Abstract
Aim
The purpose of this work is to determine if tumor-tropic neural stem cells (NSCs) can improve the tumor-selective distribution and retention of nanoparticles (NPs) within invasive brain tumors.
Materials & methods
Streptavidin-conjugated, polystyrene NPs are surface-coupled to biotinylated human NSCs. These NPs are large (798 nm), yet when conjugated to tropic cells, they are too large to passively diffuse through brain tissue or cross the blood–tumor barrier. NP distribution and retention was quantified 4 days after injections located either adjacent to an intracerebral glioma, in the contralateral hemisphere, or intravenously.
Results & conclusion
In all three in vivo injection paradigms, NSC-coupled NPs exhibited significantly improved tumor-selective distribution and retention over free-NP suspensions. These results provide proof-of-principle that NSCs can facilitate the tumor-selective distribution of NPs, a platform useful for improving intracranial drug delivery.
Keywords: cellular therapy, glioma, nanoparticles, neural stem cell, stem cell carriers, targeted delivery, tropism
Background
Selective targeting of therapeutic agents to invasive glioma could significantly improve patient prognosis. Because intravenously administered therapies do not efficiently penetrate across the blood–tumor barrier and within hypoxic tumor regions [1], selective targeting is best achieved using local drug infusions. Intratumoral infusions often involve the combined use of therapeutic nanoparticles (NPs) and convection-enhanced delivery to achieve more desirable pharmacokinetic profiles [2]. Unfortunately, NP penetration throughout the tumor remains a challenge, even when infusions incorporate digestive enzymes or hypo-osmolar solutions [3]. Furthermore, poor NP retention and off-target toxicities still occur when infused NPs overflow the intended distribution range or get lost along conductive flow paths, such as perivascular spaces or white matter tracts [4].
One innovative NP distribution strategy that may improve penetration, retention and tumor-selective distribution involves coupling NPs to the surface of tumor-tropic cells that can home to and penetrate tumors. The interest in coupling NPs to tumor-tropic cells has been broadening since a recent report demonstrating that adjuvant-loaded NPs can be surface-conjugated to T cells and hematopoietic stem cells without impairing the tropism of these cells towards tumor sites in the periphery [5]. The purpose of the present work is to evaluate the potential for tumor-tropic cells to improve the penetration, retention and tumor-selective distribution of NPs within brain tumors.
It has been shown that surface conjugation of doxorubicin-loaded NPs to mesenchymal stem cells (MSCs) does not impair MSC viability or tropism to glioma-conditioned media in vitro [6]; however, the ability of stem cells to distribute surface-conjugated NPs to tumor sites within the brain has not yet been assessed. Because MSCs can contribute to glioma progression and exhibit reduced tropism to intracranial lesions compared with neural stem cells (NSCs) [7, 8]; here, we use a human NSC line (HB1.F3) currently being evaluated in Phase I clinical studies to target a prodrug-activating enzyme to invasive glioma [9–11]. We first biotinylate the NSC surface [12] before coupling them to streptavidin-conjugated, polystyrene NPs, which were chosen for this study because of their stability, preclinical biocompatibility [13] and prior use as a generic model of intracranial NP distribution [14]. Relatively large NPs (798 nm) were selected for this study because they are too large to passively cross the blood–tumor barrier or diffuse through brain tissue, thus ensuring that observed changes in NP retention and distribution patterns are in fact NSC-mediated. These NPs are several times larger than those previously coupled to tropic cells, which may translate to increased drug-loading capacity in the future. Using 3D reconstruction and postsacrifical tissue analysis, we quantitatively assessed the ability of NSCs to improve NP retention and tumor selective distribution in vivo 4 days after injections either adjacent to the intracerebral glioma, contralaterally, or intravenously.
Material & methods
Cell culture
All cell lines were cultured in Dulbecco’s Modified Eagle’s Medium (DMEM; Invitrogen, CA, USA) supplemented with 10% fetal bovine serum (Gemini Bio, CA, USA), 1% l-glutamine (Invitrogen) and 1% penicillin–streptomycin (Invitrogen) and maintained at 37°C in a humidified incubator (Thermo Electron Corporation, CA, USA) containing 6% CO2. When the cell reached 80% confluency, they were passaged using a 0.05% trypsin/ethylenediaminetetraacetic acid solution (Invitrogen); media was changed every 2–3 days. Glioma cell lines include the firefly luciferase-expressing U251 (U251.ffluc) and U87 human glioma cell lines were obtained from American Type Culture Collection. U87 cells were used to generate glioma-conditioned media by replacing culture media with serum-free media when cells were 80% confluent, followed by a 48 h incubation. NSC lines include the human, v-myc immortalized, HB1.F3 NSC line [15], which was obtained from Seung Kim (University of British Columbia, Canada). This cell line was further transduced with lentivirus to stably expressed enhanced green fluorescent protein [16] and used to track stem cell distribution in vivo.
Biotinylation of NSCs
NSCs were biotinylated as described previously [12]. Briefly, cells were grown to 80% confluency, then culture medium was removed and cells were washed twice with phosphate-buffered saline (PBS) before incubation in cold 1 mM NaIO4/PBS solution for 20 min in the dark at 4°C. NSCs were washed with PBS (pH 6.5) at room temperature (RT). Next, NSCs were incubated in 0.5 mM biotin hydrazide (pH 6.5; Sigma, MO, USA) solution in DMEM (Invitrogen) for 90 min at RT. The cells were then washed twice with PBS (pH 7.4) before NP coupling.
Biotinylated NSC surface characterization
FACs
Freshly trypsinized biotinylated NSCs were resuspended (5 × 106 cells/ml) in staining/wash buffer (SWB; 94% PBS without Ca2+ and Mg2+, 5% fetal bovine serum and 0.001% weight/volume NaN3 [Sigma]). Cells were fixed (Fix and Perm® Cell Fixation & Cell Permeabilization Kit; Invitrogen), and rinsed. Control or biotinylated NSCs were immunostained with fluorescein-conjugated avidin (10 µg/ml) and incubated for 20 min at RT in the dark. After two final rinses with SWB, cells were resuspended to 2.5 × 104 cells/µl in SWB. The number of fluorescent cells was analyzed by flow cytometry (Guava EasyCyte™, EMD Millipore, MA, USA). A representative plot (Figure 1B) is shown, with mean ± standard deviation listed in the text (three experiments; n = 10 samples). For β1-integrin assessments, control NSCs were immunostained with anti-β1-integrin primary antibody (0.01 µg/ml; BD Biosciences, NJ, USA) or mouse IgG1-κ isotype control antibody (0.01 µg/ml; BD Pharmingen) for 40 min at RT. The samples were then washed twice with SWB, stained with goat antimouse-IgG/IgM-f luorescein isothiocyanate (10 µg/ml; BD Pharmingen) and incubated for 20 min at RT in the dark.
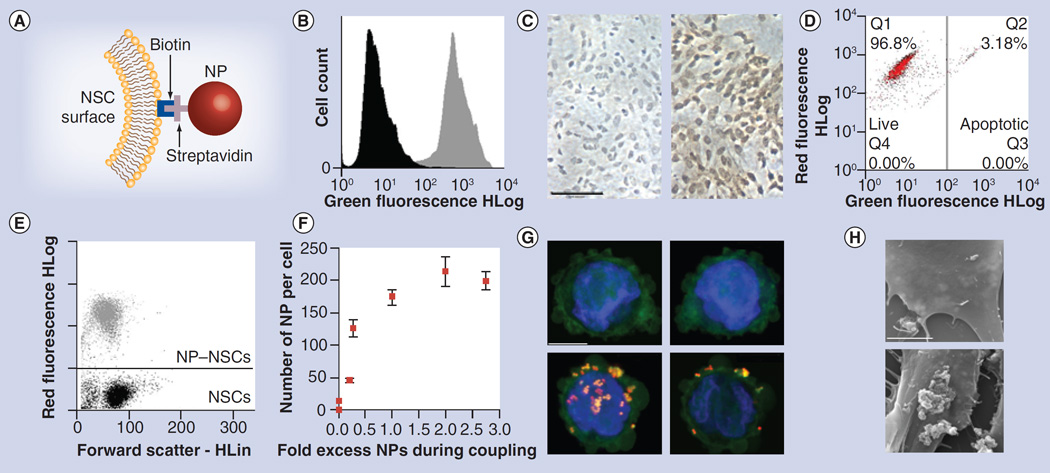
Figure 1. Nanoparticle–neural stem cell coupling.
(A) NP coupling schematic. (B) Flow analysis of control (black) and biotinylated NSCs (gray) after staining with fluorescein–avidin. (C) Brightfield images of control (left) and biotinylated NSCs (right) after staining both with avidin–horseradish peroxidise/diaminobenzidine and hematoxylin counterstain. Scale bar: 100 µm. (D) Viability of biotinylated (red) and unmodified (black) NSCs. (E) Flow analysis of biotinylated NSCs before (black) and after (gray) NP coupling. (F) Fluorometric determination of dose-dependent increase in NP binding to biotinylated NSCs. Error bars represent mean ± standard deviation. (G & H) Confocal images of control (top) and biotinylated NP–NSCs (bottom) stained with phalloidin-488 and 4-,6-diamidino-2- phenylindole. Some NPs appeared in both the green and red channel. Left: z-stack projections spanning cell thickness; right: 1-µm optical slice. Scale bar: 10 µm. (H) Standard error of the mean micrographs of NP distribution on surface of control (top) and biotinyleted (bottom) NSCs. Scale bar: 5 µm.
HLin: Linear signal amplification; HLog: Logarithmic signal amplification; NP: Nanoparticle; NSC: Neural stem cell; Q: Quadrant.
Immunohistochemistry
Control and biotinylated NSCs were rinsed then fixed in 4% paraformaldehyde and processed for immunoperoxidase-3,3β-diaminobenzidine staining after quenching endogenous peroxidases with 0.3% hydrogen peroxide/PBS for 30 min. To identify cell surface biotinylation, control and biotinylated NSCs were incubated with Vectastain® ABC Elite Kit (Vector Laboratories, CA, USA). Antibody reactivity to β1-integrins was detected using the same method after incubating serum-blocked NSCs treated with either a monoclonal anti-β1-integrin or mouse IgG control with a biotinylated anti-mouse IgG secondary antibody.
NP characterization
Nile-red-loaded, streptavidin-conjugated polystyrene NPs were commercially obtained from (SPHERO™ Fluorescent Particles FP-3056-2, Sphereotech, IL, USA). Dynamic light scattering and zeta potential measurements (n = 5) were performed in deionized water on a Brookhaven 90 Plus/BI-MAS Instrument (Brookhaven Instruments, NY, USA) at RT.
Coupling of streptavidin-conjugated NPs to biotinylated NSCs
Biotinylated cells were trypsinized, rinsed once with PBS and then resuspended in an NP:PBS suspension (pH 7.4). For coupling, the suspension was incubated (20 min, RT) with periodic trituration. The NP-coupled NSCs (NP–NSC) mixture was then centrifuged and any uncoupled particles remaining in the supernatant were removed. The cells were rinsed twice in PBS (12 ml) to encourage removal of loosely bound NPs.
Microscopic imaging of surface-associated NPs in vitro
Confocal microscopy
Suspensions of NSCs or NP–NSC hybrids (1 × 107 cells/ml) were fixed in 4% paraformaldehyde, rinsed in 0.1% Tween/PBS, then stained for 15 min at RT in the dark with a PBS solution containing AlexaFluor® 488-conjugated phalloidin (1:200; Life Technologies, CA, USA) to stain cellular filamentous-actin and 4-,6-diamidino-2-phenylindole (DAPI; 1 µg/ml) to stain cell nuclei. Cells were pelleted, rinsed and then encapsulated within 1% (weight/volume) low-melting-point agarose (Sigma) to stabilize the cells for imaging. The agarose suspension (200 µl) was placed on a glass slide where a coverslip was used to create a thin gel layer that was polymerized upon exposure to 4°C for 10 min. Images were acquired using a confocal microscope (Zeiss, Oberkochen, Germany) equipped with a 100× oil immersion objective. Each image represents a z-stack compiled from 1-µm optical slices spanning the entire thickness of the cell.
Scanning electron microscopy
NP surface localization was verified with scanning electron microscopy (SEM) after growing NSCs or NP–NSCs on glass coverslips for 12 h and then fixing cells with 1.5% glutaraldehyde in 0.1 M cacodylate buffer. Samples were then sputter coated with gold and imaged using an FEI™ Quanta 200 (OR, USA) scanning electron microscope.
Quantification of surface-associated NPs in vitro
FACS
Biotinylated NSCs with or without exposure to 0.1% (weight/volume) NPs were washed twice, then resuspended in PBS. The increase in red fluorescence as a result of NP binding was quantified using a GuavaCyte FACS cytometer and results analyzed using FlowJo™ Software (Tree Star, Inc., OR, USA; two experiments; n = 8 samples).
Fluorimeter
An NP standard curve was generated in the presence of 1 × 105 NSCs/ml in order to quantify the number of NPs bound to NSCs. Control or particle-coupled NSCs were diluted to 1 × 105 cells/ml either before or after migration, then samples in triplicate were analyzed on a SpectroMax™ M3 Microplate Reader (Molecular Devices, CA, USA) using 520 nm excitation and 605 nm emission filters. Plot of mean ± standard error of the mean is shown (three experiments; n = 12 samples).
NP–NSC in vitro viability & tumor tropism
Viability
Freshly trypsinized cells were labeled with Guava® ViaCount® (EMD Millipore), a proprietary mixture that distinguishes between viable and nonviable cells based on the differential permeability of DNA-binding dyes. The fluorescence of each dye was resolved using a Guava EasyCyte™ flow cytometer, and data analyzed using FlowJo software. A representative plot is shown, with mean ± standard deviation listed in the text (seven experiments; n = 9 samples).
Tumor tropism
Modified Boyden chamber chemotaxis assays were performed using 24-well cell culture plates with polycarbonate inserts (pore diameter: 8 µm) as described previously [17]. Glioma-conditioned media and 5% bovine serum albumin (BSA)/DMEM were added to the lower chambers as attractants (500 µl/well, triplicate samples). Suspensions of NSCs or NP–NSCs in 5% BSA/DMEM were added to the upper chambers (5 × 104 cells/250 µl). After incubation (4 h, 37°C) inserts were placed in Accutase™ (Sigma) for 10 min at 37°C. Detached cells were centrifuged (1500 rpm, 5 min), labeled with Viacount, and counted using Guava EasyCyte flow cytometer. A plot showing mean ± standard error of the mean is shown (four experiments; n = 12 samples).
In vivo glioma xenografts & NP injections
Esterase-deficient severe combined immunodeficient mice were anesthetized with an intraperitoneal injection of 132 mg/kg ketamine and 8.8 mg/kg xylazine. Mice were then immobilized in a stereotactic apparatus and received guided intracranial injections of cell suspension 2 mm lateral, 0.5 mm anterior to bregma, tracked from a depth of 2.5 to 2.25 to 2.0 mm; 0.667 µl of cell suspension was injected at each level (2 µl total). A total of 2 min elapsed before moving to the next injection level to minimize backflow through the needle track. In tumor-inoculated brains, intracranial injections contained 5 × 104 cells U251 human glioma cells, then either 3.5 × 107 free-NPs or NP–NSCs (2 × 105 NSCs, 3.5 × 107 NPs) 7 days later. In tumor-free brains, the glioma cell injections were omitted. Injections were performed with a 30-gauge 5-µl hamilton syringe over 3–5 minutes. After retracting the needle over 2–4 minutes, bone wax was used to occlude the burr hole, and skin was closed with skin glue. Intravenous injections contained 200 µl of either free NPs (3.5 × 108 NPs) or NP–NSCs (2 × 106 NSCs, 3.5 × 108 NPs). Buprenorphine analgesic (0.05 mg/kg) was administered subcutaneously to relieve postoperative pain. Results were obtained from three independent experiments that resulted in three mice per group receiving free NP injections, and 4–6 mice per group receiving NP–NSC injections. All animal protocols were approved by the City of Hope Institutional Animal care and Use Committee. Mice were euthanized consistent with the recommendations of the Panel of Euthanasia of the American Veterinary Medical Association when they appeared to be in discomfort or distress as judged by independent animal care personnel. Mice were housed in an-accredited facility and were given food and water ad libitum.
Tissue harvesting & processing
Mice were euthanized 1 or 4 days post-NP injection by CO2 asphyxiation and transcardially perfused with PBS followed by 4% paraformaldehyde (pH 7.4). The brains, liver and spleen were removed and further fixed by immersion in 4% paraformaldehyde for 24 h before sinking in 30% sucrose for 48 h. The tissues were frozen in Tissue-Tek® OCT™ (Sakura Finetek USA, Inc., CA, USA) and sectioned coronally on a Leica CM1510 S Cryostat (Leica Biosystems, Nussloch, Germany). Sections (10-µm thick) were collected on positively-charged slides (Thermo Fisher Scientific, MA, USA) for histological examination, quantification studies and immunocytochemistry.
Tissue imaging
Every fifth section was stained with DAPI (1 µg/ml, Sigma) ), rinsed, mounted with fluorescent mounting medium (Dako North America, Inc., CA, USA), then examined by fluorescence microscopy. Images of the injection and tumor site were obtained using equivalent exposure on a Nikon Eclipse TE2000-U (Nikon Corporation Instruments Company, Tokyo, Japan) microscope equipped with a SPOT™ RT Slider digital camera (SPOT™ Imaging Solutions, MI, USA). Images were recorded and stored using SPOT™ Advanced (SPOT™ Imaging Solutions) and Adobe® Photoshop® software (Adobe, CA, USA). NP volume was determined using a previously described approach [3]). The tumor was observed at multiple levels along the needle track, and the depth of the predominant engraftment site varied in different animals. The representative images were chosen with respect to the DAPI-stained sections that had the greatest amount of visible NPs rather than consistent tissue depth. Open source Reconstruct software (Boston University, MA USA) was used to construct 3D projections of NP distribution within the injection and tumor sites [18]. Image J software (NIH, MD, USA) was used to map NP distribution in representative tumor-free brains. Tumor presence in DAPI-stained slides was noted based on the presence of dense, large nuclei, and confirmed using immunohistochemistry with a rabbit polyclonal antibody against firefly luciferase protein (1:250, MBL International, MA, USA) using standard techniques. Adjacent sections were stained with hematoxylin and eosin. dark field hyperspectral imaging was performed using a CytoViva™ dark field microscope system (CytoViva Inc., AL, USA) equipped with CytoViva™ Hyperspectral Imaging System 1.2 (CytoViva Inc.). A spectral signature library was created by scanning a polystyrene NP reference sample. The library was mapped onto images of interest using the ENVI™ spectral angle mapper software (Exelis Visual Information Solutions, VA, USA).
Statistical analysis
Data are presented as mean ± standard error of the mean unless otherwise stated. Statistical significance was determined using a two-tailed students t-test (*p < 0.1; **p < 0.05; ***p < 0.01).
Results
NP–NSC coupling
NP–NSC coupling was achieved by incubating Nile-red loaded, streptavidin-conjugated polystyrene NPs with biotinylated NSCs (Figure 1A). The NPs were characterized to assess the average effective particle diameter (798 nm, 0.0137 polydispersity index value) and surface charge (−21.32 ± 3.20 mV). NSCs were biotinylated as previously described [12], which labeled 82 ± 10% of the cells with biotin moieties (Figure 1B & C) without significantly impairing cell viability (96 ± 2% live cells). Each NSC contained an average of 3.64 × 107 ± 1.1 × 107 biotin moieties, as determined using a 4´-hydroxyazobenzene-2-carboxylic acid–avidin competition assay. Biotinylated NSCs were incubated in an NP suspension at an initial coupling ratio of one NP per biotin moiety. NSCs retained their viability (97 ± 3% live cells) (Figure 1D) at all coupling ratios (data not shown), and after extensive rinsing, 98 ± 2% of NSCs contained surface-associated NPs (Figure 1E). Fluorimetric quantification was used to determine that there were 175 ± 12 NPs per NSC (Figure 1F) at a 1:1 biotin moiety:NP ratio, and that the number of NSC-coupled NPs could be modulated by adjusting this ratio (Figure 1F). Cellular NP localization was evaluated by confocal microscopy 1 h after coupling. To distinguish intracellular from extracellular located NPs, the cytoskeleton was visualized using AlexaFluor 488-phalloidin conjugate. NPs were predominantly located on the surface of NSCs (Figure 1G). This was further confirmed by SEM, which provided higher resolution images demonstrating that NPs were attached to NSC surfaces in clusters, often entangled by microvilli (Figure 1H) consistent with previous reports [19]. Further studies are needed to determine if NP clusters form before NSC binding due to aggregation of charged NPs with polymeric serum proteins, or if they form due to clustered biotin moieties on the NSC surface. Nevertheless, both visualization techniques showed surface localization of NPs, which may be suitable for NSC-mediated transport. This procedure also was an effective approach to couple NPs to nonimmortalized, nontransduced NSCs (Supplementary Figure 1; please see online at www.futuremedicine.com/doi/suppl/fon.13.217).
NP–NSCs retain tumor tropism in vitro
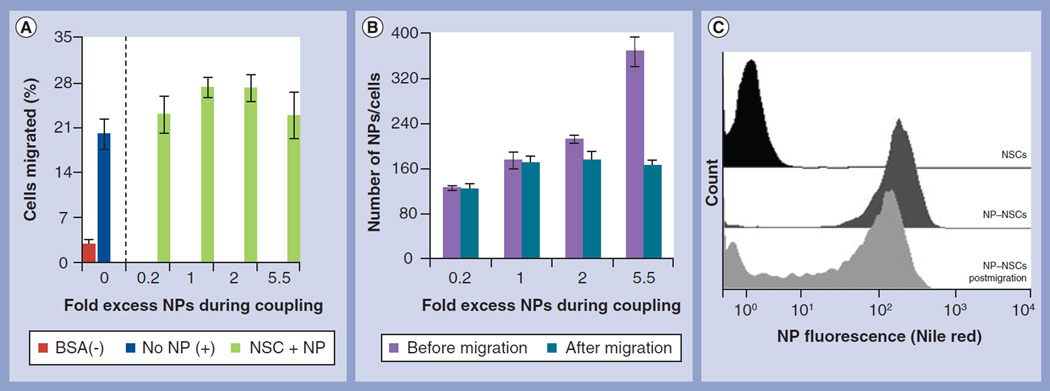
Boyden chamber transmigration assays were used to test if NP–NSCs retained their ability to migrate through a membrane toward media enriched with tumor-derived cytokines. Media containing only BSA was used as a negative control. NP–NSCs coupled using increasing NP concentrations had unaltered (p > 0.1) transmigration efficiencies compared with control NSCs (Figure 2A). After migration, NP–NSCs retained a maximum of 169 ± 11 NPs per cell, even when initially associated with more (Figure 2B). FACS analysis demonstrated that 73 ± 3% of NSCs retained NPs postmigration (Figure 2C) when coupled at a 1:1 biotin moiety:NP ratio. Preliminary data suggests that the NP binding is more complex than the biotin–streptavidin bonding scheme initially envisioned. There are potentially additional contributions from streptavidin–integrin interactions [20] and passive adsorption of polystyrene to the cell surface (Supplementary Figure 2) [13]. Having observed NSC-mediated NP transport in vitro, we proceeded to in vivo studies.
Figure 2. Nanoparticle coupling does not affect neural stem cell migration in vitro.
(A) Percentage of transmigrating control NSCs (left of dotted line) or NP–NSCs (right of dotted line) seeded in the upper well of a transwell chamber after addition of glioma-conditioned media (blue and green) or BSA-containing negative control media (red) into the lower chamber. (B) Biotinylated NSCs were exposed to increasing NP concentrations and the number of NPs per NSC before (purple) and after (teal) transmigration was assessed by quantitative fluorimetry. (C) Fluorescence profile of NP–NSCs before (black) and after (dark gray) NP coupling at a 1:1 NP:biotin moiety ratio. NP retention after transmigration is shown in light gray.
BSA: Bovine serum albumin; NP: Nanoparticle; NSC: Neural stem cell.
NSCs improve NP retention in vivo
Baseline information regarding NP retention and distribution in the absence of a tumor needed to be established first. Thus, NP distribution was observed in a tumor-free brain both 1 and 4 days after injecting either free NPs or NPs coupled to enhanced green fluorescent protein-expressing NSCs (Figure 3A & B). Fluorometric quantification was used to ensure equal amounts of NPs were injected in each case. Upon harvest, brains were cryosectioned, and every fifth section throughout the injection site imaged using fluorescence microscopy (Supplementary Figure 3). Representative images of sections that contained the largest NP surface area are shown (Figure 3C–F). To visualize cumulative particle dispersion throughout the entire injection site, the NPs in each slice were mapped then overlaid to generate a single projection (Figure 3G–J). Using images of only the red channel (NPs fluoresce red), the distance NPs distributed from the injection site and the surface area occupied by NPs in each section was quantified (Figure 3K & L). The interslice distance was used to estimate the volume of NPs retained within the brain [3], then this value was compared with the known volume of NPs initially injected to determine the percentage of NPs retained (Figure 3L).
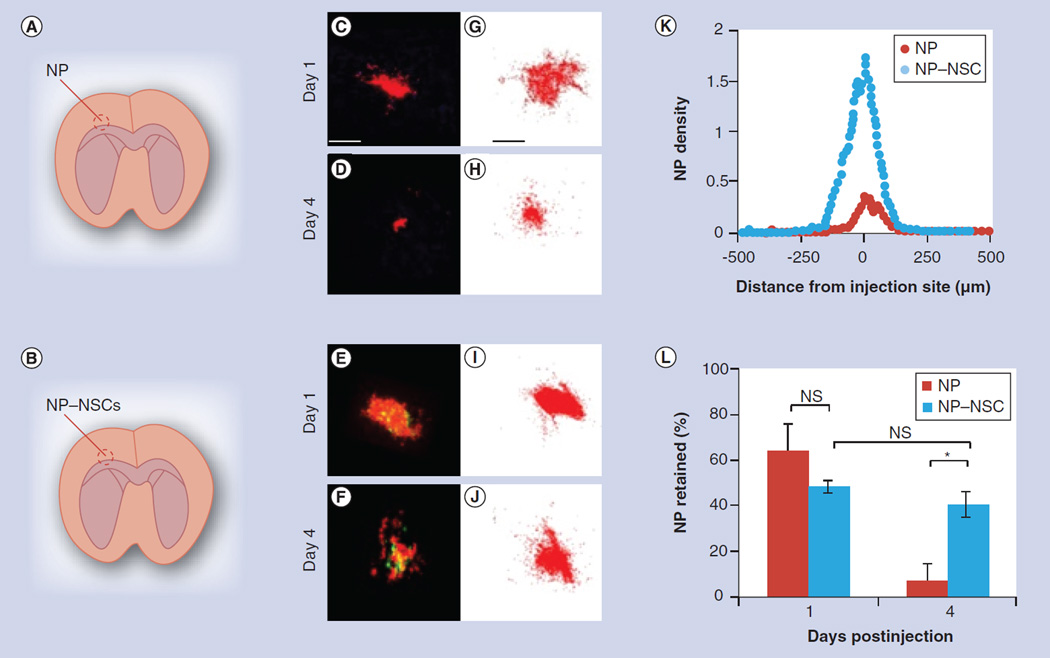
Figure 3. Comparison of free nanoparticle and nanoparticle–neural stem cell distribution in tumor-free brains.
(A & B) Depicts injection paradigm for upper (A, NPs only) and lower (B, NP–NSCs) panels. (C–F) Representative merged fluorescent images (NPs: red; NSCs: green) of brain slices that contain the greatest surface area of NPs both on day 1 (C & E) or 4 (D & F) are shown. Scale bar for (C–F) 100 µm. (G–J) Stacked image compilation showing cumulative NP distribution at representative injection sites. These compiled images were generated by overlaying the mapped NPs present in each slice throughout the injection site using Reconstruct software. Scale bar for (G–J) 100 µm. (K) Plot of NP density as a function of distance from the injection site for representative brains on day 4. (L) Percentage of NPs retained after injection into tumor-free brains. Error bars represent mean ± standard deviation.
*p < 0.05.
NP: Nanoparticle; NS: Not significant; NSC: Neural stem cell.
No significant differences in NP retention were seen 1 day postinjection; with 64 ± 12% of free NPs and 48 ± 8% of NP–NSCs retained. The NPs were initially found as a mass near the injection site, although some isolated NPs were dispersed further away. The number of dispersed NPs was twofold higher in brains that were injected with free NPs as compared with NP–NSCs; however, no other notable differences were observed. In both cases, NPs were distributed 2 mm along the injection tract. The maximum radial distance that NPs distributed was similar (free NPs: 559 ± 9 µm; NP–NSCs: 509 ± 12 µm). A total of 4 days after injection, no measurable NP diffusion or NSC mobility was observed, but the percentage of NPs present within brains that had received free NPs had decreased to 7 ± 3% (Figure 3L). A striking improvement in NP retention was observed in brains that received NP–NSCs (Figure 3K). The percentage of NPs present (40 ± 6%) on day 4 was not significantly lower (p > 0.1) than the amount observed 1 day postinjection (48 ± 8%; Figure 3L).
NSCs target NPs to tumors in vivo
To determine if NSCs can distribute NPs selectively to tumor foci, we evaluated NP distribution in mice with established U251 tumors in the left brain hemisphere. These tumors expressed firefly luciferase and grew to approximately 0.2– 0.5 mm in diameter by day 7. Tumor-bearing mice received either free NPs or NP–NSCs under three injection paradigms that involved decreasing proximity to the intracranial tumor. NPs were injected either immediately adjacent to the tumor (<200 µm away), into the contralateral hemisphere (>1 mm away) or into the tail vein. NP distribution was assessed 4 days post-NP injection, which is within the 2–10 day range when HB1.F3 NSC presence within tumors peaks [21]. Because polystyrene is difficult to identify once brought into biological systems [13], NP distribution was assessed using two complementary microscopic approaches: fluorescence visualization of Nile-red containing NPs and dark field microscopy with spectral mapping. As described in Supplementary Figure 4, we generated a polystyrene-specific spectral library that was used to confirm NP presence at tumor foci. Because of the high degree of spectral overlap that exists between tissue and polystyrene, however, our library likely underestimates the quantity of NPs present.
Paradigm 1: ispilateral NP injection
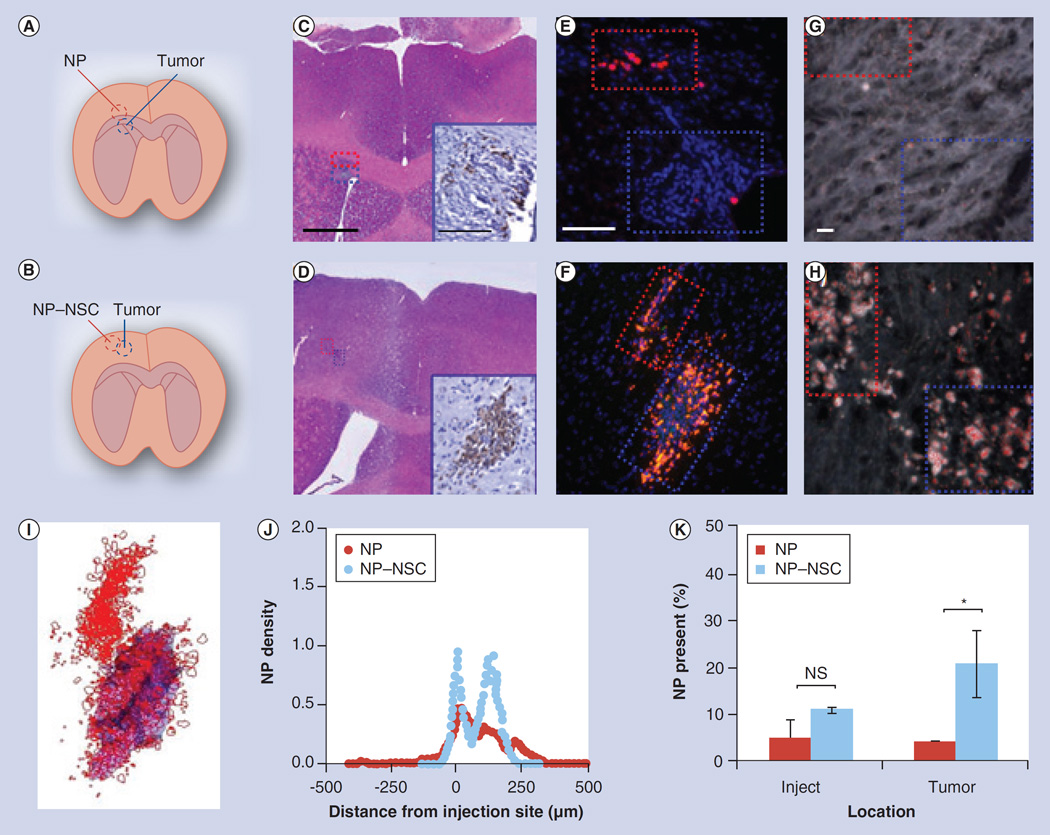
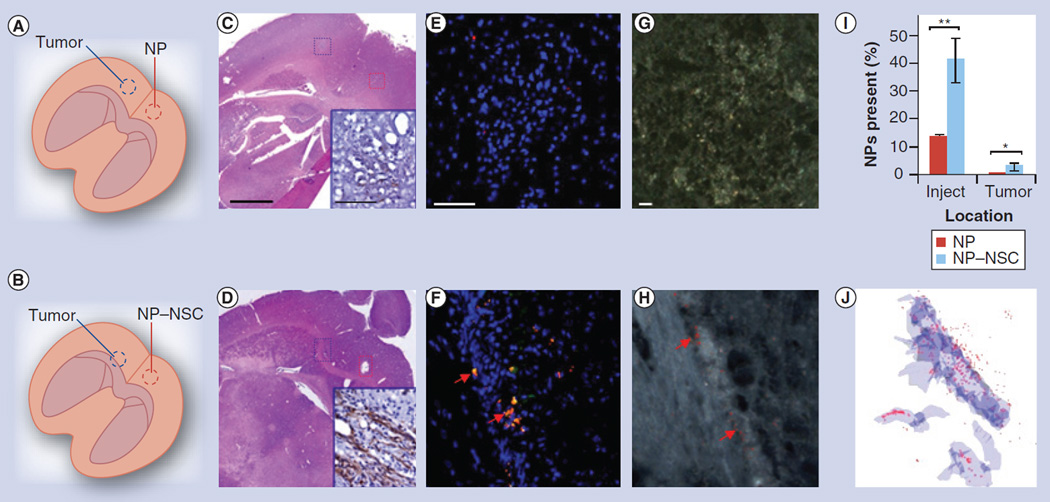
The first injection paradigm involved injecting free NPs or NP–NSCs within the narrow NP distribution range observed in the absence of a tumor, to determine if tumor presence would alter NP retention or distribution patterns (Figure 4A–D). Tumor presence did not significantly affect NP retention; at 4 days postinjection, 9 ± 4% of free-NPs and 32 ± 8% of NP–NSCs were retained (Figure 4K). By contrast, tumor presence did alter NP distribution patterns (Figure 4E, F & J). The majority of NP–NSCs (21 ± 7% of injected NPs, 66% of NPs retained in the brain) no longer remained at the injection site, but rather were redistributed to the tumor (Figure 4I & K). Aside from the injection site, no NPs were observed in nontumor regions. Spectrally mapped dark field images confirmed the presence of NPs at both the injection and the tumor sites (Figure 4G & H). To visualize the cumulative NP–NSC distribution throughout a representative brain, NP–NSCs in each slice were mapped then overlaid using 3D reconstruction software to generate a single 3D projection of the injection and tumor site (Figure 4I). By contrast, the majority of free NPs still localized to the injection site, with just a slight asymmetric distribution favoring the direction of the tumor (4 ± 0.5% passively accumulated at the tumor) (Figure 4J).
Figure 4. Comparison of free nanoparticle and nanoparticle–neural stem cell distribution when injected immediately adjacent to intracranical glioma.
(A & B) Injection paradigm for upper (A) and middle (B) panels. (C & D) A total of 4 days after NP injection, brains were sectioned throughout the entire injection and tumor site. Every fifth section was 4-,6-diamidino-2-phenylindole (DAPI)-stained and imaged using fluorescence microscopy. The slice that contained the greatest surface area of NPs was identified and adjacent sections stained with hematoxylin and eosin. Scale bar: 500 µm. Antifirefly luciferase immunohistochemistry was also performed (insets in C & D; scale bar: 100 µm) to confirm the presence of tumor cells. (E & F) Representative merged fluorescence images of tumor (blue box) and injection sites (red box; NPs: red; NSCs: green; nuclei: blue). Tumors are visible as dense clusters of DAPI-stained nuclei (blue). Scale bar: 100 µm. (G & H) Dark field micrographs of tumor (blue box) and injection (red box) sites. Polystyrene NPs were identified based on their unique spectral profile, then pseudo-colored red to ease visualization. Scale bar: 10 µm. (I) Cumulative NP dispersion throughout a representative injection and tumor site is shown. Every fifth slice throughout the injection and tumor site was imaged using only the red and blue channel, then was mapped for NPs (red) and tumor area (blue). The displayed 3D projection of all mapped images was generated using Reconstruct software. (J) NP density as a function of distance from the injection site on day 4. (K) Percentage of NPs retained at the injection (inject) and tumor sites 4 days postinjection. Error bars represent mean ± standard deviation.
*p < 0.05.
NP: Nanoparticle; NS: Not significant; NSC: Neural stem cell.
Paradigm 2: contralateral NP injection
In the second injection paradigm, free NPs or NP–NSCs were injected >1 mm away into the contralateral hemisphere (Figure 5A–D). This distance from the tumor is outside the NP distribution range observed in the absence of a tumor (Figure 3K). As expected, free NPs distributed around the injection site and no NPs were detected at the contralateral tumor site when imaged using either fluorescence (Figure 5E & I) or dark field microscopy (Figure 5G). By contrast, 3 ± 1.5% of the injected NP–NSCs (7% of those present in the brain) were redistributed selectively to the tumor site in the contralateral hemisphere (Figure 5F & I). Spectrally mapped dark field images confirmed the presence of polystyrene NPs at the tumor site (Figure 5H). 3D reconstruction software was again used to better visualize NP distribution throughout the injection and tumor sites in a representative brain (Figure 5J). Further experiments confirmed that NP–NSCs could selectively distribute to tumor foci even when the injection and tumor sites were impeded, rather than bridged by a prominent white matter tract (Supplementary Figure 5).
Figure 5. Comparison of free nanoparticle and nanoparticle–neural stem cell distribution to an intracranial glioma when injected in the contralateral hemisphere.
(A & B) Injection paradigm for upper (A) and lower (B) panels. (C & D) A total of 4 days after NP injection, brains were sectioned and every fifth section throughout the entire injection and tumor site was imaged using fluorescence microscopy. The slice that contained the greatest surface area of NPs was identified and adjacent sections stained with hematoxylin and eosin staining. Scale bar: 500 µm. Antifirefly luciferase immunohistochemistry was also performed (insets in C & D; scale bar: 100 µm) to confirm the presence of tumor cells. (E & F) Representative merged fluorescence images of tumor sites where NPs are red, NSCs are green, and nuclei are blue. Arrows indicate visible NPs. Tumors are visible as dense clusters of 4-,6-diamidino-2-phenylindole-stained nuclei (blue). Scale bar: 100 µm. (G & H) Dark field micrographs of tumor sites. Polystyrene NPs were identified based on their unique spectral profile, then pseudo-coloured red to ease visualization. Scale bar: 10 µm. (I) Percentage of NPs at the injection and tumor sites 4 days postinjection. Error bars represent mean ± standard deviation. (J) 3D reconstruction of NP (red) distribution around the tumor (blue).
*p < 0.1; **p < 0.05.
NP: Nanoparticle; NSC: Neural stem cell.
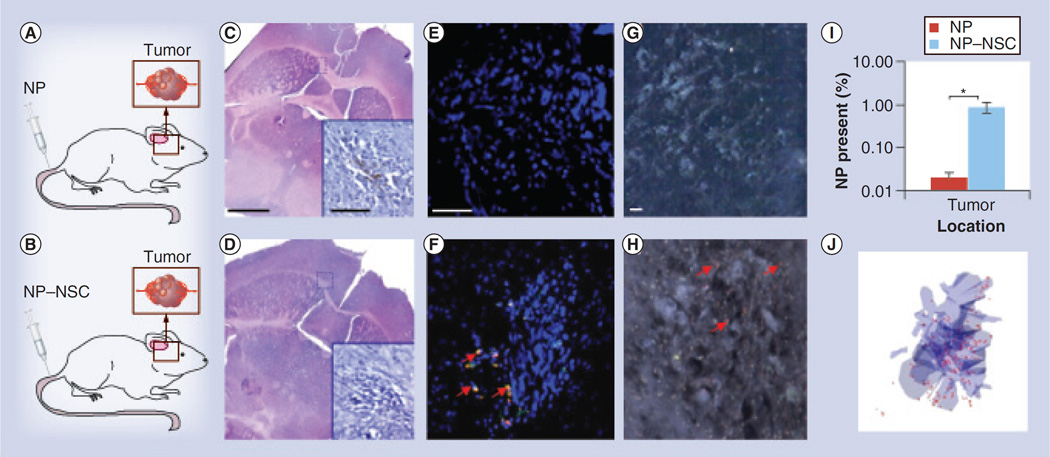
Paradigm 3: intravenous NP injection
The final paradigm involved injecting free NPs or NP–NSCs into the tail vein of mice (Figure 6A–D). As expected, negligible levels of intravenously injected free-NPs were detected at the tumor site by either fluorescence (Figure 6E & I) or dark field microscopy (Figure 6G). By contrast, 0.9 ± 0.2% of the injected NP–NSCs were observed at the intracranial tumor site, but nowhere else in the brain (Figure 6F & I). This represents a 46 ± 6-fold increase in NP intensity present within the brain when NSCs are used to distribute the NPs. Spectrally mapped dark field images confirmed the presence of polystyrene NPs at the tumor site (Figure 6H). 3D reconstruction software was again used to visualize NP–NSC distribution throughout the entire tumor site in a representative brain (Figure 6J). Note that the majority of NPs were still subject to clearance by the liver and spleen (Supplementary Figure 6), as would be expected for intravenously injected NPs in this size range [22].
Figure 6. Comparison of free nanoparticle and nanoparticle–neural stem cell distribution to intracranical glioma when injected intravenously.
(A & B) Injection paradigm for upper (A) and lower (B) panels. (C & D) A total of 4 days after NP injection, brains were sectioned and every fifth section throughout the entire injection and tumor site was imaged using fluorescence microscopy. The slice that contained the greatest surface area of NPs was identified and adjacent sections were stained with hematoxylin and eosin. Scale bar: 500 µm. Antifirefly luciferase immunohistochemistry was also performed (insets in C & D; scale bar: 100 µm) to confirm the presence of tumor cells. (E & F) Representative merged fluorescence images of tumor sites (NPs: red; NSCs: green; nuclei: blue). Arrows indicate visible NPs. Tumors are visible as dense clusters of 4-,6-diamidino-2-phenylindole-stained nuclei (blue). Scale bar: 100 µm. (G & H) Dark field micrographs of tumor sites. Polystyrene NPs were identified based on their unique spectral profile, then pseudo-coloured red to ease visualization. Arrows indicate visible NPs. Scale bar: 10 µm. (I) Percentage of NPs at the tumor sites 4 days postinjection. Error bars represent mean ± standard deviation. (J) 3D reconstruction of NP (red) distribution around the tumor (blue).
*p < 0.05.
NP: Nanoparticle; NSC: Neural stem cell.
Discussion
In this study, we demonstrate that despite the known importance of chemokine [23], integrin [24, 25] and selectin [25] surface receptors for tumor tropism, conjugating NPs to NSC surfaces had a negligible effect on viability and tropism in vitro. We demonstrate that NPs were surface localized for at least 1 h after coupling, a time point chosen to correlate with previous work showing that injected NSCs arrive at tumor foci within 50 min (<10% increase in NSC numbers at tumor foci over the next 1–2 weeks) [21]. These NP–NSC conjugates were used to test the hypothesis that NSCs can improve NP retention and tumor-specific distribution within the brain.
Following intracranial injections, NPs are typically lost due to both reflux and clearance. Previous studies report a 34–40% loss of intercerebral cell or particulate injections due to reflux [4, 26–28]. Here we show that conjugating NPs to NSCs did not improve reflux-associated loss, with approximately 40% losses in both free NP and NP–NSC injections observed after 1 day. By contrast, surface conjugating NPs to NSCs dramatically reduced NP clearance rates as assessed 4 days after injection. By this time, a dramatic (~93%) decrease in the presence of free NPs was observed, while NP–NSCs were retained at day 1 levels. The magnitude of free NP clearance observed here is within range of clearance rates reported for hydrophilic lipid nanocapsules (half-life of 7–10 h; 94% cleared 3 days postinjection) [29, 30]. Although significantly longer retention times have been achieved by altering NP surface properties (only 1% eliminated after 12 h [31]), the increased residency time is usually attributed to NP endocytosis by host brain cells [2, 32]. While our results cannot rule out the possibility that surface-conjugated NPs were endocytosed by NSCs following injection, there is reason to believe minimal endocytosis occurs because cells exhibit intrinsically low uptake of unmodified polystyrene NPs [33, 34]. This is especially true if the NPs are larger than 50–200 nm [35, 36], negatively charged [36] and anchored to cell-surface proteins [37]. Instead, we speculate that NP retention is improved because NP–NSCs are protected from perivascular clearance during the first week as postinjection edema subsides, whereas free NPs are small enough to be cleared by perivascular phagocytic cells [38–40]. Another recent study observed that MSCs can also improve intracranial retention of surface-conjugated NPs [6].
Tumor-specific distribution of NPs to tumors was investigated using both fluorescent and dark field microscopy. Although polystyrene NPs have previously been visualized using dark field microscopy [41], this is the first report to our knowledge that successfully applied spectral mapping to identify polystyrene particles within tissue slices. It is difficult given the high degree of spectral overlap that exists between polystyrene and tissue. Before assessing NSC-mediated NP distribution, we first assessed the degree to which NPs could passively diffuse throughout brain tissue. Diffusion-associated NP distribution was expected to be minimal given that the NPs are much larger than reported estimates of intracranial intercellular spaces (38–200 nm [42, 43]). If the NPs could diffuse freely, the maximum volume of distribution (Vd) should be fivefold larger than the injected volume (Vi), given that the fraction of extracellular space in the brain is approximately 20% [27]. For free NPs, the maximum radial distance that NPs distributed was 559 ± 9 µm, which corresponds to a Vd = 1.96 ± 0.06 µl; a volume less that V1 (2 µl). Most NPs distributed no more than 101–110 µm from the injection site, similar to results observed previously when injecting 100–200 nm polystyrene NPs into the brain [14].
In the absence of a tumor stimulus, NSCs did not move NPs distant from the injection site and no measureable difference was observed in the radial distribution (509 ± 12 µm) or Vd (1.63 ± 0.08 µl) of NP–NSCs relative to free NP injections. When injected adjacent to the tumor, however, NSCs distributed the majority of NPs throughout the tumor with negligible NP distribution to noninvolved brain regions. NSCs also selectively distributed NPs to tumors located >1 mm away in the contralateral hemisphere. Previous studies report that 1–10% of injected NSCs migrate to contralateral tumors [21, 38, 44–45], which is consistent with our observations that 3% of injected NP–NSCs redistributed. While our initial observations suggest that NP–NSCs relocated to tumors as efficiently as unmodified NSCs, direct comparisons are reserved for future work.
Finally, our results suggest that NSCs can even deliver surface conjugated NPs to glioma when administered intravenously, a desirable route given its less invasive nature. To our knowledge, there are no other reports of NPs greater than 200 nm penetrating the blood–tumor barrier after systemic administration [14, 43]. Numerous studies have confirmed, however, that intravenously injected NSCs can access intracranial tumors albeit at low (~0.5–1%) efficiency [38, 45]. The low efficiency is unsurprising given that delivery of cells to the brain depends on pulmonary passage, recognition of endothelial adhesion molecules, disruption of tight junctions and penetration across the basal lamina surrounding the blood vessels [25]. Further studies are needed to determine the extent to which NP-conjugation to NSCs restricts pulmonary passage, which is recognized as the major distribution challenge for larger cells. Unmodified NSCs (16 µm in diameter) pass through the lungs with 2× greater efficiency than MSCs (18 µm in diameter) [46]. When NSCs are surface conjugated to approximately 800-nm NPs, their size approaches that of a MSC and a greater portion of injected NSCs may be trapped within the lungs.
Conclusion
Here we demonstrate that NP–NSCs retain their viability and tumor tropic properties even when the NPs are significantly larger (~800nm) than those typically used for cancer therapies. This is the first study to our knowledge demonstrating that tumor-tropic cells can distribute NPs to tumors located within the brain parenchyma, whether injected adjacent to tumor (<200 µm away), into the contralateral hemisphere (>1 mm away) or into the tail vein. It was further confirmed that NSCs improve intracranial NP retention (from 7 to 40% retained on day 4). These proof-of-principle studies set the stage for future therapeutic studies in which NSCs are used to deliver large drug-loaded NPs selectively to invasive brain tumors.
Future perspective
Glioma patients are currently faced with treatment options that are not only ineffective, but can also impair the quality of their remaining few years. Such a clinical scenario underscores the importance of investing in new therapeutic approaches that, when combined with conventional therapies, are able to selectively eliminate invasive glioma [47]. Given the challenges associated with glioma-specific NP distribution, there is an emerging interest in coupling them to tumor tropic cells [47]. The present study demonstrates the feasibility of this concept using cytocompatible, polystyrene NPs that are too large to passively move through brain parenchyma to reach the tumor. We show, for the first time, that tumor-tropic NSCs can deliver NPs to intracranial tumor foci when injected at three distant locations, although the efficiency of tumor tropism decreases as distance from the tumor increases. NPs were found only at the injection site or the tumor site with negligible distribution in noninvolved brain regions. Comparative studies between antibody/aptamer- and NSC-mediated targeting are still needed, but NSC-mediated targeting may have distinct advantages over traditional NP targeting approaches when accessing tumors that are negative for, or exhibit dynamic regulation of, tumor-specific surface markers. NSCs are attracted to tumor-derived ‘cytokine storms’ and inflammatory signals [23], thus providing a potentially more robust targeting platform.
While this is not the first study surface-coupling NPs to tumor tropic cells, it does advance this emerging field in two key respects. First, earlier reports focused on conjugating smaller NPs (100–300 nm) to cells. We now demonstrate that tumor tropic cells can distribute larger NPs (~800 nm), which exponentially increases the cargo volume that each cell transports. This improved cargo capacity may facilitate the delivery of relevant drug doses [48].
Second, this represents the first study using a clinically relevant tumor tropic cell. HB1. F3 NSCs are already being tested in Phase I brain tumor trials (mediating an enzyme/ prodrug therapy). Extensive characterization studies have demonstrated the HB1.F3 line is chromosomally and functionally stable, nontumorigenic, and minimally immunogenic (HLA class II negative [49]). Differentiation of v-myc immortalized HB1.F3 NSCs has not yet been observed within a tumor setting. Although the biotin–streptavidin NP–NSC coupling scheme was sufficient for this proof-of-concept study, it may not be appropriate for clinical use given oxidative risks [50] and the immuogenicity of streptavidin [51]. Future studies will investigate the utility of coupling strategies reliant on endogenous thiols [5] or receptor–antibody combinations [6].
Together, these results raise the possibility of using tumor tropic cells to noninvasively deliver large NPs loaded with small molecule and/or siRNA-mediated therapies to intracranial tumors, as well as tumors located in the periphery. With sufficient development, tropic cell-therapeutic NP combinations may enable unprecedented control over the spatiotemporal release of therapeutics in vivo. Please also see an online supplement, available at [52].
Supplementary Material
EXECUTIVE SUMMARY.
Nanoparticle–neural stem cell coupling
A total of 175 nanoparticles (NPs) that are 798 nm in diameter can be stably coupled to the surface of HB1.F3 neural stem cells (NSCs).
NP-coupled NSCs retain tumor tropism in vitro
Conjugating NPs to the surface of clinically relevant NSCs had a negligible effect on their viability and tropism.
NSCs improve NP retention in vivo
Conjugating NPs to the surface of NSCs improves NP retention as assessed 4 days after injection.
NSCs target NPs to glioma in vivo
Using tumor topic stem cells to deliver therapeutic NPs improves tumor-selective distribution of NPs in an orthotopic glioma model, whether delivered intracranially (adjacent or contralateral to the established tumor) or intravenously.
Tumor tropic cells can target NP cargo too large to passively cross the blood–tumor barrier.
Acknowledgments
The authors gratefully acknowledge K Walker for editing assistance; Z Li, R Zerda and M Miller for technical assistance with scanning electron microscopy (funding was provided by DOD 1435-04-03GT-73134); and B Armstrong for assistance with confocal microscopy experiments.
The authors thank STOP Cancer; The Rosalinde and Arthur Gilbert Foundation; Mary Kay Foundation; California Institute of Regenerative Medicine; the Alvarez Family Foundation; and the Accelerated Brain Cancer Cures Foundation for their generous financial support. R Mooney was supported by a fellowship from the California Institute for Regenerative Medicine (Grant Number TG2- 01150). AJ Annala and KS Aboody declare an interest in TheraBiologics, Inc., an early stage biotechnology company focused on employing neural stem cells to treat cancer.
Footnotes
Disclaimer
The contents of this publication are solely the responsibility of the authors and do not necessarily represent the official views of CIRM or any other agency of the State of California.
Financial & competing interests disclosure
The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
Ethical conduct of research
The authors state that they have obtained appropriate institutional review board approval or have followed the principles outlined in the Declaration of Helsinki for all human or animal experimental investigations. In addition, for investigations involving human subjects, informed consent has been obtained from the participants involved.
References
Papers of special note have been highlighted as:
• of interest
- 1.Meikle SR, Matthews JC, Brock CS, et al. Pharmacokinetic assessment of novel anti-cancer drugs using spectral analysis and positron emission tomography: a feasibility study. Cancer Chemother. Pharmacol. 1998;42:183–193. doi: 10.1007/s002800050804. [DOI] [PubMed] [Google Scholar]
- 2.Allard E, Passirani C, Benoit JP, et al. Convection-enhanced delivery of nanocarriers for the treatment of brain tumors. Biomaterials. 2009;12:2302–2318. doi: 10.1016/j.biomaterials.2009.01.003. [DOI] [PubMed] [Google Scholar]
- 3.Neeves KB, Sawyer AJ, Foley CP, et al. Dilation and degradation of the brain extracellular matrix enhances penetration of infused polymer nanoparticles. Brain Res. 2007;1180:121–132. doi: 10.1016/j.brainres.2007.08.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brady ML, Raghavan R, Alexander A, et al. Pathways of flow for infusate loss during convection-enhanced delivery into the putamen nucleus. Stereotact. Funct. Neurosurg. 2013;91:69–78. doi: 10.1159/000342492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stephan MT, Moon JJ, Um SH, et al. Therapeutic cell engineering with surface-conjugated synthetic nanoparticles. Nat. Med. 2010;9:1035–1041. doi: 10.1038/nm.2198.• First paper to demonstrate that surface-bound nanoparticles can be stably bound to tumor-tropic cells in vivo.
- 6.Li L, Guan Y, Liu H, et al. Silica nanorattle-doxorubicin-anchored mesenchymal stem cells for tumor-tropic therapy. ACS Nano. 2011;5:7462–7470. doi: 10.1021/nn202399w.• First paper coupling chemotherapeutic nanoparticles to tumor tropic cells with the intent to improve glioma treatment efficacy.
- 7.Eskandary H, Basiri M, Nematollahi-Mahani SN, et al. The role of stem cells in tumor targeting and growth suppression of gliomas. Biologics. 2011;5:61–70. doi: 10.2147/BTT.S17838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ahmed AU, Tyler MA, Thaci B, et al. A comparative study of neural and mesenchymal stem cell-based carriers for oncolytic adenovirus in a model of malignant glioma. Mol. Pharm. 2011;8:1559–1572. doi: 10.1021/mp200161f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.A pilot feasibility study of oral 5-fluorocytosine and genetically modified neural stem cells expressing E. coli cytosine deaminase for treatment of recurrent high grade gliomas. http://clinicaltrials.gov/show/NCT01172964
- 10.Aboody K, Capela A, Niazi N, et al. Translating stem cell studies to the clinic for CNS repair: current state of the art and the need for a Rosetta stone. Neuron. 2012;70:597–613. doi: 10.1016/j.neuron.2011.05.007. [DOI] [PubMed] [Google Scholar]
- 11.Thu MS, Najbauer J, Kendall, et al. Iron labeling and pre-clinical MRI visualization of therapeutic human neural stem cells in a murine glioma model. PLoS ONE. 2009;4:e7218. doi: 10.1371/journal.pone.0007218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Krishnamachari Y, Pearce ME, Salem AK. Self-assembly of cell-microparticle hybrids. Adv. Mater. 2008;20:989–993.• Original paper surface conjugating microparticles to a cell surface.
- 13.Mailander V, Landfester K. Interaction of nanoparticles with cells. Biomacromolecules. 2009;10:2379–2400. doi: 10.1021/bm900266r. [DOI] [PubMed] [Google Scholar]
- 14.Chen MY, Hoffer A, Morrison PF, et al. Surface properties, more than size, limiting convective distribution of virus-sized particles and viruses in the central nervous system. J. Neurosurg. 2005;103:311–319. doi: 10.3171/jns.2005.103.2.0311. [DOI] [PubMed] [Google Scholar]
- 15.Kim SU, Nagai A, Nakagawa E, et al. Production and characterization of immortal human neural stem cell line with multipotent differentiation property. Methods Mol. Biol. 2008;438:103–121. doi: 10.1007/978-1-59745-133-8_10. [DOI] [PubMed] [Google Scholar]
- 16.Flax JD, Aurora S, Yang C, Simonin C, et al. Engraftable human neural stem cells respond to developmental cues, replace neurons, and express foreign genes. Nat. Biotechnol. 1998;16:1033–1039. doi: 10.1038/3473. [DOI] [PubMed] [Google Scholar]
- 17.Brown AB, Yang W, Schmidt NO, et al. Intravascular delivery of neural stem cell lines to target intracranial and extracranial tumors of neural and non-neural origin. Hum. Gene Ther. 2003;14:1777–17785. doi: 10.1089/104303403322611782. [DOI] [PubMed] [Google Scholar]
- 18.Fiala JC. Reconstruct: a free editor for serial section microscopy. J. Microscopy. 2005;218:52–61. doi: 10.1111/j.1365-2818.2005.01466.x. [DOI] [PubMed] [Google Scholar]
- 19.Lorenz MR, Kohnle MV, Dass M, et al. Synthesis of fluorescent polyisoprene nanoparticles and their uptake into various cells. Macromol. Biosci. 2008:711–727. doi: 10.1002/mabi.200700336. [DOI] [PubMed] [Google Scholar]
- 20.Alon R, Bayer EA, Wilchek M. Cell adhesion to streptavidin via RGD-dependent integrins. Eur. J. Cell Biol. 1993;60:1–11. [PubMed] [Google Scholar]
- 21.Kim JH, Lee JE, Kim SU, et al. Stereological analysis on migration of human neural stem cells in the brain of rats bearing glioma. Neurosurgery. 2010;66:333–342. doi: 10.1227/01.NEU.0000363720.07070.A8. [DOI] [PubMed] [Google Scholar]
- 22.Decuzzi P, Godin B, Tanaka T, et al. Size and shape effects in the biodistribution of intravascularly injected particles. J. Control. Release. 2010;141:320–327. doi: 10.1016/j.jconrel.2009.10.014. [DOI] [PubMed] [Google Scholar]
- 23.Kendall SE, Najbauer J, Johnston HF, et al. Neural stem cell targeting of glioma is dependent on phosphoinositide 3-kinase signaling. Stem Cells. 2008;26:1575–1586. doi: 10.1634/stemcells.2007-0887. [DOI] [PubMed] [Google Scholar]
- 24.Ziu M, Schmidt NO, Cargioli TG, et al. Glioma-produced extracellular matrix influences brain tumor tropism of human neural stem cells. J. Neurooncol. 2006;79:125–133. doi: 10.1007/s11060-006-9121-5. [DOI] [PubMed] [Google Scholar]
- 25.Karp JM, Teo GSL. Mesenchymal stem cell homing - the devil is in the details. Cell Stem Cell. 2009;4:206–216. doi: 10.1016/j.stem.2009.02.001. [DOI] [PubMed] [Google Scholar]
- 26.Hansen K, Muller FJ, Messing M, et al. A 3-dimensional extracellular matrix as a delivery system for the transplantation of glioma-targeting neural stem/progenitor cells. Neuro. Oncol. 2010;12:645–654. doi: 10.1093/neuonc/noq002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Levin VA, Fenstermacher JD, Patlak CS. Sucrose and inulin space measurements of cerebral cortex in four mammalian species. Am. J. Physiol. 1970;219:1528–1533. doi: 10.1152/ajplegacy.1970.219.5.1528. [DOI] [PubMed] [Google Scholar]
- 28.Krauze MT, Saito R, Noble C, et al. Reflux-free cannula for convection-enhanced high-speed delivery of therapeutic agents. J. Neurosurg. 2005;103:923–929. doi: 10.3171/jns.2005.103.5.0923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Allard E, Hindre F, Passirani C, et al. 188Re-loaded lipid nanocapsules as a promising radiopharmaceutical carrier for internal radiotherapy of malignant gliomas. Eur. J. Nucl. Med. Mol. Imaging. 2008;10:1838–1846. doi: 10.1007/s00259-008-0735-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.MacKay JA, Deen DF, Szoka FC., Jr Distribution in brain of liposomes after convection enhanced delivery; modulation by particle charge, particle diameter, and presence of steric coating. Brain Res. 2005;1035:139–153. doi: 10.1016/j.brainres.2004.12.007. [DOI] [PubMed] [Google Scholar]
- 31.Perlstein B, Ram Z, Daniels D, et al. Convection-enhanced delivery of maghemite nanoparticles: Increased efficacy and MRI monitoring. Neuro. Oncol. 2008;2:153–161. doi: 10.1215/15228517-2008-002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gennet N, Alexander LM, Sanchez-Martin RM, et al. Microspheres as a vehicle for biomolecule delivery to neural stem cells. N. Biotechnol. 2009;25:442–449. doi: 10.1016/j.nbt.2009.05.006. [DOI] [PubMed] [Google Scholar]
- 33.Holzapfel V, Musyanovych A, Landfester K, et al. Preparation of fluorescent carboxyl and amino functionalized polystyrene particles by miniemulsion polymerization as markers for cells. Macromol. Chem. Physics. 2005;206:2440–2449. [Google Scholar]
- 34.Lorenz MR, Holzapfel V, Musyanovych A, et al. Uptake of functionalized, fluorescent-labeled polymeric particles in different cell lines and stem cells. Biomaterials. 2006;14:2820–2828. doi: 10.1016/j.biomaterials.2005.12.022. [DOI] [PubMed] [Google Scholar]
- 35.Rejman J, Oberle V, Zuhorn I, et al. Size-dependent internalization of particles via the pathways of clathrin- and caveolae-mediated endocytosis. Biochem. J. 2004;377:159–169. doi: 10.1042/BJ20031253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Verma A, Stellacci F. Effect of surface properties on nanoparticle-cell interactions. Small. 2010;6:12–21. doi: 10.1002/smll.200901158. [DOI] [PubMed] [Google Scholar]
- 37.Stephan MT, Irvine DJ. Enhancing cell therapies from the outside: cell surface engineering using synthetic nanomaterials. Nano Today. 2011;6:309–325. doi: 10.1016/j.nantod.2011.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jackson JS, Golding JP, Chapon C, et al. Homing of stem cells to sites of inflammatory brain injury after intracerebral and intravenous administration: a longitudinal imaging study. Stem Cell Res. Ther. 2010;17:1–12. doi: 10.1186/scrt17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Carare RO, Bernardes-Silva M, Newman TA, et al. Solutes, but not cells, drain from the brain parenchyma along basement membranes of capillaries and arteries: significance for cerebral amyloid angiopathy and neuroimmunology. Neuropathol. Appl. Neurobiol. 2008;34:131–144. doi: 10.1111/j.1365-2990.2007.00926.x. [DOI] [PubMed] [Google Scholar]
- 40.MacKay JA, Deen DF, Szoka FC., Jr Distribution in brain of liposomes after convection enhanced delivery; modulation by particle charge, particle diameter, and presence of steric coating. Brain Res. 2005;1035:139–153. doi: 10.1016/j.brainres.2004.12.007. [DOI] [PubMed] [Google Scholar]
- 41.Rebner K, Schmitz M, Boldrini B, et al. Dark-field scattering microscopy for spectral characterization of polystyrene aggregates. Opt. Express. 2010;18:3116–3127. doi: 10.1364/OE.18.003116. [DOI] [PubMed] [Google Scholar]
- 42.Syková E, Nicholson C. Diffusion in brain extracellular space. Physiol. Rev. 2008;88:1277–1340. doi: 10.1152/physrev.00027.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nance EA, Woodworth GF, Sailor KA, et al. A dense poly(ethylene glycol) coating improves penetration of large polymeric nanoparticles within brain tissue. Sci. Transl. Med. 2012;4:119–149. doi: 10.1126/scitranslmed.3003594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Flexman JA, Cross DJ, Tran LN, et al. Quantitative analysis of neural stem cell migration and tracer clearance in the rat brain by MRI. Mol. Imaging Biol. 2011;13:104–111. doi: 10.1007/s11307-010-0311-3. [DOI] [PubMed] [Google Scholar]
- 45.Tang Y, Shah K, Messerli SM, et al. In vivo tracking of neural progenitor cell migration to glioblastomas. Hum. Gene Ther. 2003;14:1247–1254. doi: 10.1089/104303403767740786. [DOI] [PubMed] [Google Scholar]
- 46.Fischer UM, Harting MT, Jimenez F, et al. Pulmonary passage is a major obstacle for intravenous stem cell delivery: the pulmonary first-pass effect. Stem Cells Dev. 2009;18:683–691. doi: 10.1089/scd.2008.0253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Auffinger B, Morshed R, Tobias A, Cheng Y, Ahmed AU, Lesniak MS. Drug-loaded nanoparticle systems and adult stem cells: a potential marriage for the treatment of malignant glioma? Oncotarget. 2013;4:378–396. doi: 10.18632/oncotarget.937.• Highlights the promise of combining tumor tropic cells and nanoparticles to improve drug delivery to invasive glioma foci.
- 48.Eseonu C. Thesis. Hong Kong, China: University of Hong Kong Libraries; 2011. Intracranial drug delivery of siRNA nanoparticles to tumor cells. [Google Scholar]
- 49.Aboody KS, Najbauer J, Metz MZ, et al. Preclinical studies neural stem cell-mediated enzyme/prodrug therapy for glioma. Sci. Trans. Med. 2013;5:184ra59. doi: 10.1126/scitranslmed.3005365.• Recent report describing the safety and immunogenicity of HB1.F3 NSCs in the glioma setting.
- 50.Srinivas L, Colburn NH. Preferential oxidation of cell surface sialic acid by periodate leads to promotion of transformation in JB6 cells. Carcinogenesis. 1984;4:515–519. doi: 10.1093/carcin/5.4.515. [DOI] [PubMed] [Google Scholar]
- 51.Meyer DL, Schultz J, Lin Y, et al. Reduced antibody response to streptavidin through site-directed mutagenesis. Protein Sci. 2001;3:491–503. doi: 10.1110/ps.19901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Liver and myocardial iron overload. http://oernst.f5lvg.free.fr/liver/iron.html.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.