Abstract
The visualization of cerebral microvessels is essential for understanding brain remodeling after stroke. Injection of dyes allows for the evaluation of perfused vessels, but has limitations related either to incomplete microvascular filling or leakage. In conventional histochemistry, the analysis of microvessels is limited to 2D structures, with apparent limitations regarding the interpretation of vascular circuits. Herein, we developed a straight-forward technique to visualize microvessels in the whole ischemic mouse brain, combining the injection of a fluorescent-labeled low viscosity hydrogel conjugate with 3D solvent clearing followed by automated light sheet microscopy. We performed transient middle cerebral artery occlusion in C57Bl/6j mice and acquired detailed 3D vasculature images from whole brains. Subsequent image processing, rendering and fitting of blood vessels to a filament model was employed to calculate vessel length density, resulting in 0.922 ± 0.176 m/mm3 in healthy tissue and 0.329 ± 0.131 m/mm3 in ischemic tissue. This analysis showed a marked loss of capillaries with a diameter ≤ 10 µm and a more moderate loss of microvessels in the range > 10 and ≤ 20 µm, whereas vessels > 20 µm were unaffected by focal cerebral ischemia. We propose that this protocol is highly suitable for studying microvascular injury and remodeling post-stroke.
Keywords: Angiography, brain imaging, capillaries, focal ischemia, microcirculation
Introduction
The integrity of the cerebral microvasculature has huge importance for the successful remodeling of the ischemic brain.1 The brain parenchyma relies on the supply of nutrients, such as oxygen and glucose, and the removal of brain metabolites.2 After focal cerebral ischemia, more severely injured microvessels degenerate, with unfavorable consequences for brain plasticity and neurological recovery.3 Therefore, understanding the remodeling of microvascular circuits is essential for evaluating recovery processes in the ischemic brain and determining the potential of stroke treatments. Considering the complex architecture of the microvessel networks, a sensitive and reliable method for the three-dimensional (3D) visualization and analysis of these networks in the whole mouse brain would be highly useful, as it may allow more comprehensive insights into vascular remodeling in response to stroke and stroke treatment.
To date several techniques have been employed to visualize the cerebral vasculature of intact and ischemic rodent brains based on the optical absorption properties of intravenously delivered dyes. These include, the injection of low viscosity resin, gelatin mixed with various dyes, latex alone or in combination with carbon black, and more recently the mixture of two commercial carbon black inks.4–6 Other strategies enable the visualization of vessels using fluorescent labeled molecules such as Lycopersicon esculentum tomato-conjugated lectin,7–9 fluorescein isothiocyanate- (FITC) conjugated dextran,10–12 fluoro-turquoise-conjugated gelatin,13 FITC-conjugated albumin2 or FITC alone.14 The common idea behind these methods is the staining of vessel trees for the evaluation of vessel size, density and branching. Most of these techniques have limitations related to incomplete vascular filling or leakage of dyes into the brain parenchyma and so far vessel quantification has been restricted to superficial vessels when whole mount brains were used.4
The standard approach for the quantification of microvessels requires histological sectioning to generate two-dimensional (2D) brain sections. 3D reconstruction of the vasculature network from this material requires a large amount of high quality serial histological sections that are hard to obtain, making this methodology laborious and impractical for the quantification of large specimens.15,16 A number of optical methods have been applied for 3D imaging of the whole mouse brain that overcome the physical tissue sectioning problem. Some examples include optical projection tomography (OPT),15,17 blockface imaging,18 serial two photon tomography15,19 and ultramicroscopy.8,16,20–23 Ultramicroscopy has been combined with several clearing approaches to visualize blood vessels in different organs.8,9,22,24,25 However, to date none of these studies has systematically evaluated the 3D architecture of microvessels in the ischemic brain.
In this work, we developed a straight forward method for the visualization and analysis of cerebral microvessels, which combines a systemically injection of a gelatin hydrogel containing FITC-conjugated albumin2 with a 3D solvent-based clearing8,9 and light sheet microscopy. Image processing, segmentation, modelling and analysis of microvascular networks were performed in an automated way using a combination of readily available software tools. The utility of this procedure was demonstrated by quantifying microvessels in non-ischemic and ischemic regions of whole mouse brains allowing us to determine the length density of microvessels as a function of vessel diameter.
Materials and methods
Animals
Animal experiments were performed in accordance with the regulations of the National Institute of Health Guidelines for the Care and Use of Laboratory Animals in compliance with ARRIVE guidelines and the permission of local authorities (Bezirksregierung Düsseldorf; Az 84-02.04.2013.A194). Male C57Bl/6j mice (25–30 g body weight, 10–12 weeks; Envigo, Horst, Netherlands) were kept in a 12 h–12-h light/dark cycle with free access to food and water in groups of five animals per cage. In total 21 mice were used for the results in this study. For the optimization of the clearing procedure, sets of healthy mice were used. In a subsequent step, mice exposed to focal cerebral ischemia were evaluated. As exclusion criteria, mice were removed from the study when suffering from respiratory abnormalities or from severe motor handicaps resulting in a weight loss>20%. One out of eight ischemic mice was excluded by these criteria.
Focal cerebral ischemia
Middle cerebral artery occlusion (MCAO) using an intraluminal filament technique26 was performed as reported previously, with small modifications. Briefly, male C57Bl/6j mice were anaesthetized using 1% isoflurane (30% O2 and remainder N2O), while body temperature was maintained between 36.5 and 37.0℃ using a feedback heating system (Fluovac, Harvard Apparatus, Holliston, MA, U.S.A.). An incision at the neck midline was performed to isolate the left common and external carotid arteries. The internal carotid artery was clipped for a short time with a microvascular clip. To occlude the middle cerebral artery, a silicon resin-coated nylon monofilament was introduced through a small cut in the left common carotid and advanced to the left internal carotid artery until reaching the origin of the left middle cerebral artery (MCA) at the circle of Willis. MCAO lasted 45 min. Blood supply was restored by withdrawal of the monofilament to allow immediate reperfusion. Laser Doppler flow recordings were done to monitor the reperfusion for up to 20 min after initiation. After the surgery, wounds were sealed, the anesthesia was finished and animals were placed in a warming cabinet (37℃) for approximately 1 h to recover. All operated animals received analgetics (4 mg/kg carprofen, 0.1 mg/kg buprenorphin) before being placed back in their cages and sacrificed seven days later.
Hydrogel preparation
For accurate quantification of microvessels, we ideally required a fluorescent dye solution with low viscosity for efficient penetration of microvessels, that would retain a strong fluorescent signal with the use of organic solvents, and would not result in excessively high background fluorescence in the sample due to escape from damaged blood vessels. This technical challenge was solved by the use of a gel as reported previously2 with slight modifications. Briefly, a solution of 2% (w/v) gelatin (Sigma-Aldrich, Deisenhofen, Germany) was prepared in phosphate-buffered saline (PBS) (PBS tablets, Merck-Millipore, Darmstadt, Germany) at 60℃ and allowed to cool down to 40℃ with constant stirring. Then, FITC-conjugated albumin (Sigma-Aldrich) was added to the gelatin solution at a concentration 0.1% (w/v). The gel was filtered using filter paper (GE Whatman, Dassel, Germany) and continuously stirred at 30℃ to avoid excessive evaporation.
Animal sacrifice and hydrogel perfusion
Mice were deeply anesthetized by an intraperitoneal injection of 7% chloral hydrate and transcardially perfused with 40 mL of PBS containing 50 U/mL heparin (Ratiopharm, Ulm, Germany), followed by perfusion of 20 mL of 4% paraformaldehyde (PFA) (Merck-Millipore) in PBS. Thereafter, 10 mL of the hydrogel was perfused and each mouse immediately submerged head down into ice water for over 15 min to promote the solidification of the hydrogel. The brains were carefully removed and incubated in 4% PFA in PBS at 4℃ overnight.
Whole brain clearing
For clearing adult mouse brains perfused with hydrogel, we adapted the 3DISCO clearing technique,8,9 which combines the use of two organic solvents, i.e., tetrahydrofuran (THF) (Sigma-Aldrich) for dehydration and lipid solvation and dibenzyl ether (DBE) (Sigma-Aldrich) for matching the refractive index of the remaining dehydrated sample. We optimized THF gradients (30%, 60%, 80% and 100%) using different incubation times at room temperature with constant agitation at 300 r/min using a horizontal shaker under a laminar flow hood. To ensure complete dehydration, samples were immersed in solutions of the last THF gradient (100%) twice. Then, samples were incubated in DBE for 12 h with continued agitation and stored in this solvent until image acquisition. All incubation steps were done in 30 mL of each solvent in dark brown glass bottles.
Transmittance and brain length measurements
The light transmittance of cleared whole brains was measured from transmitted light images acquired on a BX51 upright microscope (Olympus, Tokyo, Japan) using a 1.25 × 0.04 NA Planapo N objective and a DP70 1.5 Megapixel color CCD camera. Transmittance values were calculated by dividing the transmitted light intensity from the central portion of the brains by the incident light intensity measured in the region immediately surrounding the brains.
The length of brains was measured from transmitted light images acquired on an inverted AxioObserver microscope (Zeiss, Jena, Germany) using a 1.25 ×0.04 NA Planapo N objective and Orca Flash sCMOS monochrome camera. From transmitted light images, the brains were measured along their length and across their width at the widest point, before and after clearing in order to calculate the percentage reduction in size.
Light sheet data acquisition and image processing
Cleared whole mouse brains were imaged using a commercial light sheet ultramicroscope (LaVision BioTec, Bielefeld, Germany) based around an Olympus MVX10 zoom microscope which, when combined with a 2 × 0.5 NA objective, gave a magnification range from 1.26 to 12.6. The ultramicroscope was equipped with bidirectional light sheet illumination, and an Andor Neo sCMOS camera having a 2560 × 2160 chip of 6.5 µm pixel size. We performed serial optical imaging of the brains in a ventral-dorsal direction by exciting the FITC-albumin perfused vessels using a 488 nm diode laser, and detecting via a 525/50 nm bandpass filter. Images of the entire mouse brains were acquired at 1.6 × magnification in the axial direction with 6 µm steps. On average, measurement of one brain required approximately 850 images, resulting in an acquisition time of about 10 min. For image rendering of the brains, Bitplane software (Imaris, Cologne, Germany) was used.
Vascular quantification
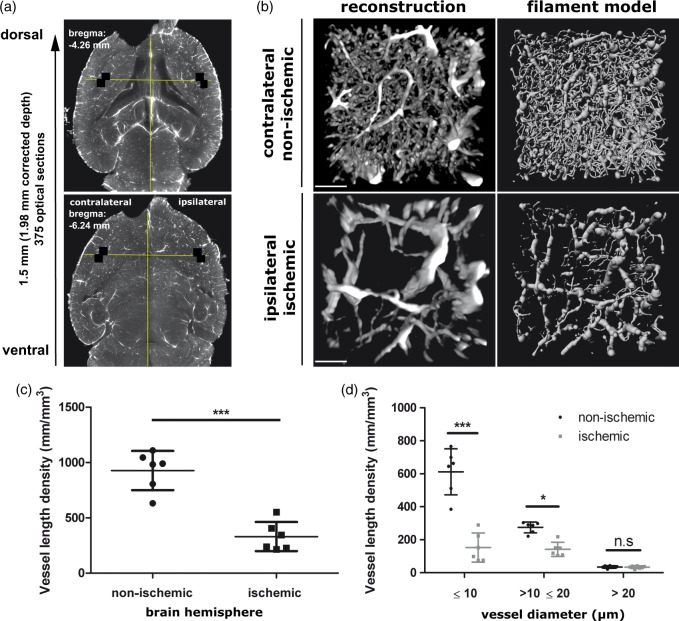
For detailed vascular quantification in stroke brains, two image stacks were taken in matching ischemic (ipsilateral) and non-ischemic (contralateral) areas within the lateral-caudal portion of the striatum, which were acquired with 6.4 × magnification and a step size of 4 µm in a ventro-dorsal direction, starting at bregma−6.24 mm and ending at bregma −4.26 mm (http://www.mbl.org/atlas232/atlas232_frame.html), resulting in 375 images that were acquired over about 3 min per image stack. From each of these image stacks, two adjacent regions of interest (ROI) measuring 508 × 508 × 1500 µm were chosen at the dorsolateral pole of the striatum at the border of the external capsule. These ROIs were defined from an overview image acquired at bregma −6.24 mm by drawing a guide line, orthogonal to the brain midline, crossing through the opening at which the lateral ventricles are connected with the third ventricle. The point, at which this line crossed the border between the striatum and external capsule, marked the position of the ROIs (see also Figure 5(a)). In the ROIs, blood vessel diameter, total vascular length and total vascular volume were measured after network modeling using the filament tracer tool of the Imaris 3D rendering software package. For the filament tracer analysis, a batch process routine was defined with each ROI requiring 31 min to compute on an imaging workstation with 8 core processors (3.3 GHz Intel Xeon E5-2643) and 128 GB RAM.
Figure 5.
3D rendering, filament modeling and quantification of blood vessels in the ischemic brain. (a) Horizontal sections across the cleared ischemic brain at −6.24 mm and −4.26 mm bregma. Between these levels, image stacks at 6.4 × magnification were acquired in the ipsilesional and contralesional striatum (i.e. the core of the middle cerebral artery territory) for a detailed quantitative analysis of microvessels. Within these image stacks the position of the ROIs (508 × 508 µm, labeled with white squares) were selected from the overview image at −6.24 mm bregma by drawing a guide line orthogonal to the midline that passed through the center of the connection of the left and right lateral ventricles with the third ventricle. The ROIs were placed in the most lateral striatum, where it faces the external capsule. (b) 3D vessel rendering and filament tracer model of blood vessels taken from the 508 × 508 × 1500 µm ROI in ipsilateral ischemic and contralateral non-ischemic brain tissue. Reconstructions show a transverse view looking down through a 250 µm slice of the rendered vessels and filament model (contrast stretched with a γ of 3.0). Effect of focal cerebral ischemia on (c) total vasculature length density, and (d) the vasculature length density stratified by vascular diameter. Note that focal cerebral ischemia most markedly affects capillaries with a diameter ≤ 10 µm. Vessels with diameter > 20 µm remain unaffected. Scale bar in (b), 100 µm. Plots in (c) and (d) show mean ± SD values. ***p < 0.001, **p < 0.01, *p < 0.05 compared with corresponding non-ischemic tissue.
Image preprocessing and data analysis
For the detailed vascular analysis, image stacks were preprocessed using open source software ImageJ (National Institutes of Health, Bethesda, MD, U.S.A.) and the Vascular Modelling Toolkit (VMTK:www.vmtk.org). In ImageJ, image stacks were first Gaussian smoothed (Sigma = 2 µm) before performing a rolling ball background subtraction (radius = 20 µm). In VMTK vessel, enhancement was applied using the multiscale Frangi vesselness filter.27 For batch preprocessing of image stacks, a script was written in the ImageJ macro language and VMTK scripts were piped together.
All the preprocessing and filament model parameters for the analysis were first optimized using image data from the contralateral region of a typical cleared mouse brain. Once the parameters were established, their values were fixed and used to analyze all remaining brain regions, both ipsi- and contralateral, in a batch process.
Both the GNU Image Manipulation Program (GIMP; open source software; https://www.gimp.org) and Inkscape (open source software; https://inkscape.org) were used for figure preparation. Illustrations were also prepared using the Servier Medical Art Powerpoint Image Bank (Servier, Suresnes, France; http://www.servier.com/Powerpoint-image-bank). Graphs were done using Graphpad Prism 5.0.
Statistical analysis
All data values are given as means ± SD. Data were analyzed using paired Student’s t tests to determine significant differences of vessel length and vascular volume between ischemic and non-ischemic brain areas and one-way ANOVA with Tukey’s test for multiple comparisons. P values of ≤ 0.05 were considered significant. Statistical analysis was performed using Graphpad Prism 5.0.
Results
Optimization of clearing technique for labeling and imaging blood vessels in the whole mouse brain
With the aim of visualizing and quantifying microvessels in whole mouse brains, we developed a straight forward technique that combined a transcardial perfusion of a low viscosity fluorescent hydrogel, composed of gelatin and FITC-conjugated albumin,2 with a slightly modified 3DISCO clearing procedure8,9 and light sheet microscopy (Figure 1). The hydrogel remains liquid at and above 30℃, which allowed the efficient filling of cerebral blood vessels during the transcardial perfusion. To prevent, or limit, leakage from blood vessels damaged by ischemia, perfused mice were submerged into ice to encourage the rapid solidification of the hydrogel within the confines of the vasculature. This was confirmed in an ischemic brain first perfused with an Alexa-647 labeled CD31 antibody, followed by a FITC-albumin hydrogel (see Supplementary Methods section). Here, in contrast to the FITC-albumin, a distinct background from CD31 leakage could be seen in the ischemic region (Supplementary Figure 1). Indeed, for all the brain samples prepared, there was no visual evidence of FITC-albumin leakage into ischemic regions. Furthermore, in this co-perfused brain, excellent co-labelling of vessel structures was observed, both in ischemic and non ischemic regions, demonstrating that the FITC-albumin hydrogel was able to perfuse the same vessel structures as the endothelial cell marker CD31 (Supplementary Figure 2 and Supplementary Table 1). In our hands, a concentration of 0.1% FITC-conjugated albumin was sufficient for a strong visual signal in microvessels, including capillaries, and a careful filtration of the hydrogel avoided the formation of aggregates that could prevent adequate vessel filling.
Figure 1.
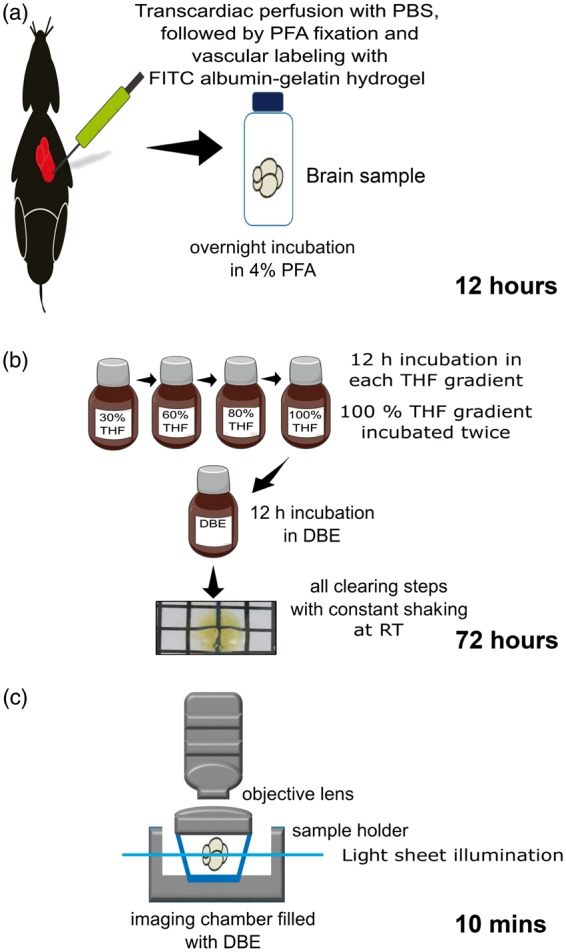
Protocol for 3D blood vessel visualization in whole mouse brains. (a) Sample preparation: During animal sacrifice mice are transcardially perfused with 40 mL of phosphate-buffered saline (PBS) containing 100 U/mL heparin, followed by 20 mL of 4% paraformaldehyde (PFA) in PBS and 10 mL hydrogel composed of 2% gelatin and 0.1% FITC-conjugated albumin in PBS. (b) Whole brain clearing: In the optimized protocol, the brain specimen is incubated in each clearing solution (tetrahydrofuran (THF) used for dehydration and lipid solvation, dibenzyl ether (DBE) for matching of refractive index) for 12 h at room temperature with constant shaking in dark bottles. To ensure complete dehydration, the 100% THF step was performed twice. (c) Light sheet microscopy: Cleared brains immersed in DBE were imaged in a dorso-ventral direction using either 1.6 × or 6.4 × magnification and corresponding step sizes of 6 µm and 4 µm in the LaVision Biotec ultramicroscope.
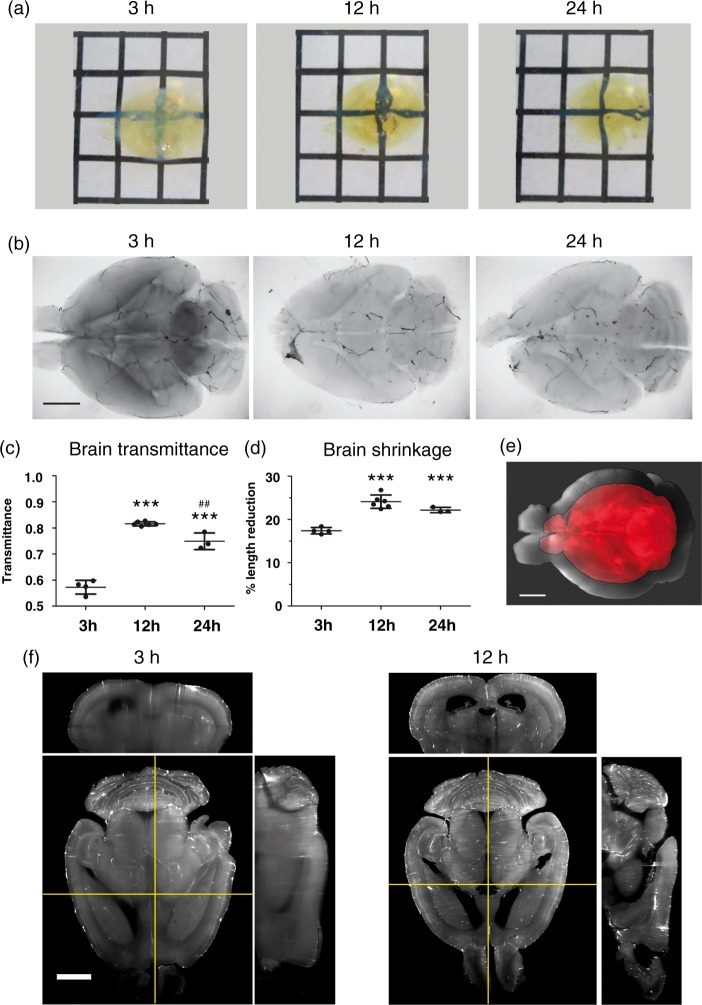
For optimizing the dehydration of brain samples and tissue clearing using the 3DISCO protocol, we systematically compared a variety of incubation times in THF gradients (30%, 60%, 80%, 100%), i.e. 3 h, 12 h or 24 h for each concentration step (Figure 1). From a visual inspection, brain transparency was much higher after incubation over 12 h or 24 h than 3 h (Figure 2(a)), as further evident in transmitted light images (Figure 2(b)). Here, the central portions of the brain were dark after 3 h relative to 12 h or 24 h incubation due to light loss by scattering in insufficiently cleared tissue portions. From the transmitted light images, the light transmittance in the most central brain regions was calculated (Figure 2(c)). Most strikingly, a relatively large and significant improvement in the transmittance by ∼40% was observed when brains were incubated over 12 h instead of 3 h. Importantly, 24 h incubation did not further increase light transmittance.
Figure 2.
Optimization of the clearing procedure of FITC-albumin gelatin hydrogel perfused whole mouse brains. (a) Cleared whole mouse brains following incubation in THF gradients for 3 h, 12 h or 24 h and DBE. Note the different degrees of transparency against the black and white grid pattern in the background. The distance between grid lines equals 5 mm. (b) Transmitted light images of representative cleared brains following incubation in THF gradients for 3 h, 12 h or 24 h and DBE. Note that transparency in the central parts of the brain is better after 12 h or 24 h than after 3 h incubation with THF. (c) Transmittance of whole brains as a function of incubation time. Transmittance was calculated from transmitted light images of brains incubated over 3 h, 12 h or 24 h of THF steps and DBE. Note that light transmittance is best after 12 h incubation. (d) Percentage brain length reduction as a function of incubation time. Note that the better transmittance after 12 h incubation compared to 3 h incubation is not achieved at the expense of major additional brain shrinkage. (e) Image of brain cleared with 12 h incubation steps (red) overlaid on the same fixed brain before clearing (grey). (f) Transverse light sheet images and orthogonal images viewed along the coronal (horizontal yellow lines, upper image panels) and median sagittal planes (vertical yellow lines, right image panels), of cleared brains. Image stacks were acquired in the Ultramicroscope with 1.6 × magnification. Note the enhanced spatial resolution in deep brain structures (e.g., lateral ventricles) after 12 h compared with 3 h incubation. Scale bars in (b), (e) and (f), 2 mm. Plots in (c) and (d) show mean ± SD values. ***p < 0.001 compared with 3-h incubation, ##p < 0.01 compared with 12-h incubation.
Since tissue deformation has previously been described in the 3DISCO protocol,8,9 we next evaluated the percentage of tissue shrinkage in the samples by comparing the measured length of each fixated brain before and after clearing (Figure 2(d) and (e)). Although there was a large improvement in the clearing efficiency after 12 h instead of 3 h incubation, there was little relative penalty in terms of tissue shrinkage. Most of the tissue shrinkage had occurred within 3 h incubation time, with only modest additional shrinkage at 12 h and no additional shrinkage at 24 h.
The increased clearing efficiency after 12 h incubation went along with a marked improvement in the resolution of fine structures in images acquired on the ultramicroscope, as shown in Figure 2(f). Differences were again most clearly observed in the central portions of the brain. Due to light scattering, structural details of larger anatomical structures in the central portions of the brain such as the brain ventricles were poorly demarcated after 3 h incubation. These structures were much more sharply resolved after 12 h incubation. Although 3 h incubation was suggested in the original 3DISCO protocols,8,23 in particular when working with fluorescent proteins, we found this insufficient for imaging of brain capillaries. Therefore, incubation times of 12 h were used for all further studies and a length correction factor of 1.318 was used to correct the metrics for the detailed vascular analysis.
3D visualization of vessels in the whole mouse brain
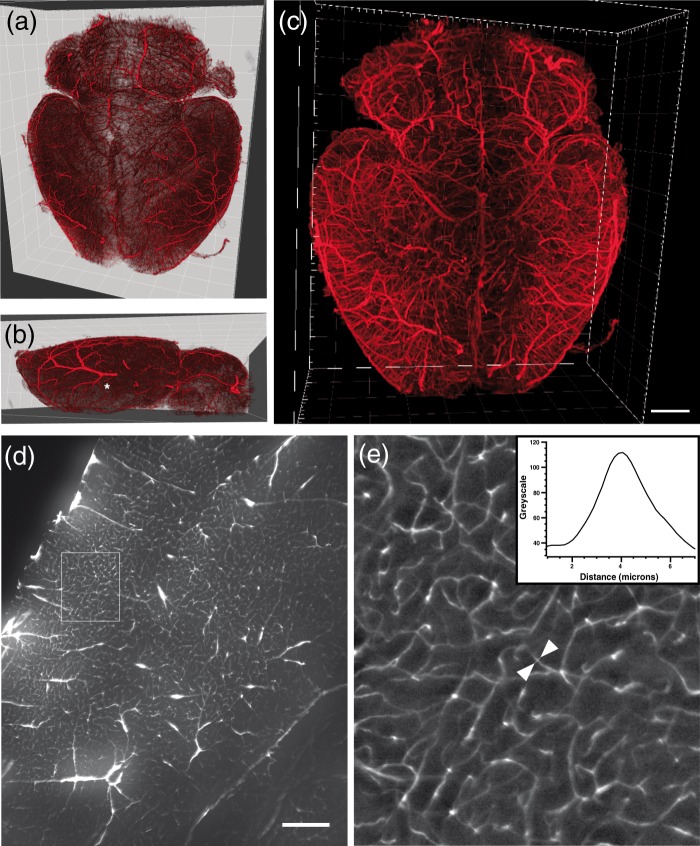
By applying the perfusion and clearing procedure described, a detailed 3D visualization of the whole brain vasculature could be performed at 1.6 × magnification from image stacks acquired in the ultramicroscope. Following volume rendering, these images are shown in blend mode, to highlight surface features, and maximum projection intensity mode in Figure 3(a) to (c). With appropriate knowledge of cerebrovascular anatomy, the anterior, middle and posterior cerebral arteries could easily be identified (MCA labeled with an asterisk (*) in Figure 3(b)). Figure 3(c) shows that detailed vascular visualization could be obtained even in the most central part of the brain. Although brain capillaries were also clearly visible at 1.6 × magnification, they were only sufficiently digitally sampled (pixel size: 1.016 µm) for subsequent analysis when imaged at 6.4 × magnification (Figure 3(d)). At closer inspection (6 × digital magnification of ROI in Figure 3(d) shown in Figure 3(e)), 5 µm was the smallest capillary diameter that could be measured and thus represents the optical resolution limit for our optical setup. Due to the high labelling efficiencies, however, all capillaries could be imaged with good signal contrast (a grey scale line profile across a typical capillary is shown in Figure 3(e), inset).
Figure 3.
Visualization of the vascular architecture of a healthy whole mouse brain. 3D volume rendering of brains are shown in blend mode (a, b), which accentuates the surface features (the middle cerebral artery in (b) is labeled with an asterisk (*)), or (c) in maximum projection intensity mode. Volume renderings were generated from image stacks acquired with 1.6 × magnification. (d) Transverse optical section of the striatum and adjacent cerebral cortex acquired at 6.4 × magnification. (e) Enlargement of insert in (d) exhibiting brain capillaries at higher magnification. The graph on the upper right displays a signal intensity profile across a typical capillary that was taken along a line connecting the two white arrows in the image. Note that capillaries with diameters of 5 µm are visualized with high contrast. Scale bar in (c), 1 mm; in (d), 300 µm.
Vascular reconstruction of the ischemic mouse brain
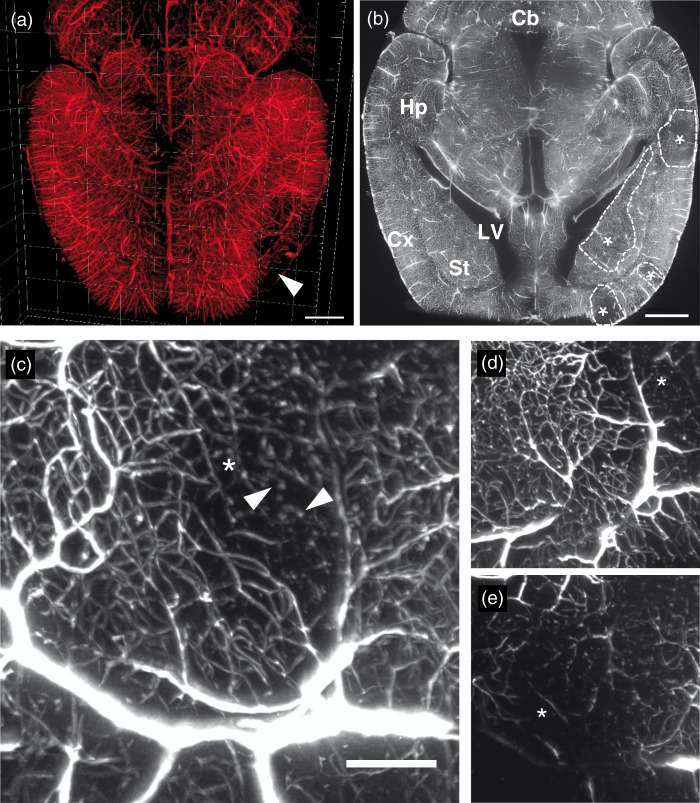
Vascular 3D reconstructions of ischemic brains were done in order to visualize the topology of brain infarcts at seven days post-stroke (Figure 4(a)). In the reconstructions, brain infarcts were characterized by a loss of FITC-albumin-stained blood vessels (Figure 4(a), white arrow). In overview images, transverse optical sections revealed better insights into the distribution of microvascular damage than 3D reconstructions, and brain structures could be identified from endogenous tissue autofluorescence (Figure 4(b); infarct boundaries outlined by dashed lines, infarcts labeled with asterisk (*)). As expected from transient MCAO, the ischemic brains showed a consistent and similar pattern of FITC-albumin signal loss centered at the lateral striatum.
Figure 4.
Visualization of the vascular architecture of an ischemic whole mouse brain. (a) 3D volume rendering of the vascular architecture in a dorsal view, showing a representative stroke that clearly affects the cortex (white arrow). Each major division in the frame of the rendered brain is equal to 1 mm. (b) Horizontal section across an ischemic brain acquired at 1.6 × optical magnification exhibiting an infarct which in this plane mainly affects the striatum. Ischemic regions are indicated with asterisks (*) with the boundaries of the stroke regions delineated by dashed lines. (c) Maximum intensity projection image of blood vessels in an ischemic brain area, taken from a 300 µm thick section in the z-direction. Areas with lack of blood vessels are shown with an asterisk (*). The white arrow indicates beads-on-a-string-like microvascular structures. Note the candelabrum-like structures with dichotomic branching in this image. (d) Maximum intensity projection of a 150 -µm z-stack section, at a position dorsal to the maximum projection image shown in (c). This region shows more intact blood vessel (asterisk shows area of the infarct). (e) Maximum intensity projection of a 150 -µm z-stack section at a position ventral to the projection in (c). In this region, the loss of blood vessels is more evident than in the previous images. Cb: cerebellum; Cx: cortex; Hp: hippocampus; LV: lateral ventricle; St: striatum. Scale bar in (a) and (b), 1 mm; contrast enhanced in (a) using γ of 3.0 and in (b) by histogram equalization; scale bar in (c), 40 µm.
Stacks of images acquired at 6.4 × magnification allowed a more detailed view of microvascular networks in and around the ischemic regions (Figure 4(c) to (e)). Ischemic tissue in the image stack revealed varying degrees of vascular damage. In particular, some microvessels exhibited candelabrum-like structures with dichotomic branching or tortuous loops (Figure 4(c)). Other microvessels filled with hydrogel appeared disconnected, as evidenced by beaded structures that followed characteristic microvascular loop contours (white arrow in Figure 4(c)). In general, however, a visual inspection of the ischemic regions suggested that the main form of vessel damage was a predominant loss of labeled microvessels.
Detailed vascular quantification in ischemic brain
To further evaluate the observation that small microvessels were most strongly affected by vascular damage, we performed a detailed analysis of vascular networks in the core of the MCA infarct, i.e. the lateral striatum, in transverse brain sections at the position where the branches of the MCA irrigate the striatum. For quantification, image stacks were acquired at 6.4 × magnification from which two ROI were selected as described in the Materials and Methods section (Figure 5(a)). The image stack acquisition was done over a distance of 1.5 mm, equivalent to a distance of 1.98 mm in an uncleared fixed mouse brain. The 3D vascular structures in each of the selected ROI (labeled with squares in Figure 5(a)) were rendered and fitted to a filament model using the filament tracer module of Imaris. Figure 5(b) shows a transverse view onto a 250 µm thick slice of the 3D rendered vasculature and the filament model from ipsilesional and contralesional ROIs, which was acquired in a typical ischemic mouse brain. Clearly from these views, the loss of vessels is preferentially confined to microvessels. From the filament models, detailed vascular metrics related to the volume, length and diameter of vessels were extracted, from which a comprehensive comparison between ischemic and contralesional non-ischemic vasculature was made in six ischemic mouse brains.
All vascular parameters reported were corrected for brain shrinkage relative to a fixed brain. After correction, the average of the vasculature length density was calculated as 0.922 ± 0.176 m/mm3 in non-ischemic brain tissue and 0.329 ± 0.131 m/mm3 in ischemic brain tissue (Figure 5(c)), corresponding to an almost 64% loss of vessel density. This loss of vessel density was also reflected in the percentage values of the vasculature volume. The total vasculature volume in non-ischemic tissue was 16.1 ± 2.5% in comparison to 9.1 ± 2.3% in ischemic tissue. Interestingly, the vessel length density for the longest connected network measured 0.915 ± 0.180 m/mm3 in non-ischemic tissue and 0.316 ± 0.132 m/mm3 in ischemic tissue, suggesting that the vast majority of the vascular volume was still covered by a single highly connected vessel network.
In order to evaluate the differential effect of ischemia on vessel patency depending on vessel size, the vessel length density was further categorized into microvessels with a diameter ≤ 10 µm (i.e. capillaries), vessels with a diameter range > 10 and ≤ 20 µm, and vessels with a diameter > 20 µm. In the non-ischemic tissue, the average length density was 0.612 ± 0.139 m/mm3 for capillaries ≤ 10 µm, 0.275 ± 0.033 m/mm3 for vessels> 10 and ≤ 20 µm, and only 0.035 ± 0.008 m/mm3 for vessels > 20 µm (Figure 5(d)), indicating that there was two times the length of capillaries ≤ 10 µm compared to larger vessels; capillaries accounting for 66.4% of the total length of all blood vessels. In ischemic tissue, the average length density was 0.153 ± 0.088 m/mm3 for capillaries ≤ 10 µm, 0.142 ± 0.043 m/mm3 for vessels measuring > 10 and ≤ 20 µm, and 0.034 ± 0.009 m/mm3 for vessels > 20 µm (Figure 5(d)), indicating an almost selective loss of capillaries, which accounted for only 46.5% of the total vessel length, with vessels > 20 µm remaining unaffected.
Discussion
We herein have developed a powerful technique for 3D imaging and automated quantification of the blood vasculature in whole mouse brains using a modified 3DISCO clearing procedure with extended clearing steps as compared to original protocols,8,9 which allowed us to visualize fine brain capillaries even in the central parts of the brain. This clearing technique was combined with an injection of FITC-albumin gelatin hydrogel, which enabled cerebral vessels and capillaries to be reliably imaged with high contrast and low background signal.
Light sheet imaging using an ultramicroscope enabled evaluation of the fine structure of cerebral microvessels and capillaries with an optical resolution of 5 µm. Due to the strong fluorescence signal of the hydrogel, even the smallest capillaries could be visualized with high signal contrast on digital images with a pixel size of 1.016 µm. Consequently, microvessels were fitted to a filament model, allowing us to perform a comprehensive analysis of the vasculature in whole mouse brain specimens. In non-ischemic striatum, we determined an average vasculature length density of 0.922 ± 0.176 m/mm3, which is in excellent agreement with previous reports using manual tracing on thin histological sections (i.e. 0.7–1.2 m/mm3)2,28 and with 3D measurements on thick histological sections (i.e. 0.88 m/mm3 in the mouse cortex)2. In the intact striatal region of the mouse brain, the vascular volume was also quantified and found to occupy 16.1 ± 2.5% of the total volume. This value is a factor of three higher than commonly reported volumes for mice using imaging techniques.29–31 A factor of three would imply a 77% overestimate of the vessel radius. This could, in part, be explained by the limited optical resolution of the light sheet, especially if a reasonable proportion of the microvessels has a diameter less than 5 µm. Indeed, the brain microvessel diameter has previously been determined by confocal microscopy in C57Bl/6 mice, where the minimum diameter was 3.97 to 4.11 µm in the cerebral cortex.2 Using FITC-dextran-perfused sections, the minimum microvasculature diameter in rat cerebral cortex was determined to be 4.15 ± 1.24 µm,12 whereas in corrosion casts stained with Indian ink, the minimum diameter of microvessels in rat cerebral cortex ranged from 3 to 8 µm.32 It should be noted that we measured a brain region particularly rich in capillaries.
We have also, for the first time, visualized and quantified the rarefication of vessels in the ischemic brain using solvent clearing combined with ultramicroscopy. In the ischemic striatum, i.e. the core of the MCA territory, the vasculature length density was reduced to 0.329 ± 0.131 m/mm3 and vascular volume ratio was 9.1 ± 2.3%. The longest connected network measured 0.316 ± 0.132 m/mm3, suggesting that the vast majority of the vascular volume was still interconnected. By categorizing vessel diameters, we were able to evaluate the loss of vasculature length density as a function of vessel size. This analysis revealed a highly selective loss of brain capillaries ≤ 10 µm that were reduced by 75%. Microvessels in the range > 10 and ≤ 20 µm were reduced to lesser extent (by 48%), while vessels > 20 µm were unaffected by focal cerebral ischemia. Our data are in line with observations of del Zoppo et al.34 after transient MCA occlusion in baboons, showing that the no reflow phenomenon mainly affects brain capillaries in focal cerebral ischemia. In their study, the number of capillaries in the striatum with a diameter of 4.0–7.5 µm was reduced by 27–39%, whereas larger vessels with a diameter of up to 60 µm were mildly decreased (not quantified). Differences of the stroke model (3 h MCAO by inflation of a balloon vs. 45 min MCAO by monofilament) or the vascular collateral density in baboons and mice might explain why slightly larger microvessels were also affected in their study.
It is well known that the blood–brain barrier is impaired following reperfusion, resulting in the leakage of blood constituents, in particular from capillaries.33 The potential consequences of this leakage for this study are twofold. Firstly, leakage should reveal itself in the form of an increased background signal around the vessels in the ischemic region. For the reliable automatic quantification of ischemic versus non ischemic vessels, the differential deterioration in the vessel signal contrast due to elevated background signal should be avoided or minimized. Secondly, leaky or non-functional vessels in the ischemic region may restrict the labeling of smaller vessels or prevent the labeling of larger parts of the vessel network. To address these issues, a cleared brain was prepared from an ischemic mouse perfused first with an Alexa-647 labeled CD31 antibody and then a FITC-albumin hydrogel. In the ischemic region of the brain, a clear and distinct background signal was observed from the CD31 endothelial cell marker, demonstrating that vessels in this region were indeed impaired and leaky to the CD31 perfusion. An equivalent background signal was not observed in the FITC-albumin hydrogel image, nor has a background been observed for FITC-albumin in any of the brains we have so far prepared. Consequently, with FITC-albumin, a strong and equivalent signal contrast, even of capillaries, can be observed in both ischemic and non-ischemic brain regions. From close analysis of the co-labeled mouse brain, we also showed that the FITC-albumin and Alexa-647 CD31 antibody were labeling the same vessel structures in the non-ischemic hemisphere of the brain, confirming that the viscosity of the hydrogel was low enough to completely fill the intact brain vasculature. This also agreed with measurements in healthy transgenic mice by Tsai et al.,2 where the FITC-albumin hydrogel was co-localized with an endogenous fluorescent protein marker of endothelial cells. In the ischemic region, where the vessels are leaky, we also observed a high degree of co-labeling of vessel structures and estimates of the vessel length densities showed CD31 label in excellent agreement with FITC-albumin, also confirming the dramatic loss of vessel density in this region. We cannot of course entirely rule out the possibility that neither the CD31 nor the FITC-albumin labels all compromised vessels in the ischemic region. However, while it could be envisaged that the perfusion of CD31 (and indeed FITC-albumin) through the vascular structure may be mostly restricted to non-compromised vessels, we would still expect to see the additional labeling of non-perfused vessels from anti-CD31 antibody leaking into the background environment. In the ischemic region, this would result in CD31 labeling more vessels than those observed with the FITC-albumin. We did not see any evidence of this in our study. Despite this, a close inspection of 3D rendering showed that in the ischemic region, some microvessels appeared filled with hydrogel in a beads-on-a string-like structure that followed characteristic loops of microvessels. We hypothesize that this phenomenon of dotted connection may represent microvessels occluded by polymorphonuclear leukocytes (PMN), which have previously been shown to provoke no reflow phenomena after focal cerebral ischemia34. Alternatively, they could also represent newly formed immature microvessels. Immunohistochemistry against markers of proliferating endothelial cells or PMN might allow the discrimination of both processes. In conclusion, with the FITC-albumin hydrogel, a comparison of functional blood vessels could be reliably made and major problems related to incomplete vascular filling or background labeling of brain parenchyma2–5,9,20,25,26 circumvented. Notably, the fluorescence of the hydrogel was not compromised after solvent clearing, and samples could be stored over months without significant loss of fluorescent signal or signal contrast.
We have shown that a modified 3DISCO protocol with 12 h incubation steps results in excellent clearing for whole mouse brains and does not impair the fluorescence signal from the FITC-albumin hydrogel, enabling the study of vascular fine structure deep into the brain. However, for combining studies of the vascular network with endogenous fluorescent protein labels a recently published update to the 3DISCO protocol could also be adapted, in particular if it is necessary to image larger specimens such as rat brains.36 Alternatively, if the toxicity of the clearing medium is a concern, another recent publication using non-toxic solvent-based dehydration and clearing agents, which also maintains fluorescent protein signal, could be explored.37
Based on the utility of light sheet microscopy for determining the characteristics of microvascular networks in whole mouse brains, we predict that light sheet microscopy will be highly valuable for evaluating vascular injury and remodeling in the stroke field. Given the non-biased nature of automated analyses, allowing detailed metrics of various morphological aspects of cerebral microvessels, this method offers itself for the analysis of responses to vascular protective and angiogenic therapies.
Funding
The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was supported by the German Research Council [HE3173/2-2 and HE3173/3-1; to D.M.H.; Priority Program “Immunobone”, to M.G; and FA159/20-1 and FA159/22-1, to A.F.] and by EU programmes (FP7 MATHIAS and EU H2020 MULTIMOT, to M.G.).
Supplementary Material
Acknowledgments
We thank Michael Wessolly for assisting us with image collection. ELH received funding from the University of Carabobo, Venezuela, and from the Wilhelm und Günter Esser Foundation.
Declaration of conflicting interests
The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Authors’ contributions
ELH, DMH, AF and MG designed the study. JS, NH and EHSM performed the animal surgeries. ELH, AB, NH and MS prepared the tissue and optimized the clearing procedure. AB and AS established the lightsheet imaging protocol. AB, AS and NH contributed to the image acquisition. AS performed the image analysis including computational analysis. ELH wrote the manuscript, and DMH, AS, AF and MG revised it. All authors read and approved the final manuscript.
Supplementary material
Supplementary material for this paper can be found at the journal website: http://journals.sagepub.com/home/jcb
References
- 1.Hermann DM, Zechariah A. Implications of vascular endothelial growth factor for postischemic neurovascular remodeling. J Cereb Blood Flow Metab 2009; 29: 1620–1643. [DOI] [PubMed] [Google Scholar]
- 2.Tsai PS, Kaufhold JP, Blinder P, et al. Correlations of neuronal and microvascular densities in murine cortex revealed by direct counting and colocalization of nuclei and vessels. J Neurosci 2009; 29: 14553–14570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hermann DM, Chopp M. Promoting brain remodelling and plasticity for stroke recovery: Therapeutic promise and potential pitfalls of clinical translation. Lancet Neurol 2012; 11: 369–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Meng H, Peng Y, Hasan R, et al. Nuclear contrast angiography: A simple method for morphological characterization of cerebral arteries. Brain Res 2009; 1261: 75–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Maeda K, Hata R, Hossmann KA. Differences in the cerebrovascular anatomy of C57black/6 and SV129 mice. Neuroreport 1998; 9: 1317–1319. [DOI] [PubMed] [Google Scholar]
- 6.Hasan MR, Herz J, Hermann DM, et al. Visualization of macroscopic cerebral vessel anatomy – A new and reliable technique in mice. J Neurosci Methods 2012; 204: 249–253. [DOI] [PubMed] [Google Scholar]
- 7.Robertson RT, Levine ST, Haynes SM, et al. Use of labeled tomato lectin for imaging vasculature structures. Histochem Cell Biol 2015; 143: 225–234. [DOI] [PubMed] [Google Scholar]
- 8.Ertürk A, Becker K, Jährling N, et al. Three-dimensional imaging of solvent-cleared organs using 3DISCO. Nat Protoc 2012; 7: 1983–1995. [DOI] [PubMed] [Google Scholar]
- 9.Ertürk A, Lafkas D, Chalouni C. Imaging cleared intact biological systems at a cellular level by 3DISCO. J Vis Exp JoVE 2014; 89: 51382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sarkar S, Schmued L. In vivo administration of fluorescent dextrans for the specific and sensitive localization of brain vascular pericytes and their characterization in normal and neurotoxin exposed brains. Neurotoxicology 2012; 33: 436–443. [DOI] [PubMed] [Google Scholar]
- 11.Hoffmann A, Bredno J, Wendland M, et al. High and low molecular weight fluorescein isothiocyanate (FITC)–dextrans to assess blood-brain barrier disruption: Technical considerations. Transl Stroke Res 2011; 2: 106–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang ZG, Zhang L, Tsang W, et al. Correlation of VEGF and angiopoietin expression with disruption of blood–brain barrier and angiogenesis after focal cerebral ischemia. J Cereb Blood Flow Metab 2002; 22: 379–392. [DOI] [PubMed] [Google Scholar]
- 13.Sarkar S, Raymick J, Paule MG, et al. In situ demonstration of Fluoro-Turquoise conjugated gelatin for visualizing brain vasculature and endothelial cells and their characterization in normal and kainic acid exposed animals. J Neurosci Methods 2013; 219: 276–284. [DOI] [PubMed] [Google Scholar]
- 14.Miyata S, Morita S. A new method for visualization of endothelial cells and extravascular leakage in adult mouse brain using fluorescein isothiocyanate. J Neurosci Methods 2011; 202: 9–16. [DOI] [PubMed] [Google Scholar]
- 15.Gleave JA, Lerch JP, Henkelman RM, et al. A method for 3D immunostaining and optical imaging of the mouse brain demonstrated in neural progenitor cells. PLoS ONE 2013; 8: e72039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dodt H-U, Leischner U, Schierloh A, et al. Ultramicroscopy: Three-dimensional visualization of neuronal networks in the whole mouse brain. Nat Methods 2007; 4: 331–336. [DOI] [PubMed] [Google Scholar]
- 17.Sharpe J, Ahlgren U, Perry P, et al. Optical projection tomography as a tool for 3D microscopy and gene expression studies. Science 2002; 296: 541–545. [DOI] [PubMed] [Google Scholar]
- 18.Weninger WJ, Geyer SH, Mohun TJ, et al. High-resolution episcopic microscopy: A rapid technique for high detailed 3D analysis of gene activity in the context of tissue architecture and morphology. Anat Embryol 2006; 211: 213–221. [DOI] [PubMed] [Google Scholar]
- 19.Ragan T, Kadiri LR, Venkataraju KU, et al. Serial two-photon tomography for automated ex vivo mouse brain imaging. Nat Methods 2012; 9: 255–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Verveer PJ, Swoger J, Pampaloni F, et al. High-resolution three-dimensional imaging of large specimens with light sheet-based microscopy. Nat Methods 2007; 4: 311–313. [DOI] [PubMed] [Google Scholar]
- 21.Santi PA, Johnson SB, Hillenbrand M, et al. Thin-sheet laser imaging microscopy for optical sectioning of thick tissues. BioTechniques 2009; 46: 287–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jährling N, Becker K, Dodt H-U. 3D-reconstruction of blood vessels by ultramicroscopy. Organogenesis 2009; 5: 227–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ertürk A, Mauch CP, Hellal F, et al. Three-dimensional imaging of the unsectioned adult spinal cord to assess axon regeneration and glial responses after injury. Nat Med 2012; 18: 166–171. [DOI] [PubMed] [Google Scholar]
- 24.Hama H, Kurokawa H, Kawano H, et al. Scale: A chemical approach for fluorescence imaging and reconstruction of transparent mouse brain. Nat Neurosci 2011; 14: 1481–1488. [DOI] [PubMed] [Google Scholar]
- 25.Susaki EA, Tainaka K, Perrin D, et al. Whole-brain imaging with single-cell resolution using chemical cocktails and computational analysis. Cell 2014; 157: 726–739. [DOI] [PubMed] [Google Scholar]
- 26.Kilic E, Spudich A, Kilic U, et al. ABCC1: A gateway for pharmacological compounds to the ischaemic brain. Brain 2008; 131: 2679–2689. [DOI] [PubMed] [Google Scholar]
- 27.Frangi AF, Niessen WJ, Vincken KL, et al. Multiscale vessel enhancement filtering. In: Wells WM, Colchester A and Delp S (eds) Medical image computing and computer-assisted interventation — MICCAI’98. Berlin: Springer Berlin Heidelberg, 1998, pp.130–137.
- 28.Boero JA, Ascher J, Arregui A, et al. Increased brain capillaries in chronic hypoxia. J Appl Physiol 1999; 86: 1211–1219. [DOI] [PubMed] [Google Scholar]
- 29.Heinzer S, Krucker T, Stampanoni M, et al. Hierarchical microimaging for multiscale analysis of large vascular networks. NeuroImage 2006; 32: 626–636. [DOI] [PubMed] [Google Scholar]
- 30.Heinzer S, Kuhn G, Krucker T, et al. Novel three-dimensional analysis tool for vascular trees indicates complete micro-networks, not single capillaries, as the angiogenic endpoint in mice overexpressing human VEGF165 in the brain. NeuroImage 2008; 39: 1549–1558. [DOI] [PubMed] [Google Scholar]
- 31.Serduc R, Vérant P, Vial J-C, et al. In vivo two-photon microscopy study of short-term effects of microbeam irradiation on normal mouse brain microvasculature. Int J Radiat Oncol 2006; 64: 1519–1527. [DOI] [PubMed] [Google Scholar]
- 32.Miyoshi Y, Date I, Ohmoto T. Three-dimensional morphological study of microvascular regeneration in cavity wall of the rat cerebral cortex using the scanning electron microscope: Implications for delayed neural grafting into brain cavities. Exp Neurol 1995; 131: 69–82. [DOI] [PubMed] [Google Scholar]
- 33.Hermann DM, ElAli A. The abluminal endothelial membrane in neurovascular remodeling in health and disease. Sci Signal 2012; 5: re4. [DOI] [PubMed] [Google Scholar]
- 34.del Zoppo GJ, Schmid-Schönbein GW, Mori E, et al. Polymorphonuclear leukocytes occlude capillaries following middle cerebral artery occlusion and reperfusion in baboons. Stroke J Cereb Circ 1991; 22: 1276–1283. [DOI] [PubMed] [Google Scholar]
- 35.Coyle P, Jokelainen PT. Dorsal cerebral arterial collaterals of the rat. Anat Rec 1982; 203: 397–404. [DOI] [PubMed] [Google Scholar]
- 36.Pan C, Cai R, Quacquarelli FP, et al. Shrinkage-mediated imaging of entire organs and organisms using uDISCO. Nat Methods 2016; 13: 859–867. [DOI] [PubMed] [Google Scholar]
- 37.Klingberg A, Hasenberg A, Ludwig-Portugall I, et al. Fully automated evaluation of total glomerular number and capillary tuft size in nephritic kidneys using lightsheet microscopy. J Am Soc Nephrol 2017; 28: 452–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.