Abstract
Mounting evidence suggests that protein methyltrans•ferases (PMTs), which catalyze methylation of histone as well as non-histone •proteins, play aa crucial role in diverse biological pathways and human• diseases. In particular, PMTs have been recognized as major players in •regulating gene expression and chromatin state. There has been an increasingly growing• interest in these enzymes as potential therapeutic targets and over the past two years tremendous progress has been made in the discovery of selective, small molecule inhibitors of protein lysine and arginine methyltransferases. Inhibitors of PMTs have been used extensively in oncology studies as tool compounds, and inhibitors of EZH2, DOT1L and PRMT5 are currently in clinical trials.
Introduction
Histone methylation was first recognized in 2000 and is one of the most actively investigated posttranslational modifications.[1] Methylation of histones as well as non-histone proteins has been implicated in various cancers and numerous other diseases, as it is a dynamic process that plays a key role in the regulation of gene expression and transcription.[2,3] Given these key functions, there has been a steadily increasing interest towards assessing the potential of these enzymes as therapeutic targets. [4–8] Therefore, the discovery of selective small-molecule inhibitors of protein methyltransferases (PMTs) has become a very active and fast growing research area (Figure 1).[9–11] In this review we focus on selective, small molecule inhibitors of PMTs that are discovered in last two years.
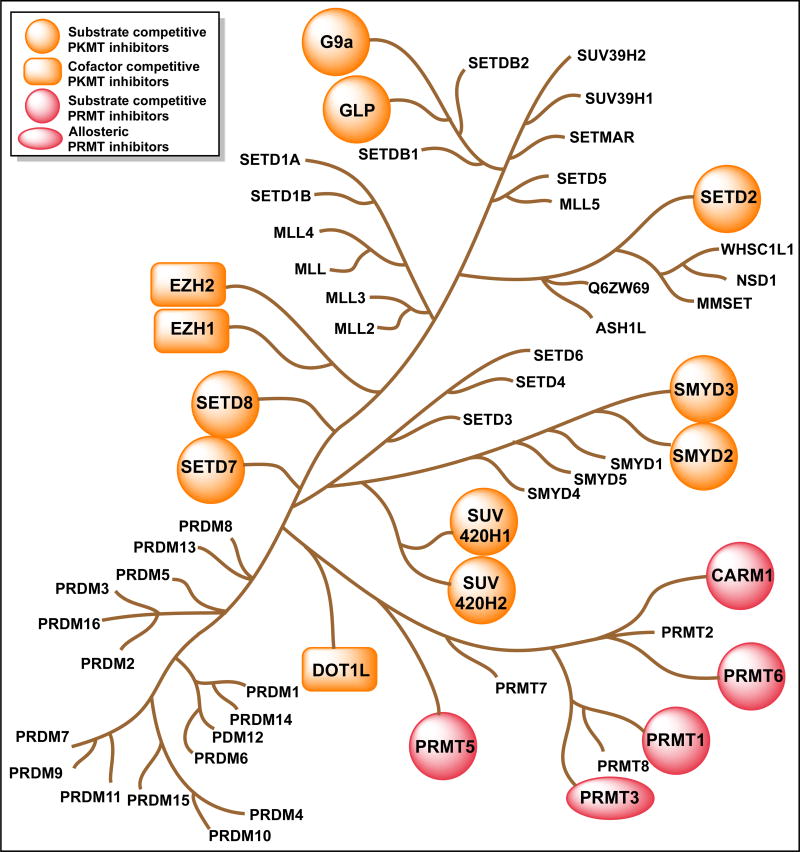
Figure 1. Phylogenetic tree of protein methyltransferases.
The methyltransferases with known inhibitors are marked and their mechanism of inhibition indicated.
Recent progress in discovery of inhibitors of PMTs
The first inhibitors of protein lysine methyltransferase (PKMT) and protein arginine methyltransferase (PRMT) were discovered in 2005 and 2004, respectively. In less than a decade, numerous PMT inhibitors with high potency and selectivity have been disclosed, some of which has entered the clinical trials emphasizing the rapid progress made in the field. For example, PKMT inhibitors, such as BIX-01294, UNC0638, UNC0642 (G9a/GLP), EPZ005687, GSK126, EI1, UNC1999, EPZ-6438, CPI-1205 (EZH2/EZH1), EPZ004777, SGC0946, EPZ-5676 (DOT1L), AZ-505 and LLY507 (SMYD2) are valuable chemical tools for further understanding biological functions of the targeted enzymes and have already been widely used in evaluating the therapeutic potential of these proteins (Table 1).[9,10] In addition, highly potent, selective, substrate-competitive PRMT inhibitors including MS023 (type I PRMTs), TP-064 (CARM1), EPZ015666 (PRMT5) and EPZ020411 (PRMT6) have been accomplished, suggesting that the substrate-binding grooves of PRMTs can also be successfully targeted (Table 1).[9,12,13] The discovery of the first allosteric PRMT3 inhibitor and the development of the PRMT3 chemical probe SGC707 have demonstrated that the allosteric binding site of PRMT3 can be exploited to yield potent, selective, and cell-active inhibitors, opening the door for discovering allosteric inhibitors of other PRMTs (Table 1).[14] Moreover, the discovery of the covalent SETD8 inhibitor MS453 has demonstrated that cysteine residues in active sites of PMTs can be selectively targeted (Table 1).[15] Furthermore, inhibitors that are disrupting the protein-protein interactions such as OICR-9429 (WDR5-MLL), MI-503 (MENIN-MLL) and A-395, EED226 (EED-PRC2) were also recently reported introducing yet another approach for the inhibition of the PMTs (Table 1).[16–19] The design, synthesis and biological studies of many of these aforementioned inhibitors have already been discussed in detail in literature.[9,10] The following sections will focus only on the potent, selective small-molecule PMT inhibitors that are discovered very recently (indicated in bold type in Table 1).
Table 1. List of known selective, small molecule inhibitors of PMTs and their mechanism of action (MOA).
Recently discovered inhibitors that are discussed in this review are shown in bold type.
| PMT | Histone targeta |
Non-histone target(s)a |
Inhibitor(s)b [ref] | MOA |
|---|---|---|---|---|
| G9a/GLP | H3K9 | p53 | BIX-01294; UNC0224; E72; UNC0638; BRD4770c; UNC0642; A-366 | Substrate competitive |
| GLP | H3K9 | p53 | MS012 [20] | Substrate competitive |
| EZH2/EZH1 | H3K27 | - | EPZ005687; GSK126; EI1; UNC1999; EPZ-6438; EPZ011989; ZLD1039; CPI-1205 [23]; Compound 1[24] | Co-factor competitive |
| EED-PRC2 | H3K27 | - | A-395 [18]; EED226 [19] | Allosteric/PPI-disruption |
| SETD7 | H3K4 | p53, p65, DNMT1 | PFI-2 | Substrate competitive |
| SMYD3 | H3K4, H4K5 | MAP3K2 | EPZ031686; EPZ030456 | Substrate competitive |
| MENIN-MLL | H3K4 | - | MI-503 | PPI-disruption |
| WDR5-MLL | H3K4 | - | OICR-9429 | PPI-disruption |
| SMYD2 | H3K4, H3K36 | p53, Rb | AZ-505; A-893; LLY507; BAY-598 [32] | Substrate competitive |
| SETD2 | H3K36 | p53 | Pr-SNF | Substrate competitive |
| SETD8 | H4K20 | p53, PCNA | Nahuoic Acid A; UNC0379 [35]; MS2177; MS453 [15] | Substrate competitive |
| SUV420H1/SUV420H2 | H4K20 | - | A-196 [36] | Substrate competitive |
| DOT1L | H3K79 | - | EPZ004777; SGC0946; EPZ-5676, Compounds 2-5 [37–39] | Co-factor competitive |
| PRMT1 | H4R3 | NPL3p, MRE11, 53BP1, ASH2L | AMI-1; NS1; A36; MS023 (Type I PRMT inhibitor) | Substrate competitive |
| PRMT3 | H4R3 | rpS2, PABPN1 | SGC707 | Allosteric |
| CARM1 | H3R17, H3R26 | CBP/p300, PABP, HuR, HuD, CA150, SAP49, SmB, U1C | MS049d [46]; Compound 7 [47]; SGC2085 [48]; TP064 [49] | Substrate competitive |
| PRMT5 | H2AR3, H4R3, H3R2, H3R8 | p53, NF-ββ | EPZ015666; GSK591; LLY-283 [50] | Substrate competitive |
| PRMT6 | H4R3, H3R2 | - | EPZ020411 | Substrate competitive |
Histone and non-histone targets of the PMTs (not a comprehensive list). Main targets of the enzymes are shown in bold type.
Known inhibitors of PMTs.[9] The inhibitors that are focus of this review are shown in bold type.
The inhibitory activity of BRD4770 against GLP was not reported and it was reported to be co-factor competitive.
MS049 is a selective, dual CARM1 and PRMT6 inhibitor.
Inhibitors of H3K9 and H3K27 methyltransferases
The mono- and dimethylation of lysine 9 residue of histone 3 (H3K9) is primarily catalyzed by G9a and GLP, which share 80% sequence identity in their SET domains. Since the identification of the first G9a and GLP inhibitor, BIX-01294 in 2007, highly potent and selective inhibitors of G9a and GLP have been published.[9] All of these inhibitors however were highly potent for both enzymes. In 2017, MS012, a potent GLP selective inhibitor MS012 was discovered (Figure 2).[20] This inhibitor while still possessing quinozoline scaffold as many of its predecessors is 140-fold selective for GLP over G9a. In addition, MS012 displayed selectivity for GLP over a broad range of other PKMTs, PRMTs, DNMTs, and RNMTs. Remarkably, X-ray structures show that this substrate-competitive inhibitor binds to GLP and G9a in virtually identical binding modes, underlining the challenges in structure-based design of selective inhibitors for these highly homologous enzymes.
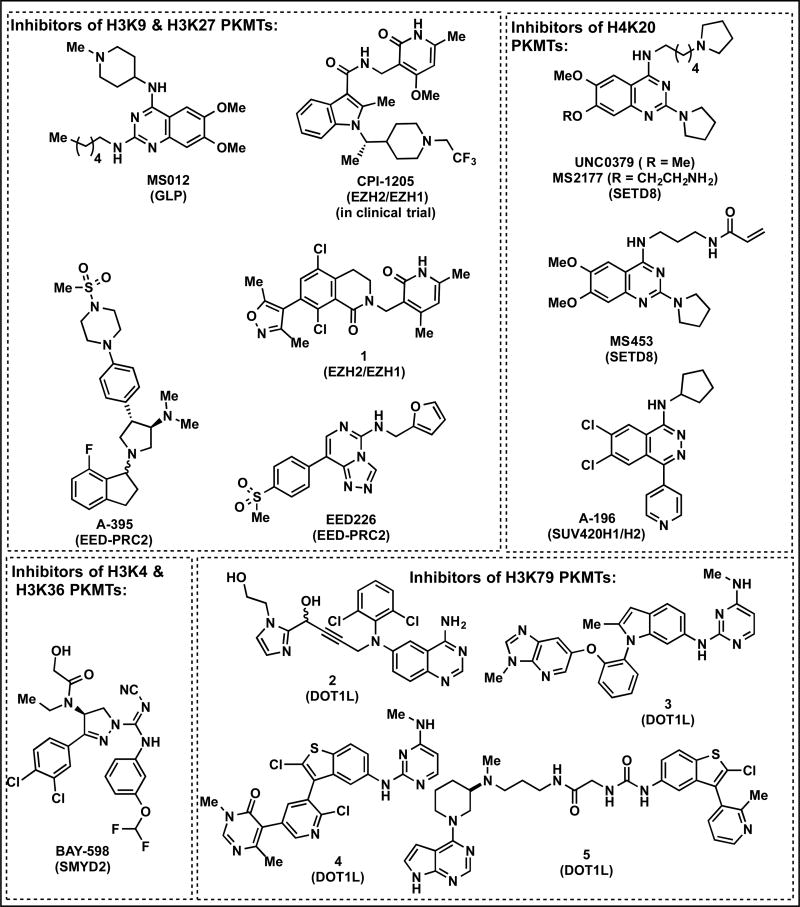
Figure 2. Structures of inhibitors of PKMTs.
The structures of recently reported inhibitors of EZH2/EZH1, EED-PRC2, SMYD2, SETD8, SUV420H1/H2 and DOT1L
Methylation of H3K27 is catalyzed by multisubunit protein complex polycomb repressive complex 2 (PRC2) and primarily functions to silence its target genes by trimethylation of H3K27.[21]. Enzymatically active PRC2 complex consists of three subunits: catalytic subunit EZH2 or EZH1, VEFS domain containing SUZ12 and WD40 repeat containing protein EED. A number of highly potent and selective inhibitors of PRC2 have been reported since 2012 (Table 1).[9] These pyridone containing and co-factor competitive EZH2 inhibitors have been used in many studies in recent years targeting various cancer types as EZH2 is highly expressed in many types of cancers.[22] Recently the discovery of CPI-1205, displaying high potency, selectivity, and cellular activity was added to the list of pyridone containing inhibitors.[23] A co-crystal structure of an analog of CPI-1205 bound to human PRC2 was also obtained.[23] In addition, very recently, new series of pyridone containing inhibitors, which led to the discovery of highly potent EZH2 inhibitor 1 (Figure 2) was reported.[24] Compound 1 displayed cellular potencies in cells that are comparable to previously reported EZH2 inhibitors. It is important to note that EPZ-6438, GSK126 and CPI-1205 are advanced into human clinical trials.
In late 2015, the first crystal structures of •an active PRC2 complex an active PRC2 complex from thee yeast Chaetomium thermophilum, which contain EZH2, EED and SUX12-VEFS in complex with inhibiting H3K27M peptide and SAH were reported.[25] Shortly after, in 2016, the structure of the human PRC2 complex was published.[26] Concurrently, the crystal structure of a small-molecule inhibitor in complex with the wild-type and Y641N-mutated PRC2 complex, consisting of human EED, human SUZ12-VEFS, and engineered American chameleon EZH2 subunits was also disclosed.[27] These crystal structures of the EZH2-EED-SUZ12 ternary complex revealed the molecular basis of the PRC2 core complex assembly and represent a landmark in this field. These structures show that the EED subunit is wrapped around by EZH2, and SUZ12 is sandwiched between EED and the SET domain of EZH2 forming a catalytically active complex. It was suggested that perhaps EED and SUZ12-VEFS allosterically activated the SET domain. The recognition of H3K27me3 by EED and the resulting increase of the methylase activity are believed to be crucial for the cellular function of PRC2 and thus, obstruction of this process would be a different approach in inhibiting PRC2 functions.
Very recently two inhibitors A-395 and EED226, which are targeting the EED H3K27me3 binding site and in turn preventing the allosteric activation of the PRC2 were reported. A-395 potently inhibited the catalytic activity of the trimeric PRC2 complex (EZH2–EED–SUZ12).[18] EED226 as well prevented the H3K27me3 binding to EED and therefore inhibited PRC2 activity when H3K27me0 peptide is used as substrate.[19] In addition both of these inhibitors reduced H3K27me2 and H3K27me3 levels in cells and inhibited proliferation of human cancer cell lines sensitive to EZH2 inhibition. Moreover they showed antitumor effects in a xenograft model in vivo and importantly, they were potent in cell lines resistant to EZH2 inhibitors. Crystal structures of these two inhibitors in complex with EED are also reported. In addition, recently peptidomimetic EED inhibitors that disrupt catalytic activity of PRC2 have been reported.[28]
Inhibitors of H3K4 and H3K36 methyltransferases
H3K4 and H3K36 methylation are hallmark of transcriptional activation.[29] SETD7, SMYD family proteins (SMYD1-3) and the MLL family proteins (MLL1-5) are some of the methyltransferases that are determined to be responsible for methylation of H3K4 in humans.[30] Studies have shown that these methyltransferases targets many non-histone proteins as well in some cases as their primary targets (Table 1). (R)-PFI-2 is the most potent, selective, and cell-active small-molecule inhibitor of SETD7 to date.[9] The major target of SMYD3 is determined to be MAP3K2 (also known as MEKK2) and highly potent, selective and cell active, small-molecule inhibitors of SYMD3, EPZ0330456 and EPZ031686 were reported and discussed elsewhere in greater detail.[9] MLL is a large multi-domain protein that is specific for H3K4 mono-, di-, and trimethylation.[30] Chromosomal rearrangements associated with MLL have been shown to cause acute myeloid, acute lymphoblastic, or mixed lineage leukemia.[31] Small molecules that perturb protein-protein interactions (PPI) of MLL with its partners, such as WDR5 (OICR-9429) and menin (MI-503) have been discovered.[9,16,17]
NSD family proteins (NSD1-3), SETD2, SETD3, ASH1L, SETMAR, and SMYD2 are responsible for H3K36 methylation.[30] SMYD2 can methylate a variety of non-histone substrates implicating effects on diverse biological processes.[30] Small molecule inhibitors of SMYD2; AZ-505, A-893 and LLY-507 have already been reported and previously reviewed.[9,10] Very recently, in 2016, a screening campaign and optimization studies yielded in the discovery of enantiomerically pure (S)-BAY-598 (Figure 2) as a cell- and in vivo-active inhibitor of SMYD2.[32] (S)-BAY-598 shows >10-fold selectivity for SMYD2 over SMYD3 and >100-fold selectivity over 31 other methyltransferases. This inhibitor is also highly selective against kinases and other primary molecular targets, including several CNS targets and competitive with the peptide substrate, but uncompetitive with cofactor.
Inhibitors of H4K20 methyltransferases
Methylation of H4K20 is catalyzed by the methyltransferases SUV420H1, SUV420H2, and SETD8.[30] SETD8 is the sole methyltransferase that catalyzes monomethylation of H4K20 and methylates many non-histone substrates, including the tumor suppressor p53 and proliferating cell nuclear antigen.[30] SETD8 has been shown to be overexpressed in various types of cancers.[33]
The first reported inhibitor of SETD8 was a marine natural product, nahuoic acid A, which was a co-factor competitive inhibitor.[34] In 2014, the first substrate-competitive, selective inhibitor of SETD8, UNC0379 was reported (Figure 2).[35]. In 2016, a more potent inhibitor, MS2177 (Figure 2), was obtained via addition of an aminoalkyl group to the 7-position of UNC0379.[15] MS2177 is competitive with the H4 peptide, but non-competitive with the cofactor S-5′-adenosyl-L-methionine (SAM). A cocrystal structure of MS2177 complexed with SETD8 revealed a cysteine residue (C311) in close proximity to the inhibitor binding site, presenting an opportunity to develop a covalent inhibitor of SETD8. Therefore, MS453 (Figure 2) was designed as a covalent inhibitor that modified C311, without effecting other cysteine residues of SETD8.[15] No covalent adduct was observed with PMTs such as PRC2, SMYD2, and SMYD3 upon incubation with MS453 suggesting specificity to SETD8. Furthermore, MS453 was highly selective for SETD8 over 29 other methyltransferases and the crystal structure of MS453 in complex with SETD8 confirmed covalent modification of C311.
SUV420H1 and SUV420H2 are highly homologous methyltransferases that di- and trimethylate H4K20.[30] A-196 (Figure 2) was recently discovered as the first potent, selective, and cell-active inhibitor of SUV420H1 and SUV420H2.[36] A-196 inhibits SUV420H1 and SUV420H2 potently in a peptide competitive manner and is >100-fold selective for SUV420H1 and SUV420H2 over other methyltransferases and a broad range of non-epigenetic targets. A-196 reduced the H4K20me3/me2 marks while elevated of H4K20me1 levels throughout the cell cycle in human cells without any observed toxicity. It inhibited 53BP1 foci formation in response to ionizing radiation, end reduced nonhomologous end joining (NHEJ) mediated DNA-break repair and it did not induce resistance/compensation effects, as H4K20me2/3 remained reduced after 20 population doublings.
Inhibitors of H3K79 methyltransferases
Highly potent, selective and cell active inhibitors of DOT1L: EPZ004777 SGC0946 and EPZ-5676 were disclosed between 2011 and 2013.[9] These inhibitors are SAM derivatives and competitive with the co-factor. In addition, a SAM derived covalent inhibitor was also reported.[9] In 2012, EPZ-5676 (also known as pinometostat) has become the first PMT inhibitor advanced to the clinic, and the first major breakthrough in the PMT inhibitor field.
In June 2016, a new series of DOT1L inhibitors that differ structurally from all previously published SAM-based inhibitors was reported.[37] These new inhibitors interact with an induced pocket adjacent to the SAM binding site, but do not bind the SAM binding site. A weak fragment-based screening hit that displayed suboptimal interactions in binding pocket was developed into a highly potent inhibitor of DOT1L, 2 (Figure 2) via careful analysis of the co-crystal structure and elegant structure-based optimization. These inhibitors were identified as SAM-competitive because, upon binding, they engage the lid loop of the SAM binding pocket and form a conformation preventing SAM binding. In a subsequent report, another structurally novel DOT1L inhibitor series that targets the same induced pocket was discovered.[38] Again a fragment based approach is utilized and resulted in the discovery of potent DOT1L inhibitors 3 and 4 (Figure 2), which displayed very high potencies in biochemical assays. The same research group realized that some of the identified fragments binding to the induced pocket extended into the methionine pocket of SAM binding site. However, the high throughput and fragment based screening methods used in the earlier studies, failed to find the fragments that occupy the SAM binding site. As already mentioned inhibitors 2-4 function in SAM-competitive manner. Interestingly, however, adenosine could bind to DOT1L in the presence of the induced pocket binders. With this observations in mind, a ternary X-ray crystal structure of adenosine and an induced pocket binder was obtained.[39] In this structure the lid loop of the SAM binding pocket in the SAM-bound state, is still folded over and collapsed onto induced pocket ligand as before. In addition, adenosine does not interact with the flexible loop in the ternary complex but otherwise forms the same contacts with DOT1L as in its binary complex. With these information in hand, the screening of a fragment-based second-site via NMR (i.e with the induced pocket already occupied) and also knowledge-based virtual screening of fragment library resembling kinase hinge binders that satisfy an adenosine-based pharmacophore were conducted. All hits discovered from both screening approaches were submitted to co-crystallization experiments with DOT1L in the absence and presence of induced pocket ligands. As a result, a pyyrolopyrimidine fragment was obtained as weak adenosine-like fragment inhibitor of DOT1L. Eventually, two ligands (adenosine-like pyyrolopyrimidine fragment and induced pocket binding fragment) that perfectly lined up for linking were identified. Linked fragments were further optimized resulting in the discovery of a highly potent inhibitor 5 with IC50 in picomolar range.[39] Impressively, inhibitor 5 performed equivalent or better than EPZ-5676 in cellular assays.
Inhibitors 2-5 displayed high selectivity against a panel of PKMTs and PRMTs, showing no inhibitory activity at up to 50 μMM. Importantly, they potently decreased H3K79me2 levels and reduced the activity of the HoxA9 promoter in cellular assays. Moreover, they efficiently inhibited the proliferation of MV4-11 cells carrying the oncogenic MLL-AF4 fusion with nanomolar IC50 values. Overall, these novel series (compounds 2, 3/4 and 5) of potent, selective, SAM-competitive DOT1L inhibitors are exciting. In addition, Compounds 3 and 4 are useful chemical tools for cellular and in vivo studies.
Inhibitors of protein arginine methyltransferases
CARM1 (PRMT4) is responsible for the asymmetric dimethylation of H3R17 and H3R26, with preference for the former. CARM1 also methylates a variety of non-histone proteins.[40] Several HTS campaigns and SAR studies resulted in the identification of CARM1 inhibitors with limited selectivity.[41–43] Cocrystal structures of the CARM1 catalytic domain in complex with these inhibitors reveal that they are anchored in the PRMT arginine-binding channel through a basic alkyl-diamino or alanine-amide tail.[44]
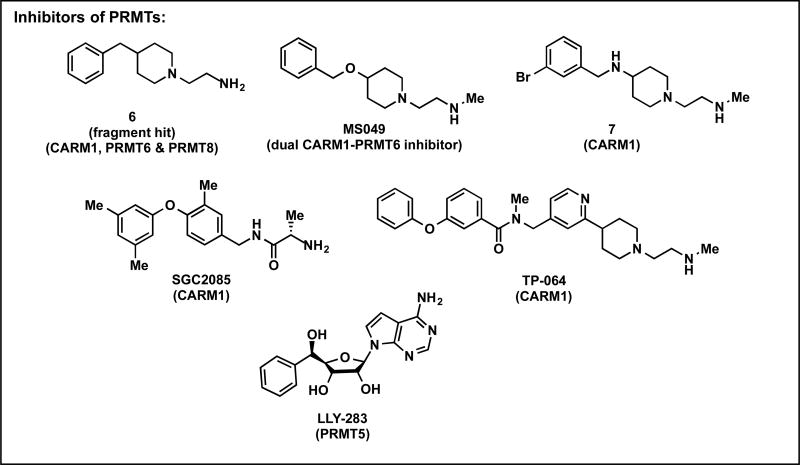
Utilizing a fragment-based approach, a commercially available diverse fragment library of compounds mimicking basic amino tails was tested against PRMT6, resulting in the discovery of fragment hit 6 that inhibits CARM1, PRMT6, and PRMT8 (Figure 3).[45] Recently, highly potent, selective, and cell-active dual CARM1 and PRMT6 inhibitor, MS049 (Figure 3), was discovered via SAR studies based on the aforementioned fragment hit 6.[46] The same research group also reported a potent and selective inhibitor of CARM1, compound 7 (Figure 3), based again on the fragment hit 6.[47] Concurrent with the two studies described above, another potent and selective CARM1 inhibitor SGC2085 displaying good selectivity was discovered via virtual screening.[48] However, this inhibitor is inactive in cell-based assays, likely due to its poor cell membrane permeability. Recently, the first potent, selective, and cell-active inhibitor of CARM1, TP-064, was discovered but detailed report of this inhibitor has not yet been published (Figure 3).[49]
Figure 3. Structures of inhibitors of PRMTs.
The structures of recently reported inhibitors of CARM1 and PRMT5.
In 2016, a potent PRMT5 inhibitor GSK3326595 (structure is not yet disclosed in literature), potently inhibits tumor growth in cellular and animal models, has entered phase I clinical trials (NCT02783300). Moreover, LLY-283, the first potent and selective SAM-competitive chemical probe for PRMT5 was recently discovered (Figure 3).[50] LLY-283 potently inhibits PRMT5, is >100-fold selective for PRMT5 over other methyltransferases and non-epigenetic targets, and shows activity in cellular assays.
Conclusion
Remarkable progress has been made in the discovery of selective, small molecule inhibitors of PMTs over the past two years. These inhibitors have been used and continue to be used extensively in studies as tool compounds to decipher biology and disease relations of these enzymes. While there has been significant progress, there is still much to be achieved in the PMT inhibitor field since, many individual targets and subgroups of targets on the PMT phylogenetic tree lack selective inhibitors, including MLL family, MMSET (NSD-2), and PRDMs (Figure 1). Potent, selective, and cell-active inhibitors of PMTs are invaluable chemical tools to better understand biological functions of PMTs and test therapeutic hypotheses concerning these proteins.
Highlights.
Inhibitors that selectively target EED component of PRC2 and GLP were discovered.
First-in-class, selective, cell active SUV420H1/H2 inhibitor was disclosed.
A covalent inhibitor of SETD8 has recently been reported.
Novel DOT1L inhibitors differing from known SAM-based inhibitors were discovered.
New inhibitors of protein arginine methyltrasferases CARM1 and PRMT5 were published.
Acknowledgments
This work was supported by Grant R01GM103893 (J.J.) and R01GM122749 (J.J.) from the National Institute of General Medical Sciences of the U.S. National Institutes of Health.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
* of particular interest
** of outstanding interest
- 1.Rea S, Eisenhaber F, O'Carroll D, Strahl BD, Sun ZW, Schmid M, Opravil S, Mechtler K, Ponting CP, Allis CD, et al. Regulation of Chromatin Structure by Site-Specific Histone H3 Methyltransferases. Nature. 2000;406:593–599. doi: 10.1038/35020506. [DOI] [PubMed] [Google Scholar]
- 2.Kouzarides T. Chromatin Modifications and their Function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 3.Bannister AJ, Kouzarides T. Regulation of Chromatin by Histone Modifications. Cell Res. 2011;21:381–395. doi: 10.1038/cr.2011.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arrowsmith CH, Bountra C, Fish PV, Lee K, Schapira M. Epigenetic Protein Families: A New Frontier for Drug Discovery. Nat. Rev. Drug Discovery. 2012;11:384–400. doi: 10.1038/nrd3674. [DOI] [PubMed] [Google Scholar]
- 5.Copeland RA, Solomon ME, Richon VM. Protein Methyltransferases as a Target Class for Drug Discovery. Nat. Rev. Drug Discovery. 2009;8:724–732. doi: 10.1038/nrd2974. [DOI] [PubMed] [Google Scholar]
- 6.Helin K, Dhanak D. Chromatin Proteins and Modifications as Drug Targets. Nature. 2013;502:480–488. doi: 10.1038/nature12751. [DOI] [PubMed] [Google Scholar]
- 7.Zagni C, Chiacchio U, Rescifina A. Histone Methyltransferase Inhibitors: Novel Epigenetic Agents for Cancer Treatment. Curr. Med. Chem. 2013;20:167–185. doi: 10.2174/092986713804806667. [DOI] [PubMed] [Google Scholar]
- 8.Schapira M. Chemical Inhibition of Protein Methyltransferases. Cell Chem. Biol. 2016;23:1067–1076. doi: 10.1016/j.chembiol.2016.07.014. [DOI] [PubMed] [Google Scholar]
- 9**.Kaniskan HÜ, Martini ML, Jin J. Inhibitors of Protein Methyltransferases and Demethylases. Chem. Rev. 2017 doi: 10.1021/acs.chemrev.6b00801. Article ASAP. A recent and comprehensive review of inhibitors of protein methyltransferases and demethylases. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schapira M, Arrowsmith CH. Methyltransferase Inhibitors for Modulation of the Epigenome and Beyond. Curr. Opin. Chem. Biol. 2016;33:81–87. doi: 10.1016/j.cbpa.2016.05.030. [DOI] [PubMed] [Google Scholar]
- 11.Pfister SX, Ashworth A. Marked for death: targeting epigenetic changes in cancer. Nat. Rev. Drug Discovery. 2017;16:241–263. doi: 10.1038/nrd.2016.256. [DOI] [PubMed] [Google Scholar]
- 12.Hu H, Qian K, Ho MC, Zheng YG. Small Molecule Inhibitors of Protein Arginine Methyltransferases. Expert Opin. Invest. Drugs. 2016;25:335–358. doi: 10.1517/13543784.2016.1144747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fuhrmann J, Clancy KW, Thompson PR. Chemical Biology of Protein Arginine Modifications in Epigenetic Regulation. Chem. Rev. 2015;115:5413–5461. doi: 10.1021/acs.chemrev.5b00003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kaniskan HÜ, Szewczyk MM, Yu Z, Eram MS, Yang X, Schmidt K, Luo X, Dai M, He F, Zang I, et al. A Potent, Selective and Cell-Active Allosteric Inhibitor of Protein Arginine Methyltransferase 3 (PRMT3) Angew. Chem. Int. Ed. 2015;54:5166–5170. doi: 10.1002/anie.201412154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15*.Butler KV, Ma A, Yu W, Li F, Tempel W, Babault N, Pittella-Silva F, Shao J, Wang J, Luo M, et al. Structure-Based Design of a Covalent Inhibitor of the SET Domain-Containing Protein 8 (SETD8) Lysine Methyltransferase. J. Med. Chem. 2016;59:9881–9889. doi: 10.1021/acs.jmedchem.6b01244. Report of a covalent inhibitor of SETD8 demonstrated that cysteine residues in active sites of PMTs can be selectively targeted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Getlik M, Smil D, Zepeda-Velazquez C, Bolshan Y, Poda G, Wu H, Dong AP, Kuznetsova E, Marcellus R, Senisterra G, et al. Structure-Based Optimization of a Small Molecule Antagonist of the Interaction Between WD Repeat-Containing Protein 5 (WDR5) and Mixed-Lineage Leukemia 1 (MLL1) J. Med. Chem. 2016;59:2478–2496. doi: 10.1021/acs.jmedchem.5b01630. [DOI] [PubMed] [Google Scholar]
- 17.Borkin D, He S, Miao H, Kempinska K, Pollock J, Chase J, Purohit T, Malik B, Zhao T, Wang J, et al. Pharmacologic Inhibition of the Menin-MLL Interaction Page 18 of 21 Blocks Progression of MLL Leukemia in vivo. Cancer Cell. 2015;27:589–602. doi: 10.1016/j.ccell.2015.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18**.He Y, Selvaraju S, Curtin ML, Jakob CG, Zhu H, Comess KM, Shaw B, The J, Lima-Fernandes E, Szewczyk MM, et al. The EED Protein-Protein Interaction Inhibitor A-395 Inactivates the PRC2 Complex. Nat. Chem. Biol. 2017;13:389–395. doi: 10.1038/nchembio.2306. A PPI disrupting/allosteric Inhibitor targeting the EED component of PRC2 complex is discovered. [DOI] [PubMed] [Google Scholar]
- 19**.Qi W, Zhao K, Gu J, Huang Y, Wang Y, Zhang H, Zhang M, Zhang J, Yu Z, Li L, et al. An Allosteric PRC2 Inhibitor Targeting the H3K27me3 Binding Pocket of EED. Nat. Chem. Biol. 2017;13:381–388. doi: 10.1038/nchembio.2304. A PPI disrupting/allosteric Inhibitor targeting the EED component of PRC2 complex is discovered. [DOI] [PubMed] [Google Scholar]
- 20*.Xiong Y, Li F, Babault N, Dong A, Zeng H, Wu H, Chen X, Arrowsmith CH, Brown PJ, Liu J, et al. Discovery of Potent and Selective Inhibitors for G9a-Like Protein (GLP) Lysine Methyltransferase. J. Med. Chem. 2017;60:1876–1891. doi: 10.1021/acs.jmedchem.6b01645. Development of a GLP inhibitor that is selective over closely related G9a as well as other methyltransferases is detailed. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Margueron R, Reinberg D. The Polycomb Complex PRC2 and its Mark in Life. Nature. 2011;469:343–349. doi: 10.1038/nature09784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim KH, Roberts CW. Targeting EZH2 in cancer. Nat. Med. 2016;22:128–134. doi: 10.1038/nm.4036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vaswani RG, Gehling VS, Dakin LA, Cook AS, Nasveschuk CG, Duplessis M, Iyer P, Balasubramanian S, Zhao F, Good AC, et al. Identification of (R)-N-((4-Methoxy-6-methyl-2-oxo-1,2-dihydropyridin-3-yl)methyl)-2-methyl-1-(1-(1 -(2,2,2-trifluoroethyl)piperidin-4-yl)ethyl)-1H-indole-3-carboxamide (CPI-1205), A Potent and Selective Inhibitor of Histone Methyltransferase EZH2, Suitable for Phase I Clinical Trials for B-Cell Lymphomas. J. Med. Chem. 2016;59:9928–9941. doi: 10.1021/acs.jmedchem.6b01315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kung PP, Rui E, Bergqvist S, Bingham P, Braganza J, Collins M, Cui M, Diehl W, Dinh D, Fan C, et al. Design and Synthesis of Pyridone-Containing 3,4-Dihydroisoquinoline-1(2H)-ones as a Novel Class of Enhancer of Zeste Homolog 2 (EZH2) Inhibitors. J. Med. Chem. 2016;59:8306–8325. doi: 10.1021/acs.jmedchem.6b00515. [DOI] [PubMed] [Google Scholar]
- 25.Jiao LY, Liu X. Structural Basis of Histone H3K27 Trimethylation by an Active Polycomb Repressive Complex 2. Science. 2015;350 doi: 10.1126/science.aac4383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Justin N, Zhang Y, Tarricone C, Martin SR, Chen S, Underwood E, De Marco V, Haire LF, Walker PA, Reinberg D, et al. Structural basis of oncogenic histone H3K27M inhibition of human polycomb repressive complex 2. Nat. Commun. 2016;7:11316. doi: 10.1038/ncomms11316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brooun A, Gajiwala KS, Deng YL, Liu W, Bolanos B, Bingham P, He YA, Diehl W, Grable N, Kung PP, et al. Polycomb Repressive Complex 2 Structure with Inhibitor Reveals A Mechanism of Activation and Drug Resistance. Nat. Commun. 2016;7:11384. doi: 10.1038/ncomms11384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Barnash KD, The J, Norris-Drouin JL, Cholensky SH, Worley BM, Li F, Stuckey JI, Brown PJ, Vedadi M, Arrowsmith CH, et al. Discovery of Peptidomimetic Ligands of EED as Allosteric Inhibitors of PRC2. ACS Comb Sci. 2017;19:161–172. doi: 10.1021/acscombsci.6b00174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Greer EL, Shi Y. Histone Methylation: A Dynamic Mark in Health, Disease and Inheritance. Nat. Rev. Genet. 2012;13:343–357. doi: 10.1038/nrg3173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Herz HM, Garruss A, Shilatifard A. SET for life: biochemical activities and biological functions of SET domain-containing proteins. Trends Biochem Sci. 2013;38:621–639. doi: 10.1016/j.tibs.2013.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kohlmann A, Schoch C, Dugas M, Schnittger S, Hiddemann W, Kern W, Haferlach T. New Insights into MLL Gene Rearranged Acute Leukemias using Gene Expression Profiling: Shared Pathways, Lineage Commitment, and Partner Genes. Leukemia. 2005;19:953–964. doi: 10.1038/sj.leu.2403746. [DOI] [PubMed] [Google Scholar]
- 32.Eggert E, Hillig RC, Koehr S, Stockigt D, Weiske J, Barak N, Mowat J, Brumby T, Christ CD, Ter Laak A, et al. Discovery and Characterization of a Highly Potent and Selective Aminopyrazoline-Based in Vivo Probe (BAY-598) for the Protein Lysine Methyltransferase SMYD2. J. Med. Chem. 2016;59:4578–4600. doi: 10.1021/acs.jmedchem.5b01890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Milite C, Feoli A, Viviano M, Rescigno D, Cianciulli A, Balzano AL, Mai A, Castellano S, Sbardella G. The emerging role of lysine methyltransferase SETD8 in human diseases. Clin. Epigenetics. 2016;8:102. doi: 10.1186/s13148-016-0268-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Williams DE, Dalisay DS, Li F, Amphlett J, Maneerat W, Chavez MAG, Wang YA, Matainaho T, Yu W, Brown PJ, et al. Nahuoic Acid A Produced by a Streptomyces sp. Isolated From a Marine Sediment Is a Selective SAM-Competitive Inhibitor of the Histone Methyltransferase SETD8. Org. Lett. 2013;15:414–417. doi: 10.1021/ol303416k. [DOI] [PubMed] [Google Scholar]
- 35.Ma A, Yu W, Li F, Bleich RM, Herold JM, Butler KV, Norris JL, Korboukh V, Tripathy A, Janzen WP, et al. Discovery of A Selective, Substrate-Competitive Inhibitor of the Lysine Methyltransferase SETD8. J. Med. Chem. 2014;57:6822–6833. doi: 10.1021/jm500871s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36**.Bromberg KD, Mitchell TR, Upadhyay AK, Jakob CG, Jhala MA, Comess KM, Lasko LM, Li C, Tuzon CT, Dai Y, et al. The SUV4-20 Inhibitor A-196 Verifies a Role for Epigenetics in Genomic Integrity. Nat. Chem. Biol. 2017;13:317–324. doi: 10.1038/nchembio.2282. First-in-class selective, inhibitor of SUV420H1 and SUV420H2 is reported and its activity in cells is displayed. [DOI] [PubMed] [Google Scholar]
- 37*.Scheufler C, Mobitz H, Gaul C, Ragot C, Be C, Fernandez C, Beyer KS, Tiedt R, Stauffer F. Optimization of a Fragment-Based Screening Hit toward Potent DOT1L Inhibitors Interacting in an Induced Binding Pocket. ACS Med. Chem. Lett. 2016;7:730–734. doi: 10.1021/acsmedchemlett.6b00168. A novel scaffold of DOT1L inhibitor differing from known SAM-derived inhibitors discovered via fragment based approach. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38*.Chen C, Zhu H, Stauffer F, Caravatti G, Vollmer S, Machauer R, Holzer P, Mobitz H, Scheufler C, Klumpp M, et al. Discovery of Novel DOT1L Inhibitors through A Structure-Based Fragmentation Approach. ACS Med. Chem. Lett. 2016;7:735–740. doi: 10.1021/acsmedchemlett.6b00167. A novel scaffold of DOT1L inhibitor differing from known SAM-derived inhibitors discovered via fragment based approach. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39*.Mobitz H, Machauer R, Holzer P, Vaupel A, Stauffer F, Ragot C, Caravatti G, Scheufler C, Fernandez C, Hommel U, et al. Discovery of Potent, Selective, and Structurally Novel Dot1L Inhibitors by a Fragment Linking Approach. ACS Med Chem Lett. 2017;8:338–343. doi: 10.1021/acsmedchemlett.6b00519. A novel scaffold of DOT1L inhibitor differing from known SAM-derived inhibitors discovered via fragment linking approach. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lee YH, Stallcup MR. Roles of Protein Arginine Methylation in DNA Damage Signaling Pathways is CARM1 A Life-or-Death Decision Point? Cell Cycle. 2011;10:1343–1344. doi: 10.4161/cc.10.9.15379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Huynh T, Chen Z, Pang SH, Geng JP, Bandiera T, Bindi S, Vianello P, Roletto F, Thieffine S, Galvani A, et al. Optimization of Pyrazole Inhibitors of Coactivator Associated Arginine Methyltransferase 1 (CARM1) Bioorg. Med. Chem. Lett. 2009;19:2924–2927. doi: 10.1016/j.bmcl.2009.04.075. [DOI] [PubMed] [Google Scholar]
- 42.Wan HH, Huynh T, Pang SH, Geng JP, Vaccaro W, Poss MA, Trainor GL, Lorenzi MV, Gottardis M, Jayaraman L, et al. Benzo[d]imidazole Inhibitors of Coactivator Associated Arginine Methyltransferase 1 (CARM1)-Hit to Lead Studies. Bioorg. Med. Chem. Lett. 2009;19:5063–5066. doi: 10.1016/j.bmcl.2009.07.040. [DOI] [PubMed] [Google Scholar]
- 43.Allan M, Manku S, Therrien E, Nguyen N, Styhler S, Robert MF, Goulet AC, Petschner AJ, Rahil G, MacLeod AR, et al. N-Benzyl-1-Heteroaryl-3-(Trifluoromethyl)-1h-Pyrazole-5-Carboxamides as Inhibitors of Co-Activator Associated Arginine Methyltransferase 1 (CARM1) Bioorg. Med. Chem. Lett. 2009;19:1218–1223. doi: 10.1016/j.bmcl.2008.12.075. [DOI] [PubMed] [Google Scholar]
- 44.Sack JS, Thieffine S, Bandiera T, Fasolini M, Duke GJ, Jayaraman L, Kish KF, Klei HE, Purandare AV, Rosettani P, et al. Structural Basis for CARM1 Inhibition by Indole and Pyrazole Inhibitors. Biochem. J. 2011;436:331–339. doi: 10.1042/BJ20102161. [DOI] [PubMed] [Google Scholar]
- 45.Ferreira de Freitas R, Eram MS, Szewczyk MM, Steuber H, Smil D, Wu H, Li F, Senisterra G, Dong A, Brown PJ, et al. Discovery of a Potent Class I Protein Arginine Methyltransferase Fragment Inhibitor. J. Med. Chem. 2016;59:1176–1183. doi: 10.1021/acs.jmedchem.5b01772. [DOI] [PubMed] [Google Scholar]
- 46.Shen Y, Szewczyk MM, Eram MS, Smil D, Kaniskan HÜ, Ferreira de Freitas R, Senisterra G, Li F, Schapira M, Brown PJ, et al. Discovery of A Potent, Selective, and Cell-Active Dual Inhibitor of Protein Arginine Methyltransferase 4 and Protein Arginine Methyltransferase 6. J. Med. Chem. 2016;59:9124–9139. doi: 10.1021/acs.jmedchem.6b01033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kaniskan HÜ, Eram MS, Liu J, Smil D, Martini ML, Shen Y, Santhakumar V, Brown PJ, Arrowsmith CH, Vedadi M, et al. Design and Synthesis of Selective, Small Molecule Inhibitors of Coactivator-Associated Arginine Methyltransferase 1 (CARM1) MedChemComm. 2016;7:1793–1796. doi: 10.1039/C6MD00342G. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ferreira de Freitas R, Eram MS, Smil D, Szewczyk MM, Kennedy S, Brown PJ, Santhakumar V, Barsyte-Lovejoy D, Arrowsmith CH, Vedadi M, et al. Discovery of a Potent and Selective Coactivator Associated Arginine Methyltransferase 1 (CARM1) Inhibitor by Virtual Screening. J. Med. Chem. 2016;59:6838–6847. doi: 10.1021/acs.jmedchem.6b00668. [DOI] [PubMed] [Google Scholar]
- 49.TP-064: A Chemical Probe For PRMT4. 2017 Jan; http://www.thesgc.org/chemical-probes/TP-064.
- 50.LLY-283: A Chemical Probe For PRMT5. 2016 Nov; http://www.thesgc.org/chemical-probes/LLY-283.