Abstract
A major challenge for the development of an effective vaccine against tuberculosis (TB) is that the attributes of protective CD4+ T cell responses are still elusive for human TB. Infection with human immunodeficiency virus-1 (HIV-1) is a major risk factor for tuberculosis (TB), and a better understanding of HIV-induced alterations of Mycobacterium tuberculosis (Mtb)-specific CD4+ T cells that leads to failed host resistance may provide insight into protective T cell immunity to TB.
Eighty-six participants from a TB-endemic setting, either HIV-infected or -uninfected and with latent or active TB, were screened using Mtb-specific MHC class II tetramers. We examined the phenotype as well as function of ex vivo Mtb-specific tetramer+CD4+ T cells using flow cytometry.
The numbers of Mtb-specific tetramer+CD4+ T cells were relatively well maintained in HIV-infected persons with active TB, despite severe immunodeficiency. However, while HIV-uninfected persons with latent TB infection exhibited ex vivo Mtb-specific CD4+ T cells predominantly of a CXCR3+CCR6+CCR4- (Th1*) phenotype; active TB or HIV infection was associated with a contraction of this subset. Nevertheless, in individuals with active TB and/or HIV infection, circulating ex vivo Mtb-specific CD4+ T cells did not display defects in exhaustion or polyfunctionality compared to healthy HIV-uninfected individuals with latent TB infection.
Collectively, these data suggest that increased susceptibility to TB disease could be related to a loss of circulating Th1* CD4+ T cells rather than major changes in the number or function of circulating CD4+ T cells.
Keywords: Mycobacterium tuberculosis, latent tuberculosis infection, active tuberculosis, HIV, MHC class II tetramers, Mtb-specific CD4+ T cells, ex vivo phenotype
Introduction
It is estimated that a third of the world's population is latently infected with Mycobacterium tuberculosis (Mtb)and in 2015, over 10 million people developed active tuberculosis (TB), of which 1.2 million (12%) were co-infected with human immunodeficiency virus (HIV) (1). While, in the majority of immunocompetent individuals, the risk of progression from latent to active TB is 2-10% in a lifetime, it increases up to an annual risk of 5-15% in HIV-infected persons (2), making HIV one of the strongest known risk factors for TB (3). Furthermore, in active TB cases, concomitant HIV infection results in accelerated TB disease progression, more severe clinical symptoms in some cases, and increased mortality (4, 5), further emphasizing the detrimental effect of HIV on Mtb immunity.
The major immune defect induced by HIV is a progressive reduction in absolute CD4+ T cells (6) that correlates with increasing TB disease risk (7), attesting to the critical role of CD4+ T cells for Mtb immunity. However, TB risk is significantly elevated even in HIV-infected persons with well-preserved CD4+ T cell counts (during the early phase of infection or after immune-restoring ART), suggesting that HIV may also induce qualitative defects in Mtb-specific CD4+ T cells. Indeed, alterations in the polyfunctional capacity (8, 9), memory profile (10) and lineage differentiation (11) of Mtb-specific CD4+ T cells have been previously reported. Moreover, HIV promotes systemic immune activation (12) and cell exhaustion (13). Altogether, these HIV-induced impairments weaken Mtb immune responses and could facilitate TB reactivation and/or promote excessive TB progression.
To date the constituents of an effective immune response to TB remaining completely understood. Indeed, although Th1 responses are the cornerstone of adaptive immunity to TB, they failed to associate with protection from infection or disease in recent clinical trials of a novel TB vaccine (14, 15). Thus, to better understand the impact of HIV on Mtb-specific responses we assessed the magnitude, phenotype and functional profile of ex vivo Mtb-specific CD4+ T cells from individuals with distinct HIV and TB clinical states, employing MHC class II tetramers. This approach allowed us to define TB disease- and HIV-induced alterations specific to Mtb-specific CD4+ T cells in their resting state. Our findings provide novel insights into cellular mechanisms of failed Mtb-specific immunity.
Materials and Methods
Study participants
Study participants (n = 86) recruited from the Ubuntu Clinic, Khayelitsha in Cape Town, South Africa, were screened for Mtb-specific MHC class II responses. To assess qualitative effects of HIV infection on Mtb-specific CD4+ T cells before profound CD4 depletion, only HIV-infected individuals with latent TB infection (LTBI) with well-maintained CD4+ T cell counts were recruited. Individuals were categorized into four groups based on their TB and HIV status: HIV-/LTBI (n=28), HIV+/LTBI (n=30), HIV-/aTB (n=14) and HIV+/aTB (n=14). LTBI was diagnosed based on a positive IFN-γ release assay (QuantiFERON®-TB Gold In-Tube, Cellestis), no symptoms of active TB disease, a negative Mtb sputum (GeneXpert) and a normal chest X-ray. Active TB disease was diagnosed based on clinical symptoms, positive chest X-ray and positive Mtb sputum. All HIV-infected individuals were antiretroviral treatment-naïve and no one had started TB treatment at the time of enrolment. The study was approved by the University of Cape Town Human Research Ethics Committee (HREC No. 158/2010 and 896/2014) and the protocol review office of the US National Cancer Institute institutional review board. All participants provided written informed consent.
CD4+ T cell counts, plasma viral load and HLA typing
Absolute blood CD4+ T cell counts were measured using a Flow-CARE PLG CD4 test (Beckman Coulter). For HIV-infected individuals, plasma HIV-1 RNA levels were quantified using Abbott m2000 RealTime HIV-1 assay. For HLA typing, DNA was extracted from peripheral blood mononuclear cells (PBMC) using the QIAamp Mini Blood kit (Qiagen). High-resolution HLA class II genotypes were determined using 454/Fluidigm HLA Typing Kits (Roche) following the manufacturer's protocols (16).
Mtb-specific MHC class II tetramers
Four custom-ordered Mtb-specific MHC class II tetramers conjugated with phycoerythrin (PE) or allophycocyanin (APC) were obtained from the NIH Tetramer Core Facility (Emory University, USA): CFP-1071-85 (EISTNIRQAGVQYSR) loaded HLA-DRB1*0401 tetramer (DRB1*0401/CFP); CFP-1051–65 (AQAAVVRFQEAANKQ) loaded HLA-DRB5*0101 tetramer (DRB5*0101/CFP); CFP-1071-85 (EISTNIRQAGVQYSR) loaded HLA-DQB1*0602 tetramer (DQB1*0602/CFP) and ESAT-631-45 (EGKQSLTKLAAAWGG) loaded HLA-DQB1*0602 tetramer (DQB1*0602/ESAT). Additionally, the human class II-associated invariant chain peptide (Clip; PVSKMRMATPLLMQA) was complexed to each of the aforementioned tetramers and used as a negative control to validate tetramer-staining specificity (Supplemental Figure 1A). The performance of PE- and APC-conjugated tetramers was compared in a subset of individuals (Supplemental Figure 1B), showing that comparable frequencies of tetramer+ cells were obtained with both reagents. Moreover, for ex vivo phenotyping, we performed a dual tetramer stain using PE- and APC-conjugated tetramers of different specificities simultaneously in a subset of samples (n=7). To validate this approach, we verified that co-staining with two different tetramers did not interfere with the detection of tetramer+ T cells (Supplemental Figure 1C).
Antigens and cell stimulation
After resting, cryopreserved PBMC were stimulated with 1 μg/mL of cognate peptide (15mers, Peptide Synthetics) from culture filtrate protein of 10 kDa (CFP-10) or early secretory antigenic target of 6 kDa (ESAT-6) proteins. Stimulations were performed in the presence of co-stimulatory antibodies: anti-CD28 and anti-CD49d (1 μg/mL; BD) for 16 hours. Brefeldin A (10 μg/mL; Sigma-Aldrich) was added at the onset of stimulation.
Cell staining
Cells were first stained with Fixable Near-IR Dead Cell Stain (Invitrogen), then with PE- and/or APC-conjugated class II tetramers (2 μg/mL and 4 μg/mL, respectively) at 37°C for 30 minutes and subsequently surface stained. When intracellular proteins were measured, cells were fixed and permeabilized using Cytofix/Cytoperm buffer (BD Biosciences) and then stained intracellularly. A summary table of the antibodies used for each panel is presented in Supplemental Table I. Samples were acquired on a LSRII flow cytometer (BD) using FACSDiva software and analysis was performed using FlowJo (v9.9.4, Treestar) and Pestle (v1.7) and Spice (v5.35) software (17). The gating strategies applied are presented in Supplemental Figure 2.
Statistical Analysis
Statistical analyses were performed using Prism (GraphPad, v5.0). Nonparametric statistical tests were used for all comparisons. The Mann-Whitney U test and Wilcoxon-matched pairs test were used for unmatched and paired samples, respectively, and the Kruskal-Wallis ANOVA using Dunn's test for multiple comparisons. Correlations were performed using the Spearman Rank test. A p value <0.05 was considered statistically significant.
Results
Ex vivo detection of Mtb-specific CD4+ T cells using MHC class II tetramers
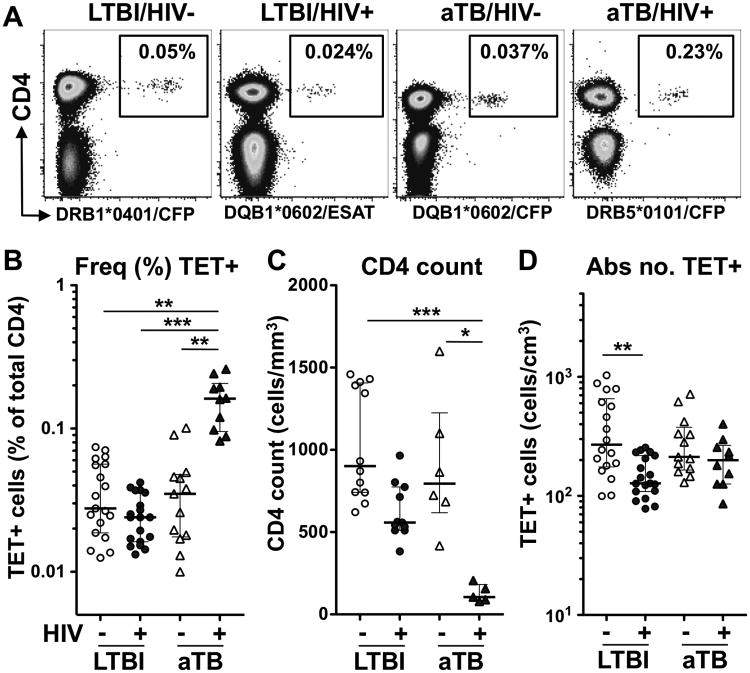
Using four different MHC class II tetramers recognizing CFP-10 or ESAT-6 epitopes from Mtb, we identified tetramer positive CD4+ T cells in 35 of the 86 participants screened. The clinical characteristics of each individual with Mtb-specific MHC class II tetramer responses are presented in Table I. The proportion of tetramer responders was similar in each group, representing approximately 40% of individuals tested (Supplemental Figure 3A). The detection rate for each tetramer (∼18% for DRB1*0401, ∼59% for DRB5*0101, ∼67% for DQB1*0602; Supplemental Figure 3B) was in accordance with the prevalence of these HLA class II types in the South African population (18). Figure 1A shows representative plots of Mtb-specific tetramer staining in one donor from each clinical group studied. While the median frequencies of tetramer+CD4+ T cells were comparable between the HIV-/LTBI, HIV+/LTBI and HIV-/aTB groups (median: 0.028%, 0.024% and 0.035%, respectively), in HIV+/aTB individuals, the median frequency of tetramer+CD4+ cells was significantly higher (0.16%, Figure 1B). To take into account variation in absolute CD4+ T cell counts between groups, particularly in persons co-infected with HIV and aTB (Figure 1C), the absolute number of Mtb-specific tetramer+CD4+ T cells was calculated (Figure 1D). The absolute number of Mtb-specific tetramer+CD4+ T cells in HIV+/LTBI individuals (median: 128 cells/cm3) was significantly lower than the HIV-/LTBI group (270 cells/cm3), representing a median fold reduction of 52%. By contrast, despite the profound CD4 depletion observed in HIV+/aTB patients, the higher frequency of Mtb-specific tetramer+CD4+ T cells observed in this group resulted in a relative maintenance of the absolute number of Mtb-specific tetramer+CD4+ T cells (median: 199 cells/cm3) to similar levels as observed in the HIV-uninfected group (Figure 1D). This suggests that notwithstanding profound lymphopenia, active bacterial replication still promotes the expansion of CD4+ T cells targeting Mtb in HIV-infected persons.
Table I. Clinical characteristics of the study cohort.
| Class II HLA type | Tetramer frequency (% CD4)a | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
| |||||||||||||
| PID | Gender | HIV Status |
TB Status |
Age | CD4 Count (cells/mm3) |
Log10 HIV viral load (copies/ml) |
DRB1 | DRB5 | DQB1 | DRB1 *0401/ CFP |
DRB5 *0101/ CFP |
DQB1 *0602/ ESAT |
DQB1 *0602/ CFP |
| Latent TB Infection | |||||||||||||
| 0018 | M | - | LTBI | 40 | nd | na | 0701/1501 | 0101 | 0202/0601 | - | 0.062 | - | - |
| 1035 | F | - | LTBI | 19 | 1459 | na | 0401/1503 | 0101 | 0302/0602 | 0.071 | 0.025 | - | nd |
| 1029 | F | - | LTBI | 22 | 1430 | na | 0301/1501 | 0101 | 0201/0602 | - | 0.055 | 0.040 | nd |
| 1052 | F | - | LTBI | 25 | 1412 | na | 0301/1503 | 0101 | 0602/0301 | - | - | 0.017 | nd |
| 1022 | F | - | LTBI | 43 | 1344 | na | 0701/1503 | 0101 | 0202/0602 | - | 0.046 | 0.014 | nd |
| 0100 | F | - | LTBI | 28 | 1157 | na | 1301/1302 | 0602/0609 | - | - | 0.056 | 0.063 | |
| 1055 | M | - | LTBI | 19 | 932 | na | 1001/1101 | 0501/0602 | - | - | 0.025 | nd | |
| 1057 | M | - | LTBI | 20 | 871 | na | 0302/1401 | 0402/0602 | - | - | 0.034 | nd | |
| 1011 | M | - | LTBI | 28 | 801 | na | 1101/1302 | 0602/0609 | - | - | - | 0.012 | |
| 1038 | F | - | LTBI | 18 | 743 | na | 0301/1101 | 0201/0602 | - | - | 0.028 | 0.014 | |
| 1072 | F | - | LTBI | 24 | 741 | na | 0102/1503 | 0101 | 0501/0602 | - | 0.019 | 0.022 | nd |
| 1061 | M | - | LTBI | 21 | 674 | na | 0804/1101 | 0319/0602 | - | - | 0.027 | nd | |
| 1066 | F | - | LTBI | 39 | 621 | na | 0401/0901 | 0302/0202 | 0.074 | - | - | nd | |
| Median [IQR] | 24 [20-34] | 902 [742-1395] | |||||||||||
| 1075 | F | + | LTBI | 31 | 965 | 3.77 | 0901/1503 | 0101 | 0202/0602 | - | 0.025 | 0.013 | nd |
| 0113 | F | + | LTBI | 41 | 803 | 4.49 | 1301/1503 | 0101 | 0604/0602 | - | 0.029 | - | - |
| 1080 | F | + | LTBI | 54 | 774 | 4.15 | 0302/1503 | 0101 | 0402/0602 | - | 0.016 | 0.014 | nd |
| 1153 | F | + | LTBI | 46 | 714 | 3.47 | 1102/1503 | 0101 | 0319/0602 | - | 0.036 | 0.024 | nd |
| 0050 | M | + | LTBI | 30 | 563 | 5.06 | 1302/1503 | 0101 | 0609/0602 | - | 0.039 | - | 0.019 |
| 0094 | F | + | LTBI | 35 | 558 | 4.99 | 0302/1503 | 0101 | 0402/0602 | - | 0.037 | 0.042 | 0.027 |
| 1076 | F | + | LTBI | 25 | 543 | 2.96 | 0401/1101 | 0302/0602 | 0.018 | - | 0.015 | nd | |
| 0131 | F | + | LTBI | 35 | 532 | 1.30 | 1302/1503 | 0101 | 0604/0602 | - | 0.024 | 0.017 | - |
| 0077 | F | + | LTBI | 38 | 511 | 3.35 | 0401/0701 | 0302/0202 | 0.024 | - | - | - | |
| 1129 | F | + | LTBI | 29 | 510 | 3.66 | 1101/1503 | 0101 | 0319/0602 | - | 0.015 | - | nd |
| 1081 | F | + | LTBI | 37 | 383 | 3.51 | 1303/1503 | 0101 | 0202/0602 | - | 0.037 | - | nd |
| Median [IQR] | 35 [30-40] | 558 [511-774] | 3.66 [3.35-4.49] | ||||||||||
|
| |||||||||||||
| Active TB infection | |||||||||||||
| 0012 | F | - | aTB | 28 | 1599 | na | 1102/1503 | 0101 | 0319/0602 | - | 0.013 | 0.045 | 0.010 |
| 0002 | M | - | aTB | 35 | 1100 | na | 1101/1101 | 0319/0602 | - | - | 0.017 | 0.026 | |
| 0014 | M | - | aTB | 26 | 865 | na | 0302/1503 | 0101 | 0402/0602 | - | 0.020 | 0.038 | - |
| 0085 | M | - | aTB | 29 | 724 | na | 0404/1454 | 0402/0602 | - | - | 0.046 | 0.018 | |
| 0133 | F | - | aTB | 30 | 685 | na | 0302/0401 | 0402/0302 | 0.090 | - | - | - | |
| 0137 | M | - | aTB | 25 | 417 | na | nd | nd | nd | 0.101 | - | 0.035 | 0.050 |
| Median [IQR] | 29 [26-31] | 795 [618-1225] | |||||||||||
| 0144 | F | + | aTB | 30 | 206 | 4.69 | 1101/1101 | 0319/0602 | - | - | 0.195 | 0.088 | |
| 0106 | F | + | aTB | 35 | 157 | 4.93 | 1503/1503 | 0101 | 0602/0602 | - | 0.163 | 0.190 | 0.098 |
| 0087 | M | + | aTB | 49 | 105 | 4.59 | 1302/1503 | 0101 | 0609/0602 | - | 0.082 | - | 0.120 |
| 0122 | F | + | aTB | 29 | 90 | 5.41 | 0302/1503 | 0101 | 0402/0602 | - | 0.242 | 0.260 | - |
| 0132 | F | + | aTB | 26 | 76 | 4.63 | 0302/1503 | 0101 | 0402/0602 | - | 0.160 | - | - |
| Median [IQR] | 30 [28-42] | 105 [83-182] | 4.69 [4.61-5.17] | ||||||||||
MHC class II tetramer frequency as a percentage of total CD4+ T cells. PID: patient identification number, F: female, M: male, IQR: interquartile range, na: not applicable, nd: not done.
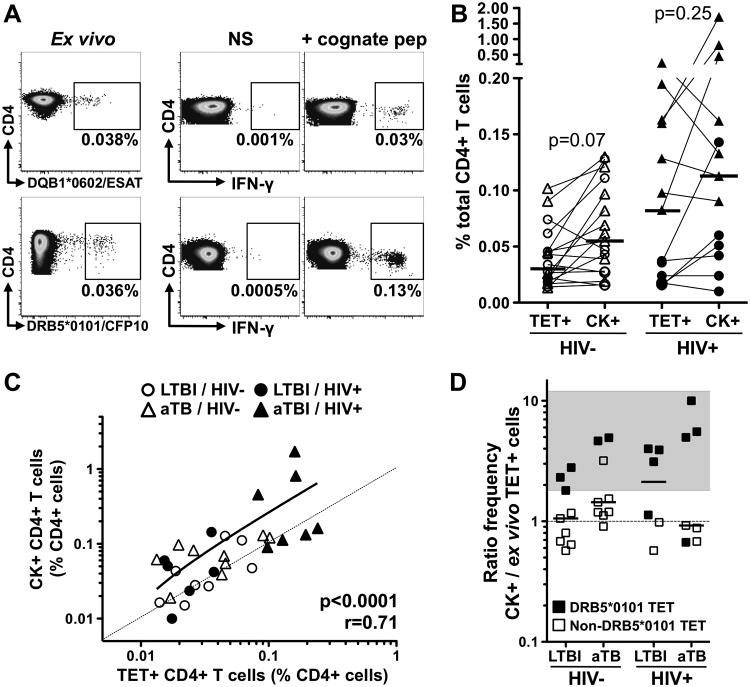
Fig. 1. Identification of Mtb-specific MHC class II tetramer (TET) responses in individuals with HIV and/or active TB.
A- Representative examples of TET+ responses in each clinical group. The frequency of TET+ cells is expressed as a percentage of total CD4+ T cells. B to D: Frequency of TET+ cells (B), absolute CD4+ T cell count (C) and absolute number of TET+ cells (D) in each clinical group (n=13 LTBI/HIV-, n=11 LTBI/HIV+, n=6 aTB/HIV- and n=5 aTB/HIV+). Bars represent the median and interquartile range. Statistical comparisons were performed using a one-way ANOVA Kruskal-Wallis test. *<0.05, **<0.01, ***<0.001.
Characterizing the phenotype of ex vivo Mtb-specific tetramer+CD4+ T cells
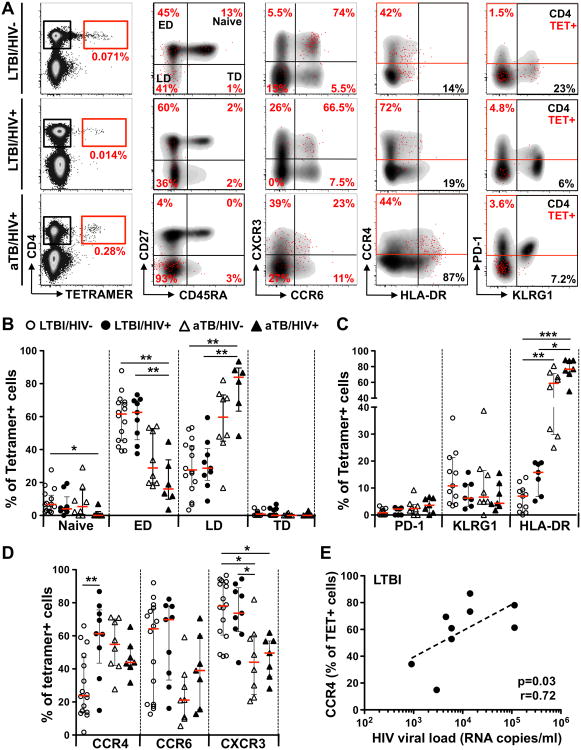
To examine the effect of HIV infection and/or TB disease on ex vivo Mtb-specific CD4+ T cells, we investigated their phenotype using flow cytometry (Figure 2A). The memory differentiation, activation status and homing potential of these cells were examined, as these are likely to be important features relating to the functional potential of T cells upon stimulation (19, 20). Due to sample availability, a subset of 29 participants was phenotyped. Based on the expression of CD27 and CD45RA, we classified four memory populations: naïve-like (CD27+CD45RA+), early differentiated (CD27+CD45RA-), late differentiated (CD27-CD45RA-) and terminally differentiated (CD27-CD45RA+). In individuals with LTBI, irrespective of HIV status, Mtb-specific CD4+ T cells were predominately early differentiated (median: 61% for HIV- and 63% for HIV+) and approximately a third of the cells exhibited a late-differentiated phenotype. Conversely, aTB individuals (regardless of their HIV status) exhibited a significantly elevated proportion of late differentiated Mtb-specific CD4+ T cells (median: 60% for HIV- and 84% for HIV+) with a concomitant reduction of early-differentiated cells (median: 29% for HIV- and 16% for HIV+; Figure 2B).
Fig. 2. Ex vivo phenotype of Mtb-specific MHC class II tetramer+CD4+ cells in individuals with HIV and/or active TB.
Due to the availability of PBMC, a subset of 29 donors (HIV-/LTBI: n=13, HIV+/LTBI: n=7, HIV-/aTB: n=4 and HIV+/aTB: n=5) was phenotyped. Ten of these 29 individuals exhibited more than one individual tetramer response (HIV-/LTBI: n=16, HIV+/LTBI: n=9, HIV-/aTB: n=8, HIV+/aTB: n=7, see Supplemental Figure 3A). A- Representative examples of memory, homing and activation profiles of TET+CD4+ T cells (red) and total CD4+ T cells (grey) in one LTBI/HIV-, one LTBI/HIV+ and one aTB/HIV+ individual. B- Comparison of the proportion of distinct memory subsets (naïve: CD27+CD45RA+, early differentiated, ED: CD27+CD45RA-, late differentiated, LD: CD27-CD45RA- and terminally differentiated, TD: CD27-CD45RA+) on ex vivo TET+CD4+ T cells from LTBI/HIV- (open circles, n=16), LTBI/HIV+ (black circles, n=9), aTB/HIV- (open triangles, n=8) and aTB/HIV+ (black triangles, n=7) individuals. C- Comparison of the expression of homing markers (CCR4, CCR6 and CXCR3) on ex vivo TET+CD4+ T cells from each clinical group. D- Comparison of the expression of activation markers (PD-1, KLRG1 and HLA-DR) on ex vivo TET+CD4+ T cells from each clinical group. Bars represent the median and interquartile range. Statistical comparisons were performed using a one-way ANOVA Kruskal-Wallis test. *<0.05, **<0.01, ***<0.001. E- Relationship between the expression of CCR4 on TET+CD4+ T cells and plasma HIV viral load in LTBI individuals. Correlations were tested by a two-tailed non-parametric Spearman Rank test.
Next, to examine the activation/exhaustion status of ex vivo Mtb-specific CD4+ T cells, the expression of PD-1, KLRG1 and HLA-DR were measured. Mtb-specific CD4+ T cells were characterized by a low expression of PD-1 (median: ∼2%) with no significant differences observed between the four groups studied (Figure 2C). Similarly, KLRG1 expression on Mtb-specific CD4+ T cells was comparable amongst the four clinical groups, with less than a quarter of the cells expressing KLRG1. In persons with active TB disease, HLA-DR expression on Mtb-specific CD4+ T cells was significantly elevated (median: 58% for HIV- and 76%for HIV+) compared to LTBI individuals (7% for HIV- and 16% for HIV+, Figure 2C). Finally, to define whether HIV or active TB alters ex vivo Mtb-specific CD4+ T cell homing potential, we measured the expression of the chemokine receptors CCR4, CCR6 and CXCR3. In the context of aTB disease (irrespective of HIV infection), Mtb-specific tetramer+CD4+ T cells were characterized by a significantly lower expression of CXCR3 (median: 44% for HIV- and 50% for HIV+) when compared to individuals with LTBI (78% for HIV- and 74% for HIV+; Figure 2D). Additionally, a trend towards lower expression of CCR6 was also observed in TB patients when compared to LTBI individuals. HIV infection per se did not significantly alter the expression of CXCR3 or CCR6 on Mtb-specific CD4+ T cells. However, specifically in the LTBI group, HIV was associated with a significant increase in the expression of CCR4 on Mtb-specific tetramer+CD4+ T cells when compared to HIV-uninfected individuals (p=0.012, median: 61% vs 24%, respectively; Figure 2D). In addition, CCR4 expression on tetramer+CD4+ T cells from HIV+/LTBI donors positively correlated with plasma HIV VL (p=0.03, r=0.72), suggesting that the increase in CCR4 expression in these cells could be driven by viral replication (Figure 2E).
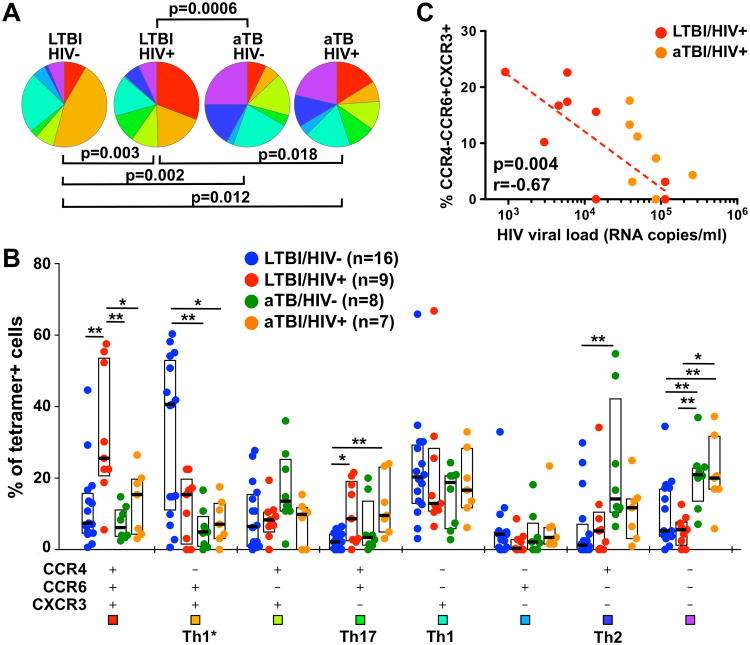
The expression pattern of the three chemokine receptors studied has previously been used to delineate T helper (Th) subsets as follows: CCR4-CCR6-CXCR3+ (Th1), CCR4+CCR6+CXCR3- (Th17), CCR4+CCR6-CXCR3- (Th2) and CCR4-CCR6+CXCR3+ (Th1*) (19, 21). This latter subset has been described as a non-conventional Th1 subset endowed with the capacity to produce IFN-γ and low levels of IL-17 (22). Detailed analysis of chemokine receptor combinations expressed by ex vivo Mtb-specific CD4+ T cells revealed that 1) these cells exhibit a broad and diverse co-expression profile of chemokine receptors; and 2) the overall distribution of these subsets was significantly different between LTBI and aTB in both HIV-uninfected and HIV-infected persons (p=0.002 and p=0.018, respectively). Furthermore, in the context of LTBI, HIV infection also significantly perturbs the global distribution of chemokine receptor expression (p=0.003) (Figure 3A). In healthy individuals, ex vivo Mtb-specific CD4+ T cells were predominately CCR4-CCR6+CXCR3+ (Th1*, median: 41%) or CCR4-CCR6-CXCR3+ (Th1, 20.4%, Figure 3B). Active TB disease induced the greatest changes on the expression pattern of chemokine receptors, where the proportion of tetramer+CD4+ T cells expressing CCR4-CCR6+CXCR3+ (Th1*) was significantly reduced (median: 5% for HIV-/aTB and 7.3% for HIV+/aTB), compared to cells from HIV-/LTBI donors (median: 40.6% for HIV-/LTBI and 15.6% for HIV+/LTBI, Figure 3B). These changes were, partly, counterbalanced by an elevated proportion of cells that did not express any of the tested chemokine receptors. The alterations induced by HIV infection, in LTBI individuals, were of a different nature compared to aTB-induced changes; tetramer+CD4+ T cells exhibited a significantly higher proportion of cells co-expressing CCR4+CCR6+CXCR3+ (median: 26%) and CCR4+CCR6+CXCR3- (Th17, 9%) when compared to the HIV-/LTBI group (7% and 2%, respectively) (Figure 3B). Although not statistically significant, these HIV-induced changes were, partly, counter balanced by a contraction of the proportion of CCR4-CCR6+CXCR3+ cells in HIV+/LTBI donors (median: 15.6%). Additionally, the proportion of Mtb-specific CCR4-CCR6+CXCR3+ CD4+ T cells negatively correlated with HIV viral load (p=0.004, r=-0.67), suggesting that HIV replication could preferentially reduce Mtb-specific Th1* responses. (Figure 3C). Of note, a lower proportion of cells expressing CCR4-CCR6+CXCR3+ in the total CD4 compartment from individuals with HIV or active TB was also observed (data not shown), indicating that the alteration of Th1* responses could be a global effect of active viral and/or bacterial replication, and not only restricted to Mtb-specific CD4+ T cells.
Fig. 3. Comparison of the chemokine receptor co-expression profile in ex vivo Mtb-specific MHC class II tetramer+CD4+ T cells in individuals with HIV and/or active TB.
A-Pie charts showing the median proportion of each possible chemokine receptor combination within ex vivo TET+CD4+ T cells. Statistical comparisons were performed using the pie statistic tool integrated in the Spice software. Each color corresponds to a different chemokine receptor combination. B- Proportions of cells expressing each possible chemokine receptor combination in ex vivo TET+CD4+ T cells using a Boolean gating strategy. LTBI/HIV- individuals are depicted with blue dots, LTBI/HIV+ individuals with red dots, aTB/HIV- individuals with green dots and aTB/HIV+ individuals with orange dots. Bars and boxes represent medians and interquartile ranges, respectively. Statistical comparisons were performed using the Student's t-test. T helper subsets assigned to known chemokine receptor combinations are indicated below. C- Relationship between the proportion of CCR4-CCR6+CXCR3+TET+CD4+ T cells and plasma HIV viral load. LTBI/HIV+ individuals are depicted with red dots and aTB/HIV+ individuals with orange dots. Correlation was tested by a two-tailed non-parametric Spearman Rank test.
Overall, these data indicate that active Mtb replication substantially altered the phenotype of ex vivo Mtb-specific CD4+ T cells, with skewing of their memory profile towards a late differentiated memory phenotype, they were highly activated and a significantly lower proportion of these cells were identified in the CCR4-CCR6+CXCR3+ (Th1*) subset, compared to individuals with LTBI. HIV-induced changes in the ex vivo Mtb-specific CD4+ T cell phenotype were more subtle, affecting primarily chemokine receptor co-expression, where CCR4+CCR6+CXCR3+ and CCR4+CCR6+CXCR3- (Th17) subsets were enriched to the detriment of CCR4-CCR6+CXCR3+ (Th1*) cells.
Characterizing the functional profile of Mtb-specific CD4+ T cells
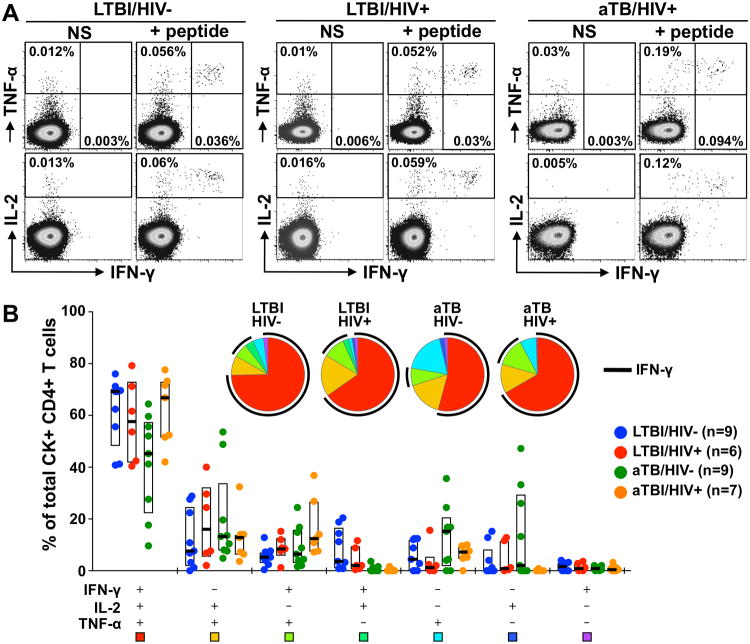
Having shown that both HIV and active TB disease impact the phenotype of ex vivo Mtb-specific CD4+ T cells, we next investigated whether the functional potential of these cells was affected by different disease states. Thus, we compared the ability of cells to secrete IFN-γ, IL-2 and TNF-α in response to CFP-10 or ESAT-6 peptide stimulation in the four clinical groups (Figure 4A). Of note, as IL-17 expression is rarely detectable in response to peptide stimulation (23, 24), this antibody was not included in our panel. Overall, despite the different phenotype observed in ex vivo Mtb-specific tetramer+CD4+ T cells in HIV or active TB, the functional capacities of Mtb-specific CD4+ T cells were comparable between the different clinical groups (Figure 4B). The majority of Mtb-responding CD4+ T cells were polyfunctional, co-expressing IFN-γ, IL-2 and TNF-α (median: 69% for HIV-/LTBI, 58% for HIV+/LTBI, 43% for HIV-/aTB and 67% for HIV+/aTB).
Fig. 4. Polyfunctional profile of Mtb-specific CD4+ T cells according to HIV and TB disease status.
A-Representative dot plots of IFN-γ, IL-2 and TNF-α production in response to cognate peptide in three individuals with distinct HIV and/or TB disease status. NS: No Stimulation. The frequencies of cytokine-producing cells are expressed as a percentage of total CD4+ T cells. B- Pie charts and graph representing the cytokine secretion profiles of Mtb-specific CD4+ T cells in response to cognate peptide stimulation. The four clinical groups are depicted as in Figure 3. Each section of the pie chart represents a specific combination of cytokines, as indicated by the color at the bottom of the graph. Horizontal bars and boxes depict the medians and interquartile ranges, respectively. The black arc on the pies corresponds to IFN-γ producing cells. Statistical comparisons were performed using a Wilcoxon rank-sum test.
As it has been shown that Mtb-specific CD4+ T cells are more permissive to HIV infection (25), we next wished to define whether such preferential targeting of Mtb-specific cells could alter their functionality by promoting cell exhaustion. Thus, we compared the frequency of ex vivo tetramer+CD4+ T cells to the frequency of Mtb-responding CD4+ T cells (i.e. cells producing IFN-γ, IL-2 or TNF-α) after cognate peptide stimulation to determine if Mtb-specific CD4+ T cells detected ex vivo using tetramers are functionally responsive to T-cell receptor (TCR) triggering (Figure 5A). The median frequency of Mtb-specific CD4+ T cells detected ex vivo using MHC class II tetramers was comparable to the median frequency of cytokine-responding CD4+ T cells in both the HIV-uninfected and HIV-infected groups (Figure 5B). In fact, there was a strong positive correlation between the frequencies of ex vivo tetramer+CD4+ T cells and cytokine-producing CD4+ T cells (p<0.0001, r=0.71, Figure 5C). Further analyses assessing the cytokine+/tetramer+ cell ratio in each clinical group revealed that, in some instances, cytokine+CD4+ T cell responses to cognate peptide were higher in magnitude (1.8- to 10-fold higher) compared to the frequency of the corresponding tetramer+CD4+ T cells (Figure 5D). Such a profile was predominantly observed for DRB5*01:01/CFP-1051–65 responses (11/12), suggesting that the CFP-1051–65 peptide is likely to be a promiscuous epitope presented by multiple HLA class II alleles. For non-DRB5*0101 restricted-epitopes, the cytokine+/tetramer+ cell ratio was close to 1 and comparable between all clinical groups (median: 0.8 for HIV-/LTBI, 0.78 for HIV+/LTBI, 1.2 for HIV-/aTB and 0.88 for HIV+/aTB, data not shown). Overall, these results suggest that neither HIV infection nor active TB promote Mtb-specific CD4+ T cell exhaustion, as most peripheral Mtb-specific CD4+ T cells detected ex vivo appear functional upon restimulation.
Fig. 5. Cytokine responsive potential of Mtb-specific MHC class II tetramer+CD4+ T cells according to HIV and TB disease status.
A-Representative examples of the frequency of TET+ cells and IFN-γ response to cognate peptide in two LTBI/HIV- individuals. NS: No Stimulation. B- Comparison of the frequencies of ex vivo TET+CD4+ T cells and cytokine+ (CK+) cells in response to cognate peptide in HIV- and HIV+ individuals (n=17 and n=13, respectively). The frequency of Mtb-specific CK+CD4+ T cells is defined as the frequency of cells expressing IFN-γ, IL-2 or TNF-α in response to cognate peptide after background (NS) subtraction. Bars represent medians. Triangles represent individuals with active TB. C- Association between the frequency of ex vivo TET+CD4+ T cells and CK+CD4+ T cells in response to cognate peptide. The dotted line represents a slope of 1. Correlations were tested by a two-tailed non-parametric Spearman Rank test. D- Ratio of the frequency of CK+CD4+ T cells/the frequency ex vivo TET+CD4+ T cells in each clinical group. Black symbols indicate DRB5*0101CFP-1051-65 responses. The grey area highlights individuals with a CK+/TET+ ratio ≥ 1.8. Statistical comparisons were performed using a Wilcoxon matched pairs test.
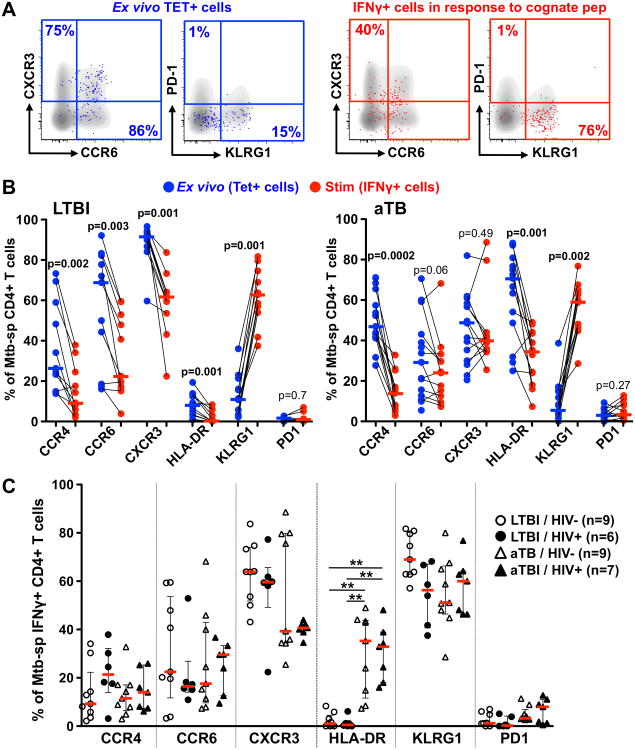
Cell stimulation alters the phenotype of ex vivo resting Mtb-specific CD4+ T cells
Finally, we compared the phenotypic profile of Mtb-specific CD4+ T cells in their resting and stimulated states (Figure 6A). Since IFN-γ expression comprised the predominate proportion of the Mtb-specific peptide response (Figure 4B), and little non-specific background for this cytokine was observed (data not shown), we focused on IFN-γ-producing cells to assess the phenotype of peptide-responding CD4+ T cells. Regardless of TB and HIV disease status, short-term TCR triggering using cognate peptide induced a significant decrease in CCR4 and HLA-DR expression, while KLRG1 was significantly elevated compared to tetramer+CD4+ T cells in their resting state (Figure 6B). Additionally, in individuals with LTBI, Mtb peptide-stimulated IFNγ+CD4+ T cells were also characterized by a significant decrease in CCR6 and CXCR3 expression compared to resting cells (p=0.003 and p=0.001, respectively, Figure 6B). These latter changes were not observed in active TB, possibly because their expression was already decreased in resting Mtb-specific CD4+ T cells (Figure 2D). Of note, these differences were not attributed to cell culture itself as the phenotypic profile of tetramer+CD4+ T cells observed ex vivo were comparable to unstimulated tetramer+CD4+ T cells after 16 h in culture (data not shown).
Fig. 6. Alteration of the phenotypic profile of ex vivo Mtb-specific MHC class II tetramer+CD4+ T cells upon short-term stimulation with cognate peptide.
A-Representative dot plots of ex vivo TET+CD4+ T cells (blue) and IFN-γ+CD4+ T cells (red) overlaid on total CD4+ T cell profile (grey) in one LTBI/HIV- individual. B- Expression of homing and activation markers on ex vivo TET+CD4+ T cells and IFN-γ+CD4+ T cells (after stimulation with cognate peptide) in LTBI individuals (n=11, left panel) and individuals with aTB (n=13, right panel). Bars represent the medians. Statistical comparisons were performed using a Wilcoxon matched pairs test. C- Comparison of the phenotypic profile of IFN-γ+CD4+ T cells between clinical groups. Statistical comparisons were performed using a one-way ANOVA Kruskal-Wallis test. *<0.05, **<0.01, ***<0.001.
When comparing the phenotypic profile of IFN-γ+CD4+ T cells between clinical groups, Figure 6C shows that chemokine receptor expression on IFN-γ+CD4+ T cells was comparable between the clinical groups. This indicates that TCR triggering-induced changes in Mtb-specific CD4+ T cells partly masks the differences in chemokine receptor expression in ex vivo tetramer+CD4+ T cells (Figure 2D). Conversely, despite the downregulation of HLA-DR upon peptide stimulation, HLA-DR on Mtb-specific IFN-γ+CD4+ T cells remained significantly higher in patients with active TB compared to LTBI (median: ∼35% vs ∼1%, respectively, Figure 6C).
These data demonstrate that the phenotypic profile of Mtb-specific CD4+ T cells was considerably altered as a result of TCR triggering, showing that the use of MHC class II tetramers permitted us to define the phenotypic nature of ex vivo Mtb-specific CD4+ T cells in a resting state; and identified differences in the context of HIV and/or active TB disease that may have been overlooked if these cells were assessed after stimulation.
Discussion
The use of MHC class II tetramers allows an unbiased quantification and characterization of antigen-specific CD4+ T cells in their resting state. In this study, we report for the first time in HIV co-infection and TB disease, the phenotypic and functional characterization ex vivo of Mtb-specific human CD4+ T cells using MHC class II tetramers. This allowed us to define the impact of HIV infection on ex vivo Mtb-specific CD4+ T cells and characterize these cells during active TB disease.
Firstly, we show comparable frequencies of ex vivo Mtb-specific CD4+ T cells between the HIV-/LTBI, HIV+/LTBI and HIV-/aTB groups and, significantly higher frequencies in HIV+/aTB compared to other groups. These results could appear inconsistent with previous reports showing that Mtb-specific CD4+ T cells are preferentially depleted during HIV infection (25, 26). However, in our study cohort, recruited from a highly TB endemic area, recurrent Mtb exposure and the relatively well-preserved CD4+ T cell count in HIV-infected individuals with LTBI (median: 558 cells/mm3) could account for the conservation of Mtb-specific CD4+ T cells. Moreover, elevated frequencies of Mtb-specific IFNγ+CD4+ T cells have been reported in severely immunocompromised HIV-infected patients with LTBI or aTB (26-29). Thus, the maintenance in the absolute number of Mtb-specific CD4+ T cells in active TB and HIV co-infection with severe lymphopenia demonstrates that bacterial replication induces the expansion of CD4+ T cells targeting Mtb, showing that memory Mtb responses are not eradicated in advanced HIV and that the residual Mtb-specific cells are not exhausted.
Secondly, assessing the phenotype of ex vivo Mtb-specific CD4+ T cells revealed that while active TB disease induced major alterations in cell profile, HIV-induced changes were of a more subtle nature. During TB disease, ex vivo Mtb-specific CD4+ T cells were highly activated, exhibited a mature memory phenotype and decreased expression of CXCR3. These specific cell features are typical of an acute infection, and suggest recent or ongoing cell stimulation (30-33). Conversely, HIV infection per sedid not induce significant changes in cell maturation or activation profile, but skewed the profile of chemokine receptor expression of ex vivo Mtb-specific CD4+ T cells. Mtb-specific cells in LTBI were previously reported to be almost exclusively CCR4-CCR6+CXCR3+ (Th1*) (34, 35). In this report, we found their chemokine receptor expression profile to be more diverse, even in the absence of HIV infection. These differences may be explained by frequent exposure to Mtb in our setting. HIV infection led to a reduction in the CCR4-CCR6+CXCR3+ subset, counterbalanced by the accumulation of CCR4+CCR6+CXCR3- cells (Th17) and a subset of cells co-expressing CCR4, CCR6 and CXCR3. This latter subset has been described in Mtb-specific CD4+ T cell clones generated from healthy individuals (22) and in peripheral blood from HIV-infected individuals on ART (36). Functionally, these cells shared characteristics with Th17 and Th1/Th17 subsets, producing IFN-γ and low amounts of IL-17 (36). The diversity and complexity in the chemokine receptor expression patterns of CD4 responses specific for a single Mtb peptide are remarkable. Our data suggest that HIV infection may bias Mtb-specific CD4+ T cell responses towards a more Th17-like phenotype. However, the manner in which skewed profiles during HIV co-infection may affect Mtb containment remains to be determined. HIV-induced alteration of chemokine receptor expression has been reported previously (37, 38). These alterations during HIV infection may alter the homing potential of Mtb-specific CD4+ T cells or affect their T helper lineage commitment, as we reported previously (11). Moreover, recent reports demonstrate that the homing potential and Th differentiation status of Mtb-specific cells can dramatically influence immune protection against TB (39, 40).
The use of Mtb-specific MHC class II tetramers allowed us to accurately probe the functionality of resting Mtb-specific CD4+ T cells. We reveal that the majority of circulating Mtb-specific CD4+ T cells appeared functional, as the frequency of cytokine-producing cells reflected the frequency of tetramer+ cells, and was comparable, regardless of HIV infection or active TB disease. We did not observe functional differences between LTBI and aTB, consistent with previous reports (41-44). This may be due to a limited number of participants analyzed in this study and CD4 responses to a single peptide not being representative of the response to the whole pathogen. However, these results further challenge the idea that polyfunctional Th1 cell responses to Mtb are protective (45). In addition, our data demonstrate that short-term TCR triggering induced considerable phenotypic changes in Mtb-specific CD4+ T cells, including substantial down-regulation in chemokine receptor expression and up-regulation of the inhibitory receptor KLRG1 (46). These phenomena have previously been reported for T cells of other specificities (30, 47). Such changes in recently activated cells could contribute to the negative regulation of T cell function. Due to the limited number of participants analyzed in this study, there was some disparity in age, gender and HIV disease severity between the studied groups. Thus, it will be of value to confirm our results in a larger cohort. Moreover, in this study, we report on HIV and aTB-associated phenotypic and functional profiles of Mtb-specific CD4+ T cells in the circulation, and it remains to de determined whether similar profiles would be observed at the site of disease (i.e. in the lung).
Overall, we describe a broader profile of Mtb-specific Th cells in individuals from a high burden setting, with major phenotypic changes induced by active TB disease, and more subtle changes during unsuppressed HIV replication. It remains to be seen whether these perturbations are normalized after treatment for HIV and/or TB. MHC class II tetramers for TB represent a useful tool for further ex vivo characterization of Mtb-specific cells, without the need for in vitro stimulation and potential modulation of expression of markers of interest. Further transcriptomic characterization of tetramer-sorted Mtb-specific cells is underway, which may identify additional phenotypic or functional differences induced by HIV and active TB.
Supplementary Material
Acknowledgments
We are grateful to the National Institutes of Health (NIH) Tetramer Core Facility (Emory University) for providing the MHC class II tetramers used in this study. We would like to thank Dr Thomas J. Scriba from the South African Tuberculosis Vaccine Initiative (SATVI) for sharing his knowledge and expertise regarding Mtb-specific MHC class II tetramer technology. We thank the study participants and staff at the Ubuntu HIV-TB clinic for their time and commitment to this project. We thank Mrs Kathryn Norman for administrative support.
Grant support. This work was funded by the National Research Foundation of South Africa (NRF, 92558 to CR)and the National Institutes of Health, the Office of the Director (OD) (NIH, R21AI115977 to CR). WAB was supported by the European and Developing Countries Clinical Trials Partnership (EDCTP; TA_08_40200_020), NRF (92755) and the Wellcome Trust (089832/Z/09/Z). NS is supported by a Clinical Infectious Diseases Research Initiative (CIDRI) postdoctoral fellowship. R.J.W. is supported by the Wellcome Trust (104803 and 203135), the European Union (FP7-Health-F3-2012-305578), Horizon 2020 under grant agreement 643381, the Francis Crick Institute which receives support from Cancer Research UK (FC00110218 to R.J.W.), the UK Medical Research Council (FC00110218 to R.J.W.), the National Institutes of Health (U01AI115940 to R.J.W.), the Medical Research Council of South Africa via its strategic health innovations partnerships, and National Research Foundation of South Africa (96841 to R.J.W.).
Footnotes
Contribution: Conceived and designed the experiments: CR, WAB and NS. Performed the experiments: NS, TLM and CR. Analyzed the data: CR and NS. Contributed reagents/materials/analysis tools: NB, RG, MC, RJW. Wrote the paper: CR, NS, WAB and RJW. All authors approved the final manuscript.
Disclosures. All authors report no potential conflicts. This project has been funded in part with federal funds from the Frederick National Laboratory for Cancer Research, under Contract No. HHSN261200800001E. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government. The HLA typing of study participants was supported in part by the Intramural Research Program of the NIH, Frederick National Lab, Center for Cancer Research.
References
- 1.WHO. Global Tuberculosis Report 2016 2016 [Google Scholar]
- 2.Aaron L, Saadoun D, Calatroni I, Launay O, Memain N, Vincent V, Marchal G, Dupont B, Bouchaud O, Valeyre D, Lortholary O. Tuberculosis in HIV-infected patients: a comprehensive review. Clin Microbiol Infect. 2004;10:388–398. doi: 10.1111/j.1469-0691.2004.00758.x. [DOI] [PubMed] [Google Scholar]
- 3.Du Bruyn E, Wilkinson RJ. The Immune Interaction between HIV-1 Infection and Mycobacterium tuberculosis. Microbiol Spectr. 2016;4:1–29. doi: 10.1128/microbiolspec.TBTB2-0012-2016. [DOI] [PubMed] [Google Scholar]
- 4.Whalen CC, Nsubuga P, Okwera A, Johnson JL, Hom DL, Michael NL, Mugerwa RD, Ellner JJ. Impact of pulmonary tuberculosis on survival of HIV-infected adults: a prospective epidemiologic study in Uganda. AIDS. 2000;14:1219–1228. doi: 10.1097/00002030-200006160-00020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kwan CK, Ernst JD. HIV and tuberculosis: a deadly human syndemic. Clin Microbiol Rev. 2011;24:351–376. doi: 10.1128/CMR.00042-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Okoye A, Picker L. CD4+ T-cell depletion in HIV infection: mechanisms of immunological failure. Immunol Rev. 2013;254:54–64. doi: 10.1111/imr.12066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sonnenberg P, Glynn JR, Fielding K, Murray J, Godfrey-Faussett P, Shearer S. How soon after infection with HIV does the risk of tuberculosis start to increase? A retrospective cohort study in South African gold miners. J Infect Dis. 2005;191:150–158. doi: 10.1086/426827. [DOI] [PubMed] [Google Scholar]
- 8.Day CL, Mkhwanazi N, Reddy S, Mncube Z, van der Stok M, Klenerman P, Walker BD. Detection of polyfunctional Mycobacterium tuberculosis-specific T cells and association with viral load in HIV-1-infected persons. J Infect Dis. 2008;197:990–999. doi: 10.1086/529048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kalsdorf B, Scriba TJ, Wood K, Day CL, Dheda K, Dawson R, Hanekom WA, Lange C, Wilkinson RJ. HIV-1 infection impairs the bronchoalveolar T-cell response to mycobacteria. Am J Respir Crit Care Med. 2009;180:1262–1270. doi: 10.1164/rccm.200907-1011OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Matthews K, Ntsekhe M, Syed F, Scriba T, Russell J, Tibazarwa K, Deffur A, Hanekom W, Mayosi BM, Wilkinson RJ, Wilkinson KA. HIV-1 infection alters CD4+ memory T-cell phenotype at the site of disease in extrapulmonary tuberculosis. Eur J Immunol. 2012;42:147–157. doi: 10.1002/eji.201141927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Riou C, Strickland N, Soares AP, Corleis B, Kwon DS, Wherry EJ, Wilkinson RJ, Burgers WA. HIV Skews the Lineage-Defining Transcriptional Profile of Mycobacterium tuberculosis-Specific CD4+ T Cells. J Immunol. 2016;196:3006–3018. doi: 10.4049/jimmunol.1502094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nasi M, Pinti M, Mussini C, Cossarizza A. Persistent inflammation in HIV infection: established concepts, new perspectives. Immunol Lett. 2014;161:184–188. doi: 10.1016/j.imlet.2014.01.008. [DOI] [PubMed] [Google Scholar]
- 13.Morou A, Palmer BE, Kaufmann DE. Distinctive features of CD4+ T cell dysfunction in chronic viral infections. Curr Opin HIV AIDS. 2014;9:446–451. doi: 10.1097/COH.0000000000000094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tameris MD, Hatherill M, Landry BS, Scriba TJ, Snowden MA, Lockhart S, Shea JE, McClain JB, Hussey GD, Hanekom WA, Mahomed H, McShane H. Safety and efficacy of MVA85A, a new tuberculosis vaccine, in infants previously vaccinated with BCG: a randomised, placebo-controlled phase 2b trial. The Lancet. 2013;381:1021–1028. doi: 10.1016/S0140-6736(13)60177-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ndiaye BP, Thienemann F, Ota M, Landry BS, Camara M, Dièye S, Dieye TN, Esmail H, Goliath R, Huygen K, January V, Ndiaye I, Oni T, Raine M, Romano M, Satti I, Sutton S, Thiam A, Wilkinson KA, Mboup S, Wilkinson RJ, McShane H. Safety, immunogenicity, and efficacy of the candidate tuberculosis vaccine MVA85A in healthy adults infected with HIV-1: a randomised, placebo-controlled, phase 2 trial. The Lancet Respiratory Medicine. 2015;3:190–200. doi: 10.1016/S2213-2600(15)00037-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Moonsamy PV, Williams T, Bonella P, Holcomb CL, Hoglund BN, Hillman G, Goodridge D, Turenchalk GS, Blake LA, Daigle DA, Simen BB, Hamilton A, May AP, Erlich HA. High throughput HLA genotyping using 454 sequencing and the Fluidigm Access Array System for simplified amplicon library preparation. Tissue Antigens. 2013;81:141–149. doi: 10.1111/tan.12071. [DOI] [PubMed] [Google Scholar]
- 17.Roederer M, Nozzi JL, Nason MC. SPICE: exploration and analysis of post-cytometric complex multivariate datasets. Cytometry A. 2011;79:167–174. doi: 10.1002/cyto.a.21015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tikly M, Rands A, McHugh N, Wordsworth P, Welsh K. Human leukocyte antigen class II associations with systemic sclerosis in South Africans. Tissue Antigens. 2004;63:487–490. doi: 10.1111/j.0001-2815.2004.00199.x. [DOI] [PubMed] [Google Scholar]
- 19.Sallusto F, Lanzavecchia A. Heterogeneity of CD4+ memory T cells: functional modules for tailored immunity. Eur J Immunol. 2009;39:2076–2082. doi: 10.1002/eji.200939722. [DOI] [PubMed] [Google Scholar]
- 20.Sallusto F, Mackay CR, Lanzavecchia A. The role of chemokine receptors in primary, effector, and memory immune responses. Annu Rev Immunol. 2000;18:593–620. doi: 10.1146/annurev.immunol.18.1.593. [DOI] [PubMed] [Google Scholar]
- 21.Acosta-Rodriguez EV, Rivino L, Geginat J, Jarrossay D, Gattorno M, Lanzavecchia A, Sallusto F, Napolitani G. Surface phenotype and antigenic specificity of human interleukin 17-producing T helper memory cells. Nat Immunol. 2007;8:639–646. doi: 10.1038/ni1467. [DOI] [PubMed] [Google Scholar]
- 22.Becattini S, Latorre D, Mele F, Foglierini M, De Gregorio C, Cassotta A, Fernandez B, Kelderman S, Schumacher TN, Corti D, Lanzavecchia A, Sallusto F. T cell immunity. Functional heterogeneity of human memory CD4(+) T cell clones primed by pathogens or vaccines. Science. 2015;347:400–406. doi: 10.1126/science.1260668. [DOI] [PubMed] [Google Scholar]
- 23.Riou C, Bunjun R, Muller TL, Kiravu A, Ginbot Z, Oni T, Goliath R, Wilkinson RJ, Burgers WA. Selective reduction of IFN-gamma single positive mycobacteria-specific CD4+ T cells in HIV-1 infected individuals with latent tuberculosis infection. Tuberculosis (Edinb) 2016;101:25–30. doi: 10.1016/j.tube.2016.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lindestam Arlehamn CS, McKinney DM, Carpenter C, Paul S, Rozot V, Makgotlho E, Gregg Y, van Rooyen M, Ernst JD, Hatherill M, Hanekom WA, Peters B, Scriba TJ, Sette A. A Quantitative Analysis of Complexity of Human Pathogen-Specific CD4 T Cell Responses in Healthy M. tuberculosis Infected South Africans. PLoS Pathog. 2016;12:e1005760. doi: 10.1371/journal.ppat.1005760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Geldmacher C, Ngwenyama N, Schuetz A, Petrovas C, Reither K, Heeregrave EJ, Casazza JP, Ambrozak DR, Louder M, Ampofo W, Pollakis G, Hill B, Sanga E, Saathoff E, Maboko L, Roederer M, Paxton WA, Hoelscher M, Koup RA. Preferential infection and depletion of Mycobacterium tuberculosis-specific CD4 T cells after HIV-1 infection. J Exp Med. 2010;207:2869–2881. doi: 10.1084/jem.20100090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Geldmacher C, Schuetz A, Ngwenyama N, Casazza JP, Sanga E, Saathoff E, Boehme C, Geis S, Maboko L, Singh M, Minja F, Meyerhans A, Koup RA, Hoelscher M. Early depletion of Mycobacterium tuberculosis-specific T helper 1 cell responses after HIV-1 infection. J Infect Dis. 2008;198:1590–1598. doi: 10.1086/593017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rangaka MX, Diwakar L, Seldon R, van Cutsem G, Meintjes GA, Morroni C, Mouton P, Shey MS, Maartens G, Wilkinson KA, Wilkinson RJ. Clinical, immunological, and epidemiological importance of antituberculosis T cell responses in HIV-infected Africans. Clin Infect Dis. 2007;44:1639–1646. doi: 10.1086/518234. [DOI] [PubMed] [Google Scholar]
- 28.Hammond AS, McConkey SJ, Hill PC, Crozier S, Klein MR, Adegbola RA, Rowland-Jones S, Brookes RH, Whittle H, Jaye A. Mycobacterial T cell responses in HIV-infected patients with advanced immunosuppression. J Infect Dis. 2008;197:295–299. doi: 10.1086/524685. [DOI] [PubMed] [Google Scholar]
- 29.Vignesh R, Kumarasamy N, Lim A, Solomon S, Murugavel KG, Balakrishnan P, Solomon SS, Mayer KH, Swathirajan CR, Chandrasekaran E, Pradeep A, Poongulali S, Benson CA, French MA. TB-IRIS after initiation of antiretroviral therapy is associated with expansion of preexistent Th1 responses against Mycobacterium tuberculosis antigens. J Acquir Immune Defic Syndr. 2013;64:241–248. doi: 10.1097/QAI.0b013e31829f6df2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sallusto F, Kremmer E, Palermo B, Hoy A, Ponath P, Qin S, Forster R, Lipp M, Lanzavecchia A. Switch in chemokine receptor expression upon TCR stimulation reveals novel homing potential for recently activated T cells. Eur J Immunol. 1999;29:2037–2045. doi: 10.1002/(SICI)1521-4141(199906)29:06<2037::AID-IMMU2037>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- 31.Petruccioli E, Petrone L, Vanini V, Sampaolesi A, Gualano G, Girardi E, Palmieri F, Goletti D. IFNgamma/TNFalpha specific-cells and effector memory phenotype associate with active tuberculosis. J Infect. 2013;66:475–486. doi: 10.1016/j.jinf.2013.02.004. [DOI] [PubMed] [Google Scholar]
- 32.Adekambi T, Ibegbu CC, Cagle S, Kalokhe AS, Wang YF, Hu Y, Day CL, Ray SM, Rengarajan J. Biomarkers on patient T cells diagnose active tuberculosis and monitor treatment response. J Clin Invest. 2015;125:1827–1838. doi: 10.1172/JCI77990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wilkinson KA, Oni T, Gideon HP, Goliath R, Wilkinson RJ, Riou C. Activation Profile of Mycobacterium tuberculosis-Specific CD4(+) T Cells Reflects Disease Activity Irrespective of HIV Status. Am J Respir Crit Care Med. 2016;193:1307–1310. doi: 10.1164/rccm.201601-0116LE. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lindestam Arlehamn CS, Gerasimova A, Mele F, Henderson R, Swann J, Greenbaum JA, Kim Y, Sidney J, James EA, Taplitz R, McKinney DM, Kwok WW, Grey H, Sallusto F, Peters B, Sette A. Memory T cells in latent Mycobacterium tuberculosis infection are directed against three antigenic islands and largely contained in a CXCR3+CCR6+ Th1 subset. PLoS Pathog. 2013;9:e1003130. doi: 10.1371/journal.ppat.1003130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Arlehamn CL, Seumois G, Gerasimova A, Huang C, Fu Z, Yue X, Sette A, Vijayanand P, Peters B. Transcriptional profile of tuberculosis antigen-specific T cells reveals novel multifunctional features. J Immunol. 2014;193:2931–2940. doi: 10.4049/jimmunol.1401151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wacleche VS, Goulet JP, Gosselin A, Monteiro P, Soudeyns H, Fromentin R, Jenabian MA, Vartanian S, Deeks SG, Chomont N, Routy JP, Ancuta P. New insights into the heterogeneity of Th17 subsets contributing to HIV-1 persistence during antiretroviral therapy. Retrovirology. 2016;13:59. doi: 10.1186/s12977-016-0293-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wacleche VS, Chomont N, Gosselin A, Monteiro P, Goupil M, Kared H, Tremblay C, Bernard N, Boulassel MR, Routy JP, Ancuta P. The colocalization potential of HIV-specific CD8+ and CD4+ T-cells is mediated by integrin beta7 but not CCR6 and regulated by retinoic acid. PLoS One. 2012;7:e32964. doi: 10.1371/journal.pone.0032964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bunders MJ, van Hamme JL, Jansen MH, Boer K, Kootstra NA, Kuijpers TW. Fetal exposure to HIV-1 alters chemokine receptor expression by CD4+T cells and increases susceptibility to HIV-1. Sci Rep. 2014;4:6690. doi: 10.1038/srep06690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sakai S, Mayer-Barber KD, Barber DL. Defining features of protective CD4 T cell responses to Mycobacterium tuberculosis. Curr Opin Immunol. 2014;29:137–142. doi: 10.1016/j.coi.2014.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sallin MA, Sakai S, Kauffman KD, Young HA, Zhu J, Barber DL. Th1 Differentiation Drives the Accumulation of Intravascular, Non-protective CD4 T Cells during Tuberculosis. Cell Rep. 2017;18:3091–3104. doi: 10.1016/j.celrep.2017.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Harari A, Rozot V, Bellutti Enders F, Perreau M, Stalder JM, Nicod LP, Cavassini M, Calandra T, Blanchet CL, Jaton K, Faouzi M, Day CL, Hanekom WA, Bart PA, Pantaleo G. Dominant TNF-alpha+ Mycobacterium tuberculosis-specific CD4+ T cell responses discriminate between latent infection and active disease. Nat Med. 2011;17:372–376. doi: 10.1038/nm.2299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pollock KM, Whitworth HS, Montamat-Sicotte DJ, Grass L, Cooke GS, Kapembwa MS, Kon OM, Sampson RD, Taylor GP, Lalvani A. T-cell immunophenotyping distinguishes active from latent tuberculosis. J Infect Dis. 2013;208:952–968. doi: 10.1093/infdis/jit265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lichtner M, Mascia C, Sauzullo I, Mengoni F, Vita S, Marocco R, Belvisi V, Russo G, Vullo V, Mastroianni CM. Multifunctional Analysis of CD4+ T-Cell Response as Immune-Based Model for Tuberculosis Detection. J Immunol Res. 2015;2015:217287. doi: 10.1155/2015/217287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kim K, Perera R, Tan DB, Fernandez S, Seddiki N, Waring J, French MA. Circulating mycobacterial-reactive CD4+ T cells with an immunosuppressive phenotype are higher in active tuberculosis than latent tuberculosis infection. Tuberculosis (Edinb) 2014;94:494–501. doi: 10.1016/j.tube.2014.07.002. [DOI] [PubMed] [Google Scholar]
- 45.Wilkinson KA, Wilkinson RJ. Polyfunctional T cells in human tuberculosis. Eur J Immunol. 2010;40:2139–2142. doi: 10.1002/eji.201040731. [DOI] [PubMed] [Google Scholar]
- 46.Voehringer D, Koschella M, Pircher H. Lack of proliferative capacity of human effector and memory T cells expressing killer cell lectinlike receptor G1 (KLRG1) Blood. 2002;100:3698–3702. doi: 10.1182/blood-2002-02-0657. [DOI] [PubMed] [Google Scholar]
- 47.McMahon CW, Zajac AJ, Jamieson AM, Corral L, Hammer GE, Ahmed R, Raulet DH. Viral and bacterial infections induce expression of multiple NK cell receptors in responding CD8(+) T cells. J Immunol. 2002;169:1444–1452. doi: 10.4049/jimmunol.169.3.1444. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.