Abstract
Histone deacetylase 8, part of a broad class of proteins responsible for regulating transcription and many other cellular processes and directly linked to a host of human disease through its mis-function, has been canonically described as a zinc-based mettalo-enzyme for many years. Recent evidence, however, has linked this protein to iron incorporation, loaded through transient interactions with the poly r(C)-binding protein 1, a metallo-chaperone and storage protein. In this report, we construct and deploy an electrospray-mass spectrometry based assay aimed at quantifying the interaction strength between these two weakly-associated proteins, as well as the zinc and iron associated form of the histone deacetylase. Despite challenges derived from artifact protein complexes derived from the electrospray process, we use carefully-constructed positive and negative control experiments, along with detailed measurements of protein ionization efficiency to validate our dissociation constant measurements for protein dimers in this size range. Furthermore, our data strongly support that complexes between histone deacetylase 8 and poly r(C)-binding protein 1 are specific, and that they are equally strong when both zinc and iron-loaded proteins are involved, or perhaps mildly promoted in the latter case, suggesting an in vivo role for the non-canonical, iron-incorporated histone deacetylase.
Keywords: Protein-protein Complex, Native Mass Spectrometry, Dissociation Constant, Metalloenzymes, Carbonic Anhydrase, Electrospray Ionization
Introduction
Histone deacetylases (HDACs) play a key role in regulating transcription and many other cellular processes by catalyzing the hydrolysis of -acetyle-lysine residues.[1, 2] Over the past decade, tremendous interest has been centered on these enzymes due to their promise as targets for therapeutic development in the context of a variety of diseases, including asthma, cancer and inflammatory lung diseases.[3–6] Understanding the fundamental role of histone acetylation and deacetylation in the basic processes surrounding gene expression is thus critically important in treating various diseases, as well as for our fundamental understanding of biochemistry.[7, 8]
There are 18 known HDAC enzymes divided phylogenetically into four classes: class I (HDAC1-3, and HDAC8), class II (HDAC4-7 and HDAC9-10), class III (sirtuins 1-7), and class IV (HDAC11).[3] Among these, histone deacetylase 8 (HDAC8) serves as our scientific focus for this work (structure shown in Figure S1). This enzyme has been directly linked to acute myeloid leukemia and the development of the actin cytoskeleton via its native enzymatic activity.[6, 9] Upon its discovery, HDAC8 was validated as a Zn(II)-dependent metalloenzyme;[10] however, in vitro activity and binding affinity assays suggest that Fe could also serve as a native metal cofactor in vivo.[11] A recent systematic investigation suggests that HDAC8 can be activated by either Zn(II) or Fe(II), depending on the local cellular environment of the enzyme.[11]
In cells, the majority of class I HDACs execute their biological function as a part of a multi-protein complex [12–14] and it is proposed that HDAC8 may operate similarly, forming complexes that alter the metal selectivity to adjust the activity of HDAC8. The mechanism by which HDAC8 recognizes and incorporates the cognate metal ions (zinc or iron) in the cell is largely unknown. Metal incorporation for a number of metallo-proteins is facilitated by metallo-chaperones [15, 16] and while there are currently no zinc-specific metallo-chaperones identified, several potential iron-specific metallo-chaperones are being investigated for roles in iron homeostasis, particularly in the assembly of Fe-S clusters.[17–20] For example, it has been reported that poly r(C)-binding protein 1 (PCBP1), a cellular iron storage protein, can function as a cytosolic iron chaperone in the delivery of iron to ferritin.[21]
The first global protein interaction network for 11 HDACs in human CEM T-cells (leukemic cell line) revealed HDAC8 interacting with multiple members of the cohesin complex [12] associated with sister chromatid segregation during mitosis.[21] Moreover, this analysis suggests that HDAC8 may also interact with the PCBP family of iron-metallo-chaperones, despite lingering controversy on the subject.[12] The PCBP family consists of four homologous RNA-binding proteins (PCBP2, PCBP3, and PCBP4) that are ubiquitously expressed in the mammalian cytosol and nucleus.[22] Human PCBP1 was recently identified as an iron chaperone for human ferritin [12] and functional assays in yeast indicate that PCBP1 facilitates the incorporation of iron into ferritin through a direct protein-protein interaction.[23] Most recently, in-vivo co-immunoprecipitation assays revealed the formation of the HDAC8-PCBP1 complex in cells, indicating that PCBP1 and HDAC8 are physically interacting independent of cellular iron concentrations, although the specificity and strength of this interaction has yet to be determined.
Over the past two decades, nano-electrospray ionization (nESI) - mass spectrometry (MS) has emerged as a key technology for the identification and quantification of protein-ligand interactions in vitro.[24–27] High throughput affinity assays of protein-small molecule complexes [26, 28–30] have been applied to the investigation of systems which are not readily accessible by conventional techniques, such as protein-glycolipid systems.[29] The major advantages of the nESI-MS approach include simplicity (no labeling or immobilization required), speed (data can be acquired in minutes), and selectivity (protein assemblies and mixtures can be further analyzed by techniques coupled to MS, for example ion mobility spectrometry).[27] The introduction of ion mobility (IM), which separates ions according to their size and charge on the millisecond timescale, in tandem with MS, further enables the acquisition of such binding information by highlighting conformer-specific small molecule interactions [31, 32], as well as offering an enhanced ability to deconvolute signals for target oligomers [33–36].
While nESI-MS has proven exceptionally useful for quantifying protein-small molecule KD values, there are many challenges to the wider application of the methodology. For example, one of the key assumptions in the interpretation of the results referenced above is that the apo and ligand-bound protein possess nearly identical nESI-MS ionization and detection efficiencies. The validity of such an assumption is strongly supported by protein-small molecule KD data currently available, as the overall accessible surface area of the binding target does significantly change upon small molecule attachment.[26, 28] In contrast, very few direct nESI-MS experiments have targeted protein-protein KD measurements. Direct nESI-MS measurements have led to accurate self-dissociation constants for both the β-lactoglobulin and hemoglobin homo-dimer,[37] as well as the complexes involved in the Hsp90 interaction network.[38] Using nESI-MS to measure the binding strength of such stable (KD < 100nM) assemblies is certainly a challenge; however, analyzing more weakly (KD > 1μM) associated proteins often results in a background of artifact protein complexes resulting from the nESI process itself. When protein concentrations are increased in order to favor the formation weaker protein-protein complexes, the ESI droplet formation process can force the formation of non-specific complexes through solvent evaporation.[39,40] As interest in weakly-associated protein interactions, and their role in formation of transient signaling complexes, intensifies, it is clear that there is a need for new nESI-MS data and strategies aimed at the detection and quantification of such assemblies.
Here we explore, for the first time, the binding affinity of HDAC8 and PCB1 using nESI-IM-MS. We conduct a series of systematic investigations aimed at three major questions: 1) Is the interaction between HDAC8 and PCBP1 relevant to HDAC8 function in vivo? 2) Can we quantify the strength of this interaction via the direct nESI-MS measurements? 3) How does metal association (Zn2+ or Fe2+) affect the binding behavior of this complex? Our nESI-MS data, which builds upon previous methods for the determination of KD values,[39] provides strong evidence that such assemblies are indeed specific, that their binding strength can indeed be quantified by nESI-MS if specific control experiments are performed, and that metal ions subtly alter the binding strength of the HDAC8:PCBP1 assembly. This data is discussed both in the specific context of HDAC8 activity, as well as the ability of the nESI-MS techniques described here to broadly quantify the binding strengths of weak protein-protein interactions.
Experimental Methods
Expression and purification of HDAC8 and PCBP1
Recombinant His6-HDAC8 was expressed in BL21 (DE3) E. coli transformed with pHD4 and purified as previously described and concentrated to 2–12 mg/mL. [11] Briefly, metal-free HDAC8 was generated by dialyzing purified HDAC8, followed by buffer exchange. Chemically competent BL21(DE3) cells were transformed with pCDF encoding His6-SUMO-tagged PCBP1, as described previously.[22] Protein fractionation following expression was carried out using a linear gradient in buffer A from 30 mM to 500 mM imidazole, to buffer B (20 mM Tris [pH 7.9], 250 mM NaCl, 500 mM imidazole, 10% glycerol, 2.5 mM TCEP), with PCBP1 eluting at 110–230 mM imidazole. The His6-SUMO tag was cleaved and the protein was buffer exchanged using dialysis with buffer A before passing over the nickel column a second time to separate the tag from the untagged protein. The protein was dialyzed overnight at 4 °C first against buffer A containing 1 mM EDTA to remove metals and then against buffer A to initially remove DTA. Finally, the protein was fractionated on a PD-10 column to remove any remaining EDTA and flash frozen for subsequent IM-MS analysis. For detailed protein expression protocols, see the associated Supporting Information document.
IM-MS experiments
All experiments were performed on a Synapt G2 ESI quadrupole-ion mobility-time-of-flight (Q-IM-ToF) mass spectrometer (Waters, Milford, MA), equipped with a nanoflow ESI source, as described previously.[41] Mass spectra were collected under positive ion mode using cesium iodide for calibration. A capillary voltage of 1.68kV was applied and sampling cone voltage and source temperature maintained at 50V and 20 °C during signal acquisition. Backing pressure was set at 7–8 mbar. During data acquisition, the quadrupole was set to dwell at m/z 2000 for 2% of the scan time, and m/z 5000 for 98% of the scan time, in order to maximize the transmission of ions in the region between 2000 and 5000 m/z. To optimize the mass resolution, trap collision voltages ranging from 30–50V were applied, with argon collision gas at a pressure of 2.56 ×10−2 mbar. IM separations were carried out using N2 buffer gas, at a pressure of 3.5 mbar. Data acquisition and processing were carried out using MassLynxV4.1 software. Protein samples were buffer exchanged into a 500 mM ammonium acetate buffer in order to produce a final concentration range of 2 – 18 μM prior to MS analysis, as measured by standard UV-Vis spectroscopy (Nanodrop, Thermo Fisher Scientific, Waltham, MA). Positive control experiments were carried out using Ferredoxin-NADP+ reductase and Ferredoxin protein (Sigma, St. Louis, MO, USA). For metal substitution experiments, either 5 μM Zn(NO3)2 or 5 μM (NH4)2Fe(SO4)2 with 250 μM ascorbic acid were used as a source of Zn2+ or Fe2+, respectively.
Binding affinity (KD) calculation by nESI-MS
The binding affinity, often used as the dissociation constant KD, for a given protein-ligand interaction, in our case is interaction between two different proteins P1 and P2, is determined from the ratio (R) of the total abundance (Ab) of bounded and free protein ions at equilibrium (eq), as measured by nESI-MS for solutions of known initial concentrations of protein (arbitrarily, the larger protein [P1]0) and ligand (the smaller protein [P2]0).[27] For a typical one-to-one protein-protein interaction as shown in equation (1), the KD value is calculated using equation (2):
| (1) |
| (2) |
Where R is given by equation (3):
| (3) |
Results and Discussion
Characterization of HDAC8 and PCBP1 interactions
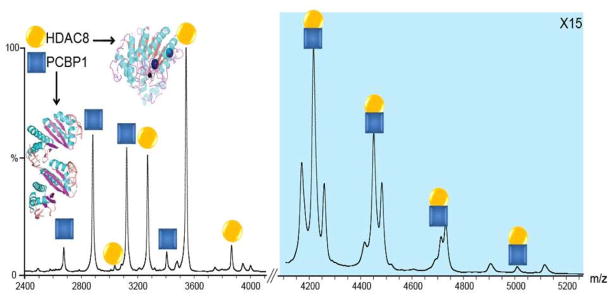
Figure 1 shows the mass spectra acquired from a solution of HDAC8 and PCBP1, and demonstrates the presence of a new species of approximately 80kDa, attributed to the formation of a HDAC8 - PCBP1 heterodimer (42.5kDa + 37.5kDa) which remains stable under conditions of mild collisional activation. The identity of the HDAC8 - PCBP1 heterodimer (H-P) was confirmed by selecting precursor ions at m/z 4452 (the putative 18+ H-P dimer) for MS/MS analysis and collision induced dissociation (CID). In addition to the H-P complex, HDAC8-HDAC8 (H-H) and PCBP1-PCBP1 (P-P) homo-dimers, and trimeric complexes comprised of both proteins are observed at higher PCBP1 to HDAC8 concentrations in solution (Figure 1, blue shaded region).
Figure 1.
Native MS data for HDAC8 incubated with PCBP1 in a 1:1 molar ratio. Signals identified as monomeric HDAC8 (orange) and PCBP1 (blue), as well as the HDAC8-PCBP1 hetero-dimer (both colors) are observed. Low levels of homo-dimer and trimer signals are also evident, but these dissociate readily upon collisional activation. Structural representations of both proteins are shown as insets. The blue shaded region of the spectrum is magnified 15 times in order to aid in visualizing the signals in that region of the spectrum.
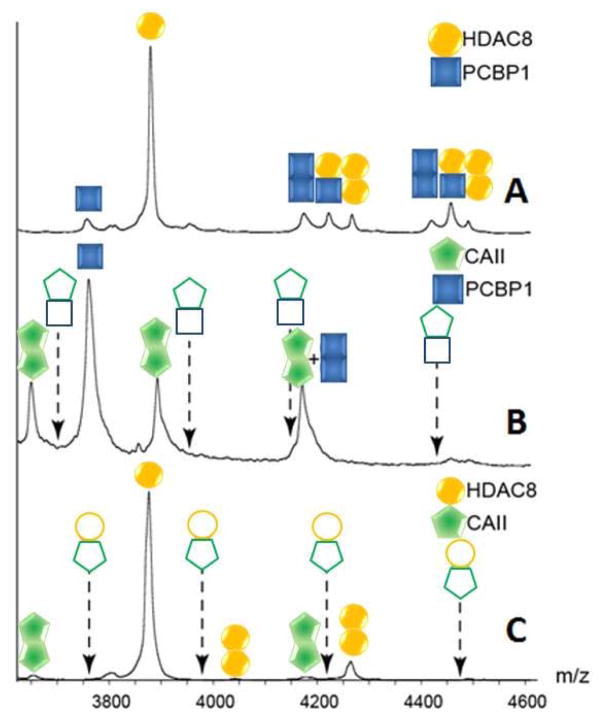
In order to rule out the possibility that the observed H-P dimer signals are due to an artifact of the nESI process, carbonic anhydrase II (CAII, molecular mass 29kDa) was employed as a negative control. CAII is a protein of a size similar to HDAC8 and PCB1, is well studied, and is not known to bind with either HDAC8 or PCBP1. [12, 42] Figure 2 shows the nESI-MS dataset acquired from a solution containing a 1:1:1 molar ratio of CAII, PCBP1 and HDAC8. We observe no evidence of CAII-PCBP1 or CAII-HDAC8 hetero-dimer complex formation under our experimental conditions, thus supporting the specificity of the H-P dimers detected. Conversely, we observe signals for homo-dimer formation throughout our dataset, which we interpret as evidence of both non-zero protein self-association constants in bulk solution for these proteins (which we attribute to signals recorded for homo-dimers at low protein concentrations) and ESI artifact complex formation (attributed to signals recorded from solutions prepared using higher protein concentrations). Metal incorporation into the proteins was verified by activity assays as described previously using samples prepared in 500 mM ammonium acetate buffer (identical to those conditions used for nESI-MS analysis), as the m/z shifts associated with metal incorporation are too small to detect directly by our nESI-MS platform. [11]
Figure 2.
(A) A region of the spectrum shown in Figure 1, showing signals for the heterodimer between HDAC8 and PCBP1. (B) A spectrum acquired under the same instrumental conditions for a sample containing equimolar amounts of CAII and PCBP1, where no heterodimers are detected (indicated by outline-only peak annotations). The peak notation that contains a “+” indicates a region of the spectrum where signal is observed, but two possible ion identities are possible. (C) A spectrum acquired identically to those above for an equimolar sample of CAII and HDAC8. As above, no hetero-dimer signals are detected. In both B and C, dashed arrows are shown representing the projected m/z values for the CAII-associated hetero-dimer assemblies.
Estimating Protein-Protein KD Values for the HDAC8:PCBP1 Complex
In order to overcome the challenges associated with quantifying weak protein KD values by nESI-MS, we worked to verify key assumptions implicit in the nESI-MS KD quantification workflow for HDAC8 and PDCB1 specifically, and also for weak protein-protein complexes in general. The first step in our workflow involves the buffer exchange of analyte protein into an ammonium acetate buffer system appropriate for native MS. Previous reports have indicated that the solution concentration of proteins can be significantly reduced during buffer exchange,[43] and as such we worked to quantify any losses experienced during this step for the proteins studied here. We find that for a typical exchange into 500mM ammonium acetate buffer using widely-available centrifugal columns (see Experimental Methods section for details), the resultant concentration of HDAC8 tends to decrease by 30–60%, and that of PCBP1 can decrease by up to 80% percent. Such significant drops in protein concentration can result in a 5 fold deviation in nESI-MS KD measurements, if not controlled. Based on this data, we built a post-buffer exchange concentration measurement step into our nESI-MS protocol for determining HDAC8:PCBP1 KD values.
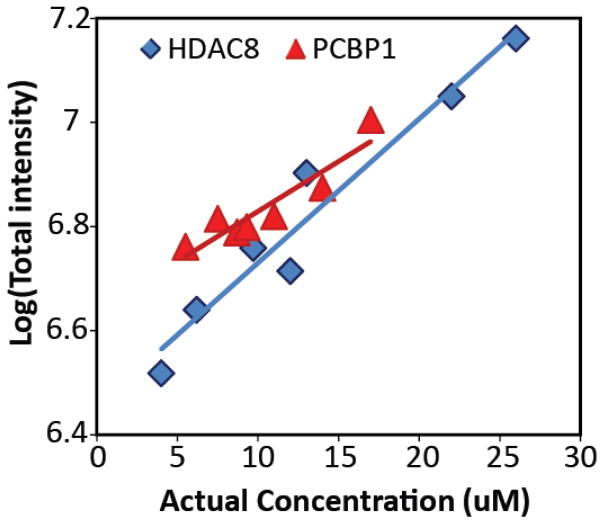
In order to control for differential nESI-MS signal intensities as a function of both PCBP1 and HDAC8 concentrations in solution, we carried out a series of carefully-controlled calibration experiments. Figure 3 shows a series of nESI-MS intensity values for the two proteins referenced above, charted against their post-buffer exchange concentrations in solution. Our results suggest that both HDAC8 and PCBP1 exhibit similar ionization, transmission and detection efficiencies, especially in within the range of concentrations from which we extract KD values (5–15 μM). As such, these results strongly suggest that both proteins are formed via similar ion formation mechanisms, most likely adhering to the well-known charge residue model for ESI protein ion formation.[44] As its solution concentration is unknown, we cannot estimate where signals for the intact H-P complex would appear on Figure 3; however, from the data shown it is highly likely the assembly follows a similar trend.
Figure 3.
A plot of the logarithm of nESI-MS intensity for HDAC8 (blue diamonds) and PCBP1 (red triangles), as a function of protein solution concentration following buffer exchange. Linear trend lines from regression analysis are shown. Taken together, these two trend-lines indicate that nESI-MS intensity can be used to confidently determine HDAC8 and PCBP1 concentrations in solution, over the range of concentrations probed, and thus the KD value associated with their complex formation. The variance values associated with the correlation coefficients from the shown trends (R2 = 0.86 for PCBP1 data and 0.78 for HDAC8 data) are carried forward into our analysis of the KD values reported here.
Low levels of collisional activation have been used previously to dissociate weakly-bound artifact complexes, thus eliminating chemical noise for clear, qualitative assessments of protein binding stoichiometries by nESI-MS.[43] Such methods, by definition, carry a risk of skewing the apparent nESI-MS KD values of detected complexes through uncontrolled CID. In order to evaluate the influence of such collisional activation on the nESI-MS signal intensities of H-P complexes, we varied the activating potential experienced by ions within the trap region of our instrument platform prior to the IM separator up to 140V, observing no evidence of significant CID, indicating that the dimer formed is highly stable in the gas phase and an unlikely source of KD error (data not shown). In addition, we altered the nESI flow rate through applying changes in the nESI backing gas in an effort to investigate the impact of altered droplet sizes and production rates on HDAC8 and PCBP1 signal intensities. These experiments resulted in no evidence of nESI artifact H-P assemblies (data not shown). In order to ensure robust nESI-MS KD values, however, we acquired multiple nESI-MS datasets over a wide range of protein concentrations, and report average values over that range (see below).
Based on the discussion and data above, herein we report KD measurements that cover four different protein molar ratios, each performed in triplicate, (Table 2) for the H-P complex, yielding an average KD value of 63 ± 30 μM (Table 1). The error in these measurements likely comes from 1) uncontrolled variations in our sample preparation, ionization, transmission and detection processes and 2) batch-to-batch sample differences that may influence protein structure, and thus binding behavior. Ultimately, advancements in nESI-IM-MS instrument design and measurement protocols should lead to even more precise KD values. Currently, measurements of protein-protein KD values that lie within the 10–1000 μM range are challenging using traditional approaches (ITC, SPR, etc). [27] As such, the nESI-MS methods described here likely provide one of the only available routes to an accurate HDAC8:PCBP1 KD value.
Table 2.
The binding affinity (KD) values for HDAC8 - PCBP1 complex measured by direct ESI-MS method.
| HDAC8 conc. (μM)* | PCBP1 conc. (μM)* | KD (μM) |
|---|---|---|
| 21 | ||
| 6 | 13 | 28 |
| 29 | ||
|
| ||
| 60 | ||
| 10 | 10 | 82 |
| 72 | ||
|
| ||
| 56 | ||
| 18 | 15 | 59 |
| 63 | ||
|
| ||
| 78 | ||
| 5 | 5.5 | 101 |
| 112 | ||
Twelve measurements (four different molar ratios, each with three replicas) are presented. Protein concentrations are measured by UV-Vis Spectroscopy directly from buffer exchanged solution.
Table 1.
The binding affinity (KD) values for (Apo/Zn/Fe) HDAC8 - PCBP1 complex.
| KD(μM) | # of replicas | |
|---|---|---|
| (Apo-Apo) | 68±30 | 4 |
| (Zn-Apo) | 60±18 | 3 |
| (Fe-Apo) | 44±29 | 5 |
ESI-MS Reveals the Role of Metal Ions in HDAC8:PCBP1 Complex Formation
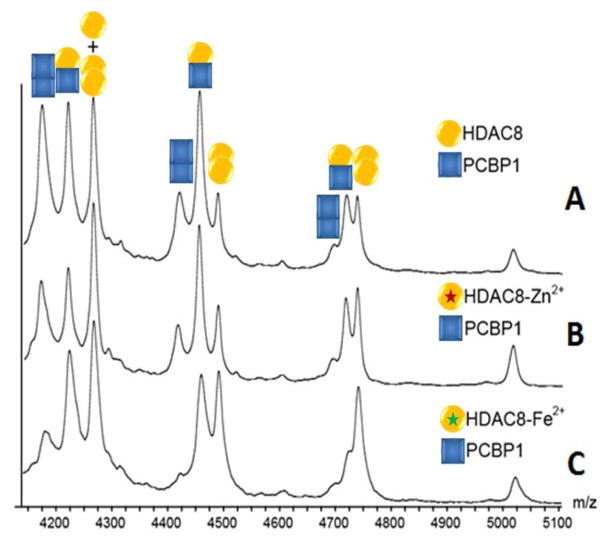
In order to investigate the influence of metal-containing HDAC8 or PCBP1 on complex formation, samples containing either Zn2+ or Fe2+ bound HDAC8 were screened for complex formation and alterations in complex collision cross-section by nESI-IM-MS. No significant changes in ion size were detected in either the intact H-P complex or its protein components by IM-MS (data not shown). HDAC8 binds tightly to zinc (KD = 5 pM) and relatively weakly to iron (KD = 0.2 μM). In the case of PCBP1, three iron binding sites are available, and ITC assays reveal a 0.9 μM affinity to the first site, and 5.8 μM for the remaining two. Our results, shown in Figure 4, suggest that, as observed in Figure 1, by using optimized buffer conditions and properly tuned instrument parameters, formation of apoHDAC8:PCBP1 complexes can be observed in significant excess relative to any homo-oligomers formed by either protein. This observation holds for metal-containing HDAC8 and PCBP1 forms.
Figure 4.
MS data of (A)apo- (B) Zn2+ bound (purple star) (C) Fe2+ bound (green star) HDAC8 incubated with PCBP1. Hetero-dimer signals for the HDAC8 - PCBP1 complex are detected in all cases, independent of metal substitution. A quantitative KD analysis suggests that, although within error, iron may subtly promote HDAC8 binding to PCBP1.
Quantitative nESI-MS KD measurements further reveal a KD value for the zinc-containing HDAC8 – PCBP1 complex of 60±18 μM, while the KD for iron-containing HDAC8 – PCBP1 assemblies is 44±29 μM (see Table 3 for details). Data from Table 1 indicates that Fe2+ mildly promotes the binding between HDAC8 and PCBP1, while noting that the complex KD values recorded for this assembly are within error of those generated for Zn2+ containing HDAC8 forms. Given that Zn2+ has long been viewed as the natural metal center for the HDAC8 active site in vivo,[10] our results strongly suggest that Fe2+ generates equivalently competent HDAC8 proteins in the context of PCBP1 binding.
Table 3.
The binding affinity (KD) values for (Zn/Fe) HDAC8 - PCBP1 complex, as well as the Ferredoxin-NADP+ reductase – Ferredoxin complex measured by direct ESI-MS method
| Protein 1 | Conc. (μM)* | Protein 2 | Conc. (μM)* | KD(μM) | Avg(μM) | Stdev(μM) |
|---|---|---|---|---|---|---|
| 9 | 8 | 77 | ||||
| (Zn)HDAC8 | 4.5 | PCBP1 | 5.5 | 71 | 60 | 18 |
| 4.5 | 5.5 | 34 | ||||
|
| ||||||
| 3 | 2 | 15 | ||||
| 4.5 | 2.3 | 21 | ||||
| (Fe)HDAC8 | 5.5 | PCBP1 | 5.5 | 28 | 44 | 29 |
| 8.5 | 8 | 70 | ||||
| 8.5 | 8 | 88 | ||||
|
| ||||||
| 2.2 | 4 | 10 | ||||
| Ferredoxin Reductase | 4.1 | Ferredoxin | 7.5 | 10 | 16 | 8 |
| 3.1 | 11.2 | 27 | ||||
Protein concentrations are measured by UV-Vis Spectroscopy directly from buffer exchanged solution. Averaged KD values and standard deviations are listed in the right-most two columns.
Validating ESI-MS Protein-Protein KD Measurements through Additional Control Experiments
As the KD values for the PCBP1: HDAC8 complexes reported here were not known previously, and the nESI-MS methodology has rarely been used to quantify the strength of such a weak protein-protein complex, we undertook additional control experiments in order to evaluate the ability of our nESI-MS methodology to measure accurately the KD of a similar protein complex of known binding strength. Following a systematic screen of the structure-based benchmark library for protein-protein binding affinity,[45] we selected the Ferredoxin-NADP+ reductase (32kDa) – Ferredoxin (10.5 kDa) complex (FNR – FDX) as our positive control system. The FNR – FDX complex has a known binding affinity of 3.57 μM, measured by difference absorption spectroscopy,[46] and is similar to HDAC8-PCBP1 complexes in terms of both its KD value and the masses of its component proteins (the intact FNR – FDX complex has a mass of 43 kDa). Our results, listed in Table 3, reveal a binding affinity of (16±8) μM, in good agreement with the literature value (i.e. within a factor of 2).[45] We also note that the absolute precision of our nESI-MS derived KD value for FNR-FDX is improved when compared with those extracted for HDAC8 – PCBP1, suggesting that the strength of the interaction observed for the latter complex may also be a quantifiable source of error in our experiments. Overall, however, these results clearly suggest the direct nESI-MS results described here, when coupled with carefully managed experimental conditions and control experiments can be used to assess the accurate binding affinities for weak protein-protein interactions.
Conclusions
In this report, we systematically investigated strength of the complexes formed between HDAC8 and PCBP1 using nESI-IM-MS. Our data suggests that HDAC8 and PCBP1 interact with each other in a specific manner, as confirmed by tandem MS experiments and comparisons with homo-oligomers also observed. Negative control experiments that tested the ability of CAII, a protein with no known affinity for either protein but possessing similar surface area to both HDAC8 and PCBP1, to form complexes with either of the two binding partners discussed above further verifies our general conclusions. Specifically, these experiments revealed no detectible CAII complexes with either HDAC8 or PCBP1, further supporting the specificity of the HDAC8-PCBP1 complex and the ability of nESI-MS to quantify the strength of weakly associated protein complexes. By optimizing buffer conditions and carefully adjusting instrument settings, we report a KD value of 63 ± 30 μM for the HDAC8-PCBP1 dimer. In order to better understand how bound metals affect the structure and binding behavior of the HDAC8 and PCBP1, Zn2+/Fe2+ doped HDAC8 were incubated with PCBP1 (with and without Fe2+), and we demonstrate that the above-noted dimer forms regardless of the metal-bound form of the component proteins used. As for the strength of the interactions detected, our measured KD values suggests that Fe(II) mildly promotes the formation of the HDAC8-PCBP1 dimer over those bound to Zn(II), strongly indicating a role for both the PCB1 Fe chaperone and Fe(II) binding in the context of HDAC8 in vivo function.
The accuracy of nESI-MS-based KD measurements in general for similar hetero-protein dimers was further evaluated by measurements on the FNR - FDX protein complex. Our nESI-MS measurements yield a KD value within error of the literature value, providing support the general accuracy and precision of this approach for such studies in general.[26, 27, 30] In order to fully evaluate the sources of error associated with the KD values reported in this work, we carried out a detailed analysis of the variances observed between protein batches, and find strong evidence to support that subtle batch-to-batch changes in protein structure likely contribute significantly the errors reported here (see Supporting Information). Future work will aim to produce a comprehensive, robust methodology with high accuracy and precision for the measurement of KD values for weak protein assemblies. For example, while we used IM separation to verify the oligomeric states of protein signals poorly resolved by MS alone in a limited number of cases in this dataset (Figure S2), IM separation could be more-broadly deployed to develop workflows aimed at resolving protein signals and binding modes that exhibit far greater overlap in m/z than those studied here. Moreover, IM offers the capability for monitoring any changes in protein conformational upon complex formation, which when coupled with accurate KD values would be a powerful approach for validating many key therapeutic targets.[32] Overall, the data presented here predicts a bright future for the application of similar nESI-MS toward weakly-associated proteins that currently represent a bottleneck in the detailed evaluation of many key processes in molecular biochemistry.
Supplementary Material
Acknowledgments
This work was supported by the National Institutes of Health, General Medical Sciences (GM095832). In addition, the authors thank Tianshuang Wu for his assistance with the statistical variance analysis of KD data.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403(6765):41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- 2.Shiio Y, Eisenman RN. Histone sumoylation is associated with transcriptional repression. Proceedings of the National Academy of Sciences. 2003;100(23):13225–13230. doi: 10.1073/pnas.1735528100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gregoretti I, Lee Y-M, Goodson HV. Molecular evolution of the histone deacetylase family: functional implications of phylogenetic analysis. Journal of molecular biology. 2004;338(1):17–31. doi: 10.1016/j.jmb.2004.02.006. [DOI] [PubMed] [Google Scholar]
- 4.Remiszewski S. Recent advances in the discovery of small molecule histone deacetylase inhibitors. Current opinion in drug discovery & development. 2002;5(4):487–499. [PubMed] [Google Scholar]
- 5.Lane AA, Chabner BA. Histone deacetylase inhibitors in cancer therapy. Journal of Clinical Oncology. 2009;27(32):5459–5468. doi: 10.1200/JCO.2009.22.1291. [DOI] [PubMed] [Google Scholar]
- 6.Barnes P, Adcock I, Ito K. Histone acetylation and deacetylation: importance in inflammatory lung diseases. European Respiratory Journal. 2005;25(3):552–563. doi: 10.1183/09031936.05.00117504. [DOI] [PubMed] [Google Scholar]
- 7.Marmorstein R. Structure of histone deacetylases: insights into substrate recognition and catalysis. Structure. 2001;9(12):1127–1133. doi: 10.1016/s0969-2126(01)00690-6. [DOI] [PubMed] [Google Scholar]
- 8.Grozinger CM, Schreiber SL. Deacetylase enzymes: biological functions and the use of small-molecule inhibitors. Chemistry & biology. 2002;9(1):3–16. doi: 10.1016/s1074-5521(02)00092-3. [DOI] [PubMed] [Google Scholar]
- 9.Minucci S, Pelicci PG. Histone deacetylase inhibitors and the promise of epigenetic (and more) treatments for cancer. Nature Reviews Cancer. 2006;6(1):38–51. doi: 10.1038/nrc1779. [DOI] [PubMed] [Google Scholar]
- 10.Finnin MS, et al. Structures of a histone deacetylase homologue bound to the TSA and SAHA inhibitors. Nature. 1999;401(6749):188–193. doi: 10.1038/43710. [DOI] [PubMed] [Google Scholar]
- 11.Gantt SL, Gattis SG, Fierke CA. Catalytic activity and inhibition of human histone deacetylase 8 is dependent on the identity of the active site metal ion. Biochemistry. 2006;45(19):6170–6178. doi: 10.1021/bi060212u. [DOI] [PubMed] [Google Scholar]
- 12.Joshi P, et al. The functional interactome landscape of the human histone deacetylase family. Molecular systems biology. 2013;9(1):672. doi: 10.1038/msb.2013.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eide DJ. Zinc transporters and the cellular trafficking of zinc. Biochimica et Biophysica Acta (BBA)-Molecular Cell Research. 2006;1763(7):711–722. doi: 10.1016/j.bbamcr.2006.03.005. [DOI] [PubMed] [Google Scholar]
- 14.Colvin RA, et al. Cytosolic zinc buffering and muffling: their role in intracellular zinc homeostasis. Metallomics. 2010;2(5):306–317. doi: 10.1039/b926662c. [DOI] [PubMed] [Google Scholar]
- 15.Fukada T, et al. Zinc homeostasis and signaling in health and diseases. JBIC Journal of Biological Inorganic Chemistry. 2011;16(7):1123–1134. doi: 10.1007/s00775-011-0797-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Beyersmann D, Hartwig A. Carcinogenic metal compounds: recent insight into molecular and cellular mechanisms. Archives of toxicology. 2008;82(8):493–512. doi: 10.1007/s00204-008-0313-y. [DOI] [PubMed] [Google Scholar]
- 17.Costello L, Franklin R. Novel role of zinc in the regulation of prostate citrate metabolism and its implications in prostate cancer. The Prostate. 1998;35(4):285–296. doi: 10.1002/(sici)1097-0045(19980601)35:4<285::aid-pros8>3.0.co;2-f. [DOI] [PubMed] [Google Scholar]
- 18.Costello LC, et al. Evidence for a zinc uptake transporter in human prostate cancer cells which is regulated by prolactin and testosterone. Journal of Biological Chemistry. 1999;274(25):17499–17504. doi: 10.1074/jbc.274.25.17499. [DOI] [PubMed] [Google Scholar]
- 19.Franklin RB, et al. hZIP1 zinc uptake transporter down regulation and zinc depletion in prostate cancer. Molecular cancer. 2005;4(1):32. doi: 10.1186/1476-4598-4-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Johnson DC, et al. Structure, function, and formation of biological iron-sulfur clusters. Annu Rev Biochem. 2005;74:247–281. doi: 10.1146/annurev.biochem.74.082803.133518. [DOI] [PubMed] [Google Scholar]
- 21.Shi H, et al. A cytosolic iron chaperone that delivers iron to ferritin. Science. 2008;320(5880):1207–1210. doi: 10.1126/science.1157643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bulteau A-L, et al. Frataxin acts as an iron chaperone protein to modulate mitochondrial aconitase activity. Science. 2004;305(5681):242–245. doi: 10.1126/science.1098991. [DOI] [PubMed] [Google Scholar]
- 23.Jensen LT, Culotta VC. Role of Saccharomyces cerevisiae ISA1and ISA2 in iron homeostasis. Molecular and cellular biology. 2000;20(11):3918–3927. doi: 10.1128/mcb.20.11.3918-3927.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Katta V, Chait BT. Observation of the heme-globin complex in native myoglobin by electrospray-ionization mass spectrometry. Journal of the American Chemical Society. 1991;113(22):8534–8535. [Google Scholar]
- 25.Loo RRO, et al. Observation of a noncovalent ribonuclease S-protein/S-peptide complex by electrospray ionization mass spectrometry. Journal of the American Chemical Society. 1993;115(10):4391–4392. [Google Scholar]
- 26.Wang W, Kitova EN, Klassen JS. Influence of solution and gas phase processes on protein-carbohydrate binding affinities determined by nanoelectrospray Fourier transform ion cyclotron resonance mass spectrometry. Analytical chemistry. 2003;75(19):4945–4955. doi: 10.1021/ac034300l. [DOI] [PubMed] [Google Scholar]
- 27.Kitova EN, et al. Reliable determinations of protein–ligand interactions by direct ESI-MS measurements. Are we there yet? Journal of the American Society for Mass Spectrometry. 2012;23(3):431–441. doi: 10.1007/s13361-011-0311-9. [DOI] [PubMed] [Google Scholar]
- 28.El-Hawiet A, et al. Quantifying labile protein–ligand interactions using electrospray ionization mass spectrometry. Journal of the American Society for Mass Spectrometry. 2010;21(11):1893–1899. doi: 10.1016/j.jasms.2010.07.008. [DOI] [PubMed] [Google Scholar]
- 29.Han L, et al. Protein–Glycolipid Interactions Studied in Vitro Using ESI-MS and Nanodiscs: Insights into the Mechanisms and Energetics of Binding. Analytical chemistry. 2015;87(9):4888–4896. doi: 10.1021/acs.analchem.5b00678. [DOI] [PubMed] [Google Scholar]
- 30.Han L, et al. Identifying carbohydrate ligands of a norovirus P particle using a catch and release electrospray ionization mass spectrometry assay. Journal of The American Society for Mass Spectrometry. 2014;25(1):111–119. doi: 10.1007/s13361-013-0752-4. [DOI] [PubMed] [Google Scholar]
- 31.Rabuck JN, et al. Activation state-selective kinase inhibitor assay based on ion mobility-mass spectrometry. Analytical chemistry. 2013;85(15):6995–7002. doi: 10.1021/ac4012655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Soper MT, et al. Amyloid-β–neuropeptide interactions assessed by ion mobility-mass spectrometry. Physical Chemistry Chemical Physics. 2013;15(23):8952–8961. doi: 10.1039/c3cp50721a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Niu S, Ruotolo BT. Collisional unfolding of multiprotein complexes reveals cooperative stabilization upon ligand binding. Protein Science. 2015;24(8):1272–1281. doi: 10.1002/pro.2699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bornschein RE, et al. Ion Mobility-Mass Spectrometry Reveals Highly-Compact Intermediates in the Collision Induced Dissociation of Charge-reduced Protein Complexes. Journal of the American Society for Mass Spectrometry. 2016;27(1):41–49. doi: 10.1007/s13361-015-1250-7. [DOI] [PubMed] [Google Scholar]
- 35.Hyung S-J, Robinson CV, Ruotolo BT. Gas-phase unfolding and disassembly reveals stability differences in ligand-bound multiprotein complexes. Chemistry & biology. 2009;16(4):382–390. doi: 10.1016/j.chembiol.2009.02.008. [DOI] [PubMed] [Google Scholar]
- 36.Ruotolo BT, et al. Evidence for macromolecular protein rings in the absence of bulk water. Science. 2005;310(5754):1658–1661. doi: 10.1126/science.1120177. [DOI] [PubMed] [Google Scholar]
- 37.Liu J, Konermann L. Protein–protein binding affinities in solution determined by electrospray mass spectrometry. Journal of the American Society for Mass Spectrometry. 2011;22(3):408–417. doi: 10.1007/s13361-010-0052-1. [DOI] [PubMed] [Google Scholar]
- 38.Ebong I-o, et al. Heterogeneity and dynamics in the assembly of the heat shock protein 90 chaperone complexes. Proceedings of the National Academy of Sciences. 2011;108(44):17939–17944. doi: 10.1073/pnas.1106261108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sun J, et al. Method for Distinguishing Specific from Nonspecific Protein–Ligand Complexes in Nanoelectrospray Ionization Mass Spectrometry. Analytical Chemistry. 2006;78(9):3010–3018. doi: 10.1021/ac0522005. [DOI] [PubMed] [Google Scholar]
- 40.Hossain BM, et al. Pulsed hydrogen/deuterium exchange MS/MS for studying the relationship between noncovalent protein complexes in solution and in the gas phase after electrospray ionization. Analytical Chemistry. 2006;78(5):1613–1619. doi: 10.1021/ac051687e. [DOI] [PubMed] [Google Scholar]
- 41.Zhong Y, et al. Characterizing the resolution and accuracy of a second-generation traveling-wave ion mobility separator for biomolecular ions. Analyst. 2011;136(17):3534–3541. doi: 10.1039/c0an00987c. [DOI] [PubMed] [Google Scholar]
- 42.Szklarczyk D, et al. STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Research. 2015;43(Database issue):D447–452. doi: 10.1093/nar/gku1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hernández H, Robinson CV. Determining the stoichiometry and interactions of macromolecular assemblies from mass spectrometry. Nature protocols. 2007;2(3):715–726. doi: 10.1038/nprot.2007.73. [DOI] [PubMed] [Google Scholar]
- 44.Kebarle P, Verkerk UH. Electrospray: From Ions in Solution to Ions in the Gas Phase, What We Know Now. Mass Spectrometry Reviews. 2009;28(6):898–917. doi: 10.1002/mas.20247. [DOI] [PubMed] [Google Scholar]
- 45.Kastritis PL, et al. A structure-based benchmark for protein–protein binding affinity. Protein Science. 2011;20(3):482–491. doi: 10.1002/pro.580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sancho J, Gómez-Moreno C. Interaction of ferredoxin-NADP+ reductase from Anabaena with its substrates. Archives of biochemistry and biophysics. 1991;288(1):231–238. doi: 10.1016/0003-9861(91)90189-p. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.