Abstract
The gene that codes for the putative dihydroorotase in the hyperthermophilic archaeon Methanococcus jannaschii was subcloned in pET-21a and expressed in Escherichia coli. A purification protocol was devised. The purity of the protein was evaluated by SDS-PAGE and the protein was confirmed by sequencing using LC-MS. The calculated molecular mass is 48104 Da. SEC-LS suggested that the protein is a monomer in solution. ICP-MS showed that there are two Zn ions per monomer. Kinetic analysis of the recombinant protein gave hyperbolic kinetics with Vmax = 12.2 µmol min−1 mg−1 and Km = 0.14 mM at 25° C. Furthermore the activity of the protein increased with temperature consistent with the hyperthermophilic nature of the organism. A homology model was constructed using the mesophilic Bacillus anthracis protein as the template. Residues known to be critical for Zn and substrate binding were conserved. The activity of the enzyme at 85° C and 90° C was found to be relatively constant over 160 min and this correlates with the temperature of optimal growth of the organism of 85° C. The amino acid sequences and structures of the two proteins were compared and this gave insight into some of the factors that may confer thermostability – more Lys and Ile, fewer Ala, Thr, Gln and Gly residues, and shorter N- and C-termini. Additional and better insight into the thermostabilization strategies adopted by this enzyme will be provided when its crystal structure is determined.
Keywords: Dihydroorotase, pyrimidine biosynthesis, Methanococcus jannaschii, enzyme kinetics, thermostability, homology modeling
1 Introduction
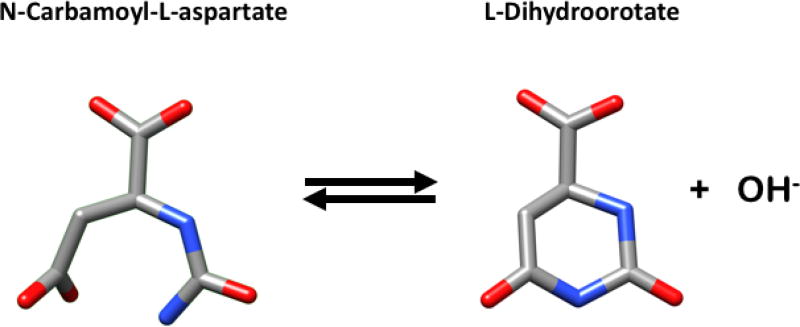
DHOase (EC 3.5.2.3) catalyzes the conversion of N-carbamoyl-L-aspartate to L-dihydroorotate (Fig. 1) in the third step of the de novo biosynthesis of pyrimidines. It is a Zinc-containing member of the amidohydrolase superfamily of metalloenzymes [1]. Historically, dihydroorotases were classified into two distinct types [2]. Long (type I) evolved first and are larger with molecular weight ~45 kDa. Short (type II) evolved more recently and are smaller with molecular weight ~38 kDa. The two classes have very low sequence identity (<20%). A more recent phylogenetic analysis [3] gave two highly divergent clusters consistent with this classification. Long DHOases include the subclasses of archaeal, bacterial type I, bacterial type III, animal CAD and CAD-like inactive DHOases from fungi. Short DHOases further subdivide in active DHOases from fungi, bacterial type II and plant DHOases. DHOase is believed to be the ancestral protein of the amidohydrolase superfamily in part because it is present in most life forms and in part because it is a biosynthetic enzyme as compared to the other members of the family that have a catabolic role [2, 4]. Other members of the superfamily acting on a cyclic amide ring include dihydropyrimidinases, D-hydantoinases, and allantoinases [1, 4].
Fig. 1.
The reversible cyclization of N-carbamoyl-L-aspartate to L-dihydroorotate catalyzed by DHOase. Colors used are dark grey for C, blue for N and red for O. Figure was prepared with UCSF Chimera.
All DHOases adopt the (βα)8-barrel fold (TIM barrel) which is characteristic of the amidohydrolase superfamily of proteins [1]. The differences between the two clusters are primarily structural [3]. Long DHOases have extra residues at the N- and C- termini forming a two-layered β stranded domain adjacent to the TIM barrel compared to short DHOases that essentially consist of the catalytic core. DHOases have two or one Zn ions in their active site as well as four conserved histidines and one aspartate that coordinate them. In type II, III and CAD DHOases the two Zn ions are bridged by a carboxylated Lys residue which is conserved in these types. In type I DHOases, the two Zn ions are bridged by an aspartate which is invariant in this type. Type II, III and CAD DHOases have a long catalytic flexible loop that covers the active site when the substrate CA is bound. The corresponding loop is short in Type I DHOases. Type I bacterial DHOases are often associated with ATCase. ATCase catalyzes the formation of CA which is the substrate of DHOase.
The basic mechanism of the DHOase reaction was determined from the crystal structure of the type II E. coli enzyme [5] and biochemically verified [6]. The CA substrate binds in the active site to the two Zn ions and to a number of invariant residues as well as main chain atoms. An invariant aspartate abstracts a proton from the amide nitrogen of CA facilitating a nucleophilic attack. This basic mechanism is maintained in all DHOases as the residues involved are invariant and the key interactions with the substrate are conserved in the known complexes [3, 5, 7, 8].
Archaeal DHOases have not been previously studied. The phylogenetic analysis [3] suggests that archaeal and bacterial type I DHOases are more closely related to each other than to the other types. Archaeal DHOases are expected to have both similarities and differences from the prokaryotic and eukaryotic DHOases. In addition, many archaea live in harsh environments and some of the differences correspond to adaptations to their physiological environment. This paper is the first study of an archaeal DHOase, that from the hyperthermophilic archaeon M. jannaschii. We are interested in studying this dihydroorotase in order to gain further insight into the variability within the dihydroorotase family and in order to understand how the structure and function of this enzyme adapt to the high temperatures that are the normal habitat of this organism. As a first step, the pyrC gene of M. jannaschii was subcloned into an expression vector, the gene product was purified, and the enzyme was characterized. A homology model was built based on the homologous mesophilic B. anthracis DHOase.
2 Materials and Methods
2.1 Plasmid Preparation
Clones of M. jannaschii genomic DNA encoding DHOase were obtained from the American Type Culture Collection (ATCC number 624773). PCR was used to amplify the open reading frame using PfuTurbo DNA polymerase (Stratagene). The forward primer was GGAATTCCATATGCTATTAAAAAACTGTAGAAT (NdeI site underlined, ORF start codon bolded) and the reverse primer was GCGGATCCTTAACATTTACATCCATAAGCTTC (BamHI site underlined, stop codon bolded). The resulting product was subcloned in pCR-Blunt II-TOPO vector (Invitrogen) and digested with NdeI and BamHI. The digested fragment was ligated into pET-21a (Novagen) using T4 DNA ligase (NEB). The ligated mix was transformed in subcloning efficiency DH5α E. coli competent cells (Invitrogen). Positive clones were identified by colony PCR and the pET-21a plasmid harboring the M. jannaschii pyrC gene was prepared using the QIAprep spin miniprep kit (Qiagen). The construct was verified by sequencing at the Genomics Core of Cleveland Clinic’s Lerner Research Institute. The plasmid was transformed in Rosetta-gami 2 (DE3) cells (Novagen) for protein expression.
2.2 Expression of the Mj-PyrC Gene Product
Typically, an overnight culture of Rosetta-gami 2 (DE3) cells harboring the pET-21a plasmid with the M. jannaschii pyrC gene in LB Lennox media supplemented with 50 µg/ml ampicillin, 34 µg/ml chloramphenicol, 50 µg/ml streptomycin and 12.5 µg/ml tetracycline was used to inoculate LB Lennox media supplemented with these antibiotics. The inoculum was 1% of the media volume. The cells were grown at 37° C, 200 rpm to an A600 of ~ 0.6 in an Innova 44 incubator shaker from New Brunswick Scientific. Protein expression was then induced by the addition of isopropyl-β-D-1-thiogalactopyranoside to a final concentration of 1 mM and the cell culture was grown for 16 additional hours at 18° C. The cells were centrifuged at 4000 rpm for 20 min in an Avanti J-26XP centrifuge and kept at −80° C until ready to use.
2.3 Purification
Before use, the frozen pellets were thawed and resuspended in 0.05 M Tris-Cl pH 7.5 buffer with 2 mM 2-mercaptoethanol (BME) and 0.05 mM zinc acetate, sonicated, and the mixture was centrifuged at 15000 rpm for 20 min. The cell supernatant was brought to 35% saturation of ammonium sulfate, stirred overnight at 4° C and centrifuged at 16000 rpm for 20 min. A heat step of the supernatant at 85° C for 15 min followed and the mixture was centrifuged at 16000 rpm for 20 min. The supernatant was dialysed in 50 mM Mes-KOH 5.8 buffer with 2 mM BME and 0.05 mM zinc-acetate followed by cation exchange chromatography using a hand packed 9 ml SP column (Lab Pack from GE Healthcare) and a gradient from 0 to 0.5 M NaCl. The fractions containing DHOase were pooled and dialyzed in 40 mM KH2PO4 pH 8 buffer with 2 mM BME and 0.05 mM zinc acetate. Ammonium sulfate was subsequently added to the protein solution to 1.3 M ammonium sulfate. The final purification step employed hydrophobic interaction chromatography using a phenyl-sepharose column [HiPrep Phenyl FF (high Sub) 16/10, GE Healthcare] that was pre-equilibrated with the same buffer as the protein. The gradient employed was 40 mM KH2PO4 pH 8, 2 mM 2-mercaptoethanol, 0.05 mM zinc acetate, 1.3 M ammonium sulfate to 40 mM KH2PO4, pH 8, 2 mM 2-mercaptoethanol, 0.05 mM zinc acetate. Fractions containing pure protein, as determined by SDS-PAGE, were pooled and concentrated.
All centrifugations were done in in an Avanti J-26XP centrifuge and the hydrophobic interaction chromatography was performed using an ÄKTAprime system (GE Healthcare). The purity of the protein throughout the purification was evaluated using SDS-PAGE.
2.4 Liquid Chromatography - Mass Spectrometry Analysis
The protein was sequenced using LC-MS by the Lerner Research Institute’s Proteomics Laboratory. The LC-MS system was a Finnigan LTQ linear ion trap mass spectrometer system and the HPLC column was a self-packed 9 cm × 75 µm Phenomenex Jupiter C18 reversed-phase capillary chromatography column. The sample used for this analysis was an SDS-PAGE gel. The bands to be analyzed were excised and digested in-gel with trypsin, chymotrypsin and endoproteinase GluC overnight at room temperature. The peptides formed in each digestion were analyzed by LC-MS. This analysis was done in a data dependent manner acquiring first a full mass scan to determine the most abundant peptide ions eluting off the LC column followed by four collision induced dissociation (CID) experiments to determine amino acid sequence in successive instrument scans. For each digest, the data were analyzed by using all CID spectra to search the NCBI non-redundant protein database with the search program Mascot (Matrix Science, London, UK) using a mammalian taxonomy filter. All matching spectra were verified by manual interpretation. The interpretation process was aided by additional searches using the programs Sequest (Thermo Fisher Scientific, San Jose, CA) and Blast (http://blast.ncbi.nlm.nih.gov). Targeted experiments were also performed involving selective reaction monitoring (SRM) for specific peptides that include residues not determined from the data dependent approach.
2.5 Determination of Dihydroorotase Activity
Dihydroorotase activity was studied in the reverse direction in 100 mM Tris-acetate pH 8.3 using the colorimetric assay of Prescott and Jones [9] and substrate concentrations to 3.15 mM. Readings were made at a wavelength of 466 nm in a SPECTRAmax PLUS 384 microplate reader from Molecular Devices using the software SoftMax Pro 6.3. The assay volume was 0.5 ml and the assay was carried out at 25° C, 45° C and 80° C. The reaction was initiated by addition of the enzyme. The background hydrolysis of dihydroorotate to carbamoyl aspartate in the absence of enzyme was monitored. All measurements were corrected for the background absorbance in the absence of substrate. In addition the enzyme kinetic data were corrected for the background reaction.
Kinetic parameters were calculated by fitting data to the Michaelis–Menten equation using the MacDaesX program by Evan Kantrowitz, Boston College. Protein concentration was measured by the Bradford assay using the Coomassie Plus Protein Assay reagent from Pierce with BSA as the standard.
2.6 Thermal Stability of Dihydroorotase
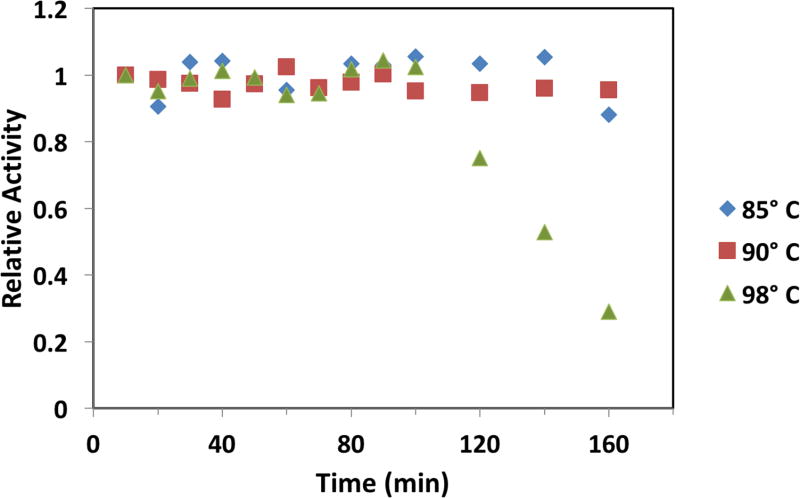
The thermal stability of the M. jannaschii DHOase was measured in the reverse direction by heating the enzyme at 85° C, 90° C and 98° C as a function of time up to 160 min. At each time point an aliquot was rapidly chilled and stored at 4° C followed by determination of enzymatic activity at 37° C using the colorimetric assay.
2.7 Zinc Inductively Coupled Plasma – Mass Spectrometry Measurements
The zinc content of the purified recombinant M. jannaschii DHOase was obtained using ICP-MS (Intertek Inc., Whitehouse, NJ). Prior to the measurements the purified protein was dialyzed against three changes of 50 mM Mes-KOH 5.8, 2 mM BME at 4° C in the presence of a dialysis bag containing Chelex-100 so that fortuitously bound metals would be removed. The protein concentration of the dialyzed sample was determined by the Bradford assay. The samples were analyzed using an Agilent 7700X ICP-MS instrument. Dublicate ICP-MS analyses of the protein and dialysis buffer solutions were carried out.
2.8 HPLC Size Exclusion Chromatography / Laser Light Scattering
High performance liquid chromatography (HPLC) size exclusion chromatography / laser light scattering was performed by the Biophysics Resource of the Keck Facility at Yale University [10]. The column used was a Superdex S-200, 10/30, HR SEC (GE Healthcare) and was connected to an HPLC system (Agilent 1200, Agilent Technologies). The elution from the column was monitored by a photodiode array UV/VIS detector (Agilent Technologies), differential refractometer (OPTILab-rEx Wyatt Corp.), static and dynamic, multiangle laser light scattering detector (HELEOS II with QELS capability, Wyatt Corp.). The SEC-UV/LS/RI system was equilibrated in a 50 mM Tris-Cl pH 8, 150 mM NaCl, 2 mM BME, 0.05 mM zinc acetate and the flow rate was 1.0 ml/min. The sample was filtered through a 0.22 µm filter before injecting to the column. Weight average molecular masses were calculated by ASTRA. During data analysis, a dn/dc value of 0.165 ml/g was used as it proved satisfactory during analyses of 3 protein standards: Aldolase (156 kDa), BSA (66 kDa) and Ovalbumin (43 kDa).
2.9 Homology Modeling
An initial homology model was generated with the I-TASSER (Iterative Threading ASSEmbly Refinement) server [11–14]. The amino acid sequence of the target was obtained from the ExPASy Bioinformatics resource portal (http://www.expasy.org) with accession number UniProtKB: Q58885. I-TASSER identifies structural templates from the Protein Data Bank using a multiple threading approach and constructs full length atomic models by iterative template fragment assembly simulations. The closest analog in the Protein Data Bank was identified by I-TASSER to be the DHOase from B. anthracis (pdb id: 3mpg). The model was examined in Chimera [15] and a few adjustments were made to its sequence alignment with the B. anthracis protein to improve the alignment with the aspartate bridging the two Zn ions in B. anthracis and known to be conserved in Type I DHOases [3]. The final model was created in Chimera using the graphical interface to Modeller [16]. Modeller models protein structure by satisfaction of spatial restraints. The structural template was chain A of the B. anthracis DHOase and the revised alignment of the two proteins was used. The Zn ions from the template were transferred to the output models. Steric clashes were identified in the accepted model and relieved using the “minimize” option in Chimera. The minimize option used Amber ff14SB [17] parameters for standard residues and Amber’s Antechamber module [18] for nonstandard residues. The stereochemistry was evaluated with PROCHECK [19].
3 Results
3.1 Expression and Purification of M. jannaschii DHOase
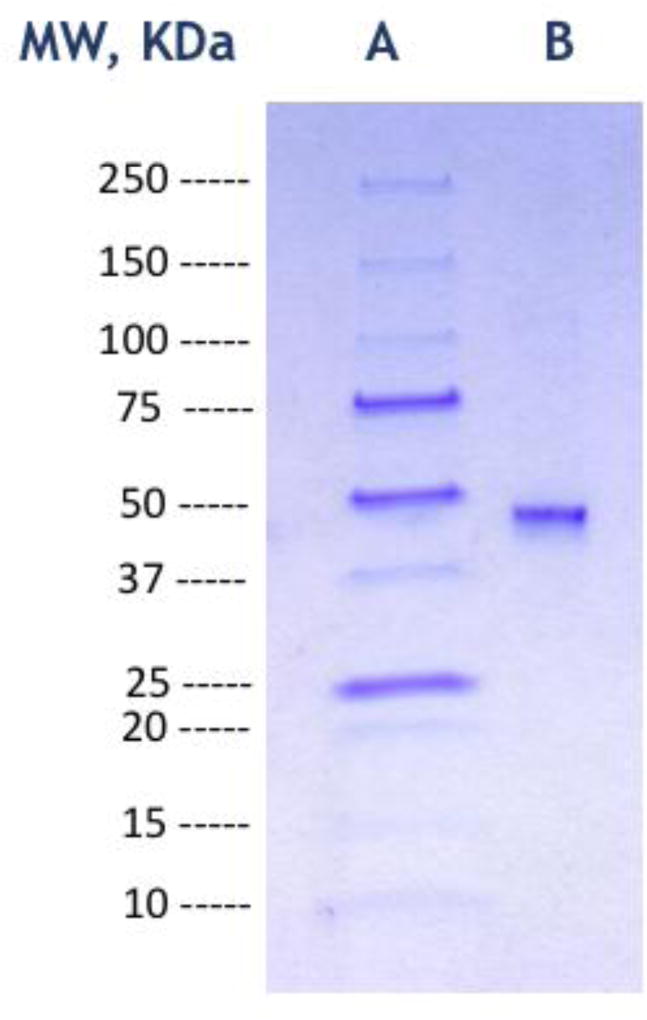
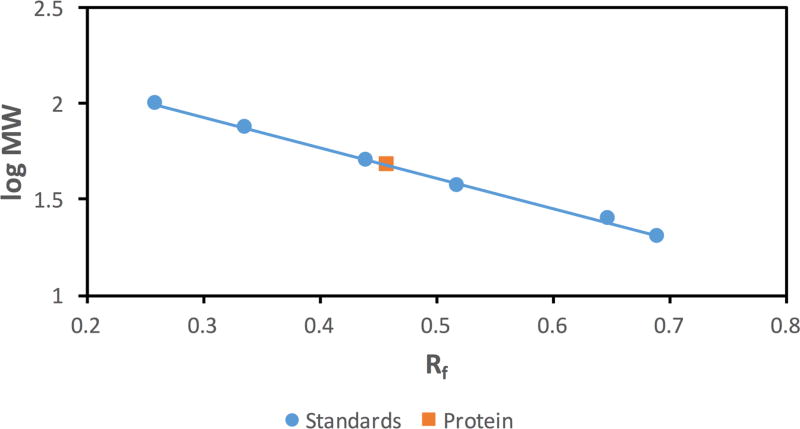
M. jannaschii DHOase was previously sequenced and cloned as part of the study of the genome of this organism [20] but it was not expressed or analyzed. In this paper we subcloned the M. jannaschii pyrC gene into the pET-21a vector, expressed it in Rosetta-gami 2 (DE3) E. coli and purified it. At the end of the purification, SDS-PAGE gave a single band (Fig. 2a). Its molecular weight was determined from a plot of the logarithm of the molecular weight of protein standards versus their relative migration distance in the SDS-PAGE gel by interpolation (Fig. 2b). The value obtained of 47503 Da is comparable to the value of 48104 Da for the M. jannaschii DHOase computed from its amino acid sequence.
Fig. 2.
(a) SDS-PAGE of purified recombinant M. jannaschii DHOase. Lane A corresponds to the molecular weight markers, lane B has the purified protein. The molecular weight markers were the Precision Plus Protein unstained standards (Biorad). (b) The corresponding plot of the logarithm of the molecular weight versus relative migration distance of the standards. Based on this plot, the molecular weight of the protein was estimated to be 47503 Da.
3.2 Identification of M. jannaschii Dihydroorotase
The purified protein was sequenced with LC-MS in order to confirm its identity as M. jannaschii DHOase and to cover as much of the protein sequence as possible.
Two bands from the SDS-PAGE gel were initially digested, one with trypsin and the other with chymotrypsin, and the digests analyzed by LC-MS in a data dependent manner. M. jannaschii DHOase was identified as the major component in both digests. A total of 49 peptides was identified in the trypsin digest covering 85% of the protein sequence. The chymotrypsin digest gave 176 peptides and covered 96% of the protein sequence. The two together covered 99% of the protein sequence. A total of five amino acids were not identified in the data dependent analysis of the tryptic and chymotryptic digests. These were the three N-terminal amino acids, Lys137, and the C-terminal amino acid Cys423. The inability to identify these areas could be due to the formation of tryptic and/or chymotryptic peptides that have poor LC-MS characteristics, the presence of a protein modification, or their low abundance precluding selection for the collision induced dissociation experiments.
A third band from the SDS-PAGE gel was digested with GluC. The digest was analyzed with LC-MS in a data dependent manner. This digest resulted in the identification of 17 peptides covering 61% of the protein sequence. The three N-terminal amino acids were identified in this analysis.
In order to determine if the Lys137, N- and C-terminal peptides were not identified in the data dependent analysis of tryptic and chymotryptic digests due to their low abundance, several SRM experiments were performed. Lys137 was identified in low abundant ions in the SRM analysis of tryptic and chymotryptic digests. The terminal Cys423 was identified from the SRM analysis of a low abundant C-terminal tryptic peptide. Several different forms of the N-terminal peptide were also targeted in the chymotryptic digest for SRM analysis but gave no indications of these three N-terminal amino acids. Fig. 3 gives a composite sequence map of the protein indicating the coverage by trypsin and chymotrypsin as well as how the remaining five residues were determined.
Fig. 3.
Composite sequence map. Peptides identified in tryptic digestion only are in red, peptides identified in chymotryptic digestion only are in blue, and peptides identified in GluC digestion only are in purple. Peptides identified in both tryptic and chymotryptic digestions are in green. Shaded areas correspond to peptides not identified in the data dependent analysis of the tryptic and chymotryptic digestions. Of them, the N-terminal tripeptide was identified in the data dependent analysis of the GluC digest, Lys137 was identified in low abundant ions in the SRM analysis of both tryptic and chymotryptic digests and Cys423 was identified in a low abundant ion in the SRM analysis of the tryptic digest.
Moreover, the ExPASy site suggested that it is possible that Lys137 is carboxylated. In ExPASy, the Zn binding site is propagated from the E. coli enzyme [5] to other DHOases using sequence alignments with the program MUSCLE [21]. E. coli [5] has a carboxylated Lys residue in the metal binding site of the enzyme at position 103 of its sequence. According to these alignments, this position corresponds to position 137 in M. jannaschii. However, this region is highly variable among DHOases [3]. If Lys137 was carboxylated, it would result in the addition of 44 Da (CO2) to the lysine residue. However, the SRM analysis of tryptic and chymotryptic digests was not able to identify Lys + 44 Da containing peptides suggesting that Lys137 is unmodified and that, in contrast to E. coli, it may not interact with the Zn ions.
3.3 Steady State Kinetics of the M. jannaschii Dihydroorotase
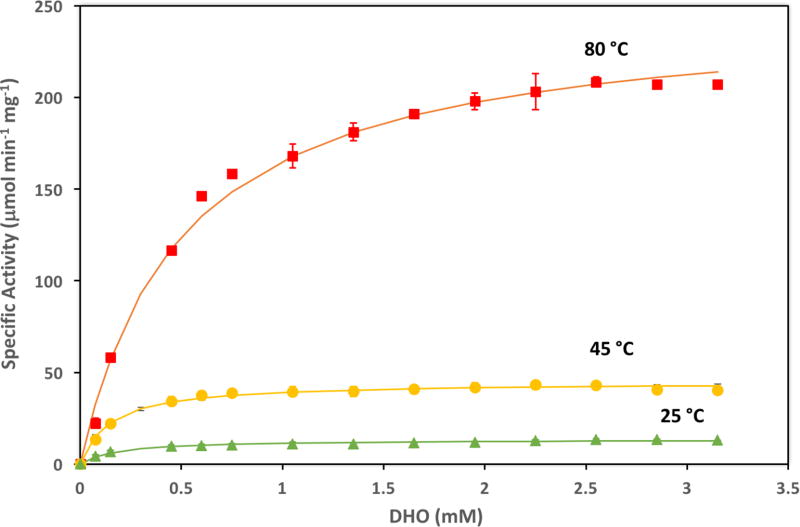
Table 1 gives the kinetic parameters of M. jannaschii DHOase in the degradative direction at pH 8.3 at 25° C, 45° C and 80° C. The saturation curves from which these parameters were determined were averages of four measurements at 25° C, three measurements at 45° C, and two measurements at 80° C and are shown in Fig. 4. At all temperatures the enzyme follows Michaelis-Menten kinetics.
Table 1.
Kinetic parameters of M. jannascii DHOase and of selected dihydroorotases in the degradative direction.
| Organism | Temperature, °C | Maximal Velocity, µmol min−1 mg−1 |
Km, mM | Type1 |
|---|---|---|---|---|
| M. jannaschii2 | 25 | 12.2 ± 0.1 | 0.140 ± 0.009 | Archaeal |
| 45 | 44.7 ± 0.6 | 0.15 ± 0.01 | ||
| 80 | 248± 5 | 0.52 ± 0.03 | ||
| B. anthracis3 | 25 | 2.4 ± 0.1 | 0.114 ± 0.016 | Bacterial type I |
| 37 | 8.6 ± 0.2 | 0.36 ± 0.06 | ||
| B. caldolyticus4 | 70 | 77.4 ± 4.5 | 0.195 ± 0.028 | Bacterial type I |
| Hamster4 | 37 | 3.9 ± 0.5 | 0.022 ± 0.005 | CAD |
| Human5 | 25 | 8.20 ± 0.24 | 0.028 ± 0.004 | CAD |
| E. coli6 | 30 | 155 ± 2 | 0.080 ± 0.001 | Bacterial type II |
Fig. 4.
Saturation curves for M. jannascii DHOase in the degradative direction at pH 8.3. Data are averages of four measurements at 25° C, three measurements at 45° C, and two measurements at 80° C.
3.4 Thermal Stability of the M. jannaschii Dihydroorotase
The thermal stability of the M. jannaschii DHOase was evaluated at 85° C, 90° C and 98° C as the optimal growth temperature of this organism is 85° C. As seen in Fig. 5, at 85° C and 90° C, the activity of the enzyme is constant over 160 min. However at 98° C, the enzyme is stable for 100 minutes after which its activity rapidly diminishes to 29 % of its initial value over the following 60 minutes. This is consistent with the observation [22] that there is no growth of M. jannaschii at 95° C in defined medium.
Fig. 5.
Thermal stability of M. jannaschii DHOase at 85° C, 90° C and 98° C. Solutions of the enzyme were heated at the indicated temperature in 0.1 M Tris-acetate pH 7.5, 150 mM NaCl, 2 mM BME. At the indicated times, samples were removed and immediately placed on ice. The colorimetric assay was used to determine enzymatic activity at 37° C in the degradative direction.
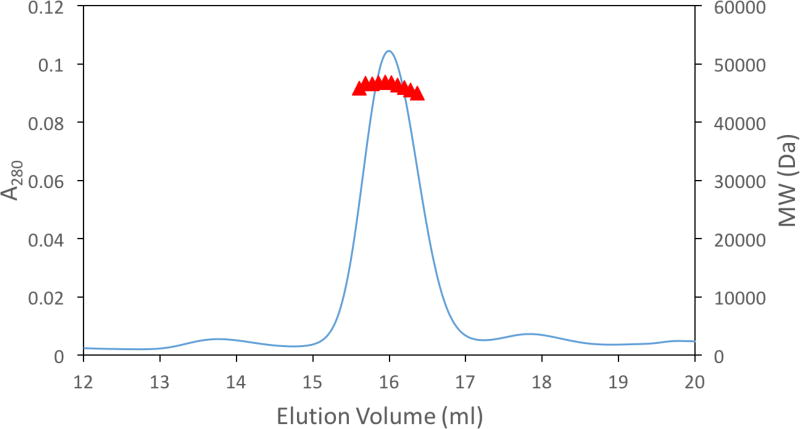
3.5 Monomeric Structure of M. jannaschii Dihydroorotase
The SEC-LS/UV/RI data were analyzed with the ASTRA software and suggested that the protein is a monomer in solution. As seen in Fig. 6, the molecular masses computed in slices of 1s intervals across the eluting peak between 15.6 and 16.3 ml are nearly constant indicating that the protein sample was monodisperse. The weight average molecular mass was 46780 Da, in agreement with the theoretical value of 48104 Da computed from the amino acid sequence, indicating that the protein is monomeric in solution.
Fig. 6.
Molecular mass distribution plot from SEC-UV/LS/RI analysis for M. jannaschii DHOase. Molecular masses are plotted as red filled triangles (for clarity only every 5th measurement of molecular mass across the eluting peak is plotted); the blue line corresponds to the UV trace of the protein eluting from the SEC column monitored at 280 nm.
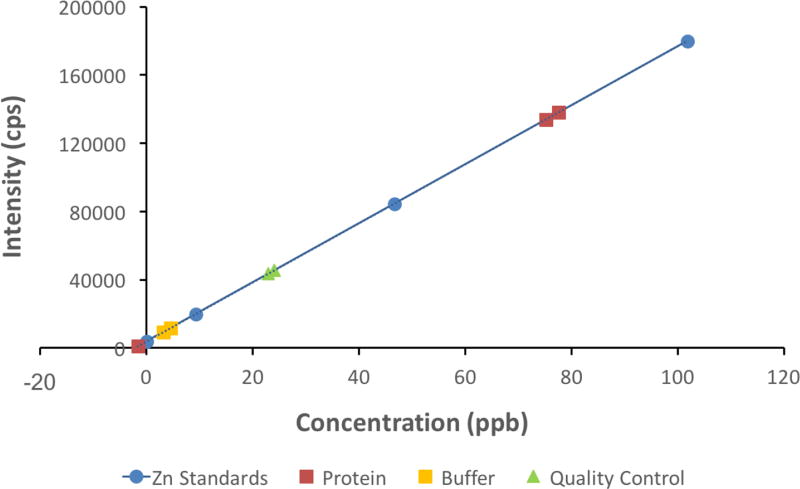
3.6 Zinc Content of Purified M. jannaschii Dihydroorotase
A set of Zn standards was prepared in an acid matrix and analyzed by ICP-MS. A calibration curve relating measured intensity (counts per second) with concentration for Zn in solution was constructed (Fig. 7). The protein and buffer samples were also prepared by acid digestion and analyzed under the same conditions as the standards. The sample intensities were measured and compared with the calibration curve, and the resulting concentrations were recorded. These concentrations (Fig. 7) were then corrected for dilution by a factor of 10. Each analysis was the average of five replicate readings and each reading was the average of 100 individual mass sweeps.
Fig. 7.
Calibration curve for Zn in ICP-MS analysis used to determine the Zn content of M. jannaschii DHOase. Blue filled circles illustrate the Zn standards used to construct this curve. Red filled squares correspond to the protein samples and orange filled squares correspond to the buffer samples. Both were diluted by a factor of 10. The green filled triangles are bracketing quality controls for the protein and buffer samples. 1ppb corresponds to 1 ug/lt.
The ICP-MS runs gave 752 ug/lt and 776 ug/lt zinc in the two protein samples of concentration 0.26 mg/ml. The Zn concentrations obtained for the buffer were 46 ug/lt and 31 ug/lt. These values give an average of 2.05 mol zinc per mol protein classifying the M. jannaschii DHOase as a two-zinc center protein.
3.7 A Model of M. jannaschii Dihydroorotase
The top I-TASSER model had a satisfactory C-score of 0.90. I-TASSER uses the C-score as the criterion for determining the best model. I-TASSER inferred the function of the protein to be a dihydroorotase, identified the B. anthracis protein as the closest structural analog in the Protein Data Bank and predicted the α Zn binding site. As the protein had two Zn ions and did not contain a carboxylated lysine, the sequence alignment with B. anthracis predicted by I-TASSER was adjusted to improve the alignment with the aspartate bridging the two Zn ions in the DHOase of B. anthracis and known to be conserved in Type I DHOases [3]. Among the 20 models generated by Modeller in Chimera, the one with the most negative zDOPE (normalized Discrete Optimized Protein Energy) score of −0.51 was selected as the best model. The zDOPE score is used by Modeller in Chimera to evaluate the plausibility of a model. The Ramachandran plot statistics [19] are satisfactory for the Chimera model with 98.4% of the residues in the most favored and additional allowed regions and only 0.5% in the disallowed regions.
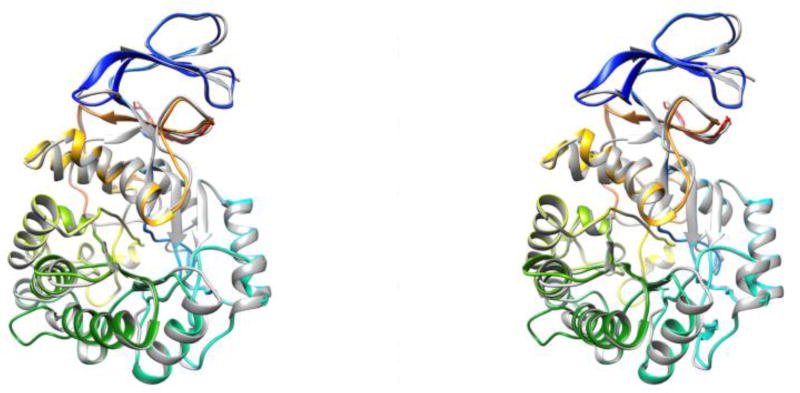
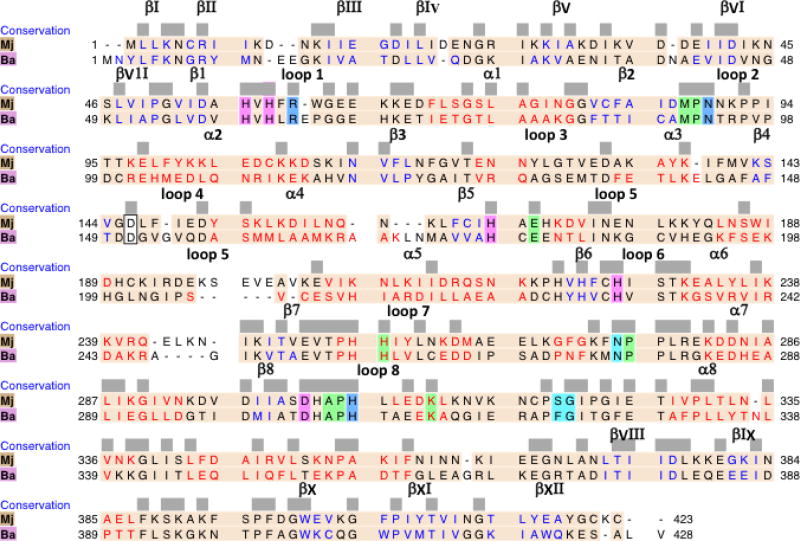
The architecture of the Mj DHOase model is closely similar to the template Ba, in particular in the secondary structural elements with differences primarily in the loops that connect them (Figs. 8a and 8b). Residues 51–340 form the catalytic (βα)8 barrel and the adjacent β stranded domain is formed by residues 1 – 50 from the N-terminal extension and residues 341 – 423 from the C- terminal extension (Figs. 8a and 8b). The outer antiparallel β sheet of the extra domain consists of the 4 strands βI, βIV, βV and βVI of the N terminal extension and the inner antiparallel β sheet consists of 6 strands, 3 from the N terminal extension, βII, βIII and βVII and 3 from the C terminal extension βVIII, βXI and βXII (Figs. 8a and 8b). The C terminal extension also contributes a β hairpin (βIX and βX) (Fig. 8b) that protrudes on one side of the TIM barrel toward the top of it (Fig. 8a). The rmsd between corresponding Cα atoms of 410 atom pairs in the two proteins within 2 Å from each other is 0.45 Å. The sequence identity between the two proteins is 32.4%.
Fig. 8.
(a) A schematic ribbons diagram illustrating the homology model of the M. jannaschii DHOase superimposed on the enzyme from B. anthracis (pdb id: 3mpg, chain A). M. jannaschii DHOase is depicted in rainbow colors, from blue at the N-terminus to red at the C-terminus. 3mpg, chain A is depicted in dark grey. In this view, the β hairpin loop described in the text for M.jannaschii (βIX and βX) is in orange in the back of the molecule. The homology model was calculated with Modeller in the UCSF chimera environment using the dihydroorotase of B. anthracis as the template. A stereo pair. Figure was drawn with the UCSF Chimera package (b)Sequence alignment of the M. jannaschii (Mj) and B. anthracis (Ba) dihydroorotases. The figure was prepared with UCSF Chimera and is based on the structural superposition of the two proteins. Identity between the two sequences is emphasized by blocks drawn at the suitable sequence positions. Invariant residues in all dihydroorotases are highlighted; those that coordinate the two Zn ions in magenta, those with side chains involved in substrate binding in blue and the rest in green. Cyan corresponds to amino acids that are not conserved but their positions are conserved and their main chain atoms interact with the substrate. Residues Asp146 in Mj and Asp151 in Ba are outlined in black. Their carboxylate oxygens bridge the two Zn ions. An aspartate at this position is conserved in Type I DHOases [3]. The coral region covers corresponding residues in the two proteins that are within 2 Å from each other. Sequence is colored according to the secondary structure: red for α helices, blue for β strands and black for loops. The secondary structural elements of the TIM barrel are labeled from β1 to α8. The β strands in the adjacent domain are labeled as βI to βXII. Helices are not labeled unless they correspond to the (βα)8 barrel. The two sequences exhibit 32% identity.
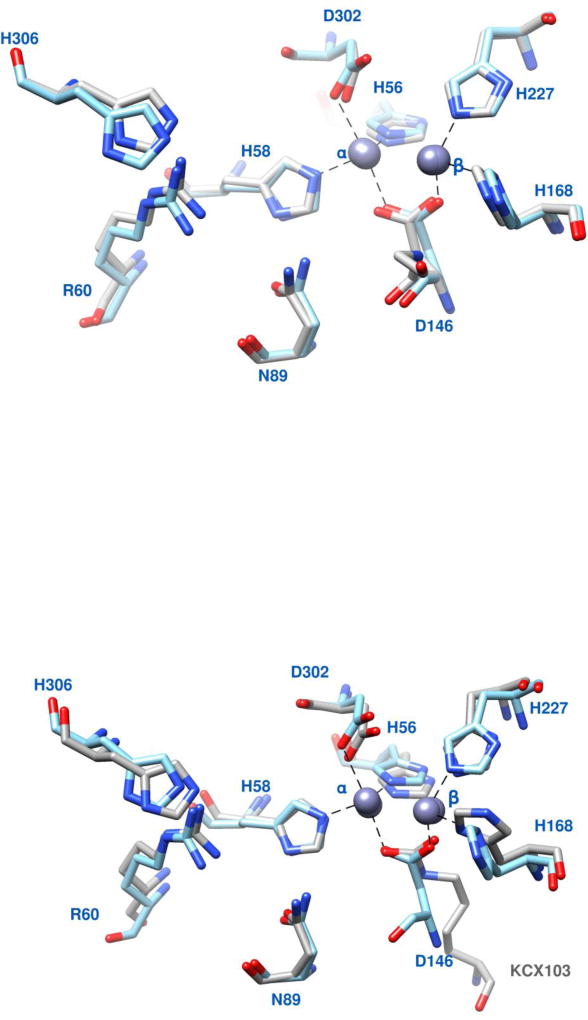
(c) Active site of the Mj model (cyan carbon atoms) superimposed on the active site of Ba dihydroorotase (pdb id: 3mpg; grey carbon atoms). Zinc atoms are grey spheres. The residues shown are those that coordinate the Zn ions and those whose side chains are involved in interactions with the substrate. D146 bridges the two Zn ions in the model. An earlier multiple sequence alignment [3] showed that this aspartate in conserved in Type I DHOases. In the superposition, the two Zn ions in Mj are slightly displaced from the Zn ions in Ba by ~0.15 Å. (d) Superposition of the active sites of the Mj model (cyan carbon atoms) and E. coli DHOase (pdb id: 1j79; grey carbon atoms). Zinc atoms are grey spheres. The residues shown are those that coordinate the Zn ions and those whose side chains are involved in interactions with the substrate. D146 bridges the two Zn ions in the model and the carboxylated lysine KCX103 bridges the two Zn ions in E. coli. An earlier multiple sequence alignment [3] showed that the carboxylated lysine is conserved in type II, III and CAD dihydroorotases and is shifted upstream relative to the conserved Aspartate in Type I dihydroorotases. In the superposition, the two Zn ions in Mj are slightly displaced from the Zn ions in Ec by ~0.22 Å
The active site on the top of the TIM barrel is closely similar to Ba (Fig. 8c) and Ec (Fig. 8d). The model has the two Zn ions coordinated to the invariant His56, His58, His227, His168, Asp302 [3] and they are bridged by the carboxylate of Asp146. Asp302 is the aspartate that abstracts a hydrogen from the amide nitrogen to facilitate the nucleophilic attack. In Ec, the two Zn ions are bridged with the carboxyl group of the carboxylated Lys103 (E. coli numbering) [5] and as seen in Fig. 8d, the carboxyl group of the carboxylated Lys103 in Ec (Ec numbering) is at the same position as the carboxyl group of the aspartate in the Mj model. Figs. 5b and 5c include also the invariant residues Arg60, Asn89, His306 that interact with the substrate in the known DHOases complexes [3, 5, 7, 8]. The substrate also interacts with the main chain at positions 275, 320 and 321 (M. jannaschii numbering) [3]. A number of other residues in the vicinity of the active site are also conserved. These are Pro88, Pro276, Pro305, Met87, Glu170, His257, Ala304, Lys311 (M. jannaschii numbering) [3] and their role may be to maintain the active site structure. All these invariant residues are highlighted in Fig. 8b.
3.8 Amino Acid Content of M. jannaschii DHOase
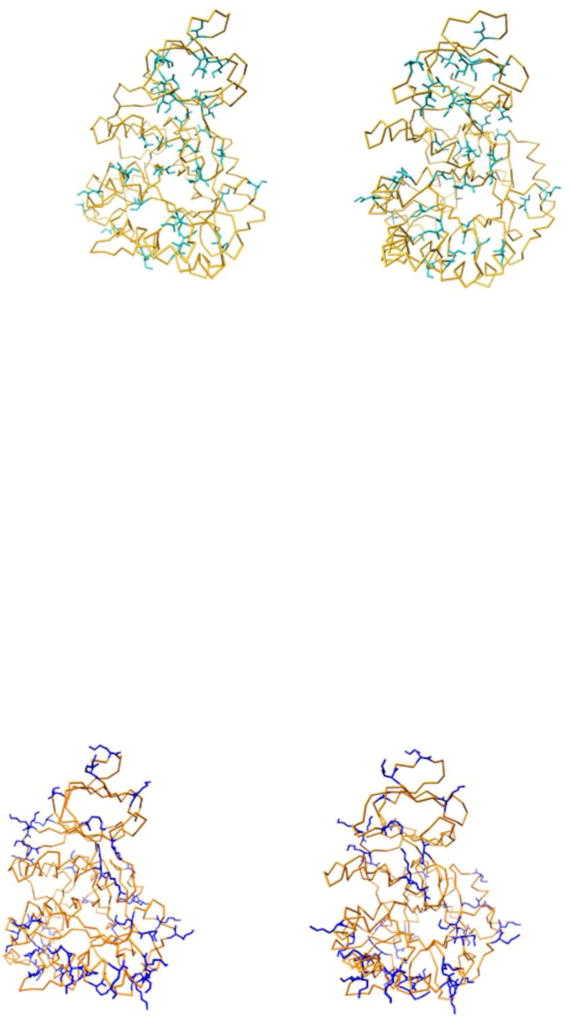
Table 2 gives the amino acid composition of Mj and Ba DHOases computed with the program protparam in ExPASy (23). As seen in this table, in Mj there is a substantial increase of charged Lys residues and large hydrophobic Ile residues and a decrease in Ala, Thr and Gly. The homology model shows that the Ile’s are mostly in the hydrophobic core concealed from the solvent (Fig. 9a) and primarily replace Val and to a lesser extent Ala, Leu and other amino acids (Fig. 8b). Lys’ are mostly on the exterior of the molecule (Fig. 9b) and replace charged and polar amino acids, more than non-polar amino acids (Fig. 8b). Table 2 shows also a decrease in Gln and an increase in Asn residues in Mj compared to the mesophilic Ba DHOase.
Table 2.
Comparison of the amino acid composition of Mj and Ba DHOases. The amino acid composition was computed with the protparam tool in ExPASy [23]. “Number” refers to the number of amino acids of a given kind in the protein, and “%” to the corresponding percentage.
| Mj | Ba | |||
|---|---|---|---|---|
| Number | % | Number | % | |
| Ala | 19 | 4.50 | 43 | 10.00 |
| Arg | 8 | 1.90 | 15 | 3.50 |
| Asn | 35 | 8.30 | 17 | 4.00 |
| Asp | 30 | 7.10 | 25 | 5.80 |
| Cys | 9 | 2.10 | 9 | 2.10 |
| Gln | 4 | 0.90 | 10 | 2.30 |
| Glu | 31 | 7.30 | 38 | 8.90 |
| Gly | 23 | 5.40 | 36 | 8.40 |
| His | 12 | 2.80 | 17 | 4.00 |
| Ile | 51 | 12.10 | 30 | 7.00 |
| Leu | 38 | 9.00 | 34 | 7.90 |
| Lys | 59 | 13.90 | 30 | 7.00 |
| Met | 4 | 0.90 | 12 | 2.80 |
| Phe | 18 | 4.30 | 14 | 3.30 |
| Pro | 15 | 3.50 | 19 | 4.40 |
| Ser | 16 | 3.80 | 10 | 2.30 |
| Thr | 12 | 2.80 | 31 | 7.20 |
| Trp | 3 | 0.70 | 3 | 0.70 |
| Tyr | 10 | 2.40 | 5 | 1.20 |
| Val | 26 | 6.10 | 30 | 7.00 |
| Total | 423 | 428 | ||
Fig. 9.
Stereo views. (a) The increased Ile content of Mj DHOase as compared to Ba. The α C backbone of Mj DHOase is shown in gold and the Ile replacements are shown in light sea green. It can be seen that most of these replacements are in the interior of the protein.
(b) The increased Lys content of Mj DHOase as compared to Ba. The α C backbone of Mj DHOase is shown in gold and the Lys replacements are shown in blue. It can be seen that most of these residues are on the surface of the protein.
4 Discussion
DHOases form a very diverse family of enzymes. Even though they catalyze the same reaction, its members differ in size, sequence, oligomeric structure, dimerization interfaces, Zn content and association or not with ATCase [3]. The current work is the first study of an archaeal DHOase and gives some understanding how this enzyme resembles and differs from other DHOases.
4.1 Structural Aspects
Archaeal DHOases are closest to bacterial type I enzymes. The latter form dimers and frequently associate with ATCase to form a stoichiometric 1:1 complex e.g. in Aa [24] and Ba [25]. It is assumed [3] that the stereochemistry of the Aa complex [24] is adopted by the other DHOases that associate with ATCase. In contrast, the Mj enzyme is a monomer in solution. In addition, a complex with ATCase is not possible as M. jannaschii has both pyrI and pyrB genes and their products associate to form a type B ATCase holoenzyme [26].
The active site has two Zn ions as most DHOases [3]. These are bridged by an aspartate Asp146 which is at the same position as the conserved aspartate in Type I bridging the two Zn ions (Fig. 8c). This differentiates Mj and Type I DHOases from types II, III and CAD that have a carboxylated lysine bridging the two Zn ions (Fig. 8d) [3]. The modified Lys is conserved within these groups. In the recent multiple sequence alignment [3], this carboxylated lysine in types II, III and CAD is shifted upstream two positions relative to the invariant aspartate in type I.
Variations from the Zn---Asp---Zn scheme in type I have been previously observed. For instance, A. aeolicus DHOase has one Zn whether complexed (pdb id: 3d6n) [24] or uncomplexed (pdb id: 1xrf and 1xrt) [27] with ATCase. The S. aureus enzyme (pdb id: 3gri) also has one Zn center bridged to the conserved Asp via a water molecule. These alternate active sites suggest analogous but somewhat different mechanisms. For instance, theoretical calculations for A. aeolicus DHOase indicate that a water in the crystal at the position of the β Zn lowers the activation energy of the reaction [8]. These findings question the invariance of a two Zn center for DHOases.
There has been recent discussion on the viability of the observed mono-zinc active site in some type I DHOases. Grande-Garcia et al. [3] suggested that all DHOases have a two-Zn center. They contend that the type I structures observed with one Zn-center in the crystalline state may represent an incomplete view of the active site where the β Zn ion is not bound or binds with partial occupancy. In these cases, it is possible that the more solvent-exposed β Zn could dissociate during purification and crystallization. This idea was supported by the fact that the β-Zn ligands are conserved in the known structures and by the incorrect estimation of the Zn content in E. coli and human DHOases. An initial analysis of both found only one zinc but further study showed two. Typically, dialysis is performed to remove fortuitously bound metal [28–30] before the analysis which could explain the loss of the second zinc. However, the existence of fully functional DHOases with a mononuclear metal center is supported by the fact that deletion of the β-site in A aeolicus DHOase [8] by replacing the two histidines binding the β Zn with Ala affected neither catalysis nor active site structure. However, when the second metal binding site in amidohydrolases with a binuclear metal center was eliminated, all catalytic activity ceased [31, 32]. Nonetheless, we have unambigiously shown that M. jannaschii DHOase contains two Zn ions. Whether other archaeal DHOases have one or two Zn centers remains to be seen.
4.2 Kinetics of M. jannaschii DHOase
The kinetic studies of Mj DHOase in Fig. 4 and Table 1 show that its specific activity increases with temperature, from 12.2 µmol min−1 mg−1 at 25° C to 248 µmol min−1 mg−1 at 80° C, which is a common feature to all enzymes. The reason is that the increase in temperature increases the number of enzyme molecules with enough energy to undergo the necessary catalytic conformational changes (33–35). The increase in Km with temperature (0.52 mM at 80° C as compared to 0.14 mM at 25° C) (Table 1, Fig. 4) in this organism is also consistent with observations in other proteins (33), and indicates an increase in the molecular flexibility of the active site. The increase in specific activity with temperature resembles the related catalytic trimer of ATCase from the same organism [26]. The fold-increase in turnover rate every 10° C increase in temperature (Q10) based on the specific activity was calculated to be 1.73 between 25° C and 80° C for the Mj DHOase. This fits nicely within the range of Q10 values observed in thermophilic enzymes of 1.2 to 2.3 based on specific activity [35]. Both Km and specific activity values in the mesophilic counterpart from Ba show also an increase with temperature (Table 1).
Close to their corresponding physiological temperatures (85° C for Mj and 37° C for Ba) the activity of Mj is much higher than the activity of its mesophilic homolog (Table 1). This is in contrast to expectations as the active sites of the two are very similar and the flexibilities of homologous proteins at their physiological temperatures are expected to be comparable (34). The large difference in the specific activities between Mj and Ba at 80° C and 37° C, could be attributed to subtle differences in the vicinity of their binding sites. This explanation has been invoked for pairs of homologous proteins that have dissimilar activities at their physiological temperatures [34]. It was also suggested [33] that these changes must alter the function by changing the mobility of the structures. Similar differences were found between the thermophilic bacterium B. caldolyticus and mesophilic hamster DHOase at their corresponding physiological temperatures in [36] (Table 1).
At 25° C, the specific activity of Mj is higher than in Ba (Table 1). This observation may be unexpected because hypethermophilic enzymes are believed to have rigid structures at ambient temperatures and catalysis requires flexibility (34). However, Elias et al (35) discuss that the rigidity of the fold of an enzyme does not necessarily affect the flexibility of its active site or its activity. Furthermore, there are reports of thermophilic enzymes that are as or more active than their mesophilic counterparts (35) as it happens in Mj. At all temperatures, the specific activity of Mj is higher than all the mesophilic proteins in Table 1.
4.3 Molecular Basis of Thermostability
The comparison of the amino acid compositions and sequences of the Mj and Ba dihydroorotases in section 3.8 (Table 2, Fig. 8b) gave insight into some of the factors that may confer stability in the Mj enzyme. The amino acid substitutions and their locations in Mj are consistent with thermostabilization strategies adopted by hyperthermophilic enzymes [34, 37–39]. Thermostable enzymes typically have more ion pairs (salt bridges), especially in networks, more hydrophobic interactions in their interior, and a larger number of bulkier hydrophobic side chains as compared to their mesophilic counterparts. It was also shown (40,41) that hyperthermophilic archaea substitute non-polar amino acids with Ile and have an increased Lys content from their corresponding mesophilic homologues. Furthermore, charged residues are overrepresented on the surface (40,41) and it was suggested (40) that a large number of exposed charged amino acids stabilize proteins at high temperatures by forming extended ion-pair networks. Such ion pairs and interconnected salt bridge networks on the surface of hyperthermophiles have been demonstrated (34). The decrease in Gln in Mj is consistent with other thermostable enzymes [37, 40). However, the Asn content in hyperthermophilic archaea is comparable to their mesophilic homologues and the reason for its increase in Mj is not clear. In addition, shorter N- and C-termini increase thermostability (34, 37) and, as seen in Figs. 8b and 8a, both N- and C-termini are shorter in M. jannaschii as compared to B. anthracis. It may be noted that the close agreement in the secondary structure of the enzymes in M. jannaschii and B. anthracis indicate that the protein core of the mesophilic protein is already optimally stable. Some of the thermostabilization strategies of DHOase in M. jannaschii are similar to those adopted by the catalytic subunit of ATCase in the same organism [42].
Additional and better insight on the stereochemistry of Mj DHOase and its thermostabilization strategies will be provided when its crystal structure is determined. In addition, more data on other archaeal DHOases are required to generalize the differences and similarities between archaeal DHOases and the other types.
Acknowledgments
This work was supported in part by grant GM071512 (JV) from the National Institutes of Health. Molecular graphics and analyses were performed with the UCSF Chimera package. Chimera is developed by the Resource for Biocomputing, Visualization, and Informatics at the University of California, San Francisco (supported by NIGMS P41-GM103311). We thank Dr. Belinda Willard of the Lerner Research Institute in Cleveland Clinic for the LC-MS, Mr. Michael Murphy of Intertek Chemicals and Pharmaceuticals for the ICP-MS and Dr. Ewa Folta-Stogniew of the Biophysics Resource of the Keck Facility at Yale for the SEC-LS. The SEC-LS/UV/RI instrumentation was supported by NIH Award Number 1S10RR023748-01. We also thank Dr. Bin Su of Cleveland State University for use of his SPECTRAmax PLUS 384 microplate reader and Drs. Barbara Zimmermann of Los Andes University and Evan Kantrowitz of Boston College for enlightening discussions.
Abbreviations
- A. aeolicus
Aquifex aeolicus
- ATCase
Aspartate transcarbamoylase
- B. anthracis, Ba
Bacillus anthracis
- BME
2-mercaptoethanol
- BSA
Bovine serum albumin
- CA
Carbamoyl aspartate
- CAD
Carbamoyl phosphate synthetase/aspartate transcarbamoylase/dihydroorotase protein
- CID
Collision induced dissociation
- DHO
Dihydroorotate
- DHOase
Dihydroorotase
- E. coli, Ec
Escherichia coli
- IPTG
Isopropyl p-D-1-thiogalactopyranoside
- LB
Luria–Bertani medium
- MES
2-(N-morpholino)ethanesulfonic acid
- M. jannaschii, Mj
Methanococcus jannaschii
- SRM
Selective reaction monitoring
- SDS-PAGE
Sodium dodecyl sulfate polyacrylamide gel electrophoresis
- S. aureus
Staphylococcus aureus
- T. thermophilus
Thermus thermophilus
- Tris
Tris(hydroxymethyl)aminomethane
Footnotes
Compliance with Ethical Standards
Conflicts of interest
The authors declare that they have no conflicts of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
References
- 1.Holm L, Sander C. An evolutionary treasure: unification of a broad set of amidohydrolases related to urease. Proteins: Structure, Function, and Bioinformatics. 1997;28:72–82. [PubMed] [Google Scholar]
- 2.Fields C, Brichta D, Shepherdson M, Farinha M, O’Donovan G. Phylogenetic analysis and classification of dihydroorotases: a complex history for a complex enzyme. Paths Pyrimidines. 1999;7:49–63. [Google Scholar]
- 3.Grande-García A, Lallous N, Díaz-Tejada C, Ramón-Maiques S. Structure, functional characterization, and evolution of the dihydroorotase domain of human CAD. Structure. 2014;22:185–198. doi: 10.1016/j.str.2013.10.016. [DOI] [PubMed] [Google Scholar]
- 4.Kim GJ, Kim HS. Identification of the structural similarity in the functionally related amidohydrolases acting on the cyclic amide ring. Biochem J. 1998;330(Pt 1):295–302. doi: 10.1042/bj3300295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Thoden JB, Phillips GN, Jr, Neal TM, Raushel FM, Holden HM. Molecular structure of dihydroorotase: a paradigm for catalysis through the use of a binuclear metal center. Biochemistry. 2001;40:6989–6997. doi: 10.1021/bi010682i. [DOI] [PubMed] [Google Scholar]
- 6.Porter TN, Li Y, Raushel FM. Mechanism of the dihydroorotase reaction. Biochemistry. 2004;43:16285–16292. doi: 10.1021/bi048308g. [DOI] [PubMed] [Google Scholar]
- 7.Rice AJ, Lei H, Santarsiero BD, Lee H, Johnson ME. Ca-asp bound X-ray structure and inhibition of Bacillus anthracis dihydroorotase (DHOase) Bioorg Med Chem. 2016;24:4536–4543. doi: 10.1016/j.bmc.2016.07.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Edwards BF, Fernando R, Martin PD, Grimley E, Cordes M, Vaishnav A, Brunzelle JS, Evans HG, Evans DR. The mononuclear metal center of type-I dihydroorotase from aquifex aeolicus. BMC Biochemistry. 2013;14:1. doi: 10.1186/1471-2091-14-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Prescott LM, Jones ME. Modified methods for the determination of carbamyl aspartate. Anal Biochem. 1969;32:408–419. doi: 10.1016/s0003-2697(69)80008-4. [DOI] [PubMed] [Google Scholar]
- 10.Folta-Stogniew E, Williams KR. Determination of molecular masses of proteins in solution: Implementation of an HPLC size exclusion chromatography and laser light scattering service in a core laboratory. J Biomol Tech. 1999;10:51–63. [PMC free article] [PubMed] [Google Scholar]
- 11.Yang J, Yan R, Roy A, Xu D, Poisson J, Zhang Y. The I-TASSER suite: protein structure and function prediction. Nat Methods. 2015;12:7–8. doi: 10.1038/nmeth.3213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yang J, Zhang Y. I-TASSER server: new development for protein structure and function predictions. Nucleic Acids Res. 2015;43:W174–81. doi: 10.1093/nar/gkv342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang Y. I-TASSER server for protein 3D structure prediction. BMC Bioinformatics. 2008;9 doi: 10.1186/1471-2105-9-40. 40-2105-9-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Roy A, Kucukural A, Zhang Y. I-TASSER: a unified platform for automated protein structure and function prediction. Nat Protoc. 2010;5:725–738. doi: 10.1038/nprot.2010.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 16.Sali A, Blundell T. Comparative protein modelling by satisfaction of spatial restraints. Protein Structure by Distance Analysis. 1994;64:C86. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- 17.Maier JA, Martinez C, Kasavajhala K, Wickstrom L, Hauser KE, Simmerling C. ff14SB: Improving the accuracy of protein side chain and backbone parameters from ff99SB. J Chem Theory Comput. 2015;11:3696–3713. doi: 10.1021/acs.jctc.5b00255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang J, Wang W, Kollman PA, Case DA. Automatic atom type and bond type perception in molecular mechanical calculations. J Mol Graph Model. 2006;25:247–260. doi: 10.1016/j.jmgm.2005.12.005. [DOI] [PubMed] [Google Scholar]
- 19.Laskowski RA, Rullmann JAC, MacArthur MW, Kaptein R, Thornton JM. AQUA and PROCHECK-NMR: programs for checking the quality of protein structures solved by NMR. J Biomol NMR. 1996;8:477–486. doi: 10.1007/BF00228148. [DOI] [PubMed] [Google Scholar]
- 20.Bult CJ, White O, Olsen GJ, Zhou L. Complete genome sequence of the methanogenic archaeon, Methanococcus jannaschii. Science. 1996;273:1058. doi: 10.1126/science.273.5278.1058. [DOI] [PubMed] [Google Scholar]
- 21.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Research. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jones W, Leigh JA, Mayer F, Woese C, Wolfe R. Methanococcus jannaschii sp. nov., an extremely thermophilic methanogen from a submarine hydrothermal vent. Arch Microbiol. 1983;136:254–261. [Google Scholar]
- 23.Gasteiger E, Hoogland C, Gattiker A, Duvaud S, Wilkins MR, Appel RD, Bairoch A. In: Protein Identification and Analysis Tools on the ExPASy Server. Walker John M., editor. The Proteomics Handbook, Humana Press; 2005. pp. 571–607. [Google Scholar]
- 24.Zhang P, Martin PD, Purcarea C, Vaishnav A, Brunzelle JS, Fernando R, Guy-Evans HI, Evans DR, Edwards BF. Dihydroorotase from the hyperthermophile Aquifiex aeolicus is activated by stoichiometric association with aspartate transcarbamoylase and forms a one-pot reactor for pyrimidine biosynthesis. Biochemistry (N Y) 2009;48:766–778. doi: 10.1021/bi801831r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kankanala R. Characterizing the oligomeric structure and catalytic activity of the dihydroorotase and aspartate transcarbamoylase from the bacterium, Bacillus anthracis. Master’s Theses and Doctoral Dissertations. 2011 Paper 339. ( http://commons.emich.edu/theses/339)
- 26.Hack ES, Vorobyova T, Sakash JB, West JM, Macol CP, Herve G, Williams MK, Kantrowitz ER. Characterization of the aspartate transcarbamoylase from Methanococcus jannaschii. J Biol Chem. 2000;275:15820–15827. doi: 10.1074/jbc.M909220199. [DOI] [PubMed] [Google Scholar]
- 27.Martin PD, Purcarea C, Zhang P, Vaishnav A, Sadecki S, Guy-Evans HI, Evans DR, Edwards BF. The crystal structure of a novel, latent dihydroorotase from Aquifex aeolicus at 1.7 Å resolution. J Mol Biol. 2005;348:535–547. doi: 10.1016/j.jmb.2005.03.015. [DOI] [PubMed] [Google Scholar]
- 28.Washabaugh MW, Collins KD. Dihydroorotase from Escherichia coli. Purification and characterization. J Biol Chem. 1984;259:3293–3298. [PubMed] [Google Scholar]
- 29.Zimmermann BH, Evans DR. Cloning, overexpression, and characterization of the functional dihydroorotase domain of the mammalian multifunctional protein CAD. Biochemistry (NY) 1993;32:1519–1527. doi: 10.1021/bi00057a016. [DOI] [PubMed] [Google Scholar]
- 30.Kelly RE, Mally MI, Evans DR. The dihydroorotase domain of the multifunctional protein CAD. Subunit structure, zinc content, and kinetics. J Biol Chem. 1986;261:6073–6083. [PubMed] [Google Scholar]
- 31.Wang C, Tsau H, Chen W, Huang C. Identification and characterization of a putative dihydroorotase, KPN01074, from Klebsiella pneumoniae. The Protein Journal. 2010;29:445–452. doi: 10.1007/s10930-010-9272-2. [DOI] [PubMed] [Google Scholar]
- 32.Ho Y, Huang Y, Huang C. Chemical rescue of the post-translationally carboxylated lysine mutant of allantoinase and dihydroorotase by metal ions and short-chain carboxylic acids. Amino Acids. 2013;44:1181–1191. doi: 10.1007/s00726-012-1451-3. [DOI] [PubMed] [Google Scholar]
- 33.Fields PA. Review: Protein function at thermal extremes: balancing stability and flexibility. Comp Biochem Physiol A Mol Integr Physiol A. 2001;129:417–431. doi: 10.1016/s1095-6433(00)00359-7. [DOI] [PubMed] [Google Scholar]
- 34.Feller G. Topical Review: Protein stability and enzyme activity at extreme biological temperatures. J Phys Condens Matter. 2010;22:323101. doi: 10.1088/0953-8984/22/32/323101. [DOI] [PubMed] [Google Scholar]
- 35.Elias M, Wieczorek G, Rosenne S, Tawfik DS. The universality of enzymatic rate– temperature dependency. Trends Biochem Sci. 2014;39:1–7. doi: 10.1016/j.tibs.2013.11.001. [DOI] [PubMed] [Google Scholar]
- 36.Huang DT, Kaplan J, Menz RI, Katis VL, Wake RG, Zhao F, Wolfenden R, Christopherson RI. Thermodynamic analysis of catalysis by the dihydroorotases from hamster and Bacillus caldolyticus, as compared with the uncatalyzed reaction. Biochemistry. 2006;45:8275–8283. doi: 10.1021/bi060595w. [DOI] [PubMed] [Google Scholar]
- 37.Vieille C, Zeikus GJ. Hyperthermophilic enzymes: sources, uses, and molecular mechanisms for thermostability. Microbiol Mol Biol Rev. 2001;65:1–43. doi: 10.1128/MMBR.65.1.1-43.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Petsko GA. Structural basis of thermostability in hyperthermophilic proteins, or ‘there’s more than one way to skin a cat’ Methods Enzymol. 2001;334:469–78. doi: 10.1016/s0076-6879(01)34486-5. [DOI] [PubMed] [Google Scholar]
- 39.Ding Y, Cai Y, Han Y, Zhao B. Comparison of the structural basis for thermal stability between archaeal and bacterial proteins Extremophiles. 2012;16:67–78. doi: 10.1007/s00792-011-0406-z. [DOI] [PubMed] [Google Scholar]
- 40.Mizuguchi K, Sele M, Cubellis MV. Environment specific substitution tables for thermophilic proteins BMC Bioinformatics. 2007;8(1):S15. doi: 10.1186/1471-2105-8-S1-S15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Saelensminde G, Halskau Ø, Helland R, Willassen NP, Jonassen I. Structure-dependent relationships between growth temperature of prokaryotes and the amino acid frequency in their proteins. Extremophiles. 2007;11:585–596. doi: 10.1007/s00792-007-0072-3. [DOI] [PubMed] [Google Scholar]
- 42.Vitali J, Colaneri MJ, Kantrowitz E. Crystal structure of the catalytic trimer of Methanococcus jannaschii aspartate transcarbamoylase. Proteins: Structure, Function, and Bioinformatics. 2008;71:1324–1334. doi: 10.1002/prot.21667. [DOI] [PubMed] [Google Scholar]