Abstract
NGR (asparagine-glycine-arginine) is a tumor vasculature-homing peptide motif widely used for the functionalization of drugs, nanomaterials and imaging compounds for cancer treatment and diagnosis. Unfortunately, this motif has a strong propensity to undergo rapid deamidation. This reaction, which converts NGR into isoDGR, is associated with receptor switching from CD13 to integrins, with potentially important manufacturing, pharmacological and toxicological implications. It is found that glycine N-methylation of NGR-tagged nanocarriers completely prevents asparagine deamidation without impairing CD13 recognition. Studies in animal models have shown that the methylated NGR motif can be exploited for delivering radiolabeled compounds and nanocarriers, such as tumor necrosis factor-α (TNF)-bearing nanogold and liposomal doxorubicin, to tumors with improved selectivity. These findings suggest that this NGR derivative is a stable and efficient tumor-homing ligand that can be used for delivering functional nanomaterials to tumor vasculature.
Keywords: deamidation, enzyme, molecular docking, N-methylglycine, peptides
1. Introduction
Tumor-homing peptides represent an important class of ligands that can be exploited for enhancing the accumulation of anticancer drugs, nanomaterials or imaging compounds in tumors, thereby improving their therapeutic or diagnostic properties. Among the various peptide motifs identified so far for this purpose, NGR (asparagine-glycine-arginine) is one of the most widely used.[1] Indeed, peptides containing this motif have proven useful for the preparation of functional materials useful for delivering a variety of compounds to tumors, such as liposomes, gold nanoparticles, DNA complexes, viral particles, anti-angiogenic compounds, chemotherapeutic drugs, imaging compounds, and proteins, such as tumor necrosis factor (TNF)-α, interferon (IFN)-γ, IFN-α-2a and tissue factor.[2] A primary mechanism underlying the tumor-homing properties of NGR relies on the recognition of aminopeptidase N (CD13, EC 3.4.11.2) expressed on the membrane of endothelial cells and other perivascular cells in tumors.[3] Remarkably, some NGR-drug conjugates are being tested in patients.[2d, 4] For example, the CNGRCG-TNFα fusion product, developed by our group and called NGR-TNF, is currently tested in Phase II and III clinical studies in patients with solid tumors, with evidence of activity and good tolerability.[4–5] The good results obtained with this drug and also with other NGR-based materials in preclinical and clinical studies highlight the usefulness and versatility of the NGR motif for the preparation of tumor-homing drugs and materials.
A major potential problem associated with the use of this motif is related to the fact that its asparagine residue has a strong propensity to undergo deamidation,[6] a spontaneous reaction that converts asparagine (N) into aspartate (D) and isoaspartate (isoD), thereby converting NGR into DGR and isoDGR (Figure 1A).[7] Remarkably, this reaction occurs in an unusually rapid manner in NGR-containing compounds (e.g., the half-life of CNGRCG is 3–5 h in physiological solutions).[7] The consequent structural changes are associated with a dramatic change-of-function, causing receptor switching from CD13 to integrins.[7a, 7b, 8] Indeed, the isoDGR-, but not NGR- or DGR-, can mimic arginine-glycine-aspartate (RGD)-motif and bind to the RGD-binding pocket of various integrins, such as αvβ3, αvβ5, αvβ6, αvβ8 and α5β1, with different affinity and selectivity depending on flanking residues and molecular scaffolds.[7b, 9] Considering the rapid kinetics of NGR deamidation, this reaction may occur during NGR-drug preparation and storage, and, potentially, also in vivo, after drug administration to animals or to patients. Thus, the poor stability of NGR may represent an important issue for drug development and manufacturing, and may also have important pharmacological and toxicological implications.
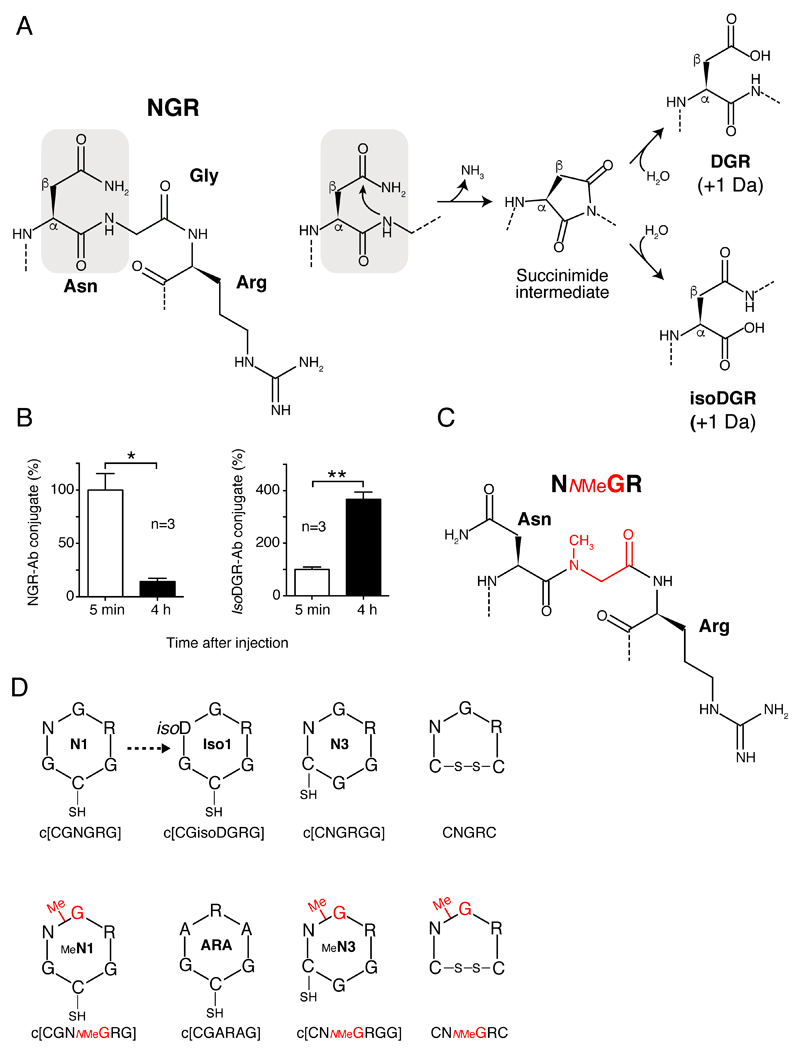
Figure 1. NGR-to-isoDGR transition and schematic representation of synthetic peptides.
A) NGR deamidation occurs by nucleophilic attack on the carbonyl group of asparagine side-chain by the adjacent peptide-bond nitrogen, leading to formation of a succinimide ring, and, after hydrolysis to isoDGR and DGR (with a gain of 1 Da).
B) NGR-to-isoDGR transition in vivo. An NGR peptide-antibody conjugate (NGR-Ab) was injected intravenous (i.v.) into mice and plasma samples, collected after 5 min and 4 h, were tested by ELISAs specific for NGR-Ab or isoDGR-Ab (see Supporting Information). *p<0.05, **p<0.01, two-tail t-test.
C) 2D representation of NNMeGR sequence and (D) of cyclic peptides containing NGR, NNMeGR, isoDGR, or ARA motifs, embedded in different scaffolds. Aminoacids are represented with the single letter code; NMeG, N-methlyglycine (sarcosine); SH, free thiol group; isoD, isoaspartate. Peptide codes are reported within each structure.
In this work, we show that the NGR-to-isoDGR transition can indeed occur in vivo, after injection into mice. Furthermore, we show that replacement of glycine with N-methylglycine (sarcosine) in a head-to-tail cyclized peptide (c[CGNGRG]) previously used for targeting TNFα-bearing gold nanoparticles to tumors,[10] completely prevents asparagine deamidation without impairing CD13 recognition. Using murine models of solid tumors, we also show that this NGR-derivative has a higher selectivity for tumor tissues than conventional NGR peptides and that it can be used for the preparation of more stable and selective drug nanocarriers, such as peptide-tagged nanogold and liposomes.
2. Results and Discussion
2.1. The NGR motif is rapidly converted to isoDGR in vivo
To assess whether the NGR-to-isoDGR transition can occur in vivo we injected in mice a conjugate consisting of a peptide encompassing the CNGRCG and TNF1-11 sequences coupled to an anti-vasostatin-1 monoclonal antibody. The plasma levels of this peptide-antibody conjugate were then analyzed at different time points using two enzyme-linked-immunosorbent assay (ELISA) based on the use of vasostatin-1 in the capture step, and antibodies against the TNF1-11 sequence (thus unable to discriminate between NGR and isoDGR), or antibodies specific for isoDGR, in the detection steps (Figure S1). A marked reduction of NGR-antibody conjugate (NGR-Ab) occurred 4 h after injection (Figure 1B, left panel), likely consequent to peptide degradation and antibody clearance. However, a marked increase of isoDGR-antibody conjugate (isoDGR-Ab) was also observed (Figure 1B, right panel, and Table S1), indicating that the NGR-to- isoDGR transition can indeed occur in vivo.
2.2. The replacement of glycine with N-methylglycine in NGR peptides prevents asparagine deamidation
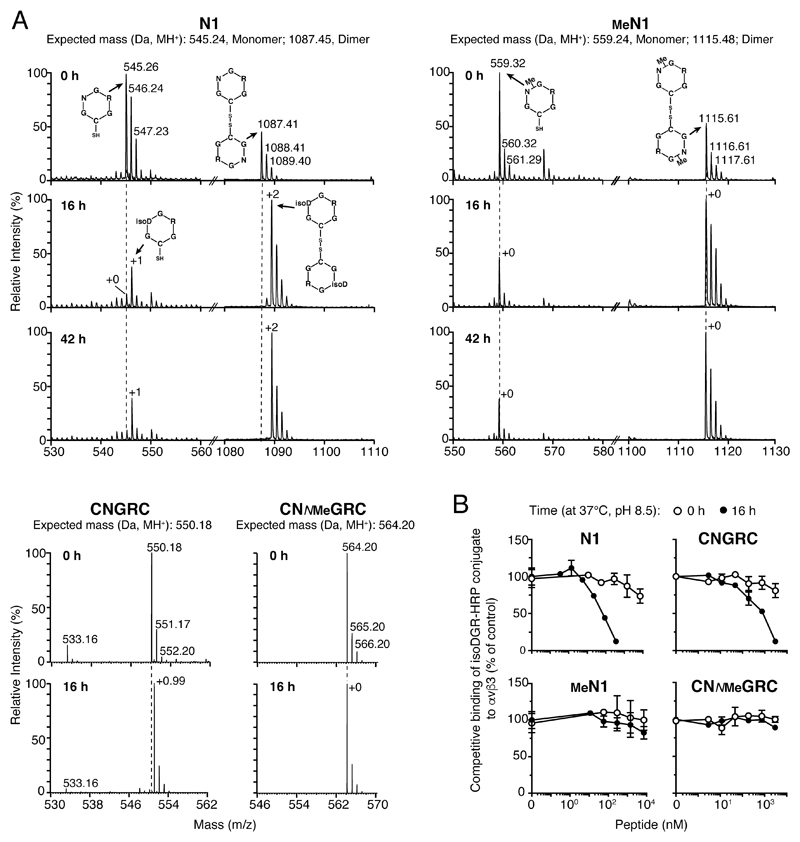
In the attempt to develop a more stable peptide, we hypothesized that glycine N-methylation in NGR (Figure 1C and D) can prevent asparagine deamidation and, consequently, improve peptide stability. To test this hypothesis, we synthetized various cyclic peptides containing NGR and their N-methylated derivatives (NNMeGR) embedded in different scaffolds (see Figure 1D and Table S2). Mass spectrometry (MS) analysis of the head-to-tail cyclized peptide c[CGNGRG] (called N1) before and after incubation for 16 or 42 h in 0.1 ammonium bicarbonate, pH 8.5 (a condition known to favor NGR deamidation), showed a gain of 1 Da, pointing to complete deamidation (Figure 2A, upper panel, left). In contrast, no change of molecular mass was observed with the methylated peptide c[CGNNMeGRG] (called MeN1) (Figure 2A, upper panel, right). Glycine N-methylation blocked deamidation also in other NGR peptides, such as CNGRC (disulfide-bridged) (Figure 2A, bottom panel) and c[CNGRGG] (head-to-tail cyclized, called N3) (Figure S2A). Integrin binding assays showed that N1, N3 and CNGRC, but not MeN1, MeN3 and CNMeGRC, could bind αvβ3 integrin after 16 h of incubation (Figure 2B and Figure S2B). Thus, glycine N-methylation in NGR peptides completely prevents asparagine deamidation and isoDGR formation.
Figure 2. Glycine N-methylation in NGR peptides prevents asparagine deamidation.
A) MS analysis (deconvoluted molecular weight spectrum) of N-methylated and non-methylated NGR peptides before and after incubation at 37 °C in 0.1 M ammonium bicarbonate buffer, pH 8.5 for 16 or 42 h. The molecular weights of monomers and disulfide-bridged dimers, formed during incubation and analysis, are shown. The +2 Da observed for N1 after 16-42 h likely accounts for complete dimer deamidation.
B) Binding of peptides to αvβ3 coated-microtiter plates before (0 h) and after (16 h) of incubation, as measured by competitive ELISA.
2.3. Glycine N-methylation of N1 does not impair CD13 recognition
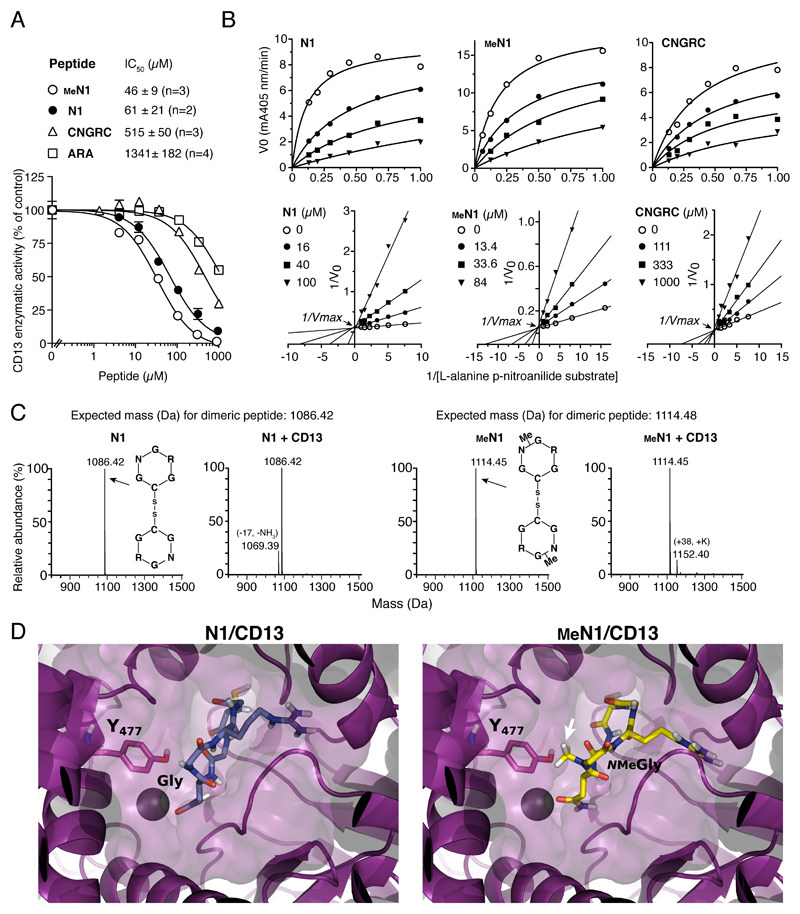
To assess whether glycine N-methylation affects CD13 recognition we performed enzyme inhibition assays with methylated and non-methylated peptides. Both N1 and MeN1 could inhibit CD13 (aminopeptidase N) enzymatic activity (Figure 3A and Table S3). Steady-state enzyme kinetic analyses (Figure 3B) showed that these peptides could inhibit the enzymatic activity of CD13 with similar potency (N1, Ki = 12.4 ± 5.4 μM; MeN1, Ki = 8.9 ± 1.8 µM). Interestingly, MeN1 was about 15-fold more potent than the reference CNGRC peptide (CNGRC, Ki = 134 ± 28 μM). MeN1, N1, and CNGRC affected the Michaelis constant (Km), but not the maximum velocity (Vmax) of CD13 (Figure 3B, bottom panels). These data are consistent with the competitive inhibition model and suggest that N1, MeN1 and CNGRC can bind within, or close to, the active site of the enzyme. Interestingly, while N-methylation of N1 did not impair CD13 binding, a reduction of inhibitory potency was observed after modification of N3, CNGRC and NGRAHA (Table S3). Thus, the overall effect of peptide methylation on CD13 recognition depends on the molecular scaffold of NGR. No change in the molecular mass of N1 and MeN1 were observed upon 40 min incubation with CD13 (Figure 3C), indicating that no peptide cleavage occurred and, therefore, that these peptides were not CD13 substrates.
Figure 3. N1 glycine N-methylation does not prevent CD13 recognition.
A) Effect of different peptides on CD13 enzymatic activity. IC50, peptide concentration that inhibit 50% of activity; n, number of independent experiments (each in duplicate). A representative experiment is shown (mean ± standard error, SE).
B) Steady state kinetic analysis of CD13 in the presence of various amounts of substrate and peptides. Representative plots of initial velocity (V0) versus substrate concentration (upper panels) and double reciprocal plot (lower panels). The inhibitory constant (Ki) values reported in each plot are the result of two independent experiments (mean ± SE).
C) MS analysis of N1 and MeN1 (30 µM) after incubation with or without CD13. No changes in the molecular mass were observed, suggesting that both peptides are resistant to degradation by CD13.
D) Representative decoy poses of N1 (left panel) and MeN1 (right panel) with CD13. CD13 is shown as purple cartoon and surface. The Y477 of CD13 interacting with G or NMeG is highlighted in sticks and labeled with the one-letter code. The arrow indicates the N-methylation. Zn2+ is represented as a sphere; peptides are represented as sticks.
2.4. Molecular docking of N1 and MeN1 on CD13 reveals that both peptides can accommodate into the same enzyme cavity
To provide a structural rationale for this hypothesis, N1 and MeN1 were docked into the CD13 crystallographic structure (PDB code: 4fyr),[11] as described in Supporting Information. Both peptides could adopt a binding mode reminiscent of that displayed by CNGRCG in complex with CD13 (PDB code: 4ou3)[12] (Figure 3D and Figure S3). Analysis of the docking poses of MeN1 and N1 revealed that the asparagine is involved in the coordination of the zinc ion and in stable interactions with the GXMEN catalytic motif of CD13. Moreover, the cysteine and the arginine establish polar interactions with the enzyme IV domain. Finally, the glycine and its methylated form interact with Y477 of CD13, an important residue involved in the catalytic transition state,[13] through polar and hydrophobic interactions, respectively. No other significant contacts were detected with residues of the large internal cavity of the enzyme. These results suggest that both peptides can accommodate into the enzyme cavity and that N-methylation is well tolerated (Figure 3D, Figure S3, Figure S4 and Table S5).
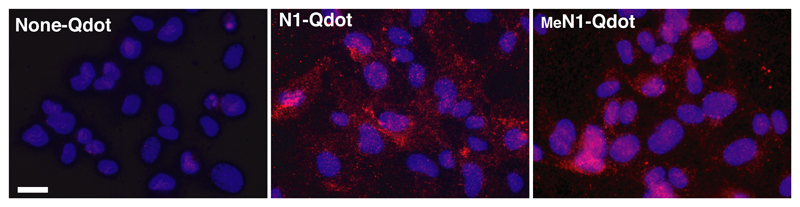
2.5. MeN1 and N1 bind endothelial cells
N1 and MeN1, coupled to Qdot fluorescent nanoparticles, could bind CD13+ endothelial cells (HUVEC) with similar potencies (Figure 4), suggesting that the methyl-group of MeN1 does not impair the recognition of CD13, on these cells.
Figure 4. N1 glycine N-methylation does not prevent binding to CD13+ human umbilical vein cells (HUVEC).
Binding of N1 and MeN1 coupled to fluorescent Qdot605 nanoparticles (6 nM, 2h) to cultured HUVEC (see Supporting Information). None-Qdot corresponds to nanoparticles activated with sulfosuccinimidyl 4-(N-maleimidomethyl)cyclohexane-1-carboxylate (sulfo-SMCC), but lacking the peptide.
Magnification, 400X; bar, 20 µm; red, Qdots; blue, nuclear staining with 4′,6-diamidine-2′-phenylindole dihydrochloride (DAPI).
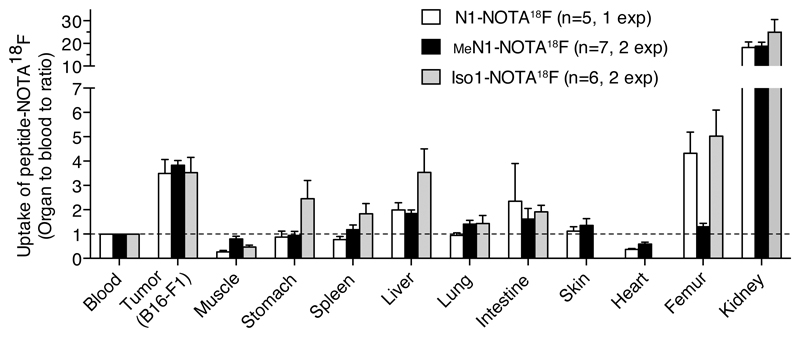
2.6. Radio-labeled MeN1 homes to tumors with a selectivity higher than that of N1
To compare the tumor-homing properties of N1 and MeN1 we prepared 18F-radiolabeled N1-and MeN1–2,2′,2 ″-(1,4,7-triazacyclononane-1,4,7-triyl)triacetic acid (MeN1-NOTA) conjugates (Figure S5) and analyzed their biodistribution in B16-F1 melanoma-bearing mice, 3 h after injection. In parallel, to assess the contribution of deamidated products, we performed a similar study with c[CGisoDGRC] (called Iso1, see Figure 1D and Fig S5). The results showed 4-fold accumulation of MeN1-NOTA18F in tumors compared to blood and other tissues (Figure 5). Similar accumulation was observed with N1- and Iso1-NOTA18F. However, in these cases a certain degree of accumulation was observed also in other organs (e.g. stomach, spleen, liver, intestine, femur), pointing to a lower specificity of N1 and Iso1. Co-administration of labeled peptides with an excess of the corresponding free peptide-NOTA showed a significant inhibition of tumor uptake in the case of MeN1 and Iso1, but not in the case of N1 (Figure S6). One possible explanation for the lower specificity of N1-NOTA18F and the lack of inhibition by N1 is that a small amount of Iso1 was formed in this conjugate, which is less specific and which could not be competed by free N1. These results suggest that glycine N-methylation does not impair the tumor-homing properties of this NGR peptide, and, remarkably, that it increases its tumor selectivity (likely by preventing isoDGR formation).
Figure 5. Radio-labeled MeN1 homes to tumors with a selectivity higher than that of N1 and Iso1.
Biodistribution of radiolabeled N1-, MeN1-, and Iso1-NOTA18F in B16-F1 melanoma-bearing mice (3 h after administration). Cumulative results of 1 or 2 independent experiments (bars, mean ± SE; n=5-7 mice/group).
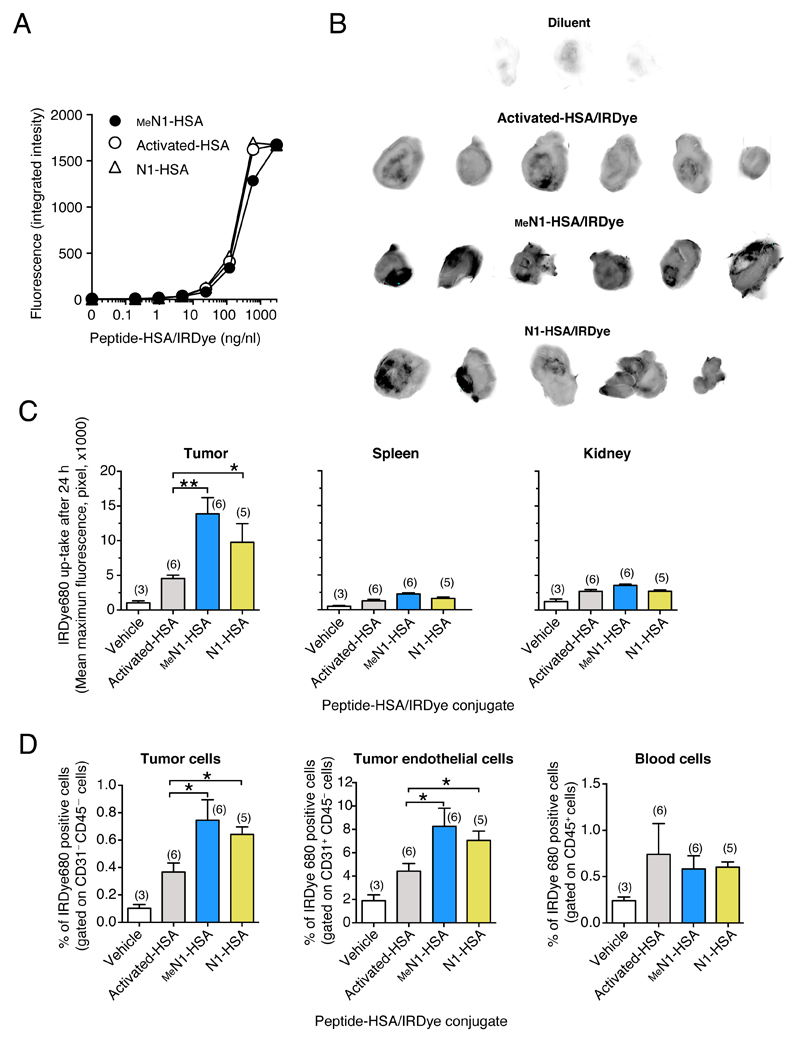
2.7. The MeN1-albumin conjugate accumulates on tumor and endothelial cells, but not on blood cells
Other in vivo assays performed with peptide-albumin conjugates and fluorescence-labeled albumin (HSA) (Figure 6A and Figure S7) showed that N1 and MeN1 could increase the accumulation of albumin in tumors, but not in spleen and kidney, to a similar extent (Figure 6B and 6C). Interestingly, fluorescence activated cell sorting (FACS) analysis of cells obtained from these tumors showed that N1 and MeN1 increased albumin accumulation on tumor and endothelial cells, but not on blood cells (Figure 6D). These data further support the concept that glycine N-methylation in N1 does not abrogate its capability to target the tumor and endothelial cells. This also suggests that MeN1 may recognize with high affinity a CD13 form expressed by the endothelium of tumor vessels, and not (or less efficiently) CD13 expressed by other tissues. In line with this hypothesis, we did not observe binding of fluorescent MeN1-HSA at low (nanomolar) concentration to human acute T lymphoblastic leukemia cells (MOLT-4) transfected with human CD13 cDNA (CD13/MOLT-4) (Table S6).
Figure 6. N1 and MeN1-tagged albumin home to CD13+ tumors.
A) Fluorescence intensity of HSA tagged with N1 and MeN1-and labeled with IRDye680, as analyzed using the Odyssey scanner. “Activated-HSA/IRDye” corresponds to the negative control (lacking the peptide).
B-C) Tumor-homing properties of N1-HSA/, MeN1-HSA/, or activated-HSA/IRDye conjugates in the WEHI-164 fibrosarcoma model. Tumor-bearing mice were injected, i.v., with 30 μg of N1-, MeN1-, activated-HSA/IRDye. After 24 h, tumors, spleen, and kidneys were explanted and analyzed with an Odyssey CLx scanner to quantify the fluorescence uptake. Tumor fluorescence images (black-and-white) (B) and tissue fluorescence quantification (histograms) are shown (C). Each tumor was subdivided into small squared areas of equivalent size and fluorescence intensity (pixel) of each area was evaluated using the in-build software of the scanner. Bars represent the mean ± SE intensity values of the 15 areas with the highest intensity (mean maximum fluorescence); the number of mice/group is indicated on each bar.
D) In vivo binding of N1-HSA/, MeN1-HSA/, or Activated-HSA/IRDye to tumor cells, tumor endothelial cells and blood cells, as measured by FACS. The blood and the tumors of mice described in (B) were analyzed, after tissue disaggregation, by FACS using anti-CD31 and anti-CD45 antibodies. The percentage (mean ± SE) of IRDye680 positive cells gated on CD31–CD45– (tumor cells), CD31+CD45+ (endothelial cells), CD45+ (blood cells) are shown. The number of mice/group is indicated on each bar.
Considering that cyclic isoDGR peptides with different scaffolds are selective for different integrin subtypes, [7b, 9] it is possible that changes in the NGR scaffold might also affect CD13 selectivity for different CD13 isoforms. The above results suggest that N1 methylation does not cause loss of selectivity for the endothelial form of CD13, which is an important pre-requisite for efficient and selective tumor vascular targeting.
2.8. MeN1 can be exploited as a ligand for delivering functional nanomaterials to the tumor vasculature
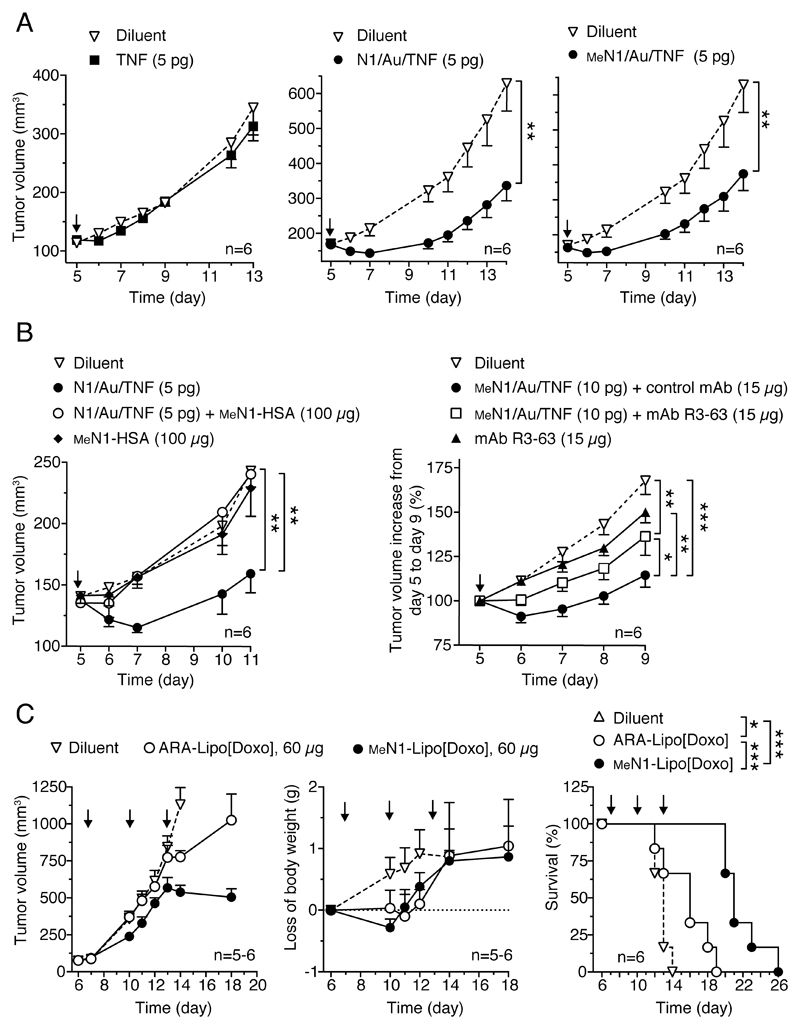
We then investigated the potential of MeN1 to deliver drug-loaded nanoparticles, such as TNF-bearing nanogold and liposomal doxorubicin, to tumors. We have shown previously that the therapeutic properties of TNF-bearing nanogold (Au/TNF) can be enhanced after functionalization with N1 (N1/Au/TNF).[10] Notably, systemic administration of N1/Au/TNF bearing 5 pg of bioactive TNF are sufficient to induce anti-tumor effects in the WEHI-164 fibrosarcoma model, whereas 5 pg of non-targeted Au/TNF or free TNF were almost totally inactive.[10] To assess whether also MeN1can be exploited for nanodrug delivery to tumors we prepared N1/ and MeN1/Au/TNF, and analyzed their physicochemical properties, their cytotoxic activity in vitro and their anti-tumor activity in vivo. In vitro studies showed that these products were characterized by similar physicochemical and biological properties (Figure S8 and Table S7). In vivo studies showed that doses equivalent to 5 pg of TNF of both nanodrugs induced similar effects in the WEHI-164 model (Figure 7A). Mechanistic studies showed a significant reduction of tumor perfusion at 48-72 h after 5 pg dose administration, as evaluated by contrast-enhanced ultrasound (CEUS) imaging technique (Figure S9), presumably related to the vasculature damaging activity of TNF.[14]
Figure 7. Anti-tumor effects of nanodrugs functionalized with MeN1.
A) Anti-tumor effects of TNF and TNF-coated nanoparticles (Au/TNF) tagged with N1 or MeN1 alone or in combination (B) with MeN1-HSA, anti-CD13 mAb (R3-63) or control mAb (IgG2a), in the WEHI-164 fibrosarcoma model. MeN1-HSA was co-administered with N1/Au/TNF, whereas antibodies were given 2.5 h before the MeN1/Au/TNF.
C) Effect of MeN1– and ARA–liposomal doxorubicin (–Lipo[Doxo]) on tumor growth, animal weight and survival in the B16-F1 melanoma model.
Arrows, time of treatment; tumor volume (6 mice/group, mean ± SE). * (P < 0.05); ** (P < 0.01); *** (P< 0.001), by two-tail t-test (A-B) and log-rank (Mantel-Cox) test (C).
Interestingly, the anti-tumor activity of N1/Au/TNF was completely inhibited by an excess of MeN1-HSA (Figure 7B, left panel), suggesting that N1 and MeN1can bind the same receptor in vivo, likely CD13, and that NGR deamidation (and integrin targeting) was not necessary for activity. Furthermore, the anti-tumor activity of MeN1/Au/TNF was significantly inhibited by the anti-CD13 mAb R3-63 (Figure 7B, right panel), supporting the hypothesis that CD13 is an important receptor of MeN1 in vivo.
Next, we compared the anti-tumor activity MeN1-tagged liposomal doxorubicin (MeN1-Lipo[doxo], 60 µg, three times) with that of liposomal doxorubicin tagged with a negative control peptide (ARA-Lipo[doxo]) in the B16-F1 melanoma model. Treatment with MeN1-Lipo[doxo] significantly prolonged the survival of mice compared to ARA-Lipo[doxo], with no evidence of increased toxicity, as judged from animal loss of body weight (Figure 7C). These results suggest that MeN1can be exploited as vehicle for delivering TNF-bearing gold nanoparticles and liposomal doxorubicin to tumors, thereby enhancing their anti-tumor activity.
3. Conclusion
The results of in vitro and in vivo studies show that replacement of glycine with N-methylglycine (sarcosine) in N1 peptide prevents deamidation without impairing CD13 recognition. The observation that MeN1 promoted the selective accumulation of NOTA18F and fluorescence-labeled HSA in tumors and that the anti-tumor activities of TNF-bearing gold nanoparticles and liposomal doxorubicin were significantly improved after coupling with this peptide support the validity of MeN1 as a ligand for delivering functional nanomaterials to the tumor vasculature.
Thus, MeN1 represents an efficient tumor-homing ligand that overcomes a major problem encountered with conventional NGR peptides, i.e. their poor asparagine stability. Glycine N-methylation may also contribute to enhance the stability of NGR to proteolytic degradation in vivo.[15]
MeN1 might be used, in principle, for the preparation of a variety of tumor-homing compounds endowed with high stability and selectivity, including nanoparticles, liposomes, viral particles, chemotherapeutic drugs, DNA complexes, anti-angiogenic compounds, cytokines, imaging compounds, and others, as previously done with NGR peptides.
4. Experimental Section
For experimental details see Supporting Information.
Supporting Information
Supplementary material (regarding experimental methods and additional results) is available in the online version of this article at link http://onlinelibrary.wiley.com/doi/10.1002/adfm.201701245/abstract
Acknowledgements
This work was supported by grants from Worldwide Cancer Research (formerly known as AICR, GR:14-0066), Ministero della Salute of Italy (No. RF-2011-02350836), and Associazione Italiana Ricerca sul Cancro (No. AIRC: IG-14338 and IG-9965 to A.C.; IG-17468 to G.M.; IG-14231 and IG-18474 to M.P.). F.P. was supported by Fondazione Umberto Veronesi and Istituto G. Gaslini Awards.
The authors thank the Protein Microsequencing Facility (ProMiFa) for mass spectrometry analyses and the Preclinical Imaging Facility (Experimental Imaging Center) for CEUS-experiments at the San Raffaele Scientific Institute.
Footnotes
The final publication is available at link http://onlinelibrary.wiley.com/doi/10.1002/adfm.201701245/abstract
References
- [1].Corti A, Curnis F. Curr Pharm Biotechnol. 2011;12:1128. doi: 10.2174/138920111796117373. [DOI] [PubMed] [Google Scholar]
- [2].a) Curnis F, Sacchi A, Borgna L, Magni F, Gasparri A, Corti A. Nat Biotechnol. 2000;18:1185. doi: 10.1038/81183. [DOI] [PubMed] [Google Scholar]; b) Curnis F, Gasparri A, Sacchi A, Cattaneo A, Magni F, Corti A. Cancer Res. 2005;65:2906. doi: 10.1158/0008-5472.CAN-04-4282. [DOI] [PubMed] [Google Scholar]; c) Meng J, Yan Z, Wu J, Li L, Xue X, Li M, Li W, Hao Q, Wan Y, Qin X, Zhang C, et al. Cytotherapy. 2007;9:60. doi: 10.1080/14653240601094322. [DOI] [PubMed] [Google Scholar]; d) Bieker R, Kessler T, Schwoppe C, Padro T, Persigehl T, Bremer C, Dreischaluck J, Kolkmeyer A, Heindel W, Mesters RM, Berdel WE. Blood. 2009;113:5019. doi: 10.1182/blood-2008-04-150318. [DOI] [PubMed] [Google Scholar]
- [3].a) Pasqualini R, Koivunen E, Kain R, Lahdenranta J, Sakamoto M, Stryhn A, Ashmun RA, Shapiro LH, Arap W, Ruoslahti E. Cancer Res. 2000;60:722. [PMC free article] [PubMed] [Google Scholar]; b) Bhagwat SV, Lahdenranta J, Giordano R, Arap W, Pasqualini R, Shapiro LH. Blood. 2001;97:652. doi: 10.1182/blood.v97.3.652. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Curnis F, Arrigoni G, Sacchi A, Fischetti L, Arap W, Pasqualini R, Corti A. Cancer Res. 2002;62:867. [PubMed] [Google Scholar]; d) Di Matteo P, Arrigoni GL, Alberici L, Corti C, Gallo-Stampino C, Traversari C, Doglioni C, Rizzardi GP. J Histochem Cytochem. 2011;59:47. doi: 10.1369/jhc.2010.956644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Corti A, Curnis F, Rossoni G, Marcucci F, Gregorc V. BioDrugs. 2013;27:591. doi: 10.1007/s40259-013-0048-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].a) Gregorc V, Zucali PA, Santoro A, Ceresoli GL, Citterio G, De Pas TM, Zilembo N, De Vincenzo F, Simonelli M, Rossoni G, Spreafico A, et al. J Clin Oncol. 2010;28:2604. doi: 10.1200/JCO.2009.27.3649. [DOI] [PubMed] [Google Scholar]; b) Gregorc V, Citterio G, Vitali G, Spreafico A, Scifo P, Borri A, Donadoni G, Rossoni G, Corti A, Caligaris-Cappio F, Del Maschio A, et al. Eur J Cancer. 2010;46:198. doi: 10.1016/j.ejca.2009.10.005. [DOI] [PubMed] [Google Scholar]
- [6].a) Robinson NE, Robinson AB. Proc Natl Acad Sci U S A. 2001;98:944. doi: 10.1073/pnas.98.3.944. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Robinson NE. Proc Natl Acad Sci U S A. 2002;99:5283. doi: 10.1073/pnas.082102799. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Geiger T, Clarke S. J Biol Chem. 1987;262:785. [PubMed] [Google Scholar]; d) Stephenson RC, Clarke S. J Biol Chem. 1989;264:6164. [PubMed] [Google Scholar]; e) Tyler-Cross R, Schirch V. J Biol Chem. 1991;266:22549. [PubMed] [Google Scholar]
- [7].a) Curnis F, Longhi R, Crippa L, Cattaneo A, Dondossola E, Bachi A, Corti A. J Biol Chem. 2006;281:36466. doi: 10.1074/jbc.M604812200. [DOI] [PubMed] [Google Scholar]; b) Curnis F, Cattaneo A, Longhi R, Sacchi A, Gasparri AM, Pastorino F, Di Matteo P, Traversari C, Bachi A, Ponzoni M, Rizzardi GP, et al. J Biol Chem. 2010;285:9114. doi: 10.1074/jbc.M109.044297. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Corti A, Curnis F. J Cell Sci. 2011;124:515. doi: 10.1242/jcs.077172. [DOI] [PubMed] [Google Scholar]
- [8].a) Curnis F, Sacchi A, Gasparri A, Longhi R, Bachi A, Doglioni C, Bordignon C, Traversari C, Rizzardi GP, Corti A. Cancer Res. 2008;68:7073. doi: 10.1158/0008-5472.CAN-08-1272. [DOI] [PubMed] [Google Scholar]; b) Corti A, Curnis F, Arap W, Pasqualini R. Blood. 2008;112:2628. doi: 10.1182/blood-2008-04-150862. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Takahashi S, Leiss M, Moser M, Ohashi T, Kitao T, Heckmann D, Pfeifer A, Kessler H, Takagi J, Erickson HP, Fassler R. J Cell Biol. 2007;178:167. doi: 10.1083/jcb.200703021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].a) Spitaleri A, Mari S, Curnis F, Traversari C, Longhi R, Bordignon C, Corti A, Rizzardi GP, Musco G. J Biol Chem. 2008;283:19757. doi: 10.1074/jbc.M710273200. [DOI] [PubMed] [Google Scholar]; b) Frank AO, Otto E, Mas-Moruno C, Schiller HB, Marinelli L, Cosconati S, Bochen A, Vossmeyer D, Zahn G, Stragies R, Novellino E, et al. Angew Chem. 2010;122:9465. doi: 10.1002/anie.201004363. [DOI] [PubMed] [Google Scholar]; Angew Chem Int Ed Engl. 2010;49:9278. [Google Scholar]; c) Curnis F, Sacchi A, Longhi R, Colombo B, Gasparri A, Corti A. Small. 2013;9:673. doi: 10.1002/smll.201202310. [DOI] [PubMed] [Google Scholar]; d) Bochen A, Marelli UK, Otto E, Pallarola D, Mas-Moruno C, Di Leva FS, Boehm H, Spatz JP, Novellino E, Kessler H, Marinelli L. J Med Chem. 2013;56:1509. doi: 10.1021/jm301221x. [DOI] [PubMed] [Google Scholar]; e) Kapp TG, Rechenmacher F, Neubauer S, Maltsev OV, Cavalcanti-Adam EA, Zarka R, Reuning U, Notni J, Wester HJ, Mas-Moruno C, Spatz J, et al. Sci Rep. 2017;7:39805. doi: 10.1038/srep39805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Curnis F, Fiocchi M, Sacchi A, Gori A, Gasparri A, Corti A. Nano Res. 2016;9:1393. doi: 10.1007/s12274-016-1035-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Wong AH, Zhou D, Rini JM. J Biol Chem. 2012;287:36804. doi: 10.1074/jbc.M112.398842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Liu C, Yang Y, Chen L, Lin YL, Li F. J Biol Chem. 2014;289:34520. doi: 10.1074/jbc.M114.566802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Luciani N, Marie-Claire C, Ruffet E, Beaumont A, Roques BP, Fournie-Zaluski MC. Biochemistry. 1998;37:686. doi: 10.1021/bi971705p. [DOI] [PubMed] [Google Scholar]
- [14].Stam TC, Jongen-Lavrencic M, Eggermont AM, Swaak AJ. Eur J Clin Investig. 1996;26:1085. doi: 10.1046/j.1365-2362.1996.480599.x. [DOI] [PubMed] [Google Scholar]
- [15].Chatterjee J, Rechenmacher F, Kessler H. Angew Chem Int Ed Engl. 2013;52:254. doi: 10.1002/anie.201205674. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.