Abstract
Acute myeloid leukemia (AML) and the myelodysplastic syndromes (MDS) are initiated and sustained by self-renewing malignant stem cells; thus, eradication of AML and MDS stem cells is required for cure. We identified CD99 as a cell surface protein frequently overexpressed on AML and MDS stem cells. Expression of CD99 allows for prospective separation of leukemic stem cells (LSCs) from functionally normal hematopoietic stem cells (HSCs) in AML, and high CD99 expression on AML blasts enriches for functional LSCs as demonstrated by limiting dilution xenotransplant studies. Monoclonal antibodies (mAbs) targeting CD99 induce the death of AML and MDS cells in a SRC-family kinase dependent manner in the absence of immune effector cells or complement, and administration of anti-CD99 mAbs exhibit anti-leukemic activity in AML xenografts. These data establish CD99 as a novel marker of AML and MDS stem cells, as well as a promising therapeutic target in these disorders.
Introduction
Acute myeloid leukemia (AML) and the myelodysplastic syndromes (MDS) are disorders arising in the hematopoietic system that share common molecular origins, with MDS patients showing a propensity to progress to AML. AML arises from immature hematopoietic cells and is composed of leukemic blasts organized in a developmental hierarchy reminiscent of normal hematopoiesis. At the apex of this hierarchy are leukemic stem cells (LSCs) that possess the capacity to self-renew and differentiate into non-self renewing progeny that comprise the vast majority of leukemic blasts(1, 2). LSCs appear to be largely resistant to conventional chemotherapy and are thought to serve as the reservoir of minimal residual disease (MRD) that is responsible for disease relapse following initial treatment(3). Xenografts and mouse models of AML suggest that fully transformed LSCs arise not from hematopoietic stem cells (HSCs), but rather more committed myeloid progenitors that acquire aberrant self-renewal(4–6). In contrast, MDS is initiated by neoplastic HSCs that fail to give rise to sufficient numbers of mature hematopoietic cells, leading to bone marrow failure(7–9). True to their identity as disease stem cells, when transplanted into immunodeficient animals MDS HSCs give rise to long-term grafts that recapitulate features of MDS(7, 10). Similar to AML LSCs, MDS HSCs are also highly resistant to standard therapies(8, 11). Thus, despite the likely differing cellular origins of AML and MDS, curative therapies for these malignancies must eliminate disease stem cells (LSCs or MDS HSCs, respectively), as they are likely the only self-renewing disease cells in the bone marrow (BM).
In efforts to identify potential therapeutic targets in AML, a number of groups have identified cell surface proteins preferentially expressed on AML LSCs compared to normal HSCs, including CD47(12), CD44(13), CD96(14), TIM3(15, 16), and CD123(17). While these antigens are present on LSCs, their relative levels of expression among AML cells have not been shown to enrich for functional LSCs. In the context of MDS, aberrant HSC cell surface protein expression has not been carefully characterized, and therefore no therapies targeting MDS stem cells based on differentially expressed cell surface antigens have been described or evaluated.
Here, we demonstrate that CD99 is expressed at increased levels at a high frequency on immunophenotypic AML LSCs and MDS HSCs compared to their normal hematopoietic counterparts, hematopoietic stem and progenitor cells (HSPCs). Furthermore, high CD99 expression can be used to identify and prospectively separate leukemic cells from non-leukemic cells in AML patient BM specimens, as well as to selectively enrich for functional LSC activity, as demonstrated by xenotransplantation assays. Ligation of AML and MDS stem cells with a monoclonal antibody (mAb) recognizing CD99 directly induces cytotoxicity in vitro. Moreover, anti-CD99 mAbs exhibit anti-leukemic activity in AML xenografts, demonstrating the potential of anti-CD99 mAbs as therapeutic agents.
Results
MDS HSCs and AML blasts frequently express high levels of CD99
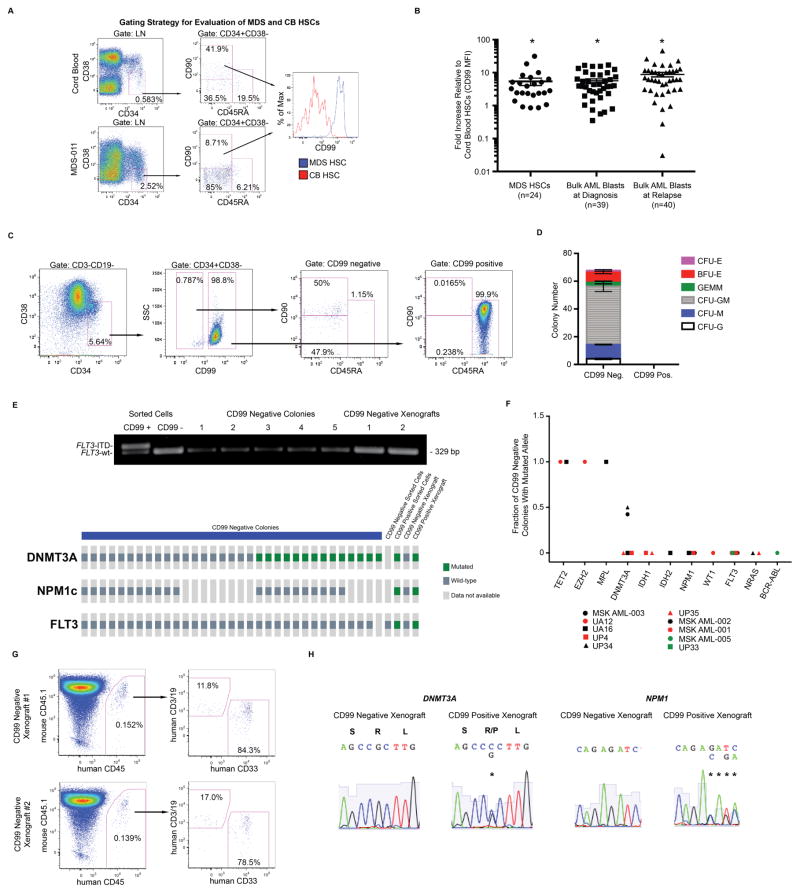
To identify candidate cell surface proteins differentially expressed on MDS HSCs, we evaluated the transcriptomes of purified HSCs (lineage negative [LN] CD38+CD34−CD90+CD45RA−) from MDS patients and age-matched controls(18), identifying 25 dysregulated transcripts encoding cell surface antigens. We used flow cytometry (FC) to validate cell surface expression of these antigens on MDS HSCs from 24 MDS patient specimens (Supp. Table 1) as compared with cord blood (CB) HSC controls, identifying CD99 as the most frequently overexpressed among the antigens tested (83% of cases as defined by a >50% increase in mean fluorescence intensity [MFI], mean 6.35-fold increase, Fig. 1A–B). Normal adult BM HSCs expressed similar levels of cell surface CD99 as compared with CB HSCs (Supp. Fig. 1). We previously demonstrated that CD99 transcripts are more highly expressed in LSC-enriched CD34+CD38− AML cells compared with normal bone marrow CD34+CD38− cells(14), suggesting that CD99 protein is also upregulated on AML LSCs. Comparison of cell surface CD99 expression on unfractionated bulk leukemic blasts (CD45[low]SSC[low], representative gating shown in Supp. Fig. 2) from 79 paired diagnosis/relapse AML samples (Supp. Table 2) with expression on normal CB HSCs by FC confirmed elevated levels of CD99 in 82% of diagnostic samples and 90% of relapse samples (average 8.34-fold increase, Fig. 1B).
Fig. 1. Myelodysplastic syndrome (MDS) and acute myeloid leukemia (AML) stem cells express high levels of CD99.
(A) Hematopoietic stem cells (HSCs; lineage negative [LN] CD34+CD38−CD90+CD45RA−) from patients with MDS (n=24) and normal cord blood (CB) controls (n=24) were analyzed by flow cytometry (FC) for CD99 expression, (B) shown as fold-change CD99 mean fluorescence intensity (MFI) on MDS HSCs compared with CB HSCs. Unfractionated blasts (CD45[low]SSC[low]) from diagnostic (n=39) or relapse (n=40) AML specimens and normal CB HSCs (n=18) were analyzed by FC for CD99 expression, shown as fold-change CD99 MFI on AML blasts compared with CB HSCs. Error bars represent ±SEM. *P<0.0001 (paired t-test). (C) CD99 expression was evaluated by FC on CD3-CD19-CD34+CD38− cells from AML specimen MSK AML-003. By immunophenotype, CD99 negative cells include HSCs and multipotent progenitors (MPPs; LN CD34+CD38−CD90−CD45RA−), while CD99 positive cells are almost entirely composed of lymphoid-primed multipotent progenitor-like cells (LMPP-like; LN CD34+CD38−CD90−CD45RA+). (D) When these populations were plated in methylcellulose (750 cells in triplicate), normal myeloid/erythroid colonies formed only from the CD99 negative fraction. Error bars represent ±SEM of triplicates. (E) All CD99 negative derived methylcellulose colonies lacked the heterozygous FLT3-ITD and NPM1 abnormalities present in MSK AML-003, while 14 out of 33 sequenced colonies harbored a heterozygous DNMT3A R882P abnormality. CD99 negative sorted cells and xenografts lacked DNMT3A, NPM1, and FLT3 mutations, while all three abnormalities were present in CD99 positive sorted cells and xenografts. (F) Summary of the percentage of CD99 negative cell derived colonies from ten AML specimens that were positive for disease associated mutations in the indicated alleles (which were confirmed to be present in the bulk fraction of each corresponding AML). Each data point represents a summary of results for each AML harboring a particular mutation (shown in detail in Supp. Fig. 4). (G) CD99 negative cells (2,500 cells) were transplanted into sublethally irradiated NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ (NSG) mice (n=3), with mice demonstrating human lymphomyeloid engraftment in the BM 10 months after transplantation. Representative FACS-plots are shown from two CD99 low xenografts. (H) Sanger sequencing traces reveal the absence of DNMT3A R882P and NPM1 W288fs mutations in CD99 negative cell derived xenografts, and the presence of both of these abnormalities in CD99 positive cell derived xenografts.
CD99 distinguishes between leukemic and non-leukemic hematopoietic cells
To determine whether differential CD99 expression can be used to identify and prospectively isolate LSCs from pre-leukemic or residual normal HSCs(19), we measured CD99 expression in the LSC-enriched CD34+CD38− fraction of AML (Fig. 1C, Supp. Fig. 3A). Within this fraction, CD99 positive cells expressed an antigenic profile consistent with lymphoid-primed multipotent progenitors (LMPPs, CD34+CD38−CD90−CD45RA+), the immunophenotype most highly enriched for LSCs in the majority of human AML(5), while CD99 negative cells exhibited an antigenic profile consistent with a mixture of HSCs and multipotent progenitors (MPPs) (CD34+CD38−CD90+/−CD45RA−) present in proportions similar to those observed in normal hematopoiesis(20). CD34+CD38− CD99 negative cells from ten independent AML specimens were FACS-purified and grown in methylcellulose and demonstrated the robust myeloid colony formation characteristic of normal HSCs, but not observed with LSCs (Fig. 1D, Supp. Fig. 3B–C, Supp. Fig. 4)(21). These colonies lacked the full complement of molecular genetic abnormalities identified in the corresponding bulk AML blasts, consistent with their derivation from residual normal or pre-leukemic HSCs(19, 22, 23) (Fig. 1E–F, Supp. Fig. 3B, Supp. Fig. 4). Moreover, transplantation of FACS-purified CD99 negative cells into sublethally irradiated NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ (NSG) mice (n=3) led to lymphomyeloid human engraftment (Fig. 1G, Supp. Fig. 5C), with engrafted cells lacking all leukemia associated mutations (Fig. 1E, H), further confirming the utility of CD99 in separating LSCs from functionally normal pre-leukemic HSCs. In contrast, transplantation of FACS-purified CD99 positive cells into NSG mice (n=4) led to engraftment of a rapidly lethal myeloid leukemia (Supp. Fig. 5A–C). This capability to prospectively separate leukemic from non-leukemic hematopoiesis may eventually allow for effective purging of LSCs from autologous BM grafts that may be used clinically for therapeutic HSC transplantation.
CD99 enriches for functional leukemic stem cells
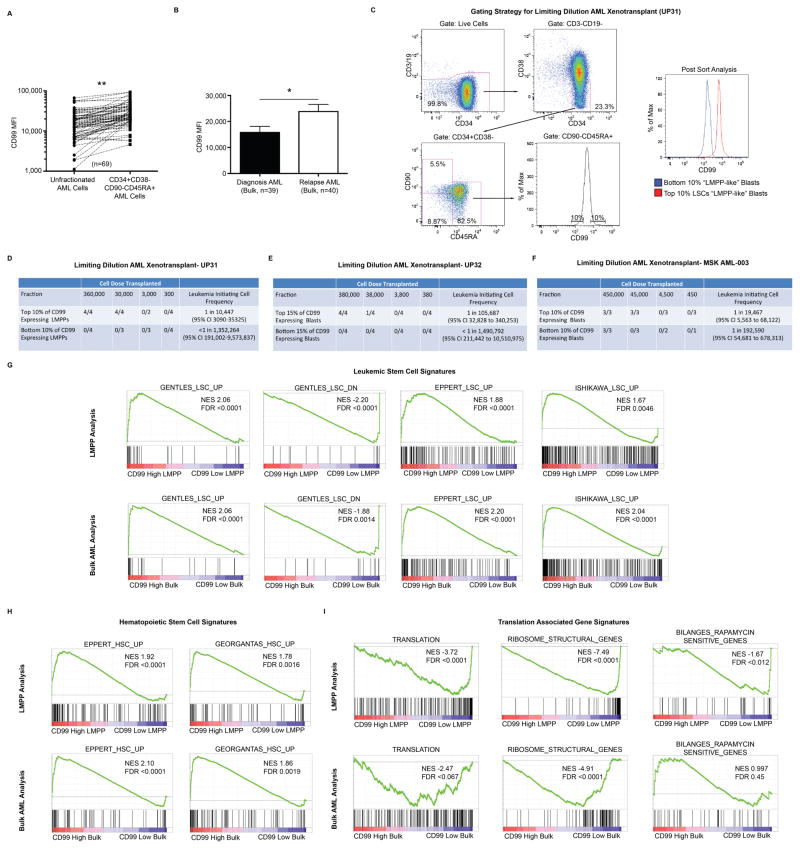
In AML specimens with an identifiable LMPP-like population (CD34+CD38−CD90−CD45RA+, n=69) within the CD99 positive leukemic fraction, this LSC-enriched population consistently expressed higher levels of CD99 compared with more differentiated blasts, demonstrating intra-leukemic heterogeneity of CD99 expression and suggesting that LSCs exhibit the highest levels of CD99 (Fig. 2A, Supp. Fig. 2). Consistent with the chemoresistant nature of LSCs, CD99 expression was significantly higher on leukemic blasts at relapse when compared directly with paired specimens from the time of diagnosis (Fig. 2B). To test whether CD99 enriches for LSC function among CD99 positive leukemic blasts, we transplanted the highest and lowest CD99 expressing leukemic cells (top and bottom 10%, respectively) within the LSC-enriched LMPP-like fraction of a CD34 positive primary AML specimen into sublethally irradiated NSG mice. Leukemia-initiating cell (L-IC) activity was only observed in mice transplanted with the top 10% of CD99 expressing LMPP-like cells, indicating that CD99 is not only highly expressed in AML, but is also expressed at the highest levels on functional LSCs (Fig. 2C–D). Similar results were obtained when transplanting the highest and lowest CD99 expressing leukemic cells (top and bottom 15%, respectively) from a CD34 negative AML (Fig. 2E, gating strategy shown in Supp. Fig. 6A–B), which represents a subset of AMLs for which a strategy to enrich for functionally defined LSCs has not been reported. To test whether CD99 can enrich for functional LSCs irrespective of CD34 and CD38 expression, we transplanted the highest and lowest CD99 expressing leukemic cells (top and bottom 10%, respectively) among bulk unfractionated leukemic blasts (CD45[low]SSC[low]) from an AML with variable CD34 and CD38 expression, demonstrating a 10-fold increase in L-IC activity in the CD99 high fraction (Fig. 2F, gating strategy shown in Supp. Fig. 6C–D). When the CD99 high and low populations from both CD34+ and CD34− AMLs were cultured in vitro in liquid culture supplemented with cytokines, CD99 high cells demonstrated improved viability compared with CD99 low cells (Supp. Fig. 7), a previously described characteristic of LSC-enriched blasts in AML(24, 25).
Fig. 2. CD99 expression enriches for functional leukemic stem cells (LSCs).
(A) In AML specimens with an identifiable LMPP-like (LN CD34+CD38−CD90−CD45RA+) LSC-enriched population (n=69), CD99 expression was higher in LMPP-like blasts compared with bulk unfractionated blasts. **P<0.0001 (paired t-test). (B) CD99 expression was higher on bulk unfractionated AML blasts from relapse samples (n=40) as compared with diagnostic samples (n=39). Error bars represent ±SEM. *P=0.01 (paired t-test). (C) Gating strategy for purifying LSC-enriched LMPP-like cells from AML specimen UP31. From this fraction, the highest and lowest CD99 expressing blasts (top and bottom 10%, respectively) were FACS-purified to >95% purity and transplanted at limiting dilution into sublethally irradiated NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ (NSG) mice. Sort gates were drawn based on normal HSC, MPP, and LMPP populations in CB as depicted in Fig. 1A. (D) Leukemic engraftment (defined as >0.1% detectable human cells in the bone marrow >12 weeks after transplantation) was only observed in mice transplanted with “top 10%” CD99 expressing LMPP-like blasts. (E) The highest and lowest CD99 expressing blasts (top and bottom 15%, respectively) from the CD34 negative AML specimen UP32 were FACS-purified to >95% purity and transplanted at limiting dilution into NSG mice. Leukemic engraftment was only observed in mice transplanted with “top 15%” CD99 expressing blasts. (F) The highest and lowest CD99 expressing blasts (top and bottom 10%, respectively) from the bulk (CD45[low] SSC[low] CD45RA+) fraction of AML specimen MSK AML-003 were FACS-purified to >95% purity and transplanted at limiting dilution into NSG mice. Leukemia-initiating cell activity was estimated to be 10-fold higher in the “top 10%” CD99 expressing blasts. (G–I) The top 10% and bottom 10% of CD99 expressing blasts within the LMPP-like fraction of eight independent primary AML specimens, as well as the bulk fraction from six of these AML specimens, were FACS-purified and RNA-sequencing was performed. Gene set enrichment analysis revealed in “top 10%” CD99 expressing blasts enrichment for LSC (27–29) and HSC gene signatures (28, 30), as well depletion of ribosomal gene transcripts (KEGG)(54) and gene signatures associated with translation (REACTOME)(55, 56).
RNA-sequencing of the top 10% and bottom 10% of CD99 expressing blasts within the LMPP-like fraction of seven primary AML specimens and the bulk fraction of a CD34− AML (post-sort data shown in Fig. 2C, Supp. Fig. 6, Supp. Fig. 8) revealed 1558 differentially expressed genes (p<0.05). Gene set enrichment analysis(26) revealed enrichment for LSC(27–29) and HSC(28, 30) gene signatures in CD99 high blasts (Fig. 2G–H). Furthermore, CD99 high blast associated transcriptomes were depleted for ribosomal gene transcripts and gene signatures associated with translation (Fig. 2I), consistent with recent reports that both normal and malignant hematopoietic stem cells exhibit highly regulated levels of translation(31, 32). Since functional LSCs have been demonstrated to exist in populations outside of the LMPP fraction of AML, we performed RNA-sequencing of the top 10% and bottom 10% of CD99 expressing cells among bulk leukemic blasts from six of the eight primary AMLs initially tested (excluding one AML that was entirely CD34− and one that was entirely CD34+, post-sort data shown in Supp. Fig. 9) and found enrichment for the same LSC and HSC gene signatures, as well as depletion of translation associated gene signatures (Fig. 2G–I).
In six of the specimens confirmed to harbor recurrent AML-associated somatic mutations, we were able to assess for the burden of mutant transcripts in the CD99 high and low fractions. In three of the six specimens, the variant transcript frequency (VTF) was higher in the CD99 high fractions (MSK AML-002 [particularly in the LMPP fraction], MSK AML-005, and MSK AML-006), while there was no difference in two specimens (MSK AML-003, UP31) and a decrease in VTF in one specimen (MSK AML-006) (Supp. Fig. 10). Thus, in a subset of AMLs, CD99 expression may correlate in part with clonal heterogeneity.
Collectively, these data provide functional and transcriptomal evidence that CD99 is preferentially expressed on LSCs, identifying CD99 as the first LSC specific cell surface marker that can be used to selectively enrich for LSC activity in AML. Other previously identified LSC markers do not appear likely to share this feature, as they are expressed at equal or decreased levels on LSC-enriched fractions compared to more differentiated blasts (Supp. Fig. 11)(12–15, 17). Thus, these data validate CD99 expression as a novel method for the purification of LSCs, which will allow for more refined studies of this biologically and clinically significant cell population.
Anti-CD99 mAbs are directly cytotoxic to AML and MDS cells
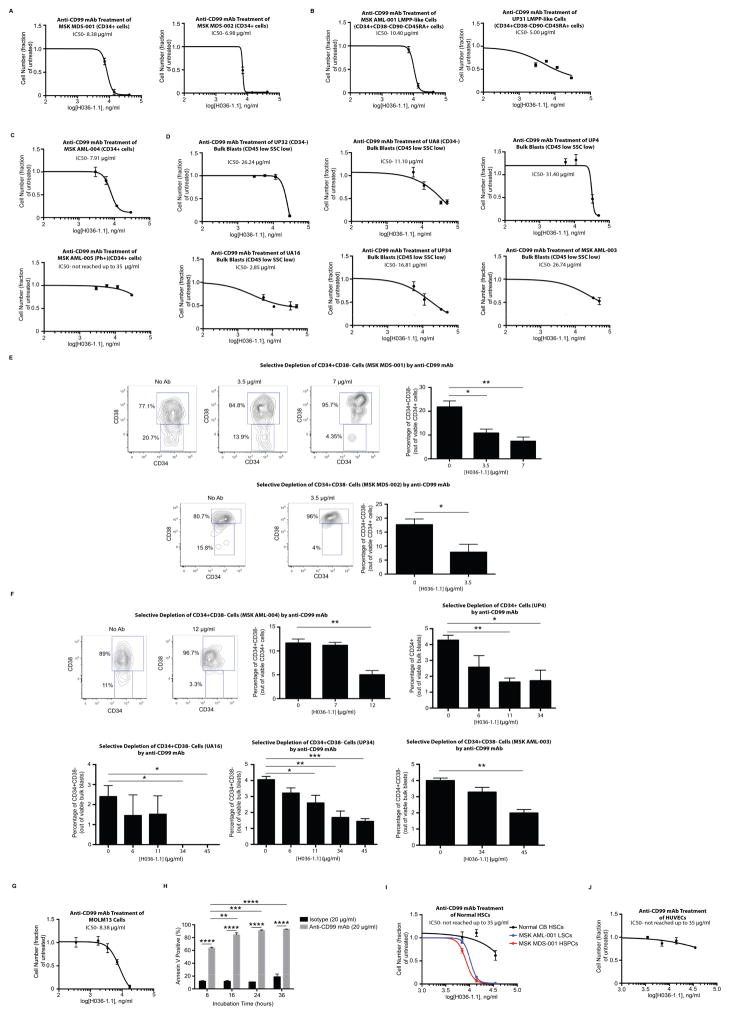
As therapeutic targeting of CD99 with mAbs has shown promise in other human cancers such as Ewing’s sarcoma(33), we tested the ability of anti-CD99 mAbs to induce cytotoxicity in AML and MDS stem/progenitor cells in vitro. The anti-CD99 mAb clone H036-1.1 was cytotoxic to purified primary MDS CD34+ cells (Fig. 3A) as well as LMPP-like AML blasts (Fig. 3B), AML CD34+ blasts (Fig. 3C), and bulk leukemic blasts (Fig. 3D). Of note, the only primary AML specimen we tested that was resistant to anti-CD99 mAb treatment harbored a BCR-ABL translocation (Fig. 3C). In addition, when treated with lower concentrations of anti-CD99 mAb, MDS HSPCs and AML blasts with at least partial CD34 expression exhibited selective depletion of the least mature populations (either CD34+CD38− or CD34+) (Fig. 3E–F, Supp. Fig. 12). Anti-CD99 mAbs were also cytotoxic to myeloid leukemia cell lines such as the AML-derived cell line MOLM13 (Fig. 3G). Anti-CD99 mAbs induced apoptosis as confirmed by annexin V and activated caspase 3 staining (Fig. 3H, Supp. Fig. 13) in the absence of immune effector cells or complement, consistent with a direct cytotoxic effect.
Fig. 3. An anti-CD99 monoclonal antibody (mAb) is cytotoxic to MDS and AML cells.
(A) Incubation of purified CD34+ cells from MSK MDS-001 and MDS-002 with anti-CD99 mAb (clone H036-1.1) for 48 hours led to a significant decrease in cell number. Similar results were obtained with (B) CD34+CD38−CD90−CD45RA+ LMPP-like cells from AML specimens MSK AML-001 and UP31, as well as (C) CD34+ cells from MSK AML-004. CD34+ cells from the BCR-ABL positive AML MSK AML-005 were incubated with anti-CD99 mAb (H036-1.1) for 48 hours. The IC50 was not reached using mAb concentrations up to 35 μg/ml. (D) Incubation of bulk blasts (CD45[low]SSC[low]) purified from UP32, UA8, UP4, UA16, UP34, and MSK AML-003 with anti-CD99 mAb (clone H036-1.1) for 48 hours led to a significant decrease in cell number. (E) At the end of the incubation of MSK MDS-001 and MSK MDS-002 with anti-CD99 mAb, CD34 and CD38 expression was measured on remaining viable cells, demonstrating selective depletion of CD34+CD38− cells. Representative FACS-plots are shown. *P<0.05, **P<0.01 (unpaired t-test). (F) At the end of incubation of MSK AML-004, UP4, UA16, UP34, and MSK AML-003 with anti-CD99 mAb, CD34 and CD38 expression was measured on remaining viable cells, demonstrating selective depletion of CD34+CD38− cells (or CD34+ cells in UP4). Representative FACS-plots are shown here and in Supp. Fig. 12. *P<0.05, **P<0.01 (unpaired t-test). (G) MOLM13 cells were incubated with anti-CD99 mAb (H036-1.1) for 48 hours, leading to a marked decrease in cell number. (H) Incubation of MOLM13 cells with anti-CD99 mAb (H036-1.1, 20 μg/ml) led to induction of apoptosis over the course of 36 hours. **P<0.01, ***P<0.001, ****P<0.0001 (unpaired t-test). (I) 700 HSCs (LN CD34+CD38−CD90+CD45RA−) purified from CB were incubated with anti-CD99 mAb (H036-1.1) for 48 hours. The IC50 was not reached using mAb concentrations up to 35 μg/ml. Juxtaposed for comparison is the sensitivity of MSK MDS-001 and MSK AML-001 to anti-CD99 mAb as shown in panels A and B. (J) Similar results were obtained when human umbilical vein endothelial cells (HUVECs) were incubated with anti-CD99 mAb (H036-1.1) for 48 hours. For panels A–J, error bars represent ±SEM of biological triplicates.
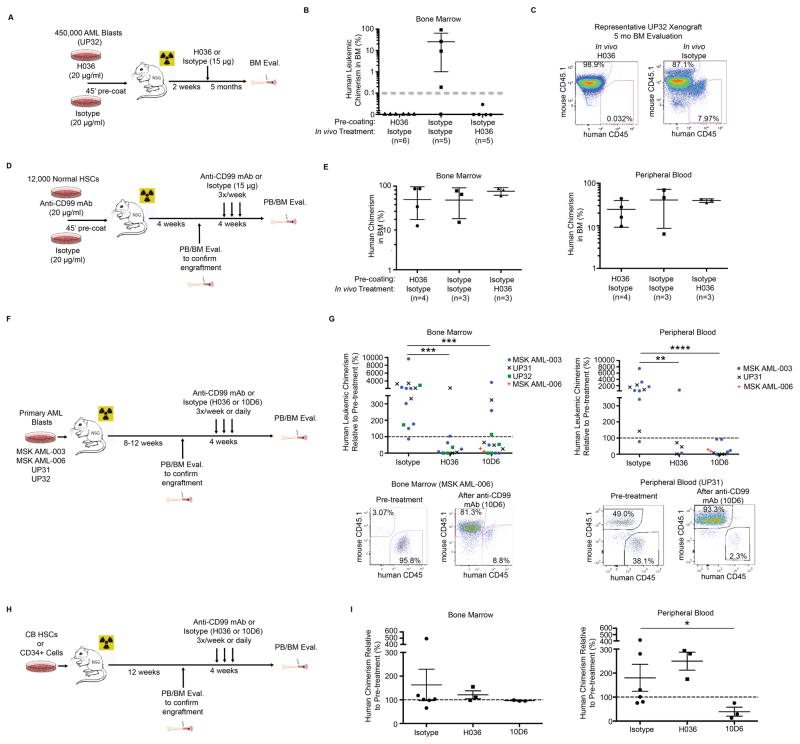
Since normal HSCs and endothelial cells express low and intermediate levels of CD99, respectively (Supp. Fig. 1), we tested whether cytotoxic anti-CD99 mAbs would affect their growth or survival. At concentrations toxic to AML and MDS cells, anti-CD99 mAbs had minimal effects on HSC growth in liquid culture supplemented with cytokines (Fig. 3I) and exerted no significant toxicity to human umbilical vein endothelial cells (HUVECs) (Fig. 3J). Ex vivo incubation of leukemic blasts from an AML patient previously shown to engraft NSG mice with anti-CD99 mAb (clone H036-1.1) for 45 minutes prior to xenotransplantation completely abolished their engraftment capability (Fig. 4A–C). To rule out a possible effect of anti-CD99 mAbs on LSC homing and to determine whether anti-CD99 mAbs can eradicate LSCs in vivo, NSG mice transplanted with leukemic blasts from the same AML specimen were allowed to engraft for two weeks prior to treatment with 15 μg of anti-CD99 mAb (clone H036-1.1) or isotype control (Fig. 4A). One treatment with anti-CD99 mAb was sufficient to completely abolish AML engraftment as measured in the BM at five months post-transplantation (Fig. 4C, Supp. Fig. 14). When followed out to 11 months, none of the mice transplanted with AML cells treated with anti-CD99 mAb ex vivo or in vivo demonstrated engraftment. Conversely, treatment of normal CB HSCs with anti-CD99 mAb (H036-1.1) ex vivo or in vivo in the same manner did not have any significant effect on engraftment in the BM or PB (Fig. 4D–E). The eradication of LSCs in vivo by anti-CD99 mAbs is likely due to the direct cytotoxic effect of the antibody, since IgM antibodies such as H036-1.1 have minimal capability to induce antibody-dependent cellular cytotoxicity (ADCC)(34), and NSG mice are deficient in hemolytic complement(35).
Fig. 4.
(A) Schematic for combined ex vivo and in vivo anti-CD99 mAb (H036-1.1) treatment of AML specimen UP32. (B) Summary of engraftment in UP32 xenografts five months after transplantation (>0.1% threshold for human engraftment demarcated with dotted gray line). Error bars represent ±SD. (C) Representative FACS plots of human engraftment levels in anti-CD99 mAb and isotype control treated animals. (D) Schematic for combined ex vivo and in vivo anti-CD99 mAb (H036-1.1) treatment of normal CB HSCs (LN CD34+CD38−CD90+CD45RA−). (E) Summary of engraftment based on assessment of human CD45+ cells two months after transplantation and one month after antibody treatment. (F) Schematic for engraftment of primary AML specimens and in vivo treatment with anti-CD99 mAbs (H036-1.1 or 10D6). (G) Human leukemic chimerism in xenografts relative to pre-treatment chimerism after four weeks of antibody treatment with the indicated anti-CD99 mAb clone or isotype. **P<0.01, ***P<0.001, ****P<0.0001 (Mann-Whitney U test). Representative FACS plots of human engraftment levels in the BM and PB before and after anti-CD99 mAb treatment. (H) Schematic for in vivo treatment of mice engrafted with normal CB HSCs with anti-CD99 mAb (H036-1.1 or 10D6). (I) Summary of CB HSC derived human engraftment in the BM and PB after four weeks of antibody treatment with the indicated anti-CD99 mAb clone or isotype. *P<0.05 (Mann-Whitney U test).
To determine whether anti-CD99 mAbs have anti-leukemic activity against established AML, we transplanted NSG mice with four independent AML specimens and confirmed human leukemic engraftment in both the BM and PB (with the exception of specimen UP32, which demonstrated detectable engraftment in the BM, but not PB) (Fig. 4F). Engrafted mice were treated for four weeks with the anti-CD99 mAb H036-1.1 (15 μg/ml three times weekly) or isotype control. Additionally, we generated a novel anti-CD99 mAb (clone 10D6, IgG1κ isotype) that demonstrates direct cytotoxicity to AML cells (Supp. Fig. 15), and we also treated engrafted mice with this mAb for four weeks (40 μg daily). Treatment with either anti-CD99 mAb (clone H036-1.1 or 10D6) led to a significant reduction in leukemic burden in both the BM and PB (Fig. 4G). In contrast, treatment of NSG mice engrafted with normal CB HSCs with anti-CD99 mAbs (H036-1.1 or 10D6) had minimal effects on engraftment, with the exception of a modest reduction in PB chimerism with clone 10D6 (Fig. 4H–I). Together, these results indicate that anti-CD99 mAbs preferentially induce death in AML disease stem cells, possess a potentially large therapeutic window, and represent a promising AML stem cell-directed therapeutic strategy.
Anti-CD99 monoclonal antibodies activate SRC-family kinases
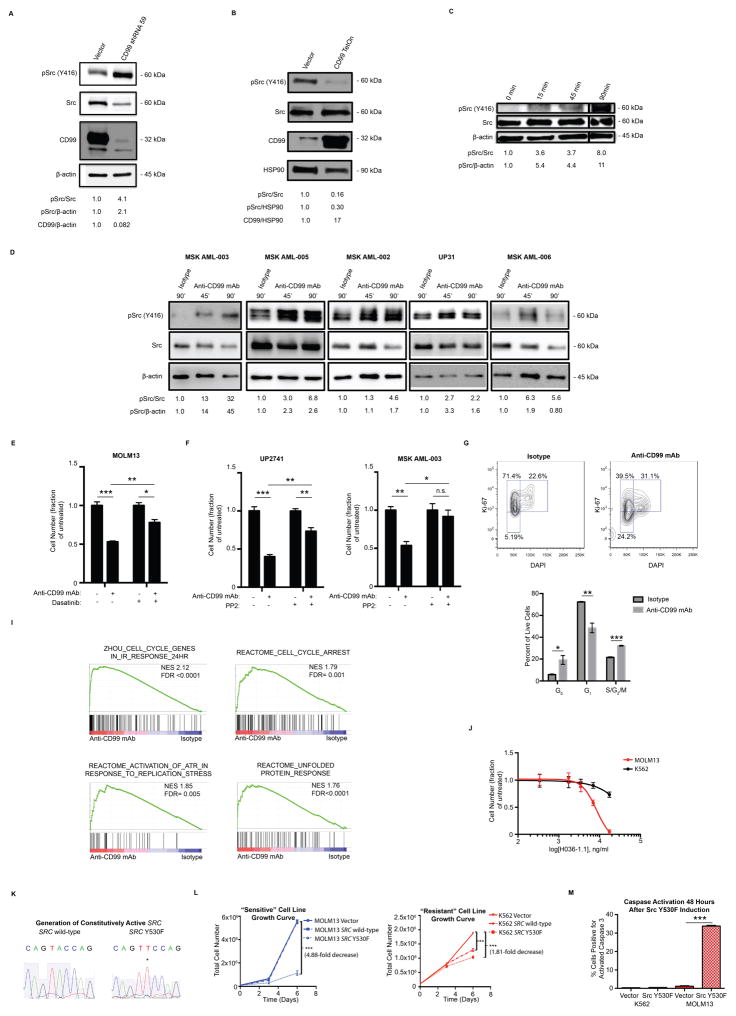
To determine the mechanism of anti-CD99 mAb induced cell death, we assessed the effect of anti-CD99 mAbs on SRC-family kinase (SFK) activation, since CD99 has been shown to negatively regulate SFK activation in osteosarcoma cells(36). We confirmed that CD99 negatively regulates SFK activation in AML cell lines, as shRNA-mediated knockdown of CD99 in the AML cell line MOLM13 induced SFK activation, while overexpression repressed SFK activation (Fig. 5A–B). Cytotoxic anti-CD99 mAbs recapitulated the effects of CD99 knockdown, inducing rapid and robust SFK activation in both AML cell lines (Fig. 5C) as well as primary AML blasts (Fig. 5D). Pharmacologic inhibition of SFKs with the small molecule inhibitors PP2 or dasatinib significantly attenuated anti-CD99 mAb induced cytotoxicity in both AML cell lines (Fig. 5E) and primary AML blasts (Fig. 5F), confirming that anti-CD99 mAbs promote cell death in part by inducing SFK activation. Consistent with the deleterious effects of rapid SFK activation, we observed cell cycle arrest (Fig. 5G), as well as enrichment for gene expression signatures associated with DNA damage response, replication stress, and the unfolded protein response (Fig. 5I). The chronic myeloid leukemia blast crisis derived cell line K562 was highly resistant to the cytotoxic effects of anti-CD99 mAbs (Fig. 5J, Supp. Fig. 15). Similar to the resistant primary sample shown in Fig. 3C, this cell line harbors the constitutively active tyrosine kinase BCR-ABL, which promotes high basal levels of SFK activation(37). Thus, resistance to anti-CD99 mAb induced cytotoxicity is likely due to the ability of K562 cells to better tolerate acute SFK activation. To determine whether SFK activation is sufficient to induce cell death, we generated a constitutively active SRC mutant (Y530F) (Fig. 5K) and overexpressed it in anti-CD99 mAb sensitive and resistant cell lines (MOLM13 and K562, respectively). While MOLM13 cells exhibited a marked decrease in cell growth (Fig. 5L), cell cycle arrest (Supp. Fig. 16), and increased apoptosis (Fig. 5M) in response to constitutive SRC activation, thereby phenocopying the effects of treatment with anti-CD99 mAb, K562 cells demonstrated a modest decrease in growth and no increase in apoptosis (Fig. 5L–M) or cell cycle arrest (Supp. Fig. 16). Together, these findings suggest that anti-CD99 mAbs promote cell death by inducing SFK activation and oncogenic stress in myeloid leukemias that do not exhibit constitutive activation of SFKs.
Fig. 5. Anti-CD99 mAbs induce apoptosis by promoting SRC-family kinase (SFK) activation.
(A) MOLM13 cells were transduced with an shRNA targeting CD99. Western blot confirmed knockdown of CD99, which was accompanied by an increase in SFK activation, as measured by pSRC (Y416). (B) MOLM13 cells were transduced to overexpress CD99 under a doxycycline inducible promoter, and 1 μg/ml doxycycline was added to the media. Western blot confirmed overexpression of CD99 24 hours after addition of doxycycline, which was accompanied by a decrease in pSRC (Y416). (C) Incubation of MOLM13 cells with anti-CD99 mAb (clone H036-1.1, 20 μg/ml) induced rapid SFK activation. (D) Incubation of the indicated primary AML specimens with anti-CD99 mAb (clone H036-1.1, 36 μg/ml) induced rapid SFK activation. (E) MOLM13 cells were incubated with dasatinib (1 μM) or DMSO for eight hours prior to incubation with anti-CD99 mAb (H036-1.1, 5 μg/ml) for 48 hours. Anti-CD99 mAb induced a significant reduction in cell number that was partially rescued by dasatinib treatment. *P<0.05, **P<0.01, ***P<0.001 (unpaired t-test). Error bars represent ±SEM of biological triplicates. (F) Primary AML blasts were incubated with PP2 (20 μM) or DMSO immediately prior to incubation with anti-CD99 mAb (H036-1.1, 36 μg/ml) for 48 hours. Anti-CD99 mAb induced a significant reduction in cell number that was partially rescued by dasatinib treatment. *P<0.05, **P<0.01, ***P<0.001 (unpaired t-test). Error bars represent ±SEM of biological triplicates. (G) Incubation of MOLM13 cells with anti-CD99 mAb (H036-1.1, 20 μg/ml) for 36 hours leads to a marked redistribution of cells from the G1 to the G0 or S/G2/M phases of the cell cycle. *P<0.05, **P<0.01, ***P<0.001 (unpaired t-test). (I) MOLM13 cells were incubated with anti-CD99 mAb (H036-1.1, 36 μg/ml) and after 24 hours live cells (propidium iodide negative) were FACS-purified and RNA-sequencing was performed. Gene set enrichment analysis revealed enrichment for gene signatures associated with DNA-damage response(57), cell cycle arrest, replication stress, and the unfolded protein response(55, 58). (J) K562 cells were incubated with anti-CD99 mAb (H036-1.1) for 48 hours. The IC50 was not reached using mAb concentrations up to 17.4 μg/ml. The sensitivity of MOLM13 cells to anti-CD99 mAb, as shown in Fig. 3G, is juxtaposed for comparison. Error bars represent ±SEM of biological triplicates. (K) A constitutively active SRC (Y530F) mutant was generated by site directed mutagenesis. (L) MOLM13 and K562 cells were transduced to overexpress wild-type SRC or mutant SRC (Y530F) using a doxycycline-inducible system; SRC (Y530F) expression induced a greater decrease in cell growth in MOLM13 as compared with K562 cells. Error bars represent ±SEM of biological triplicates. **P<0.01, ***P<0.001 (unpaired t-test). (M) SRC (Y530F) expression induced apoptosis, as measured by activated caspase 3, in MOLM13 but not K562 cells. Error bars represent ±SEM of biological triplicates. ***P<0.001 (unpaired t-test).
Discussion
Both AML and MDS fulfill central tenets of the cancer stem cell hypothesis, whereby disease-initiating stem cells display the ability to self-renew and differentiate into non-self-renewing progeny that comprise the majority of neoplastic cells(1, 7–9). As disease-initiating cells represent the only self-renewing cells in these diseases, therapies aimed at inducing durable remissions or cures must target these populations, which are resistant to current standard therapies(3, 8, 11). Prior studies have identified a number of antigens differentially expressed on AML LSCs(12–17), but these markers are not specific for, or enriched on, functional LSCs (Supp. Fig. 11). Examples of therapies that target cell surface antigens in AML include antibodies against CD47 to enhance phagocytic clearance of blasts(12), against CD44 to disrupt blast:niche interactions(13), and against CD123 to impair cytokine signaling(17). Numerous separate efforts to target LSCs with small molecule inhibitors of self-renewal or survival pathways are also underway(38, 39). Despite these efforts, strategies to exploit unique AML LSC surface antigens have yet to be successfully translated to the clinic. Similarly, therapies directly targeting MDS stem cells have yet to be described, which is not surprising since very little information is available regarding aberrant antigen expression on MDS HSCs.
By examining a large number of primary AML and MDS patient specimens, we have identified CD99 as a cell surface antigen expressed at increased levels on disease-initiating cells compared with normal HSPCs in the vast majority AML and MDS cases. Similar to a subset of previously defined AML associated cell surface markers present on LSCs(12, 14–16), differences in CD99 expression allow for the prospective separation of leukemic blasts from residual normal or pre-leukemic hematopoietic cells. However, CD99 also exhibits the unique capability to enrich for functional LSCs within the leukemic blast population as demonstrated by limiting dilution xenograft assays, even when assessing the LN CD34+CD38−CD90−CD45RA+ LMPP-like fraction of AML, which is most highly enriched for LSC activity(5). This characteristic is not shared by any other previously described LSC markers and will allow for more refined studies of the functional and molecular features of LSCs in the future.
Given the high frequency of aberrant CD99 expression in our series of AML and MDS cases and the fact that these cases were not selected with prior knowledge of their cytogenetic and mutational profiles, our studies suggest that CD99 is likely to be an exploitable therapeutic target in the vast majority of AML and MDS patients. Although we do not expect all myeloid leukemias to be sensitive to this single strategy, we speculate that it may be possible to enhance the therapeutic efficacy of anti-CD99 mAbs by combining their direct cytotoxic activity with other strategies including conjugation with cytotoxic adducts (40, 41) or the derivation of bi-specific antibodies to simultaneously target other leukemia associated antigens or recruit cytotoxic T-cells (42, 43). The robust and rapid activation of SFKs following anti-CD99 mAb binding suggests a mechanism of cytotoxicity via induction of oncogenic stress, as has been described for a number of oncogenes such as RAS(44), c-MYC(45), BCR-ABL(46), and SYK(47). Accordingly, overexpression of constitutively active SRC is sufficient to induce apoptosis in anti-CD99 mAb sensitive MOLM13 cells, but not in K562 cells, which are relatively resistant to anti-CD99 mAbs and exhibit high basal levels of SFK activation. One possible explanation for the relative resistance of K562 cells may be their lack of functional CDKN2A and TP53(48), both critical mediators of oncogene-induced cell cycle arrest(44). Thus, while prior studies have suggested inhibiting the SFK pathway in AML as a potential therapeutic strategy (49), our studies demonstrate that activation of SFKs may represent a novel therapeutic vulnerability in myeloid malignancies.
Although CD99 appears to be a robust marker of disease stem cells and a promising therapeutic target in AML, a number of unresolved issues remain. First, it is not clear whether CD99 is only upregulated in fully transformed LSCs, or whether it is also upregulated on a subset of pre-leukemic HSCs. A number of recent studies have identified pre-leukemic HSCs which functionally give rise to normal hematopoiesis but which may harbor a subset of the full complement of somatic mutations which are present in fully transformed leukemia(19, 22, 23). In our studies, the CD99 negative fraction of a number of specimens completely lacked mutations previously described to be pre-leukemic (e.g. DNMT3A and IDH1/2)(19, 22, 23). Thus, it appears that CD99 may be upregulated on pre-leukemic HSCs in some AMLs. Further characterization of CD99 positive and negative populations by comprehensive targeted sequencing and other approaches will be necessary to fully characterize the stage of leukemogenesis during which CD99 is upregulated, as well as the mechanisms of its upregulation. Identifying the spectrum of pre-leukemic HSCs that upregulate CD99 will be important to assess whether anti-CD99 mAbs may also target this potential reservoir of pre-neoplastic cells that may re-initiate disease.
Second, although we demonstrate that immunophenotypically defined disease initiating MDS HSCs (7, 9) express high levels of CD99, it is unclear whether these high levels of CD99 expression are sufficient to prospectively separate MDS HSCs from residual normal HSCs. We were unable to assess this directly due to the limited cellularity of the MDS samples we evaluated. Additionally, given the high burden of involvement of the HSC compartment by the MDS clone at diagnosis (>95–99% based on prior studies)(7, 8, 11), addressing this question may require evaluation of CD99 expression in the post-therapy and/or minimal residual disease setting, at which time MDS HSCs persist at significant, but likely lower levels(8, 11). Single cell transcriptomal/exome sequencing approaches may also have the potential to address this question by correlating differences in CD99 expression with somatic mutational status. Finally, it is unclear whether high CD99 expression enriches for disease initiating function among MDS HSCs. We were unable to address this question experimentally since current MDS xenograft models require transplantation of relatively large numbers of cells(7, 10), thereby not allowing for limiting dilution analyses using samples with limited cellularity.
In sum, our evaluation of primary patient specimens and xenograft models reveals that CD99 is a novel AML LSC and MDS HSC marker that also identifies cells with functional disease-initiating capability, particularly in AML. Moreover, anti-CD99 mAbs effectively target AML LSCs, directly inducing their death in part by activating SFKs. These studies establish the use of anti-CD99 mAbs as a promising and novel disease stem cell directed therapeutic strategy and identifies a previously unappreciated molecular vulnerability in myeloid malignancies.
Materials and Methods
Experimental Design
Research objectives
The primary objectives of this study were to characterize the frequency of expression of CD99 on disease initiating stem cells in MDS and AML, and to determine the potential of anti-CD99 mAbs as potential therapeutic agents. Our initial data confirmed our pre-specified hypotheses that CD99 is frequently expressed on disease initiating stem cells in MDS and AML, and that anti-CD99 mAbs exhibit direct cytotoxicity to disease cells. Based on our initial data, we hypothesized that CD99 may also enrich for functional leukemic stem cells, and that anti-CD99 mAbs may exert their cytotoxic effects by modulating cell-signaling pathways regulated by CD99. Our secondary objectives were thus to validate CD99 as a cell surface marker which can be used to enrich for functional leukemic stem cells, and to characterize cell-signaling pathways perturbed by anti-CD99 mAbs.
Research subjects and study design
For validation of CD99 expression, 24 MDS specimens and 79 AML specimens were analyzed (39 diagnostic specimens and 40 relapse specimens- for one patient, relapse specimens were available from two different time points). No patient samples were excluded from the analysis or inclusion in results. Human AML and MDS specimens were obtained from patients at Memorial Sloan Kettering Cancer Center, the University of Pennsylvania, the University of Adelaide, and the University of Rochester with informed consent under IRB-approved protocols. Umbilical cord blood (CB) specimens were purchased from the New York Blood Center (New York, New York). Normal human BM specimens were purchased from AllCells Inc. (Emeryville, California).
For limiting dilution analyses, at least 21 xenografts were analyzed for each AML specimen to ensure a large enough sample size for statistical comparison. Experimental groups (e.g. top 10% and bottom 10% of CD99 expressing LSCs) were transplanted into NSG mice in a randomized manner, with age and sex of animals balanced between groups. Animals that died within one week of transplantation were censored, but no other animals were excluded from the analysis. Outcome assessments were performed in a blinded manner (animals were assigned a random numerical code prior to analysis). In vitro cytotoxicity assays were performed on consecutive primary MDS and AML specimens based on availability and adequate cellularity, with no samples excluded from analysis or inclusion in results.
Experimental Methods
Flow cytometry and fluorescence activated cell sorting
All FACS-sorting and flow cytometry analysis was performed on a FACSAria II cell sorter (BD Biosciences, San Jose, California). Analysis of leukemic blasts and LSCs in human AML specimens was performed using the following antibodies: CD19 (HIB29), CD3 (HIT3a), CD34 (581), CD45RA (HI100), CD47 (CC2C6), and CD44 (BJ18) from Biolegend, San Diego, California, CD38 (HIT2), CD99 (3B2/TA8), CD123 (6H6), and TIM3 (F38-2E2) from eBioscience, San Diego, California, and CD90 (5E10) from BD Biosciences, San Jose, California. Given the expression of aberrant differentiation antigens on leukemic cells, an abbreviated lineage stain of CD3/CD19 was used, with lineage negative (LN) CD34+CD38−CD90+CD45RA− cells designated HSCs, LN CD34+CD38−CD90−CD45RA− cells MPPs, and LN CD34+CD38−CD90−CD45RA+ cells LMPPs.
Analysis of HSPCs in human MDS specimens and cord blood was performed using a full lineage stain including: CD2 (RPA-2.10), CD3 (HIT3a), CD4 (RPA-T4), CD7 (M-T701), CD8 (RPA-T8), CD10 (HI10a), CD11b (ICRF44), CD14 (TuK4), CD19 (CC2C6), CD20 (2H7), GPA (HIR2), and CD56 (B159), all from BD Biosciences, San Jose, California. Definitions of HSCs, MPPs, and LMPPs/LSCs remained as described above.
Analysis of human and murine cells in xenograft experiments was performed using the following antibodies: Mouse Ter119 (TER-119) and CD45.1 (A20) from eBioscience, San Diego, California, human CD45 (2D1), CD99 (3B2/TA8), CD38 (HIT2), and CD33 (WM53) from eBioscience, San Diego, California and human CD34 (581) from Biolegend, San Diego, California. Engrafted human cells were identified based on their immunophenotype: mouse Ter119 and mouse CD45 negative, human CD45 positive, with confirmation of CD34, CD38, CD99, CD45RA, and CD33 expression depending on the immunophenotype of the transplanted AML.
Methylcellulose colony assays
HSPCs (CD3-CD19-CD34+CD38−) from primary AML specimens were double FACS-sorted to >95% purity and seeded in methylcellulose with myeloid/erythroid-promoting cytokines (MethoCult H4435; Stem Cell Technologies, Vancouver, Canada). Colony number and lineage were scored after 14 days with an Olympus BX41 microscope.
Genotyping of methylcellulose colonies
Methylcellulose colonies were aspirated, and cells were resuspended in sterile water and heated to 95°C for 10 minutes. FLT3 mutational status was determined by PCR using primers that yield a product of 329 bp for wild-type alleles, and products of varying larger sizes for ITD alleles. Reactions were performed in 50 μl total volume containing 5 μl template, 5 μl each of 10 μM forward primer 11F (5′-GCAATTTAGGTATGAAAGCCAGC-3′) and reverse primer 12R (5′-CTTTCAGCATTTTGACGGCAACC-3′), 1.25 U Taq DNA Polymerase, 50 mM KCl, 30 mM Tris-HCl, 1.5 mM Mg, and 200 μM of each dNTP. Thermocycling conditions were as follows: 30 sec at 95°C, 30 sec at 62°C, and 30 sec at 72°C for 40 cycles. The presence of the BCR-ABL (e6a2) translocation present in MSK AML-005 was detected using the same reaction mixture with forward primer (5′-GACTTCATTATCAGCTCAGAATGCACC-3′) and reverse primer (5′-AGATACTCAGCGGCATTGCGG-3′). Thermocycling conditions were as follows: 30 sec at 95°C, 30 sec at 56°C, and 60 sec at 72°C for 40 cycles. The mutational status of other genes was determined by Sanger sequencing of PCR fragments amplified from genomic DNA using the primers listed in Table 3.
In vitro antibody incubation assays
Cell lines were plated at a concentration of 2,500 cells per 20 μl of RPMI 1640, 10% heat-inactivated fetal calf serum (Thermo Fisher, Rockford, Illinois), and Penicillin (5000 U/ml)/Streptomycin (5000 μg/ml)(Life Technologies, Carlsbad, California) in a flat-bottom 384-well tissue culture treated plate (Corning, Corning, New York). Primary MDS, AML, and CB specimens were plated at a concentration of 700–2,500 cells per 20 μl of Stemspan SFEM media (Stem Cell Technologies, Vancouver, Canada) supplemented with 40 μg/ml human LDL (Sigma-Aldrich, St. Louis, Missouri) and the following cytokines (Peprotech, Rocky Hill, New Jersey): 100 ng/ml Flt-3 ligand, 100 ng/ml SCF, 50 ng/ml TPO, 20 ng/ml IL-3, and 20 ng/ml IL-6. Human umbilical vein endothelial cells (HUVECs) were plated at a concentration of 5,000 cells per 20 ul of M199 medium (Thermo Fisher) containing 20% heat-inactivated fetal calf serum (Thermo Fisher, Rockford, Illinois), 15 mM HEPES (Sigma-Aldrich, St. Louis, Missouri), 50 mg/L EC mitogen (Biomedical Technologies Inc., Stoughton, Massachusetts), 50 mg/ml heparin (Sigma-Aldrich, St. Louis, Missouri), 2mM L-Glutamine (Cellgro, Manassas, Virginia), and Penicillin (100 IU/ml)/Streptomycin (100 μg/ml)/Amphotericin B (250 ng/ml)(Life Technologies, Carlsbad, California). For in vitro antibody incubation assays, we utilized anti-CD99 mAb clone H036-1.1 (Abcam, Cambridge, United Kingdom) in sterile PBS. H036-1.1 was added at the indicated concentrations up to 35 μg/ml, and cells were incubated for 48 hours at 37°C, 5% CO2. At the end of the treatment time course, cells were resuspended in buffer (phosphate buffered saline with 2% FCS) containing 100 ng/ml propidium iodide (Sigma-Aldrich, St. Louis, Missouri) and quantified using flow cytometry counting beads (BD Biosciences, San Jose, California). The SFK inhibitor dasatinib (Selleck Chemicals, Houston, Texas) was resuspended in DMSO and used at a final concentration of 1 μM. The SFK inhibitor PP2 (EMD Millipore) was resuspended in DMSO and used at a final concentration of 20 μM.
Apoptosis assays
Annexin V and caspase 3 staining was performed according to the manufacturer’s instructions (BD Biosciences, San Jose, California). For cell cycle analysis, cells were stained with a LIVE/DEAD fixable dead cell stain (Life Technologies, Carlsbad, California) according to the manufacturer’s instructions, followed by fixation and permeabilization (BD cytofix/cytoperm). Cells were then stained with Ki-67 (SolA15) from eBioscience (San Diego, California) for 30 minutes at room temperature followed by DAPI (2 μg/ml, Sigma-Aldrich, St. Louis, Missouri) at room temperature for 10 minutes.
Site-directed mutagenesis
Mutant SRC (Y530F) was generated from a human SRC cDNA (CCSB-Broad Lentiviral Expression Library, Open Biosystems, Lafayette, Colorado) using a QuikChange II XL site-directed mutagenesis kit according to the manufacturer’s instructions (Agilent Technologies, Santa Clara, California) and the following primers: sense (5′-CCACCGAGCCCCAGTTCCAGCCCG-3′) and anti-sense (5′-CGGGCTGGAACTGGGGCTCGGTGG-3′). Successful mutagenesis was confirmed by Sanger sequencing, and the mutant SRC cDNA was cloned into a lentiviral expression vector (pLentiLox 3.7, pLL3.7) containing a doxycycline-inducible promoter. Lentiviral particles were generated using standard techniques(50). To induce ectopic wild-type SRC or SRC (Y530F) expression, 50 ng/ml of doxycycline was added to the media.
Western blotting
5–10 × 106 MOLM13 cells were incubated with H036-1.1 or isotype control (20 μg/ml) for a 90-minute time course. At each time point, cells were pelleted, washed in PBS containing sodium orthovanadate, and lysed using Pierce IP lysis buffer (Thermo Pierce, Rockford, Illinois) containing FOCUS-Protease Arrest (GBiosciences, Houston, Texas) and Phosphatase Inhibitor Cocktail II (EMD Millipore, Billerica, Massachusetts). Western blotting was performed using anti-CD99 mAb clone EPR3097Y from Abcam (Cambridge, United Kingdom), and phospho-SRC family kinase (Y416) mAb clone D49G4, total SRC family kinase mAb clone 32G6, β-actin mAb clone 13E5, and HSP90 mAb clone C45G5 from Cell Signaling Technology (Danvers, Massachusetts). Electrochemiluminescent reagent (EMD Millipore, Billerica, Massachusetts) was applied to Western blots and images were acquired using a GE ImageQuant LAS 4000 instrument (GE Life Sciences, Pittsburgh, Pennsylvania). Densitometry analysis was performed using GelQuant.NET image analysis software (http://biochemlabsolutions.com/).
Xenotransplantation assays
All mice were housed in Memorial Sloan Kettering Cancer Center (MSKCC) animal facilities. All animal procedures were conducted in accordance with the Guidelines for the Care and Use of Laboratory Animals and were approved by the Institutional Animal Care and Use Committees (IACUCs) at MSKCC. NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ (NSG) mice 8–10 weeks of age were used for xenogeneic transplantation assays. FACS-sorted human cells were transplanted into sublethally irradiated NSG mice (185 cGy) via retro-orbital injection. Hu-CD34-NSG™ mice engrafted with human cord blood derived HSCs (Jackson Laboratories, Bar Harbor, Maine) were also utilized for experiments shown in Fig. 4D. Peripheral blood and BM analysis was performed by flow cytometry at the indicated times using the antibodies described above. Anti-CD99 mAb clone H036-1.1 or IgM isotype control was administered by retro-orbital injection, and.anti-CD99 mAb clone 10D6 or IgG isotype control was administered by intraperitoneal injection.
RNA sequencing
Primary AML specimens were FACS-sorted into Trizol LS (Thermo Fisher, Rockford, Illinois). cDNA libraries were prepared using a SMARTer mRNA amplification kit (Clontech, Mountain View, California) and sequencing was performed using the Hi-Seq platform (Illumina, San Diego, California) with 40 million paired end reads per sample.
Statistical and data analysis
All flow cytometry data were analyzed using Flowjo (TreeStar, Ashland, Oregon). L-IC frequency estimations were calculated using a Poisson distribution probability calculator (L-Calc, http://www.stemcell.com/en/Products/All-Products/LCalc-Software.aspx, Stem Cell Technologies, Vancouver, Canada). RNA-seq data was aligned by STAR(51) to human genome hg37 and reads for each gene were counted by HTSeq(52). Differentially expressed genes were identified by DESeq2(53), followed by gene set enrichment analyses(26). P-values were calculated using two-tailed t-tests (Prism 7, GraphPad Inc., La Jolla, California). Estimated variation was taken into account for each group of data and is indicated as standard error or standard deviation in each figure legend.
Supplementary Material
Fig. S1. CD99 is expressed at comparable levels on adult bone marrow hematopoietic stem cells (HSCs) as compared with cord blood (CB) HSCs.
Fig. S2. CD99 is highly expressed on bulk acute myeloid leukemia (AML) blasts, and its level of expression is increased further on leukemic stem cell (LSC) enriched populations.
Fig. S3. CD99 expression distinguishes leukemic from residual normal or pre-leukemic hematopoietic stem and progenitor cells (HSPCs).
Fig. S4. CD99 negative CD34+CD38− cells from AML specimens lack the full complement of mutations present in bulk AML cells.
Fig. S5. CD99 positive CD34+CD38− AML cells engraft a lethal myeloid leukemia.
Fig. S6. CD99 expression enriches for functional leukemic stem cells (LSCs).
Fig. S7. LSC-enriched AML cells with high levels of CD99 expression demonstrate improved survival in vitro.
Fig. S8. FACS-purification of CD99 high and low lymphoid primed multipotent progenitor-like (LMPP-like) cells from primary AML specimens.
Fig. S9. FACS-purification of CD99 high and low bulk unfractionated AML blasts.
Fig. S10. SNV Analysis of CD99 high and low AML blasts.
Fig. S11. Other described leukemic stem cell markers are not enriched on LSC-enriched “LMPP-like” AML cells.
Fig. S12. Selective depletion of CD34+CD38− or CD34+ primary AML cells by anti-CD99 mAbs.
Fig. S13. Time course of anti-CD99 mAb induced apoptosis in MOLM13 cells.
Fig. S14. Confirmation of expression of human acute myeloid leukemia (AML) cell surface markers on xenografted leukemic blasts.
Fig. S15. Anti-CD99 mAb clone 10D6 is directly cytotoxic to AML cell lines.
Fig. S16. Expression of constitutively active SRC leads to cell cycle arrest in anti-CD99 mAb sensitive but not resistant cells.
Supp. Table 1. Clinical characteristics of primary MDS specimens.
Supp. Table 2. Clinical characteristics of primary AML specimens.
Supp. Table 3. Primers used for Sanger sequencing of CD99 negative methylcellulose colonies.
Acknowledgments
We thank F. Zhao, C. Miller, G. Kone, C. Li, J. Morowitz, L. Yu, and S. Zheng for technical support. Next generation sequencing protocols and sequencing were performed by the Integrated Genomics Operation at Memorial Sloan Kettering Cancer Center.
Funding: This work was supported by a Young Investigator Award from the Conquer Cancer Foundation of the American Society of Clinical Oncology, a U.S. Department of Defense Postdoctoral Fellow Award in Bone Marrow Failure Research (BM120096), and a Fellow Scholar Award from the American Society of Hematology, all to S.S.C., as well as a Clinical Scientist Award from the Doris Duke Charitable Foundation, a Translational Research Program Award from the Leukemia and Lymphoma Society, and a Geoffrey Beene Grant from the Geoffrey Beene Cancer Research Center, all to C.Y.P.
Footnotes
Author contributions: S.S.C. and C.Y.P. designed the study. F.E.G, R.L.L., V.K., and A.M.M. accrued primary human MDS specimens. M.C. provided primary AML specimens with known engraftment potential. S.S.C. performed FACS-sorting of human samples and in vitro and xenograft assays. S.S.C. performed antibody treatment and biochemical studies. W.H. analyzed RNA-sequencing data. W.S.E. and M.T. performed biochemical studies. S.S.C. and C.Y.P. analyzed the data. S.S.C. and C.Y.P. prepared the manuscript with input from the other authors.
Competing interests: None of the authors have any financial conflicts of interest to report.
Data and materials availability: RNA-sequencing data has been deposited into the Gene Expression Omnibus (GSE86506).
References and Notes
- 1.Bonnet D, Dick JE. Human acute myeloid leukemia is organized as a hierarchy that originates from a primitive hematopoietic cell. Nature medicine. 1997;3:730–737. doi: 10.1038/nm0797-730. [DOI] [PubMed] [Google Scholar]
- 2.Lapidot T, Sirard C, Vormoor J, Murdoch B, Hoang T, Caceres-Cortes J, Minden M, Paterson B, Caligiuri MA, Dick JE. A cell initiating human acute myeloid leukaemia after transplantation into SCID mice. Nature. 1994;367:645–648. doi: 10.1038/367645a0. [DOI] [PubMed] [Google Scholar]
- 3.Ishikawa F, Yoshida S, Saito Y, Hijikata A, Kitamura H, Tanaka S, Nakamura R, Tanaka T, Tomiyama H, Saito N, Fukata M, Miyamoto T, Lyons B, Ohshima K, Uchida N, Taniguchi S, Ohara O, Akashi K, Harada M, Shultz LD. Chemotherapy-resistant human AML stem cells home to and engraft within the bone-marrow endosteal region. Nature biotechnology. 2007;25:1315–1321. doi: 10.1038/nbt1350. [DOI] [PubMed] [Google Scholar]
- 4.Krivtsov AV, Twomey D, Feng Z, Stubbs MC, Wang Y, Faber J, Levine JE, Wang J, Hahn WC, Gilliland DG, Golub TR, Armstrong SA. Transformation from committed progenitor to leukaemia stem cell initiated by MLL-AF9. Nature. 2006;442:818–822. doi: 10.1038/nature04980. [DOI] [PubMed] [Google Scholar]
- 5.Goardon N, Marchi E, Atzberger A, Quek L, Schuh A, Soneji S, Woll P, Mead A, Alford KA, Rout R, Chaudhury S, Gilkes A, Knapper S, Beldjord K, Begum S, Rose S, Geddes N, Griffiths M, Standen G, Sternberg A, Cavenagh J, Hunter H, Bowen D, Killick S, Robinson L, Price A, Macintyre E, Virgo P, Burnett A, Craddock C, Enver T, Jacobsen SE, Porcher C, Vyas P. Coexistence of LMPP-like and GMP-like leukemia stem cells in acute myeloid leukemia. Cancer cell. 2011;19:138–152. doi: 10.1016/j.ccr.2010.12.012. [DOI] [PubMed] [Google Scholar]
- 6.Jamieson CH, Ailles LE, Dylla SJ, Muijtjens M, Jones C, Zehnder JL, Gotlib J, Li K, Manz MG, Keating A, Sawyers CL, Weissman IL. Granulocyte-macrophage progenitors as candidate leukemic stem cells in blast-crisis CML. The New England journal of medicine. 2004;351:657–667. doi: 10.1056/NEJMoa040258. [DOI] [PubMed] [Google Scholar]
- 7.Pang WW, Pluvinage JV, Price EA, Sridhar K, Arber DA, Greenberg PL, Schrier SL, Park CY, Weissman IL. Hematopoietic stem cell and progenitor cell mechanisms in myelodysplastic syndromes. Proc Natl Acad Sci U S A. 2013;110:3011–3016. doi: 10.1073/pnas.1222861110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Will B, Zhou L, Vogler TO, Ben-Neriah S, Schinke C, Tamari R, Yu Y, Bhagat TD, Bhattacharyya S, Barreyro L, Heuck C, Mo Y, Parekh S, McMahon C, Pellagatti A, Boultwood J, Montagna C, Silverman L, Maciejewski J, Greally JM, Ye BH, List AF, Steidl C, Steidl U, Verma A. Stem and progenitor cells in myelodysplastic syndromes show aberrant stage-specific expansion and harbor genetic and epigenetic alterations. Blood. 2012;120:2076–2086. doi: 10.1182/blood-2011-12-399683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Woll PS, Kjallquist U, Chowdhury O, Doolittle H, Wedge DC, Thongjuea S, Erlandsson R, Ngara M, Anderson K, Deng Q, Mead AJ, Stenson L, Giustacchini A, Duarte S, Giannoulatou E, Taylor S, Karimi M, Scharenberg C, Mortera-Blanco T, Macaulay IC, Clark SA, Dybedal I, Josefsen D, Fenaux P, Hokland P, Holm MS, Cazzola M, Malcovati L, Tauro S, Bowen D, Boultwood J, Pellagatti A, Pimanda JE, Unnikrishnan A, Vyas P, Gohring G, Schlegelberger B, Tobiasson M, Kvalheim G, Constantinescu SN, Nerlov C, Nilsson L, Campbell PJ, Sandberg R, Papaemmanuil E, Hellstrom-Lindberg E, Linnarsson S, Jacobsen SE. Myelodysplastic syndromes are propagated by rare and distinct human cancer stem cells in vivo. Cancer cell. 2014;25:794–808. doi: 10.1016/j.ccr.2014.03.036. [DOI] [PubMed] [Google Scholar]
- 10.Medyouf H, Mossner M, Jann JC, Nolte F, Raffel S, Herrmann C, Lier A, Eisen C, Nowak V, Zens B, Mudder K, Klein C, Oblander J, Fey S, Vogler J, Fabarius A, Riedl E, Roehl H, Kohlmann A, Staller M, Haferlach C, Muller N, John T, Platzbecker U, Metzgeroth G, Hofmann WK, Trumpp A, Nowak D. Myelodysplastic Cells in Patients Reprogram Mesenchymal Stromal Cells to Establish a Transplantable Stem Cell Niche Disease Unit. Cell stem cell. 2014 doi: 10.1016/j.stem.2014.02.014. [DOI] [PubMed] [Google Scholar]
- 11.Tehranchi R, Woll PS, Anderson K, Buza-Vidas N, Mizukami T, Mead AJ, Astrand-Grundstrom I, Strombeck B, Horvat A, Ferry H, Dhanda RS, Hast R, Ryden T, Vyas P, Gohring G, Schlegelberger B, Johansson B, Hellstrom-Lindberg E, List A, Nilsson L, Jacobsen SE. Persistent malignant stem cells in del(5q) myelodysplasia in remission. The New England journal of medicine. 2010;363:1025–1037. doi: 10.1056/NEJMoa0912228. [DOI] [PubMed] [Google Scholar]
- 12.Majeti R, Chao MP, Alizadeh AA, Pang WW, Jaiswal S, Gibbs KD, Jr, van Rooijen N, Weissman IL. CD47 is an adverse prognostic factor and therapeutic antibody target on human acute myeloid leukemia stem cells. Cell. 2009;138:286–299. doi: 10.1016/j.cell.2009.05.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jin L, Hope KJ, Zhai Q, Smadja-Joffe F, Dick JE. Targeting of CD44 eradicates human acute myeloid leukemic stem cells. Nature medicine. 2006;12:1167–1174. doi: 10.1038/nm1483. [DOI] [PubMed] [Google Scholar]
- 14.Hosen N, Park CY, Tatsumi N, Oji Y, Sugiyama H, Gramatzki M, Krensky AM, Weissman IL. CD96 is a leukemic stem cell-specific marker in human acute myeloid leukemia. Proc Natl Acad Sci U S A. 2007;104:11008–11013. doi: 10.1073/pnas.0704271104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kikushige Y, Shima T, Takayanagi S, Urata S, Miyamoto T, Iwasaki H, Takenaka K, Teshima T, Tanaka T, Inagaki Y, Akashi K. TIM-3 is a promising target to selectively kill acute myeloid leukemia stem cells. Cell Stem Cell. 2010;7:708–717. doi: 10.1016/j.stem.2010.11.014. [DOI] [PubMed] [Google Scholar]
- 16.Jan M, Chao MP, Cha AC, Alizadeh AA, Gentles AJ, Weissman IL, Majeti R. Prospective separation of normal and leukemic stem cells based on differential expression of TIM3, a human acute myeloid leukemia stem cell marker. Proc Natl Acad Sci U S A. 2011;108:5009–5014. doi: 10.1073/pnas.1100551108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jin L, Lee EM, Ramshaw HS, Busfield SJ, Peoppl AG, Wilkinson L, Guthridge MA, Thomas D, Barry EF, Boyd A, Gearing DP, Vairo G, Lopez AF, Dick JE, Lock RB. Monoclonal antibody-mediated targeting of CD123, IL-3 receptor alpha chain, eliminates human acute myeloid leukemic stem cells. Cell Stem Cell. 2009;5:31–42. doi: 10.1016/j.stem.2009.04.018. [DOI] [PubMed] [Google Scholar]
- 18.McGowan KA, Pang WW, Bhardwaj R, Perez MG, Pluvinage JV, Glader BE, Malek R, Mendrysa SM, Weissman IL, Park CY, Barsh GS. Reduced ribosomal protein gene dosage and p53 activation in low-risk myelodysplastic syndrome. Blood. 2011;118:3622–3633. doi: 10.1182/blood-2010-11-318584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shlush LI, Zandi S, Mitchell A, Chen WC, Brandwein JM, Gupta V, Kennedy JA, Schimmer AD, Schuh AC, Yee KW, McLeod JL, Doedens M, Medeiros JJ, Marke R, Kim HJ, Lee K, McPherson JD, Hudson TJ, Brown AM, Yousif F, Trinh QM, Stein LD, Minden MD, Wang JC, Dick JE. Identification of pre-leukaemic haematopoietic stem cells in acute leukaemia. Nature. 2014;506:328–333. doi: 10.1038/nature13038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Majeti R, Park CY, Weissman IL. Identification of a hierarchy of multipotent hematopoietic progenitors in human cord blood. Cell Stem Cell. 2007;1:635–645. doi: 10.1016/j.stem.2007.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Griffin JD, Lowenberg B. Clonogenic cells in acute myeloblastic leukemia. Blood. 1986;68:1185–1195. [PubMed] [Google Scholar]
- 22.Jan M, Snyder TM, Corces-Zimmerman MR, Vyas P, Weissman IL, Quake SR, Majeti R. Clonal evolution of preleukemic hematopoietic stem cells precedes human acute myeloid leukemia. Science translational medicine. 2012;4:149ra118. doi: 10.1126/scitranslmed.3004315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Corces-Zimmerman MR, Hong WJ, Weissman IL, Medeiros BC, Majeti R. Preleukemic mutations in human acute myeloid leukemia affect epigenetic regulators and persist in remission. Proc Natl Acad Sci U S A. 2014;111:2548–2553. doi: 10.1073/pnas.1324297111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Montesinos JJ, Sanchez-Valle E, Flores-Figueroa E, Martinez-Jaramillo G, Flores-Guzman P, Miranda-Peralta E, Gutierrez-Romero M, Mayani H. Deficient proliferation and expansion in vitro of two bone marrow cell populations from patients with acute myeloid leukemia in response to hematopoietic cytokines. Leuk Lymphoma. 2006;47:1379–1386. doi: 10.1080/10428190500465424. [DOI] [PubMed] [Google Scholar]
- 25.Ito S, Barrett AJ, Dutra A, Pak E, Miner S, Keyvanfar K, Hensel NF, Rezvani K, Muranski P, Liu P, Melenhorst JJ, Larochelle A. Long term maintenance of myeloid leukemic stem cells cultured with unrelated human mesenchymal stromal cells. Stem cell research. 2015;14:95–104. doi: 10.1016/j.scr.2014.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gentles AJ, Plevritis SK, Majeti R, Alizadeh AA. Association of a leukemic stem cell gene expression signature with clinical outcomes in acute myeloid leukemia. JAMA: the journal of the American Medical Association. 2010;304:2706–2715. doi: 10.1001/jama.2010.1862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Eppert K, Takenaka K, Lechman ER, Waldron L, Nilsson B, van Galen P, Metzeler KH, Poeppl A, Ling V, Beyene J, Canty AJ, Danska JS, Bohlander SK, Buske C, Minden MD, Golub TR, Jurisica I, Ebert BL, Dick JE. Stem cell gene expression programs influence clinical outcome in human leukemia. Nature medicine. 2011;17:1086–1093. doi: 10.1038/nm.2415. [DOI] [PubMed] [Google Scholar]
- 29.Saito Y, Kitamura H, Hijikata A, Tomizawa-Murasawa M, Tanaka S, Takagi S, Uchida N, Suzuki N, Sone A, Najima Y, Ozawa H, Wake A, Taniguchi S, Shultz LD, Ohara O, Ishikawa F. Identification of therapeutic targets for quiescent, chemotherapy-resistant human leukemia stem cells. Science translational medicine. 2010;2:17ra19. doi: 10.1126/scitranslmed.3000349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Georgantas RW, 3rd, Tanadve V, Malehorn M, Heimfeld S, Chen C, Carr L, Martinez-Murillo F, Riggins G, Kowalski J, Civin CI. Microarray and serial analysis of gene expression analyses identify known and novel transcripts overexpressed in hematopoietic stem cells. Cancer research. 2004;64:4434–4441. doi: 10.1158/0008-5472.CAN-03-3247. [DOI] [PubMed] [Google Scholar]
- 31.Signer RA, Magee JA, Salic A, Morrison SJ. Haematopoietic stem cells require a highly regulated protein synthesis rate. Nature. 2014;509:49–54. doi: 10.1038/nature13035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cai X, Gao L, Teng L, Ge J, Oo ZM, Kumar AR, Gilliland DG, Mason PJ, Tan K, Speck NA. Runx1 Deficiency Decreases Ribosome Biogenesis and Confers Stress Resistance to Hematopoietic Stem and Progenitor Cells. Cell Stem Cell. 2015;17:165–177. doi: 10.1016/j.stem.2015.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Scotlandi K, Baldini N, Cerisano V, Manara MC, Benini S, Serra M, Lollini PL, Nanni P, Nicoletti G, Bernard G, Bernard A, Picci P. CD99 engagement: an effective therapeutic strategy for Ewing tumors. Cancer research. 2000;60:5134–5142. [PubMed] [Google Scholar]
- 34.Rumpold H, Wiedermann G, Scheiner O, Kraft D, Stemberger H. Lack of evidence for IgM-induced ADCC: studies with monoclonal and polyclonal antibodies. Immunology. 1981;43:161–170. [PMC free article] [PubMed] [Google Scholar]
- 35.Shultz LD, Schweitzer PA, Christianson SW, Gott B, Schweitzer IB, Tennent B, McKenna S, Mobraaten L, Rajan TV, Greiner DL, et al. Multiple defects in innate and adaptive immunologic function in NOD/LtSz-scid mice. J Immunol. 1995;154:180–191. [PubMed] [Google Scholar]
- 36.Scotlandi K, Zuntini M, Manara MC, Sciandra M, Rocchi A, Benini S, Nicoletti G, Bernard G, Nanni P, Lollini PL, Bernard A, Picci P. CD99 isoforms dictate opposite functions in tumour malignancy and metastases by activating or repressing c-Src kinase activity. Oncogene. 2007;26:6604–6618. doi: 10.1038/sj.onc.1210481. [DOI] [PubMed] [Google Scholar]
- 37.Danhauser-Riedl S, Warmuth M, Druker BJ, Emmerich B, Hallek M. Activation of Src kinases p53/56lyn and p59hck by p210bcr/abl in myeloid cells. Cancer research. 1996;56:3589–3596. [PubMed] [Google Scholar]
- 38.Guzman ML, Rossi RM, Karnischky L, Li X, Peterson DR, Howard DS, Jordan CT. The sesquiterpene lactone parthenolide induces apoptosis of human acute myelogenous leukemia stem and progenitor cells. Blood. 2005;105:4163–4169. doi: 10.1182/blood-2004-10-4135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Goff DJ, Court Recart A, Sadarangani A, Chun HJ, Barrett CL, Krajewska M, Leu H, Low-Marchelli J, Ma W, Shih AY, Wei J, Zhai D, Geron I, Pu M, Bao L, Chuang R, Balaian L, Gotlib J, Minden M, Martinelli G, Rusert J, Dao KH, Shazand K, Wentworth P, Smith KM, Jamieson CA, Morris SR, Messer K, Goldstein LS, Hudson TJ, Marra M, Frazer KA, Pellecchia M, Reed JC, Jamieson CH. A Pan-BCL2 inhibitor renders bone-marrow-resident human leukemia stem cells sensitive to tyrosine kinase inhibition. Cell Stem Cell. 2013;12:316–328. doi: 10.1016/j.stem.2012.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sievers EL. Efficacy and safety of gemtuzumab ozogamicin in patients with CD33-positive acute myeloid leukaemia in first relapse. Expert opinion on biological therapy. 2001;1:893–901. doi: 10.1517/14712598.1.5.893. [DOI] [PubMed] [Google Scholar]
- 41.Sutherland MK, Yu C, Lewis TS, Miyamoto JB, Morris-Tilden CA, Jonas M, Sutherland J, Nesterova A, Gerber HP, Sievers EL, Grewal IS, Law CL. Anti-leukemic activity of lintuzumab (SGN-33) in preclinical models of acute myeloid leukemia. mAbs. 2009;1:481–490. doi: 10.4161/mabs.1.5.9288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Topp MS, Gokbuget N, Stein AS, Zugmaier G, O’Brien S, Bargou RC, Dombret H, Fielding AK, Heffner L, Larson RA, Neumann S, Foa R, Litzow M, Ribera JM, Rambaldi A, Schiller G, Bruggemann M, Horst HA, Holland C, Jia C, Maniar T, Huber B, Nagorsen D, Forman SJ, Kantarjian HM. Safety and activity of blinatumomab for adult patients with relapsed or refractory B-precursor acute lymphoblastic leukaemia: a multicentre, single-arm, phase 2 study. The Lancet Oncology. 2015;16:57–66. doi: 10.1016/S1470-2045(14)71170-2. [DOI] [PubMed] [Google Scholar]
- 43.Arndt C, von Bonin M, Cartellieri M, Feldmann A, Koristka S, Michalk I, Stamova S, Bornhauser M, Schmitz M, Ehninger G, Bachmann M. Redirection of T cells with a first fully humanized bispecific CD33-CD3 antibody efficiently eliminates AML blasts without harming hematopoietic stem cells. Leukemia: official journal of the Leukemia Society of America, Leukemia Research Fund, U.K. 2013;27:964–967. doi: 10.1038/leu.2013.18. [DOI] [PubMed] [Google Scholar]
- 44.Serrano M, Lin AW, McCurrach ME, Beach D, Lowe SW. Oncogenic ras provokes premature cell senescence associated with accumulation of p53 and p16INK4a. Cell. 1997;88:593–602. doi: 10.1016/s0092-8674(00)81902-9. [DOI] [PubMed] [Google Scholar]
- 45.Evan GI, Wyllie AH, Gilbert CS, Littlewood TD, Land H, Brooks M, Waters CM, Penn LZ, Hancock DC. Induction of apoptosis in fibroblasts by c-myc protein. Cell. 1992;69:119–128. doi: 10.1016/0092-8674(92)90123-t. [DOI] [PubMed] [Google Scholar]
- 46.Dengler MA, Staiger AM, Gutekunst M, Hofmann U, Doszczak M, Scheurich P, Schwab M, Aulitzky WE, van der Kuip H. Oncogenic stress induced by acute hyper-activation of Bcr-Abl leads to cell death upon induction of excessive aerobic glycolysis. PloS one. 2011;6:e25139. doi: 10.1371/journal.pone.0025139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chen Z, Shojaee S, Buchner M, Geng H, Lee JW, Klemm L, Titz B, Graeber TG, Park E, Tan YX, Satterthwaite A, Paietta E, Hunger SP, Willman CL, Melnick A, Loh ML, Jung JU, Coligan JE, Bolland S, Mak TW, Limnander A, Jumaa H, Reth M, Weiss A, Lowell CA, Muschen M. Signalling thresholds and negative B-cell selection in acute lymphoblastic leukaemia. Nature. 2015;521:357–361. doi: 10.1038/nature14231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rui HB, Su JZ. Co-transfection of p16(INK4a) and p53 genes into the K562 cell line inhibits cell proliferation. Haematologica. 2002;87:136–142. [PubMed] [Google Scholar]
- 49.Dos Santos C, McDonald T, Ho YW, Liu H, Lin A, Forman SJ, Kuo HY, Bhatia R. The Src and c-Kit kinase inhibitor dasatinib enhances p53-mediated targeting of human acute myeloid leukemia stem cells by chemotherapeutic agents. Blood. 2013;122:1900–1913. doi: 10.1182/blood-2012-11-466425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Naldini L, Blomer U, Gallay P, Ory D, Mulligan R, Gage FH, Verma IM, Trono D. In vivo gene delivery and stable transduction of nondividing cells by a lentiviral vector. Science. 1996;272:263–267. doi: 10.1126/science.272.5259.263. [DOI] [PubMed] [Google Scholar]
- 51.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Anders S, Pyl PT, Huber W. HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Milacic M, Haw R, Rothfels K, Wu G, Croft D, Hermjakob H, D’Eustachio P, Stein L. Annotating cancer variants and anti-cancer therapeutics in reactome. Cancers. 2012;4:1180–1211. doi: 10.3390/cancers4041180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Croft D, Mundo AF, Haw R, Milacic M, Weiser J, Wu G, Caudy M, Garapati P, Gillespie M, Kamdar MR, Jassal B, Jupe S, Matthews L, May B, Palatnik S, Rothfels K, Shamovsky V, Song H, Williams M, Birney E, Hermjakob H, Stein L, D’Eustachio P. The Reactome pathway knowledgebase. Nucleic Acids Res. 2014;42:D472–477. doi: 10.1093/nar/gkt1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhou T, Chou J, Mullen TE, Elkon R, Zhou Y, Simpson DA, Bushel PR, Paules RS, Lobenhofer EK, Hurban P, Kaufmann WK. Identification of primary transcriptional regulation of cell cycle-regulated genes upon DNA damage. Cell Cycle. 2007;6:972–981. doi: 10.4161/cc.6.8.4106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Fabregat A, Sidiropoulos K, Garapati P, Gillespie M, Hausmann K, Haw R, Jassal B, Jupe S, Korninger F, McKay S, Matthews L, May B, Milacic M, Rothfels K, Shamovsky V, Webber M, Weiser J, Williams M, Wu G, Stein L, Hermjakob H, D’Eustachio P. The Reactome pathway Knowledgebase. Nucleic Acids Res. 2016;44:D481–487. doi: 10.1093/nar/gkv1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. CD99 is expressed at comparable levels on adult bone marrow hematopoietic stem cells (HSCs) as compared with cord blood (CB) HSCs.
Fig. S2. CD99 is highly expressed on bulk acute myeloid leukemia (AML) blasts, and its level of expression is increased further on leukemic stem cell (LSC) enriched populations.
Fig. S3. CD99 expression distinguishes leukemic from residual normal or pre-leukemic hematopoietic stem and progenitor cells (HSPCs).
Fig. S4. CD99 negative CD34+CD38− cells from AML specimens lack the full complement of mutations present in bulk AML cells.
Fig. S5. CD99 positive CD34+CD38− AML cells engraft a lethal myeloid leukemia.
Fig. S6. CD99 expression enriches for functional leukemic stem cells (LSCs).
Fig. S7. LSC-enriched AML cells with high levels of CD99 expression demonstrate improved survival in vitro.
Fig. S8. FACS-purification of CD99 high and low lymphoid primed multipotent progenitor-like (LMPP-like) cells from primary AML specimens.
Fig. S9. FACS-purification of CD99 high and low bulk unfractionated AML blasts.
Fig. S10. SNV Analysis of CD99 high and low AML blasts.
Fig. S11. Other described leukemic stem cell markers are not enriched on LSC-enriched “LMPP-like” AML cells.
Fig. S12. Selective depletion of CD34+CD38− or CD34+ primary AML cells by anti-CD99 mAbs.
Fig. S13. Time course of anti-CD99 mAb induced apoptosis in MOLM13 cells.
Fig. S14. Confirmation of expression of human acute myeloid leukemia (AML) cell surface markers on xenografted leukemic blasts.
Fig. S15. Anti-CD99 mAb clone 10D6 is directly cytotoxic to AML cell lines.
Fig. S16. Expression of constitutively active SRC leads to cell cycle arrest in anti-CD99 mAb sensitive but not resistant cells.
Supp. Table 1. Clinical characteristics of primary MDS specimens.
Supp. Table 2. Clinical characteristics of primary AML specimens.
Supp. Table 3. Primers used for Sanger sequencing of CD99 negative methylcellulose colonies.