Abstract
Purpose
We created a novel inducible mouse line Keratocan-rtTA (KeraRT) that allows specific genetic modification in corneal keratocytes and tenocytes during development and in adults.
Methods
A gene-targeting vector (Kera- IRES2-rtTA3) was constructed and inserted right after the termination codon of the mouse Kera allele via gene targeting techniques. The resulting KeraRT mouse was crossed to tet-O-Hist1H2B-EGFP (TH2B-EGFP) to obtain KeraRT/TH2B-EGFP compound transgenic mice, in which cells expressing Kera are labeled with green fluorescence protein (GFP) by doxycycline (Dox) induction. The expression patterns of GFP and endogenous Kera were examined in KeraRT/TH2B-EGFP. Moreover, KeraRT was bred with tet-O-TGF-α to generate a double transgenic mouse, KeraRT/tet-O-TGF-α, to overexpress TGF-α in corneal keratocytes upon Dox induction.
Results
Strong GFP-labeled cells were detected in corneal stroma, limbs, and tail when KeraRT/TH2B-EGFP mice were fed Dox chow. There was no GFP in any single transgenic KeraRT or TH2B-EGFP mouse. Histological analysis showed that GFP in the cornea was limited to stromal keratocytes of KeraRT/TH2B-EGFP, which is consistent with Kera expression. Induction of GFP occurred in 24 hours and reached a plateau by 7 days after Dox induction. GFP could be detected 3-months after induction of KeraRT/TH2B-EGFP. Ectopic expression of TGF-α in corneal keratocytes caused hyperplasia in the corneal epithelium and stroma.
Conclusions
The novel Dox inducible KeraRT driver mouse line is a useful genetic tool for gene manipulation and elucidating gene functions in corneal stroma and tendons of limbs and tail during embryonic development, homeostasis and pathogenesis.
Keywords: knock-in mouse model, GFP, corneal stroma, Dox-inducible, keratocan
Genetically modified mice generated by transgenesis and gene targeting techniques provide important experimental animal models for elucidating the function of genes during development and homeostasis in the adult as well as the pathophysiology of human disease.1,2 However, embryonic lethality and congenital defects observed in many of these experimental mice preclude their use for studying the consequences of genetic modification in adult animals. To circumvent these difficulties, the tetracycline (tet)-inducible transgenic system was developed that allows tissue/cell type–specific manipulation of gene deletion and ectopic expression at specific time points.3–5 This system uses double and triple transgenic mouse lines: one line serves as a driver mouse carrying a tTA (tet-off) or rtTA (tet-on) transcription factor fusion gene, whose expression is under the control of a cell type–specific promoter/enhancer unit of choice, such as Kera as used in this study. Another transgenic mouse serves as the effecter or reporter, which consists of a tet responsive element (TRE) ligated to a cytomegalovirus (CMV) minimal promoter and a gene of interest, such as green fluorescent protein (GFP) and TGF-α in this study. The binding of tTA or rtTA to the tet-operon depends on the absence or presence of doxycycline (Dox, a tetracycline derivative), respectively. The gene attached to the CMV minimal promoter-tetO elements can be activated or inactivated in a Dox-dependent manner.5,6 This strategy has been used widely to produce mouse models to manipulate genes in a temporal-spatial fashion.3,7
The mouse Kera gene expression tracks the morphogenesis of cornea and tendon during mouse development and homeostasis in the adult,8,9 making it a suitable candidate for a mouse driver to study gene function during cornea and tendon development and homeostasis. Several transgenic mouse lines driven by the Kera promoter, such as Kera-Bgn,10 Kera-Cre (KC),11 and Kera-rtTA (KR),12 which ectopically express biglycan, Cre, and rtTA, respectively, have been generated for studies of keratocyte-specific genetic modification. Among these mouse lines, KR is the only driver mouse line established by taking advantage of the Dox-inducible transgenic strategy.12 Interesting data have been obtained from these mouse lines by crossing with effecter mouse lines; however, some limits and disadvantages have been recognized due to either leakage (such as promiscuous ablation of floxed genes during gametogenesis by the Kera-Cre transgene11) or inefficient manipulation of gene expression in the adult, such as KR (our unpublished data). Therefore, establishing a better driver mouse line is required to overcome the pitfalls found in the KR and KC mouse lines.
We previously have generated the Krt12rtTA knock-in mouse line, an excellent corneal epithelium-specific mouse driver in which a IRES-rtTA minigene was introduced following the stop codon of the mouse Krt12 allele.6 This strategy was adapted to produce a novel keratocyte-specific driver mouse line. Efforts were made in this study to generate a knock-in mouse line called KeraRT by inserting the IRES-rtTA minigene immediately behind the stop codon of the mouse Kera allele. Keratocan and rtTA proteins will be produced simultaneously in the corneal keratocytes of this new driver mouse due to the IRES element. This new driver was crossed with the reporter/effector mouse lines TH2B-EGFP and tet-O-TGF-α to characterize this new mouse line functionally.
Materials and Methods
Preparation of Kera-IRES-rtTA Targeting Vector
Conventional cloning techniques were used to construct a Kera-IRES2-rtTA3 targeting vector as shown in Figure 1A. Briefly, to make the pgkDTA backbone, FRT-PGKneo-FRT cassette amplified by PCR was fused with pgkNeolox2DTA.2 (Addgene 13449) by using In-Fusion HD cloning kit (Clontech Laboratories, Mountain View, CA, USA) according to the manufacturer's instructions. Then, the 2.9-kb 3′ and 3.3-kb 5′ homology arms of the C57BL/6N Kera gene, IRES2, and rtTA3 fragments were generated by PCR with 15 base pairs (bp) overlap primers. The 5′ homology arm of Kera gene, IRES2, and rtTA3 fragments were inserted into the EcoRV site of pFRT-PGKneo-FRT-pgkDTA backbone. The obtained DNA plasmid containing the reversed pgkDTA-3.3-kb 5′ homology of Kera-IRES2-rtTA3-reversed FRT-PGKneo-FRT was linearized by SmaI and fused with the 2.9-kb 3′ homology arm of Kera. All cloned sequences were confirmed by an automatic sequencing machine (3500 Genetic Analyzer; Thermo Fisher Scientific, Waltham, MA, USA). PCR primers used in construction of the targeting vector are summarized in Supplementary Table S1.
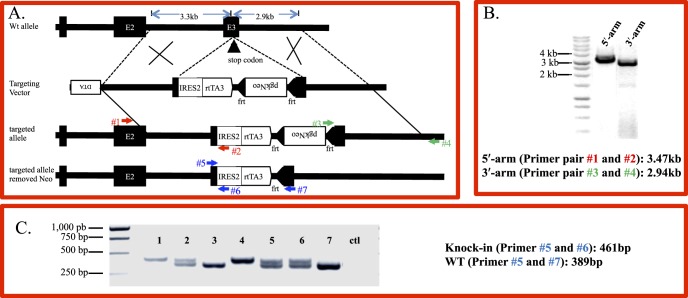
Figure 1.
Generation of keratocyte-specific knock-in mouse line KeraRT via gene targeting. (A) To generate the targeting vector the IRES-rtTA cassette followed by a positive selection marker pgk-Neo minigene was inserted after the Kera exon 3 stop codon. The final targeting vector containing the 3.3 kb 5′-arm, 2.9 kb 3′-arm, and a negative selection marker DTA, was linearized by SacII before being transfected into a C57/BL ES cell line via electroporation. (B) Targeted ES clones were identified by PCR. To identify targeted ES cells, two specific PCR primer pairs detecting the 5′-arm and 3′-arm, respectively, are shown as numbered arrows. Lane 1 is the PCR product using primer #1 and #2 detecting the 5′-arm of targeted clones. Lane 2 is the PCR product using primer #3 and #4 detecting the 3′-arm of targeted clones. (C) Genotyping by PCR of offspring from heterozygous mating. The PCR primer pairs used for identification of the knock-in allele also are shown as numbered arrows. Shown is the expected size of the PCR product amplified by three primers included in one single reaction to distinguish homo-, heterozygous and WT. Lanes 1 and 4 are homozygous knock-in mice. Lane 2, 5, and 6 are heterozygous; while lanes 3 and 7 are WT.
Generation of Knock-In KeraRT Mice
The targeting vector Kera-IRES2-rtTA3 linearized by SacII (Fig. 1A, Supplementary Fig. S1) was introduced into C57BL/6 ES cells by the University of Cincinnati Gene Targeted Mouse Service Core Facility. ES cell clones that stably incorporated the targeting vector were selected by growth medium containing geneticin (225 μg/mL) for 8 days. The homologous recombinant ES cell clones were identified by a double-PCR–based method using two primer pairs (Fig. 1A): KeraE2-F1(primer #1), 5′ CCTGCAGCACCTTCACCTTGATCACAAC (located in Kera exon2, beyond the targeting vector) and reverse primer: IRES-R1(primer #2), 5′ TTATTCCAAGCGGCTTCGGCCAGTAACG produced a 3473 bp PCR product; and KeraE3-F1 (primer #3), 5′ CGGATCCACTAGTTCTAGAGCGGCCTAGTTTA and reverse Kera3-R1(primer #4), 5′ GATTGCAAAATTCTTCGCAGGACCACACAGTCTT (the 3′ flanking polyadenylation signal of Kera allele, which is beyond the targeting vector) produced a 2943 bp PCR product (Figs. 1A, 1B). The identified ES clones were verified further by sequencing and restriction enzyme digestion of the junction regions of the PCR products (Supplementary Figs. S2A, S2B). Correctly targeted ES cells were expanded and were used for blastocyst injection by the Gene Targeted Mouse Service Core Facility at the University of Cincinnati. The germline chimeric mice were bred with Swiss Black females to obtain heterozygous KeraRT mice. The heterozygous mice then were mated with FLP mice (stock number: 003946;13 Jackson Laboratories, Bar Harbor, ME, USA) to genetically remove the Neo gene, which were identified by tail DNA PCR. PCR primer pair: 5′-CACTGATATTGTAAGTAGTTTGC-3′; 5′-CTAGTGCGAAGTAGTGATCAGG-3′ was used to identify the FLP allele. PCR primers: 5′-CATCGAGCGAGCACGTACTCG-3′; 5′-GAGCGGCGATACCGTAAAGCAC-3′ were used to detect the Neo gene (Supplementary Fig. S2).
Generation of Double Transgenic Mice KeraRT/TH2B-EGFP and KeraRT/tet-O-TGF-α
The knock-in KeraRT mice were crossed with an enhanced GFP (EGFP) reporter mouse line, tet-o-Hist1H2B-EGFP (TH2B-EGFP, Stock number: 005104;14 Jackson Laboratories) to obtain the double transgenic mice KeraRT/TH2B-EGFP. The knock-in mice were bred with transgenic mouse line tet-O-TGF-α15 to generate double transgenic mice KeraRT/tet-O-TGF-α. All mice were bred at the Animal Facility at the University of Cincinnati Medical Center and School of Optometry, Indiana University. Experimental procedures for handling the mice were approved by the Institutional Animal Care and Use Committee, University of Cincinnati/College of Medicine and Indiana University/School of Optometry. Animal care and use conformed to the ARVO Statement for the Use of Animals in Ophthalmologic and Vision Research.
Genotyping of Double Transgenic Mice by PCR
The knock-in KeraRT mice were identified by PCR using the following PCR primers (Fig. 1C): Kera-F1 (primer #5), 5′-TGGTGGCTTGCTTCAAGCTTCTTC-3′; reverse primer Kera-R1 (primer #6), 5′-TATCCAACTCACAACGTGGCACTG-3′, and Kera-R2 (primer #7), 5′-GGAGTCTGCACTACCAGTACTCAT-3′ (Figs. 1A, 1C). The identification of reporter mice TH2B-EGFP also were performed by PCR using tail DNA using specific primer pairs (Jackson Laboratories).
Administration of Dox Chow
Mice were subjected to systemic induction by Dox chow (1 g/kg; Custom Animal Diets, Bangor, PA, USA) for different periods of time. To induce mice from the embryonic stage, pregnant dams were given an intraperitoneal (IP) injection of Dox (80 μg/g body weight in PBS, pH7.4; Clontech Laboratories) then fed Dox chow (ad libitum). Control animals were littermates containing a single transgene.
Hematoxylin and Eosin Staining and Immunohistochemistry Fluorescence Staining
Enucleated eyes were fixed overnight in 4% paraformaldehyde (PFA) in PBS at 4°C, followed by paraffin or cryo embedding. Deparaffinized sections (5 μm) were stained with hematoxylin and eosin and examined by a Nikon ECLIPSE E800 microscope. Cryo-sections (10 μm) used in this study were blocked with 3% BSA in PBS containing 0.05% NP-40 for 1 hour at room temperature, then incubated overnight at 4°C with goat anti-keratocan antibody (homemade antibody16) diluted in the same buffer. After three washes in PBS/0.1% Tween-20 (PBST), slides were incubated at room temperature for 1 hour with the Alexa-Fluor 555-conjugated secondary antibodies (Life Technologies, Carlsbad, CA, USA) and 1 ng/mL 4′,6-diamidino-2-phenylendole (DAPI) as a nuclear counterstain. Following incubation, the slides were washed with PBST and mounted with Mowiol (Sanofi-Aventis, Bridgewater, NJ, USA). Sections were examined and photographed using a Zeiss microscope equipped with a camera (Axiocam Mrm). For data acquisition, we used the Axiovision 4.6 software (Carl Zeiss Meditec, Dublin, CA, USA).
Statistical Analysis
Six continuous paraffin sections from each mouse eye collected at P1, P2, and P3 after Dox induction were used to count the stromal cell numbers of the central cornea under the full view fields of the microscope. A 2-tailed Student's t-test (Excel; Microsoft, Redmond, WA, USA) was used to analyze the significance of difference; P < 0.05 was considered statistically significant.
Results
Generation of KeraRT Knock-In Mouse Line
To generate a Dox-inducible stromal keratocyte-specific knock-in mouse line, a targeting vector Kera-IRES2-rtTA3 containing the IRES2-rtTA3 cassette was constructed (Fig. 1A, Supplementary Fig. S1A) and the SacII linearized construct (Supplementary Fig. S1B) was introduced into C57/BL ES cells. Six ES clones were identified as homologous recombinants by a double-PCR–based method (Fig. S1B; Supplementary Table S1), which were verified further by restriction enzyme digestion (Supplementary Fig. S2A) and junction area sequencing of the PCR product (Supplementary Fig. 2B). These targeted clones were expanded and injected into blastocysts from which 10 chimera mice were obtained (Supplementary Table S2). The heterozygous knock-in mice produced from mating germline chimeras with albino C57 mice were further bred with FLP mice to remove the Neo gene from the recombinant allele (Supplementary Fig. S3). It was noted that the Neo gene was removed only in some of the double transgenic mice containing KeraRT and FLP. For example, mouse # 4 and mouse #8 are double transgenic mice from the same litter, but the Neo gene was removed in #8 but not #4 (Supplementary Fig. S3). Heterozygous knock-in mice that had the Neo gene removed were subjected to natural breeding to get homozygous mice, which were identified by tail DNA PCR (Fig. 1C).
Functional Analysis of the Knock-In Mice KeraRT
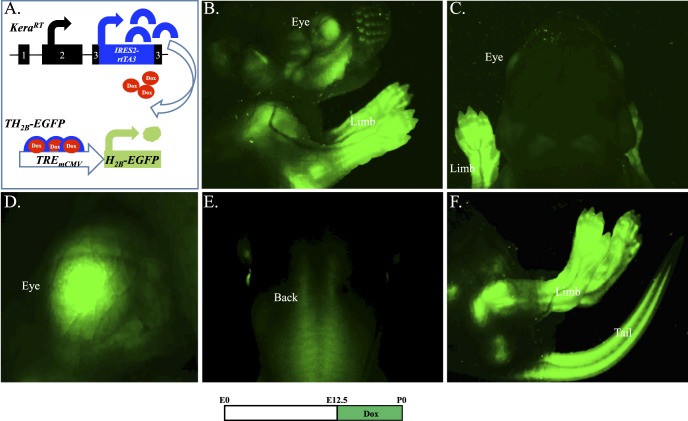
To test the functionality of the rtTA3 minigene in the modified Kera allele, KeraRT driver mice were bred with reporter TH2B-EGFP mice. After Dox induction, the resulting double transgenic mice should show GFP signal in tissues where the endogenous keratocan protein is expressed, such as the corneal stroma (Fig. 2A). As expected, double transgenic offspring induced from embryonic day 12.5 (E12.5) to the date of birth (P0) had strong GFP signals in the eye, limbs, and tail, but lacked signal in other tissues/organs (Fig. 2B–2F). This pattern is consistent with our previous published results in Kera-β-Geo transgenic mice.17 Single transgenic littermates (KeraRT or TH2B-EGFP) never displayed any GFP fluorescence (data not shown). This indicated that the green fluorescent signals detected in the double transgenic mice are real signals resulting from the GFP fusion protein produced as a consequence of Dox induction.
Figure 2.
Functional analysis of KeraRT knock-in driver mouse line. (A) Dox-dependent expression of GFP. The double transgenic mouse KeraRT/TH2B-EGFP generated by crossing the driver KeraRT with the reporter TH2B-EGFP will express GFP in keratocan-positive tissues and cells once administered with Dox chow. Images from a stereomicroscope show the GFP (green) expression pattern in a Dox-treated double transgenic mouse at P0. (B) Cephalic and limb. (C) Back of head. (D) The eye region. (E) Back of the body. (F) Hind legs and tail.
Stromal Keratocyte-Specific Induction of GFP During Development
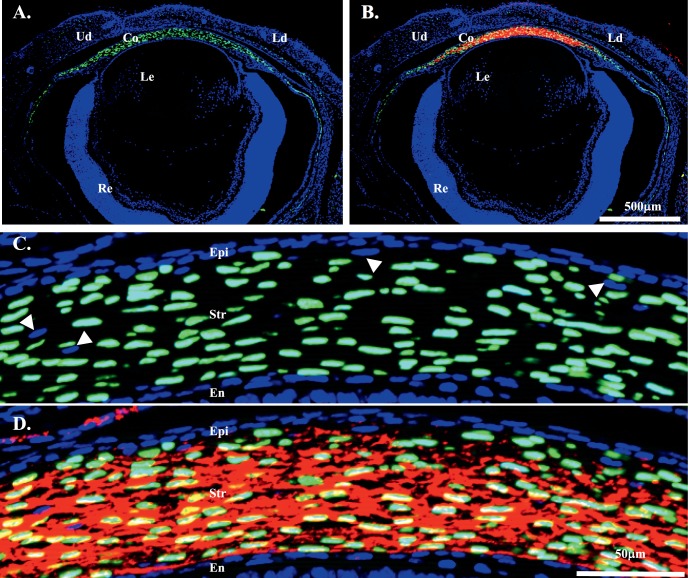
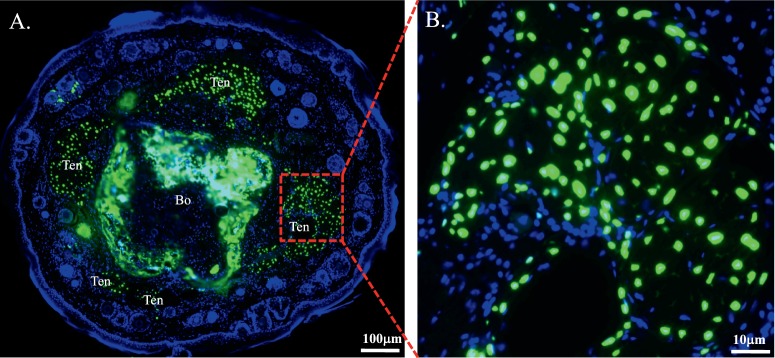
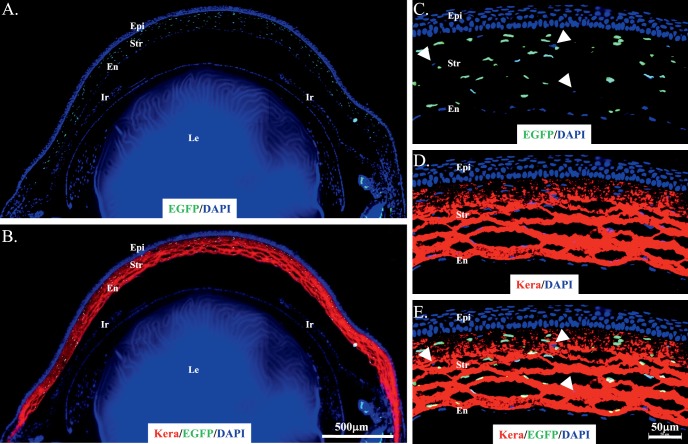
At the cellular level, GFP fluorescence was present in neural crest–derived mesenchymal cells of the stroma of cornea, limbus, and sclera of double transgenic mice, but not in epithelial cells (Figs. 3A, 3C). The GFP expression pattern in the cornea is in agreement with the results obtained with the transgenic mouse line KR.12 However, only a few mesenchymal cells in the eyelid have GFP expression in KeraRT mice, which is different from the transgenic mouse line KR showing strong GFP signals in the eyelid.12 In addition, IHC revealed that keratocan protein was expressed in the corneal stroma of KeraRT mice (Figs. 3B, 3D), which suggests that the functionality of the Kera allele in KeraRT is not affected by the simultaneous expression of rtTA3 in the keratocytes of KeraRT mice. In addition, the tails of double transgenic mice contained nuclear-bound GFP fluorescence in the tendon cells, which express keratocan protein (Fig. 4, Supplementary Fig. S5).
Figure 3.
The double transgenic mouse KeraRT/TH2B-EGFP shows GFP expression in the eye at P0. (A, B) GFP was present in neural crest–derived mesenchymal cells of the cornea, limbus, and some of the sclera. It also appears in a few stromal cells of the eyelid. Immunostaining shows keratocan was expressed in the corneal stroma (red in [B]). (C, D) GFP and keratocan were present exclusively in the central corneal stroma cells of the above eye section, but not in epithelium or endothelium. Note that approximately 10% of corneal stroma keratocytes lacked expression of GFP (arrows). Ud, upper lid; Ld, lower lid; Co, cornea; Epi, corneal epithelium; Str, stroma; En, endothelium; Le, lens.
Figure 4.
GFP expression in the tail of double transgenic mice KeraRT/TH2B-EGFP after Dox induction. Strong GFP fluorescence is present in the tenocytes and bone mesenchymal cells of the induced double transgenic mice (A). Higher magnification shows clear GFP signals located in the nucleus of tail tenocytes (B).
Stromal Keratocyte-Specific Induction of GFP in Adult Mice
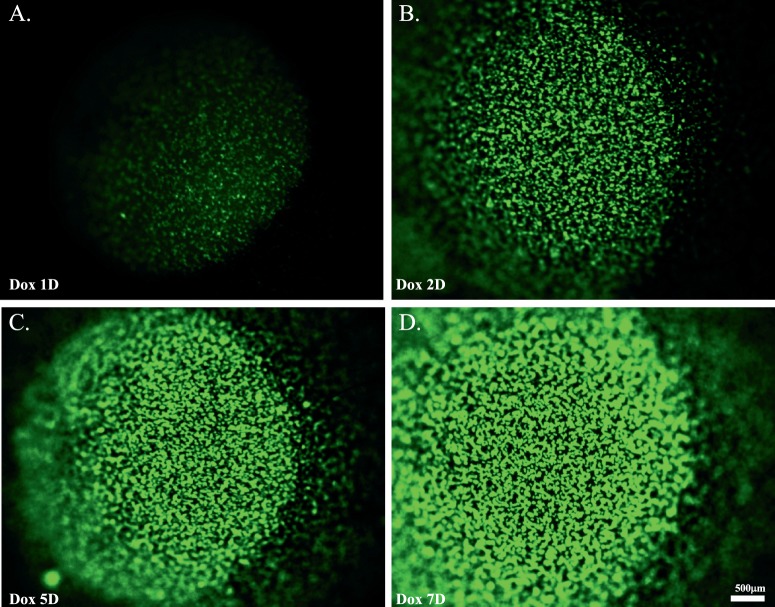
To determine the temporal and spatial GFP expression pattern of adult KeraRT/TH2B-EGFP mice, 3-month old double transgenic mice were Dox-induced for different time points. Before induction, no GFP signal was identified in the double transgenic mice (Supplementary Figs. S4A, S4B). After 1 day Dox induction, weak GFP signal was detected in the cornea, which increased in intensity during a 7-day induction period (Fig. 5). These data suggested that adult KeraRT mice can be induced specifically and efficiently by Dox, which is consistent with the keratocan protein expression pattern. This is quite different from our previously reported KR mice generated by transgenesis that led to random insertion of the rtTA transgene in the mouse chromosome. We also found that the GFP signal was quite stable in the double transgenic KeraRT/TH2B-EGFP mice. Figure 6 shows strong GFP signals were detected 3 months after the cessation of Dox-induction of double transgenic mice that were induced for 48 hours at 2 months of age. At the cellular level, GFP was detected only in stromal keratocytes but not in the epithelium or endothelium (Figs. 6C–E), which is consistent with the pattern seen during embryonic induction (Fig. 3) or adolescent induction (data not shown). Figures 6D and 6E also indicated that the endogenous Kera expression pattern was not altered in the double transgenic mice at the adult stage.
Figure 5.
Dox-induction of a 3-month-old double transgenic mouse for different time periods. (A–D) Images showing GFP expression in double transgenic mice induced for 1 day (A), 2 days (B), 5 days (C), and 7 days (D). After 1-day Dox induction, a weak GFP signal was detected in the cornea, which increased in intensity during a 7-day induction period.
Figure 6.
GFP was detected 3 months after induction of 2-month-old double transgenic mice. (A, B) GFP was detected in corneal stroma cells of double transgenic mice after 3 months of Dox induction from P60 to P62. (C–E) Images taken from the central cornea of the same section shown in (A) and (B). Red fluorescence showing keratocan expression in the stromal cells of the double transgenic mice (B, D, E). Note that a very low ratio of corneal stroma keratocytes lacked GFP (C–E, arrows). Ir, iris.
Ectopic Expression of TGF-α in KeraRT/tet-O-TGF-α Caused Hyperplasia in Corneal Stroma and Epithelium
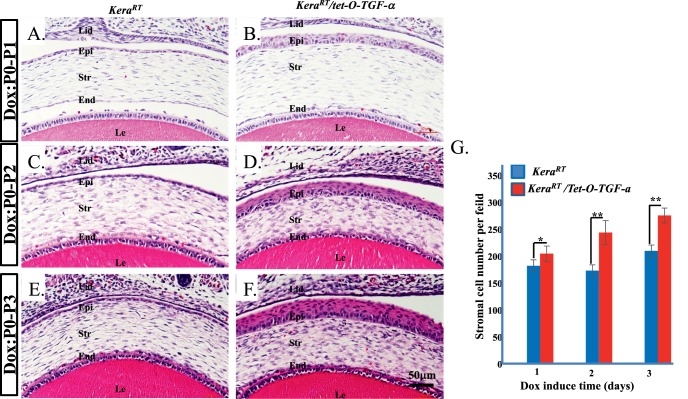
To functionally characterize this novel mouse line further, double transgenic mice KeraRT/tet-O-TGF-α were generated by mating KeraRT with tet-O-TGF-α and were administered Dox from P0 to P1, P2, and P3, respectively. The mice did not have significant changes in general appearance between control and double transgenic littermates (data not shown). Eyeballs collected from induced mice were subjected to hematoxylin and eosin staining, which revealed hyperplasia in the corneal stoma and in corneal epithelium. The corneal stromal cells significantly increased after Dox induction (Fig. 7G). Notably, even though TGF-α was overexpressed in corneal stromal keratocytes of the KeraRT/tet-O-TGF-α mouse, the cell layers of the corneal epithelium in double transgenic mice continually increased to 7 to 8 layers at P3, while the single transgenic control mice maintained 1 to 2 corneal epithelial cell layers. Even at P1 after 1-day induction, corneal epithelial cells amplified to 4 to 5 layers. At P2, after 2-day induction, it became 5 to 6 layers (Figs. 7A–F). The molecular mechanism underlying this phenotype will be investigated and reported in the future. These data indicated that this novel mouse line is a useful model for gene manipulation to elucidate the biological functions in corneal stroma during embryonic development.
Figure 7.
Ectopic expression of TGF-α in the keratocytes of KeraRT/tet-O-TGF-α double transgenic mice caused hyperplasia in corneal epithelium and stroma. (A–F) Hematoxylin and eosin staining of mouse neonates that were induced with Dox P0 to P1, P2, and P3, respectively. (G) Statistical analysis of the stromal cell number present in the central cornea under each microscopic field of view. The cell layers of the corneal epithelium in Dox-induced double transgenic mice continually increased to 7 to 8 layers at P3, while the single transgenic control mice maintained 1 to 2 corneal epithelial layers. Even at P1, after 1 day of induction, corneal epithelial cells amplified to 4 to 5 layers. At P2, after 2 days of induction, it became 5 to 6 layers. Likewise, corneal stromal cells significantly increased after 2 and 3 days of Dox induction. Lid, eyelid. *P < 0.05, **P < 0.01.
Discussion
We established a novel inducible genetic mouse tool that allows for spatial and temporal gene modification in Kera-expressing cells. These cells represent a subset of neural crest–derived cells destined to become keratocytes of the corneal stroma and tenocytes of the limb tendons during embryonic development. This novel mouse driver will allow for the study of the biological role of genes of interest by taking advantage of gain and/or loss of function strategies. For gain-of-function studies, this driver mouse can be crossed with an effecter transgenic mouse containing the gene of interest driven by the TRE, such as tet-O-TGF-α as used in this study. The resulting double transgenic mice will express the gene of interest in the corneal stroma and tendon cells upon Dox induction. For loss-of-function studies, the driver KeraRT must be mated with the tet-O-Cre18 mouse to generate a double transgenic mouse, KeraRT/tet-O-Cre, which then is bred with a flox mouse containing loxP sites flanking the essential exon(s) of the gene of interest. In this scenario, the gene of interest can be ablated in the corneal stroma and tendon cells upon Dox induction. In our laboratory, with the motivation to examine the roles of Wnt canonical signaling pathway in corneal keratocytes during corneal development and homeostasis, the mouse line Ctnnb1flox19 currently is being bred with the double transgenic mouse, KeraRT/tet-O-Cre to generate the triple transgenic mouse line, KeraRT/tet-O-Cre/Ctnnb1flox for specifically knocking out of the key mediator of Wnt canonical signaling pathway, β-catenin, in corneal stromal kratocytes and limb tendon cells after Dox induction. An interesting phenotype after deletion of β-catenin is revealed specifically in the mouse eyes, which will be reported later. Accordingly, KeraRT is an efficient and valuable tool to manipulate the expression of gene interested. In addition, this novel KeraRT driver mouse can complement other neural crest cell lineage drivers, such as Wnt1-Cre,20 P0-Cre,21 and Kera-Cre11 mice due to the fact that none of the previous lines is inducible, which may cause embryonic lethality of the experimental mice when knocking out a gene of interest as early as the Wnt1 or P0 promoter become active during development. Therefore, this inducible driver mouse line is useful for regulating gene expression in a temporal, spatial specific manner during and after embryonic development.
In addition, for experiments using adult mice, this new driver mouse line is advantageous to the previously reported inducible mouse line KR12 in that the expression of rtTA transgene functions in cornea of adult mice. Although the KR driver is a useful keratocyte-specific mouse line to modulate gene expression during embryonic development, it is inefficient at the adult stage (our unpublished observation). Thus, KR is not beneficial to study gene functions in the adult. Our newly generated KeraRT line can fill this gap and is a valuable tool to efficiently perform genetic modification in the adult (Fig. 5).
Comparing the results from embryonic induction to those from adult induction, we noticed an interesting phenomenon, which is that most of the stromal keratocytes show strong GFP signal after Dox induction, while approximately 10% of them do not (arrows pointed to cells in Figs. 3C, 3D; Figs. 6C–E; Supplementary Fig. S6). There are several possible explanations: one is that there is allele-specific expression like that seen in our Krt12rtTA driver mice.6 Another possibility is epigenetically inactivated expression of GFP in those cells.22 Thirdly, the cells lacking GFP may arise from another origin, such as mesoderm, although most of the keratocytes originate from periocular mesenchymal cells of neural crest origin23 and residential dendritic cells of hematopoietic cell lineage. This hypothesis will need to be investigated in our laboratory by using mesodermal CreER driver like Col1a2-Cre/ERT2-ALPP (Jax mice: 01623724 and tamoxifen-inducible effecter mice with red fluorescence crossed with the KeraRT/TH2B-EGFP background. The compound mice induced by Dox and tamoxifen will show GFP fluorescence in neural-crest derived keratocytes and red fluorescence in mesoderm-derived keratocytes in the corneal stroma.
In our Dox-induced KeraRT/TH2B-EGFP mice, GFP appears in some of the sclera cells in addition to the stroma while keratocan can be detected only in the corneal stroma (Fig. 3). This inconsistency may be due to the stability of GFP, which means that during early development keratocan may be expressed in this part of the sclera and then expressed more selectively in the corneal stroma.8 Keratocan may not be detected because of degradation, but at an early stage GFP was produced that is quite stable and still detected at P1.
In Dox-induced KeraRT/TH2B-EGFP transgenic adult mice, tendons in the limb and tail also were GFP-labeled (Fig. 4, Supplementary Fig. S5). This implies that KeraRT adult mice express rtTA protein in tendon tissues of the limbs and tail. This phenomenon is in agreement with the keratocan protein expression pattern, which is found in the corneal stroma and tendon of mouse tails in the adult. Keratocan protein has been detected by immunofluorescent staining of tendons in the limbs and tail of adult mice (Supplementary Fig. S5), which is consistent with data reported.9
Taken together, this newly generated inducible keratocyte-specific driver KeraRT mouse line will be a valuable tool for gene manipulation and elucidating gene function in the corneal stroma during embryonic development, maintenance of homeostasis and pathogenesis.
Supplementary Material
Acknowledgments
Supported by grants from the National Institutes of Health/National Eye Institute (NIH/NEI, Bethesda, MD, USA) R01 EY21501, EY23086 (to CYL), EY011845 (to WWYK), Research Prevent Blindness and Ohio Lions Foundation for Eye Research.
Disclosure: Y. Zhang, None; W.W.-Y. Kao, None; Y. Hayashi, None; L. Zhang, None; M. Call, None; F. Dong, None; Y. Yuan, None; J. Zhang, None; Y.-C. Wang, None; O. Yuka, None; A. Shiraishi, None; C.-Y. Liu, None
References
- 1. Kao WW-Y,, Liu C-Y. The use of transgenic and knock-out mice in the investigation of ocular surface cell biology. Ocul Surf. 2003; 1: 5–19. [DOI] [PubMed] [Google Scholar]
- 2. Lewandoski M. Conditional control of gene expression in the mouse. Nat Rev Genet. 2001; 2: 743–755. [DOI] [PubMed] [Google Scholar]
- 3. Kao WW-Y. Ocular surface tissue morphogenesis in normal and disease states revealed by genetically modified mice. Cornea. 2006; 25 (Suppl 1): S7–S19. [DOI] [PubMed] [Google Scholar]
- 4. Cho A,, Haruyama N,, Kulkarni AB. Generation of transgenic mice. Curr Protoc Cell Biol. 2009; Chapter 19:Unit 19.11. [DOI] [PMC free article] [PubMed]
- 5. Gossen M,, Freundlieb S,, Bender G,, Müller G,, Hillen W,, Bujard H. Transcriptional activation by tetracyclines in mammalian cells. Science. 1995; 268: 1766–1769. [DOI] [PubMed] [Google Scholar]
- 6. Chikama T-I, Hayashi Y, Liu C-Y, et al. . Characterization of tetracycline-inducible bitransgenic Krt12rtTA/+/tet-O-LacZ mice. Invest Ophthalmol Vis Sci. 2005; 46: 1966–1976. [DOI] [PubMed] [Google Scholar]
- 7. Sakamoto K,, Gurumurthy CB,, Wagner KU. Generation of conditional knockout mice. Methods Mol Bio. 2014; 1194: 21–35. [DOI] [PubMed] [Google Scholar]
- 8. Liu CY, Shiraishi A, Kao CW, et al. . The cloning of mouse keratocan cDNA and genomic DNA and the characterization of its expression during eye development. J Biol Chem. 1998; 273: 22584–22588. [DOI] [PubMed] [Google Scholar]
- 9. Rees SG, Waggett AD, Kerr BC, et al. . Immunolocalisation and expression of keratocan in tendon. Osteoarthr Cartil. 2009; 17: 276–279. [DOI] [PubMed] [Google Scholar]
- 10. Hayashi Y, Liu C-Y, Jester JJ, et al. . Excess biglycan causes eyelid malformation by perturbing muscle development and TGF-alpha signaling. Dev Biol. 2005; 277: 222–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Weng DY, Zhang Y, Hayashi Y, et al. . Promiscuous recombination of LoxP alleles during gametogenesis in cornea Cre driver mice. Mol Vis. 2008; 20: 562–571. [PMC free article] [PubMed] [Google Scholar]
- 12. Zhang Y,, Kao WW-Y,, Pelosi E,, Schlessinger D,, Liu C-Y. Notch gain of function in mouse periocular mesenchyme downregulates FoxL2 and impairs eyelid levator muscle formation, leading to congenital blepharophimosis. J Cell Sci. 2011; 124: 2561–2572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Farley FW,, Soriano P,, Steffen LS,, Dymecki SM. Widespread recombinase expression using FLPeR (flipper) mice. Genesis. 2000; 28: 106–110. [PubMed] [Google Scholar]
- 14. Kanda T,, Sullivan KF,, Wahl GM. Histone–GFP fusion protein enables sensitive analysis of chromosome dynamics in living mammalian cells. Curr Biol. 1998; 8: 377–385. [DOI] [PubMed] [Google Scholar]
- 15. Hardie WD,, Le Cras TD,, Jiang K,, Tichelaar JW,, Azhar M,, Korfhagen TR. Conditional expression of transforming growth factor-α in adult mouse lung causes pulmonary fibrosis. Am J Physiol - Lung Cell Mol Physiol. 2004; 286: L741–L749. [DOI] [PubMed] [Google Scholar]
- 16. Liu C-Y,, Birk DE,, Hassell JR,, Kane B,, Kao WW-Y. Keratocan-deficient mice display alterations in corneal structure. J Biol Chem. 2003; 278: 21672–21677. [DOI] [PubMed] [Google Scholar]
- 17. Liu C,, Arar H,, Kao C,, Kao WW. Identification of a 3.2 kb 5′-flanking region of the murine keratocan gene that directs beta-galactosidase expression in the adult corneal stroma of transgenic mice. Gene. 2000; 250: 85–96. [DOI] [PubMed] [Google Scholar]
- 18. Perl A-KT,, Wert SE,, Nagy A,, Lobe CG,, Whitsett JA. Early restriction of peripheral and proximal cell lineages during formation of the lung. Proc Natl Acad Sci U S A. 2002; 99: 10482–10487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Brault V, Moore R, Kutsch S, et al. . Inactivation of the beta-catenin gene by Wnt1-Cre-mediated deletion results in dramatic brain malformation and failure of craniofacial development. Development. 2001; 128: 1253–1264. [DOI] [PubMed] [Google Scholar]
- 20. Danielian PS,, Muccino D,, Rowitch DH,, Michael SK,, McMahon AP. Modification of gene activity in mouse embryos in utero by a tamoxifen-inducible form of Cre recombinase. Curr Biol. 1998; 8: 1323–1326. [DOI] [PubMed] [Google Scholar]
- 21. Feltri ML,, D'Antonio M,, Previtali S,, Fasolini M,, Messing A,, Wrabetz L. P0-Cre transgenic mice for inactivation of adhesion molecules in Schwann cells. Ann N Y Acad Sci. 1999; 883: 116–123. [PubMed] [Google Scholar]
- 22. Cui C,, Wani MA,, Wight D,, Kopchick J,, Stambrook PJ. Reporter genes in transgenic mice. Transgenic Res. 1994; 3: 182–194. [DOI] [PubMed] [Google Scholar]
- 23. Gage PJ,, Rhoades W,, Prucka SK,, Hjalt T. Fate maps of neural crest and mesoderm in the mammalian eye. Invest Ophthalmol Vis Sci. 2005; 46: 4200–4208. [DOI] [PubMed] [Google Scholar]
- 24. Zheng B,, Zhang Z,, Black CM,, de Crombrugghe B,, Denton CP. Ligand-dependent genetic recombination in fibroblasts: a potentially powerful technique for investigating gene function in fibrosis. Am J Pathol. 2002; 160: 1609–1617. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.