Abstract
While the combined antiretroviral therapy has resulted in a significant decrease in HIV-1 related morbidity and mortality, the HIV-1 pandemic has not been substantially averted. To curtail the 2.4 million new infections each year, a prophylactic HIV-1 vaccine is urgently needed. This review first summarizes four major completed clinical efficacy trials of prophylactic HIV-1 vaccine and their outcomes. Next, it discusses several other approaches that have not yet advanced to clinical efficacy trials, but provided valuable insights into vaccine design. Among them, live-attenuated vaccines (LAVs) provided excellent protection in a non-human primate model. However, safety concerns have precluded the current version of LAVs from clinical application. As the major component of this review, two synthetic biology approaches for improving the safety of HIV-1 LAVs through controlling HIV-1 replication are discussed. Particular focus is on a novel approach that uses unnatural amino acid-mediated suppression of amber nonsense codon to generate conditionally replicating HIV-1 variants. The objective is to attract more attention towards this promising research field and to provoke creative designs and innovative utilization of the two control strategies.
Keywords: HIV-1 vaccine, amber suppression, unnatural amino acid, live-attenuated vaccine, virus engineering
Introduction
Human immunodeficiency virus type 1 (HIV-1) has caused one of the greatest global public health crises. It is estimated that the HIV/AIDS pandemic has already claimed 40 million human lives, 35 million people are living with HIV-1 infection and approximately 2.4 million people are newly infected with HIV-1 worldwide each year.1 A safe and effective HIV-1 vaccine is urgently needed to prevent HIV-1 transmission and to curtail the HIV-1 pandemic. Nevertheless, the development of such vaccines may still have a long way to go.
Up to now, four major types of HIV-1 vaccines have been evaluated in clinical Phase IIB or Phase III efficacy trials (Table 1).2,3 The first one is a protein subunit HIV-1 vaccine based on monomeric Env protein. However, this vaccine failed to elicit protective humoral immune responses. To address this issue, the focus of vaccine research shifted to the second vaccine development strategy – elicitation of cellular immune response by using adenovirus type 5 vector encoding gag, pol, and nef genes of HIV-1. This approach, exemplified by the ‘Merck STEP trial’, also failed.4 The third vaccine strategy employs a DNA prime followed by a recombinant adenovirus vector boost, aiming at generating both cellular and humoral responses, but it failed, too. The fourth vaccine strategy uses a canarypox vector prime followed by a protein subunit boost, i.e. the HIV-1 vaccine trial (RV144) conducted in Thailand in 2009.5 This vaccine approach was designed to elicit both cellular and humoral immune responses and to enhance the immune response by several prime and boost. The results showed that a poxvirus-protein prime-boost provided about 31% protection against HIV-1 acquisition.5 While the modest protection may not be significant enough for clinical uses, the landmark RV144 trial implicated that an HIV-1 vaccine was possible, and brought renewed energy to the field. With a successful initial clinical trial, a large clinical Phase III trial (HVTN 702) will be conducted in South Africa to determine whether the regimen is safe, tolerable, and effective at preventing HIV-1 infection.
Table 1. Summary of HIV-1 vaccine strategies.
| Vaccine strategies | Description | Outcome |
|---|---|---|
| Completed clinical efficacy trials | ||
| Protein subunits | Express bivalent HIV-1 Env gp120 | No efficacy7–10 |
| Recombinant adenovirus vector | Express HIV-1 Gag/Pol/Nef | No efficacy, increased risk11–15 |
| DNA Prime-Adenovirus vector boost | DNA prime expressing HIV-1 Gag, Pol, Nef, Env, boost with adenovirus vector expressing Gag, Pol, Env | No efficacy17–21 |
| Canarypox vector prime and protein subunit boost | Canarypox vector expressing Env, Gag, Pro, boost with Env gp120 | 31% protection5,22–24 |
| Representative strategies undergoing clinical trials | ||
| Protein subunits with improved immunogenic properties | Employ soluble Env gp140 | Elicit moderate systemic and mucosal antibodies25,26 |
| Other immunogens | Use peptides and lipopeptides | Elicit specific T-cell responses27,28 |
| Broadly neutralizing antibodies | Generate or passively administer broadly neutralizing antibodies | Provide complete protection in non-human primates29–33 |
| Novel strategies by employing synthetic biology tools | ||
| Doxycycline-dependent HIV-1 | Doxycycline-dependent expression of HIV-1 proteome | Observe protection in SIV-rtTA vaccinated animals56–64 |
| Amber suppression-dependent HIV-1 (single-cycle) | Unnatural amino acid-mediated translation of HIV-1 proteome using exogenously provided aaRS-tRNACUA pair | Single-cycle replication of HIV-1 in vitro65 |
| Amber suppression-dependent HIV-1 (multi-cycle) | Unnatural amino acid-mediated translation of HIV-1 proteome using genomic copy of aaRS-tRNACUA pair | Multi-cycle replication of HIV-1 in vitroa |
From a manuscript that is currently under revision.
The following discussions first summarize the lessons from these vaccine trials and then state-of-art HIV-1 live-attenuated vaccines (LAVs) are discussed. Among all the HIV-1 vaccine modalities developed thus far, HIV-1 LAVs, such as nef-deleted LAV, confer the best protection against HIV-1 acquisition in simian immunodeficiency virus (SIV)/rhesus macaque model of HIV-1 infection.6 However, owing to safety concerns, current versions of HIV-1 LAVs cannot enter clinical trials. Given their strong protective efficacy in animal models, HIV-1 LAVs are still of great interest. This review mainly discusses strategies aiming to improve the safety of HIV-1 LAVs, particularly the strategy based on controlling HIV-1 replication through the unnatural amino acid-mediated suppression of amber nonsense codon.
Four HIV-1 Vaccine Strategies from Completed Phase IIB or III Clinical Trials
HIV-1 vaccine based on protein subunits
When researchers started the development of HIV-1 vaccine in the late 1980s, vaccine paradigms were brought from successful vaccine strategies against other pathogens. Several clinical trials focused on multiple recombinant envelope proteins derived from different HIV-1 strains. VAX004 and VAX003 employed recombinant bivalent subtype B/B and B/E GP120 and went as far as Phase III clinical trials.7,8 These early approaches were able to successfully induce binding antibodies, but had limited success in eliciting neutralizing antibodies against HIV-1. In addition, the antibodies produced lacked the breadth of neutralization to other HIV-1 strains.9,10 The failure to induce neutralizing antibody responses led to poor or even no efficacy of these vaccines in early trials. With the goal of developing an efficacious HIV-1 vaccine, alternative strategies were pursued.
HIV-1 vaccine based on recombinant adenovirus vectors
In the 2000s, a vaccine strategy for eliciting cytotoxic T lymphocytes (CTLs) response was emphasized. The rationale was that CTL response would be effective against different HIV-1 strains by targeting the conserved region of different HIV-1. Thus eliciting CTL response became the major trend of vaccine design afterwards. In this strategy, HIV-1 genes were inserted into viral vectors in order to deliver these antigens into MHC class I antigen-presenting route. In the initial clinical trial (HVTN502 or STEP trial), gag, pol, and/or nef genes of HIV-1 subtype B were used as antigens and inserted into a recombinant adeno 5 (Ad5) vector. This vaccine contained no Env antigen, and the main focus was on inducing the CTL response in order to lyse infected cells post viral entry. Although the vaccine did not replicate well in vivo,11 it conferred effective immunogenicity and was capable of reducing viremia after infection, especially in the non-human primate study.12 However, the human clinical trials failed to elicit protection, instead enhanced virus acquisition was observed in comparison with the control group.11,13,14 One plausible explanation is that the pre-existing immunity to Ad5 increased the availability of HIV-1 target cells at the portal of virus entry.15 Another possible reason is that the CTL responses targeted the variable regions rather than the conserved regions, even though durable and considerable CTL responses recognizing both HIV-1 clade B and C were obtained.16
HIV-1 vaccine based on DNA prime and adenovirus vector boost
To overcome the breadth issue of CTL responses and to combine the humoral immunity responses, a third strategy was developed. It employed HIV-1 DNA prime (envA, envB, envC, gagB, polB, nefB) and boosted by a recombinant Ad5 vector harboring envA, envB, envC, and gag-polB. This novel prime-boost strategy was demonstrated to be protective against the low-dose challenge in non-human primate study.17 However, the human clinical trials, including HVTN204 and HVTN505, showed no efficacy, no decrease in viral load, and little neutralizing antibody activities.18,19 Certain data indicated that most of the binding antibodies were cross-reactive to an E. coli antigen,20 which may explain why this strategy was unsuccessful. In addition, some other studies indicated that the failure might be caused by a viral escape mechanism derived from neutralizing antibody resistance.21
Canarypox vector prime and protein subunit boost HIV-1 vaccine
A fourth vaccine strategy was developed between 2004 and 2009, which employed the same prime and boost concept as that of strategy three. This vaccine used a canarypox vector for priming and viral proteins for boosting. By employing this strategy, the RV144 Thai HIV-1 vaccine trial (referred to as Thai Trial vaccine hereafter) showed modest but significant protection against HIV-1 acquisition.5 This HIV-1 vaccine employed ALVAC expressing Env from HIV-1 clade E and Gag and Pol from HIV-1 clade B as the prime, followed by AIDSVAX GP120 from clade B and E as the boost and alum as the adjuvant. For the first time in the history of HIV-1 vaccine development, the Thai Trial vaccine demonstrated that a vaccine could be possible to protect people from HIV-1 infection. The Thai Trial vaccine had 60.5% efficacy after one year and had more than 31% efficacy three and half years post vaccination. The RV144 Thai Trial is one of the most visible achievements so far in HIV-1 vaccine development. All other HIV-1 vaccines that entered clinical efficacy trials either ended up lacking efficacy or even enhanced HIV-1 transmission. Currently, the protection of the Thai Trial vaccine is correlated with antibodies of the V1V2 region of gp120 (in particular the IgG1 and IgG3 subclass)-mediated antibody-dependent cell-mediated cytotoxicity (ADCC).22 However, the exact protective mechanisms of Thai Trial vaccine are still not fully understood.
Although the level of protection elicited by Thai Trial vaccine is modest and not sufficient for clinical use, it laid the foundation for further improvement of its protection. Based on the RV144 Thai Trial vaccine, a few other HIV-1 vaccine forms, such as HVTN097, HVTN100, and HVTN702,23,24 were constructed and examined. A better understand of the underlying mechanisms of Thai Trial vaccine-induced protection is therefore critical for further improving its efficacy. With the lessons learned from above clinical trials and the identification and characterization of HIV-1 broadly neutralizing antibodies, designs of novel and efficacious HIV vaccines are expected.
Other strategies
Beyond the above four clinically evaluated concepts, great efforts have been continuously made to develop a range of different approaches (Table 1), which could probably shed light on the development of preventive HIV-1 vaccines. Among them, protein subunits that exhibit improved immunogenic properties are of great interests. For example, Env gp140, which represents a better mimicry of the native trimeric insoluble Env gp160 than the traditional Env gp120, was able to elicit moderate systemic and mucosal antibodies in clinical trials.25,26 In addition to protein subunits, other potential immunogens, such as peptide and lipopeptides, have been proven to elicit HIV-1 specific CD8 and CD4 T-cell responses in clinical trials.27,28 To generate protective antibodies against diverse HIV-1 strains, mosaic immunogens have been designed by in silico tools that identify and optimize the coverage of global HIV-1 epitopes.29,30 In non-human primates, expanded cellular and humoral immune responses have been observed.29,30 The generation of HIV-1 immunogens that can elicit broadly neutralizing antibodies (bNAbs) is also more promising than ever before. Several approaches have been pursued to elicit bNAbs. One approach aims to design immunogens that could better mimic bNAb epitopes.31,32 Another approach uses B-cell lineage vaccine strategy to design specific immunogens that target the desired precursors of bNAb-producing cells, allowing the maturation of B-cells through uncommon pathways.33 In a third approach, passive administration of bNAbs, such as VRC01, has provided complete protection in rhesus macaques34 and proven to be safe in human.35
Live-attenuated vaccines (LAVs)
By mimicking naturally occurring infections, LAVs have successfully prevented a series of viral diseases, such as measles, mumps, yellow fever, and chickenpox by eliciting broad and long-lasting protective immune responses.36 Researchers have developed a number of SIV based live-attenuated vaccines.6,37 The attenuation is generally achieved by the deletion of accessory gene(s) from the viral genome, either individually or in combination.38–41 The majority of monkeys that are vaccinated with such deletion mutants of SIV are able to efficiently protect against pathogenic SIV challenge. One notable example was the nef-deleted SIV mutants, which provided the vaccinated monkeys with a strong protection (the average protection rate was 95% versus the average of all the other type of vaccines 7%) from homologous and heterologous SIV challenge.42,43 Although these nef-deleted SIV mutants could protect some vaccinated monkeys from challenge, they led to the establishment of a life-long and persistent viral infection and caused AIDS in some animals especially the infant – monkeys.44–46 Moreover, restoration of deletion in nef has been revealed by genetic analysis. This raised significant safety concerns that the LAVs could revert back into virulent forms and cause dis ease over time in vaccinated animals.47–49
Multiple deletions in both accessory genes and regulatory elements on the virus genome can be used to increase vaccine safety by decreasing viral replication capacity, but often led to the loss of vaccine efficacy.42 In fact, a reported evidence showed an inverse correlation between the attenuation of virus replication capacity and the degree of protection.42 In addition, an HIV-1 variant with deletions in the accessory regulatory protein-encoding genes, including vpr, nef , and LTR sequences, could still regain substantial replication capacity through viral evolution in long-term cell culture infections.50
The above results highlight the genetic instability and evolutionary capacity of attenuated SIV/HIV-1 strains. An uncontrolled replication of attenuated virus can lead it to regain virulence, which poses a serious safety risk for any future experimentation with HIV-1 LAVs in humans. As an alternative strategy, a virus that can execute only a single round of replication has been used as a vaccine.51–54 However, because of the limited replication, such a single-cycle virus vaccine is less potent and does not provide the necessary duration for the induction of protective immunity against HIV-1. Therefore, novel strategies are needed to improve the safety of HIV-1 LAVs. With unsuccessful attempts based on other vaccine modalities,55 HIV-1 LAVs represent a promising approach if the safety concerns can be resolved.
Development of HIV-1 Vaccine Using Conditionally Replicating Virus
Research efforts have been made to construct conditionally replicating HIV-1 or SIV variants.56–65 One representative example is the use of doxycycline (dox)-inducible gene expression system (the Tet-On system66), which allows the virus to replicate in the presence of dox (Table 1).61 The other strategy to control HIV-1 replication entails the manipulation of essential HIV-1 protein biosynthesis through unnatural amino acid (unAA)-mediated suppression of genome-encoded amber nonsense codon (Table 1).
Doxycycline-dependent HIV-1
Bekhout et al. constructed a number of HIV-rtTA and SIV-rtTA variants (Fig. 1(A)) in which the transcription activation can be switched on and off using a doxycycline (dox)-inducible gene expression system.62 In their constructs, the natural Tat-TAR transcription control element was replaced with the dox-inducible Tet-On gene expression system.
Figure 1.
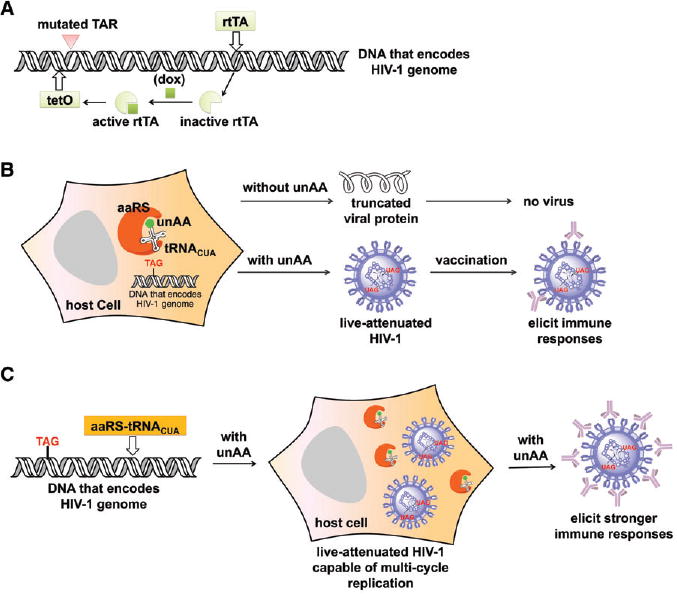
Synthetic biology approaches for the development of conditionally replicating HIV-1 variants. (A) Doxycycline-dependent HIV-1. (B) First generation of amber suppression-dependent HIV-1. In this design, HIV-1 mutant can only complete a single infection cycle in the host in the absence of exogenously provided tRNACUA-aaRS pair. (C) Second generation of amber suppression-dependent HIV-1. In this design, HIV-1 mutant with a genomic copy of the tRNACUA-aaRS pair can complete multiple infection cycles in the host. Abbreviations: TAR, trans-activating response element; rtTA, reverse tetracycline-controlled transactivator; dox, doxycycline; tetO, tetracycline operator; aaRS, aminoacyl-tRNA synthetase; unAA, unnatural amino acid.
In HIV-1, the LTR (long terminal repeat) acts as the viral promoter. The expression of viral genes is dependent on the Tat (trans-activator of transcription; a transactivation factor) protein and the TAR (trans-activating response element) region of the LTR. Tat induces transcriptional elongation by directly interacting with the bulge of TAR region and by recruiting cellular transcriptional co-activators, such as the positive transcription elongation complex (P-TEFb). In the absence of Tat, RNA polymerases initiating transcription mostly stall near the promoter and only short RNAs are produced. In order to abolish the Tat-TAR transcription control mechanism, mutations were introduced into both Tat and TAR.62 Specifically, Tat was inactivated by the Tyr26Ala mutation that hinders activation of the HIV-1 LTR promoter. Several mutations were introduced into the bulge and loop regions of TAR, which prevented TAR from binding to Tat and subsequent transcription activation.
In order to restore HIV-1 gene expression in a dox-dependent manner, a set of dox-associated control elements, including the tet-operator (tetO) sequence and the reverse tetracycline– controlled transactivator (rtTA) protein-encoding gene, were inserted into the viral genome. Multiple copies of tetO were inserted between NFκB and SP1 transcription factor binding sites in the U3 promoter of the 5′-LTR. The nef gene was replaced by the reverse rtTA gene.62 With the administration of dox, the dox-bound rtTA protein was able to bind exclusively to the tetO and initiated viral gene expression.62 On the other hand, the removal of dox resulted in the inactive form of rtTA and switched off viral gene expression.62
To further improve viral replication in the presence of dox, the Nef protein was restored in an HIV-rtTA-Ub-nef variant using the Ub fusion protein system.62 This modification led to the expression of the complete proteome of HIV-1. In comparison with the Nef-lacking HIV-rtTA variants, HIV-rtTA-Ub-nef replicated more efficiently in primary T cells and HIS mice. The conditionally replicating HIV variants could also be further optimized by using the safety-lock rtTA variants as well as the improved tetO elements. The use of this optimized rtTA–tetO system can potentially reduce the chance of conditional-live HIV-1 variants from losing the dox-control.63,64
While the tetO-rtTA system is powerful at generating conditionally replicating viruses, the long-term in vivo evolution of tetO–rtTA along with the acquired mutations within the virus genome could cause problems. This might lead to inactivated or constitutively active transcription of the vaccine virus, which exposes the vaccination hosts to the danger of receiving no protection or life-long infections.
Amber suppression-dependent HIV-1
Amber suppression is a generally applicable method to incorporate unnatural amino acids (unAAs) with desirable functions into a protein of interest in bacteria, yeast, mammalian cells, and – even in animals.67–69 However, few applications were reported to incorporate unAAs into proteins in live viruses, which is probably due to the complicated life-cycle of human viruses.65 Chen and co-workers first demonstrated a successful incorporation of unAAs into a surface protein of hepatitis D without compromising its viability or infectivity.70 Rather than manipulating the structural protein of viruses, we are interested in developing an amber suppression-dependent approach to stringently turn on/off the replication of HIV-1 strains (Fig. 1(B)), which can be potentially used as LAVs. This approach (Fig. 1(B), (C)) entails the manipulation of essential HIV-1 protein biosynthesis through amber suppression that is precisely controlled by three mutually dependent exogenous regulatory components, a unique amber suppressor tRNACUA (component 1) that can decode a blank codon (e.g. amber nonsense codon67–69 or a quadruplet codon71,72) and a special aminoacyl-tRNA synthetase (aaRS, component 2) that charges the suppressor tRNACUA with an unnatural amino acid (unAA; component 3).
Introduction to the approach
In the amber suppression strategy, TAG amber nonsense codons were introduced into the essential viral genes of HIV-1. An orthogonal aaRS-tRNACUA pair that specifically recognizes an unAA was included during virus replication. In the absence of the unAA, the full-length essential HIV-1 proteins could not be synthesized and led to no production of infectious HIV-1. On the other hand, the amber nonsense codons could be decoded and rendered the assembly and replication of HIV-1 in the presence of unAA. The key control components of the amber suppression system include an aaRS that specifically recognizes the small molecule switch, an unAA of one's choice, and charges it onto an amber suppressor tRNACUA. To examine the feasibility of using this approach in HIV-1 vaccine development, it was first demonstrated that an overexpression of aaRS and tRNACUA from an exogenously provided plasmid exhibited no apparent influence on the assembly of wild-type HIV-1 virus (pSUMA; the infectious molecular clone of a founder/transmitter HIV-1 virus, catalog #11748, NIH AIDS Reagent Program) by in vitro infectivity assays.65 Next, it was demonstrated that unAA was absolutely required for the assembly of live HIV-1 variants containing amber mutations in their genomes.65
The Choice of Amber Mutation Sites on HIV-1 Genome
HIV-1 has a highly compact and efficient genome. The HIV-1 genome contains three major viral structural protein-encoding genes gag, pol and env, two essential regulatory elements tat and rev, and four accessory regulatory protein-encoding genes, nef , vpr, vif and vpu. The gag (group-specific antigen) gene encodes the precursor Gag protein that is subsequently processed by the HIV protease (PR) into matrix protein p17 (MA), capsid protein p24 (CA), spacer peptide 1 (SP1), nucleocapsid protein (NC), spacer peptide 2 (SP2), and P6 protein. The pol gene encodes important viral enzymes, including reverse transcriptase (RT), RNase H, integrase (IN), and protease. The env gene encodes gp160 that is post-translationally cleaved to gp120 and gp41. In addition to the coding genes, HIV-1 also contains several essential genomic structural elements such as long terminal repeat (LTR) and trans-activating response element (TAR). Furthermore, HIV-1 uses a sophisticated splicing system to produce over 40 different mRNA species, both completely and incompletely spliced.
Thus, it is critical to choose proper amber mutation sites on the HIV-1 genome so that the assembly and function of HIV-1 are not affected by amber mutations. While a large amount of genetic information of HIV is available, the choice of amber mutation sites mostly relied on a trial and error approach. Multiple HIV-1 mutants were initially constructed that contained amber mutations at a range of different sites, including SUMA-Tyr132 (amber mutation at position Tyr132 of gag), SUMA-Ala119 (amber mutation at position Ala119 of gag), SUMA-Tyr59 (amber mutation at position Tyr59 of pol), and SUMA-Leu365 (amber mutation at position Leu365 of pol). Among these four mutants, only the SUMA-Tyr59 mutant was confirmed to contain a proper amber mutation that did not affect viral assembly and function.65
The Choice of UNAA
Another important consideration in applying amber suppression strategy to HIV-1 vaccine development is the choice of unAA. First, the introduction of an unAA should not affect the function of HIV-1 proteins. Ideally, the structure and chemical property of unAA should resemble the natural amino acid that it replaces. For example, Tyr132 in SUMA-Tyr132 should be substituted with a close analog of tyrosine. To this end, three tyrosine analogs were examined, including p-acetylphenylalanine (AcF), p-iodophenylalanine (IodoF), and p-azidophenylalanine (AzF).65 Although IodoF is structurally very similar to Tyr, it could not be functionally introduced into Gag protein to replace Tyr132. Presumably, the hydrophobic iodo substituent might interfere with the translation of Gag, which led to the translational termination at position Tyr132 and the production of truncated proteins. On the other hand, AcF could be efficiently introduced at the position of Tyr132 of Gag. However, the assembled HIV-1 mutants did not show any infectivity. In this case, replacing Tyr with AcF probably interfered with proteolytic processing of Gag or abolished the function of p24 (a processed product from Gag). Finally, AzF was shown to be a proper unAA for the construction of amber suppression-dependent HIV-1 mutants.65
The fidelity of unAA incorporation is critical for the generation of safe and conditionally replicating HIV-1 vaccine using the amber suppression strategy. Ideally, the full-length essential HIV-1 proteins should only be synthesized in the presence of the unAA of one's choice. This requires that the special aaRS-tRNACUA pair does not recognize any of the endogenous natural amino acids in the host. To this end, the fidelity of the tRNACUA-AzFRS (an evolved aaRS that specifically charge the tRNACUA with AzF73) pair was thoroughly examined. This was conducted by co-transfecting plasmid pAzFRS (a plasmid containing AzFRS and tRNACUA) with a reporter plasmid encoding an enhanced green fluorescent protein (EGFP) with an amber nonsense codon at position 40 (pEGFP-TAG40).65 The full length EGFP was observed only in the presence of 1mM AzF. On the other hand, no fluorescent signal was detected in the absence of AzF. The site-specific incorporation of AzF at position 40 was further confirmed by tandem mass spectrometry.65 Therefore, the AzFRS-tRNACUA pair can be used to incorporate AzF into proteins in response to amber nonsense codon with excellent fidelity. Besides fidelity, the efficiency of the tRNACUA-aaRS pair is also important. Optimization of the tRNACUA-AzFRS pair can be performed using a previously reported strategy by fine tuning the interaction between tRNACUA and AzFRS.74
Construction and testing of amber suppression-dependent HIV-1 variants
To demonstrate the feasibility of amber suppression-dependent virus replication, the assembly and function of SUMA-Tyr59 mutant were examined in the presence and in the absence of AzF (Fig. 2). The AzFRS-tRNACUA pair was provided exogenously on a plasmid. The experimental results showed that live and functional SUMA-Tyr59 mutant could be assembled in the presence of AzF. On the other hand, no detectable functional virus was produced in the absence of AzF. The infectivity of SUMA-AzF59 is roughly 1.7% of the wild-type SUMA according to the tissue culture infectious dose 50 (TCID50) values.
Figure 2.
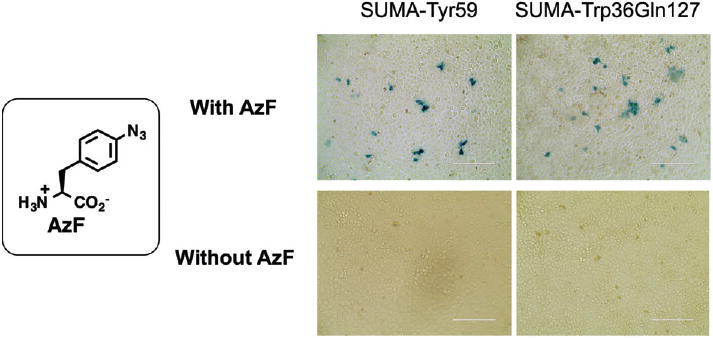
Infection assays with HIV-1 mutants in the presence and the absence of AzF. SUMA-Tyr59 contains an amber mutation at position Tyr59 of pol. SUMA-Trp36Gln127 contains two amber mutations in matrix protein domain of Gag. The assay was conducted with TZM-bl cells, which harbors a Tat-driven bacterial lacZ gene. A positive X-gal staining (to detect the expression of Tat) indicates that HIV-1 virus is active. AzF, p-azidophenylalanine.
Based on theoretical calculations, if we take the HIV mutation rate as approximately 3 × 10−5 per nucleotide base per cycle of replication75 and the virus life cycle as one day, the possibility of mutating amber codon(s) back to sense codon(s) is approximately 1%, 0.01%, and 0.0001%, respectively, in the human life span (100 years) when one, two, and three amber codons are used. Therefore, introducing one amber mutation into the HIV genome may not sufficiently prevent the virus regaining virulence through mutations. Two or more amber mutations are needed in the vaccine development. To this end, a HIV mutant (SUMA-Trp36Gln127) was constructed to contain two amber mutations in the matrix protein section of Gag. To our delight, similar infectivity was observed with the SUMA-Trp36Gln127 mutant in comparison with HIV-1 mutant containing only single amber mutation. As a control, no live SUMA-Trp36Gln127 virus was produced in the absence of AzF during viral assembly in 293T cells according to the infection assay (Fig. 2).
In the above experiments, the AzFRS-tRNACUA pair was provided exogenously on a plasmid, and cannot be inherited by the progeny virus. Once injected in the vaccination host, the assembled live viruses can only survive one-cycle since the host does not have either the AzFRS-tRNACUA pair or the unAA. As previously reported, transient replication might not be sufficient to elicit potent protective immune responses,76 HIV-1 mutants that are capable of multi-cycle infection of host cells are needed as HIV-1 LAV. To address this problem, an HIV-1 mutant that contains a genomic copy of the aaRS-tRNACUA pairs was recently constructed (the manuscript is currently under revision) and examined. In this case, the regulatory components are always co-expressed with the viral proteins to ensure the replication capacity of attenuated HIV-1. The ability to control multi-cycle HIV-1 replication in mammalian cells represents an important step towards the generation of a safe and efficacious vaccine to control the worldwide HIV-1 epidemic.
Conclusion
In this review, we first summarized four Phase IIB or III clinical trials of HIV-1 vaccines (Table 1). Although only the RV144 Thai vaccine trial demonstrated a moderate protection, the results from all of those strategies have provided valuable information and lessons for future vaccine development. Since none of the prior vaccine strategies yielded an effective HIV-1 vaccine in clinical applications, new and more effective approaches are highly desirable, such as the use of conditionally replicating HIV-1 strains (Table 1). The current frontiers of the development of conditionally replicating HIV-1 mutants were discussed, including the dox-dependent and the amber suppression-dependent systems. While the dox-dependent strategy has been extensively reviewed,62 this article represents the first overview of the technical details and perspectives of the newly developed amber suppression-dependent system. In addition, a closer comparison of the two systems is provided. While both of them could potentially be used to increase the safety of HIV-1 LAVs, the two approaches differ in their level (transcription or translation) of control. The dox-dependent strategy controls HIV-1 replication at the DNA transcription level. One individual event of losing the dox-dependent control would lead to the synthesis of one copy of mRNA, which serves as the template for the synthesis of multiple copies of protein. On the other hand, the amber suppression-dependent approach controls virus replication at the protein translation level, one undesirable read-through of amber codons would lead to the synthesis of one copy of protein. In this sense, the amber suppression-dependent approach may give a lower level of basal replication of HIV-1 in comparison with the dox-dependent strategy. It may also be possible to further improve the safety of conditionally replicating HIV-1 variants by a combination of the above two strategies and/or the inclusion of additional control mechanisms. The intention of this review is to attract more attention towards this promising research field and to provoke creative designs and innovative utilization of the two control strategies.
In summary, the two control strategies for the construction of conditionally replicating HIV-1 variants laid a solid foundation for the future development of replication-competent but controllable HIV-1 LAVs. There are still considerable technical challenges that need to be overcome to yield an effective and safe HIV-1 vaccine. Nevertheless, the two conditionally replicating HIV-1 strategies represent elegant combinations of chemical/synthetic biology and virology, which is likely to open up new avenues in HIV-1 vaccine research. In addition, these control strategies can also be potentially applied to the development of novel LAVs against other pathogens or to improve the safety of vaccines currently being used.
Acknowledgments
This work was supported by Grant 1R01AI111862 (to J. Guo, Q. Li, and W. Niu) from DHHS-NIH-NIAID.
References
- 1.UNAIDS Fact sheet. 2016;2016 [Google Scholar]
- 2.Sheets RL, Zhou T, Knezevic I. Review of efficacy trials of HIV-1/AIDS vaccines and regulatory lessons learned: a review from a regulatory perspective. Biologicals. 2016;44:73–89. doi: 10.1016/j.biologicals.2015.10.004. [DOI] [PubMed] [Google Scholar]
- 3.Cohen YZ, Dolin R. Novel HIV vaccine strategies: overview and perspective. Ther Adv Vaccines. 2013;1114:99–112. doi: 10.1177/2051013613494535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Watkins DI, Burton DR, Kallas EG, Moore JP, Koff WC. Nonhuman primate models and the failure of the Merck HIV-1 vaccine in humans. Nat Med. 2008;14:617–621. doi: 10.1038/nm.f.1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rerks-Ngarm S, Pitisuttithum P, Nitayaphan S, Kaewkungwal J, Chiu J, Paris R, et al. Vaccination with ALVAC and AIDSVAX to prevent HIV-1 infection in Thailand. N Eng J Med. 2009;361:2209–2220. doi: 10.1056/NEJMoa0908492. [DOI] [PubMed] [Google Scholar]
- 6.Koff WC, Johnson PR, Watkins DI, Burton DR, Lifson JD, Hasenkrug KJ, et al. HIV vaccine design: insights from live attenuated SIV vaccines. Nat Immunol. 2006;7:19–23. doi: 10.1038/ni1296. [DOI] [PubMed] [Google Scholar]
- 7.Flynn NM, Forthal DN, Harro CD, Judson FN, Mayer KH, Para MF, et al. Placebo-controlled phase 3 trial of a recombinant glycoprotein 120 vaccine to prevent HIV-1 infection. J Infect Dis. 2005;191:654–665. doi: 10.1086/428404. [DOI] [PubMed] [Google Scholar]
- 8.Pitisuttithum P, Gilbert P, Gurwith M, Heyward W, Martin M, van GF, et al. Randomized, double-blind, placebo-controlled efficacy trial of a bivalent recombinant glycoprotein 120 HIV-1 vaccine amonginjec-tion drug users in Bangkok, Thailand. J Infect Dis. 2006;194:1661–1671. doi: 10.1086/508748. [DOI] [PubMed] [Google Scholar]
- 9.Mascola JR, Snyder SW, Weislow OS, Belay SM, Belshe RB, Schwartz DH, et al. Immunization with envelope subunit vaccine products elicits neutralizing antibodies against laboratory-adapted but not primary isolates of human immunodeficiency virus type 1. J Infect Dis. 1996;173:340–348. doi: 10.1093/infdis/173.2.340. [DOI] [PubMed] [Google Scholar]
- 10.Gilbert PB, Peterson ML, Follmann D, Hudgens MG, Francis DP, Gurwith M, et al. Correlation between immunologic responses to a recombi-nant glycoprotein 120 vaccine and incidence of HIV-1 infection in a phase 3 HIV-1 preventive vaccine trial. J Infect Dis. 2005;191:666–677. doi: 10.1086/428405. [DOI] [PubMed] [Google Scholar]
- 11.Zak DE, Andersen-Nissen E, Peterson ER, SatoA, Hamilton MK, Borgerd- ing J, et al. Merck Ad5/HIV induces broad innate immune activation that predicts CD8+ T-cell responses but is attenuated by preexisting Ad5 immunity. Proc Nat Acad Sci USA. 2012;109:E3503–E3512. doi: 10.1073/pnas.1208972109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schmitz JE, Kuroda MJ, Santra S, Sasseville VG, Simon MA, Lifton MA, et al. Control of viremia in simian immunodeficiency virus infection by CD8+ lymphocytes. Science. 1999;283:857–860. doi: 10.1126/science.283.5403.857. [DOI] [PubMed] [Google Scholar]
- 13.Moodie Z, Metch B, Bekker LG, Churchyard G, Nchabeleng M, Mlisana K, et al. Continued follow-up of phambili phase 2b randomized HIV-1 vaccine trial participants supports increased HIV-1 acquisition among vaccinated men. PLoS One. 2015;10:e0137666. doi: 10.1371/journal.pone.0137666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gray GE, Allen M, Moodie Z, Churchyard G, Bekker LG, Nchabeleng M, et al. Safety and efficacy of the HVTN 503/phambili study of a clade-B-based HIV-1 vaccine in South Africa: a double-blind, randomised, placebo-controlled test-of-concept phase2bstudy. Lancet Infect Dis. 2011;11:507–515. doi: 10.1016/S1473-3099(11)70098-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hu H, Eller MA, Zafar S, Zhou Y, Gu M, Wei Z, et al. Preferential infection of human Ad5-specific CD4 T cells by HIV in Ad5 naturally exposed and recombinant Ad5-HIV vaccinated individuals. Proc Nat Acad Sci USA. 2014;111:13439–13444. doi: 10.1073/pnas.1400446111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li F, Finnefrock AC, Dubey SA, Korber BTM, Szinger J, Cole S, et al. Mapping HIV-1 vaccine induced T-cell responses: bias towards less-conserved regions and potential impact on vaccine efficacy in the Step study. PLoS One. 2011;6:e20479. doi: 10.1371/journal.pone.0020479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Letvin L, Rao SS, Montefiori DC, Seaman MS, Sun Y, Lim SY, et al. Immune and genetic correlates of vaccine protection against mucosal infection by SIV in monkeys. Sci Trans Med. 2011;3:81ra36. doi: 10.1126/scitranslmed.3002351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Churchyard GJ, Morgan C, Adams E, Hural J, Graham BS, Moodie Z, et al. A phase IIA randomized clinical trial of a multiclade HIV-1 DNA prime followed by a multiclade rAd5 HIV-1 vaccine boost in healthy adults (HVTN204) PLoS One. 2011;6:e21225. doi: 10.1371/journal.pone.0021225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hammer SM, Sobieszczyk ME, Janes H, Karuna ST, Mulligan MJ, Grove D, et al. Efficacy trial of a DNA/rAd5 HIV-1 preventive vaccine. N Eng J Med. 2013;369:2083–2092. doi: 10.1056/NEJMoa1310566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Williams WB, Liao HX, Moody MA, Kepler TB, Alam SM, Gao F, et al. Diversion of HIV-1 vaccine–induced immunity by gp41-microbiota cross-reactive antibodies. Science. 2015;349 doi: 10.1126/science.aab1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Roederer M, Keele BF, Schmidt SD, Mason RD, Welles HC, Fischer W, et al. Immunological and virological mechanisms of vaccine-mediated protection against SIV and HIV. Nature. 2014;505:502–508. doi: 10.1038/nature12893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim JH, Excler JL, Michael NL. Lessons from theRV144 Thai phase IIIHIV-1 vaccine trial and the search for correlates of protection. Annu Rev Med. 2015;66:423–437. doi: 10.1146/annurev-med-052912-123749. [DOI] [PubMed] [Google Scholar]
- 23.Gray GE, Andersen-Nissen E, Grunenberg N, Huang Y, Roux S, Laher F, et al. HVTN 097: evaluation of the RV144 vaccine regimen in HIV uninfected South African adults. AIDS Res Hum Retroviruses. 2014;30:A33–A34. [Google Scholar]
- 24.Gray GE, Laher F, Lazarus E, Ensoli B, Corey L. Approaches to preventative and therapeutic HIV vaccines. Curr Opin Virol. 2016;17:104–109. doi: 10.1016/j.coviro.2016.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hurwitz JL, Lockey TD, Jones B, Freiden P, Sealy R, Coleman J, et al. First phase I clinical trial of an HIV-1 subtype D gp140 envelope protein vaccine: immune activity induced in all study participants. Aids. 2008;22:149–151. doi: 10.1097/QAD.0b013e3282f174ed. [DOI] [PubMed] [Google Scholar]
- 26.Cosgrove CA, Lacey CJ, Cope AV, Bartolf A, Morris G, Yan C, et al. Comparative immunogenicity of HIV-1 gp140 vaccine delivered by parenteral, and mucosal routes in female volunteers; MUCOVAC2, a randomized two centre study. PLoS One. 2016;11:e0152038. doi: 10.1371/journal.pone.0152038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Salmon-Ceron D, Durier C, Desaint C, Cuzin L, Surenaud M, Hamouda NB, et al. Launay O and group AVt, Immunogenicity and safety of an HIV-1 lipopeptide vaccine in healthy adults: a phase 2 placebo-controlled ANRS trial. AIDS. 2010;24:2211–2223. doi: 10.1097/QAD.0b013e32833ce566. [DOI] [PubMed] [Google Scholar]
- 28.Cobb A, Roberts LK, Palucka AK, Mead H, Montes M, Ranganathan R, et al. Development of a HIV-1 lipopeptide antigen pulsed therapeutic dendritic cell vaccine. J Immunol Methods. 2011;365:27–37. doi: 10.1016/j.jim.2010.11.002. [DOI] [PubMed] [Google Scholar]
- 29.Barouch DH, Stephenson KE, Borducchi EN, Smith K, Stanley K, McNally AG, et al. Protective efficacy of a global HIV-1 mosaic vaccine against heterologous SHIV challenges in rhesus monkeys. Cell. 2013;155:531–539. doi: 10.1016/j.cell.2013.09.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ondondo B, Murakoshi H, Clutton G, Abdul-Jawad S, Wee EG, Gatanaga H, et al. Novel conserved-region T-cell mosaic vaccine with high global HIV-1 coverage is recognized by protective responses in untreated infection. Mol Ther. 2016;24:832–842. doi: 10.1038/mt.2016.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Graziani GM, Angel JB. HIV-1 immunogen: an overview of almost 30 years of clinical testing of a candidate therapeutic vaccine. Expert Opin Biol Ther. 2016;16:953–966. doi: 10.1080/14712598.2016.1193594. [DOI] [PubMed] [Google Scholar]
- 32.Mann JK, Ndung'u T. HIV-1 vaccine immunogen design strategies. Virol J. 2015;12:3. doi: 10.1186/s12985-014-0221-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Haynes BF, Kelsoe G, Harrison SC, Kepler TB. B-cell-lineage immunogen design in vaccine development with HIV-1 as a case study. Nat Biotechnol. 2012;30:423–433. doi: 10.1038/nbt.2197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pegu A, Yang ZY, Boyington JC, Wu L, Ko SY, Schmidt SD, et al. Neutralizing antibodies to HIV-1 envelope protect more effectively in vivo than those to the CD4 receptor. Sci Trans Med. 2014;6:243ra288. doi: 10.1126/scitranslmed.3008992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ledgerwood JE, Coates EE, Yamshchikov G, Saunders JG, Holman L, Enama ME, et al. Safety, pharmacokinetics and neutralization of the broadly neutralizing HIV-1 human monoclonal antibody VRC01 in healthy adults. Clin Exp Immunol. 2015;182:289–301. doi: 10.1111/cei.12692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Desrosiers RC. HIV with multiple gene deletions as a live attenuated vaccine for AIDS. AIDS Res Hum Retroviruses. 1992;8:411–421. doi: 10.1089/aid.1992.8.411. [DOI] [PubMed] [Google Scholar]
- 37.Daniel MD, Kirchhoff F, Czajak SC, Sehgal PK, Desrosiers RC. Protective effects of a live attenuated SIV vaccine with a deletion in the nef gene. Science. 1992;258:1938–1941. doi: 10.1126/science.1470917. [DOI] [PubMed] [Google Scholar]
- 38.Johnson RP, Desrosiers RC. Protective immunity induced by live-attenuated simian immunodeficiency virus. Curr Opin Immunol. 1998;10:436–443. doi: 10.1016/s0952-7915(98)80118-0. [DOI] [PubMed] [Google Scholar]
- 39.Mills J, Desrosiers R, Rud E, Almond N. Live-attenuated HIV vaccines: a proposal for further research and development. AIDS Res Hum Retroviruses. 2000;16:1453–1461. doi: 10.1089/088922200750005976. [DOI] [PubMed] [Google Scholar]
- 40.Whitney JB, Ruprecht RM. Live attenuated HIV vaccines: pitfalls and prospects. Curr Opin Infect Dis. 2004;17:17–26. doi: 10.1097/00001432-200402000-00004. [DOI] [PubMed] [Google Scholar]
- 41.Koff WC, Johnson PR, Watkins DI, Burton DR, Lifson JD, Hasenkrug KJ, et al. HIV vaccine design: insights from live attenuated SIV vaccines. Nat Immunol. 2006;7:19–23. doi: 10.1038/ni1296. [DOI] [PubMed] [Google Scholar]
- 42.Ruprecht RM. Live attenuated AIDS viruses as vaccines: promise or peril? Immunol Rev. 1999;170:135–149. doi: 10.1111/j.1600-065x.1999.tb01335.x. [DOI] [PubMed] [Google Scholar]
- 43.Li Q, Zeng M, Duan L, Voss JE, Smith AJ, Pambuccian S, et al. Live simian immunodeficiency virus vaccine correlate of protection: local antibody production and concentration on the path of virus entry. J Immunol. 2014;193:3113–3125. doi: 10.4049/jimmunol.1400820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Baba TW, Jeong YS, Pennick D, Bronson R, Greene MF, Ruprecht RM. Pathogenicity of live, attenuated SIV after mucosal infection of neonatal macaques. Science. 1995;267:1820–1825. doi: 10.1126/science.7892606. [DOI] [PubMed] [Google Scholar]
- 45.Wyand MS, Manson KH, Lackner AA, Desrosiers RC. Resistance of neonatal monkeys to live attenuated vaccine strains of simian immunodeficiency virus. Nat Med. 1997;3:32–36. doi: 10.1038/nm0197-32. [DOI] [PubMed] [Google Scholar]
- 46.Baba TW, Liska V, Khimani AH, Ray NB, Dailey PJ, Penninck D, et al. Live attenuated, multiply deleted simian immunodeficiency virus causes AIDS in infant and adult macaques. Nat Med. 1999;5:194–203. doi: 10.1038/5557. [DOI] [PubMed] [Google Scholar]
- 47.Whatmore AM, Cook N, Hall GA, Sharpe S, Rud EW, Cranage MP. Repair and evolution of nef in vivo modulates simian immunodeficiency virus virulence. J Virol. 1995;69:5117–5123. doi: 10.1128/jvi.69.8.5117-5123.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dittmer U, Nisslein T, Bodemer W, Petry H, Sauermann U, Stahl-Hennig C, et al. Cellular immune response of rhesus monkeys infected with a partially attenuated nef deletion mutant of the simian immunodeficiency virus. Virology. 1995;212:392–397. doi: 10.1006/viro.1995.1496. [DOI] [PubMed] [Google Scholar]
- 49.Stahl-Hennig C, Dittmer U, Nisslein T, Pekrun K, Petry H, Jurkiewicz E, et al. Attenuated SIV imparts immunity to challenge with pathogenic spleen-derived SIV but cannot prevent repair of the nef deletion. Immunol Lett. 1996;51:129–135. doi: 10.1016/0165-2478(96)02567-9. [DOI] [PubMed] [Google Scholar]
- 50.Berkhout B, Verhoef K, Van WJLB, Back NKT. Genetic instability of live, attenuated human immunodeficiency virus type 1 vaccine strains. J Virol. 1999;73:1138–1145. doi: 10.1128/jvi.73.2.1138-1145.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Evans DT, Bricker JE, Desrosiers RC. A novel approach for producing lentiviruses that are limited to a single cycle of infection. J Virol. 2004;78:11715–11725. doi: 10.1128/JVI.78.21.11715-11725.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Evans DT, Bricker JE, Sanford HB, Lang S, Carville A, Richardson BA, et al. Immunization of macaques with single-cycle simian immunodeficiency virus (SIV) stimulates diverse virus-specific immune responses and reduces viral loads after challenge with SIVmac239. J Virol. 2005;79:7707–7720. doi: 10.1128/JVI.79.12.7707-7720.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Falkensammer B, Rubner B, Hiltgartner A, Wilflingseder D, Stahl HC, Kuate S, et al. Role of complement and antibodies in controlling infection with pathogenic simian immunodeficiency virus (SIV) in macaques vaccinated with replication-deficient viral vectors. Retro-virology. 2009;6:60. doi: 10.1186/1742-4690-6-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kuate S, Stahl-Hennig C, ten HP, Heeney J, Uberla K. Single-cycle immunodeficiency viruses provide strategies for uncoupling in vivo expression levels from viral replicative capacity and for mimicking live-attenuated SIV vaccines. Virology. 2003;313:653–662. doi: 10.1016/s0042-6822(03)00388-x. [DOI] [PubMed] [Google Scholar]
- 55.Berkhout B, Paxton WA. HIV vaccine: it may take two to tango, but no party time yet. Retrovirology. 2009;6:88. doi: 10.1186/1742-4690-6-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Verhoef K, Marzio G, Hillen W, Bujard H, Berkhout B. Strict control of human immunodeficiency virus type 1 replication by a genetic switch: Tet for Tat. J Virol. 2001;75:979–987. doi: 10.1128/JVI.75.2.979-987.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Smith SM, Khoroshev M, Marx PA, Orenstein J, Jeang KT. Constitutively dead, conditionally live HIV-1 genomes: ex vivo implications for a live virus vaccine. J Biol Chem. 2001;276:32184–32190. doi: 10.1074/jbc.M101604200. [DOI] [PubMed] [Google Scholar]
- 58.Berkhout B, Marzio G, Verhoef K. Control over HIV-1 replication by an antibiotic; a novel vaccination strategy with a drug-dependent virus. Virus Res. 2002;82:103–108. doi: 10.1016/s0168-1702(01)00399-9. [DOI] [PubMed] [Google Scholar]
- 59.Das AT, Zhou X, Vink M, Klaver B, Berkhout B. Conditional live virus as a novel approach towards a safe live attenuated HIV vaccine. Expert Rev Vaccines. 2002;1:293–301. doi: 10.1586/14760584.1.3.293. [DOI] [PubMed] [Google Scholar]
- 60.Das AT, Verhoef K, Berkhout B. A conditionally replicating virus as a novel approach toward an HIV vaccine. Methods Enzymol. 2004;388:359–379. doi: 10.1016/S0076-6879(04)88028-5. [DOI] [PubMed] [Google Scholar]
- 61.Marzio G, Verhoef K, Vink M, Berkhout B. In vitro evolution of a highly replicating, doxycycline-dependent HIV for applications in vaccine studies. Proc Nat Acad Sci USA. 2001;98:6342–6347. doi: 10.1073/pnas.111031498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Das AT, Berkhout B. Conditionally replicating HIV and SIV variants. Virus Res. 2016;216:66–75. doi: 10.1016/j.virusres.2015.05.004. [DOI] [PubMed] [Google Scholar]
- 63.Zhou X, Vink M, Klaver B, Verhoef K, Marzio G, Das AT, et al. The genetic stability of a conditional live HIV-1 variant can be improved by mutations in the Tet-On regulatory system that restrain evolution. J Biol Chem. 2006;281:17084–17091. doi: 10.1074/jbc.M513400200. [DOI] [PubMed] [Google Scholar]
- 64.Zhou X, Vink M, Berkhout B, Das AT. Modification of the Tet-On regulatory system prevents the conditional-live HIV-1 variant from losing doxycycline-control. Retrovirology. 2006;3:82. doi: 10.1186/1742-4690-3-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wang N, Li Y, Niu W, Sun M, Cerny R, Li Q, et al. Construction of a live-attenuated HIV-1 vaccine through genetic code expansion. Angew Chem Int Ed. 2014;53:4867–4871. doi: 10.1002/anie.201402092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Baron U, Bujard H. Tet repressor-based system for regulated gene expression in eukaryotic cells: principles and advances. Methods Enzymol. 2000;327:401–421. doi: 10.1016/s0076-6879(00)27292-3. [DOI] [PubMed] [Google Scholar]
- 67.Wu X, Schultz PG. Synthesis at the interface of chemistry and biology. J Am Chem Soc. 2009;131:12497–12515. doi: 10.1021/ja9026067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Davis L, Chin JW. Designer proteins: applications of genetic code expansion in cell biology. Nat Rev Mol Cell Biol. 2012;13:168–182. doi: 10.1038/nrm3286. [DOI] [PubMed] [Google Scholar]
- 69.Niu W, Guo J. Expanding the chemistry of fluorescent protein biosensors through genetic incorporation of unnatural amino acids. Mol BioSyst. 2013;9:2961–2970. doi: 10.1039/c3mb70204a. [DOI] [PubMed] [Google Scholar]
- 70.Lin S, Yan H, Li L, Yang M, Peng B, Chen S, et al. Site-specific engineering of chemical functionalities on the surface of live hepatitis D virus. Angew Chem Int Ed. 2013;52:13970–13974. doi: 10.1002/anie.201305787. [DOI] [PubMed] [Google Scholar]
- 71.Niu W, Schultz PG, Guo J. An expanded genetic code in mammalian cells with a functional quadruplet codon. ACS Chem Biol. 2013;8:1640–1645. doi: 10.1021/cb4001662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wang N, Shang X, Cerny R, Niu W, Guo J. Systematic evolution and study of UAGN decoding tRNAs in a genomically recoded bacteria. Sci Rep. 2016;6:21898. doi: 10.1038/srep21898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Chin JW, Santoro SW, Martin AB, King DS, Wang L, Schultz PG. Addition of p-azido-L-phenylalanine to the genetic code of Escherichia coli. J Am Chem Soc. 2002;124:9026–9027. doi: 10.1021/ja027007w. [DOI] [PubMed] [Google Scholar]
- 74.Wang N, Ju T, Niu W, Guo J. Fine-tuning interaction between aminoacyl-tRNA synthetase and tRNA for efficient synthesis of proteins containing unnatural amino acids. ACS Synth Biol. 2014;4:207–212. doi: 10.1021/sb500195w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Rambaut A, Posada D, Crandall KA, Holmes EC. The causes and consequences of HIV evolution. Nat Rev Genet. 2004;5:52–61. doi: 10.1038/nrg1246. [DOI] [PubMed] [Google Scholar]
- 76.Wyand MS, Manson KH, Garcia-Moll M, Montefiori D, Desrosiers RC. Vaccine protection by a triple deletion mutant of simian immunodeficiency virus. J Virol. 1996;70:3724–3733. doi: 10.1128/jvi.70.6.3724-3733.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]


