Abstract
Maternal smoking during pregnancy (MSDP) has detrimental effects on fetal development and on the health of the offspring into adulthood. Energy homeostasis through ATP production via the mitochondria (mt) plays a key role during pregnancy. This study aimed to determine if MSDP resulted in differences in DNA methylation to the placental mitochondrial chromosome at the transcription and replication control region, the D-Loop, and if these differences were also present in an alternate neonatal tissue (foreskin) in an independent birth cohort. We investigated mtDNA methylation by bisulfite-pyrosequencing in two sections of the D-Loop control region and in long interspersed nuclear element-1 (LINE-1) genomic sequences in placenta from 96 mother–newborn pairs that were enrolled in a Rhode Island birth cohort along with foreskin samples from 62 infants from a Kentucky birth cohort. In both placenta and foreskin, mtDNA methylation in the light chain D-Loop region 1 was positively associated with MSDP in placenta (difference +2.73%) ( P = 0.001) and foreskin (difference + 1.22%) ( P = 0.08). Additionally, in foreskin, a second segment of the D-Loop-heavy chain region 1 showed a small but significant change in methylation with MSDP (+ 0.4%, P = 0.04). No methylation changes were noted in either tissue at the LINE-1 repetitive element. We identified a similar pattern of epigenetic effect to mitochondria arising in cells from different primordial lineages and in different populations, associated with MSDP. These robust and consistent results build evidence that MSDP may impact mt D-Loop methylation, as one mechanism through which this exposure affects newborn health.
Keywords: DNA methylation, mitochondria, maternal smoking, pregnancy, placenta
Introduction
Understanding the role of environmental and lifestyle exposures on inter-generational health has been the subject of many recent studies [ 1–6 ]. Cigarette smoking, specifically maternal smoking during pregnancy (MSDP), is one such environmental exposure that warrants further study. For > 50 years, investigators have known that MSDP alters infant birth weight [ 7 , 8 ]. However, the exact cellular mechanisms driving this physiological difference remain elusive. Exposure to maternal cigarette smoking during pregnancy is associated with offspring morbidity and mortality, including increased risks for miscarriage, stillbirth, low birth weight, preterm birth, asthma, obesity, altered neurobehavior, and other conditions [ 9 ]. Maternal cigarette smoking during pregnancy can interfere with placental growth and functioning, and it has been proposed that this may occur through the disruption of normal and necessary placental epigenetic patterns [ 10 ].
Epigenetics represents a broad range of mechanisms involved in gene expression control that are not associated with modifications to the primary DNA sequence. These include DNA methylation, histone post-translational modifications, and regulatory non-coding RNA sequences (i.e micro-RNA). Epigenetic regulation during the in utero period has been suggested to be the mechanism by which fetal programming occurs. Epigenetic variations have important regulatory roles within the placenta and have been associated with a number of developmental outcomes [ 11 ]. The most well-characterized epigenetic modification in human epidemiologic studies is DNA methylation, which can alter the transcription or transcriptional potential of genes and when occurring in gene promoters are usually associated with transcriptional repression.
Mitochondrial epigenetics is an emerging field [ 12 ] with several recent reports examining mitochondrial methylation related to environmental exposures [ 13 , 14 ].
The mitochondria is the “powerhouse” of the cell and is involved in many cellular processes, including oxidative phosphorylation and ATP production, β-fatty acid oxidation, the citric acid cycle, and regulation of apoptosis (intrinsic pathway).
Mitochondrial (mt) DNA is a closed-circular, double-stranded molecule 16.6 kb in size and containing 37 genes (2 ribosomal RNAs, 22 transfer RNAs, and 13 protein-coding mRNAs) all of which are essential for normal mitochondrial function. The strands of the DNA duplex can be distinguished on the basis of G + T base composition, which results in different buoyant densities of each strand (“heavy” and “light”) in denaturing cesium chloride gradients [ 15 ]. Most information is encoded on the heavy (H) strand, with genes for 2 rRNAs, 14 tRNAs, and 12 polypeptides. The light (L) strand codes for eight tRNAs and a single polypeptide. The D-loop region is the major control site for mtDNA expression, containing the heavy strand origin of replication and the major promoters for transcription for both H and L strands ( Fig. 1A ) [ 16 ]. Transcription proceeds via a full-length polycistronic precursor that is cleaved to release individual tRNAs, rRNAs, and mRNAs [ 17 ].
Figure 1.
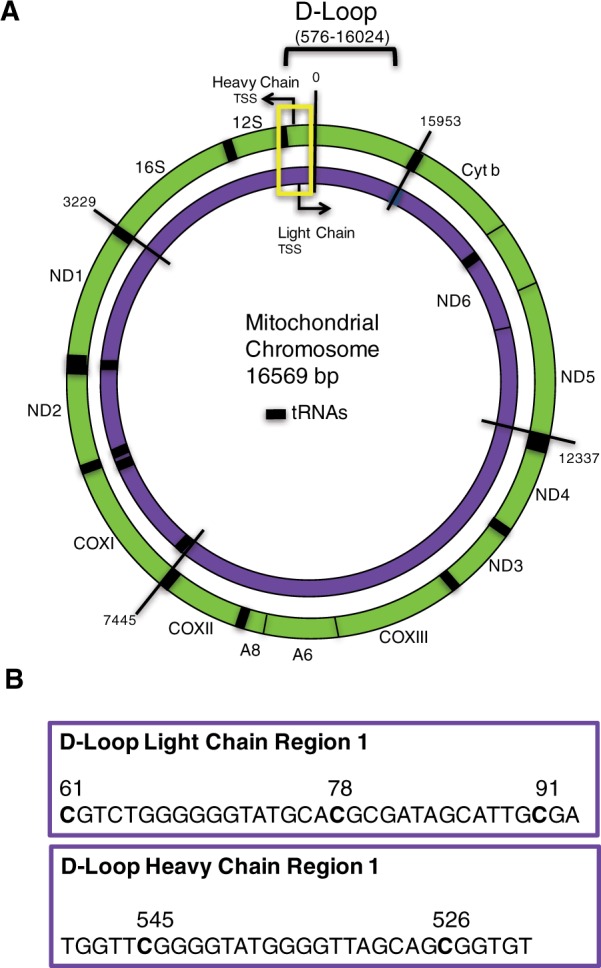
Map of mitochondrial genome and sequences on the MT chromosome used for methylation analysis. The outer circle (green) represents the heavy chain MT chromosome and the inner circle (purple) being the MT light chain ( A ). The 16,569 base chromosome is numbered at various segments for orientation. Coding genes are labeled on the H & L chains and the 12S and 16S ribosomal RNA genes. tRNAs are represented by thick black bars. The D-Loop control region is contained within bases 576–16,024. Yellow rectangle outlines the section of the mitochondrial chromosome within which the methylation assays were designed. The specific MT-chromosomal sequences and CpG sites analyzed in D-Loop are detailed ( B )
In this study, we examined the association between mtDNA methylation from newborns with MSDP as a prototypical deleterious environmental/lifestyle exposure, both in placenta, derived from the trophoectoderm lineage, as well as in foreskin tissue, a neonatal tissue comprising cells of various primordial lineages. We hypothesized that if we could quantify differences in mtDNA methylation related to smoking and that these differences may be identifiable in multiple tissues. To our knowledge, this is the first study to examine mitochondrial epigenetics related to maternal smoking during pregnancy.
Methods
Rhode Island Child Health Study Population
Study subjects involved in the placenta analyses were part of the Rhode Island Child Health Study (RICHS), which enrolled healthy mother and infant pairs following delivery at the Women and Infants Hospital of Rhode Island (Providence, RI, USA). Term infants born small for gestational age (lowest 10th percentile), or large for gestational age (highest 10th percentile), based on birth weight and gestational age and calculated from the Fenton growth chart [ 18 ], were selected; infants appropriate for gestational age matched on gender, gestational age (±3 days), and maternal age (±3 years) are also enrolled. Only singleton, viable infants were included in the study. Other exclusion criteria were maternal age <18 or >40 years, a life-threatening medical complication of the mother, and congenital or chromosomal abnormality of the infant. A structured chart review was used to collect information from the maternal inpatient medical record from delivery. While still in the hospital after delivery but prior to discharge, mothers participated in an interviewer-administered structured questionnaire to obtain information on demographics, prenatal health behaviors, medical history, information on various lifestyle exposures (including smoking and recreational drug use), occupational histories, and personal care product usage. Smoking was retrospectively assessed as over the lifetime, 3 months prior to pregnancy, and during each trimester of pregnancy, asking participants about the types of cigarettes used (filtered or nonfiltered, menthol, or nonmenthol) and the average number of packs of cigarette smoked per day during these periods. Between September 1, 2009, and July 31, 2013, 1150 eligible infants were identified and 721 enrolled (63%). Study protocols were approved by the Institutional review boards for Women and Infants’ Hospital and Dartmouth College. Mothers provided written informed consent for participation and also for participation of her infant. Based on the administered maternal questionnaire, all full-term smokers (FT) ( n = 36) and first trimester-only smokers (1T) ( n = 23) were identified and included in this study along with a randomly selected non-smoker (NS) comparison group ( n = 37).
Placental Sample Collection
For each subject and within 2 hours of delivery, 12 samples of placenta tissue, three from each of four quadrants (totaling approximately 8–10 g of tissue) were excised. All samples were taken from the maternal side of the placenta, 2 cm from the umbilical cord insertion site, free of maternal decidua. The samples were placed immediately in RNAlater (Life Technologies, Grand Island, NY) and stored at 4 °C. At least 72 hours later, placenta samples were removed from RNAlater, blotted dry, snap-frozen in liquid nitrogen, homogenized by pulverization using a stainless steel cup and piston unit (Cellcrusher, Cork, Ireland) to create a uniform sample, and stored at −80 °C until needed for examination.
Kentucky Study Population and Sample Collection
This was a study of male neonates born at the University of Kentucky from June 2012 to March 2013 ( n = 38) and July to October 2015 ( n = 24). Inclusion criteria were English-speaking mothers, ≥ 18 years old following the delivery of a non-anomalous, singleton infant that would be circumcised within 72 hours of birth. The study was approved by the University of Kentucky Institutional Review Board, and all the mothers provided the clinical and surgical consents required for the operative procedure and for the collection of the circumcised foreskin tissue for research purposes. All procedures were performed either directly by or under supervision of an MD Board Certified in Obstetrics and Gynecology with dedicated hospital privileges for performing the circumcision procedure. Neonatal circumcision was performed in the standard manner using sterile aseptic precautions. We did not note any intra-/postoperative complications in our study cohort. The excised foreskin tissue was dissected immediately after collection to separate the dartos (hypodermis) from the epidermis/dermis layers. A portion of the epidermis/dermis was snap-frozen in liquid nitrogen for DNA isolation, while the remainder of the samples was used for separate experiments. The DNA from the epidermis/dermis (referred to as foreskin in this article) was isolated and analyzed together as described below because the two layers are difficult to separate mechanically in fresh tissues. All neonates enrolled in the study were healthy. One neonate per group was pre-term, with the remaining neonates born at ≥ 37 weeks. Demographic and clinical data were abstracted from a thorough review of the maternal and neonatal electronic medical records (EMR). Smoking status before pregnancy was ascertained at the first prenatal visit, as documented in the intake questionnaire in the outpatient maternal EMR system. Smoking status during pregnancy was verified based on the documentation of the same in the maternal EMR during the course of ongoing prenatal care and the status at the time of delivery was similarly confirmed using EMR at the time of presentation for delivery. Patients who were noted to have been smoking at the beginning of pregnancy (1st prenatal visit) but had quit smoking during the ongoing course of prenatal care or at the time of delivery—were considered as “smoking quitters,” while the remainder were deemed to have been “smokers” for the entire pregnancy time-course (the population of smoking quitters was too low to analyze as part of this study).
Nucleic Acid Extraction and Bisulfite Modification
DNA was extracted from the placenta and foreskin samples using the Qiagen DNAeasy Blood and Tissue Kit or QIAprep Spin Miniprep Kit (Qiagen, Inc., Valencia, CA). Purified DNA was quantified using a Qubit 3.0 Fluorometer (Thermo Fisher Scientific Inc., Waltham, MA), and DNA samples (500 ng) were bisulfite modified using the EZ DNA Methylation Kit (Zymo Research, Irvine, CA, USA) and stored at −20 °C. Samples were randomized for analysis.
Bisulfite Pyrosequencing DNA Methylation Analysis
Quantitative bisulfite pyrosequencing was used to assess DNA methylation status for specific CpG sites within mitochondrial D-Loop (NCBI Reference Sequence: NC_012920.1), along with previously described long interspersed nuclear element-1 (LINE-1) for global methylation [ 19 , 20 ]. We have included LINE-1 analysis in our study as this assay provides us with two important internal controls. LINE-1 sequences are highly repeated human retrotransposon sequences and constitute about 15% of the human genome. Approximately one-third of genomic DNA methylation occurs in repetitive elements. The DNA derived from both birth cohorts’ bio-specimens comprised a mix of both mitochondrial and genomic DNA. Therefore, analysis of these repetitive elements can serve as a surrogate marker for global genomic DNA methylation [ 19 ]. In this way, we can compare mtDNA methylation to genomic DNA methylation within the same tissue and note any differences related to MSDP. Second, we used LINE-1 pyrosequencing assay as a measure of DNA bisulfite conversion efficiency. A ratio was generated using the LINE-1-based conversion control as follows: high peak height/(high + low) peaks (i.e T/(T + C)) [ 21 ]. Only samples with DNA bisulfite conversion efficiency ≥ 95% were used in analysis.
Taking into account the issue of nuclear mtDNA (NUMTs) [ 22 ], mitochondrial D-Loop primer sets were designed using Qiagen PyroMark CpG Assay Design software (V2.0) with specificity of mitochondrial polymerase chain reaction (PCR) product confirmed via the on-line primer design and search tool: BiSearch ( http://bisearch.enzim.hu/ ) and agarose gel electrophoresis.
Detailed information regarding primer sequences is provided in online Supplementary material, Table S1 . Percent DNA methylation at each CpG site was quantified using the PyroMark MD instrument and the PyroMark Q-CpG software, version 1.0.11. (Qiagen). Samples were run in triplicate, and data points were rejected if they were greater or less than two times the standard deviation of the mean of the triplicate group. DNA methylation across all CpG sites within each assay was averaged to obtain average percent methylation for analysis.
mtDNA Content Analysis
Relative mtDNA content was measured from placental tissue by a quantitative real-time PCR assay by determining the ratio of mitochondrial copy number to a single copy gene [ 23 , 24 ]. The reference single copy gene used in this study was human globin (beta). Primers used for mtDNA content were identical to those previously described [ 23 ].
Statistical Analysis
For each region of interest, Pearson correlation indicated that values across loci within the region were strongly correlated ( r > 0.75). Due to this, we took the average methylation value for each individual across the loci measured here. A Shapiro–Wilk test was performed for both the RICHS and Kentucky cohorts in order to determine whether methylation values were normally distributed for LDLR1, HDLR1, and LINE1. In both the RICHS and Kentucky cohorts LINE1 methylation was normally distributed while LDLR1 and HDLR1 methylation was not normally distributed.
Associations between maternal smoking status during pregnancy and DNA methylation for RICHS were determined by 1-way analysis of variance (ANOVA) or by Kruskal–Wallis one-way ANOVA for normally and non-normally distributed data, respectively. A Dunn’s multiple comparisons test was performed post hoc in order to determine the relationship between smoking status and methylation. Within the Kentucky cohort, the extent of methylation was associated with smoking status by Student’s t -test or by Mann–Whitney U test for normally and non-normally distributed data, respectively. Descriptive statistics were calculated for each smoking status grouping, and to determine whether there were demographic differences within our groupings, we used a Fisher’s exact test and one-way ANOVA to compare count and continuous data, respectively. All data were analyzed using the R statistical environment v3.2.4. Data underlying these analyses can be provided for purposes of verification by contacting the authors.
Results
RICHS study subject characteristics are listed in Table 1 . Overall, mothers in the study were on average 28.1 years old, with no differences in maternal age depending on smoking status. Maternal body mass index also did not differ based on smoking status. Non-smoking mothers tended to have the highest level of maternal education with 55.6% FT, 43.5% 1T, and 29.7% of NS mothers having attained a high-school education or less. Infant sex did not differ across the maternal smoking groups, but as expected, FT pregnancy smokers had a greater proportion of infants consider small for gestational age (50%) compared to first term only (17%) or non-smokers (NS) (16%). Infants of non-smoking mothers showed the highest overall birth weight average at 3666.0 g, with the 1T and FT pregnancy smokers showing a 7.4% (3378.48 g) and 17.85% (3011.5 g) reduction in birth weight, respectively. The average gestational age for each group was similar. Finally, pack years ((cigarettes/day/20)/years of smoking) was used to evaluate differences in lifetime primary smoke exposures between the 1T smokers and FT pregnancy smokers. No differences were noted ( P = 0.56).
Table 1.
The characteristics of RICHS study subjects
| NS (n = 37) | 1st trimester-only smokers (n = 23) | FT pregnancy smokers (n = 36) | P value | |
|---|---|---|---|---|
| Maternal | ||||
| Age, mean (range) | 29.18 (18–40) | 27.91 (20–39) | 27.14 (20–38) | 0.35 |
| BMI | ||||
| Normal | 17 (45.95%) | 9 (39.13%) | 18 (50%) | 0.45 |
| Overweight | 7 (18.92%) | 8 (34.78%) | 10 (27.78%) | |
| Obese | 12 (32.43%) | 6 (26.09%) | 6 (16.67%) | |
| Maternal education | ||||
| < 11th grade | 2 (5.41%) | 0 | 11 (30.56%) | 0.002 |
| High school graduate | 9 (24.32%) | 10 (43.48%) | 9 (25%) | |
| Junior college | 10 (27.03%) | 7 (30.43%) | 12 (33.33%) | |
| College graduate | 10 (27.03%) | 4 (17.39%) | 4 (11.11%) | |
| Post graduate school | 6 (16.22%) | 2 (8.7%) | ||
| Gender | ||||
| Male | 13 (35.14%) | 11 (47.83%) | 16 (44.44%) | 0.62 |
| Female | 24 (64.86%) | 12 (52.17%) | 20 (55.56%) | |
| Birthweight group | ||||
| AGA | 19 (51.35%) | 13 (56.52%) | 15 (41.67%) | 0.006 |
| LGA | 12 (32.43%) | 6 (26.09%) | 3 (8.33%) | |
| SGA | 6 (16.22%) | 4 (17.39%) | 18 (50%) | |
| Birthweight (g) | 3666.0 ± 650.68 | 3378.48 ± 683.37 | 3011.5 ± 598.58 | 0.0001 |
| Range (g) | 2510–4920 | 1920–4530 | 2280–4380 | |
| Gestational weeks | 39.22 (37–41) | 39.28 (37–41) | 39.04 (37–41) | 0.54 |
AGA: Appropriate for Gestational Age
LGA: Large for Gestational Age
SGA: Small for Gestational Age
Pyrosequencing assays were designed to measure DNA methylation of the mitochondrial chromosome on the light chain in D-Loop region (LDLR) as shown in Figure 1 . As a mark of genomic global methylation, LINE-1 repetitive region was also quantified. Median methylation was evaluated in both cohorts in LDLR1 and HDLR1 assays as the data were not normally distributed; however, LINE-1 assay data were normally distributed; therefore, mean methylation was reported.
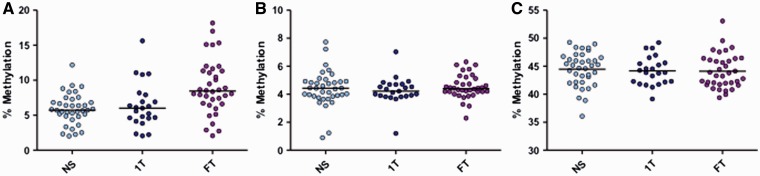
The three CpGs of LDLR1 showed strong correlation among each other (online Supplementary material, Fig. S1 ); therefore, the median percent methylation across all three CpGs was used for further analysis. A significant difference was seen when comparing the median methylation across groups ( P = 0.001, Kruskal–Wallis) most notably when comparing NS 5.72% to smokers 8.45% (difference +2.73%) ( P = 0.001, Dunn’s post hoc)( Fig. 2A ). To examine the specificity of the D-Loop methylation assay (LDLR1), we used two additional DNA regions as surrogate measurements for methylation selectivity: a second location in the D-Loop (HDLR1) and global methylation in genomic DNA (LINE-1). We found no significant methylation differences among smoking groups at loci HDLR1 in placenta ( P = 0.66) ( Fig. 2B ) nor LINE-1 ( P = 0.87) ( Fig. 2C ). In addition, no significant differences in mitochondrial copy number were found between the smoking group and the non-smoking group ( P = 0.95) (online Supplementary material, Fig. S2 ).
Figure 2.
Mitochondrial and global methylation associated with maternal smoking during pregnancy in placenta . Median methylation % was measured for each pyrosequencing assay from placenta of mothers comparing NS ( n = 37), 1T smokers ( n = 23) and FT pregnancy smokers ( n = 36). Mitochondrial LDLR1 showed significantly increased methylation in full pregnancy smokers ( P = 0.001, Kruskal-Wallis, NS to FT P = 0.001, Dunn’s post hoc) ( A ). HDLR1 ( B ) showed no differences in methylation among the smoking groups ( P = 0.66). Horizontal lines represent the median. Global methylation measured via LINE-1 (C) showed no differences between the groups ( P = 0.87). Horizontal lines represent the mean (LINE-1)
Next, we sought to determine if the differences in D-Loop methylation in placenta could also be observed in a neonatal tissue and in an independent population. The Kentucky Birth Cohort subject characteristics are listed in Table 2 .
Table 2.
The characteristics of Kentucky study subjects
| NS (n = 41) | FT pregnancy smokers (n = 21) | P value | |
|---|---|---|---|
| Maternal | |||
| Age, mean (years) | 28.12 (20–39) | 25.71 (21–38) | 0.047 |
| BMI | |||
| Normal | 25 (60.98%) | 9 (42.86%) | 0.015 |
| Overweight | 7 (17.07%) | 7 (33.33%) | |
| Obese | 9 (21.95%) | 5 (23.81%) | |
| Maternal education | |||
| < 11th grade | 4 (9.76%) | 3 (14.29%) | 0.061 |
| High school grad | 12 (29.27%) | 14 (66.67%) | |
| Junior college | 1 (2.44%) | 0 | |
| College graduate | 16 (39.02%) | 4 (19.05%) | |
| Post graduate school | 8 (19.51%) | 0 | |
| Infant | |||
| Birthweight group | |||
| AGA | 38 (92.68%) | 15 (71.43%) | 0.015 |
| LGA | 1 (2.44%) | 0 | |
| SGA | 2 (4.88%) | 6 (28.57%) | |
| Birthweight (g) | 3437 ± 451.87 | 3145 ± 372.65 | 0.009 |
| Range (g) | 2469–4550 | 2380–3890 | |
| Gestational weeks | 38.80 (36–41) | 39.14 (35–41) | 0.284 |
This cohort included two groups: FT pregnancy smoking mothers versus non-smoking mothers. Overall, mothers in the study were on average 27.31 ± 4.95 years old, with FT pregnancy smokers mean age at 25.71 ± 3.86 years and NS at 28.12 ± 5.28 years old. Infants of non-smoking mothers showed a birth weight average at 3437.46 ± 451.87 g, while FT pregnancy smoker infants had a mean birth weight of 3145 ± 372.65 g, representing a significant 8.5% reduction in birth weight ( P = 0.009). The average gestational age for each group was similar with FT at 39.14 and NS at 38.80 weeks.
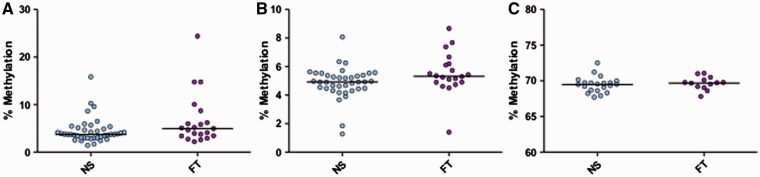
Although not statistically significant, D-Loop assay LDLR1 revealed a trend similar to that observed in placenta when comparing the median methylation between NS and smokers (3.75% vs. 4.97%, P = 0.08, Mann–Whitney, Fig. 3A ). Moreover, we have tested the relationship between LDLR1 mean methylation and smoking status within the Kentucky cohort removing a single highest data point as seen in Figure 3A . The resulting methylation difference between NS and smokers is in the same direction (+0.73%; P = 0.14) and similar magnitude to our original findings.
Figure 3.
Mitochondrial and global methylation associated with maternal smoking during pregnancy in foreskin. Median methylation % was measured for each pyrosequencing assay from foreskin of infants of the Kentucky birth cohort. Mitochondrial LDLR1 showed a trend of increased methylation in FT pregnancy smokers ( n = 21) vs. NS ( n = 41) ( P = 0.08, Mann–Whitney) (A). Mitochondrial HDLR1 showed significantly increased methylation in FT pregnancy smokers ( n = 20) vs. NS ( n = 38) ( P = 0.04, Mann–Whitney) (B) (four individual samples failed pyrosequencing assay for HDLR1). Horizontal lines represent the median. Global methylation measured via LINE-1 (C) showed no differences between the groups ( P = 0.58, t -test), (FT n = 13, NS n = 21). Horizontal lines represent the mean (LINE-1)
Additionally, in foreskin, a second segment of the D-Loop region—HDLR1—showed a small but significant change in methylation with MSDP (+ 0.4%; P = 0.04) ( Fig. 3B ). No methylation changes were noted in foreskin at the genomic global methylation mark: LINE-1 repetitive element ( P = 0.58) ( Fig. 3C ).
In sum, the range of median methylation levels in LDLR1 in placenta and foreskin was (5.72–8.45%) and (3.75–4.97%), respectively. The range of median methylation levels in HDLR1 in placenta and foreskin was (4.1–4.5%) and (4.5–5.0%), respectively.
Discussion
Environmental exposures are increasingly recognized as having a crucial influence on fetal development and physiology, as well as long-term health. During pregnancy, the placenta plays a key role in regulating infant growth and metabolic homeostasis. Energy production in the form of ATP through oxidative phosphorylation in placental mitochondria impacts many aspects of fetal development and growth, particularly by altering function of the placenta. Mitochondrial epigenetics is an emerging field that examines how mtDNA methylation may affect not only development but also health and disease later in life. A number of novel studies have examined how airborne particulate matter including cigarette smoke, influences genomic, and/or mtDNA methylation [ 3 , 13 , 25–27 ]. Additionally, it should also be considered that, as mitochondria are posited as of solely maternal inheritance, the study of mtDNA methylation inheritance and its effects on the risk of the disease could be of great benefit [ 28 ]. The goal of our study was to broaden the above findings by focusing on in utero effects of maternal smoking during pregnancy. This is the first study to observe similar mtDNA methylation changes in two different tissues (i.e. placenta and foreskin), from two different birth cohorts, associated with maternal smoking during pregnancy.
Our initial strategy was to develop a mitochondrial specific methylation assay that was contained within the D-Loop control region of the mitochondrial chromosome, as this region contains the transcription start sites for both the light and heavy chains and the heavy chain DNA replication start site, and we observed a positive association between MSDP and mitochondrial D-Loop methylation. Several previous studies have demonstrated methylation at the mitochondrial D-Loop from a variety of cell types and tissues [ 13 , 29 ] including the work of Bianchessi et al. [ 30 ] in endothelial cells at region LSR4 that overlaps with our assay, but none have demonstrated the methylation differences we see comparing smoking to non-smoking mothers. Janssen et al. [ 13 ] investigated methylation changes in placenta in a Belgian birth cohort associated with airborne particulates and found that placental mtDNA methylation is positively associated with particulate matter exposure during gestation. Other work that addresses mitochondrial D-Loop methylation includes Feng et al. [ 31 ] who investigated the correlation between ND2 expression and D-Loop methylation in colorectal cancer and Pirola et al. [ 32 ] examined D-Loop methylation in the setting of nonalcoholic fatty liver disease; however, these latter two studies are difficult to directly compare to our results as their approach uses methylation specific PCR and either ratios of methylated/unmethylated in study participants or percent of total samples methylated across the study population, and not the extent of methylation across the regions examined.
Consistent with a prior study that found that placental genomic DNA methylation changes with smoking at specific CpG dinucleotides [ 27 , 33 ], we found methylation increases at specific D-Loop mtDNA CpGs (LDLR1) in placenta and at two D-Loop segments—LDLR1 and HDLR1—in a second tissue, foreskin. However, no significant changes were seen in genomic methylation of LINE-1 sequence. Several previous studies have reported global methylation changes in infant DNA associated with maternal smoking [ 2 , 25 , 33–35 ]; however, neither placenta nor foreskin were examined in these studies. This global methylation discrepancy may have to do with fact that each tissue has a unique epigenetic signature that likely reflects differential functions [ 20 ]. Additionally, methylation of the D-Loop region itself may be a cell/tissue type-specific or even gender-specific phenomenon. Further studies are needed to address and clarify this issue.
Mitochondria certainly contain the machinery required to epigenetically modify mtDNA [ 36 ]. Consequently, it is plausible to hypothesize that epigenetic alterations at specific targeted locations on the mitochondrial chromosome may impact transcription and/or replication of the mitochondrial DNA, subsequent mitochondrial gene expression and perhaps even regulation of oxidative phosphorylation. Such changes could affect the proper functioning of placental trophoblasts, and thus could have various effects on placental activity and fetal development.
Previous studies have found both positive [ 37 ] or negative [ 13 ] association between mtDNA count number (mtDNAcn) and environmental exposures. And both higher [ 38 ] and lower [ 39 ] placental mtDNAcn have been reported associated with cigarette smoking during pregnancy. We did not see differences in placental mtDNAcn associated with maternal smoking in our study (online Supplementary material, Fig. S2 ). It is possible that our small sample size does not afford us enough power to discern a clear trend.
To determine if the D-Loop methylation differences associated with MSDP in placenta were specific to this tissue, we examined newborn foreskin from an independent birth cohort. Foreskin, a tissue that is rich in various cell types including fibroblasts, epidermal keratinocytes, T cells, and Langerhans cells, has been used in vitro studies for >50 years [ 40 ]. More recently, however, genome-wide DNA methylation profiling has been reported from foreskin [ 41 ] and comparative analysis of human mitochondrial methylomes [ 42 ]. Nonetheless, our study is the first that we are aware of to examine infant mitochondrial methylation in relation to smoking during pregnancy. The similarity between D-Loop % methylation increase comparing smoking vs. non-smoking in the RICHS birth cohort vs. Kentucky birth cohort is remarkable at 2.73% vs. 1.22%, respectively. We do note, however, that there may be one influential point in the Kentucky placental LDLR1 assay and we have tested the relationship between LDLR1 mean methylation and smoking status within the Kentucky cohort removing the single data point resulting in a difference of +0.73% and P = 0.14. Considering that cigarette smoke toxicity depends not only on the intrinsic characteristics of the toxic components in the environmental stress but also on the individual susceptibility of the subjects exposed to the cigarette smoke [ 36 ], the fact that such similarities could be observed in these distinct populations and in two different infant tissue types, is notable. Additionally, the observation of significant mtDNA methylation in a second segment of the D-Loop (HDLR1) in one tissue but not in the other may again reflect the type of epigenetic tissue or cell-specificity as seen in other studies [ 20 , 43 , 44 ] or even a gender-specific phenomenon. Further studies are needed to address and clarify this issue.
Historically, the observed methylation of the mtDNA has been very low (1–6%) [ 37 , 45 ]. Our observations are mostly consistent with previous results, with the exception of our smokers placental group higher at 8.45%. With the placenta being a physical and functional connection between mother and developing infant, fulfilling not only metabolic and endocrine functions but also serving a protective role, namely, helping to reduce the transfer of potentially toxic substances to the developing embryo/fetus [ 46 ], it might not be surprising with environmental exposures to observe higher levels of mtDNA methylation in the placenta itself as opposed to lower mtDNA methylation in an alternate infant tissue, as we observe here.
Furthermore, the exact temporal influence of environmental exposures remains to be investigated; however, Kobayashi et al. [ 47 ] have reported that mitochondrial methylation differs by developmental stage in mice where mtDNA is unmethylated in blastocysts and embryonic stem cells but only partially methylated in germ cells, suggesting a possible role for mtDNA methylation in early developmental processes and that earlier exposures may lead to more consistent effects across all tissues.
This study has a number of limitations. Like other work in this area, we cannot address the specific mechanisms linking this environmental exposure to mtDNA methylation. Also, we acknowledge that we are limited in our analysis of bio-specimens as we had available only one infant tissue-type per birth cohort to measure mtDNA methylation. A follow-up study might consider multiple bio-specimens such as placenta, umbilical cord blood, and foreskin from each study participant. Approximately 25% of the pregnancy non-smoking mothers in the RICHS cohort self-report exposure to secondhand smoke in the home or workplace, although no quantitative assessment of exposure was available. Future work should consider a larger study including more quantitative assessments of second-hand smoke exposure. Additionally, maternal cotinine was not measured in these birth cohorts. Further studies should consider that serum cotinine levels could provide a more accurate measure of maternal smoking habits versus a self-reported questionnaire. Moreover, we have analyzed DNA methylation via pyrosequencing at CpG sites only. It is beyond the scope of this study to examine non-CpG-associated cytosine methylation [ 29 ] or 5-hydroxymethylation in the mitochondrial chromosome [ 48 ]. As we have reported effects on only two mitochondrial DNA regions, we are not able to address potential epigenetic alterations at other locations within the mitochondrial chromosome. We also note that the analyses performed, due to the limited sample size, are univariate, and that some of the associations may be affected by uncontrolled confounding. Larger studies where appropriate multivariable modeling can be implemented would be necessary. Finally, to this point, a number of studies have identified gender-specific epigenetic alterations associated with environmental exposures. Sen et al. [ 49 ] found both male-specific and female-specific changes in DNA methylation profiles from dried blood spots in young children related to lead exposure. Also, Hall et al. [ 50 ] have examined sex differences in genome-wide DNA methylation patterns as it relates to insulin secretion and metabolism. Moreover, related to cigarette smoking, Campesi et al. [ 51 ] observed gender-specific changes in global DNA methylation and a wide range of plasma inflammation biomarkers. We cannot rule out that the absolute differences we observe between the RICHS vs. Kentucky birth cohorts may be related to comparing a mixed gender birth cohort to a male-only cohort.
In summary, this is the first study to report comparable mitochondrial D-Loop methylation changes associated with maternal smoking in two independent birth cohorts and alternate tissue types. The exact mechanisms responsible for changes in mitochondrial methylation and ultimately the effects on fetal development and growth remain to be determined but areas of potential interest for investigation may include intrauterine hypoxia or specific chemical constituents of tobacco smoke. In addition, it will be important to examine if these differences in mtDNA methylation can be linked to long-term outcome in these infants, and so follow-up in longitudinal studies is warranted.
Funding
This work was supported by the U.S. National Institutes of Health (NIH), National Institute of Mental Health (NIMH) R01MH094609, National Institutes for Environmental Health Sciences (NIEHS) R01ES022223, and P01 ES022832 and by the US Environmental Protectional Agency (EPA) grant RD83544201. Its contents are solely the responsibility of the grantee and do not necessarily represent the official views of the US EPA. Furthermore, the US EPA does not endorse the purchase of any commercial products or services mentioned in the presentation.
Supplementary data
Supplementary data is available at EnvEpig online.
Conflict of interest statement . None declared.
Authorship
CJM and KJP conceived the study. CJM, KJP, and DAA designed the study. DAA, DJG, BAB, NRC, and JFL performed the experiments and acquired the data. BBG, NRC, and KJP analyzed the data. DAA wrote the article. All authors edited and approved the article.
Supplementary Material
Supplementary Data
References
- 1. Appleton AA, Armstrong DA, Lesseur C, Lee J, Padbury JF, Lester BM, Marsit CJ. Patterning in placental 11-B hydroxysteroid dehydrogenase methylation according to prenatal socioeconomic adversity . PLoS One 2013. ; 8 : e74691 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Breton CV, Byun HM, Wenten M, Pan F, Yang A, Gilliland FD. Prenatal tobacco smoke exposure affects global and gene-specific DNA methylation. Am J Respir Crit Care Med 2009. ; 180 : 462 – 467 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Breton CV, Salam MT, Wang X, Byun HM, Siegmund KD, Gilliland FD. Particulate matter, DNA methylation in nitric oxide synthase, and childhood respiratory disease. Environ Health Perspect 2012. ; 120 : 1320 – 1326 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Conradt E, Lester BM, Appleton AA, Armstrong DA, Marsit CJ. The roles of DNA methylation of NR3C1 and 11beta-HSD2 and exposure to maternal mood disorder in utero on newborn neurobehavior. Epigenetics 2013. ; 8 : 1321 – 1329 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Parle-McDermott A, Ozaki M. The impact of nutrition on differential methylated regions of the genome. Adv Nutr 2011. ; 2 : 463 – 471 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Marsit CJ. Influence of environmental exposure on human epigenetic regulation. J Exp Biol 2015. ; 218(Pt 1) : 71 – 79 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Abernathy JR, Greenberg BG, Wells HB, Frazier TM. Smoking as an independent variable in a multiple regression analysis upon birth weight and gestation. Am J Public Health Nations Health 1966. ; 56 : 626 – 633 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lowe CR. Effect of mothers' smoking habits on birth weight of their children. Br Med J 1959. ; 2 : 673 – 676 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Maccani JZ, Maccani MA. Altered placental DNA methylation patterns associated with maternal smoking: current perspectives. Adv Genomics Genet 2015. ; 2015 : 205 – 214 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Knopik VS, Maccani MA, Francazio S, McGeary JE. The epigenetics of maternal cigarette smoking during pregnancy and effects on child development. Dev Psychopathol 2012. ; 24 : 1377 – 1390 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Paquette AG, Marsit CJ. The developmental basis of epigenetic regulation of HTR2A and psychiatric outcomes. J Cell Biochem 2014. ; 115 : 2065 – 2072 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ghosh S, Singh KK, Sengupta S, Scaria V. Mitoepigenetics: The different shades of grey. Mitochondrion 2015. ; 25 : 60 – 66 . [DOI] [PubMed] [Google Scholar]
- 13. Janssen BG, Byun HM, Gyselaers W, Lefebvre W, Baccarelli AA, Nawrot TS. Placental mitochondrial methylation and exposure to airborne particulate matter in the early life environment: An ENVIRONAGE birth cohort study. Epigenetics 2015. ; 10 : 536 – 544 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Byun HM, Motta V, Panni T, Bertazzi PA, Apostoli P, Hou L, Baccarelli AA. Evolutionary age of repetitive element subfamilies and sensitivity of DNA methylation to airborne pollutants. Part Fibre Toxicol 2013. ; 10 : 28.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Babykin MM, Zinchenko VV. Rapid separation of DNAs by buoyant density in three-layer CsCl gradients. Anal Biochem 1984. ; 137 : 175 – 181 . [DOI] [PubMed] [Google Scholar]
- 16. Clayton DA. Transcription and replication of mitochondrial DNA. Hum Reprod 2000. ; 15 Suppl 2 : 11 – 17 . [DOI] [PubMed] [Google Scholar]
- 17. Mercer TR, Neph S, Dinger ME, Crawford J, Smith MA, Shearwood AM, Haugen E, Bracken CP, Rackham O, Stamatoyannopoulos JA , et al. . The human mitochondrial transcriptome. Cell 2011. ; 146 : 645 – 658 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Fenton TR. A new growth chart for preterm babies: Babson and Benda’s chart updated with recent data and a new format. BMC Pediatrics 2003. ; 3 : 13.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Yang AS, Estécio MR, Doshi K, Kondo Y, Tajara EH, Issa JP. A simple method for estimating global DNA methylation using bisulfite PCR of repetitive DNA elements. Nucleic Acids Res 2004. ; 32 : e38.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Armstrong DA, Lesseur C, Conradt E, Lester BM, Marsit CJ. Global and gene-specific DNA methylation across multiple tissues in early infancy: implications for children's health research. Faseb J 2014. ; 28 : 2088 – 2097 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Bassil CF, Huang Z, Murphy SK. Bisulfite pyrosequencing. Methods Mol Biol 2013. ; 1049 : 95 – 107 . [DOI] [PubMed] [Google Scholar]
- 22. Hazkani-Covo E, Zeller RM, Martin W. Molecular poltergeists: mitochondrial DNA copies (numts) in sequenced nuclear genomes. PLoS Genet 2010. ; 6 : e1000834.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hou L, Zhu ZZ, Zhang X, Nordio F, Bonzini M, Schwartz J, Hoxha M, Dioni L, Marinelli B, Pegoraro V , et al. . Airborne particulate matter and mitochondrial damage: a cross-sectional study. Environ Health 2010. ; 9 : 48.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lee HC, Yin PH, Lu CY, Chi CW, Wei YH. Increase of mitochondria and mitochondrial DNA in response to oxidative stress in human cells. Biochem J 2000. ; 348 Pt 2 : 425 – 432 . [PMC free article] [PubMed] [Google Scholar]
- 25. Breton CV, Siegmund KD, Joubert BR, Wang X, Qui W, Carey V, Nystad W, Håberg SE, Ober C, Nicolae D , et al. . Prenatal tobacco smoke exposure is associated with childhood DNA CpG methylation. PLoS One 2014. ; 9 : e99716.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ladd-Acosta C, Shu C, Lee BK, Gidaya N, Singer A, Schieve LA, Schendel DE, Jones N, Daniels JL, Windham GC , et al. . Presence of an epigenetic signature of prenatal cigarette smoke exposure in childhood. Environ Res 2016. ; 144(Pt A) : 139 – 148 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Suter M, Ma J, Harris A, Patterson L, Brown KA, Shope C, Showalter L, Abramovici A, Aagaard-Tillery KM. Maternal tobacco use modestly alters correlated epigenome-wide placental DNA methylation and gene expression. Epigenetics 2011. ; 6 : 1284 – 1294 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lambertini L, Byun HM. Mitochondrial epigenetics and environmental exposure. Curr Environ Health Rep 2016. ; 3 : 214 – 24 . [DOI] [PubMed] [Google Scholar]
- 29. Bellizzi D, D'Aquila P, Scafone T, Giordano M, Riso V, Riccio A, Passarino G.. The control region of mitochondrial DNA shows an unusual CpG and non-CpG methylation pattern. DNA Res 2013. ; 20 : 537 – 547 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Bianchessi V, Vinci MC, Nigro P, Rizzi V, Farina F, Capogrossi MC, Pompilio G, Gualdi V, Lauri A. Methylation profiling by bisulfite sequencing analysis of the mtDNA Non-Coding Region in replicative and senescent Endothelial Cells. Mitochondrion 2016. ; 27 : 40 – 47 . [DOI] [PubMed] [Google Scholar]
- 31. Feng S, Xiong L, Ji Z, Cheng W, Yang H. Correlation between increased ND2 expression and demethylated displacement loop of mtDNA in colorectal cancer. Mol Med Rep 2012. ; 6 : 125 – 130 . [DOI] [PubMed] [Google Scholar]
- 32. Pirola CJ, Gianotti TF, Burgueño AL, Rey-Funes M, Loidl CF, Mallardi P, Martino JS, Castaño GO, Sookoian S. Epigenetic modification of liver mitochondrial DNA is associated with histological severity of nonalcoholic fatty liver disease. Gut 2013. ; 62 : 1356 – 1363 . [DOI] [PubMed] [Google Scholar]
- 33. Joubert BR, Felix JF, Yousefi P, Bakulski KM, Just AC, Breton C, Reese SE, Markunas CA, Richmond RC, Xu CJ , et al. . DNA methylation in newborns and maternal smoking in pregnancy: genome-wide consortium meta-analysis. Am J Hum Genet 2016. ; 98 : 680 – 696 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Joubert BR, Håberg SE, Nilsen RM, Wang X, Vollset SE, Murphy SK, Huang Z, Hoyo C, Midttun Ø, Cupul-Uicab LA , et al. . 450K epigenome-wide scan identifies differential DNA methylation in newborns related to maternal smoking during pregnancy. Environ Health Perspect 2012. ; 120 : 1425 – 1431 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Richmond RC, Simpkin AJ, Woodward G, Gaunt TR, Lyttleton O, McArdle WL, Ring SM, Smith AD, Timpson NJ, Tilling K , et al. . Prenatal exposure to maternal smoking and offspring DNA methylation across the lifecourse: findings from the Avon Longitudinal Study of Parents and Children (ALSPAC). Hum Mol Genet 2015. ; 24 : 2201 – 2217 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Pirini F, Guida E, Lawson F, Mancinelli A, Guerrero-Preston R. Nuclear and mitochondrial DNA alterations in newborns with prenatal exposure to cigarette smoke. Int J Environ Res Public Health 2015. ; 12 : 1135 – 1155 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Byun HM, Panni T, Motta V, Hou L, Nordio F, Apostoli P, Bertazzi PA, Baccarelli AA. Effects of airborne pollutants on mitochondrial DNA methylation. Part Fibre Toxicol 2013. ; 10 : 18.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Garrabou G, Hernàndez AS, Catalán García M, Morén C, Tobías E, Córdoba S, López M, Figueras F, Grau JM, Cardellach F. Molecular basis of reduced birth weight in smoking pregnant women: mitochondrial dysfunction and apoptosis. Addict Biol 2016. ; 21 : 159 – 170 . [DOI] [PubMed] [Google Scholar]
- 39. Bouhours-Nouet N, May-Panloup P, Coutant R, de Casson FB, Descamps P, Douay O, Reynier P, Ritz P, Malthièry Y, Simard G. Maternal smoking is associated with mitochondrial DNA depletion and respiratory chain complex III deficiency in placenta. Am J Physiol Endocrinol Metab 2005. ; 288 : E171 – E177 . [DOI] [PubMed] [Google Scholar]
- 40. Frost P, Weinstein GD, Hsia SL. Metabolism of estradiol-17beta and estrone in human skin. J Invest Dermatol 1966. ; 46 : 584 – 585 . [DOI] [PubMed] [Google Scholar]
- 41. Choudhry S, Deshpande A, Qiao L, Beckman K, Sen S, Baskin LS. Genome-wide DNA methylation profiling of CpG islands in hypospadias. J Urol 2012. ; 188(4 Suppl) : 1450 – 1455 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Ghosh S, Sengupta S, Scaria V. Comparative analysis of human mitochondrial methylomes shows distinct patterns of epigenetic regulation in mitochondria. Mitochondrion 2014. ; 18 : 58 – 62 . [DOI] [PubMed] [Google Scholar]
- 43. Wan J, Oliver VF, Wang G, Zhu H, Zack DJ, Merbs SL, Qian J. Characterization of tissue-specific differential DNA methylation suggests distinct modes of positive and negative gene expression regulation. BMC Genomics 2015. ; 16 : 49.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Huang YT, Chu S, Loucks EB, Lin CL, Eaton CB, Buka SL, Kelsey KT. Epigenome-wide profiling of DNA methylation in paired samples of adipose tissue and blood. Epigenetics 2016. ; 11 : 227 – 236 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Maresca A, Zaffagnini M, Caporali L, Carelli V, Zanna C. DNA methyltransferase 1 mutations and mitochondrial pathology: is mtDNA methylated? Front Genet 2015. ; 6 : 90.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Gude NM, Roberts CT, Kalionis B, King RG. Growth and function of the normal human placenta. Thromb Res 2004. ; 114 : 397 – 407 . [DOI] [PubMed] [Google Scholar]
- 47. Kobayashi H, Sakurai T, Sato S, Nakabayashi K, Hata K, Kono T. Imprinted DNA methylation reprogramming during early mouse embryogenesis at the Gpr1-Zdbf2 locus is linked to long cis-intergenic transcription. FEBS Lett 2012. ; 586 : 827 – 833 . [DOI] [PubMed] [Google Scholar]
- 48. Ghosh S, Sengupta S, Scaria V. Hydroxymethyl cytosine marks in the human mitochondrial genome are dynamic in nature. Mitochondrion 2016. ; 27 : 25 – 31 . [DOI] [PubMed] [Google Scholar]
- 49. Sen A, Heredia N, Senut MC, Hess M, Land S, Qu W, Hollacher K, Dereski MO, Ruden DM. Early life lead exposure causes gender-specific changes in the DNA methylation profile of DNA extracted from dried blood spots. Epigenomics 2015. ; 7 : 379 – 393 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Hall E, Volkov P, Dayeh T, Esguerra JL, Salö S, Eliasson L, Rönn T, Bacos K, Ling C. Sex differences in the genome-wide DNA methylation pattern and impact on gene expression, microRNA levels and insulin secretion in human pancreatic islets. Genome Biol 2014. ; 15 : 522.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Campesi I, Carru C, Zinellu A, Occhioni S, Sanna M, Palermo M, Tonolo G, Mercuro G, Franconi F. Regular cigarette smoking influences the transsulfuration pathway, endothelial function, and inflammation biomarkers in a sex-gender specific manner in healthy young humans. Am J Transl Res 2013. ; 5 : 497 – 509 . [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Data