Abstract
The translocator protein (TSPO) is a mitochondrial membrane protein, of as yet uncertain function. Its purported high expression on activated macrophages, has lent utility to TSPO targeted molecular imaging in the form of positron emission tomography (PET), as a means to detect and quantify inflammation in vivo. However, existing literature regarding TSPO expression on human activated macrophages is lacking, mostly deriving from brain tissue studies, including studies of brain malignancy, and inflammatory diseases such as multiple sclerosis. Here, we utilized three human sources of monocyte derived macrophages (MDM), from THP-1 monocytes, healthy peripheral blood monocytes and synovial fluid monocytes from patients with rheumatoid arthritis, to undertake a detailed investigation of TSPO expression in activated macrophages. In this work, we demonstrate a consistent down-regulation of TSPO mRNA and protein in macrophages activated to a pro-inflammatory, or ‘M1’ phenotype. Conversely, stimulation of macrophages to an M2 phenotype with IL-4, dexamethasone or TGF-β1 did not alter TSPO expression, regardless of MDM source. The reasons for this are uncertain, but our study findings add some supporting evidence for recent investigations concluding that TSPO may be involved in negative regulation of inflammatory responses in macrophages.
Introduction
Translocator protein (TSPO) is an 18kDa outer mitochondrial membrane protein, as yet of uncertain function, although roles in steroidogenesis, apoptosis and cholesterol transport have been proposed [1]. It is increasingly being utilized as a molecular target for the in vivo imaging of inflammation using Positron Emission Tomography (PET), based upon its known high expression on macrophages in a variety of inflammatory diseases, including multiple sclerosis [1–3], and more recently, rheumatoid arthritis (RA) [4–5].
Macrophages are a key cellular component of the inflammatory response, having roles in phagocytosis, antigen presentation, as well as potentiation of inflammation through the production of numerous pro-inflammatory cytokines, such as tumour necrosis factor-⍺ (TNF-⍺) and interleukin-23 (IL-23) [6].
Macrophages exhibit plasticity in response to their milieu [7]. Indeed, the paradigm of two opposing ends of a spectrum of macrophage phenotypes has long been established. In this model, pro-inflammatory (or ‘classically activated’) macrophages are designated the term ‘M1’, having been activated by ‘M1’ stimuli, such as lipopolysaccharide (LPS) and interferon-ɣ (IFN- ɣ) [8]. Such macrophages produce TNF-α, and reactive oxygen species, and are associated with Th-1 cell responses, potentiating inflammation [9]. In contrast, monocytes in the presence of macrophage-colony stimulating factor (M-CSF), interleukin-4 (IL-4), glucocorticoids (such as dexamethasone) or tumour growth factor-β (TGF-β), are thought to activate to an anti-inflammatory (or ‘alternatively-activated’) M2 macrophage phenotype [8,10]. Such macrophages produce chemokines such as CCL17, and are associated with Th-2 cell responses [10].
As our knowledge of macrophage phenotype develops, assisted by transcriptional studies, there is increasing realization that ‘M1’ and ‘M2’ likely only represent two ends of a spectrum of multiple, complex macrophage phenotypes that exist in vivo, dependent on the combination of stimuli macrophages encounter [10,11].
There is scant literature investigating the expression of TSPO on macrophages of different phenotype. Such an investigation would not only enable more accurate interpretation of TSPO targeted in vivo imaging, but also potentially provide insight into any role of TSPO in macrophage biology.
The majority of investigations into TSPO expression on macrophages of different phenotype have been carried out on murine models in vitro [12,13]. Bonsack et al. undertook a histological study of microglia in a murine model of intracerebral haemorrhage. This failed to demonstrate any significant difference in TSPO expression on microglia expressing the M1 marker (CD16/32), and those expressing the M2 marker (CD206) [12]. However, immunohistochemistry may not be a truly accurate means of TSPO quantification on macrophages, since TSPO is ubiquitous [1], and further, using a single macrophage marker (e.g. CD16/32 vs CD206) may not be a precise means of classifying M1 or M2 phenotype. An additional in vitro study of the murine macrophage cell line, RAW 264.7 [13], demonstrated a significant increase in TSPO protein levels after 24 hours stimulation with LPS, in a dose-dependent manner as determined by an increase in the binding of the TSPO specific radio-ligand [11C]PBR28 [13].
However, caution is needed in translating findings of such studies to human macrophages, given the debate surrounding potential fundamental differences between human and murine macrophages [14–16]. For example, much evidence suggests the enzyme arginase 1 is well known to be expressed in murine but not human macrophages activated by the same stimuli [17–19], and there is further controversy surrounding differences in production of inducible nitric oxide synthase (iNOS) and nitric oxide (NO) on activation of human and murine macrophages by the same stimuli [14–16, 20].
Here, we undertake an investigation of TSPO expression in human monocyte derived macrophages (MDM), activated to opposing ends of the currently accepted spectrum of macrophage phenotypes [21]. We demonstrate a significant difference in TSPO expression between M1 phenotype macrophages activated by the M1 ‘pro-inflammatory’ stimuli, LPS and IFN-γ, and macrophages activated by M2 ‘reparative-state’ stimuli (IL-4, dexamethasone and TGF-β1). Human macrophage/monocytes used for the study were derived from: (i) the human monocytic cell line THP-1, originating from a paediatric patient with acute myelomonocytic leukaemia [22]; (ii) peripheral blood monocytes from healthy volunteers and (iii) RA synovial fluid derived monocytes.
Materials and methods
Cell culture
All cells were cultured and maintained in RPMI 1640 medium (Lonza, Biowhittaker®, Belgium) supplemented with 10% endotoxin free heat-inactivated foetal bovine serum (FBS), 1% penicillin/streptomycin (Lonza, Biowhittaker®, Belgium). Cells were maintained in a humidified incubator at 37°C and 5% CO2 (New Brunswick Scientific Innova™ CO-170) unless otherwise stated.
THP-1 monocytes
Human monocytic THP-1 cells were purchased from the American Type Culture Collection (ATCC, Maryland, USA) and were cultivated in 75cm2 flasks, in culture media (stated above). THP-1 cells were passaged every 4 days at a ratio of between 1:2 and 1:3, to ensure cell density was not more than 0.8–1.2x106 cells/mL. Cells were used from passage numbers 4–6.
Peripheral blood monocytes from healthy volunteers
Peripheral blood monocytes were isolated from blood cones of five healthy donors purchased from the National Blood Service (Colindale, Edgware, London, UK). Here, the peripheral blood mononuclear cell fraction was extracted using density gradient centrifugation with Histopaque (Lonza, Whittaker), and continuous counter-flow elutriation to isolate the monocyte fraction. Counter-flow elutriation is a well-established process for isolating cells of different size and has been established as a technique for isolating monocytes for decades [23–26]. It combines centrifugation (the process of sedimentation under the influence of a centrifugal force), and elutriation (the process of separation by washing), to generate fractions of cells of desired size from peripheral blood [27].
Cells isolated from the blood cone underwent centrifugal elutriation in a Beckman JE6 Elutriator (Beckman Coulter, High Wycombe, UK) to obtain monocyte-rich fractions. The centrifuge was run at room temperature and a constant speed of 2200rpm, as described in Banfalvi et al. [27]. In brief, cells were loaded into the elutriation chamber at an increasing flow rate, and subjected to an opposing centrifugal force compared to the flow velocity. Since the centrifugal force is constant, separation of cells according to size and granularity occurs by altering the flow of the media into the centrifuge, done via a pump. Increasing the low rate in gradual steps yields successive fractions of increasingly large or dense cells being collected. Monocytes were ejected when the flow rate was increased to 18-22mLs/minute. The percentage of monocytes in the fraction being ejected was determined by concomitant analysis of cell size and granularity using flow cytometry.
Only fractions with monocyte content over 85% (as determined by flow cytometry) were utilised in this work. Cells were counted using a Neubauer chamber haemocytometer and adjusted to a concentration of 1x106 cells/mL.
Rheumatoid Arthritis-derived synovial fluid monocytes
Aspirated synovial fluid samples from the knee joints of five patients; aged 38–65, 2 males, 3 females- with established RA (as per American College of Rheumatology updated diagnostic criteria [28]) were added into 10mL lithium heparin tubes. Full ethics approval was granted for collection of synovial fluid from NRES Committee London (reference: 07/H0706/81). Synovial fluid was diluted 1:1 with sterile phosphate buffered saline (PBS), layered over Histopaque®, and centrifuged. The intermediate cell layer was removed, labelled with CD14+ magnetic beads (Miltenyi Biotech, Germany) and sorted using Magnetic Activated Cell Sorting (MACS) as per manufacturer’s instructions (Miltenyi Biotech). The resultant monocytes were washed and resuspended in 1mL of MACS buffer. The cells were counted and adjusted to 1X106 cells/mL.
Differentiation of monocytes to macrophages
THP-1 MDM
To differentiate THP-1 MDM, THP-1 cells were plated at a density of 0.5x106/cm2 (CELLSTAR®, Greiner Bio-One, UK), and treated with 10ng/mL of phorbol 12-myristate 13-acetate (PMA, Sigma-Aldrich, Poole, UK), for 72 hours to generate a macrophage phenotype as previously described [22].
Healthy peripheral blood and RA synovial fluid MDM
Monocytes from healthy peripheral blood or synovial fluid mononuclear fractions were counted and resuspended to a density of 1x106 cells/mL in RPMI 1640 (Lonza, Whittaker). Macrophages were differentiated from elutriated healthy monocytes and synovial fluid-derived monocytes, by culturing cells with 100ng/mL of recombinant human M-CSF (Peprotech Inc., UK) at a concentration of 1x106 cells/mL, for 7 days [29]. Thereafter, fluorescence activated cell sorting (FACS) was used to confirm monocytes had been differentiated to monocyte-derived macrophages (MDM), i.e. CD68-positive cells [30] (see S1 Fig).
On day 7, cells were treated with 10ng/mL LPS (Peprotech Inc. London) and 20ng/mL recombinant human IFN-γ (Peprotech Inc., London), for 2, 4, 6 or 24 hours, to generate M1 phenotype macrophages. To generate M2 phenotype macrophages, cells were treated with either IL-4 20ng/mL, 500nM dexamethasone, or 3ng/mL TGF-β1 for 24 hours as described in previous studies [29,31–33].
Immunofluorescence staining
After the specified incubation period, MDM were lifted using EDTA (Sigma-Aldrich), followed by gentle scraping. Cells were washed in PBS, counted and incubated at 1 million cells/mL in 1.6% paraformaldehyde solution (Thermo-Fischer) at 4°C for 20 minutes. Using a cyto-centrifuge (Cytospin, Shandon Southern Products, Runcorn, UK), cells were spun onto slides, fixed for 15 minutes in ice cold acetone, and stored at -20°C until required. For multiple antibody immunofluorescence staining and image acquisition, protocols were adapted from Dakin et al. [34], using the primary antibodies rabbit anti-human TSPO at 1:1000 (LSbio), and mouse anti-human CD68 at 1:600 (Dako). Isotype control antibodies were a cocktail of mouse immunoglobulin G (IgG1), and rabbit immunoglobulin fraction of serum from non-immunized rabbits, solid-phase absorbed (Dako). Immunofluorescence images were acquired on a Zeiss LSM 710 confocal microscope as described previously [34].
Real-time quantitative Polymerase Chain Reaction (qPCR)
RNA was isolated using the E.Z.N.A. EaZy RNA extraction kit, and complementary DNA (cDNA) was synthesized using 500 ng of RNA, random primers (Invitrogen), and Moloney murine leukemia virus (Promega). Quantitative PCR was performed using SYBR Green I JumpStart (Sigma-Aldrich). List and details of the primers used is provided in Table 1.
Table 1. Primers used for real-time PCR.
| GENE | SENSE PRIMER | ANTI-SENSE PRIMER |
|---|---|---|
| 18S ribosomal RNA (18S) | GTAACCCGTTGAACCCCA | CCATCCAATCGGTAGTAGCG |
| Translocator protein(TSPO) | GCGGCCTGGCTAACTCCTGC | AAAGCGGGAGCCCACGAAGC |
| Sterol-27-hydroxylase (CYP27A1) | ACTGCACCAGTTACAGGTGCTTTACA | CCATGTCGTTCCGTACTGGGTACT |
| ATP binding cassette transporter (ABCA1) | TTCCCGCATTATCTGGAAAGC | CAAGGTCCATTTCTTGGCTGT |
|
Tumour necrosis factor-alpha (TNF-α) |
GGCCAAGCCCTGGTATGAG | TAGTCGGGCCGATTGATCTC |
| Prostaglandin endoperoxide synthase 2 (PTGS2) | ATGCTGACTATGGCTACAAAAGC | TCGGGCAATCATCAGGCAC |
| Interleukin 12 beta (IL-12β) | AGAAGATGGTATCACCTGGACC | GAACCTCGCCTCCTTTGTGAC |
| Interleukin 10 (IL-10) | TACGGCGCTGTCATCGATT | GGCTTTGTAGATGCCTTTCTCTTG |
|
Transglutaminase 2 (TGM2) |
GCCACTTCATTTTGCTCTTCAA | TCCTCTTCCGAGTCCAGGTACA |
|
Mannose receptor C type-1 (MRC1) |
AAGGCGGTGACCTCACAAG | AAAGTCCAATTCCTCGATGGTG |
| CD200 Receptor 1 (CD200R1) | GACCAGAGAGGGTCTCACCA | TTGAAGCGGCCACTAAGAAG |
In order to provide evidence that macrophages had been activated to an appropriate phenotype by the stimuli, mRNA expression of multiple markers were assessed by qPCR. TNF-α, prostaglandin endoperoxide synthase 2 (PTGS2), and interleukin-12 β (IL-12β) mRNA are thought to be more highly expressed in M1 compared to M2 phenotype macrophages [9]. In contrast, interleukin-10 (IL-10), Transglutaminase 2 (TGM2), Mannose receptor C type-1 (MRC1), and CD200 Receptor 1 (CD200R1) mRNA have all been reported as M2 macrophage markers, demonstrated to be more highly expressed in M2 compared to M1 macrophages [35–37].
Enzyme-Linked Immunosorbent Assay (ELISA)
It is recognized that TNF-α production in macrophages may serve as an indicator of macrophage phenotype, being higher in M1 compared to M2 phenotype macrophages [35]. Therefore, TNF-α ELISA was undertaken, to provide additional evidence of macrophage phenotype, at protein level.
The concentrations of soluble human TNF-α in cell culture media were measured using commercially-available ELISA (Quantikine human TNF-α ELISA Kits, R&D Systems Europe) according to the manufacturer’s instructions. Each reaction was run in triplicate. The optical density measurements were carried out using ELISA plate reader (Multiskan Ascent Thermo Labsystems, Finland) at 450 nm wavelength. The lower and higher detection limits of the kits were 15.6pg/mL and 1000pg/mL, respectively.
Western blots
Total cellular protein extracts were prepared using denaturating lysis buffer [8M urea, 1% sodium dodecyl sulfate, 1% glycerol, 10 mM Tris (pH 6.8), 0.5 mM protease inhibitor cocktail (Sigma-Aldrich, containing AEBSF (4-(2-aminoethyl) benzenesulfonyl fluoride hydrochloride), aprotinin, bestatin, E-64, leupeptin and pepstatin A), 1mM dithiothreitol (Sigma-Aldrich)]. Protein extracts were separated on NuPAGE Tris-Acetate gels (4–12%; Invitrogen) and blotted onto polyvinylidene difluoride (PVDF) membranes. The membranes were incubated overnight with goat anti-human TSPO monoclonal antibody (GeneTex) at a titre of 1:10,000, or mouse anti-β-actin at 1:10,000 (Sigma-Aldrich). Following incubation with secondary polyclonal donkey anti-goat or rabbit anti-mouse antibodies (1:500 and 1:3000, respectively) coupled with horseradish peroxidase (Dako) the resultant bands were visualized using ECL Plus (GE Healthcare). Optical densitometry analysis was carried out utilizing ImageJ software (NIH), as previously described [38].
Radio-ligand saturation binding assay
[3H]PBR28 is a radioligand with high selectivity and affinity for TSPO [39]. PBR28 has been used successfully in multiple imaging studies as a means of detecting TSPO protein both in vitro and in vivo [13,40,41]. To ensure all donors were high affinity binders to [3H]PBR28, DNA extraction and genotyping was performed for all cell donors as previously described by Owen et al. [42].
Due to the large volume of protein required to undertake saturation binding (minimum 2.4mg cytoplasmic protein per condition for each donor), only healthy donor peripheral blood monocytes and MDM were used for radioligand binding work. Three conditions of MDM were generated; unstimulated, and stimulated with 10ng/mL LPS plus 20ng/mL IFN-γ or with 20ng/mL IL-4, for 24 hours. Cells were pelleted, and membrane preparation, along with homogenate and competition binding assays carried out as described previously [43].
Analysis of saturation binding data
All saturation and competition data were analysed using the iterative nonlinear regression curve-fitting software supplied with GraphPad Prism 7.0. Single-site and two-site models were compared using the least-squares algorithm. The null hypothesis, that the data fitted a single-site model, was rejected if the p-value was less than 0.05. Amount of tracer required (fmol/mg protein) to occupy all TSPO sites (Bmax) was calculated. Bmax value was multiplied by the total amount of protein in each cell pellet used, and then divided by number of cells in each pellet, to generate a value of fmol of [3H]PBR28 required to bind to all TSPO present in the cell pellet being assayed [44].
Statistical analysis
All experiments for mRNA and Western blots were carried out in triplicate. Results are expressed as mean ± standard error of the mean (SEM). Graph Pad Prism 7.0 software was used for all data analyses. To evaluate effects of stimuli (M-CSF, LPS, IFN-γ, IL-4, TGF-β1, dexamethasone), analysis of variance (ANOVA), followed by a Bonferroni’s Multiple Comparison Test was used if there were more than two groups to compare, or Student’s t-test applied if two groups were analysed. P values less than 0.05 were considered significant (*p<0.05, **p <0.01, ***p≤0.001).
Results
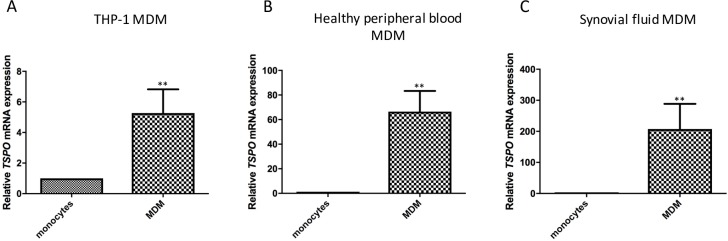
TSPO mRNA is up-regulated in macrophages compared to monocytes
TSPO is up-regulated on MDM compared to monocytes, even without further addition of stimuli to generate an activated macrophage phenotype (Fig 1). For THP-1 monocytes, addition of 10ng/mL PMA for 72 hours, resulted in an average 6-fold increase of TSPO expression (6.52 ± 1.28 fold change) (Fig 1A). For both healthy human peripheral blood MDM and synovial fluid MDM, monocytes were differentiated with 100ng/mL M-CSF for 7 days. Although all groups of MDM demonstrated a significant increase in TSPO expression compared to their monocyte counterparts (p<0.01), the magnitude of TSPO up-regulation was greater in MDM derived from the synovial fluid of arthritis patients (mean fold change of 284.6 ± 27.88 on synovial fluid MDM compared to synovial fluid monocytes, versus mean fold change 65.4 ± 16.88 for healthy peripheral blood MDM compared to healthy monocytes; Fig 1B and 1C).
Fig 1. TSPO mRNA expression of monocytes compared to MDM.
(A) THP-1 monocytes and THP-1 MDM; (B) healthy peripheral blood monocytes and corresponding MDM (C) synovial fluid monocytes and corresponding MDM. Results are expressed as fold change mRNA expression (compared to 18s RNA). Each experiment was run in triplicate, with data expressed as mean ± SEM of five independent experiments. Relative TSPO mRNA expression in MDM was compared to their respective monocyte counterpart whereby statistically significant differences are noted as **p<0.001 using Student’s t-test.
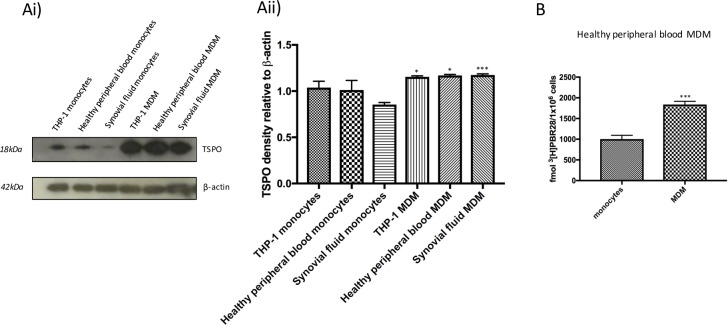
TSPO protein is up-regulated on macrophages compared to monocytes
Western blot data demonstrated increased TSPO protein expression in MDM from all sources of human macrophages compared to monocytes (Fig 2A), with a small (p = 0.05), but significant increase in TSPO densitometry on monocyte to macrophage differentiation (Fig 2A). In order to provide a quantitative assessment of TSPO protein expression in monocytes compared to macrophages, [3H]PBR28 radioligand binding studies were undertaken. This demonstrated a significant (p<0.001) up-regulation of TSPO at protein level in healthy human peripheral blood MDM compared to monocytes (mean 1838 ± 45.37 fmol [3H]PBR28 per 1x106 MDM, compared to mean 1004 ± 52.61 fmol [3H]PBR28 per 1x106 monocytes) (Fig 2B).
Fig 2. TSPO protein expression is higher in MDM than in monocytes.
TSPO protein was assessed by Western blot with β-actin as loading control (Ai), with accompanying densitometry normalised to β-actin (Aii). Results shown are for THP-1 monocytes, healthy peripheral blood monocytes, synovial fluid monocytes, and corresponding MDM. (B) [3H]PBR28 tracer binding per 1x106 cells in healthy human peripheral blood monocytes and MDM. Each experiment was run in triplicate, and data expressed as mean± SEM for five independent experiments. *** p<0.001 using Student’s t-test, comparing data from each MDM with that of their counterpart monocytes.
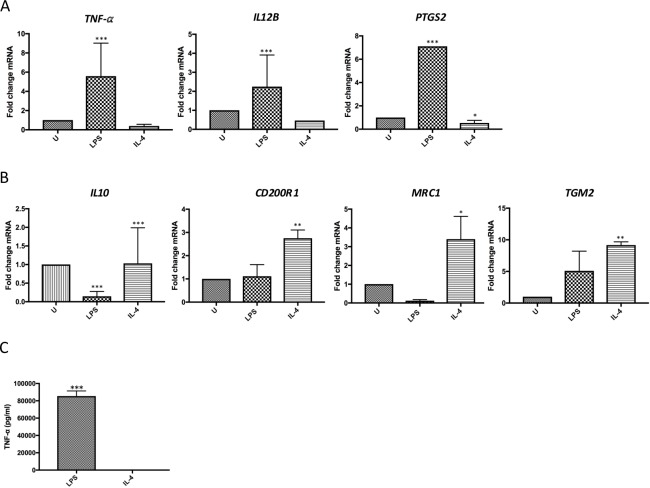
Changes in M1- & M2-specific mRNA markers in response to LPS plus IFN-γ or IL-4 stimulation
Transcriptional studies are increasingly used to accurately differentiate between macrophage phenotypes, with the expression of mRNA of TNF-α, IL-12β and PTGS2 being higher in M1 macrophages than M2 macrophages [9,34]. In contrast, mRNA of IL-10, CD200R1, MRC1 and TGM2 are well-recognised to be upregulated in M2 compared to M1 macrophages [9,35–37].
Analysis of the M1 macrophage markers, TNF-α, IL-12β and PTGS2, revealed significantly higher expression of all these M1 markers in healthy human peripheral blood MDM treated with LPS plus IFN-γ, compared to MDM treated with IL-4 (p<0.01) (Fig 3A). Analysis of mRNA expression of M2 markers, IL-10, CD200R1, MRC1 and TGM2, showed a significantly greater expression in healthy human peripheral blood MDM treated with IL-4 compared to MDM treated with LPS and IFN-γ (Fig 3B). Table 2 demonstrates the ratio of fold change mRNA expression of each marker in MDM treated with LPS plus IFN-γ to those treated with IL-4. In addition, stimulation of human peripheral blood MDM treated with LPS and IFN-γ resulted in a significant increase in TNF-α secretion as compared to IL-4-treated cultures (85,481 ± 3452 pg/mL vs. 27.16 ± 4.17 pg/mL) (Fig 3C).
Fig 3. Expression of M1 & M2 markers in M1 and M2 MDM compared to untreated MDM.
Expression of M1 & M2 markers relative to untreated healthy human peripheral blood MDM (U) in MDM treated with 10ng/mL LPS plus 20ng/mL IFN-γ (LPS) (M1 stimuli), or 20ng/mL IL-4 for 24 hours (IL-4) (M2 stimulus). (A) mRNA expression of M1 macrophage markers; TNF-α, IL-12β and PTGS2, in LPS plus IFN-γ or IL-4 treated MDM, compared to untreated MDM. (B) M2-specific mRNA expression of M2 macrophage markers; IL-10, CD200R1, MRC1, TGM2, in LPS plus IFN-γ- or IL-4-treated MDM, compared to untreated MDM. (C) TNF-α in supernatant of healthy human peripheral blood MDM stimulated with 10ng/mL LPS plus 20ng/mL IFN-γ, or 20ng/mL IL-4 for 24 hours, were measured using an ELISA Quantikine assay. Each experiment was performed in triplicate, with data expressed as mean ± SEM of five independent experiments. (*p<0.05, **p <0.01, ***p≤0.001).
Table 2. M1:M2 ratio of mean fold change of macrophage mRNA markers.
| M1 markers | M1:M2 ratio |
| Tumour necrosis factor alpha (TNF-α) | 18.12 |
| Interleukin 12β (IL-12β) | 9.66 |
| prostaglandin-endoperoxide synthase 2 (PTGS2, also known as Cox2) | 13.40 |
| M2 markers | M1:M2 ratio |
| Interleukin-10 (IL-10) | 0.16 |
| CD200R | 0.41 |
| C-type mannose receptor 1 (MRC-1, also known as CD206) | 0.04 |
| Transglutaminase 2 (TGM-2) | 0.34 |
M1: M2 ratio of mean fold change of each M1 or M2 specific macrophage marker in healthy human peripheral blood MDM. M1 phenotype MDM were generated by stimulation of MDM with 10ng/mL LPS plus 20ng/mL IFNγ for 24 hours, and M2 phenotype MDM generated by stimulation of MDM with 20ng/mL IL-4 for 24 hours (M2). Values are expressed to nearest 2 decimal places and presented as mean fold change for 5 independent experiments.
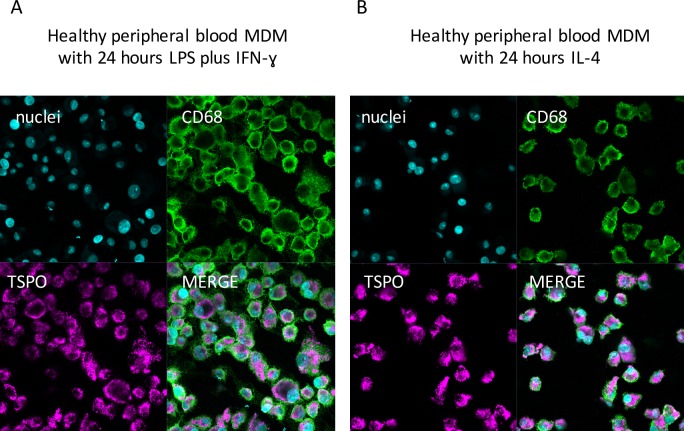
Immunofluorescence demonstrates no difference in staining for TSPO on MDM treated with LPS and IFN-γ and MDM treated with IL-4
Immunofluorescence staining of MDM treated with LPS and IFN-γ (Fig 4A), and MDM treated with IL-4 (Fig 4B) indicated that all cells were CD68 positive, confirming their macrophage phenotype. Furthermore, staining for TSPO demonstrated that the CD68+ cells expressed TSPO protein irrespective if they were stimulated with LPS and IFN-γ or IL-4 (Fig 4).
Fig 4. Representative immunofluorescence staining images of cytospin slides of healthy peripheral blood M1 and M2 MDM.
Immunofluorescence staining of cytospin slides of healthy peripheral blood MDM with the nuclear stain POPO-1 (to confirm presence of cells), TSPO, the macrophage marker CD68 and merging of the three stains. (A) staining of MDM stimulated with LPS plus IFN-γ for 24 hours; (B) staining of MDM stimulated with IL-4 for 24 hours. x400 magnification. Scale bar = 20μm.
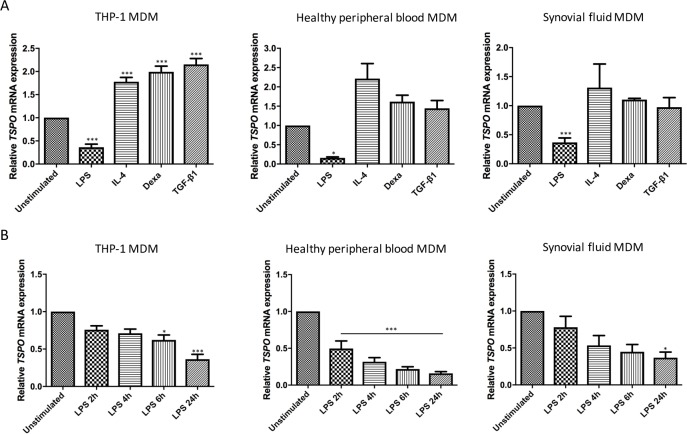
TSPO mRNA expression is reduced in M1 macrophages
For MDM derived from all sources (THP-1 monocytes, healthy human peripheral blood monocytes, and synovial fluid monocytes), there was a statistically significant down-regulation of TSPO mRNA expression in MDM stimulated with LPS and IFN-γ, compared to both unstimulated MDM (p<0.01 in THP-1 MDM, p<0.05 in healthy human peripheral blood MDM, and p<0.01 in synovial fluid MDM; Fig 5A). Likewise, there was a reduction in TSPO mRNA expression between MDM stimulated with LPS plus IFN-γ, compared to those stimulated with IL-4, dexamethasone or TGF-β1 (p<0.01 for THP-1 MDM, healthy human MDM and synovial fluid MDM) (Fig 5A).
Fig 5. Relative TSPO mRNA expression in MDM from THP-1, healthy human peripheral blood, and RA synovial fluid.
MDMs were stimulated with: (A) 10ng/mL LPS plus 20ng/mL IFN-γ, 20ng/mL IL-4, 500nM dexamethasone or 3ng/mL TGF-β1 for 24 hours, or (B) LPS plus IFN-γ for 2, 4, 6 and 24 hours. Each experiment was run in triplicate, with data expressed as mean of five independent experiments ± SEM and compared to unstimulated control cultures. (*p<0.05, **p <0.01, ***p≤0.001).
In contrast, there was no statistically significant difference between TSPO mRNA expression in unstimulated MDM versus MDM treated with M2 stimuli (IL-4, dexamethasone, or TGF-β1) in healthy human peripheral blood MDM or synovial fluid MDM. In THP-1 MDM, there was a statistically significant increase in TSPO mRNA expression in MDM stimulated with any M2 stimuli (IL-4, dexamethasone or TGF-β1) compared to unstimulated MDM (p<0.001).
In order to ensure there is no increase in TSPO mRNA expression in LPS plus IFN-γ stimulated MDM prior to 24 hours, a series of different stimulation durations were employed; 2,4,6 and 24 hours (Fig 5B). This demonstrated a reduction in TSPO expression over time in MDM stimulated with LPS plus IFNγ, with a statistically significant reduction in TSPO expression (relative to unstimulated MDM) being apparent as early as 2 hours for healthy human peripheral blood MDM (p<0.001), by 6 hours for THP-1 MDM (p<0.05), and by 24 hours in synovial fluid MDM (p<0.05).
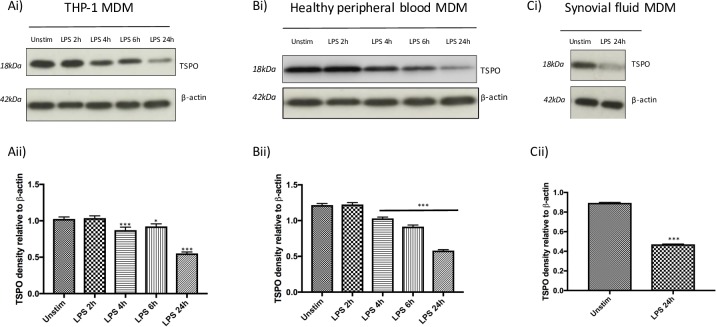
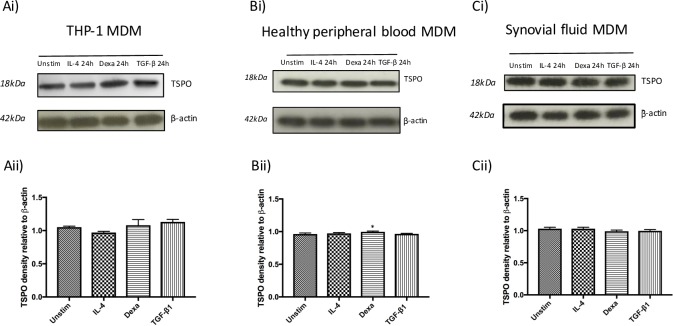
TSPO protein expression in human M1 and M2 macrophages
Results of Western blots indicated a down-regulation of TSPO protein expression in MDM stimulated with LPS plus IFN-γ at 24 hours in THP-1 MDM, healthy peripheral blood MDM, and synovial fluid MDM (Fig 6A–6C). Densitometry measurements of bands on western blot confirmed a significant reduction in TSPO band density on stimulation with LPS plus IFN-γ in MDM of all sources, as early as 2 hours (Fig 6A–6C). In contrast, protein expression of TSPO in healthy peripheral blood MDM stimulated with M2 stimuli did not appear to differ from unstimulated MDM, with only a small significant increase (p = 0.05) in TSPO band density in healthy peripheral blood MDM on stimulation with dexamethasone 500nM for 24 hours (mean ratio of 0.96 ± 0.01 in unstimulated healthy peripheral blood MDM compared to 1.00 ± 0.01 in healthy peripheral blood MDM treated with dexamethasone) (Fig 7).
Fig 6. Western blotting indicates TSPO protein expression decreases on treatment of MDM with M1 stimuli LPS plus IFN-γ.
(i) representative western blot of TSPO protein with β-actin acting as loading control and (ii) densitometry normalized to β-actin in (A) THP-1 MDM, (B) healthy peripheral blood MDM and (C) synovial fluid MDM either unstimulated (unstim) or treated with 10ng/mL LPS plus 20ng/mL IFN-γ (LPS) for 2,4,6 and 24 hours. Densitometry data is expressed as the mean of five independent experiments ± SEM. (*p<0.05, **p <0.01, ***p≤0.001).
Fig 7. Western blotting indicates TSPO protein expression is largely unchanged in MDM treated with M2 stimuli.
(i) representative Western blots of TSPO protein with β-actin acting as loading control and (ii) densitometry normalized to β-actin in (A) THP-1 MDM, (B) healthy peripheral blood MDM and (C) synovial fluid MDM either unstimulated (Unstim) treated with 20ng/mL IL-4, 500nM dexamethasone or 3ng/mL TGF-β1 for 24 hours. Densitometry data is expressed as the mean of five independent experiments ± SEM. (*p<0.05, **p <0.01, ***p≤0.001).
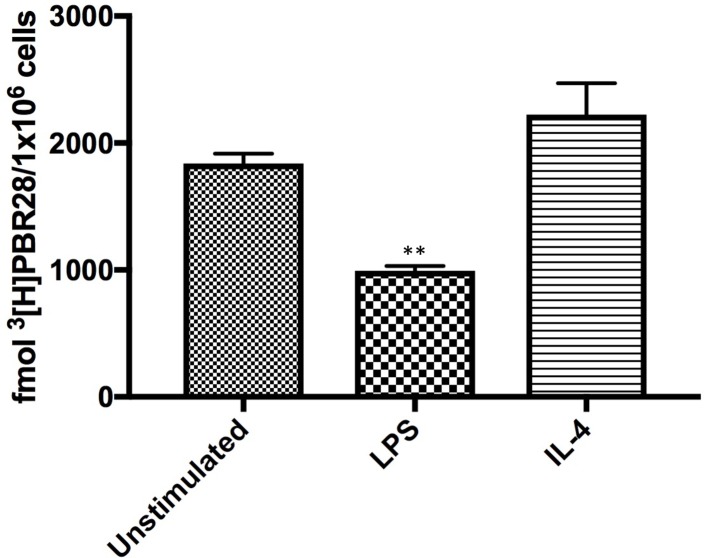
To provide a quantitative measure of TSPO protein expression, radio-ligand binding was undertaken using the TSPO specific radio-ligand [3H]PBR28 (Fig 8). Radioligand binding demonstrated significantly less [3H]PBR28 binding in healthy human peripheral blood MDM stimulated with LPS plus IFN-γ compared to unstimulated MDM (p<0.01, with mean binding of 994.3 fmol [3H]PBR28 per 1x106 MDM treated with LPS plus IFN-γ, compared to 1838 fmol [3H]PBR28 per 1x106 unstimulated MDM). There was no significant difference between unstimulated MDM and MDM treated with IL-4 (mean binding of 2223 fmol [3H]PBR28 per 1x106 MDM stimulated with IL-4). There was likewise a statistically significant (p<0.001) reduction of fmol [3H]PBR28/1x106 cells between MDM stimulated with LPS plus IFN-γ compared to those stimulated with IL-4.
Fig 8. Binding of the TSPO specific radio-ligand [3H]PBR28 in unstimulated, M1 and M2 healthy human peripheral blood MDM.
Binding of [3H]PBR28 in unstimulated healthy peripheral blood MDM, in MDM stimulated with LPS plus IFN-γ, and MDM stimulated with IL-4 for 24 hours. Each experiment was performed in triplicate, with data expressed as mean ± SEM of five independent experiments. ** p<0.01 compared to unstimulated.
Discussion
Macrophages are cells of the innate immune system and long established as key players in inflammatory diseases [45,46]. Indeed, in RA, the importance of macrophages in disease pathogenesis is well known, with infiltration of macrophages in joint tissue correlating with disease activity and joint damage [47].
The high expression of TSPO on macrophages has led to its use as a cellular target for the imaging of multiple inflammatory diseases; indeed, in the case of RA, TSPO-targeted imaging in vivo has been demonstrated to detect joint inflammation even before clinical signs of arthritis are apparent [48,49].
A better understanding of the expression of TSPO on macrophages of different phenotype would aid more accurate interpretation of in vivo TSPO targeted imaging in humans with inflammatory disease. Further, since the function of TSPO is as yet undetermined, further knowledge of TSPO expression according to macrophage phenotype, may lend insight into the potential role of TSPO in macrophage biology.
There has been scant assessment of TSPO expression on different phenotypes of human macrophages. Sandiego et al. attempted to ascertain the impact of the M1 stimulus, LPS, on microglia TSPO expression, by administering Escherichia coli LPS to healthy human volunteers, via peripheral intravenous injection [50]. TSPO PET radio-ligand [11C]PBR28 imaging of the brain was performed prior to, and two hours post LPS administration into peripheral blood. The authors’ data demonstrated a significant increase (nearly 2-fold on average) in [11C]PBR28 binding in the brain, post LPS treatment, reflecting increased TSPO expression [50]. These findings imply that LPS increases TSPO expression in the brain, but does not necessarily imply that it increases TSPO expression in microglia. Indeed, multiple cell types other than microglia exist in the brain (neurons, astrocytes and oligodendrocytes [51]) and circulating leukocytes may infiltrate brain tissue during a systemic inflammatory response [52]. Since TSPO is known to be fairly ubiquitous [53–55], it cannot therefore be assumed that the increased TSPO tracer signal on brain imaging was solely attributable to the resident microglia, and not due to contribution from other neural cells, or infiltrating cells. Further, since LPS was administered peripherally, it cannot be assumed that LPS directly up-regulated TSPO expression on microglia; indeed, imaging was undertaken at 2 hours post LPS administration, a far shorter time interval than has been previously reported for LPS to penetrate the blood brain barrier, and further LPS may not penetrate this directly [56].
Our study not only confirmed that TSPO is up-regulated on MDM compared to monocytes, but further provides the first in vitro data on TSPO mRNA and protein expression in human macrophages of known M1 or M2 phenotype, as defined by mRNA assessment of multiple M1 and M2 markers, as well as MDM production of TNF-α. Our data also indicate that TSPO mRNA and protein was down-regulated on MDM activated to an M1 phenotype (regardless of source), by stimulation with LPS plus IFN-γ at 24 hours. Down-regulation of TSPO mRNA expression was noted as early as 2 hours of treatment. The magnitude of down-regulation of TSPO protein on M1 stimulation of MDM was such that there was no significant difference between [3H]PBR28 binding in healthy human monocytes and healthy human M1 MDM, indicating that, the up-regulation of TSPO protein that occurs on monocyte-to-macrophage differentiation, is lost on exposure to M1 stimuli.
In contrast, there was no significant change in TSPO expression at mRNA or protein level in healthy peripheral MDM, or synovial fluid MDM activated to an M2 phenotype, compared to unstimulated MDM. Our immunofluorescence staining demonstrated TSPO expression on both M1 and M2 macrophages, in keeping with the fact that TSPO was still detectable on both macrophage phenotypes, but reinforces the point that histological staining may not be the optimal means of quantifying TSPO protein expression.
It must be noted that M1 and M2 stimuli applied in this study may not exist in isolation in vivo, where a combination of stimuli may act on monocytes and macrophages at any one time within a specific tissue [11,57], thus macrophage phenotypes in vivo may be more complex. Further study of in vivo human MDM harvested from different tissues, and disease states, will clarify macrophage phenotypes in vivo.
The reasons for a difference in TSPO expression between M1 and M2 phenotype human MDM are uncertain, given that the function of TSPO is yet to be conclusively demonstrated. However, our findings could suggest that TSPO is involved in the negative regulation of inflammation, by promoting an M2 macrophage phenotype. This hypothesis is supported by an investigation by Bae et al. who engineered the murine BV2 microglia cell line to overexpress TSPO [58]. This study concluded that TSPO overexpression was associated with reduced production of pro-inflammatory cytokines on LPS treatment of immortalized microglia BV2 cells, with increased expression of ‘M2’ murine macrophage markers IL-10 and MRC1, compared to microglia that did not overexpress TSPO [58].
One potential hypothesis by which TSPO expression could be associated with skewing macrophage phenotype away from the pro-inflammatory end of the spectrum, could be due to the postulated role of TSPO in cholesterol efflux [59]. A downregulation of cholesterol efflux could promote intra-cellular lipid accumulation, potentiating a pro-inflammatory (M1) macrophage phenotype, through promoting lipid raft receptor signaling, previously recognized to be an important component of pro-inflammatory signaling pathways [60,61]. Additionally, a downregulation of cholesterol efflux could promote intra-cellular cholesterol crystals accumulation, activating the inflammasome [62]. The importance of TSPO in cholesterol efflux is indicated by the fact that THP-1 derived macrophages transfected to overexpress TSPO, exhibit an increase in cholesterol efflux, whilst ligation of TSPO increased cholesterol efflux further in macrophages [63]. Silencing TSPO expression in human MDM, and investigating cholesterol efflux in in vitro macrophages of known M1 or M2 phenotype, would provide further confirmatory evidence to support this hypothesis.
The data presented herein have implications for the field of TSPO-targeted PET imaging, where TSPO specific radio-ligands are increasingly being used to image a range of inflammatory disease states in vivo [4,64–67]. In particular, in RA, TSPO-targeted PET has already been demonstrated to have potential utility for diagnosis of subclinical RA, prediction of disease flare (44, 66), and may well have utility as a tool for assessing response to therapy. With the increasing sensitivity and specificity of next generation TSPO targeted radio-ligands [5,39], it may be important to consider differences in TSPO expression in macrophages of M1 polarity compared to M2 when interpreting TSPO PET images. Indeed, given our study findings, TSPO PET itself may provide insight into the M1 or M2 polarization of macrophages in synovial tissue in RA, for which there is much conflicting data currently [46].
Supporting information
(A) SSC-A/FSC of harvested healthy peripheral blood monocytes. (B) SSC-A/FSC of unstimulated healthy peripheral blood MDM, generated after differentiation of monocytes with 100ng/mL M-CSF for 7 days. (C) SSC-A/PE-Cy7-A -CD68 staining of monocytes. (D) SSC-A/PE-Cy7-A -CD68 staining of unstimulated healthy human MDM. SSC-A = side scatter area (measure of cell granularity), FSC-A = forward scatter area (measure of cell size).
S1A and B Figs demonstrate clear increase in granularity of cells, consistent with monocyte differentiation to macrophages. S1C and D Figs demonstrate a clear increase in CD68 expression in macrophages (D) compared to monocytes (C), along with an increase in cell granularity, consistent with successful monocyte to macrophage differentiation.
(TIF)
Acknowledgments
The authors gratefully acknowledge patients from Nuffield Orthopaedic Centre Oxford Rheumatology Outpatients Department, for donation of synovial fluid.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
NN was supported by the NIHR BRC at the University of Oxford through an unrestricted grant (OXF141232).
References
- 1.Batarseh A, Papadopoulos V. Regulation of translocator protein 18 kDa (TSPO) expression in health and disease states. Mol Cell Endocrinol. 2010;327(1–2):1–12. doi: 10.1016/j.mce.2010.06.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Papadopoulos V, Lecanu L, Brown RC, Han Z, Yao ZX. Peripheral-type benzodiazepine receptor in neurosteroid biosynthesis, neuropathology and neurological disorders. Neuroscience. 2006;138(3):749–56. doi: 10.1016/j.neuroscience.2005.05.063 [DOI] [PubMed] [Google Scholar]
- 3.Vowinckel E, Reutens D, Becher B, Verge G, Evans A, Owens T, et al. PK11195 binding to the peripheral benzodiazepine receptor as a marker of microglia activation in multiple sclerosis and experimental autoimmune encephalomyelitis. J Neurosci Res. 1997;50(2):345–53. doi: 10.1002/(SICI)1097-4547(19971015)50:2<345::AID-JNR22>3.0.CO;2-5 [DOI] [PubMed] [Google Scholar]
- 4.van der Laken CJ, Elzinga EH, Kropholler MA, Molthoff CF, van der Heijden JW, Maruyama K, et al. Noninvasive imaging of macrophages in rheumatoid synovitis using 11C-(R)-PK11195 and positron emission tomography. Arthritis Rheum. 2008;58(11):3350–5. doi: 10.1002/art.23955 [DOI] [PubMed] [Google Scholar]
- 5.Gent YY, Weijers K, Molthoff CF, Windhorst AD, Huisman MC, Kassiou M, et al. Promising potential of new generation translocator protein tracers providing enhanced contrast of arthritis imaging by positron emission tomography in a rat model of arthritis. Arthritis Res Ther. 2014;16(2):R70 doi: 10.1186/ar4509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Navegantes KC, de Souza Gomes R, Pereira PAT, Czaikoski PG, Azevedo CHM, Monteiro MC. Immune modulation of some autoimmune diseases: the critical role of macrophages and neutrophils in the innate and adaptive immunity. J Transl Med. 2017;15:36 doi: 10.1186/s12967-017-1141-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hume DA. The Many Alternative Faces of Macrophage Activation. Front Immunol. 2015;6:370 doi: 10.3389/fimmu.2015.00370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sica A, Mantovani A. Macrophage plasticity and polarization: in vivo veritas. J Clin Invest. 2012;122(3):787–95. doi: 10.1172/JCI59643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Murray PJ, Wynn TA. Protective and pathogenic functions of macrophage subsets. Nat Rev Immunol. 2011;11(11):723–37. doi: 10.1038/nri3073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Murray PJ, Allen JE, Biswas SK, Fisher EA, Gilroy DW, Goerdt S, et al. Macrophage activation and polarization: nomenclature and experimental guidelines. Immunity. 2014;41(1):14–20. doi: 10.1016/j.immuni.2014.06.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Martinez FO, Gordon S. The M1 and M2 paradigm of macrophage activation: time for reassessment. F1000prime Rep. 2014;6:13 doi: 10.12703/P6-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bonsack Ft, Alleyne CH Jr., Sukumari-Ramesh S. Augmented expression of TSPO after intracerebral hemorrhage: a role in inflammation? J Neuroinflammation. 2016;13(1):151 doi: 10.1186/s12974-016-0619-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shao X, Wang X, English SJ, Desmond T, Sherman PS, Quesada CA, et al. Imaging of carrageenan-induced local inflammation and adjuvant-induced systemic arthritis with [(11)C]PBR28 PET. Nucl Med Biol. 2013;40(7):906–11. doi: 10.1016/j.nucmedbio.2013.06.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Murray PJ, Wynn TA. Obstacles and opportunities for understanding macrophage polarization. J Leukoc Biol. 2011;89(4):557–63. doi: 10.1189/jlb.0710409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schneemann M, Schoeden G. Macrophage biology and immunology: man is not a mouse. J Leukoc Biol. 2007;81(3):579; discussion 80. doi: 10.1189/jlb.1106702 [DOI] [PubMed] [Google Scholar]
- 16.Mestas J, Hughes CC. Of mice and not men: differences between mouse and human immunology. J Immunol. 2004;172(5):2731–8. [DOI] [PubMed] [Google Scholar]
- 17.Thomas AC, Mattila JT. “Of Mice and Men”: Arginine Metabolism in Macrophages. Front Immunol. 2014;5:479 doi: 10.3389/fimmu.2014.00479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Raes G, Van den Bergh R, De Baetselier P, Ghassabeh GH, Scotton C, Locati M, et al. Arginase-1 and Ym1 are markers for murine, but not human, alternatively activated myeloid cells. J Immunol. 2005;174(11):6561; author reply -2. [DOI] [PubMed] [Google Scholar]
- 19.Lawrence T, Natoli G. Transcriptional regulation of macrophage polarization: enabling diversity with identity. Nat Rev Immunol. 2011;11(11):750–61. doi: 10.1038/nri3088 [DOI] [PubMed] [Google Scholar]
- 20.Schneemann M, Schoedon G. Species differences in macrophage NO production are important. Nat Immunol. 2002;3(2):102 doi: 10.1038/ni0202-102a [DOI] [PubMed] [Google Scholar]
- 21.Mosser DM, Edwards JP. Exploring the full spectrum of macrophage activation. Nat Rev Immunol. 8(12):958–969. doi: 10.1038/nri2448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Auwerx J. The human leukemia cell line, THP-1: a multifacetted model for the study of monocyte-macrophage differentiation. Experientia. 1991;47(1):22–31. [DOI] [PubMed] [Google Scholar]
- 23.Stroncek DF, Fellowes V, Pham C, Khuu H, Fowler DH, Wood LV, et al. Counter-flow elutriation of clinical peripheral blood mononuclear cell concentrates for the production of dendritic and T cell therapies. J Transl Med. 2014;12:241 doi: 10.1186/s12967-014-0241-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Berger TG, Strasser E, Smith R, Carste C, Schuler-Thurner B, Kaempgen E, et al. Efficient elutriation of monocytes within a closed system (Elutra) for clinical-scale generation of dendritic cells. J Immunol Methods. 2005;298(1–2):61–72. doi: 10.1016/j.jim.2005.01.005 [DOI] [PubMed] [Google Scholar]
- 25.Yasaka T, Mantich NM, Boxer LA, Baehner RL. Functions of human monocyte and lymphocyte subsets obtained by countercurrent centrifugal elutriation: differing functional capacities of human monocyte subsets. J Immunol. 1981;127(4):1515–8. [PubMed] [Google Scholar]
- 26.Schwanke U, Nabereit A, Moog R. Isolation of monocytes from whole blood-derived buffy coats by continuous counter-flow elutriation. J Clin Apher. 2006;21(3):153–7. doi: 10.1002/jca.20077 [DOI] [PubMed] [Google Scholar]
- 27.Banfalvi G. Cell cycle synchronization of animal cells and nuclei by centrifugal elutriation. Nat Protoc. 2008;3(4):663–73. doi: 10.1038/nprot.2008.34 [DOI] [PubMed] [Google Scholar]
- 28.Aletaha D, Neogi T, Silman AJ, Funovits J, Felson DT, Bingham CO 3rd, et al. 2010 Rheumatoid arthritis classification criteria: an American College of Rheumatology/European League Against Rheumatism collaborative initiative. Arthritis Rheum. 2010;62(9):2569–81. doi: 10.1002/art.27584 [DOI] [PubMed] [Google Scholar]
- 29.Martinez FO. Analysis of gene expression and gene silencing in human macrophages. Curr Protoc Immunol. 2012;Chapter 14:Unit 14.28.1–3. [DOI] [PubMed] [Google Scholar]
- 30.Liu H, Shi B, Huang C-C, Eksarko P, Pope RM. Transcriptional diversity during monocyte to macrophage differentiation. Immunol Lett. 2008;117(1):70–80. doi: 10.1016/j.imlet.2007.12.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Frede S, Stockmann C, Freitag P, Fandrey J. Bacterial lipopolysaccharide induces HIF-1 activation in human monocytes via p44/42 MAPK and NF-kappaB. Biochem J. 2006;396(3):517–27. doi: 10.1042/BJ20051839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Krausgruber T, Blazek K, Smallie T, Alzabin S, Lockstone H, Sahgal N, et al. IRF5 promotes inflammatory macrophage polarization and TH1-TH17 responses. Nat Immunol. 2011;12(3):231–8. doi: 10.1038/ni.1990 [DOI] [PubMed] [Google Scholar]
- 33.Han J, Hajjar DP, Tauras JM, Feng J, Gotto AM Jr., Nicholson AC. Transforming growth factor-beta1 (TGF-beta1) and TGF-beta2 decrease expression of CD36, the type B scavenger receptor, through mitogen-activated protein kinase phosphorylation of peroxisome proliferator-activated receptor-gamma. J Biol Chem. 2000;275(2):1241–6. [DOI] [PubMed] [Google Scholar]
- 34.Dakin SG, Martinez FO, Yapp C, Wells G, Oppermann U, Dean BJF, et al. Inflammation activation and resolution in human tendon disease. Sci Transl Med. 2015;7(311):311ra173–311ra173. doi: 10.1126/scitranslmed.aac4269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Martinez FO, Gordon S, Locati M, Mantovani A. Transcriptional profiling of the human monocyte-to-macrophage differentiation and polarization: new molecules and patterns of gene expression. J Immunol. 2006;177(10):7303–11. [DOI] [PubMed] [Google Scholar]
- 36.Roszer T. Understanding the mysterious M2 macrophage through activation markers and effector mechanisms. Mediators Inflamm. 2015;2015:816460 doi: 10.1155/2015/816460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Koning N, van Eijk M, Pouwels W, Brouwer MS, Voehringer D, Huitinga I, et al. Expression of the inhibitory CD200 receptor is associated with alternative macrophage activation. J Innate Imm. 2010;2(2):195–200. [DOI] [PubMed] [Google Scholar]
- 38.Taylor SC, Berkelman T, Yadav G, Hammond M. A defined methodology for reliable quantification of Western blot data. Mol Biotechnol. 2013;55(3):217–26. doi: 10.1007/s12033-013-9672-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chauveau F, Boutin H, Van Camp N, Dolle F, Tavitian B. Nuclear imaging of neuroinflammation: a comprehensive review of [11C]PK11195 challengers. Eur J Nucl Med Mol Imaging. 2008;35(12):2304–19. doi: 10.1007/s00259-008-0908-9 [DOI] [PubMed] [Google Scholar]
- 40.Oh U, Fujita M, Ikonomidou VN, Evangelou IE, Matsuura E, Harberts E, et al. Translocator protein PET imaging for glial activation in multiple sclerosis. J Neuroimmune Pharmacol. 2011;6(3):354–61. doi: 10.1007/s11481-010-9243-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nair A, Veronese M, Xu X, Curtis C, Turkheimer F, Howard R, et al. Test-retest analysis of a non-invasive method of quantifying [(11)C]-PBR28 binding in Alzheimer's disease. EJNMMI Res. 2016;6(1):72 doi: 10.1186/s13550-016-0226-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Owen DR, Yeo AJ, Gunn RN, Song K, Wadsworth G, Lewis A, et al. An 18-kDa translocator protein (TSPO) polymorphism explains differences in binding affinity of the PET radioligand PBR28. J Cereb Blood Flow Metab. 2012;32(1):1–5. doi: 10.1038/jcbfm.2011.147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Owen DR, Howell OW, Tang SP, Wells LA, Bennacef I, Bergstrom M, et al. Two binding sites for [3H]PBR28 in human brain: implications for TSPO PET imaging of neuroinflammation.J Cereb Blood Flow Metab. 2010;30(9):1608–18. doi: 10.1038/jcbfm.2010.63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Canat X, Carayon P, Bouaboula M, Cahard D, Shire D, Roque C, et al. Distribution profile and properties of peripheral-type benzodiazepine receptors on human hemopoietic cells. Life Sci. 1993;52(1):107–18. [DOI] [PubMed] [Google Scholar]
- 45.Wynn TA, Chawla A, Pollard JW. Origins and Hallmarks of Macrophages: Development, homeostasis, and disease. Nature. 2013;496(7446):445–55. doi: 10.1038/nature12034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kennedy A, Fearon U, Veale DJ, Godson C. Macrophages in synovial Inflammation. Front Immunol 2011;2:52 doi: 10.3389/fimmu.2011.00052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mulherin D, Fitzgerald O, Bresnihan B. Synovial tissue macrophage populations and articular damage in rheumatoid arthritis. Arthritis Rheum. 1996;39(1):115–24. [DOI] [PubMed] [Google Scholar]
- 48.Gent YY, Voskuyl AE, Kloet RW, van Schaardenburg D, Hoekstra OS, Dijkmans BA, et al. Macrophage positron emission tomography imaging as a biomarker for preclinical rheumatoid arthritis: findings of a prospective pilot study. Arthritis Rheum. 2012;64(1):62–6. doi: 10.1002/art.30655 [DOI] [PubMed] [Google Scholar]
- 49.Gent YY, Ahmadi N, Voskuyl AE, Hoetjes N, van Kuijk C, Britsemmer K, et al. Detection of subclinical synovitis with macrophage targeting and positron emission tomography in patients with rheumatoid arthritis without clinical arthritis. J Rheumatol. 2014;41(11):2145–52. doi: 10.3899/jrheum.140059 [DOI] [PubMed] [Google Scholar]
- 50.Sandiego CM, Gallezot JD, Pittman B, Nabulsi N, Lim K, Lin SF, et al. Imaging robust microglial activation after lipopolysaccharide administration in humans with PET. Proc Natl Acad Sci U S A. 2015;112(40):12468–73. doi: 10.1073/pnas.1511003112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Garcia-Cabezas MA, John YJ, Barbas H, Zikopoulos B. Distinction of Neurons, Glia and Endothelial Cells in the Cerebral Cortex: An Algorithm Based on Cytological Features. Front Neuroanat. 2016;10:107 doi: 10.3389/fnana.2016.00107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ransohoff RM, Kivisakk P, Kidd G. Three or more routes for leukocyte migration into the central nervous system. Nat Rev Immunol. 2003;3(7):569–81. doi: 10.1038/nri1130 [DOI] [PubMed] [Google Scholar]
- 53.Scarf AM, Kassiou M. The translocator protein. J Nucl Med. 2011; 52(5):677–80. doi: 10.2967/jnumed.110.086629 [DOI] [PubMed] [Google Scholar]
- 54.Morin D, Musman J, Pons S, Berdeaux A, Ghaleh B. Mitochondrial translocator protein (TSPO): From physiology to cardioprotection. Biochem Pharmacol. 2016;105:1–13. doi: 10.1016/j.bcp.2015.12.003 [DOI] [PubMed] [Google Scholar]
- 55.Ching AS, Kuhnast B, Damont A, Roeda D, Tavitian B, Dolle F. Current paradigm of the 18-kDa translocator protein (TSPO) as a molecular target for PET imaging in neuroinflammation and neurodegenerative diseases. Insights Imaging. 2012;3(1):111–9. doi: 10.1007/s13244-011-0128-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Banks WA, Gray AM, Erickson MA, Salameh TS, Damodarasamy M, Sheibani N, et al. Lipopolysaccharide-induced blood-brain barrier disruption: roles of cyclooxygenase, oxidative stress, neuroinflammation, and elements of the neurovascular unit. J Neuroinflamm. 2015;12:223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Davies LC, Jenkins SJ, Allen JE, Taylor PR. Tissue-resident macrophages. Nat Immunol. 2013;14(10):986–95. doi: 10.1038/ni.2705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bae KR, Shim HJ, Balu D, Kim SR, Yu SW. Translocator protein 18 kDa negatively regulates inflammation in microglia. J Neuroimmune Pharmacol. 2014;9(3):424–37. doi: 10.1007/s11481-014-9540-6 [DOI] [PubMed] [Google Scholar]
- 59.Allen AM, Taylor JM, Graham A. Mitochondrial (dys)function and regulation of macrophage cholesterol efflux. Clin Sci. 2013;124(8):509–15. doi: 10.1042/CS20120358 [DOI] [PubMed] [Google Scholar]
- 60.Fessler MB, Parks JS. Intracellular lipid flux and membrane microdomains as organizing principles in inflammatory cell signaling. J Immunol. 2011;187(4):1529–35. doi: 10.4049/jimmunol.1100253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chowdhury SM, Zhu X, Aloor JJ, Azzam KM, Gabor KA, Ge W, et al. Proteomic Analysis of ABCA1-Null Macrophages Reveals a Role for Stomatin-Like Protein-2 in Raft Composition and Toll-Like Receptor Signaling. Mol Cell Proteomics. 2015;14(7):1859–70. doi: 10.1074/mcp.M114.045179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Niyonzima N, Samstad EO, Aune MH, Ryan L, Bakke SS, Rokstad AM, et al. Reconstituted High-Density Lipoprotein Attenuates Cholesterol Crystal-Induced Inflammatory Responses by Reducing Complement Activation. J Immunol. 2015;195(1):257–64. doi: 10.4049/jimmunol.1403044 [DOI] [PubMed] [Google Scholar]
- 63.Taylor JM, Allen AM, Graham A. Targeting mitochondrial 18 kDa translocator protein (TSPO) regulates macrophage cholesterol efflux and lipid phenotype. Clin Sci. 2014;127(10):603–13. doi: 10.1042/CS20140047 [DOI] [PubMed] [Google Scholar]
- 64.Lamare F, Hinz R, Gaemperli O, Pugliese F, Mason JC, Spinks T, et al. Detection and quantification of large-vessel inflammation with 11C-(R)-PK11195 PET/CT. J Nucl Med. 2011;52(1):33–9. doi: 10.2967/jnumed.110.079038 [DOI] [PubMed] [Google Scholar]
- 65.Park E, Gallezot JD, Delgadillo A, Liu S, Planeta B, Lin SF, et al. (11)C-PBR28 imaging in multiple sclerosis patients and healthy controls: test-retest reproducibility and focal visualization of active white matter areas. Eur J Nucl Med Mol Imaging. 2015;42(7):1081–92. doi: 10.1007/s00259-015-3043-4 [DOI] [PubMed] [Google Scholar]
- 66.Schweitzer PJ, Fallon BA, Mann JJ, Kumar JS. PET tracers for the peripheral benzodiazepine receptor and uses thereof. Drug Discov Today. 2010;15(21–22):933–42. doi: 10.1016/j.drudis.2010.08.012 [DOI] [PubMed] [Google Scholar]
- 67.Bernards N, Pottier G, Theze B, Dolle F, Boisgard R. In vivo evaluation of inflammatory bowel disease with the aid of muPET and the translocator protein 18 kDa radioligand [18F]DPA-714. Mol Imaging Biol. 2015;17(1):67–75. doi: 10.1007/s11307-014-0765-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) SSC-A/FSC of harvested healthy peripheral blood monocytes. (B) SSC-A/FSC of unstimulated healthy peripheral blood MDM, generated after differentiation of monocytes with 100ng/mL M-CSF for 7 days. (C) SSC-A/PE-Cy7-A -CD68 staining of monocytes. (D) SSC-A/PE-Cy7-A -CD68 staining of unstimulated healthy human MDM. SSC-A = side scatter area (measure of cell granularity), FSC-A = forward scatter area (measure of cell size).
S1A and B Figs demonstrate clear increase in granularity of cells, consistent with monocyte differentiation to macrophages. S1C and D Figs demonstrate a clear increase in CD68 expression in macrophages (D) compared to monocytes (C), along with an increase in cell granularity, consistent with successful monocyte to macrophage differentiation.
(TIF)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.