Abstract
“Omics” research poses acute challenges regarding how to enhance validation practices and eventually the utility of this rich information. Several strategies may be useful, including routine replication, public data and protocol availability, funding incentives, reproducibility rewards or penalties, and targeted repeatability checks.
The exponential growth of the “omics” fields (genomics, transcriptomics, proteomics, metabolomics, and others) fuels expectations for a new era of personalized medicine. However, clinically meaningful discoveries are hidden within millions of analyses (1). Given this immense biological complexity, separating true signals from red herrings is challenging, and validation of proposed discoveries is essential.
Some fields already employ stringent replication criteria. For example, in genomics, genome-wide association studies demand high statistical significance (P values < 5 × 10−8) and perform large-scale replication efforts within international consortia (2). Conversely, other fields continue to perform “mile-long, inch-thick” research (3), in which many factors are tested once (“discovered”) but are rarely further validated. Studies in gene expression profiling and transcriptomics sometimes try to validate the results using different assays within single populations as well as statistical techniques such as cross-validation, which do not require the evaluation of additional, independent samples. However, such methods do not guarantee good performance across different populations. Moreover, very often cross-validation overestimates classifier performance, probably because biases are introduced in the process (4, 5). Independent external validation usually yields more conservative results, but may also be inflated because of optimism, selective reporting, and other biases (5, 6). Independent external validation by completely different teams remains rare.
Even strong replication of omics results does not automatically imply the potential for successful adoption in clinical or public health practice. Demonstrating clinical validity requires evaluation of the predictive value in real-practice populations, whereas clinical utility requires evaluation of the balance of benefits and harms associated with the adoption of these technologies for different intended uses (7). Ideally, randomized clinical trials are needed to assess whether omics information improves patient outcomes. Long-term, large-scale trials, such as those under way for OncotypeDX (a diagnostic test that analyzes a panel of 21 genes within a breast tumor to assess the likelihood of disease recurrence and/or patient benefit from chemotherapy) and MammaPrint (a breast cancer signature of 70 genes) also require careful consideration of design issues (8, 9), because information on available classifiers constantly changes and new classifiers are proposed. There is at least one recent unfortunate example, where gene signatures were moved into clinical trial experimentation with insufficient previous validation. Three trials of gene signatures to predict outcomes of chemotherapy in treating non–small-cell lung cancer and breast cancer were suspended in 2011 after the realization that their supporting published evidence was nonreproducible (10).
Many scientists now demand reproducible omics research (11). This requires access to the full data, protocols, and analysis codes for published studies so that other scientists can repeat analyses and verify results. Fortunately, several public data repositories exist, such as the Gene Expression Omnibus, ArrayExpress, and the Stanford Microarray Database. There have also been many calls for diverse comprehensive study registries, such as for tumor biomarkers, a field riddled with uncertainty because of suboptimal study design and data quality, and a poor replication record (12, 13). Many leading journals are now working to adopt policies to make public deposition of data and protocols a prerequisite for publication (14). However, the practice of making this information accessible is applied inconsistently; furthermore, it is challenging to verify that complete data and protocols are indeed deposited, files are usable, and results repeatable. An empirical assessment of 18 published papers of microarray studies showed that independent analysts could perfectly reproduce the results of only two of the studies, and it sometimes took over a month to reproduce a single figure (15). Moreover, in some fields that started with good prospects for public data deposition, such as genome-wide association studies, many investigators have become more resistant to sharing data because of perceived confidentiality issues (16).
Data, protocol, and code deposition for access by qualified investigators should be more widely adopted. Possible steps in this direction include making data-protocol-algorithm deposition a prerequisite for publication in all journals, not just select ones, and mandating this procedure for all future funded proposals and for all data submitted to regulatory agencies. Some resources are needed to support the development and expansion of such repositories, but hopefully once the infrastructure is set up and streamlined, maintenance should not be expensive. Moreover, federal funding agencies could offer financial bonuses to investigators whose publicly available data have demonstrably been used by multiple other investigators, with citations made to the publicly available data sets. Actual repeatability exercises require considerable effort, and it is currently not feasible to have a “police force” of analysts repeating all published and deposited data and analyses. However, targeted repeatability checks are possible by specialists in bioinformatics/biostatistics (or by other field experts), with a priority for postulated discoveries that have potentially high clinical impact. For example, a repeatability check of all relevant data can become a prerequisite to clinical trial experimentation. Given that few discoveries (currently much less than 1%) progress that far in the translation process, such a repeatability check is logistically manageable without an inordinate cost. Such checks can be competitively outsourced to bioinformatics experts, for example, who have not been involved in any of the original work. Funding agencies may also outsource spot checks of repeatability in a small number of randomly selected funded omics projects and may offer bonuses to investigators with repeatable results and penalize the future funding of investigators with nonrepeatable results. In this fashion, the penalities accrued from instances of nonreproducible results may help to cover the costs of the validation exercises.
Another possibility is to make collections of massive amounts of omics data available to all investigators who are interested in analyzing them. Rewards (funding) could go to those who succeed in producing the best models (for example, models that achieve the best predictive discrimination for cancer), which can also be validated by other, independent data sets. This is a mechanism similar to what Netflix has used. The movie rental company launched an open public competition to generate models that would best predict movie ratings based on available collected data on movie preferences (17). Such an approach makes data available on an egalitarian basis to all potentially interested investigators, maximizes data transparency, and rewards investigators that use the best methods and get the most-reproducible results.
Some fields have recognized the need to reevaluate even earlier stages of measurement validity. For example, analytic validity remains a challenge in proteomics. One study (18) tested the ability of 27 laboratories to evaluate standardized samples containing 20 highly purified recombinant human proteins with mass spectrometry, a simple challenge compared to the thousands of proteins involved in clinical samples. Only seven laboratories reported all 20 proteins correctly, and only one lab captured all tryptic peptides of 1250 daltons. This exemplifies how factors such as missed identifications, contaminants, and poor database matching can create havoc. In such fields, efforts to improve inter-laboratory reproducibility of data should precede efforts to promote independent validation in large samples from different data sets (19, 20). Several efforts such as the recently launched Human Proteome Project (21) and other Human Proteome Organisation (www.hupo.org) initiatives are currently under way to create networks of laboratories; interactions that will hopefully enhance reproducibility.
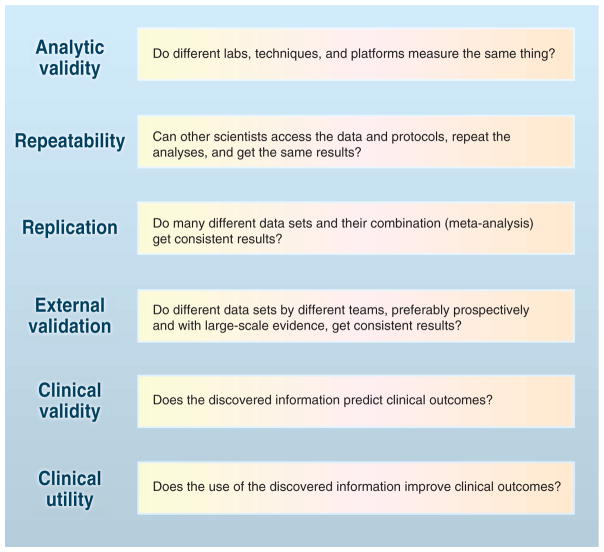
The validation of large-scale omics discoveries involves multiple steps (Fig. 1). Some steps are easier to accomplish in specific fields than others—for example, genotyping of common gene variants has reached almost-perfect analytical validity, with measurement error <0.01%, and the genome community continues to pursue the development of reference materials for use in validating whole-genome sequencing and variant calling; on the other hand, the field of proteomics is struggling with measurement error basics. Given that new measurement platforms continuously emerge, and information is increasingly combined from many of them, one has to decide which approaches are more reproducible and potentially informative. Empirical replication with large-scale studies is therefore increasingly necessary. One may argue that this is not easy because of technical and cost considerations. However, similar arguments were made for fields such as human genome epidemiology, which then saw the cost of DNA sequencing decrease over a billionfold over the past 20 years and the amount of information increase proportionally. Costs could decrease for other technologies as technologies attract the attention of many investigators, especially in large consortia, thereby driving data reproducibility in a field. Funding incentives, reproducibility rewards and/or non-reproducibility penalties, and targeted requirements for repeatability checks may enhance the public availability of useful data and valid analyses.
Fig. 1.
The validation of omics research for use in medicine and public health requires fulfilling multiple steps. [Adapted from (7)]
Acknowledgments
The opinions expressed in this paper are those of the authors and do not reflect an official position of the Department of Health and Human Services.
References and Notes
- 1.Ioannidis JPA. PLoS Med. 2005;2:e124. doi: 10.1371/journal.pmed.0020124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chanock SJ, et al. Nature. 2007;447:655. doi: 10.1038/447655a. [DOI] [PubMed] [Google Scholar]
- 3.Riley RD, et al. Health Technol Assess. 2003;7:1. doi: 10.3310/hta7050. [DOI] [PubMed] [Google Scholar]
- 4.Dupuy A, Simon RM. J Natl Cancer Inst. 2007;17:99. doi: 10.1093/jnci/djk018. [DOI] [PubMed] [Google Scholar]
- 5.Castaldi PJ, Dahabreh IJ, Ioannidis JP. Brief Bioinform. 2011;12:189. doi: 10.1093/bib/bbq073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jelizarow M, Guillemot V, Tenenhaus A, Strimmer K, Boulesteix AL. Bioinformatics. 2010;26:1990. doi: 10.1093/bioinformatics/btq323. [DOI] [PubMed] [Google Scholar]
- 7.Teutsch SM, et al. EGAPP Working Group. Genet Med. 2009;11:3. doi: 10.1097/GIM.0b013e318184137c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sparano JA, Paik S. J Clin Oncol. 2008;26:721. doi: 10.1200/JCO.2007.15.1068. [DOI] [PubMed] [Google Scholar]
- 9.Cardoso F, et al. J Clin Oncol. 2008;26:729. doi: 10.1200/JCO.2007.14.3222. [DOI] [PubMed] [Google Scholar]
- 10.Baggerly K, Coombes KR. Ann Appl Stat. 2009;3:1309. [Google Scholar]
- 11.Baggerly K. Nature. 2010;467:401. doi: 10.1038/467401b. [DOI] [PubMed] [Google Scholar]
- 12.Andre F, et al. Nat Rev Clin Oncol. 2011;8:171. doi: 10.1038/nrclinonc.2011.4. [DOI] [PubMed] [Google Scholar]
- 13.Ioannidis JPA, Panagiotou OA. JAMA. 2011;305:2200. doi: 10.1001/jama.2011.713. [DOI] [PubMed] [Google Scholar]
- 14.Al-Sheikh-Ali AA, Qureshi W, Al-Mallah MH, Ioannidis JP. PLoS ONE. 2011;6:e24357. doi: 10.1371/journal.pone.0024357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ioannidis JPA, et al. Nat Genet. 2009;41:149. doi: 10.1038/ng.295. [DOI] [PubMed] [Google Scholar]
- 16.Johnson AD, Leslie R, O’Donnell CJ. PLoS Genet. 2011;7:e1002269. doi: 10.1371/journal.pgen.1002269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ransohoff DF. Clin Chem. 2010;56:172. doi: 10.1373/clinchem.2009.126698. [DOI] [PubMed] [Google Scholar]
- 18.Bell AW, et al. HUPO Test Sample Working Group. Nat Methods. 2009;6:423. doi: 10.1038/nmeth.1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mischak H, et al. Sci Transl Med. 2010;2:46ps42. doi: 10.1126/scitranslmed.3001249. [DOI] [PubMed] [Google Scholar]
- 20.Mann M. Nat Methods. 2009;6:717. doi: 10.1038/nmeth1009-717. [DOI] [PubMed] [Google Scholar]
- 21.Legrain P, et al. Mol Cell Proteom. 2011;10:M111.009993. doi: 10.1074/mcp.M111.009993. [DOI] [PMC free article] [PubMed] [Google Scholar]