Introduction
Tree species compositions are altered in forest ecosystems as a result of species interactions, natural or artificial disturbances and human interferences (Frehlich, 2016). The issue of local species changes has recently received increased attention in the context of climate change, but human interferences cannot be dismissed as they can hardly be disentangled from climatic disturbances and species interactions. There are numerous reviews suggesting that modifications of temperature and precipitation reshape tree distributions (Ruis-Labourdette et al., 2012; Pucko et al., 2012; Ozolinčius et al., 2014), or change forest succession (Laflower et al., 2016). These concerns have triggered research in modeling approaches aiming at predicting species distributions under contrasting climatic scenarios (Iverson, 2008; Cheaib et al., 2012; Wang et al., 2016). In contrast to the comprehensive research going on in modeling approaches, little evidence is available from field observations of tree species composition. Indeed, in situ assessment of recent shifts of tree distributions are very limited under lowland conditions (Walther et al., 2005; Delzon et al., 2012) while a few more studies focused on elevational gradients (Kullman, 2002; Peñuelas & Boada, 2003; Lenoir et al., 2008; Bodin et al., 2013). Such observations are usually based on the comparison of vegetation inventories over several decades, which do not allow to disentangle the various causes of differential migration of species (but see Monleon & Lintz, 2015). The inherent ecological processes driving species changes can only be dissected if observations are conducted at the spatiotemporal scale at which they are acting. Following this reasoning, we monitored species composition at the single tree level in a mixed stand of Quercus petraea (Matt.) Liebl. and Quercus robur L., and targeted in our analysis the changes in species occupancy from one generation to the next. Historical changes in distribution and growth of the two species has been highly debated in recent years (Bobiec et al., 2011; Hlásny et al., 2014). Tree-ring analysis indicated that there was a long-term increase of radial growth in both species over the last century, but that Q. petraea maintained higher growth than Q. robur particularly during dry years (Becker et al., 1994; Friedrichs et al., 2009). Together with observations regarding the ecological requirements of the two species (Levy et al., 1992; Eaton et al., 2016), these authors predicted a steady retreat of Q. robur and an expansion of Q. petraea if current climatic trends will continue. Indeed, it has been repeatedly observed in mixed oak stands that the former species was more prone to decline than the latter following the severe summer droughts that occurred from the mid-seventies onward (Durand et al., 1983; Lévy et al., 1994). To date no assessments have been made on the recruitment success of the two species during the critical phase of natural regeneration. The difficulty to reliably identify the two species at young stages has probably precluded such studies. The two species have also different successional status which may alter their spatial distribution in mixed stands. It is well known that Q. robur is more a pioneer type species colonizing open areas, while Q. petraea is rather a late successional species establishing in areas already occupied by Q. robur (Levy et al., 1992). Here we employ succession in its broadest meaning as the processes by which a community changes its composition over time. We examine how hybridization and backcrossings may be acting as a genetic process reinforcing succession. Earlier observations of reproduction under natural conditions suggested indeed that interspecific pollination is asymmetric with pollen of Q. petraea more frequently pollinating Q. robur than vice versa (Chybicki & Burczyck, 2013; Lagache et al., 2014). Interspecific sexual barriers may facilitate the progressive invasion of Q. petraea, the late successional species into Q. robur, the early successional species (Petit et al., 2004). While asymmetric reproduction has now been observed in natura by parentage analysis (Bacilieri et al., 1996), no demographic or genetic survey in mixed stands has confirmed the progressive invasion due to hybridization and backcrossing, and the contribution of these succession dynamics to species changes. To sum up, there are pending issues regarding the regeneration dynamics and species distribution in mixed Q. petraea/Q. robur stands. It is unclear whether species abundance changes can be depicted at a fine spatiotemporal scale. And the likely underlying drivers of these changes have not been disentangled. We assessed the species reproductive success and recruitment over two successive generations in a mixed Q. petraea/Q. robur stand at the individual tree level using genetic fingerprints. These investigations were conducted in a selected stand where long-term research has been implemented in population and evolutionary genetics (Bacilieri et al., 1996; Streiff et al., 1998; Lagache et al., 2013; Gerber et al., 2014). The mixed composition of the stand is representative of the ecological preferences of the two species in western Europe. Our approach was driven by the following three questions:
-
(1)
Are there detectable demographic and spatial changes of Q. petraea/Q. robur in mixed stands over short and recent time spans?
-
(2)
Is there evidence of asymmetric hybridization and backcrossing in extant mixed stands as predicted by earlier theory and mating studies?
-
(3)
What are the likely factors driving species changes? Is it reproduction, environmental changes, species interactions, human interferences, or succession dynamics?
Material and methods
Study stand and sampling
The study stand is part of the Petite Charnie State Forest located in western France (latitude: 48.086°N; longitude: 0.168°W). The two species are sympatric in western part of France (Fig. S1b), where they are at the edge of the optimum of their bioclimatic niche (Fig. S1c). The State Forest extends over 712 ha and is mainly composed of broad-leaved trees (Q. petraea, Q. robur) with a slight predominance of Q. petraea (Fig. S1a). The study stand is situated in the centre of the Petite Charnie State Forest and covers 5.19 ha (square of 230 x 226m). At the beginning of our investigation in 1989 it comprised Q. petraea and Q. robur in even proportions (Bacilieri et al., 1995). In 1989, when the trees were 90 years old, a regeneration felling left 426 seed trees on the site. This seed cut was followed by a removal cut in 1992 and 1993 leaving 298 trees. In 2001 a final clear cut was implemented. Seedlings of the natural regeneration resulted from all the successful mating events that occurred between 1989 and 2001 (see Methods S1 and Table 1 for details of the silvicultural and regeneration operations). In summer 2014, a systematic sampling of 2510 saplings was made in the regeneration, corresponding to the selection of 1 seedling every 3 to 6 meters. The present study is based on data collected in three cohorts (Table 1):
The 426 adult mature trees in 1989. This is an exhaustive sampling of all seed trees remaining after the regeneration cut (cohort 1a). Variation of leaf morphology of these trees was analysed in an earlier study aiming at species assignment (Kremer et al., 2002).
The 298 adult mature trees in 1994. This is also an exhaustive sampling of the trees remaining after the two removal cuts in 1992 and 1993 (cohort 1b), among which 260 were analysed in this study (Methods S1).
The 2510 seedlings, systematically sampled in the regeneration in 2014 (cohort 2).
Table 1.
History of silvicultural operations, assessments and sampling conducted in the study stand of Quercus petraea and Q. robur.
| Year | Silviculture operation | Census Number | Sampling size | Assessments and operations for this study |
|---|---|---|---|---|
| 1989 | Regeneration felling | 426 (cohort 1a) | 426 (422)* trees | Leaf morphology in cohort 1a |
| 1992 | Botanical survey | 34 plots | Ecological mapping of the study stand | |
| 1992-1993 | Removal cut | 298 (cohort 1b) | ||
| 1995 to 2001 | 298 | Scion collection for grafting | ||
| 1995 to 2001 | 298 | 298 (260)* trees | Collection of bud and leaf tissue for DNA extraction in cohort 1b | |
| 1998 to 2001 | Final clear cut | |||
| 2013 | Mechanical systematic cleaning | |||
| 2014 | > 6000/ha (cohort 2) | 2510 (2490) * saplings | Collection of bud and leaf tissue for DNA extraction | |
| 2015 | 2490 saplings | SNP genotyping of cohort 1b and 3 | ||
| 2016 | 49 plots | Assessment of sapling densities in cohort 2 |
Numbers between brackets indicate the ultimate sample sizes used for the analysis of the data, after discarding individuals due to technical constraints and assessment difficulties during phenotyping (cohort 1a) or genotyping (cohort 1b and 2)
For cohort 1b and 2, leaf or bud tissues were collected for DNA extraction and SNP genotyping for species assignment and parentage analysis. Species assignment of trees of cohort 1a removed in 1992/1993 was based only on morphological traits (Kremer et al., 2002). All trees of all cohorts were mapped by recording their GPS coordinates, using post processed differential corrections.
Botanical survey, fine scale ecological mapping and demographic inventory
In July 1992, a floristic survey was conducted within 34 plots (circular plots of 64 m2 each) systematically distributed throughout the study stand (Bacilieri et al., 1995; Methods S2). The botanical survey (119 plant species) data were used to infer key soil characteristics. Bioindication of soil variables were drawn from large databases of species indicator values established for temperate western European forests (Gégout et al., 2003, 2005; Ellenberg et al., 1992, Methods S2). We calculated mean indicator values for the following soil attributes: pH, soil moisture, ratio of carbon to nitrogen (C/N) and organic matter content for each sampling plot. These variables were then further downscaled to a single tree level after kriging, which is an interpolation method used in geostatistics (Methods S2).
In July 2016, a demographic survey was conducted to assess sapling densities in cohort 2 to derive census estimates (Methods S2). The survey was based on a systematic sampling of 49 square survey plots distributed according to a grid system throughout the study stand. The area of each plot was 25 m2 and all saplings present in a given plot were counted.
DNA extraction and SNP genotyping
DNA of parental trees of cohort 1b had been previously extracted by Lagache et al. (2013) and Mariette et al. (2002). DNA of seedlings of cohort 2 was isolated from 5 punches of leaves using the Invisorb DNA plant HTS 96 kit (Invitek GmbH, Berlin, Germany), according to manufacturer’s recommendations.
Four medium-throughput SNP genotyping assays were developed using a MassARRAY® System (Agena Bioscience™) and iPLEX® chemistry. Three multiplexed assays were designed (W1 and W2 with 40 SNPs and W3 with 29 SNPs) from a collection of oak SNPs previously validated by Lepoittevin et al. (2015). We selected the SNPs according to their MAF (Minor Allele Frequency) > 30 % and to their map position. This procedure resulted in a distribution of one SNP every 5 to 10 cM throughout the genome (Bodénès et al., 2015). Our earlier estimates of LD (Linkage Disequilibrium) clearly showed that LD in natural oak populations is negligible between markers separated by such genetic distances (Alberto et al. 2013, Figure S2) Another multiplex of 17 SNPs (W4) selected for their interspecific differentiation between Q. petraea and Q. robur in Guichoux et al. (2013) was added to the SNP panel. Overall, a total of 126 SNPs distributed in four multiplexes were finally used to genotype all individuals (260 parents of cohort 1b and 2490 offspring of cohort 2). Twenty samples of cohort 2 had to be discarded before extraction due to the poor quality of the leaf material collected. In the genotyping assay, nucleotide base calls for SNPs were exported and assessed in MassARRAY® TYPER 4.0 genotyping software. Base calls were automatically determined and then all plots were manually validated. Each SNP locus was recorded as successful after visual inspection of the scatter plots (Methods S3). This validation step resulted in the selection of 82 SNPs (Table S1).
Species assignment
We assigned trees of cohorts 1b and 2 to their respective species (Q. robur or Q. petraea) using version 2.3.3 of STRUCTURE (Pritchard et al., 2000). The analysis was conducted over the whole data set comprising trees of cohort 1b and 2. The number of groups tested was K = 1 to 8. The admixture model with correlated allele frequencies was used with the default parameter settings (initial value of α =1, maximum value of α =10, λ = 1). A burn-in of 250 000 steps was followed by a Markov chain Monte Carlo repetition of 500 000 steps, with 30 iterations. The most likely number of populations (K) was estimated using the Ln probability of the data according to Pritchard et al. (2000) and the Delta-K method by Evanno et al. (2005) as implemented in STRUCTURE HARVESTER (Earl & von Holdt, 2012). The most probable number of populations was 2 according to the Delta-K value and the mean Ln probability of the data. Runs generated at K=2 were clustered and averaged using CLUMPAK (Kopelman et al., 2015).
Individual trees were assigned to the two species according to the value of the admixture coefficient (q). Assignment was made in three groups according to different threshold values of q: Q. petraea pure species (q ≥0.9), admixed trees (q varying between 0.1 and 0.9) and Q. robur pure species (q ≤ 0.1). The choice of the threshold for q was based on the results of a simulation study specifically designed for species assignment in interspecific oak mixtures (Neophytou, 2014). This study showed that the performance of assignment (efficiency and accuracy) was highest with STRUCTURE when the threshold of q was set to 0.90. The study indeed indicated that 99% of purebreds were correctly assigned to their taxonomic groups (either Q. petraea or Q. robur), and 85 % of the admixed individuals were also correctly assigned (Neophytou, 2014). Although the simulations were based on allele frequencies of different markers than ours, but with similar levels of interspecific differentiation to ours, we assume that the chosen threshold level (q = 0.9 and q = 0.1) would provide similar levels of performance in our study. Considering the admixed individuals, q values varying between 0.375 and 0.625 would be expected for F1 hybrids and values lower than 0.375 or larger than 0.625 for backcrossings. However the simulation study also indicated that q values of first generation hybrids and backcrossed individuals largely overlapped (Neophytou, 2014). Therefore, in what follows, these individuals (hybrids + backcrossed) will be designated as “admixed” individuals.
A retrospective assignment to the two species was also conducted on cohort 1a using leaf morphological traits, after comparing the leaf morphological variation of trees of cohort 1b with their admixture coefficient (Methods S4). However leaf morphological data did not allow to assign trees to the admixed group.
Parentage analysis
Parentage analysis was conducted using CERVUS 3.0.7 (Marshall et al., 1998) between adult trees of cohort 1b and offspring saplings in cohort 2. CERVUS uses simulations to evaluate the confidence in assignment of parentage to the most likely candidate parent. In addition to using observed allele frequencies, the simulation takes into account the number of candidate parents (1 000), the proportion of candidate parents sampled (30%), completeness of genetic typing (94%) and estimated frequency of typing error (0.01) when generating genotypes. The number of simulated offspring was set at 10000 and the minimum number of loci typed by individual was set at 40.
Parentage analyses were conducted with stringent parameters assuming no errors in genotyping (a strict exclusion analysis: 0.0 error rate) and a high confidence level (95%). Results of the parentage analysis were then used to calculate the reproductive success of each parent tree and to investigate hybridization and backcrossings. The method does not allow in our case to differentiate between seed and pollen parent. As parentage analysis was done on a systematic sampling of saplings, the number of saplings assigned to a given tree does not correspond to the absolute reproductive success of that tree (i.e. the total number of saplings the tree produced). In what follows we will therefore call it the relative reproductive success.
Data resource
Leaf morphology data and SNP genotypic data were stored in the TreePop database, available at http://treepop.pierroton.inra.fr/. To access data, go to the “Publication data” section at the top of the home page, using pc26_treepop as username and pc26_treepop as password.
Results
SNP diversity and species assignment
Four multiplexes containing 126 SNPs were used for genotyping 260 and 2490 parental and offspring trees, respectively. After analyzing each SNP profile separately (Methods S3), we selected 82 SNP loci (23 for W1, 25 for W2, 18 for W3 and 16 for W4) for the final analyses. Forty-four SNPs have been entirely discarded after visual inspection of the scatter plots because of failed clustering (low intensity magnitude, more than 3 clusters, too weak or no amplification, Methods S3). On average, 80 loci were genotyped for parental trees (Min: 45 / Max: 82) and 77 loci for offspring (Min: 32 / Max: 82). The call rate (ratio of number of assigned genotypes to the total number of genotypes) for parental and offspring samples was 97% and 94% respectively.
Results of the Bayesian clustering analysis STRUCTURE indicated that individuals clustered mostly in the two species groups (Table 2, Fig. S2). We identified in cohort 1b, 135 Quercus robur (51.9%), 110 Quercus petraea (42.3%) and 15 admixed individuals (5.8%). In cohort 2, the 2490 saplings were subdivided into 820 Q. robur (32.9%), 1570 Q. petraea (63.1%) and 100 admixed (4%) offspring (Table 2, Fig. S2). Admixed trees were equally distributed below and above the q=0.5 value in cohort 1b and 2 (Fig. S2b and S2d). We compared species assignment for each tree across the 30 iterations of STRUCTURE. The results were consistent across all iterations. All trees assigned to Q. petraea had q values larger than 0.9 in all 30 iterations. This was also the case for all trees assigned to Q. robur which had q values lower than 0.1 in all 30 iterations. Finally q values of all admixed trees ranged between 0.9 and 0.1 in all 30 iterations. Species assignment based on morphological traits (“morpho groups”) was also done in cohort 1a, after checking for congruence of assignments methods based on leaf morphological traits and SNP fingerprints (Methods S4). By using a threshold value of the first principal component (PCA1) of 1, 196 (46.4%) trees were assigned to Q. petraea and 226 (53.6%) to Q. robur (Table 2).
Table 2.
Number of trees (in percentage) assigned to the different taxonomic groups in the three cohorts.
| Taxonomic group | Cohort 1a | Cohort 1b | Cohort 2 |
|---|---|---|---|
| Q. petraea | 196 (46.4%) | 110 (42.3%) | 1570 (63.1%) |
| Q. robur | 226 (53.6%) | 135 (51.9%) | 820 (32.9%) |
| Admixed | NA | 15 (5.8%) | 100 (4.0%) |
NA: number of admixed individuals could not be estimated given the species assignment method based on leaf morphological traits (see text and Methods S4). Hence the two “morpho groups” of cohort 1a also comprised admixed individuals. Assuming that admixed individuals were not preferentially selected during the removal cut in 1992-1993 and equally distributed between the two “morpho groups”, the corrected numbers of Q. petraea and Q. robur in cohort 1a are 185 (43.8%) and 213 (50.4%). Admixed trees would have been 24 (5.8%) in cohort 1a
Species assignment was based either on SNP data using Bayesian clustering analysis (STRUCTURE) for cohort 1b and 2 or leaf morphological traits using principal component analysis for cohort 1a (Methods S4).
Spatial distribution of the species
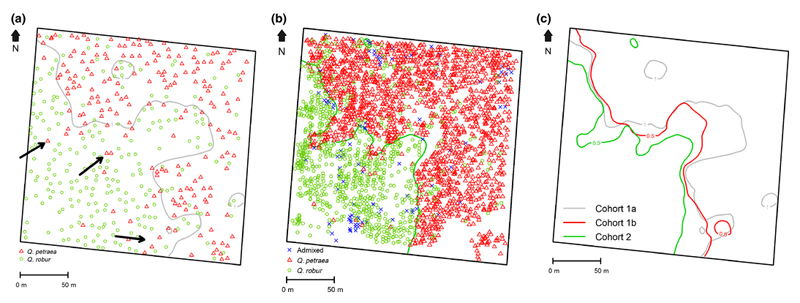
We used the threshold value of PCA1=1 in cohort 1a (Methods S4) and the threshold value of q=0.5 in cohort 1b and cohort 2 to map the species distribution in the study stand by plotting contour lines. The plotting of the contour line separating the two species ignores the admixed individuals. The plotting was done for comparative purposes separately within the three cohorts. Mapping of the contour lines was done by plotting kriging interpolated values of PCA1=1 and q=0.5 (Fig. 1a, b, c). Using this procedure, areas occupied by the two species were estimated, and were designated as Q. petraea zone and Q. robur zone. Overall there was a clear shift from cohort 1a to cohort 2 resulting in the increase of the Q. petraea zone from 49.8% to 66.5% of the whole area (Table 3). Particularly the central indentation of Q. robur into Q. petraea is reducing in size from cohort 1a to cohort 1b. Given the census numbers in both zones (Table 2) and their areas (Table 3), the tree densities in cohort 1a were 79 stems/ha for the Q. petraea zone and 84 stems/ha for the Q. robur zone. For cohort 1b these figures were 44 stems/ha and 58 stems/ha respectively.
Fig.1.
Spatial distribution of the trees in the three cohorts and contour lines of the two species.
All trees of all cohorts were mapped by recording their GPS coordinates, using post processed differential corrections.
a Cohort 1a
Mapping of the contour line between Q. petraea and Q. robur was done by plotting kriging interpolated values of PCA1=1 (First principal component of the leaf morphology analysis, Methods S4). Admixed trees were not considered for the calculation of the contour line. Arrows indicate hotspots where Q. petraea expanded.
b Cohort 2
Mapping of the contour line between Q. petraea and Q. robur was done by plotting kriging interpolated values of q=0.5 (q: value of the admixture coefficient). Admixed trees were not considered for plotting the contour line.
c Contour lines between Q. petraea and Q. robur in the three cohorts.
Contour lines of cohort 1a and cohort 2 are reproduced from Fig.1a and Fig. 1b. Mapping of the contour line in cohort 1b was done by plotting kriging interpolated values of q=0.5 in cohort 1b (q: value of the admixture coefficient). Admixed trees were not considered for plotting the contour line.
Table 3.
Absolute (m2) and relative (%) areas occupied by the two species in the three cohorts and estimated total and relative (%) number of saplings in cohort 2 (in italics).
| Species (number of inventory plots in cohort 2 ) | Cohort 1a | Cohort 1b | Cohort 2 |
|---|---|---|---|
| Q. petraea zone (34) | 25 848 (49.8%) | 29 028 (55.9%) | 34 548 (66.5%) 29 421 (73.2%) |
| Q. robur zone (15) | 26 084 (50.2%) | 22 904 (44.1%) | 17 384 (33.5%) 10 794 (26.8%) |
Absolute areas of each species were estimated based on the contours lines plotted in Fig. 1. These calculations do not take into account the presence of admixed trees.
After plotting contour lines between the two species, one can notice the presence of “outlier” trees, i.e. trees of one species that are located in the zone of the other species. Because of their neighborhood, “outlier” trees are more exposed to hybridization than others. It is worthwhile noticing that the density of “outlier” Q. petraea within the Q. robur zone is lower than the reciprocal in both adult cohorts 1a and 1b (11.5/ha vs 14.3/ha in cohort 1a and 4.4/ha vs 7.9/ha in cohort 1b). These results further highlight the higher recruitment success in cohort 2 of Q. petraea, as the presence of “outlier” Q. robur parental trees did not constrain the expansion of Q. petraea. On the contrary “outlier” Q. petraea trees, despite less frequent, were clearly instrumental to the expansion of Q. petraea as can be seen by comparing Fig. 1a and 1b (arrows on Fig. 1a). Indeed, substantial expansion of Q. petraea from cohort 1a to cohort 2 occurred around the Q. petraea “outlier” trees of cohort 1a.
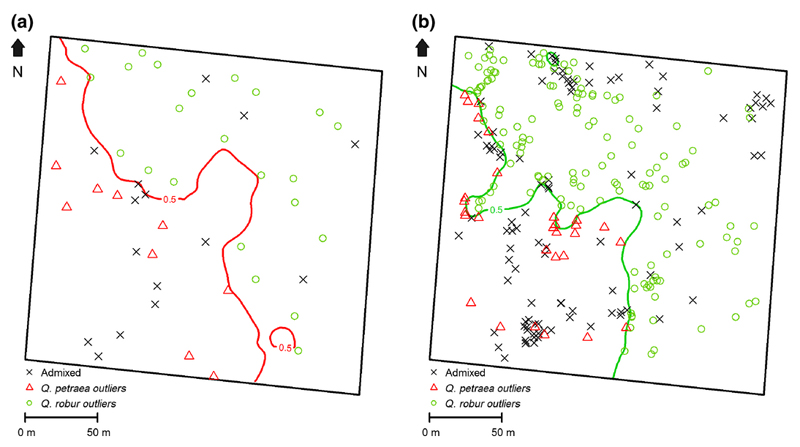
Finally, although admixed trees were not taken into account for plotting the contour line, we positioned the admixed individuals within the Q. petraea and Q. robur zones for cohort 1b (Fig. 2a) and cohort 2 (Fig. 2b). The proportion of admixed trees is higher in the Q. robur zone than in the Q. petraea zone (difference not significant in cohort 1b, and significant in cohort 2, Table 4a). Expressed in terms of tree densities there are 2.54 times more admixed trees in the Q. robur zone than in the Q. petraea zone in cohort 1b, and this figure amounts to 2.63 in cohort 2 (Table 4a; Fig. 2). While these figures were calculated on the basis of the areas occupied by the two species zones, we did similar calculations on a single tree basis in order to take into account also the “outlier” trees. To do so we considered the neighborhood area of all trees of cohort 1b, and counted the presence of admixed saplings of cohort 2 in their neighborhood. The neighborhood of a tree was defined as a circle of 10 m around the tree based on earlier assessments of acorn dispersion conducted in the same stand (Gerber et al., 2014). Admixed saplings in cohort 2 are more frequently located in the neighborhood of adult Q. robur trees (cohort 1b) than in the neighborhood of adult Q. petraea trees (significant differences, Table 4a). The same trend is also present in the Q. petraea zone of cohort 2, but the differences are not significant (Table 4b).
Fig. 2.
Distribution of admixed and “outlier” trees in cohort 1b (Fig. 2a) and in cohort 2 (Fig. 2b).
Contour lines of cohort 1b and cohort 2 are reproduced from Fig. 1c and Fig. 1b. Admixed trees were not considered for plotting the contour line. Admixed were located on the map once the contour line was plotted (see text).
a Distribution of admixed and “outlier” trees in cohort 1b.
b Distribution of admixed and “outlier” saplings in cohort 2.
Table 4a. Demographic statistics of admixed trees in cohort 1b and cohort 2.
a Number (and %) of admixed trees in the Q. petraea and Q. robur zones in cohort 1b and cohort 2.
| Q. petraea zone | Q. robur zone | p value of chi square test | |
|---|---|---|---|
| Cohort 1b | |||
| Number of admixed trees | 5 (3.9%) | 10 (7.6%) | p= 0.316 |
| Number of non admixed trees | 123 (96.1%) | 122 (92.4%) | |
| Density of admixed trees (nb trees /ha) | 1.72 | 4.37 | |
| Cohort 2 | |||
| Number of admixed saplings | 43 (2.5%) | 57 (7.5 %) | p= 1.177e-08 |
| Number of non admixed saplings | 1682 (97.5%) | 708 (92.5%) | |
| Density of admixed saplings (nb saplings /ha) | 12.45 | 32.79 | |
Table 4b. Demographic statistics of admixed trees in cohort 1b and cohort 2.
b Statistics of the number of saplings in cohort 2 according to the neighborhood of trees of cohort 1b.
| Q. petraea neighborhood | Q. robur neighborhood | p value of chi square test | |
|---|---|---|---|
| Whole cohort 2 (Q. petraea zone + Q. robur zone) | |||
| Number of admixed trees* | 39 (3.0%) | 70 (5.5%) | p= 0.002 |
| Number of non admixed trees | 1243 (97.0%) | 1194 (94.5%) | |
|
Q. petraea zone in
cohort 2 In this zone Q. robur parental trees are “outlier” trees | |||
| Number of admixed trees | 36 (3.2 %) | 16 (4.1%) | p= 0.505 |
| Number of non admixed trees | 1075 (97.8%) | 371 (95.9%) | |
|
Q. robur zone in
cohort 2 In this zone Q. petraea parental trees are “outlier” trees | |||
| Number of admixed trees | 3 (1.7 %) | 54 (6.0%) | p= 0.027 |
| Number of non admixed trees | 178 (98.3%) | 843 (94.0%) | |
the total count of admixed saplings in cohort 2 can be larger than 100 (Table 2), as a given sapling can be assigned to more than one neighborhood.
The neighborhood of a tree in cohort 1b was defined as a circle of 10 m around the tree based on earlier assessments of acorn dispersion conducted in the same stand (Gerber et al., 2014). The counts in the table correspond to the number of saplings occurring either in the neighborhood of a Q. petraea adult tree or a Q. robur neighborhood.
Demographic and reproductive monitoring
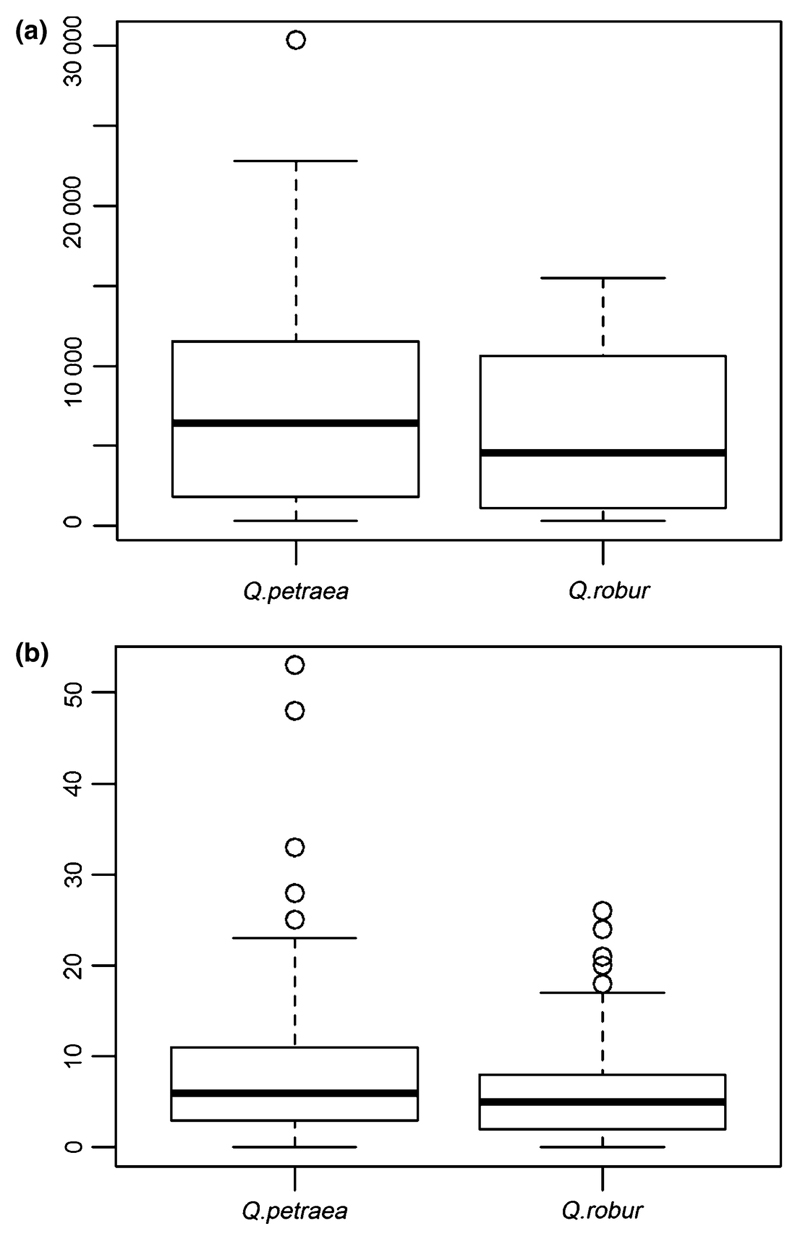
We used the data of the demographic survey to estimate sapling densities in cohort 2 for each species. As species assignment was not feasible for all saplings, we assumed that all saplings within a survey plot belonged to the same species. Species assignment for a plot was determined by the zone in which the demographic plot was located. We derived average estimates of densities by bulking data over all survey plots present in the Q. petraea zone, and those present in the Q. robur zone. Mean sapling density of Q. petraea was higher than in Q. robur (8516 saplings/ha vs 6209 saplings/ha, Fig. 3a). These differences were not significant (Wilcoxon rank sum test, p=0.43) because of the extreme variation in sapling densities among inventory plots in both species (from 285/ha to 30 330/ha in Q. petraea vs 327/ha to 15 490/ha in Q. robur), with no significant differences in the variance of sapling densities between both species. Pooling the data of sapling densities and distribution areas of the two species (Table 3) resulted in a total census number of 29 421 (73.2%) saplings of Q. petraea and 10 794 (26.8%) saplings of Q. robur in cohort 2 (Table 3)
Fig. 3.
Box plot of sapling density and relative reproductive success in Q. petraea and Q. robur.
a Sapling density (cohort 2).
Sapling density was assessed in 49 square survey plots distributed according to a grid system throughout the study stand. The area of each plot was 25 m2 on average and all saplings present in a given plot were counted. Fig. 3a shows the variation of sapling density across the survey plots. The bold horizontal line in the box indicates the median value of the densities, the bottom and top of the box correspond to the 1st and 3rd quartile of the densities. Horizontal lines at the extremities delimit 1.5 times the interquartile range above the 3rd quartile and below the 1st quartile. Circles correspond to outlier densities.
b Relative reproductive success of parental trees in cohort 1b.
Relative reproductive success was calculated for each parent tree using parentage analysis comparing saplings of cohort 2 and parental trees of cohort 1b (see text). Fig 3b shows the variation of the number of offspring between the parental trees (these numbers are relative to the sampling strategy, and are not absolute numbers). The bold horizontal line in the box indicates the median value of the reproductive success, the bottom and top of the box correspond to the 1st and 3rd quartile. Horizontal lines at the extremities delimit 1.5 times the interquartile range above the 3rd quartile and below the 1st quartile. Circles correspond to outlier values of reproductive success.
Similar results were also obtained for relative reproductive success of parental trees in cohort 1b estimated by parentage analysis (strict exclusion and 0 error rate) based on the systematic sampling of 2490 saplings in cohort 2. Parentage analysis was conducted for 2487 saplings as 3 were discarded because less than 40 SNPs could be reliably genotyped (Table 5). A total of 1453 saplings were assigned to at least one parent (58.4%). Only one parent was identified for 45.3% of the offspring (n = 1126), while two parents were found for 13.1% of offspring (n = 327). Only 17 adult trees (6.5%) among the 260 did not contribute to any offspring among the 2487 saplings investigated (Table 5; Fig. 4)
Table 5.
Number of parents and offspring assigned by parentage analysis.
| Q. robur | Q. petraea | Admixed** | Total*** | |
|---|---|---|---|---|
| Parents (cohort 1b) | ||||
| Number of trees which have at least one offspring | 125 (92.6%) | 107 (97.3%) | 11 (73.3%) | 243 (93.5%) |
| Number of trees with no offspring | 10 (7.4%) | 3 (2.7%) | 4 (26.7%) | 17 (6.5%) |
| Offspring (cohort 2) | ||||
| Minimum / maximum number of offspring* (per parental tree) | 0 /26 | 0 /53 | 0 /14 | |
| Number of offspring assigned to two parents | 193 (23.6%) | 122 (7.8%) | 12 (12.1%) | 327 (13.1%) |
| Number of offspring assigned to only one parent | 443 (54.1%) | 636 (40.5%) | 47 (47.5%) | 1126 (45.3%) |
| Number of offspring assigned to no parent | 183 (22.3%) | 811 (51.7%) | 40 (40.4%) | 1034 (41.6%) |
| Mean number of offspring per parental tree * | 6.2 | 8.1 | 3.5 | _ |
These numbers are not absolute numbers. They are relative to the sapling sampling.
Parentage could not be obtained for one admixed sapling in cohort 2, due to a limited number of successfully genotyped SNPs. Total number of admixed trees is 99.
Parentage could not be obtained for three saplings in cohort 2, due to a limited number of successfully genotyped SNPs. Total number of saplings is 2487.
Parentage analyses (CERVUS) were conducted with stringent parameter settings, assuming no errors in genotyping (a strict exclusion analysis: 0.0 error rate) and a high confidence level (95%).
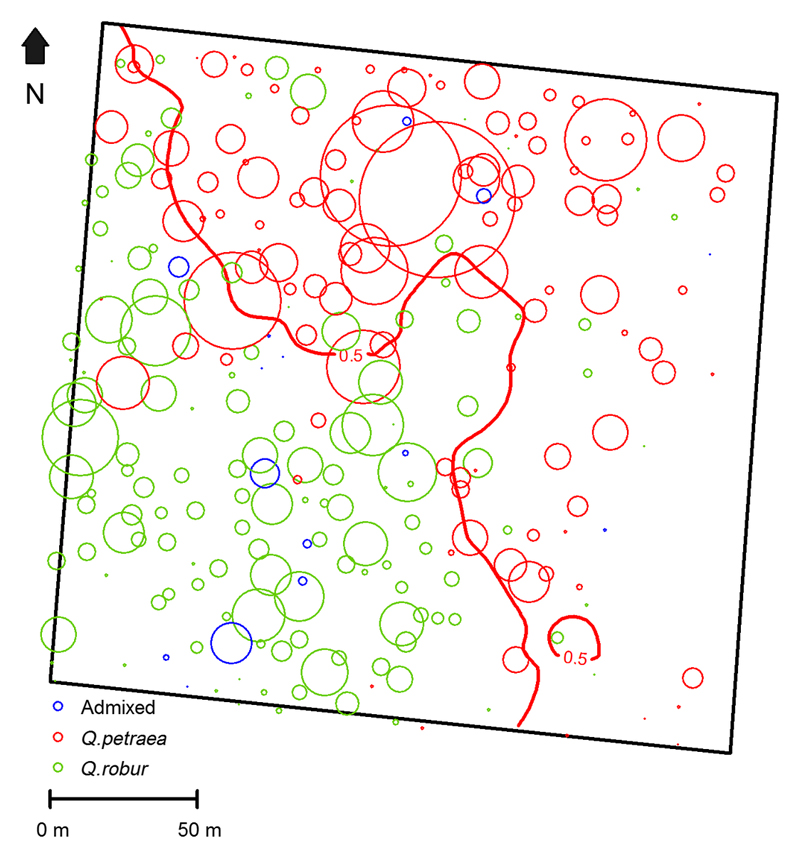
Fig. 4.
Distribution of the relative reproductive success.
Centers of circles indicate the position of parental trees in cohort 1b, while the size of the circle is proportional to the relative reproductive success of the parental trees (largest circle: 53 and smallest circle: 0). The thick line represents species separation line in cohort 1b.
Relative reproductive success of Q. robur was on average lower than in Q. petraea (6.2 versus 8.1 offspring per parent (Wilcoxon rank sum test, p=0.09) and its distribution was skewed in both species towards larger values (Table 5; Fig. 3b). The maximum number of offspring per parent varied from 0 to 26 for Q. robur and from 0 to 53 for Q. petraea. Admixed parent trees exhibited less reproductive success then the pure species trees (3.5 offspring on average per parent, varying from 0 to 14).
Hybridization and backcrossings
Among the 327 saplings of cohort 2, for which both parents could be identified in cohort 1b, 36.4% originated from intraspecific Q. petraea crosses, and 56.9% from intraspecific Q. robur crosses. None was assigned to F1 Q. petraea x Q. robur hybrids (Table 6). The remaining 6.7% came from backcrosses between pure species and admixed parents, either Q. petraea x admixed crosses (2.7%) or Q. robur x admixed crosses (4%). Among the 100 admixed saplings of cohort 2, 12 were assigned to two parents of cohort 1b and corresponded mainly to backcrosses (5 to Q. petraea x admixed crosses, and 6 to Q. robur x admixed crosses), whereas one came from an intraspecific Q. petraea cross (notice that the admixture coefficient of that individual was 0.892, close to the threshold 0.90) (Table 7). Finally, half of the backcrosses resulted in pure species offspring. Indeed among the 9 Q. petraea x admixed reported offspring (Table 6), 5 were assigned as admixed in cohort 2 (Table 7), and the remaining 4 became pure Q. petraea. Similarly among the 13 Q. robur x admixed reported offspring (Table 6), 6 were assigned as admixed in cohort 2 (Table 7), and the remaining 7 became pure Q. robur.
Table 6.
Number of saplings of cohort 2 of known biparental parentage.
| Parent species | Range of q values of parents | Number | Percentage | Range of q values of saplings |
|---|---|---|---|---|
| Q. petraea x Q. petraea | [0.959/0.999] | 119 | 36.4% | [0.890/0.999] |
| Q. robur x Q. robur | [0.001/0.046] | 186 | 56.9% | [0.001/0.058] |
| admixed x admixed | 0 | 0% | - | |
| Q. petraea x Q. robur | 0 | 0% | - | |
| Q. petraea x admixed | [0.528/0.998] | 9 | 2.7% | [0.690/0.997] |
| Q. robur x admixed | [0.001/0.710] | 13 | 4.0% | [0.006/0.420] |
| 327 | 100% |
Parent trees and species were obtained from parentage analysis using CERVUS, whereas species assignment resulted from Bayesian clustering analysis (STRUCTURE). This table provides data for only those saplings for which parentage analysis allowed to identify the two parents in the stand (327 trees, see Table 5).
Table 7.
Number of admixed saplings of cohort 2 subdivided according to their parentage.
| Range of q values parents | Number | Percentage | Range of q values admixed saplings | |
|---|---|---|---|---|
| Q. petraea x Q. petraea | [0.982/0.994] | 1 | 1.0% | 0.892 |
| Q. robur x Q. robur | - | 0 | 0% | - |
| admixed x admixed | - | 0 | 0% | - |
| Q. petraea x Q. robur | - | 0 | 0% | - |
| Q. petraea x admixed | [0.528/0.997] | 5 | 5.1% | [0.693/0.873] |
| Q. robur x admixed | [0.001/0.710] | 6 | 6.1% | [0.120/0.419] |
| Q. petraea x unknown | [0.907/0.997] | 15 | 15.1% | [0.398/0.897] |
| Q. robur x unknown | [0.001/0.016] | 18 | 18.2% | [0.113/0.606] |
| admixed x unknown | [0.124/0.554] | 14 | 14.1% | [0.103/0.760] |
| unknown x unknown | - | 40 | 40.4% | - |
| 99 | 100% |
Admixed individuals resulted from Bayesian clustering analysis (STRUCTURE), whereas parent species were obtained from parentage analysis using CERVUS. This table provides a breakdown of the 99 admixed individuals of Table 2 (parentage could not be obtained for one tree, due to a limited number of successfully genotyped SNPs).
Ecological preferences of the species and admixed individuals
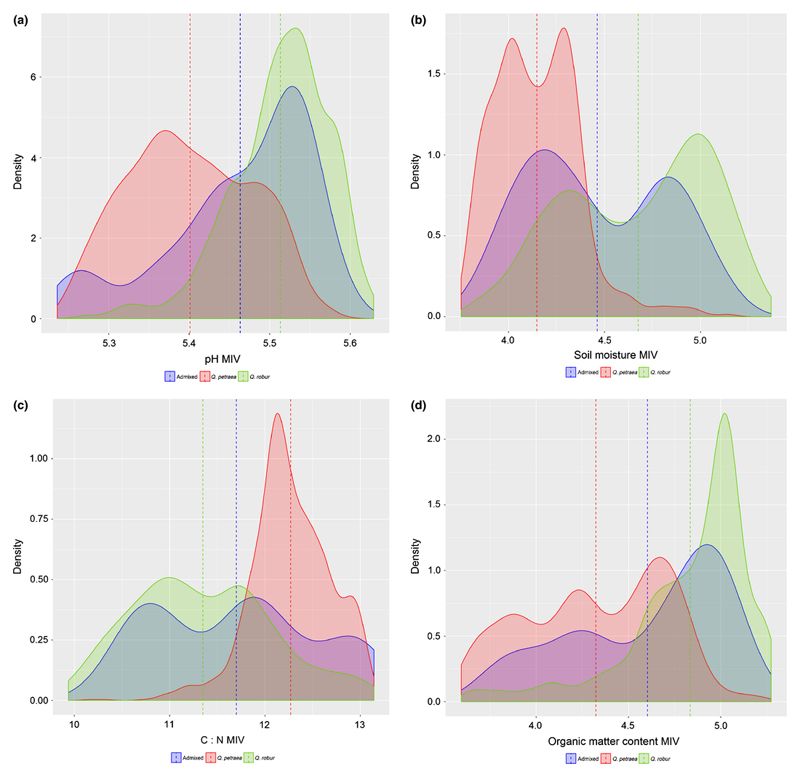
Floristic indicators of pH, soil moisture, C/N ratio and organic matter showed statistically significant differences between the two species, while the admixed group was usually intermediate between both species. We illustrate these results for cohort 2 where the sample sizes were the largest (Fig. 5) but similar results were also obtained in cohort 1a and 1b (data not shown). The range of ecological indicators of the admixed trees overlapped generally with the range of ecological indicators of Q. robur (Fig. 5). These data corroborate the spatial and demographic observations that admixed individuals occur more frequently under the Q. robur canopy than under the Q. petraea canopy. Finally we compared also changes of ecological indicators across all three cohorts. Differences among cohorts were statistically significant for all ecological indicators for Q. robur (Fig. S3a). C/N decreased from cohort 1a to cohort 2, whereas pH values, soil moisture and organic matter increased from cohort 1a to cohort 2. A similar trend was observed in Q. petraea for C/N and organic matter but differences among cohorts were only significant for organic matter (Fig. S3b). In order to provide an interpretable meaning of the floristic derived indicator values, we projected their observed values at la Petite Charnie within their distribution in France after compiling floristic surveys in Q.petraea and Q. robur stands across the country (Fig. S4). The floristic survey at La Petite Charnie indicates, in comparison to the range of variation observed in France, average values for the soil C/N ratio and the pH, but drier conditions and a more organic soil.
Fig. 5.
Probability density of ecological indicator values in the two species and admixed group of cohort 2.
Indicator values of soil variables were inferred from a systematic botanical survey conducted in 34 plots (64m2 surface each). These values were drawn from large databases of species indicator values established for temperate western European forests (see text). The y-axis is the probability density within the three groups (Q. petraea, Q. robur, admixed), in each class associated with a specific indicator value (x axis). Vertical lines indicate mean values.
Probability densities were plotted using ggplot2 (Wickham, 2009).
a pH
b Soil moisture
c C/N ( carbon/nitrogen) ratio
d Organic matter
Discussion
This survey provides unprecedented results on the changes of species occupancy along two generations in a mixed oak stand (Q. petraea/Q. robur). Our results clearly show that Q. petraea has expanded at the local scale, both numerically and spatially (Table 2; Fig. 1). Multiple factors may lead to different recruitment success: differences in reproduction, differential intraspecific response to environmental conditions, interspecific competition during the recruitment phase, human interferences during past or present silvicutural practices. We add to these factors the contribution generated by asymmetric reproduction that has been advocated –at least theoretically- to drive succession in mixed Q. petraea/ Q. robur forests over longer terms (Petit et al., 2004). Although our methods and results do not allow to disentangle the effects of these factors, we discuss their likely contributions to the recruitment successes that we observed in the two oak species.
Differential reproduction
It is important to recall that the regeneration phase cumulated 12 years of reproduction (1989-2001). Thus the results obtained are free from year to year variations that can impact the flowering, pollination, or seed crop. There is no background information on the frequency of flowering, but reasonable good acorn crops from a silvicultural point of view occur every three years on average in this part of France (Jarret, 2004). We cannot exclude that there might have been species differences in seed crop, as expected by the lower densities of Q. petraea zone especially in cohort 1b (44/ha in the Q. petraea zone vs 58/ha in the Q. robur zone). While on a mean tree basis, acorn production is correlated to light availability, the canopy was already very open during the regeneration phase (less than 60 trees /ha in cohort 1b), thus reducing the limiting factor of light availability. Furthermore on the whole area basis the overall seed crop depends also on the total number of seed trees, especially under low densities, and higher overall seed crops would have been expected for Q. robur. Finally, given the long period of regeneration (12 years), we argue that species differences, if they occurred, likely averaged out over years as a result of yearly fluctuations of species seed crop differences in opposite directions.
Interspecific competition and/ or differential intraspecific response to environmental conditions
Repeated drought events occurred during the last decades (1989-2010) which resulted in documented decline, growth losses and tree mortalities in western European forests (Bréda & Badeau, 2008; Carnicer et al., 2011). Locally, droughts in 1989, 1990, 1996 and 2005 were exceptional for the whole period (Methods S5). Overall, we recorded an increase of 20% of the annual soil water deficit during the period 1989-2010, in comparison to the period 1955-1988 (Methods S5). We also noticed that soil moisture at La Petite Charnie is at the drier margin within France (Fig S4).We therefore suspect that differential survival may have occurred due to either intra- or interspecific competition for water. Differential survival of Q. petraea and Q. robur may be caused by competitive exclusion, particularly at the frontiers between the two species, where the expansion of Q. petraea was most visible (Fig. 1b). Competition experiments show indeed contrasting responses between the two species. Généré and Le Bouler (1996) and Guibert and Généré (2000) conducted a long lasting experiment where acorns of the two species were sown in controlled mixtures in nursery beds under high densities. After one year in the nursery, the seedlings were transferred to the forest in densities mimicking natural regeneration while maintaining the same mixtures between species as in the nursery. The whole experiment was replicated in three different forests and height growth and survival was assessed over four successive years in the field. During the nursery step, the density of seedlings of Q. robur increased after one year in all mixture modalities. In the field experiments, regardless of the year and the forest, mortality was systematically higher in Q. petraea than in Q. robur, in pure and in mixed conditions. Cumulative height growth of the surviving seedlings was also higher in Q. robur than in Q. petraea. But more interestingly, the height growth performance did not change in Q. petraea between pure and mixed conditions, while growth systematically increased in Q. robur under mixed conditions in all three forests. Superior juvenile growth of Q. robur was also observed in seedling by seedling mixtures with Q. petraea (Landergott et al., 2012). Clearly the consistent outcome of these experiments was a demographic increase and an increase of growth of Q. robur in mixed plantations, thus demonstrating the higher competitive ability of this species at least at a juvenile stage (Guibert & Généré, 2000).
How can these observations be reconciled with our own data that indicated an opposite trend? On the one hand, one may advocate that the superior growth and competitive ability of Q. robur is transient and more pronounced at the early juvenile stage (Ponton et al., 2002). Support for this hypothesis comes from the comparative tree-ring analysis in older stands, which show larger ring width in Q. petraea than Q. robur (Becker et al., 1994; Friedrichs et al., 2009; Levy et al., 1992) and from the observation of adult tree survival (Ponton et al., 2002). These time trends in growth suggest a tipping point where the growth curves of both species cross each other, and Q. petraea becomes a stronger competitor than Q. robur. Our results suggest that this tipping point would need to occur sometime between age 15 and 25. A second hypothesis is that the growth and competitive ability of Q. robur in the juvenile phase can be reduced under drought conditions (Fonti et al., 2013). As stated and shown experimentally by these authors Q. robur is more competitive under favorable growing conditions than under severe exposition to drought. Earlier experiments comparing the two species under different levels of drought confirm indeed that Q. petraea sustained stress better than Q. robur in terms of tolerance to drought (Vivin et al., 1993; Arend et al., 2013) and to higher temperature (Hu et al., 2015). Differential response under drought conditions may result in differences of recruitment success in pure species settings (better survival of Q. petraea versus Q. robur) as well as under mixtures of both species, where it would actually enhance competitive exclusion. Both species competitive interactions or intraspecific processes as differential survival under drought conditions may be acting separately or concurrently.
Species shifts due to human interferences
Both oak species have been cultivated over centuries in the western part of France, and one unavoidable question is whether the shift we observed in one generation was the result of present or past human action. If past silvicultural practices have driven the two species out of their central environmental envelope, the shift we observe today may simply correspond to a backwards move to their natural preferences. There is no strong historical clue that can support this hypothesis. History suggests that the Petite Charnie Forest was growing as high forest up to the 18th century (Dufour, 1984) undergoing recurrently natural regeneration. At that time, important and very active ironworks were established in the area and the Petite Charnie Forest was intensively treated as short-term coppice to maintain high levels of wood production for feeding the forge industry during the 18th and 19th century (Dufour, 1984; Pesche, 1829). Switching back from coppice to high forest came later at the end of the 19th and beginning of the 20th century. Although Q. robur exhibits a stronger ability for resprouting, coppicing is unlikely to foster the spatial expansion of Q. robur, as coppicing is a clonal reproduction system that maintains trees in place and does not produce new propagules that are dispersed. Thus it is likely that the feedback move of the species (contemporary expansion of Q. petraea or retraction of Q. robur) to their ecological niche is of minor importance in the shift we observed.
Asymmetric reproduction and succession
We found evidence of hybridization and backcrossings between Q. petraea and Q. robur as suggested by the persistence of admixed trees in the two cohorts. The proportion of admixed trees remained remarkably similar over the two generations (Table 2), and corresponded mostly to backcrossed trees as shown by the parentage analysis (Table 6). Surprisingly we did not identify any F1 hybrid between the two species; such hybrids most likely do exist but could not be identified because they were present in such low densities that our sampling did not allow to capture them. A second reason might be that their parents were not part of the subset of trees that were genotyped (either trees of cohort 1a that were cut during the removal cut, or trees from outside of the study stand). A third reason could be that hybridization between the two species mostly involves more intermediate parent individuals, that is, adults that show more intermediate allele frequencies. Parentage analysis did neither allow to detect asymmetric hybridization and backcrossings, as we found equal numbers of backcrosses with the two pure species, and we could not identify male and female parents. Again, such asymmetric hybridization or backcrossing could hardly be detected by parentage analysis given our sampling. While parentage analysis was not resolutive enough to depict asymmetric reproduction, the demographic and ecological survey provided however indirect evidence that it is actually going on. These conclusions are based on the distribution of admixed individuals within the mixed stand that occur more frequently within the Q. robur zone than within the Q. petraea zone, as shown by the demographic and ecological survey (Table 4). In other related studies, genetic surveys aiming at inventorying admixed individuals in mixed stands confirm that admixture proportions may provide a relevant clue to the succession and colonization dynamics (Beatty et al., 2016; Neophytou et al., 2015). There is one reported diachronic study that aimed to track hybridization and backcrossing triggering succession dynamics in a mixed Q. petraea/ Q. robur stand (Boratyński et al., 2010). Using a similar approach than ours but based on morphological traits, these authors found that the proportion of hybrids was larger under a Q. robur canopy than under a Q. petraea canopy. Furthermore their demographic investigations showed also a higher recruitment success at age 17 of Q. petraea. While our observations suggest that admixed individuals actually persist and can contribute to succession, they also indicate that this process remains overall limited, as only 5.8% of admixed saplings were present in cohort 1b and 4% in cohort 2 (Table 2). We suspect that some of the admixed individuals were eliminated by natural selection or competition with pure species during earlier stages. Indeed hybridization rates estimated at the acorn stage were on average much higher (sometimes up to 40%) in different European stands (Gerber et al., 2014), but much lower (0.1 to 3%) in one single year monitoring in our study stand (Lagache et al., 2013). Nevertheless the proportion of admixed individuals is similar to values recorded in other stands in central Europe using microsatellites (3% in the Rhine Valley, Neophytou et al., 2015; 5.7% in the South of France, Lepais et al., 2009). If this limited number of admixed trees become ultimately Q. petraea once the backcrossings are completed, they will only contribute little to the Q. petraea expansion, in comparison to the recruitment success that was discussed earlier.
In conclusion, we suspect that species differential response to current environmental conditions and interspecific competition are the predominant factors of the recruitment success of Q. petraea, while human interferences, differential reproduction and succession dynamics are likely of more limited importance. We anticipate that in mixed even-aged Q. petraea/Q. robur stands, under current ongoing environmental change, Q. petraea will potentially replace Q. robur in the western part of their distribution particularly in sites that become drier and that were formely prone to Q. robur. This replacement will not only operate by catastrophic events of oak decline and mortality as it is often predicted (Bréda & Badeau, 2008; Urli et al., 2015), but also by a gradual progress of Q. petraea in the regeneration phase of mixed stands, as we showed here.
Supplementary Material
Summary.
Large scale tree distribution changes have received considerable attention but underlying demo-genetic mechanisms are less documented. We used a diachronic approach to track species shifts in a mixed oak stand (Quercus petraea/Q. robur) at a fine spatiotemporal scale.
Species assignment was made using Single Nucleotide Polymorphism (SNP) fingerprints employing clustering and parentage analysis. Mating patterns and reproductive success were assessed by parentage analysis. Plot-based inventories of soil parameters and sapling densities provided ecological and demographic information respectively.
Sapling density and reproductive success was higher in Q. petraea than in Q. robur, and were correlated with a spatial expansion of Q. petraea (50% to 67% of the area). Admixed trees resulting from hybridization and backcrossing between both species were more frequent under the Q. robur canopy.
We suspect that species differential response to ongoing environmental changes and interspecific competition are the predominant factors of the recruitment success of Q. petraea, while human interferences, differential reproduction and hybridization (and backcrossings) are likely of more limited importance. We anticipate in mixed Q. petraea/ Q. robur stands, under current ongoing environmental change, these processes will be enhanced, at least in the western part of the distribution of the two species.
Acknowledgements
This research was supported by the European Research Council through the Advanced Grant Project TREEPEACE (#FP7-339728). We acknowledge the assistance of the local ONF (Office National des Forêts) staff for the management of the Petite Charnie oak experimental site. We are grateful to Benjamin Dencausse, Véronique Gaillard, Sophie Gerber, Frédéric Lagane, Thibault Leroy, Jean Marc Louvet, Nastasia Merceron, Pierre Montpied, Patrick Reynet for their contribution during field collections, DNA extraction, and data analysis during various steps of the project. Genotyping was performed at the Genome-Transcriptome facility at the Functional Genomic Center of Bordeaux. The UMR 1137 and 1202 are supported by a grant overseen by the French National Research Agency (ANR) as part of the “Investissements d’Avenir” program respectively Labex ARBRE and Labex COTE.
Footnotes
Author contributions
A.K. designed the study. L.T. carried out the field sampling, and demographic survey with contributions of the technical staff. E.C. designed and implemented the genotyping procedure. F.E. contributed to the data flow and management. A.D. installed the study plot at the beginning and carried out the ecological monitoring. L.T did the demographic and ecological analysis with the help of J.L.D. and V.B. E.C. did the species assignment and parentage analysis. A.K., L.T. and E.C. wrote the manuscript, and all other authors reviewed and amended the complete manuscript.
References
- Alberto FJ, Derory J, Boury C, Frigério JM, Zimmermann NE, Kremer A. Imprints of natural selection along environmental gradients in phenology-related genes of Quercus petraea. Genetics. 2013;195:495–512. doi: 10.1534/genetics.113.153783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arend M, Brem A, Kuster TM, Günthardt-Goerg MS. Seasonal photosynthetic responses of European oaks to drought and elevated daytime temperature. Plant Biology. 2013;15(Suppl. 1):169–176. doi: 10.1111/j.1438-8677.2012.00625.x. [DOI] [PubMed] [Google Scholar]
- Bacilieri R, Ducousso A, Kremer A. Genetic, morphological, ecological and phenological differentiation between Quercus petraea (Matt.) Liebl. and Quercus robur L. in a mixed stand in the northwest of France. Silvae Genetica. 1995;44:1–10. [Google Scholar]
- Bacilieri R, Ducousso A, Petit RJ, Kremer A. Mating system and asymmetric hybridization in a mixed stand of European oaks. Evolution. 1996;50:900–908. doi: 10.1111/j.1558-5646.1996.tb03898.x. [DOI] [PubMed] [Google Scholar]
- Beatty GE, Montgomery WI, Spaans F, Tosh DG, Provan J. Pure species in a continuum of genetic and morphological variation: sympatric oaks at the edge of their range. Annals of Botany. 2016;117:541–549. doi: 10.1093/aob/mcw002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker M, Nieminen TM, Gérémia F. Short-term variations and long-term changes in oak productivity in northeastern France. The role of climate and atmospheric CO2. Annals of Forest Science. 1994;51:477–492. [Google Scholar]
- Becker M, Lévy G. Le point sur l’écologie comparée du chêne sessile et du chêne pédonculé. Revue Forestière Française. 1982;42:148–154. [Google Scholar]
- Bobiec A, Jaszcz E, Wojtunik K. Oak (Quercus robur L.) regeneration as a response to natural dynamics of stands in European hemiboreal zone. European Journal of Forest Research. 2011;130:785–797. [Google Scholar]
- Bodénès C, Chancerel E, Ehrenmann F, Kremer A, Plomion C. High-density linkage mapping and distribution of segregation distortion regions in the oak genome. DNA Research. 2016;23:115–124. doi: 10.1093/dnares/dsw001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodin J, Badeau V, Bruno E, Cluzeau C, Moisselin JM, Walther GR, Dupouey JL. Shifts of forest species along an elevational gradient in Southeast France: climate change or stand maturation? Journal of Vegetation Science. 2013;24:269–283. [Google Scholar]
- Boratyński A, Marcysiak K, Lewandowska A, Jasińska A, Iszkuło G. Interrelations among con-generic and co-occurring tree species: asymmetric hybridization and the high success of Quercus petraea (Matt.) Liebl. regeneration in mixed Q. petraea Q. robur L. stands. Polish Journal of Ecology. 2010;58:273–283. [Google Scholar]
- Bréda N, Badeau V. Forest tree responses to extreme drought and some biotic events: Towards a selection according to hazard tolerance? Comptes Rendus Géosciences. 2008;340:651–662. [Google Scholar]
- Carnicer J, Coll M, Ninyerola M, Pons X, Sanchez G, Peñuelas J. Widespread crown condition decline, and amplified tree mortality with increased climate change-type drought. Proceedings of the National Academy of Sciences USA. 2011;108:1474–1478. doi: 10.1073/pnas.1010070108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheaib A, Badeau V, Boe J, Chuine I, Delire C, Dufrêne E, François C, Gritti ES, Legay M, Pagé C, et al. Climate change impacts on tree ranges: model intercomparison facilitates understanding and quantification of uncertainty. Ecology Letters. 2012;15:533–544. doi: 10.1111/j.1461-0248.2012.01764.x. [DOI] [PubMed] [Google Scholar]
- Chybicki IJ, Burczyk J. Seeing the forest through the trees: comprehensive inference on individual mating patterns in a mixed stand of Quercus robur and Q. petraea. Annals of Botany. 2013;112:561–574. doi: 10.1093/aob/mct131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delzon S, Urli M, Samalens JC, Lamy JB, Lischke H, Sin F, Zimmermann NE, Porté AJ. Field evidence of colonisation by Holm oak, at the northern margin of its distribution range, during the Anthropocene period. PLoS One. 2013;8:e80443. doi: 10.1371/journal.pone.0080443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dufour J. Les forges et la forêt dans la Sarthe. Revue géographique des Pyrénées et du Sud Ouest. 1984;55:219–230. [Google Scholar]
- Durand P, Gelpe J, Lemoine B, Riom J, Timbal J. Le dépérissement du chêne pédonculé dans les Pyrénées atlantiques. Revue Forestière Française. 1983;35:357–368. [Google Scholar]
- Earl DA, vonHoldt BM. STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conservation Genetics Resources. 2012;4:359–361. [Google Scholar]
- Eaton E, Caudullo G, Oliveira S, de Rigo D. Quercus robur and Quercus petraea in Europe: distribution, habitat, usage and threats. In: San-Miguel-Ayanz J, de Rigo D, Caudullo G, Houston Durrant T, Mauri A, editors. European Atlas of Forest Tree Species. Luxembourg: Publication Office of the European Union; 2016. pp. 160–163. [Google Scholar]
- Ellenberg H, Weber HE, Düll R, Wirth V, Werner W, Paulißen D. Zeigerwerte von Pflanzen in Mitteleuropa. Scripta Geobotanica. 1992;18:1–248. [Google Scholar]
- Evanno G, Regnaut S, Goudet J. Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Molecular Ecology. 2005;14:2611–2620. doi: 10.1111/j.1365-294X.2005.02553.x. [DOI] [PubMed] [Google Scholar]
- Fonti P, Heller O, Cherubini P, Rigling A, Arend M. Wood anatomical responses of oak saplings exposed to air warming and soil drought. Plant Biology. 2013;15(Suppl. 1):210–219. doi: 10.1111/j.1438-8677.2012.00599.x. [DOI] [PubMed] [Google Scholar]
- Frelich L. Forest dynamics. F1000Research. 2016;5(F1000 Faculty Rev):183. doi: 10.12688/f1000research.7412.1. [DOI] [Google Scholar]
- Friedrichs DA, Buentgen U, Frank DC, Esper J, Neuwirth B, Loeffler J. Complex climate controls on 20th century oak growth in Central-West Germany. Tree Physiology. 2009;29:39–51. doi: 10.1093/treephys/tpn003. [DOI] [PubMed] [Google Scholar]
- Gégout JC, Hervé JC, Houllier F, Pierrat JC. Prediction of forest soil nutrient status using vegetation. Journal of Vegetation Science. 2003;14:55–62. [Google Scholar]
- Gégout JC, Coudun C, Bailly G, Jabiol B. Ecoplant: A forest site database linking floristic data with soil and climate variables. Journal of Vegetation Science. 2005;16:257–260. [Google Scholar]
- Généré B, Le Bouler H. Etude des performances en pépinière de lots de glands constitués d’un mélange variable en chênes sessile et pédonculé. Revue Forestière Française. 1996;48:21–30. [Google Scholar]
- Gerber S, Chadœuf J, Gugerli F, Lascoux M, Buiteveld J, Cottrell J, Dounavi A, Fineschi S, Forrest L, Fogelqvist J, Goicoechea PG, et al. High rates of gene flow by pollen and seed in oak populations across Europe. PlosOne. 2014;9:e85130. doi: 10.1371/journal.pone.0085130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guibert M, Généré B. Evaluation en jeunes plantations de lots mélangés de chênes sessile (Quercus petraea) et pédonculé (Quercus robur). (Early assessment of mixed sessile and pedunculate stands at three plantation sites in France) Revue Forestière Française. 2000;52:303–315. [Google Scholar]
- Guichoux E, Garnier-Géré P, Lagache L, Lang T, Boury C, Petit RJ. Outlier loci highlight the direction of introgression in oaks. Molecular Ecology. 2013;22:450–462. doi: 10.1111/mec.12125. [DOI] [PubMed] [Google Scholar]
- Hlásny T, Mátyás C, Seidl R, Kulla L, Merganičová K, Trombik J, Dobor L, Barcza Z, Konôpka B. Climate change increases the drought risk in Central European forests: what are the options for adaptation? Lesnicky Casopis Forestry Journal. 2014;60:5–18. [Google Scholar]
- Hu B, Simon J, Günthardt-Goerg MS, Arend M, Kuster TM, Rennenberg H. Changes in the dynamics of foliar metabolites in oak saplings by drought and air warming depend on species ad soil type. Plos One. 2015;10:e0126701. doi: 10.1371/journal.pone.0126701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iverson LR, Prasad AM, Matthews SN, Peters M. Estimating potential habitat for 134 eastern US tree species under six climate scenarios. Forest Ecology and Management. 2008;254:390–406. [Google Scholar]
- Jarret P. Guide des sylvicultures. Chênaie atlantique. Lavoisier; Paris, France: 2004. p. 333. [Google Scholar]
- Kopelman NM, Mayzel J, Jakobsson M, Rosenberg NA, Mayrose I. CLUMPAK: a program for identifying clustering modes and packaging population structure inferences across K. Molecular Ecology Resources. 2015;15:1179–1191. doi: 10.1111/1755-0998.12387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kremer A, Dupouey JL, Deans JD, Cottrell J, Csaikl U, Finkeldey R, Espinel S, Jensen J, Kleinschmit J, Van Dam B, et al. Leaf morphological differentiation between Quercus robur and Quercus petraea is stable across western European mixed oak stands. Annals of Forest Science. 2002;59:777–787. [Google Scholar]
- Kullman L. Rapid recent range-margin rise of tree and shrub species in the Swedish Scandes. Journal of Ecology. 2002;90:68–77. [Google Scholar]
- Laflower DM, Hurteau MD, Koch GW, North MP, Hungate BA. Climate-driven changes in forest succession and the influence of management on forest carbon dynamics in the Puget Lowlands of Washington State, USA. Forest Ecology and Management. 2016;362:194–204. [Google Scholar]
- Lagache L, Klein EK, Ducousso A, Petit RJ. Distinct male reproductive strategies in two closely related oak species. Molecular Ecology. 2014;23:4331–4343. doi: 10.1111/mec.12766. [DOI] [PubMed] [Google Scholar]
- Lagache L, Klein EK, Guichoux E, Petit RJ. Fine-scale environmental control of hybridization in oaks. Molecular Ecology. 2013;22:423–436. doi: 10.1111/mec.12121. [DOI] [PubMed] [Google Scholar]
- Landergott U, Gugerli F, Hoebee S, Finkeldey R, Holderegger R. Effects of seed mass on seedling height and competition in European white oaks. Flora. 2012;207:721–725. [Google Scholar]
- Lenoir J, Gegout JC, Marquet PA, De Ruffray P, Brisse H. A significant upward shift in plant species optimum elevation during the 20th century. Science. 2008;320:1768–1771. doi: 10.1126/science.1156831. [DOI] [PubMed] [Google Scholar]
- Lepais O, Petit RJ, Guichoux E, Lavabre E, Alberto F, Kremer A, Gerber S. Species relative abundance and direction of introgression in oaks. Molecular Ecology. 2009;18:2228–2242. doi: 10.1111/j.1365-294X.2009.04137.x. [DOI] [PubMed] [Google Scholar]
- Lepoittevin C, Bodénès C, Chancerel E, Villate L, Lang T, Lesur I, Boury C, Ehrenmann F, Zelenica D, Boland A, et al. Single-nucleotide polymorphism discovery and validation in high density SNP array for genetic analysis in European white oaks. Molecular Ecology Resources. 2015;15:1446–1459. doi: 10.1111/1755-0998.12407. [DOI] [PubMed] [Google Scholar]
- Levy G, Becker M, Duhamel D. A comparison of the ecology of pedunculate and sessile oaks: radial growth in the centre and Northwest of France. Forest Ecology and Management. 1992;55:51–63. [Google Scholar]
- Levy G, Delatour C, Becker M. Le dépérissement du chêne des années 1980 dans le centre de la France, point de départ d’une meilleure compréhension de l’équilibre et de la productivité de la chênaie. Revue Forestière Française. 1994;46:995–503. [Google Scholar]
- Mariette S, Cottrell J, Csaikl UM, Goikoechea P, Konig A, Lowe AJ, Van Dam DC, Barreneche T, Bodénès C, Streiff R, et al. Comparison of levels of genetic diversity detected with AFLP and microsatellite markers within and among mixed Q. petraea (Matt.) Liebl. and Q. robur L. stands. Silvae Genetica. 2002;51:72–79. [Google Scholar]
- Marshall TC, Slate J, Kruuk LEB, Pemberton JM. Statistical confidence for likelihood-based paternity inference in natural populations. Molecular Ecology. 1998;7:639–655. doi: 10.1046/j.1365-294x.1998.00374.x. [DOI] [PubMed] [Google Scholar]
- Monleon VJ, Lintz HE. Evidence of tree species’ range shifts in a complex landscape. Plos One. 2015;10:e0118069. doi: 10.1371/journal.pone.0118069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neophytou C. Bayesian clustering analyses for genetic assignment and study of hybridization in oaks: effects of asymmetric phylogenies and asymmetric sampling schemes. Tree Genetics & Genomes. 2014;10:273–285. [Google Scholar]
- Neophytou C, Gärtner SM, Vargas-Gaete R, Michiels HG. Genetic variation of Central European oaks: shaped by evolutionary factors and human intervention? Tree Genetics & Genomes. 2015;11:79. doi: 10.1007/s11295-015-0905-7. [DOI] [Google Scholar]
- Ozolinčius R, Lekevičius E, Stakėnas V, Galvonaitė A, Samas A, Valiukas D. Lithuanian forest and climate change: possible effects on tree composition. European Journal Forest Research. 2015;133:51–60. [Google Scholar]
- Peñuelas J, Boada M. A global change-induced biome shift in the Montseny mountains (NE Spain) Global Change Biology. 2003;9:131–140. [Google Scholar]
- Pesche JR. Dictionnaire topographique, historique et statistique de la Sarthe. Vol. 1. Bachelier; Paris: 1829. p. 390. [Google Scholar]
- Petit RJ, Bodénès C, Ducousso A, Roussel G, Kremer A. Hybridization as a mechanism of invasion in oaks. New Phytologist. 2004;161:151–164. [Google Scholar]
- Ponton S, Dupouey JL, Breda N, Dreyer E. Comparison of water-use efficiency of seedlings from two sympatric oak species: genotype × environment interactions. Tree Physiology. 2002;22:413–422. doi: 10.1093/treephys/22.6.413. [DOI] [PubMed] [Google Scholar]
- Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pucko C, Beckage B, Perkins T, Keeton WS. Species shifts in response to climate change: Individual or shared responses? Journal of the Torrey Botanical Society. 2011;138:156–176. [Google Scholar]
- Ruiz-Labourdette D, Schmitz MF, Pineda FD. Changes in tree species composition in Mediterranean mountains under climate change: Indicators for conservation planning. Ecological Indicators. 2013;24:310–323. [Google Scholar]
- Streiff R, Labbe T, Bacilieri R, Steinkellner H, Gloessl J, Kremer A. Within-population genetic structure in Quercus robur L. and Quercus petraea (Matt.) Liebl. assessed with isozymes and microsatellites. Molecular Ecology. 1998;7:317–328. [Google Scholar]
- Urli M, Lamy JB, Sin F, Burlett R, Delzon S, Porté AJ. The high vulnerability of Quercus robur to drought at its southern margin paves the way for Quercus ilex. Plant Ecology. 2015;216:177–187. [Google Scholar]
- Vivin P, Aussenac G, Levy G. Differences of drought resistance among 3 deciduous oak species grown in large boxes. Annals of Forest Science. 1993;50:221–233. [Google Scholar]
- Walther GR, Berger S, Sykes MT. An ecological “footprint” of climate change. Proc R Soc B. 2005;272:1427–1432. doi: 10.1098/rspb.2005.3119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang WJ, He HS, Thompson FR, III, Fraser JS, Dijak WD. Landscape- and regional-scale shifts in forest composition under climate change in the Central Hardwood Region of the United States. Landscape Ecology. 2016;31:149–163. [Google Scholar]
- Wickham H. ggplot2: Elegant Graphics for Data Analysis. Springer-Verlag; New York: 2009. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.