Abstract
Background
Enteric fever remains a major public health problem in low resource settings and antibiotic resistance is increasing. In Asia, an increasing proportion of infections is caused by Salmonella enterica serovar Paratyphi A, which for a long time was assumed to cause a milder clinical syndrome compared to Salmonella enterica serovar Typhi.
Methodology
A retrospective chart review study was conducted of 254 unique cases of blood culture confirmed enteric fever who presented at a referral adult hospital in Phnom Penh, Cambodia between 2008 and 2015. Demographic, clinical and laboratory data were collected from clinical charts and antibiotic susceptibility testing was performed. Whole genome sequence analysis was performed on a subset of 121 isolates.
Results
One-hundred-and-ninety unique patients were diagnosed with Salmonella Paratyphi A and 64 with Salmonella Typhi. In the period 2008–2012, Salmonella Paratyphi A comprised 25.5% of 47 enteric fever cases compared to 86.0% of 207 cases during 2013–2015. Presenting symptoms were identical for both serovars but higher median leukocyte counts (6.8 x 109/L vs. 6.3 x 109/L; p = 0.035) and C-reactive protein (CRP) values (47.0 mg/L vs. 36 mg/L; p = 0.034) were observed for Salmonella Typhi infections. All but one of the Salmonella Typhi isolates belonged to haplotype H58 associated with multidrug resistance (MDR) (i.e. resistance to ampicillin, chloramphenicol and co-trimoxazole).;42.9% actually displayed MDR compared to none of the Salmonella Paratyphi A isolates. Decreased ciprofloxacin susceptibility (DCS) was observed in 96.9% (62/64) of Salmonella Typhi isolates versus 11.5% (21/183) of Salmonella Paratyphi A isolates (all but one from 2015). All isolates were susceptible to azithromycin and ceftriaxone.
Conclusions
In Phnom Penh, Cambodia, Salmonella Paratyphi A now causes the majority of enteric fever cases and decreased susceptibility against ciprofloxacin is increasing. Overall, Salmonella Typhi was significantly more associated with MDR and DCS compared to Salmonella Paratyphi A.
Author summary
Enteric fever is a bloodstream infection caused by the bacteria Salmonella Typhi or Salmonella Paratyphi A, B, or C. It is common in low resource settings and linked to poor water quality and sanitation. The disease is also endemic in Cambodia and since 2013 there has been a sharp increase in the number of Salmonella Paratyphi A infections. We sought to compare the clinical phenotypes and antibiotic susceptibilities of Salmonella Paratyphi A infections with those of Salmonella Typhi infections in this setting. We retrospectively collected demographic, clinical and laboratory data from clinical charts of 254 patients with a blood culture positive for enteric fever. We also assessed antibiotic susceptibility patterns and sequenced the genome of a subset of isolates. We found that since 2013 the majority of enteric fever cases are caused by Salmonella Paratyphi A which increasingly shows decreased susceptibility to the antibiotic ciprofloxacin, the current first line treatment. In contrast, in Salmonella Typhi a re-emergence of susceptibility for the former first line antibiotics of ampicillin, co-trimoxazole and chloramphenicol was observed. Presenting symptoms of Salmonella Typhi and Salmonella Paratyphi A were identical, minor differences were observed in laboratory parameters.
Introduction
Salmonella enterica serovar Typhi (Salmonella Typhi) and Salmonella enterica serovars Paratyphi (Salmonella Paratyphi) A, B, and C are Gram-negative bacteria which can invade the bloodstream and cause typhoid and paratyphoid fever respectively (also jointly known as ‘enteric fever’). They are confined to the human host and are transmitted via the fecal-oral route. Enteric fever poses a serious disease burden in low resource settings where the infection is linked to poor sanitation and limited access to safe drinking water [1]. Although enteric fever has become rare in Western countries it continues to affect international travelers returning from endemic countries [2].
Patients with enteric fever typically present with acute fever and non-specific symptoms. For a long time, Salmonella Paratyphi A was thought to cause milder disease than Salmonella Typhi but several studies have contradicted this [1–4].
For both serovars, antibiotic resistance is increasingly reported and there is now widespread presence of co-resistance against the former first line treatment options of ampicillin, co-trimoxazole and chloramphenicol (known as ‘multidrug resistance’) and decreased susceptibility to ciprofloxacin, the current first line drug [5, 6]. Resistance to ciprofloxacin is also increasingly reported [7, 8]. Worrisome are recent reports on emerging resistance against third-generation cephalosporins and azithromycin, the current alternative treatment options [9, 10].
Although historically the majority of enteric fever cases were caused by Salmonella Typhi, the proportion of Salmonella Paratyphi A infections has been increasing steadily since the turn of the century, in particular on the Asian continent [11].
In 2013, a significant increase in Salmonella Paratyphi A infections was also observed in Cambodia, a country where enteric fever remains one of the most common clinical and blood culture-confirmed diseases [12]. The increase was described in local residents as well as in travelers returning from Cambodia to Europe, New Zealand, Japan and the United States [13–16].
Surprisingly, little is known about the clinical and microbiological characteristics of Salmonella Typhi and Salmonella Paratyphi A infections in Cambodian adults.
This study therefore aims to assess the clinical and microbiological aspects of enteric fever in patients attending an adult hospital in Phnom Penh, Cambodia, during 2008–2015. More specifically it aims to assess differences between infections caused by Salmonella Typhi as compared to Salmonella Paratyphi A.
Materials and methods
Study setting & population
Sihanouk Hospital Center of HOPE (SHCH) is a 40-bed non-governmental referral hospital for adults in Phnom Penh, Cambodia. Since July 2007, SHCH and the Institute of Tropical Medicine (ITM) in Antwerp, Belgium, have been jointly organising the surveillance of bloodstream infections at this hospital and its associated clinics. For this study, all data collected between 2008–2015 were analyzed. Blood cultures were systematically sampled in all patients presenting at SHCH who were suspected of having sepsis according to the Systemic Inflammatory Response Syndrome (SIRS) criteria [17]. Recently, new definitions and criteria for sepsis have been proposed such as the Sequential [Sepsis-related] Organ Failure Assessment (SOFA) score [18]. Over the 8-year period, 18,927 blood cultures were sampled from mostly, but not exclusively, adults [19]. Of these cultures 1,654 (8,7%) yielded clinically significant organisms.
Clinical review
From all patients whose blood was drawn for culture, basic demographic and clinical data were registered in a surveillance logbook. A medical doctor verified missing data with patients during a routine phone call one week after discharge from the hospital which was part of standard care. In addition, for this study, the available medical charts of all patients with blood culture-confirmed enteric fever were reviewed retrospectively by a second medical doctor for additional symptoms and signs.
Laboratory methods
Hematology parameters were analyzed using a Sysmex KX-21 and T-1800i analyzer (Sysmex Corporation, Kobe, Japan) and CRP values were measured using a TEMIS Linear Analyzer (Linear Chemicals sl, Montgat, Spain).
Blood cultures were sampled and worked-up as previously described [20]. Isolates biochemically identified as Salmonella spp. were stored at -70°C on porous beads in cryopreservative (Microbank, Pro-Lab Diagnostics, Richmond Hill, Canada) and eventually shipped to the ITM in Belgium. At ITM, the isolates were serotyped using commercial antisera (Sifin, Berlin, Germany) following the White-Kauffmann-Le Minor scheme. A selection of 91 isolates were sent to the Institut Pasteur in Paris for confirmation and whole genome sequencing.
At the ITM, antibiotic susceptibility was determined for all available isolates by disk diffusion on Mueller-Hinton II agar in accordance with the CLSI 2016 guidelines [21]. The following antimicrobial drugs (Neo-Sensitabs, Rosco, Taastrup, Denmark) were tested: ampicillin, sulfamethoxazole-trimethoprim, chloramphenicol, nalidixic acid, pefloxacin, gentamicin, tetracycline, ceftriaxone, ceftazidime, meropenem and ertapenem. Nalidixic acid and pefloxacin served as predictors for ciprofloxacin non-susceptibility.
In addition, for all available isolates, minimal inhibitory concentration (MIC) values for ciprofloxacin and azithromycin were determined by the E-test macro method (bioMérieux, Marcy L'Etoile, France).
Quality control was performed using Escherichia coli (ATCC 25922) and Staphylococcus aureus (ATCC 29213).
Multidrug resistance (MDR) was defined as co-resistance to ampicillin, chloramphenicol and trimethoprim-sulfamethoxazole [22]. For comparison with previously published literature, we used the superseded term ‘decreased ciprofloxacin susceptibility (DCS)’, defined as MIC-values of ≥0.12 mg/L and ≤0.5 mg/L, i.e. currently classified as ‘intermediate susceptibility' but associated with treatment failures or delayed treatment response [21].
Assessment of antimicrobial resistance genes and Salmonella Typhi H58 typing
Whole genome sequencing was carried out on all 65 Salmonella Typhi isolates and a selection of 26 Salmonella Paratyphi A isolates at the Plateforme de microbiologie mutualisée (P2M) from the Pasteur International Bioresources network (PIBnet, Institut Pasteur, Paris, France). Short-read sequences from 30 previously published Salmonella Paratyphi A genomes were also included [23]. The run accession numbers and related metadata are detailed in S1 Table. Short-read sequences have been deposited to the European Nucleotide Archive (ENA) (http://www.ebi.ac.uk/ena) (accession number PRJEB19906).
The MagNAPure 96 system (Roche Diagnostics, Indianopolis, IN, USA) was used for DNA extraction, libraries were prepared using the Nextera XT kit (Illumina, San Diego, CA, USA) and sequencing was done with the NextSeq 500 system (Illumina). Read alignment, single nucleotide polymorphism (SNP) detection and maximum-likelihood phylogeny were carried out as described previously [23]. Sequence assembly was performed using SPAdes v. 3.6.0 [24].
Salmonella Typhi isolates were categorized as belonging to haplotype H58 based on the presence of the H58 specific single SNP (T at nucleotide 252 on the gene glpA corresponding to STY2513 from GenBank accession no. AL513382, Salmonella Typhi CT18) [25]. Genotyphi (https://github.com/katholt/genotyphi) was also used to classify Salmonella Typhi [26]. Salmonella Paratyphi A isolates were categorized as belonging to clade C5 (the dominant clade in Cambodia) based on the presence of the C5-specific SNP (G to A at position 2 381 607 within the SPA_RS11495 gene) [23]. The presence of antibiotic resistance genes was determined with ResFinder version 2.1 (https://cge.cbs.dtu.dk/services/ResFinder/) [27] and plasmids with PlasmidFinder version 1.3 (https://cge.cbs.dtu.dk/services/PlasmidFinder/) and pMLST version 1.4 (https://cge.cbs.dtu.dk/services/pMLST-1.4/) [27, 28]. The presence of mutations in the quinolone-resistance determining region of the DNA gyrase and topoisomerase IV genes (gyrA, gyrB, parC and parE) was assessed by the visual examination of sequences.
Data registration, statistical analysis
Demographic, clinical and microbiological data were entered encoded into an Excel database that was created for this study (Microsoft, Redmond, WA, USA). The code referring to the patient identity was only known by two medical doctors. Access to the database was restricted to these two medical doctors and patient identifiers were removed prior to analysis.
Only the first isolate and associated clinical data for each unique patient was considered. Isolates recovered from a second blood culture drawn within two weeks after the initial one were considered as duplicates, whereas isolates recovered from a repeat blood culture more than two weeks after the initial one were considered as recurrences (either relapse or repeat infections). Statistical analysis was done with Stata 12 (Stata Corp., College Station, TX, USA). Continuous variables are described by a median and interquartile range (IQR). Comparisons between Salmonella Typhi and Salmonella Paratyphi were performed using a Mann-Whitney U test for continuous values and a Chi square test or Fisher exact test for proportions. A p-value of < 0.05 was considered significant.
Ethics
The study was conducted according to the principles expressed in the Declaration of Helsinki and involves use of information that was previously collected in the course of routine care. Ethical approval for the Microbiological Surveillance Study was granted by the Institutional Review Board of the ITM, the Ethics Committee of Antwerp University, and the National Ethics Committee for Health Research in Cambodia. This study and approval includes retrospective review of demographic and clinical data which are part of routine clinical history taking as recorded in the clinical chart.
Results
Surveillance of invasive Salmonella infections
Between 1 January 2008 and 31 December 2015 193 Salmonella Paratyphi A isolates were retrieved from 190 patients and 65 Salmonella Typhi isolates from 64 patients. There were no Salmonella Paratyphi B or C isolates; sixty-two non-typhoidal Salmonella isolates were retrieved from 49 patients. The combined annual proportion of Salmonella Typhi and Salmonella Paratyphi A among all clinically significant organisms varied between 2.8% (2008) and 31.7% (2014).
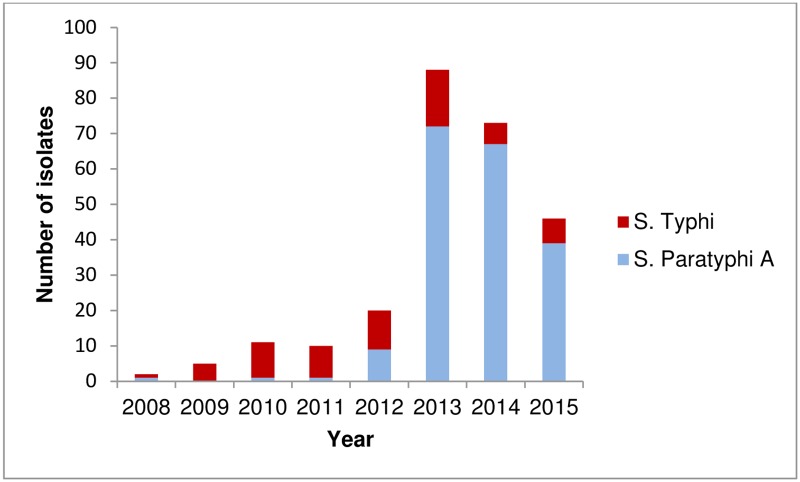
During 2008–2012, enteric fever was caused mostly by Salmonella Typhi (35 cases) and only 12 cases of Salmonella Paratyphi A infection were identified (Fig 1). In 2013 however, a sharp increase in the number of Salmonella Paratyphi A cases was observed with a total of 72 unique cases. In 2014 and 2015, the absolute annual number of Salmonella Paratyphi A cases decreased, but remained higher than for the period preceding 2013. During this period, the number of Salmonella Typhi cases remained relatively stable.
Fig 1. Number of unique confirmed blood-culture Salmonella enterica serovar Paratyphi A and Salmonella enterica serovar Typhi isolates at the Sihanouk Hospital Center of HOPE, Phnom Penh, Cambodia, 2008–2015.
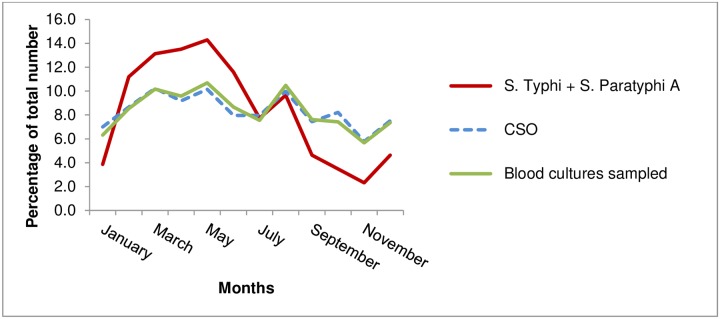
The majority of Salmonella Paratyphi A (64.7%; 123/190) and Salmonella Typhi infections (59.4%; 38/64) cases occurred during the dry season (months November—April) while there was an overall decreasing trend during the rainy season (months June-October) (Fig 2). Compared to the monthly percentage of total blood cultures sampled and clinically significant organisms found, the monthly combined percentage of Salmonella Typhi and Salmonella Paratyphi A was higher during the hot and dry season (March—May) and lower during the rainy season (June-October).
Fig 2. Cumulative monthly percentages of total number of blood culture bottles sampled, clinically significant organisms (CSO) and Salmonella Paratyphi A and Salmonella Typhi isolated from blood cultures at the Sihanouk Hospital Center of HOPE, Phnom Penh, Cambodia.
Recurrent infections
There were four cases of recurrent infections (37–48 days interval between first and recurrent infection), three with Salmonella Paratyphi A and one with Salmonella Typhi; whole genome sequence data was available for three of the four pairs. SNP analysis of the paired isolates revealed that they differed by only two or three SNPs and the isolate pairs formed discrete clusters within the trees (S1 and S2 Figs).
Epidemiological and clinical features
Available epidemiological, clinical and radiographic findings of all enteric fever patients are listed in Table 1.
Table 1. Epidemiological, clinical and radiographic findings for patients presenting with enteric fever, according to pathogen (Salmonella Typhi versus Salmonella Paratyphi A).
| N | Salmonella Typhi | N | Salmonella Paratyphi A | Significant p-values | |
|---|---|---|---|---|---|
| Epidemiology (numbers (% of total)) | |||||
| Male | 64 | 32 (50) | 190 | 93 (48.9) | |
| Phnom Penh resident | 63 | 38 (60.3) | 189 | 154 (81.5) | 0.001 |
| Hospitalization | 63 | 11 (17.5) | 188 | 19 (10.1) | |
| Previous antibiotic exposurea | |||||
| Epidemiology (median (IQR)) | |||||
| Median age, years | 64 | 23 (18–30) | 190 | 26 (22–31) | 0.015 |
| Median duration of symptoms, days | 40 | 4 (3–7) | 112 | 4 (3–7) | |
| Median duration of hospitalization, days | 10b | 5.5 (3–6) | 19 | 4 (2–5) | |
| Symptom and signs (numbers (% of total)) | |||||
| Fever | 48 | 48 (100) | 182 | 181 (99.5) | |
| Abdominal pain | 48 | 30 (62.5) | 182 | 113 (62.1) | |
| Diarrhea | 48 | 12 (25.0) | 182 | 40 (22.0) | |
| Constipation | 48 | 4 (8.3) | 182 | 10 (5.5) | |
| Vomiting | 48 | 11 (22.9) | 182 | 27 (14.8) | |
| Headache | 43 | 24 (55.8) | 178 | 114 (64.0) | |
| Cough | 40 | 15 (37.5) | 168 | 46 (27.4) | |
| Physical findings (median (IQR)) | |||||
| Temperature, °C | 46 | 38.4 (37.3–39.0) | 179 | 38.0 (37.0–38.8) | |
| Pulse, beats/min. | 46 | 106 (95–119) | 179 | 105 (91–117) | |
| Systolic blood pressure, mm Hg | 46 | 107 (100–117) | 179 | 113 (103–123) | 0.036 |
| Diastolic blood pressure, mm Hg | 46 | 67.5 (60–77) | 179 | 70 (64–78) | |
| Physical findings (numbers (% of total)) | |||||
| Fever (Temperature >38°C) | 46 | 26 (56.5) | 179 | 88 (49.2) | |
| Tachycardia (Pulse >100 beats/min)c | 46 | 27 (58.7) | 179 | 109 (60.9) | |
| Abdominal tenderness | 39 | 15 (38.5) | 164 | 56 (34.1) | |
| Radiographic signs | |||||
| Ultrasound: Splenomegaly | 11 | 3 (27.3) | 38 | 5 (13.2) | |
| Ultrasound: Hepatomegaly | 11 | 0 (0) | 38 | 6 (15.8) | |
| Chest X-ray: Normal | 10 | 9 (90.0) | 32 | 31 (96.9) | |
For statistical analysis the X2 test or Fisher exact test was used for proportions and the Mann-Whitney U test for continuous variables. Only statistically significant p-values are listed
IQR = Interquartile range
a = Two weeks before presentation
b = Data on duration of hospitalization was missing for one Salmonella Typhi patient
c = For children aged 5–12 years (N = 4) tachycardia was defined as >120 beats/min
Eleven out of 189 (5.8%) Salmonella Paratyphi A patients had a known co-morbidity, i.e. HIV (n = 7), Diabetes Mellitus type 1 or 2 (n = 3) and leukemia (n = 1). Of 63 Salmonella Typhi patients also nine (14.3%) had a known co-morbidity, i.e. HIV (n = 7) and Diabetes Mellitus type 1 (n = 2). All but one HIV patient were on antiretroviral therapy at time of presentation. There were 11 patients known to have hepatitis B-positive serology (8 Salmonella Paratyphi A, 3 Salmonella Typhi). At least five could be classified as inactive chronic carriers and two had signs of chronic liver disease on ultrasound.
Patients with a Salmonella Paratyphi A infection were more likely to be living in Phnom Penh compared to Salmonella Typhi patients (81.5% (154/189) vs. 60.3% (38/63); p = 0.001) but the median duration of illness at presentation was the same (four days).
Eleven (17.5%) patients with typhoid fever and 19 (10.1%) with paratyphoid fever were hospitalized, with no statistically significant difference between the two groups (p = 0.119). Reasons for hospitalization included sepsis, persistent fever despite antibiotic therapy, dizziness due to low blood pressure, suspicion of dengue hemorrhagic fever (thrombocytopenia) and dysregulated diabetes mellitus. There were no deaths nor complications noted.
The most frequently reported symptoms in all enteric fever patients together were fever in 229 patients (99.6%), headache in 138 (62.4%) and abdominal pain in 143 (62,2%). Presence or absence of classic enteric fever signs such as a coated tongue and rose spots were infrequently mentioned in clinical files and therefore not evaluated.
Despite the non-specific symptoms, physicians noted typhoid fever in their differential diagnosis upon admission in 67.7% (136/201) of the cases.
There were no statistically significant differences in individual symptoms between typhoid and paratyphoid fever patients, but patients infected with Salmonella Typhi had a slightly but significantly lower median systolic blood pressure (107 mm Hg vs. 113 mm Hg; p = 0.036).
Treatment was not systematically recorded for all patients as many were lost to follow-up. Various antimicrobial regimens were used, but ceftriaxone (2g I.V., once daily) was given most frequently as empirical treatment and as monotherapy, normally for 10–14 days. In case of de-escalation to oral antibiotics, this concerned mostly ciprofloxacin (500 mg, twice daily) and next amoxicillin/clavulanate (625 mg, three times a day). In case of persistent fever while awaiting blood culture results, amikacin was occasionally added to ceftriaxone.
Laboratory parameters
The laboratory parameters of enteric fever patients on admission are summarized in Table 2. Common laboratory abnormalities for enteric fever patients included moderately risen transaminase levels in 133 patients (70.7%), an elevated CRP in 53 patients (94.6%) and eosinopenia in 49 patients (90.7%). Hematological abnormalities were uncommon; the leukocyte count was normal in 88.1% of all patients. Compared to Salmonella Paratyphi A infected patients, Salmonella Typhi patients had slightly but significantly higher median values for leukocytes (6.8 x 109/L vs. 6.3 x 109/L; p = 0.035) and C-reactive protein (CRP) (47.0 mg/L vs. 36 mg/L; p = 0.034), with more presence of leukocytosis (10.0% vs. 2.2% p = 0.015). Salmonella Paratyphi A infection was associated with a higher monocyte count compared to Salmonella Typhi (0.48 x 109/L vs. 0.33 x 109/L), but this difference did not reach statistical significance (p = 0.069).
Table 2. Laboratory parameters at presentation for enteric fever patients attending the Sihanouk Hospital Center of HOPE in Phnom Penh, Cambodia.
| N | Salmonella Typhi | N | Salmonella Paratyphi A | Significant p-values | |
|---|---|---|---|---|---|
| Values expressed as median (IQR) | |||||
| Hemoglobin, g/L | 60 | 13.1 (12.1–14.2) | 186 | 13.2 (12.2–14.5) | |
| Leukocyte count, cells x 109/L | 60 | 6.8 (5.6–8.8) | 186 | 6.3 (5.1–7.6) | 0.035 |
| Monocyte count, cells x 109/L | 12 | 0.33 (0.28–0.55) | 41 | 0.48 (0.37–0.72) | |
| Eosinophil count, cells x 109/L | 12 | 0 (0–0.01) | 41 | 0 (0–0.03) | |
| Basophil count, cells x 109/L | 12 | 0.02 (0.01–0.03) | 40b | 0.01 (0.01–0.02) | |
| Thrombocyte count, cells x 109/L | 52 | 205 (166–251) | 185 | 205 (165–247) | |
| CRP, mg/L | 13 | 47.0 (38.6–79.2) | 43 | 36.0 (25.2–54.7) | 0.034 |
| AST, units/L | 40 | 47.0 (33.5–81.5) | 148 | 43.0 (30.5–72.5) | |
| ALT, units/L | 40 | 38.5 (26–88) | 148 | 39.0 (24.5–72.5) | |
| Sodium mmol/L | 33 | 132 (130–135) | 105 | 135 (132–137) | 0.024 |
| Potassium mmol/L | 33 | 3.9 (3.7–4.2) | 105 | 3.9 (3.6–4.1) | |
| Chloride mmol/L | 32 | 99 (96–101) | 105 | 100 (97–102) | |
| Values expressed as numbers (% of total) | |||||
| Leukocytosis (leukocytes >11 x109/L) | 60 | 6 (10) | 186 | 4 (2.2) | 0.015 |
| Leukopenia (leukocytes <4x109/L) | 60 | 4 (6.7) | 186 | 17 (9.1) | |
| Eosinopenia (eosinophils <0.08x109/L) | 13 | 13 (100) | 41 | 36 (87.8) | |
| Thrombocytopenia (<150x109L) | 52 | 11 (21.2) | 185 | 31 (16.8) | |
| Elevated AST and/or ALT (units/L)a | 40 | 28 (70.0) | 148 | 105 (70.9) | |
For statistical analysis the X2 test or Fisher exact test was used for proportions and the Mann-Whitney U test for continuous variables. Only statistically significant p-values are listed
ALT = Alanine transaminase
AST = Aspartate transaminase
CRP = C-reactive protein
IQR = Interquartile range
a = ≥ 18 years: Males: AST ≥35U/L and ALT ≥45U/L; Females: AST ≥31U/L and ALT ≥34U/L; 13–17 years: AST ≥42U/L and ALT ≥45U/L; 7–12 years: AST ≥48U/L and ALT ≥44U/L
b = Basophil count not measurable in 1 patient
Microbiological features
Both anaerobic and aerobic blood cultures showed signs of growth after a median of two days (IQR 2–3) for all enteric fever patients. In 221 enteric fever patients a pair of one aerobic bottle and one anaerobic bottle was sampled, and in 180 of those cases (81.4%) both bottles grew. In the other cases (growth in only a single bottle), it was the aerobic bottle which grew in nearly two-thirds (65.9%; 27/41) of pairs.
Reported antibiotic exposure in the two weeks before blood culture sampling was not associated with a difference in the median days to growth for both aerobic bottles and anaerobic bottles.
In total, 183 out of 190 (96.3%) unique Salmonella Paratyphi A isolates and all 64 unique Salmonella Typhi isolates recovered during the study period were available for antibiotic susceptibility testing (Table 3).
Table 3. Antibiotic resistance rates of 64 Salmonella Typhi isolates and 183 Salmonella Paratyphi A isolates, Sihanouk Hospital Center of HOPE, Phnom Penh, Cambodia.
| 2008–2012 | 2013–2015 | 2008–2012 | 2013–2015 | |
|---|---|---|---|---|
|
Salmonella Typhi (N = 35) % resistant |
Salmonella Typhi (N = 29) % resistant |
Salmonella Paratyphi A (N = 12) % resistant |
Salmonella Paratyphi A (N = 171) % resistant |
|
| MDR | 62.9 | 17.2 | 0 | 0 |
| Nalidixic acid | 97.1a | 93.1 | 0 | 12.3 |
| DCS | 100 | 93.1 | 0 | 12.3 |
| MDR + DCS | 62.9 | 17.2 | 0 | 0 |
| Tetracycline | 62.9 | 24.1 | 0 | 0 |
| Gentamicin | 0 | 0 | 0 | 0 |
| Ceftriaxone | 0 | 0 | 0 | 0 |
| Azithromycin | 0 | 0 | 0 | 0 |
| Meropenem | 0 | 0 | 0 | 0 |
| Ertapenem | 0 | 0 | 0 | 0 |
MDR = Multidrug resistance
DCS = Decreased ciprofloxacin susceptibility. Defined as MIC value ≥0.12 mg/L and ≤0.5 mg/L
a = The remaining isolate displayed intermediate resistance to nalidixic acid
For Salmonella Typhi, there was a significant decrease (p = <0.001) in the proportion of isolates that were MDR over the 8-year period (62.9% vs. 17.2%) while decreased susceptibility to ciprofloxacin remained at nearly 100% (96.9%; 62/64) during the entire period. For Salmonella Paratyphi A the emergence of DCS was noted as of 2015 (S1 Table). In this year 19 out of 36 unique isolates (52.8%) showed DCS.
Overall, Salmonella Typhi was significantly more likely to be MDR and more likely to display DCS than was Salmonella Paratyphi A (42.2% vs. 0.0%; p = <0.001 and 96.9% vs. 11.5%; p = <0.001 respectively).
Of note, for both serovars no ciprofloxacin resistance was reported and the presence of nalidixic acid and pefloxacin resistance were excellent predictors of DCS except in case of one isolate with a single gyrB mutation (Table 4 and S1 Table). Furthermore, no resistance against third-generation cephalosporins, carbapenems or azithromycin was found.
Table 4. Mutations found in the DNA gyrase genes in 62 Salmonella enterica serovar Typhi isolates and 21 Salmonella enterica serovar Paratyphi A isolates.
| Serovar | Resistance phenotype | MIC-value ciprofloxacin (mg/L) | gyrA | gyrB |
|---|---|---|---|---|
|
Salmonella Typhi (n = 62) |
NaR, PefR, DCS | 0.19–0.38 | Ser83Phe (n = 60) | - |
| NaR, PefR, DCS | 0.38 | Asp87Asn + Ser83Phe (n = 1) | - | |
| NaI, PefR, DCS | 0.094 | - | Ser464Phe (n = 1) | |
|
Salmonella Paratyphi A (n = 21) |
NaR, PefR, DCS | 0.25–0.38 | Ser83Phe (n = 20) | - |
| NaR, PefR, DCS | 0.125 | Asp87Gly (n = 1) | - |
NaR = Nalidixic acid resistant. NaI = Nalidixic acid intermediate susceptible. PefR = Pefloxacin resistant. DCS = Decreased Ciprofloxacin Susceptibility. MIC = Minimum Inhibitory Concentration.
Molecular analysis of subtypes and resistance mechanisms
All but one of the Salmonella Typhi isolates were confirmed to be of the H58 haplotype (recently reclassified as the 4.3.1 genotype), with only 27 out of 63 (42.9%) unique H58 isolates displaying MDR but 98.4% (62/63) displaying DCS. The only non-H58 isolate belonged to genotype 3.2.1 and was pan-susceptible. All of the Salmonella Paratyphi A isolates with DCS belonged to the C5 clade.
Most frequently observed in both Salmonella Typhi and Salmonella Paratyphi A with DCS was the gyrA mutation leading to serine-to-phenylalanine substitution at codon 83 (Ser83Phe) (Table 4). These isolates showed DCS and resistance to pefloxacin and nalidixic acid. There was one Salmonella Typhi isolate with a double gyrA mutation and one with a gyrB mutation. The latter mutation (serine-to-phenylalanine substitution at codon 464 (Ser464Phe)) was associated with intermediate susceptibility to nalidixic acid and DCS. No mutations in ParC or ParE were observed.
Various resistance genes (blaTEM-1B, catA1, sul1, sul2, dfrA7, tet(B), strAB) were detected in MDR Salmonella Typhi which were associated with the presence of an incHI1 PST6 plasmid (S1 Table).
Discussion
The present study describes the clinical and microbiological aspects of enteric fever in a large group of patients attending an adult hospital in Cambodia with several interesting results.
First, and in line with some other Asian countries, a clear increase was noted in the proportion of Salmonella Paratyphi A infections. This can largely be explained by a community outbreak which occurred in 2013, but the number of cases has remained high also in succeeding years. A recent genetic study on the Cambodian Salmonella Paratyphi A outbreak isolates showed that these isolates belong to a clade that has been circulating in the South-East Asian region already for decades [23]. Further, no indications were found for significant genetic changes within the Cambodian isolates suggesting that environmental and/or behavioral factors are more likely to play a role.
Patients with paratyphoid fever were significantly more likely than typhoid fever patients to be residents of Phnom Penh, which suggests that exposure to the bacterium is more common in the city. Previous studies from Nepal and Indonesia have linked paratyphoid fever to recent immigration into the capital and consumption of street food [29, 30]. Increased dependency on street food has been linked to urbanization, and Phnom Penh is rapidly expanding.
As part of urban expansion, some of the city’s peri-urban lakes have been filled with sand to reclaim land [31]. These lakes are estimated to receive 80% of the city’s (untreated) waste water and act as a natural sewage treatment through aquatic cultivation of vegetables of which some are consumed raw [32]. Reductions in the size of these lakes could have led to higher concentrations of fecal sludge and bacteria in the remaining water and increased flooding in the city [33].
The majority of enteric fever cases occurred in the dry season. During this season more vegetables are harvested and it is also known for an increased availability of snails and clams (bivalve shellfish), due to low water levels in rivers. Shellfish are known to be able to concentrate micro-organisms from water. They are popular snacks which are dried outside rather than boiled during the dry season. In addition, this season coincides with the two most important festivities of the year, the Chinese and Khmer New Year which are associated with increased migration in and out of the city and longer storage duration of food. Last, high daily temperatures may lead to more indiscriminate intake of water and ice cubes. These factors are currently being explored more in-depth.
Second, based on individual symptoms at presentation, infections caused by Salmonella Typhi vs. Salmonella Paratyphi A were similar and indistinguishable, which is in line with other studies from Asia [3, 4]. For both serovars, the median pulse rates at presentation (106 and 105 beats/minute) were high. This has been noted before in children with enteric fever [34].
No complications or deaths occurred which could be ascribed to a prompt start of antibiotic therapy and an early presentation. The latter can also explain the absence of relative bradycardia and the low rate of diarrhea observed which are typically seen in later stages of the disease [35].
In general, laboratory abnormalities were non-specific and leukocyte counts were normal in 87.4% of all enteric fever patients which was also found by others [36]. Higher leukocyte counts and CRP values were found among Salmonella Typhi-infected patients, suggesting a more severe infection. This is in line with a recent human challenge study, which found that a challenge with Salmonella Paratyphi A in healthy volunteers resulted in a milder disease profile (high rates of afebrile bacteremia) than that observed following typhoid challenge [37].
Some differences in presentation were noted when comparing these results to travelers infected with the same Salmonella Paratyphi A C5 strain returning from Cambodia to France. In the latter study, the majority of patients did have diarrhea (70.6%) and were hospitalized (86%) [14]. This difference may be due to other waterborne or oral-fecal infections travelers frequently contract and/or less financial constrains related to hospital admission [2]. Clinical presentation in the present study also differed from typhoid fever patients in African countries where higher rates of severe complications and mortality are observed [38]. As the same H58 haplotype of Salmonella Typhi is dominant in Asia and in eastern Africa, differences could perhaps be explained by timely access to health care and adequate treatment as well as to host-related factors including underlying co-morbidities like malnutrition.
It has been suggested that isolates of the H58 lineage and MDR strains in general are associated with increased virulence and pathogenicity [39–41]. Therefore the results regarding the clinical presentation and severity of cases as described here, might not be applicable to areas where other lineages dominate.
As a third observation, antibiotic resistance trends were very different for the two serovars. While 42.2% of the unique Salmonella Typhi isolates displayed MDR, none of the Salmonella Paratyphi A isolates did. DCS was present in nearly all Salmonella Typhi isolates, but only emerged in Salmonella Paratyphi A from 2015. The rapid increase of DCS in Salmonella Paratyphi during that year is of concern as ciprofloxacin is the treatment of choice for uncomplicated enteric fever.
Fourth, although all but one Salmonella Typhi isolates were found to belong to the globally dominant H58 haplotype, more than half were not associated with MDR and the proportion of (plasmid-mediated) MDR Salmonella Typhi significantly decreased during the study period. This trend has previously been noted in India, Nepal and neighboring country Vietnam [42–44], but contrasts with a recent study on Salmonella Typhi isolates from rural Cambodia where 89% of the H58 isolates displayed the MDR phenotype [45]. The re-emergence of susceptibility might result from a lack of antibiotic pressure since fluoroquinolones have become the preferred treatment both in community and hospital settings in Phnom Penh.
No resistance against ceftriaxone nor against azithromycin was observed. However, reports on extended spectrum beta-lactamases (ESBL) positive and azithromycin resistant Salmonella spp. isolates are emerging globally including one from the same hospital on Salmonella enterica serovar Choleraesuis [20, 46, 47] underlining the importance of continued microbiological surveillance.
Last, molecular analysis of isolates from three patients with a recurrent infection showed relapse was more likely than re-infection with isolate pairs differing only 2–3 SNPs which can occur during the period of persistence within the human body and suggests relapse rather than re-infection [48]. Relapse is estimated to occur in around 5–10% of enteric fever cases usually two to three weeks after the resolution of fever [5]. In our study, a blood culture confirmed recurrence was witnessed only in 4 out of 254 cases (1.6%). It is likely that other recurrent infections have been missed, partly due to the different medical systems that co-exist in Cambodia in which patients readily switch from one healthcare provider to another, especially if symptoms persist.
This study has several limitations. First, the hospital-based setting precluded generalization to patients whose symptoms were not severe enough to seek medical care in a hospital or clinic. Second, the study concerned mostly adults and therefore findings might not be equally applicable to a pediatric population. Third, the study was retrospective in nature; not all clinical charts were available for review and clinical record keeping was variable among different clinicians. It was not possible to reliably estimate time to defervescence.
Despite these limitations, this is one of the largest and most comprehensive descriptive studies on Salmonella Paratyphi A infections so far which is relevant given the global increase in Salmonella Paratyphi A infections. The data do not represent one single hospital, but several clinics located in different districts of the city. Some of these clinics have reduced rates for the poor and during the study period all blood cultures were provided for free. This helped to overcome some of the bias associated with a hospital based study.
The high proportion of Salmonella Typhi and Paratyphi A recovered from blood cultures indicated that enteric fever is a very frequent disease in Phnom Penh. While efforts are made to increase the microbiological diagnostic capacity in the country, a rapid test for invasive Salmonella infections would be a welcome tool for fast and reliable diagnosis. It could increase knowledge on the burden of disease in the community and could replace the flawed Widal test that is still frequently used.
As the current Salmonella Typhi vaccine provides no to very little protection against Salmonella Paratyphi A, the development of an effective Salmonella Paratyphi A vaccine should be promoted, pending improved water quality and sanitation [49].
Conclusion
Enteric fever is frequent in Phnom Penh and the proportion of cases due to Salmonella Paratyphi A has increased. Studies to investigate risk factors and possible transmission routes are urgently needed to advise public health interventions. No MDR was observed for Salmonella Paratyphi A but DCS increased rapidly. DCS remained highly prevalent in Salmonella Typhi while MDR rates have declined. Ceftriaxone and azithromycin remain highly active in vitro but continued surveillance is imperative to monitor resistance.
Supporting information
(XLSX)
Maximum Likelihood (ML) tree of 185 Salmonella Paratyphi A genomes including 159 previously published genomes (Kuijpers & Le Hello et al., 2016 [23] and Zhou et al., 2014 [50]). Fifty-six genomes represent isolates collected at the Sihanouk Hospital Center of HOPE, Phnom Penh, Cambodia between 2008–2015. For readability, only the position of the reference genome (Salmonella Paratyphi A ATCC 9150) and the paired isolates are shown. Only clades C1-C5 are indicated. The big arrow indicates the paired isolates (ID 6778 and 6748, 2 SNPs difference; ID 6610 and 6670; 3 SNPs difference).
(PPTX)
Maximum Likelihood (ML) tree of 66 Salmonella Typhi genomes including the Salmonella Typhi CT18 reference genome (AL513382) and 65 genomes of Salmonella Typhi isolates collected at the Sihanouk Hospital Center of HOPE, Phnom Penh, Cambodia between 2008–2015. For readability, only the position of the reference genome (Salmonella Typhi CT18) and of the paired isolates (ID 4764 and 4855, 3 SNPs difference) are indicated (with a big arrow).
(PPTX)
(PDF)
Acknowledgments
We would like to thank Barbara Barbé, Marleen Verlinden and Marjan Peeters for technical assistance at ITM. We would also like to thank all colleagues involved in blood culture surveillance and clinical care at the Sihanouk Hospital Center of HOPE and Laura Brinas, Andrea Alexandru, Maud Vanpeene, Sobhy Wilhame, and Vincent Enouf (Institut Pasteur, Pasteur International Bioresources network (PIBnet), Plateforme de microbiologie mutualisée (P2M), Paris, France, for performing the sequencing experiments.
Data Availability
All relevant data are within the paper and its Supporting Information files. Short-read sequences have been deposited to the European Nucleotide Archive (ENA) (http://www.ebi.ac.uk/ena), under study accession number PRJEB19906 (http://www.ebi.ac.uk/ena/data/view/PRJEB19906).
Funding Statement
The surveillance of bloodstream infections at the Sihanouk Hospital Center of HOPE is supported by the Belgian Directorate of Development Cooperation (DGD) (https://diplomatie.belgium.be/en/policy/development_cooperation/who_we_are/our_organisation/dgd) through project 2.08 of the Third Framework Agreement between the Belgian DGD (Ministry of Development Cooperation) and the Institute of Tropical Medicine (ITM). This study was further supported by the Strategic Network Laboratory Quality Management (LQM) project (http://www.labquality.be/) (Belgian Development Cooperation), the Institut Pasteur and the French government’s Investissement d’Avenir programme, Laboratoire d’Excellence ‘Integrative Biology of Emerging Infectious Diseases’ (http://www.agence-nationale-recherche.fr/?ProjetIA=10-LABX-0062) (grant number ANR-10-LABX-62-IBEID). LMFK is supported by the Flemish Ministry of Sciences (EWI, SOFI project IDIS) (http://www.ewi-vlaanderen.be/en) and received additional travel grants (grant numbers K2.060.16N and K2.065.17N) from the Fund for Scientific Research Flanders (F.W.O.-Vlaanderen, Belgium) (http://www.fwo.be/). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Bhan MK, Bahl R, Bhatnagar S. Typhoid and paratyphoid fever. Lancet. 2005;366(9487):749–62. doi: 10.1016/S0140-6736(05)67181-4 [DOI] [PubMed] [Google Scholar]
- 2.Meltzer E, Sadik C, Schwartz E. Enteric fever in Israeli travelers: a nationwide study. J Travel Med. 2005;12(5):275–81. [DOI] [PubMed] [Google Scholar]
- 3.Maskey AP, Day JN, Phung QT, Thwaites GE, Campbell JI, Zimmerman M, et al. Salmonella enterica serovar Paratyphi A and S. enterica serovar Typhi cause indistinguishable clinical syndromes in Kathmandu, Nepal. Clin Infect Dis. 2006;42(9):1247–53. doi: 10.1086/503033 [DOI] [PubMed] [Google Scholar]
- 4.Vollaard AM, Ali S, Widjaja S, Asten HA, Visser LG, Surjadi C, et al. Identification of typhoid fever and paratyphoid fever cases at presentation in outpatient clinics in Jakarta, Indonesia. Trans R Soc Trop Med Hyg. 2005;99(6):440–50. doi: 10.1016/j.trstmh.2004.09.012 [DOI] [PubMed] [Google Scholar]
- 5.Parry CM, Hien TT, Dougan G, White NJ, Farrar JJ. Typhoid fever. N Engl J Med. 2002;347(22):1770–82. doi: 10.1056/NEJMra020201 [DOI] [PubMed] [Google Scholar]
- 6.Wain J, Kidgell C. The emergence of multidrug resistance to antimicrobial agents for the treatment of typhoid fever. Trans R Soc Trop Med Hyg. 2004;98(7):423–30. doi: 10.1016/j.trstmh.2003.10.015 [DOI] [PubMed] [Google Scholar]
- 7.Medalla F, Sjolund-Karlsson M, Shin S, Harvey E, Joyce K, Theobald L, et al. Ciprofloxacin-resistant Salmonella enterica Serotype Typhi, United States, 1999–2008. Emerg Infect Dis. 2011;17(6):1095–8. doi: 10.3201/eid/1706.100594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chiou CS, Lauderdale TL, Phung DC, Watanabe H, Kuo JC, Wang PJ, et al. Antimicrobial resistance in Salmonella enterica Serovar Typhi isolates from Bangladesh, Indonesia, Taiwan, and Vietnam. Antimicrob Agents Chemother. 2014;58(11):6501–7. doi: 10.1128/AAC.03608-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pokharel BM, Koirala J, Dahal RK, Mishra SK, Khadga PK, Tuladhar NR. Multidrug-resistant and extended-spectrum beta-lactamase (ESBL)-producing Salmonella enterica (serotypes Typhi and Paratyphi A) from blood isolates in Nepal: surveillance of resistance and a search for newer alternatives. Int J Infect Dis. 2006;10(6):434–8. doi: 10.1016/j.ijid.2006.07.001 [DOI] [PubMed] [Google Scholar]
- 10.Hassing RJ, Goessens WH, van Pelt W, Mevius DJ, Stricker BH, Molhoek N, et al. Salmonella subtypes with increased MICs for azithromycin in travelers returned to The Netherlands. Emerg Infect Dis. 2014;20(4):705–8. doi: 10.3201/eid2004.131536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ochiai RL, Wang X, von Seidlein L, Yang J, Bhutta ZA, Bhattacharya SK, et al. Salmonella paratyphi A rates, Asia. Emerg Infect Dis. 2005;11(11):1764–6. doi: 10.3201/eid1111.050168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vlieghe ER, Phe T, De Smet B, Veng HC, Kham C, Lim K, et al. Bloodstream infection among adults in Phnom Penh, Cambodia: key pathogens and resistance patterns. PLoS One. 2013;8(3):e59775 doi: 10.1371/journal.pone.0059775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vlieghe E, Phe T, De Smet B, Veng CH, Kham C, Sar D, et al. Increase in Salmonella enterica serovar Paratyphi A infections in Phnom Penh, Cambodia, January 2011 to August 2013. Euro Surveill. 2013;18(39). [DOI] [PubMed] [Google Scholar]
- 14.Tourdjman M, Le Hello S, Gossner C, Delmas G, Tubiana S, Fabre L, et al. Unusual increase in reported cases of paratyphoid A fever among travellers returning from Cambodia, January to September 2013. Euro Surveill. 2013;18(39). [DOI] [PubMed] [Google Scholar]
- 15.Saitoh T, Morita M, Shimada T, Izumiya H, Kanayama A, Oishi K, et al. Increase in paratyphoid fever cases in Japanese travellers returning from Cambodia in 2013. Epidemiol Infect. 2016;144(3):602–6. doi: 10.1017/S0950268815001648 [DOI] [PubMed] [Google Scholar]
- 16.Judd MC, Grass JE, Mintz ED, Bicknese A, Mahon BE. Salmonella enterica Paratyphi A Infections in Travelers Returning from Cambodia, United States. Emerg Infect Dis. 2015;21(6):1089–91. doi: 10.3201/eid2106.150088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bone RC, Balk RA, Cerra FB, Dellinger RP, Fein AM, Knaus WA, et al. Definitions for sepsis and organ failure and guidelines for the use of innovative therapies in sepsis. The ACCP/SCCM Consensus Conference Committee. American College of Chest Physicians/Society of Critical Care Medicine. Chest. 1992;101(6):1644–55. [DOI] [PubMed] [Google Scholar]
- 18.Seymour CW, Liu VX, Iwashyna TJ, Brunkhorst FM, Rea TD, Scherag A, et al. Assessment of Clinical Criteria for Sepsis: For the Third International Consensus Definitions for Sepsis and Septic Shock (Sepsis-3). JAMA. 2016;315(8):762–74. doi: 10.1001/jama.2016.0288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Phe T VE, Lim K, Veng CH, Thai S, Leng L, Kham C, Jacobs J. Surveillance of bloodstream infection and antibiotic resistance in Phnom Penh, Cambodia (2007–2014). Poster presented at: 17th International Congress on Infectious Diseases; 2016 March 2–5; Hyderabad, India.
- 20.Vlieghe ER, Phe T, De Smet B, Veng CH, Kham C, Bertrand S, et al. Azithromycin and ciprofloxacin resistance in Salmonella bloodstream infections in Cambodian adults. PLoS Negl Trop Dis. 2012;6(12):e1933 doi: 10.1371/journal.pntd.0001933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Clinical and Laboratory Standards Institute (CLSI). Performance standards for antimicrobial susceptibility testing. 26th informational supplement. CLSI Document M100-S26. Wayne, PA: CLSI 2016.
- 22.World Health Organization (WHO). Background document: the diagnosis, treatment and prevention of typhoid fever. 2003. [Google Scholar]
- 23.Kuijpers LMF, Le Hello S, Fawal N, Fabre L, Tourdjman M, Dufour M, Sar D, Kham C, Phe T, Vlieghe E, Bouchier C, Jacobs J, Weill FX. Genomic analysis of Salmonella enterica serotype Paratyphi A during an outbreak in Cambodia, 2013–2015. Microbial Genomics. 2016; doi: 10.1099/mgen.0.000092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 2012;19(5):455–77. doi: 10.1089/cmb.2012.0021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Roumagnac P, Weill FX, Dolecek C, Baker S, Brisse S, Chinh NT, et al. Evolutionary history of Salmonella typhi. Science. 2006;314(5803):1301–4. doi: 10.1126/science.1134933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wong VK, Baker S, Connor TR, Pickard D, Page AJ, Dave J, et al. An extended genotyping framework for Salmonella enterica serovar Typhi, the cause of human typhoid. Nat Commun. 2016;7:12827 doi: 10.1038/ncomms12827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zankari E, Hasman H, Cosentino S, Vestergaard M, Rasmussen S, Lund O, et al. Identification of acquired antimicrobial resistance genes. J Antimicrob Chemother. 2012;67(11):2640–4. doi: 10.1093/jac/dks261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Carattoli A, Zankari E, Garcia-Fernandez A, Voldby Larsen M, Lund O, Villa L, et al. In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob Agents Chemother. 2014;58(7):3895–903. doi: 10.1128/AAC.02412-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vollaard AM, Ali S, van Asten HA, Widjaja S, Visser LG, Surjadi C, et al. Risk factors for typhoid and paratyphoid fever in Jakarta, Indonesia. JAMA. 2004;291(21):2607–15. doi: 10.1001/jama.291.21.2607 [DOI] [PubMed] [Google Scholar]
- 30.Karkey A, Thompson CN, Tran Vu Thieu N, Dongol S, Le Thi Phuong T, Voong Vinh P, et al. Differential epidemiology of Salmonella Typhi and Paratyphi A in Kathmandu, Nepal: a matched case control investigation in a highly endemic enteric fever setting. PLoS Negl Trop Dis. 2013;7(8):e2391 doi: 10.1371/journal.pntd.0002391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Halim H, Muong, V. Capital’s remaining freshwater lake a sinking ship. Phnom Penh Post. 2017 Feb 23. http://www.phnompenhpost.com/post-property/capitals-remaining-freshwater-lake-sinking-ship
- 32.Sar S, Chervier C, Lim P, Warrender C, Warrender GW, Gilbert RG. Seasonal Direct-Use Value of Cheung Ek Peri-Urban Lake, Phnom Penh, Cambodia. Int J Environmental & Rural Development. 2010;1:113–8 [Google Scholar]
- 33.Yeap C. Faecal build-up a threat: study. The Phnom Penh Post. 2012 Apr 25. http://www.phnompenhpost.com/national/faecal-build-threat-study
- 34.Davis TM, Makepeace AE, Dallimore EA, Choo KE. Relative bradycardia is not a feature of enteric fever in children. Clin Infect Dis. 1999;28(3):582–6. doi: 10.1086/515143 [DOI] [PubMed] [Google Scholar]
- 35.Cunha BA. Osler on typhoid fever: differentiating typhoid from typhus and malaria. Infect Dis Clin North Am. 2004;18(1):111–25. doi: 10.1016/S0891-5520(03)00094-1 [DOI] [PubMed] [Google Scholar]
- 36.Caumes E, Ehya N, Nguyen J, Bricaire F. Typhoid and paratyphoid fever: a 10-year retrospective study of 41 cases in a Parisian hospital. J Travel Med. 2001;8(6):293–7. [DOI] [PubMed] [Google Scholar]
- 37.Dobinson HC, Gibani MM, Jones C, Thomaides-Brears HB, Voysey M, Darton TC, et al. Evaluation of the Clinical and Microbiological Response to Salmonella Paratyphi A Infection in the First Paratyphoid Human Challenge Model. Clin Infect Dis. 2017;64(8):1066–73. doi: 10.1093/cid/cix042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Otegbayo JA, Daramola OO, Onyegbutulem HC, Balogun WF, Oguntoye OO. Retrospective analysis of typhoid fever in a tropical tertiary health facility. Trop Gastroenterol. 2002;23(1):9–12. [PubMed] [Google Scholar]
- 39.Wong VK, Baker S, Pickard DJ, Parkhill J, Page AJ, Feasey NA, et al. Phylogeographical analysis of the dominant multidrug-resistant H58 clade of Salmonella Typhi identifies inter- and intracontinental transmission events. Nat Genet. 2015;47(6):632–9. doi: 10.1038/ng.3281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bhutta ZA. Impact of age and drug resistance on mortality in typhoid fever. Arch Dis Child. 1996;75(3):214–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wain J, Pham VB, Ha V, Nguyen NM, To SD, Walsh AL, et al. Quantitation of bacteria in bone marrow from patients with typhoid fever: relationship between counts and clinical features. J Clin Microbiol. 2001;39(4):1571–6. doi: 10.1128/JCM.39.4.1571-1576.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shrestha KL, Pant ND, Bhandari R, Khatri S, Shrestha B, Lekhak B. Re-emergence of the susceptibility of the Salmonella spp. isolated from blood samples to conventional first line antibiotics. Antimicrob Resist Infect Control. 2016;5:22 doi: 10.1186/s13756-016-0121-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Menezes GA, Harish BN, Khan MA, Goessens WH, Hays JP. Antimicrobial resistance trends in blood culture positive Salmonella Typhi isolates from Pondicherry, India, 2005–2009. Clin Microbiol Infect. 2012;18(3):239–45. doi: 10.1111/j.1469-0691.2011.03546.x [DOI] [PubMed] [Google Scholar]
- 44.Le TA, Fabre L, Roumagnac P, Grimont PA, Scavizzi MR, Weill FX. Clonal expansion and microevolution of quinolone-resistant Salmonella enterica serotype typhi in Vietnam from 1996 to 2004. J Clin Microbiol. 2007;45(11):3485–92. doi: 10.1128/JCM.00948-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pham Thanh D, Thompson CN, Rabaa MA, Sona S, Sopheary S, Kumar V, et al. The Molecular and Spatial Epidemiology of Typhoid Fever in Rural Cambodia. PLoS Negl Trop Dis. 2016;10(6):e0004785 doi: 10.1371/journal.pntd.0004785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nair S, Ashton P, Doumith M, Connell S, Painset A, Mwaigwisya S, et al. WGS for surveillance of antimicrobial resistance: a pilot study to detect the prevalence and mechanism of resistance to azithromycin in a UK population of non-typhoidal Salmonella. J Antimicrob Chemother. 2016;71(12):3400–3408. doi: 10.1093/jac/dkw318 [DOI] [PubMed] [Google Scholar]
- 47.Kalonji LM, Post A, Phoba MF, Falay D, Ngbonda D, Muyembe JJ, et al. Invasive Salmonella Infections at Multiple Surveillance Sites in the Democratic Republic of the Congo, 2011–2014. Clin Infect Dis. 2015;61 Suppl 4:S346–53. [DOI] [PubMed] [Google Scholar]
- 48.Okoro CK, Kingsley RA, Quail MA, Kankwatira AM, Feasey NA, Parkhill J, et al. High-resolution single nucleotide polymorphism analysis distinguishes recrudescence and reinfection in recurrent invasive nontyphoidal Salmonella typhimurium disease. Clin Infect Dis. 2012;54(7):955–63. doi: 10.1093/cid/cir1032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Simanjuntak CH, Paleologo FP, Punjabi NH, Darmowigoto R, Soeprawoto, Totosudirjo H, et al. Oral immunisation against typhoid fever in Indonesia with Ty21a vaccine. Lancet. 1991;338(8774):1055–9. [DOI] [PubMed] [Google Scholar]
- 50.Zhou Z, McCann A, Weill FX, Blin C, Nair S, Wain J, et al. Transient Darwinian selection in Salmonella enterica serovar Paratyphi A during 450 years of global spread of enteric fever. Proc Natl Acad Sci U S A. 2014;111(33):12199–204. doi: 10.1073/pnas.1411012111 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(XLSX)
Maximum Likelihood (ML) tree of 185 Salmonella Paratyphi A genomes including 159 previously published genomes (Kuijpers & Le Hello et al., 2016 [23] and Zhou et al., 2014 [50]). Fifty-six genomes represent isolates collected at the Sihanouk Hospital Center of HOPE, Phnom Penh, Cambodia between 2008–2015. For readability, only the position of the reference genome (Salmonella Paratyphi A ATCC 9150) and the paired isolates are shown. Only clades C1-C5 are indicated. The big arrow indicates the paired isolates (ID 6778 and 6748, 2 SNPs difference; ID 6610 and 6670; 3 SNPs difference).
(PPTX)
Maximum Likelihood (ML) tree of 66 Salmonella Typhi genomes including the Salmonella Typhi CT18 reference genome (AL513382) and 65 genomes of Salmonella Typhi isolates collected at the Sihanouk Hospital Center of HOPE, Phnom Penh, Cambodia between 2008–2015. For readability, only the position of the reference genome (Salmonella Typhi CT18) and of the paired isolates (ID 4764 and 4855, 3 SNPs difference) are indicated (with a big arrow).
(PPTX)
(PDF)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files. Short-read sequences have been deposited to the European Nucleotide Archive (ENA) (http://www.ebi.ac.uk/ena), under study accession number PRJEB19906 (http://www.ebi.ac.uk/ena/data/view/PRJEB19906).