene to the tetrad stage. Chromosomes appearing as single threads were observed at the pachytene stage (Figure 5C). At diplotene, abnormal chromosomes were linked together, and we observed abnormally shaped chromosomes, instead of well-defined bivalents (Figures 5D,E). During diakinesis, irregularly shaped chromosomes, chromosome bridges, and entangled chromosomes were visualized (Figures 5F,G). In metaphase I, most of the entangled chromosomes were aligned on the equatorial plate, such as those found in the wild-type, while other chromosomes did not align at all on the equatorial plate (Figure 5I). Chromosome bridges also were found in Osfignl1 at metaphase I (Figure 5H). At anaphase, the chromosomes separated and moved to opposite sides of the cell, because of these chromosome fragments, the numbers of chromosomes were difficult to distinguish as separate chromosomes (Figure 5J). Moreover, multiple chromosomes and several chromosome fragments were observed at this stage (Figure 5K). During the second meiotic division, lagging chromosomes and chromosome fragments were still observed in the cell, and resulted in formation of the tetrads with nuclei of different sizes from wild-type (Figures 5L–O). These results indicated that unrepaired ectopic recombination may occur between non-homologous chromosomes, finally leading to chromosome breakage and ultimately abortion of microspore development. These results suggest that meiotic defects cause complete male sterility of Osfignl1 plants.
FIGURE 5.
4′,6-diamidino-phenylindole-stained chromosome spreads of male meiosis in Osfignl1 plants. (A) Leptotene. (B) Zygotene. (C) Pachytene. (D,E) Diplotene. Arrows indicate entangled chromosomes. (F,G) Diakinesis. Arrows indicate multivalent chromosome and the arrowheads indicate chromosome bridges. (H,I) Metaphase I, showing chromosome bridges (arrowhead). Chromosomes did not align on the equatorial plate (arrow). (J,K) Anaphase I, showing chromosome fragments (arrowheads) and entangled chromosome (arrows). (L) Metaphase II. (M) Anaphase II. (N) Telophase II. (O) Tetrad. From Metaphase II (L) to tetrad (O), showing lagging chromosome (arrowheads). Bars = 5 μm.
We compared meiosis progression between the wild-type and Osfignl1 male meiocytes based on the lengths of spikelets. In Osfignl1 mutant plants, male meiocytes progressed normally before the pachytene phase as wild-type did. In the wild-type, 52% of microsporocytes were found in 5.6–5.8 mm spikelets during the diplotene stage, while 90% of Osfignl1 mutant microsporocytes were still in the pachytene stage (Supplementary Figure S3). When most of wild-type meiocytes (94%, n = 62) in relatively longer (6.3–6.5 mm) spikelets had progressed from pachytene into late prophase I, the Osfignl1 mutant meiocytes (100%, n = 187) remained in the pachytene phase (Supplementary Figure S3). When spikelet length reached 6.7–6.8 mm, a majority of wild-type meiocytes completed meiosis, however, 5% of Osfignl1 male meiocytes were still observed at the pachytene stage. These observations suggest that a slowdown of the pachytene stage of meiosis in Osfignl1 meiocytes leads to a lengthening of the progression of male meiosis.
Map-Based Cloning of OsFIGNL1 and Its Protein Sequence Analysis
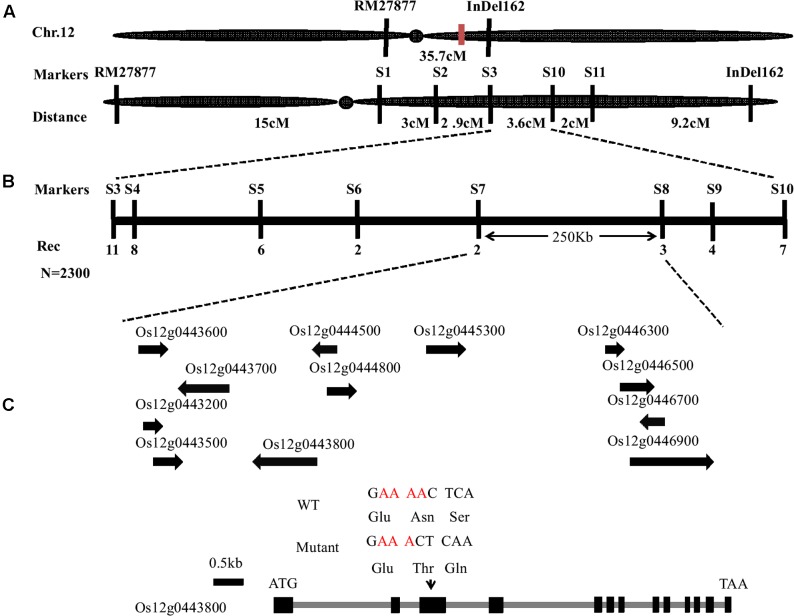
To isolate the OsFIGNL1 gene, a map-based cloning strategy was employed. We generated an F2 population by crossing the Osfignl1 mutant with Zhonghua11 (japonica rice) for linkage analysis. The mutant gene was primarily mapped to a genetic distance of 22.9 and 12.8 cM with RM27877 and InDel162 markers on chromosome 12, respectively (Figure 6A). To finely map the OsFIGNL1 gene, a total of 2,300 F2 population individuals exhibiting the male-sterile phenotype were used for genetic analysis. Finally, we mapped the candidate gene to a 250 kb between markers S7 and S8, which contained 12 predicted open reading frames, annotated by the Rice Annotation Project Database (RAP-DB5) (Figure 6B). We sequenced the predicted genes of this 250-kb region, and found that the Osfignl1 mutant carries a single nucleotide Adenine deletion in the third exon of the annotated gene Os12g0443800.1, resulting in a premature stop codon at the 187th amino acid residue (Figure 6C). Based on the annotations of the RAP-DB, we found that Os12g0443800.2 and Os12g0443800.1 were annotated as a P-loop containing nucleoside triphosphate hydrolase superfamily protein with putative, overlapping transcribed sequences (Supplementary Figure S4A), suggesting that these ORFs may be a single, transcriptional unit. As expected, RT-PCR showed that Os12g0443800 containing Os12g0443800.1 and Os12g0443800.2 represented a single mRNA, and the sequence of Os12g0443800.2 revealed a mis-annotation in the RAP-DB. OsFIGNL1 consisted of 13 exons and 12 introns (Figure 6C), comprising a 2,085-bp cDNA region, encoding a predicted protein of 694 amino acids (Supplementary Figure S4B).
FIGURE 6.
Map-based cloning of OsFIGNL1 gene on chromosome 12. (A) Primary mapping was performed using markers RM27877 and InDel162. (B) The OsFIGNL1 locus was finally mapped to a 250-kb region between the molecular markers S7 and S8. Names and positions of the markers are noted. (C) A schematic representation of the gene structure of OsFIGNL1. The mutant sequence has a single nucleotide deletion in the third exon. ATG and TAA represent the start and stop codon, respectively. Black boxes indicate exons, and intervening gray lines indicate introns. cM, Centimorgan; Rec, Recombinants; N, Total number of F2 plants used for fine mapping.
To confirm that the male sterility was caused by the mutation in Os12g0443800, we amplified an 11.8-kb genomic DNA fragment containing the full-length ORF of Os12g0443800 from Zh8015, and transformed it into calli induced from young panicles of homozygous Osfignl1 mutant plants. The transgenic plants had high seed-setting rate, similar to those of wild-type (Supplementary Figure S5A). The complemented lines accumulated abundant starch granules and displayed yellow anthers (Supplementary Figures S5B–E), indicating that the 1 bp deletion in Os12g0443800 was responsible for the pollen abortion phenotype of Osfignl1. To further confirm OsFIGNL1 function is responsible for the phenotype observed in the Osfignl1 mutant, we generated CRISPR/Cas9 mutation lines of OsFIGNL1 in the Zh8015 genetic background. Among the 12 CRISPR T0 lines obtained, two lines were found to harbor a homozygous mutation in OsFIGNL1 with significantly reduced transcription level (Supplementary Figures S6A,E). As expected, the two homozygous lines phenotypically mimicked Osfignl1 with shorter, thinner anthers and shriveled pollens, while the remaining heterozygous or negative lines grew normally as wild-type (Supplementary Figures S6B,C). Similar to the Osfignl1 mutants, aberrant chromosomes and chromosomal entanglement also can be found in the homozygous mutant lines during male meiosis (Supplementary Figure S6D). Taken together, the results above demonstrated that Os12g0443800 was the rice OsFIGNL1 gene.
An interProScan search found that OsFIGNL1 was predicted to encode a protein with an ATPase domain (453–583) and a VPS4 domain (656–690)6 (Supplementary Figure S4B). Further alignment of the protein sequences of FIGNL1 homologs from Arabidopsis thaliana (Genbank accession KM055500), O. sativa (Os12g0443800), Homo sapiens (NP_001036227.1), Mus musculus (NP_001156832.1), and C. elegans (NP_504197.1) indicated that all five proteins contained a VPS4 domain and an ATPase domain (Supplementary Figure S7). We constructed a phylogenetic tree from the OsFIGNL1 full-length protein sequences of 23 orthologs from different species acquired by BLASTP (NCBI) search using rice OsFIGNL1 as the query. OsFIGNL1-related sequences were classified into two clades: clade I is comprised of dicot proteins, while the clade II members belong to monocotyledon species including the Brachypodium distachyon, Hordeum vulgare, Zea mays, Setaria italica, and Sorghum bicolor (Supplementary Figure S8). The BLASTP search also revealed that OsFIGNL1 was closely related to Brachypodium distachyon with 80% identity. Taken together, these analyses imply that OsFIGNL1 is conserved in land plants.
Subcellular Localization OsFIGNL1 Protein and Expression Pattern of Rice OsFIGNL1 Gene
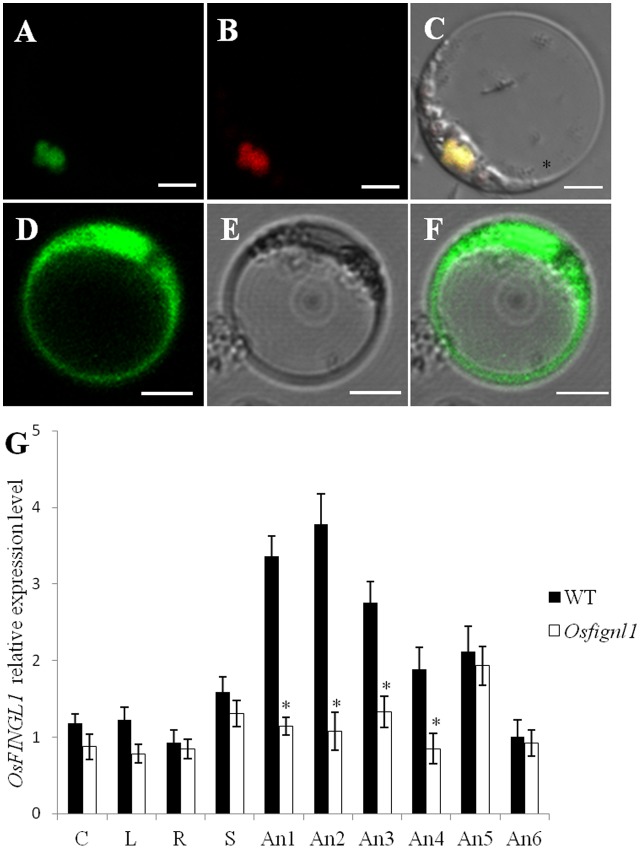
In order to determine the subcellular localization of OsFIGNL1 protein, we constructed a vector expressing OsFIGNL1-GFP (green fluorescent protein) fusion protein driven by the 35S promoter. After introduction into rice protoplast cells, the OsFIGNL1-GFP fusion protein was detected exclusively in the nucleus (Figures 7A–C), whereas the GFP empty vector control was found throughout the cell (Figures 7D–F), indicating that OsFIGNL1 encodes a nuclear-localized protein. qRT-PCR analysis was conducted to examine the expression patterns of OsFIGNL1 in various tissues and growth stages. The results suggested that OsFIGNL1 was constitutively expressed in all the tested issues, including mature root, mature sheath, mature leaf, mature culm, relatively higher levels detected in anthers. Particularly, OsFIGNL1 expressed predominantly in anthers at the meiosis stage, and progressively reduced as anther development proceeded (Figure 7G). This expression pattern well supported the compromised meiosis that we observed in Osfignl1.
FIGURE 7.
Subcellular localization and expression pattern analysis of rice OsFIGNL1. (A) Subcellular localization of full-length OsFIGNL1 fused with GFP is detected in nucleus. (B) OsMADS3-mCherry was used as a nuclear marker. (C) A merged image of (A,B). (D–F) GFP alone is detected throughout the rice protoplast cells. Bars = 5 μm. (G) Expression pattern analysis of rice OsFIGNL1. R, mature root; C, mature culm; L, mature leaves; S, mature sheath. An1, spikelet lengths from 4 to 5 mm; An2, spikelet lengths from 5 to 6 mm; An-3, spikelet lengths from 6 to 7 mm; An-4, spikelet lengths from 7 to 8 mm; An-5, spikelet lengths from 8 to 9 mm; An-6, spikelet lengths > 10 mm. OsActin1 was used a control for normalizing expression in all assays. ∗Indicate significant difference at P < 0.01.
OsFIGNL1 Interacts with Rice RAD51 and DMC1
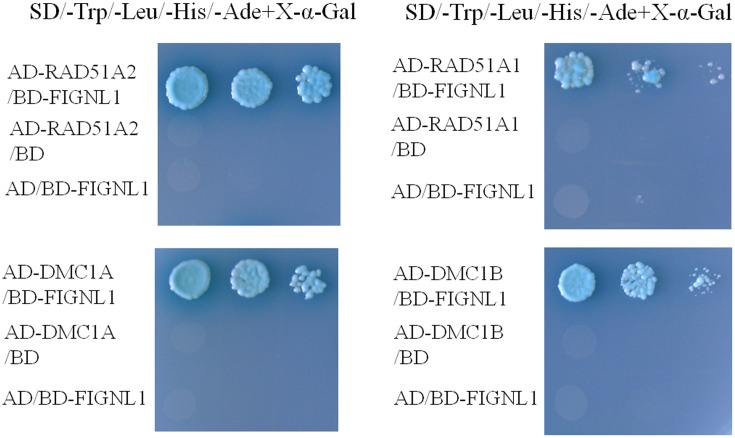
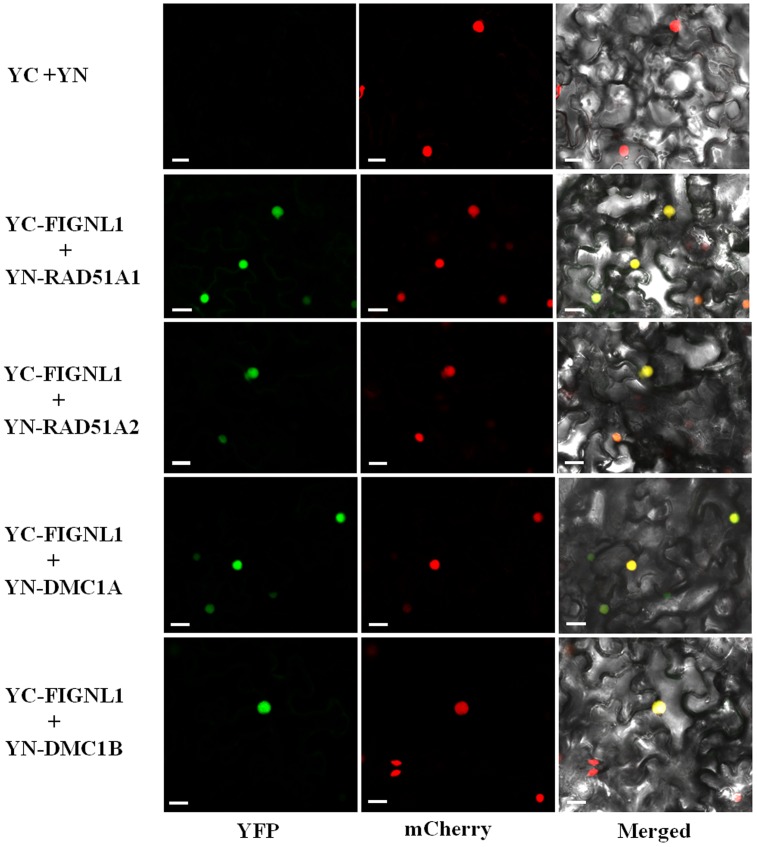
A previous study showed that human FIGNL1 specifically interacts with RAD51 protein (Yuan and Chen, 2013). Rice genome contains two highly similar RAD51 proteins, RAD51A1 and RAD51A2, both of which are DNA-binding and participate in homologous chromosome pairing, though the specific activity may differ from each other (Morozumi et al., 2013). To investigate whether OsFIGNL1 interacts with RAD51A1, RAD51A2, DMC1A, and DMC1B in rice, we cloned full-length RAD51A1, RAD51A2, DMC1A, and DMC1B into the prey vector pGADT7, while OsFIGNL1 was cloned into the bait vector pGBKT7. Yeast two-hybrid analysis demonstrated that rice OsFIGNL1 could physically interact with the rice RAD51A1, RAD51A2, DMC1A, and DMC1B, but not with RAD51B and RAD51D (data not shown) (Figure 8). To prove the interactions further, a BiFC assay demonstrated that OsFIGNL1 interacted with RAD51A1, RAD51A2, DMC1A, and DMC1B in Nicotiana benthamiana leaves cells and interacting proteins had a nuclear localization. These results suggest that rice OsFIGNL1 interacts with rice RAD51A1, RAD51A2, DMC1A, and DMC1B in vivo (Figure 9).
FIGURE 8.
Yeast two-hybrid interactions between rice OsFIGNL1 and RAD51A1 RAD51A2, DMC1A and DMC1B. Y2H assay to test interactions between OsFIGNL1, RAD51A1, RAD51A2, DMC1A and DMC1B. OsFIGNL1 was cloned into the bait vector pGBKT7 (BD), and rice RAD51A1, RAD51A2, DMC1A, and DMC1B were inserted in the pray vector pGADKT7 (AD). The empty pGBKT7 and pGADKT7 vectors were used as negative controls.
FIGURE 9.
Bimolecular fluorescence complementation (BiFC) assays of rice OsFIGNL1 interaction between RAD51A1, RAD51A2, DMC1A, and DMC1B in vivo. The expression of pSPYNE (YN)/pSPYCE(YC) was used as negative control. OsMADS3-mCherry was used as a nuclear marker. Bars = 20 μm.
Discussion
OsFIGNL1 Is a Member of AAA Family and Controls Male Fertility in Rice
In the present study, using map-based cloning, we isolated the rice fertility gene OsFIGNL1. The results of this mutagenesis and complementation study strongly indicated that OsFIGNL1 is a critical gene for male fertility in rice. The morphological and cytologic analyses suggest that OsFIGNL1 plays a key role in anther development, and is essential for male meiosis. Here, we report that the rice OsFIGNL1 gene encodes a nuclear protein which belongs to a group of meiosis-related AAA proteins (Frickey and Lupas, 2004). FIDGETIN is the first reported AAA protein, which contains a conserved ATPase domain spanning 200–250 amino acid residues. Sequence alignment showed that the FIGNL1 protein consists of several motifs including a VPS4 domain (Vajjhala et al., 2006), FIGNL1-RAD51-Binding Domain (FRBD) (Yuan and Chen, 2013), as well as the conserved Walker A, Walker B, and Arg fingers domains of the ATPase domain (Yakushiji et al., 2004; Peng et al., 2013), indicating that these functional domains were evolutionarily conserved and likely critical for its function in these species. However, despite of the high level of conservation in the ATPase region, the N-termini were not conserved among the different species of OsFIGNL1 homologs (Supplementary Figure S7). Furthermore, human FIGNL1’s RAD51 Binding Domain (FRBD), a region that is highly specific for binding RAD51, is also found in O. sativa (Supplementary Figure S7) and Arabidopsis (Girard et al., 2015). Several FIGNL1 proteins have been reported to be functional in regulating diverse cellular processes among different species. Although there may exist essential and conserved function domains among FIGNL1s, molecular and biological functions of the AAA family members may have diverged among different species. For example, Zhao et al. (2016) recently found that human FIGNL1 localizes at centrosomes, and is involved in ciliogenesis. Previous reports showed that C. elegans FIGNL1 binds to microtubules and controls mitotic progression in the germ line (Luke-Glaser et al., 2007). In our study, the results revealed that the loss function of OsFIGNL1 does not affect female fertility, but leads to complete male sterility. This conclusion was supported by backcrossing between the Osfignl1 mutant and wild-type plants using a mutant as maternal recipient, which also can produce hybrid seeds. The phenotypic characterization of embryo sac development of differential stages showed that Osfignl1 embryo sacs had no obvious differences compared with wild-type plants, suggesting that OsFIGNL1 is only essential for male meiosis but not indispensable for female reproductive development. In contrast to most of the previously reported meiotic mutants which exhibited complete male and female sterility, the Osfignl1 mutant is male sterile with normal female fertility. These findings suggest that the meiosis in rice megagametogenesis and male gametogenesis are regulated by different genes. Since OsFIGNL1 is most highly expressed in meiosis stage anthers, though its expression is not limited to the anther, we hypothesize that OsFIGNL1 plays a minor role in female meiosis aside from its important role in male meiosis. This role may be associated with regulation of gene expression in different tissues. Here, the present results demonstrate that the rice OsFIGNL1 mainly functions in male fertility and is essential for male meiosis.
OsFIGNL1 May Be Involved in the Interplay between the Rice RAD51 and DMC1 for Repair Meiotic DSBs
RAD51 and DMC1, both highly conserved in eukaryotes, load onto single-stranded DNA and catalyze the strand exchange between homologous DNA molecules (Baumann and West, 1998). RAD51 protein was shown to mediate the search for homologous sequences and catalyze strand exchange, which plays an import role in DSBs repair (Baumann et al., 1996; Sheridan et al., 2008; Kurzbauer et al., 2012). S. cerevisiae RAD51, similar to Escherichia coli RecA protein, plays an essential role in mitotic and meiotic homologous recombination (Shinohara et al., 1992; Krogh and Symington, 2004). In Arabidopsis, the loss of function of AtRAD51 causes severe abnormalities in meiosis and complete sterility, although the mutant exhibits normal vegetative and flower development (Li et al., 2004). Arabidopsis Atrad51 null mutant is deficient in DSBs repair and exhibits extensive chromosome fragmentation and chromosome bridges during meiosis, whereas Atdmc1 single mutant shows 10 univalents at metaphase I (Pradillo et al., 2012). The phenotypic differences between Atdmc1 and Atrad51 mutants suggest that the specific role of AtRAD51 and AtDMC1 in plant meiosis may be different. Previous studies suggest that AtRAD51 seem to be more involved in intersister recombination, while AtDMC1 could be primarily repair meiotic DSBs using the homologous chromosomes as templates (Pradillo et al., 2012). Alternatively, Kurzbauer et al. (2012) proposed that AtRAD51 and AtDMC1 localize to opposite DNA ends at a meiotic DSB, and AtDMC1 functions independently and spatially separated from AtRAD51 during meiosis. Recently, Da Ines et al. (2013) have reported that AtDMC1 would be chiefly responsible for catalyzing strand invasion, while AtRAD51 would just play a supporting role for promoting AtDMC1 activity in meiotic homologous recombination. In rice, Osdmc1a Osdmc1b mutant displays univalent chromosomes at diakinesis and synapsis is seriously disrupted (Wang H.J. et al., 2016), resulting in complete sterility. However, the meiotic phenotype has not been reported in rice rad51a1, rad51a2, and rad51a1 rad51a2 double mutant to date. A BLASTP (NCBI) search revealed that the rice OsFIGNL1 was similar to the Arabidopsis FIGNL1 with 60% identity (Supplementary Figure S7). So far, only Arabidopsis FIGNL1 has been well characterized in plants (Girard et al., 2015). In wild-type Arabidopsis meiosis, FIGNL1 limits the formation of COs by regulating AtRAD51 and AtDMC1 foci dynamics, while mutation of FIGNL1 increases the number of AtRAD51 foci, but not AtDMC1 foci. In Arabidopsis, mutation of FIGNL1 would restore the level of CO formation in CO-deficient mutants. In addition, human FIGNL1 is found to interact with RAD51, and is necessary for homologous recombination repair (Yuan and Chen, 2013). In this study, using a yeast two-hybrid system and BiFC assay, we revealed strong interactions of OsFIGNL1 with rice RAD51A1, RAD51A2, DMC1A, and DMC1B. Given their crucial roles in strand exchange activities during meiosis, OsFIGNL1 is likely to be involved in homologous recombination mediated by the rice RAD51 and DMC1 proteins. In vivo data have shown that AtBRCA2 protein could bind to AtDMC1 and AtRAD51 (Dray et al., 2006). AtBRCA2 mediates the recruitment of AtRAD51 and AtDMC1 for strand invasion and plays an important role for both somatic and meiotic homologous recombination (Seeliger et al., 2012). It is proposed that OsFIGNL1 may work as a complex with DMC1 and RAD51 for recruitment to DSBs at the initiation of meiotic recombination and thus participates in meiotic homologous recombination in rice. It seems likely that OsFIGNL1 may act as an accessory factor involved in DNA strand invasion by cooperating with DMC1/RAD51 regulating male meiosis. However, cytological studies and biochemical data are needed to establish the molecular mechanism of the effect of OsFIGNL1 on DMC1/RAD51, which load onto chromosomes for DNA strand invasion in accurate meiotic recombination.
The Loss of OsFIGNL1 Function Disturbs Meiotic Chromosome Behavior and Affects Meiotic Cell Cycle Progression
In Arabidopsis, several meiotic mutants, such as Atrad51, Atrad51c, and Atxrcc3, have been found to show observable chromosome fragments or chromosome entanglement during meiosis (Pradillo et al., 2014). Previous studies found that Arabidopsis Atrad51 mutants failed to synapse and instead developed an entangled mass of chromosomes, interconnected by chromatin bridges at metaphase I (Li et al., 2004; Pradillo et al., 2012). The abnormal DSB repair mutants that exhibit various meiotic defects of chromosome fragmentation also have been reported in rice. The Osmre11 mutant contained an entangled mass of chromosomes at diakinesis, and formed large chromosome aggregates at metaphase I (Ji et al., 2013). The rice XRCC3 and OsRAD51C have been reported to be essential for DSB repair in meiosis (Tang et al., 2014; Zhang et al., 2015), and their mutants exhibit chromosome fragments and chromosome entanglements during the process of meiosis. In the Osrad51c mutant, chromosome fragments were first observed at early metaphase I (Kou et al., 2012; Tang et al., 2014). In the rice xrcc3-1 meiocytes, chromosomes with irregular shape and chromosome entanglements were observed from diplotene to metaphase I (Zhang et al., 2015). A previous study has shown that chromosomal fragmentation is thought to be caused by the accumulation of unrepaired DSBs of DNA (Schommer et al., 2003). In the Osfignl1 mutant, the entangled chromosomes and chromosome bridges were observed at diakinesis and metaphase I, possibly due to the defects in homologous recombination or improper repair of DSBs. Compared with the previously reported Osrad51c mutants and the xrcc3 mutant (Tang et al., 2014; Zhang et al., 2015), the abnormal chromosome behavior phenotype in the Osfignl1 mutant, in our study, was less severe from metaphase I to the end of the meiotic stage. However, it is obvious that the chromosome morphology among these mutants was not the same. PMCs of Osfignl1 mutant displayed obvious defects in meiotic chromosome behavior, which resulted in complete male sterility due to pollen abortion. The meiotic defects were visible in Osfignl1 male meiocytes starting from pachytene stage. We also noted that some chromosomes were trapped as single threads at the pachytene stage, suggesting that homolog juxtaposition and synapsis may be deficient. In the diakinesis stage, chromosome bridges between two chromosomes were observed in the PMCs of Osfignl1 plants, which might be associated with unresolved homologous/non-homologous intermediates during the first meiotic division. We also observed chromosome fragmentation and lagging chromosomes in Osfignl1 male gametocytes from late prophase I to meiosis II, which suggests that mutation of OsFIGNL1 causes broken chromosomes or incorrect segregation of homologous chromosomes.
Loss of OsFIGNL1 function in mice leads to an impaired progression of the germ cells through lengthening the first wave of pachytene stages, thus affecting meiotic cell cycle progression (L’Hote et al., 2011). The extension of the pachytene during the meiosis I in the Osfignl1 male meiocytes is consistent with the role of FIGNL1 in the regulation of male meiosis in mice (L’Hote et al., 2011). However, in Arabidopsis, different mutation sites of the FIGNL1 gene allow formation of bivalents without affecting meiotic cell cycle progression (Girard et al., 2015), which is inconsistent with the observation that Osfignl1 stayed longer at the pachytene stage than those in wild-type meiocytes. A reason for abnormal progression of the meiotic cell cycle in Osfignl1 male meiocytes may be that our mutagenesis of the rice OsFIGNL1 gene created a premature stop at the 187th amino acid residue, leading to a severe alteration of the protein structure, which differs from previously studied mutations of Arabidopsis FIGNL1. In Arabidopsis, loss of function of the synaptonemal complex protein ZYP1 significantly delays prophase I progression, indicating the existence of surveillance mechanisms that monitor progression through prophase I (Higgins et al., 2005). It is possible that prolonged arrest in the pachytene stage is the result of the existence of a recombination checkpoint in Osfignl1 mutant. The pachytene checkpoint was found to be activated in mouse (Wang et al., 2011), C. elegans (Gartner et al., 2000) and Drosophila (Ghabrial and Schupbach, 1999). However, meiotic mutants that invoke the pachytene checkpoint control mechanisms were less studied in plants. The observation that male meiocytes in Osfignl1 can proceed through late prophase I to the end of meiosis II after a brief arrest. Taken together, these results suggest that OsFIGNL1 participates in the regulation of meiotic cell cycle progression and plays an important role in recombination events during male meiosis in rice.
Conclusion
This report presents a functional characterization of OsFIGNL1 in rice. Our results show that OsFIGNL1 encodes a conserved AAA-ATPase domain and participates in control of microspore and anther development in rice. Our results provide evidence that disruption of OsFIGNL1 function leads to extension of meiotic cell cycle progression and abnormalities in chromosome behavior during male meiosis, thus resulting in pollen abortion. This work provides novel insight into the function of OsFIGNL1 in rice meiosis.
Author Contributions
Study conception and design: PZ, YZ, LC, and SC; acquisition of data: PZ, LS, SS, and WW; analysis and interpretation of data: PZ, YZ, ZL, DX, and PY; drafting of manuscript: PZ, YZ, ZY, BS, KW, and LC; critical revision: PZ, YZ, SC, and LC. All authors have read and approved to submit it to your journal.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We would like to thank Prof. Jian Zhang (China National Rice Research Institute) for critical proof reading, feedback, and editing of the manuscript. This work was supported by grants from the National Key Transform Program (#2016ZX08001-002), the National Natural Science Foundation of China (#31501290), the Zhejiang Provincial Natural Science Foundation of China (Grant #LQ14C130003), and the Super Rice Breeding Innovation Team and Rice Heterosis Mechanism Research Innovation Team of the Chinese Academy of Agricultural Sciences Innovation Project (CAAS-ASTIP-2013-CNRRI).
Footnotes
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2017.01639/full#supplementary-material
References
- Baumann P., Benson F. E., West S. C. (1996). Human Rad51 protein promotes ATP-dependent homologous pairing and strand transfer reactions in vitro. Cell 87 757–766. 10.1016/S0092-8674(00)81394-X [DOI] [PubMed] [Google Scholar]
- Baumann P., West S. C. (1998). Role of the human RAD51 protein in homologous recombination and double-stranded break repair. Trends Biochem. Sci. 23 247–251. 10.1016/S0968-0004(98)01232-8 [DOI] [PubMed] [Google Scholar]
- Bleuyard J. Y., White C. I. (2004). The Arabidopsis homologue of Xrcc3 plays an essential role in meiosis. EMBO J. 23 439–449. 10.1038/sj.emboj.7600055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bugreev D. V., Huang F., Mazina O. M., Pezza R. J., Voloshin O. N., Camerini-Otero R. D., et al. (2014). HOP2-MND1 modulates RAD51 binding to nucleotides and DNA. Nat. Commun. 5:4198 10.1038/ncomms5198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cloud V., Chan Y. L., Grubb J., Budke B., Bishop D. K. (2012). Rad51 is an accessory factor for Dmc1-mediated joint molecule formation during meiosis. Science 337 1222–1225. 10.1126/science.1219379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couteau F., Belzile F., Horlow C., Grandjean O., Vezon D., Doutriaux M. P. (1999). Random chromosome segregation without meiotic arrest in both male and female meiocytes of a dmc1 mutant of Arabidopsis. Plant Cell 11 1623–1634. 10.1105/tpc.11.9.1623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox G. A. (2000). The mouse fidgetin gene defines a new role for AAA family proteins in mammalian development. Nat. Genet. 26 383–383. 10.1038/79923 [DOI] [PubMed] [Google Scholar]
- Da Ines O., Degroote F., Goubely C., Amiard S., Gallego M. E., White C. I. (2013). Meiotic recombination in Arabidopsis is catalysed by DMC1, with RAD51 playing a supporting role. PLOS Genet. 9:e1003787 10.1371/journal.pgen.1003787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dray E., Siaud N., Dubois E., Doutriaux M. P. (2006). Interaction between Arabidopsis Brca2 and its partners Rad51, Dmc1, and Dss1. Plant Physiol. 140 1059–1069. 10.1104/pp.105.075838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edlinger B., Schlogelhofer P. (2011). Have a break: determinants of meiotic DNA double strand break (DSB) formation and processing in plants. J. Exp. Bot. 62 1545–1563. 10.1093/jxb/erq421 [DOI] [PubMed] [Google Scholar]
- Filippo J. S., Sung P., Klein H. (2008). Mechanism of eukaryotic homologous recombination. Annu. Rev. Biochem. 77 229–257. 10.1146/annurev.biochern.77.061306.125255 [DOI] [PubMed] [Google Scholar]
- Frickey T., Lupas A. N. (2004). Phylogenetic analysis of AAA proteins. J. Struct. Biol. 146 2–10. 10.1016/j.jsb.2003.11.020 [DOI] [PubMed] [Google Scholar]
- Gartner A., Milstein S., Ahmed S., Hodgkin J., Hengartner M. O. (2000). A conserved checkpoint pathway mediates DNA damage-induced apoptosis and cell cycle arrest in C. elegans. Mol. Cell 5 435–443. 10.1016/S1097-2765(00)80438-4 [DOI] [PubMed] [Google Scholar]
- Ghabrial A., Schupbach T. (1999). Activation of a meiotic checkpoint regulates translation of Gurken during Drosophila oogenesis. Nat. Cell. Biol. 1 354–357. 10.1038/14046 [DOI] [PubMed] [Google Scholar]
- Girard C., Chelysheva L., Choinard S., Froger N., Macaisne N., Lehmemdi A., et al. (2015). AAA-ATPase FIDGETIN-LIKE 1 and helicase FANCM antagonize meiotic crossovers by distinct mechanisms. PLOS Genet. 11:e1005369 10.1371/journal.pgen.1005369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins J. D., Sanchez-Moran E., Armstrong S. J., Jones G. H., Franklin F. C. H. (2005). The Arabidopsis synaptonemal complex protein ZYP1 is required for chromosome synapsis and normal fidelity of crossing over. Gene Dev. 19 2488–2500. 10.1101/gad.354705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji J. H., Tang D., Wang M., Li Y. F., Zhang L., Wang K. J., et al. (2013). MRE11 is required for homologous synapsis and DSB processing in rice meiosis. Chromosoma 122 363–376. 10.1007/s00412-013-0421-1 [DOI] [PubMed] [Google Scholar]
- Keeney S., Giroux C. N., Kleckner N. (1997). Meiosis-specific DNA double-strand breaks are catalyzed by Spo11, a member of a widely conserved protein family. Cell 88 375–384. 10.1016/S0092-8674(00)81876-0 [DOI] [PubMed] [Google Scholar]
- Kou Y. J., Chang Y. X., Li X. H., Xiao J. H., Wang S. P. (2012). The rice RAD51C gene is required for the meiosis of both female and male gametocytes and the DNA repair of somatic cells. J. Exp. Bot. 63 5323–5335. 10.1093/jxb/ers190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogh B. O., Symington L. S. (2004). Recombination proteins in yeast. Annu. Rev. Genet. 38 233–271. 10.1146/annurev.genet.38.072902.091500 [DOI] [PubMed] [Google Scholar]
- Kurzbauer M. T., Uanschou C., Chen D., Schlogelhofer P. (2012). The recombinases DMC1 and RAD51 are functionally and spatially separated during meiosis in Arabidopsis. Plant Cell 24 2058–2070. 10.1105/tpc.112.098459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- L’Hote D., Vatin M., Auer J., Castille J., Passet B., Montagutelli X., et al. (2011). Fidgetin-Like1 is a strong candidate for a dynamic impairment of male meiosis leading to reduced testis weight in mice. PLOS ONE 6:e27582 10.1371/journal.pone.0027582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Pinot F., Sauveplane V., Werck-Reichhart D., Diehl P., Schreiber L., et al. (2010). Cytochrome P450 family member CYP704B2 catalyzes the ω-hydroxylation of fatty acids and is required for anther cutin biosynthesis and pollen exine formation in rice. Plant Cell 22 173–190. 10.1105/tpc.109.070326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M. R., Li X. X., Zhou Z. J., Wu P. Z., Fang M. C., Pan X. P., et al. (2016). Reassessment of the four yield-related genes Gn1a, DEP1, GS3, and IPA1 in rice using a CRISPR/Cas9 system. Front. Plant Sci. 7:377 10.3389/Fpls.2016.00377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li N., Zhang D. S., Liu H. S., Yin C. S., Li X. X., Liang W. Q., et al. (2006). The rice tapetum degeneration retardation gene is required for tapetum degradation and anther development. Plant Cell 18 2999–3014. 10.1105/tpc.106.044107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W. X., Chen C. B., Markmann-Mulisch U., Timofejeva L., Schmelzer E., Ma H., et al. (2004). The Arabidopsis AtRAD51 gene is dispensable for vegetative development but required for meiosis. Proc. Natl. Acad. Sci. U.S.A. 101 10596–10601. 10.1073/pnas.0404110101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W. X., Ma H. (2006). Double-stranded DNA breaks and gene functions in recombination and meiosis. Cell Res. 16 402–412. 10.1038/sj.cr.7310052 [DOI] [PubMed] [Google Scholar]
- Li W. X., Yang X. H., Lin Z. G., Timofejeva L., Xiao R., Makaroff C. A., et al. (2005). The AtRAD51C gene is required for normal meiotic chromosome synapsis and double-stranded break repair in Arabidopsis. Plant Physiol. 138 965–976. 10.1104/pp.104.058347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin J. Z., Zhou B., Yang Y. Z., Mei J., Zhao X. Y., Guo X. H., et al. (2009). Piercing and vacuum infiltration of the mature embryo: a simplified method for Agrobacterium-mediated transformation of indica rice. Plant Cell Rep. 28 1065–1074. 10.1007/s00299-009-0706-2 [DOI] [PubMed] [Google Scholar]
- Luke-Glaser S., Pintard L., Tyers M., Peter M. (2007). The AAA-ATPase FIGL-1 controls mitotic progression, and its levels are regulated by the CUL-3MEL-26 E3 ligase in the C. elegans germ line. J. Cell Sci. 120 3179–3187. 10.1242/jcs.015883 [DOI] [PubMed] [Google Scholar]
- Luo Q., Tang D., Wang M., Luo W., Zhang L., Qin B., et al. (2013). The role of OsMSH5 in crossover formation during rice meiosis. Mol. Plant 6 729–742. 10.1093/mp/sss145 [DOI] [PubMed] [Google Scholar]
- McCormick S. (2004). Control of male gametophyte development. Plant Cell 16 S142–S153. 10.1105/tpc.016659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milman N., Higuchi E., Smith G. R. (2009). Meiotic DNA Double-Strand Break repair requires two nucleases, MRN and Ctp1, to produce a single size class of Rec12 (Spo11)-oligonucleotide complexes. Mol. Cell. Biol. 29 5998–6005. 10.1128/Mcb.01127-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morozumi Y., Ino R., Ikawa S., Mimida N., Shimizu T., Toki S., et al. (2013). Homologous pairing activities of two rice RAD51 proteins, RAD51A1 and RAD51A2. PLOS ONE 8:e75451 10.1371/journal.pone.0075451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray M. G., Thompson W. F. (1980). Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res. 8 4321–4325. 10.1093/nar/8.19.4321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neale M. J., Pan J., Keeney S. (2005). Endonucleolytic processing of covalent protein-linked DNA double-strand breaks. Nature 436 1053–1057. 10.1038/nature03872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nonomura K. L., Nakano M., Fukuda T., Eiguchi M., Miyao A., Hirochika H., et al. (2004). The novel gene HOMOLOGOUS PAIRING ABERRATION IN RICE MEIOSIS1 of rice encodes a putative coiled–coil protein required for homologous chromosome pairing in meiosis. Plant Cell 16 1008–1020. 10.1105/tpc.020701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onitake A., Yamanaka K., Esaki M., Ogura T. (2012). Caenorhabditis elegans fidgetin homolog FIGL-1 a nuclear-localized AAA ATPase, binds to SUMO. J. Struct. Biol. 179 143–151. 10.1016/j.jsb.2012.04.022 [DOI] [PubMed] [Google Scholar]
- Peng W. T., Lin Z. J., Li W. R., Lu J., Shen Y. Q., Wang C. G. (2013). Structural insights into the unusually strong ATPase activity of the AAA domain of the Caenorhabditis elegans Fidgetin-like 1 (FIGL-1) Protein. J. Struct. Biol. 288 29305–29312. 10.1074/jbc.M113.502559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pezza R. J., Camerini-Otero R. D., Bianco P. R. (2010). Hop2-Mnd1 condenses DNA to stimulate the synapsis phase of DNA strand exchange. Biophys. J. 99 3763–3772. 10.1016/j.bpj.2010.10.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pradillo M., Lopez E., Linacero R., Romero C., Cunado N., Sanchez-Moran E., et al. (2012). Together yes, but not coupled: new insights into the roles of RAD51 and DMC1 in plant meiotic recombination. Plant J. 69 921–933. 10.1111/j.1365-313X.2011.04845.x [DOI] [PubMed] [Google Scholar]
- Pradillo M., Varas J., Oliver C., Santos J. L. (2014). On the role of AtDMC1, AtRAD51 and its paralogs during Arabidopsis meiosis. Front. Plant Sci. 5:23 10.3389/Fpls.2014.00023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajanikant C., Melzer M., Rao B. J., Sainis J. K. (2008). Homologous recombination properties of OsRad51, a recombinase from rice. Plant Mol. Biol. 68 479–491. 10.1007/s11103-008-9385-6 [DOI] [PubMed] [Google Scholar]
- Sakane I., Kamataki C., Takizawa Y., Nakashima M., Toki S., Ichikawa H., et al. (2008). Filament formation and robust strand exchange activities of the rice DMC1A and DMC1B proteins. Nucleic Acids Res. 36 4266–4276. 10.1093/nar/gkn405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schommer C., Beven A., Lawrenson T., Shaw P., Sablowski R. (2003). AHP2 is required for bivalent formation and for segregation of homologous chromosomes in Arabidopsis meiosis. Plant J. 36 1–11. 10.1046/j.1365-313X.2003.01850.x [DOI] [PubMed] [Google Scholar]
- Seeliger K., Dukowic-Schulze S., Wurz-Wildersinn R., Pacher M., Puchta H. (2012). BRCA2 is a mediator of RAD51- and DMC1-facilitated homologous recombination in Arabidopsis thaliana. New Phytol. 93 364–375. 10.1111/j.1469-8137.2011.03947.x [DOI] [PubMed] [Google Scholar]
- Shen Y., Tang D., Wang K. J., Wang M., Huang J., Luo W. X., et al. (2012). ZIP4 in homologous chromosome synapsis and crossover formation in rice meiosis. J. Cell Sci. 125 2581–2591. 10.1242/jcs.090993 [DOI] [PubMed] [Google Scholar]
- Shen Y. J., Jiang H., Jin J. P., Zhang Z. B., Xi B., He Y. Y., et al. (2004). Development of genome-wide DNA polymorphism database for map-based cloning of rice genes. Plant Physiol. 135 1198–1205. 10.1104/pp.103.038463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheridan S. D., Yu X., Roth R., Heuser J. E., Sehorn M. G., Sung P., et al. (2008). A comparative analysis of Dmc1 and Rad51 nucleoprotein filaments. Nucleic Acids Res. 36 4057–4066. 10.1093/nar/gkn352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinohara A., Ogawa H., Ogawa T. (1992). Rad51 protein involved in repair and recombination in S. cerevisiae is a RecA-like protein. Cell 69 457–470. 10.1016/0092-8674(92)90447-K [DOI] [PubMed] [Google Scholar]
- Tamura K., Stecher G., Peterson D., Filipski A., Kumar S. (2013). MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30 2725–2729. 10.1093/molbev/mst197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang D., Miao C. B., Li Y. F., Wang H. J., Liu X. F., Yu H. X., et al. (2014). OsRAD51C is essential for double-strand break repair in rice meiosis. Front. Plant Sci. 5:167 10.3389/Fpls.2014.00167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao J., Zhang L., Chong K., Wang T. (2007). OsRAD21-3, an orthologue of yeast RAD21, is required for pollen development in Oryza sativa. Plant J. 51 919–930. 10.1111/j.1365-313X.2007.03190.x [DOI] [PubMed] [Google Scholar]
- Uanschou C., Ronceret A., Von Harder M., De Muyt A., Vezon D., Pereira L., et al. (2013). Sufficient amounts of functional HOP2/MND1 complex promote interhomolog DNA repair but are dispensable for intersister DNA repair during meiosis in Arabidopsis. Plant Cell 25 4924–4940. 10.1105/tpc.113.118521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vajjhala P. R., Wong J. S., To H. Y., Munn A. L. (2006). The beta domain is required for Vps4p oligomerization into a functionally active ATPase. FEBS J. 273 2357–2373. 10.1111/j.1742-4658.2006.05238.x [DOI] [PubMed] [Google Scholar]
- Waadt R., Schmidt L. K., Lohse M., Hashimoto K., Bock R., Kudla J. (2008). Multicolor bimolecular fluorescence complementation reveals simultaneous formation of alternative CBL/CIPK complexes in planta. Plant J. 56 505–516. 10.1111/j.1365-313X.2008.03612.x [DOI] [PubMed] [Google Scholar]
- Wang C. L., Wang Y., Cheng Z. J., Zhao Z. G., Chen J., Sheng P. K., et al. (2016). The role of OsMSH4 in male and female gamete development in rice meiosis. J. Exp. Bot. 67 1447–1459. 10.1093/jxb/erv540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H. J., Hu Q., Tang D., Liu X. F., Du G. J., Shen Y., et al. (2016). OsDMC1 is not required for homologous pairing in rice meiosis. Plant Physiol. 171 230–241. 10.1104/pp.16.00167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Chang C. Y., Wu J. F., Tung K. S. (2011). Nuclear localization of the meiosis-specific transcription factor Ndt80 is regulated by the pachytene checkpoint. Mol. Biol. Cell 22 1878–1886. 10.1091/mbc.E10-12-1011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu W., Zheng X. M., Lu G., Zhong Z., Gao H., Chen L., et al. (2013). Association of functional nucleotide polymorphisms at DTH2 with the northward expansion of rice cultivation in Asia. Proc. Natl. Acad. Sci. U.S.A. 110 2775–2780. 10.1073/pnas.1213962110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z., Ji J., Tang D., Wang H., Shen Y., Shi W., et al. (2015). OsSDS is essential for DSB formation in rice meiosis. Front. Plant Sci. 6:21 10.3389/fpls.2015.00021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yakushiji Y., Yamanaka K., Ogura T. (2004). Identification of a cysteine residue important for the ATPase activity of C. elegans fidgetin homologue. FEBS Lett. 578 191–197. 10.1016/j.febslet.2004.11.009 [DOI] [PubMed] [Google Scholar]
- Yang Y., Mahaffey C. L., Berube N., Nystuen A., Frankel W. N. (2005). Functional characterization of fidgetin, an AAA-family protein mutated in fidget mice. Exp. Cell Res. 304 50–58. 10.1016/j.yexcr.2004.11.014 [DOI] [PubMed] [Google Scholar]
- Yoo S. D., Cho Y. H., Sheen J. (2007). Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat. Protoc. 2 1565–1572. 10.1038/nprot.2007.199 [DOI] [PubMed] [Google Scholar]
- Yuan J. S., Chen J. J. (2013). FIGNL1-containing protein complex is required for efficient homologous recombination repair. Proc. Natl. Acad. Sci. U.S.A. 110 10640–10645. 10.1073/pnas.1220662110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B. W., Wang M., Tang D., Li Y. F., Xu M., Gu M. H., et al. (2015). XRCC3 is essential for proper double-strand break repair and homologous recombination in rice meiosis. J. Exp. Bot. 66 5713–5725. 10.1093/jxb/erv253 [DOI] [PubMed] [Google Scholar]
- Zhang D. B., Luo X., Zhu L. (2011). Cytological analysis and genetic control of rice anther development. J. Genet. Genomics 38 379–390. 10.1016/j.jgg.2011.08.001 [DOI] [PubMed] [Google Scholar]
- Zhang D. S., Liang W. Q., Yin C. S., Zong J., Gu F. W., Zhang D. B. (2010). OsC6, encoding a lipid transfer protein, is required for postmeiotic anther development in rice. Plant Physiol. 154 149–162. 10.1104/pp.110.158865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X., Jin M., Wang M., Sun L., Hong X., Cao Y., et al. (2016). Fidgetin-like 1 is a ciliogenesis-inhibitory centrosome protein. Cell Cycle 15 2367–2375. 10.1080/15384101.2016.1204059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou S. R., Wang Y., Li W. C., Zhao Z. G., Ren Y. L., Wang Y., et al. (2011). Pollen Semi-Sterility1 encodes a Kinesin-1-Like protein important for male meiosis, anther dehiscence, and fertility in rice. Plant Cell 23 111–129. 10.1105/tpc.109.073692 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.