Abstract
Studies in recent years have uncovered a diverse set of eukaryotic receptors that recognize lipopolysaccharide (LPS), the major outer-membrane component of gram-negative bacteria. Indeed, Toll-like Receptors, G-protein coupled receptors, Integrins, Receptor-like kinases, and caspases have emerged as important LPS-interacting proteins. In this Review, the mammalian receptors that detect LPS are described. I highlight how no host protein is involved in all LPS responses, but a single lipid (phosphatidylinositol 4,5-bisphosphate) regulates many LPS responses, including endocytosis, phagocytosis, inflammation and pyroptosis. I further describe LPS response systems that operate specifically in plants, and discuss potentially new LPS response systems that await discovery. This diversity of receptors for a single microbial product underscores the importance of host-microbe interactions in multiple kingdoms of life.
Academic and therapeutic benefits of a cross-species analysis of innate immunity
Multicellular organisms come in many forms and have many needs. These forms can range from the gigantic elephant to the diminutive house mouse, and these needs range from the desire of humans for companionship to the desire of tigers for solitude. Despite this diversity of features and lifestyles, there are fundamental features of all multicellular organisms. One of these features is the need to identify potentially infectious microorganisms, which can pose an existential threat to the host. Among these infectious threats is that posed by encounters with bacteria. Throughout history, bacterial infections have been a threat to human longevity, and one of the triumphs of our times has been the production of vaccines and antibiotics that prevent these infections. For example, in the year 1900 the life expectancy of humans in the United States was 47.3 years, with infectious diseases being the leading cause of lethality [1]. In the years following 1924, widespread vaccination and antibiotic use has been estimated to prevent over 100 million infections, and shift non-infectious disease to emerge as the leading cause of death [1]. Today, the life expectancy in the United States has experienced a considerable rise, and is approaching 80 years of age [2]. These remarkable medical advances underscore the value of basic microbiology and immunology to individuals, and society. It is worth noting, however, that much of our knowledge of host-microbe interactions has derived from the desire to protect ourselves, not the ecosystem in which we reside. Despite our interests in protecting ourselves, all multicellular organisms face the threat of bacterial infection. Only through the study of all organisms can a true systems-wide understanding of immune defense be achieved.
An interest in understanding host-microbe interactions in all organisms is not merely academic, as infections of many non-humans can influence our way of life. For example, infections of livestock and sea life have considerable effects on food availability [3], and in some instances, infections that originate in farmed animals can cause deadly disease when transmitted to humans. An example of this threat comes from epidemiological studies of influenza outbreaks, as the infectious virus often originates from a farm animal [4]. As our knowledge of human biology improves, so has our ability to target and eliminate the threat of infections. Based on this premise, it stands to reason that similar benefits could be achieved through a greater understanding of the immune systems of a wide range of organisms that occupy diverse ecological niches. Indeed, genetic and bioinformatic studies have suggested that the principles established to explain the detection of bacteria by mice and humans may not apply to all multicellular organisms [5, 6]. As such, species-specific solutions to the problem of bacterial infection may exist. Herein, I will discuss the mechanisms involved in the recognition of lipopolysaccharide (LPS), the major component of the cell wall of gram-negative bacteria. LPS is composed of three moieties, lipid A, a core oligosaccharide and the O-antigen (Figure 1). Of note, the structures of lipid A and the core oligosaccharide are conserved across bacterial species (with some exceptions), whereas the O-antigen is significantly more variable [7]. The mechanisms by which LPS is detected by the innate immune system will be used as a frame of reference for a discussion of the evolutionary aspects of host-microbe interactions. I will first describe the current knowledge of how LPS is detected in land animals, such as mice and humans. I will then discuss recent studies that have revealed novel LPS detection mechanisms that may operate in plants, invertebrates and in primitive single celled eukaryotes. This knowledge illustrates the diversity of strategies that exist in nature to combat bacterial infections, and raises the question of whether a better understanding of these strategies can be used for therapeutic purposes.
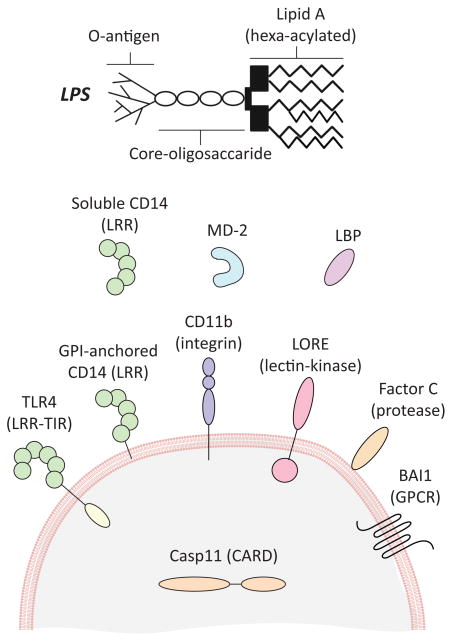
Figure 1.
Diverse LPS receptors are found in nature.
Depiction of the types of LPS receptors in plants and animals. Receptors include representatives from various families of proteins, including Toll-like receptors (TLRs), integrins, G-protein coupled receptors (GPCRs), proteases and kinases. Receptors can be membrane proteins, soluble cytosolic proteins or soluble extracellular proteins (as indicated). Of note, Factor C is secreted but lipid binding, hence its positioning at the membrane. This diversity illustrates the complexity of the natural LPS response systems, and raises the question of how many others await discovery.
Abbreviations used: LRR: Leucine-rich repeats, LBP: LPS-binding protein, GPI: Glycosylphosphatidylinositol, LORE: Lipooligosaccharide-specific reduced elicitation, BAI1: Brain-specific angiogenesis inhibitor 1, CARD: caspase activation and recruitment domain
LPS detection in mammals: diverse pathways are coordinated by a common host lipid
Perhaps due to the central role of LPS in inducing inflammation during bacterial infections [8], much of our knowledge of LPS responses has derived from studies in humans and mice. In these organisms, the detection of LPS is first achieved through the actions of LPS binding protein (LBP), which is present in the extracellular fluids [9]. LBP makes physical contacts with micelles of free LPS and with gram-negative bacteria [10]. These interactions facilitate the extraction of a monomer of LPS by CD14, the first membrane-bound LPS receptor identified [10, 11]. CD14 exists as a glycosylphosphatidylinositol (GPI)-anchored protein at the plasma membrane of many mammalian cells, most notably phagocytes, but can also be secreted from cells in a soluble (GPI-free) state [12]. LPS-CD14 complexes are critical for subsequent inflammatory responses for three reasons. First, the LPS present in complex with CD14 can be transferred to two sets of heterodimers of MD-2 and Toll-like receptor 4 (TLR4) [10, 13, 14]. Of these LPS-binding proteins, TLR4 is the unique in that it exists as a transmembrane protein, thereby serving as a conduit to transmit the information of LPS detection to the cytosol [15]. Mechanistically, CD14-mediated transfer of LPS to two sets of TLR4/MD-2 complexes results in the dimerization of the cytosolic Toll/IL-1R homology (TIR) domain present in these TLR4 molecules, and the accumulation of this complex in plasma membrane microdomains with features similar to lipid rafts [16]. The dimerized TLR4 TIR domains are detected by a protein TIRAP (also known as Mal) [17, 18], which is a promiscuous phosphoinositide-binding protein [19, 20]. Interactions between TIRAP and plasma membrane-localized phosphatidylinositol 4,5-bisphosphate (PI(4,5)P2) position this adaptor at the cell surface prior to microbial encounters. This positioning allows TIRAP to rapidly detect dimerized TLR4, an event that subsequently leads to the assembly of a cytosolic supramolecular organizing center (SMOC) called the myddosome [19, 21]. The myddosome has attracted much attention in recent years, as it serves as the principal subcellular site of TLR signal transduction [19, 22–24]. This SMOC consists minimally of TIRAP, its related adaptor protein MyD88 and several IRAK family kinases that initiate downstream signaling [19]. Upon assembly, the proteins within the myddosome promote the NF-κB and AP-1 dependent inflammatory responses that typify TLR4 signal transduction. The specific details of each step in the TLR4 pathway has been reviewed expertly elsewhere [25]. The second activity of LPS-bound CD14 is to promote the internalization of itself and dimerized TLR4 from the plasma membrane [26]. This process moves TLR4 into endosomes, and promotes a second wave of NF-κB and AP-1 activation that reinforces the responses induced from the plasma membrane [26, 27]. TLR4 signaling from endosomes also induces transcriptional responses that are distinct from those initiated at the cell surface, with the best-defined being those mediated by the transcription factor IRF3 [26, 27]. IRF3 activation is mediated by the actions of the adaptor proteins TRAM and TRIF, which are thought to only engage endosome-localized TLR4. IRF3 acts along with NF-κB and AP-1 to drive the expression of type I interferon (IFN) genes, which are important for various cell intrinsic antibacterial responses (e.g. autophagy and pyroptosis) [28, 29]. In dendritic cells, TLR4-induced type I IFN expression also promotes the activation of natural killer cells, which then release the highly antibacterial cytokine IFNγ [30, 31]. Importantly, while TLR4 is a cargo for this CD14-dependent endocytosis pathway, TLR4 signaling does not regulate this process. Rather, CD14 (independently of TLR4 signaling) promotes the endocytosis process that is important to position TLR4 in a subcellular location where IRF3-dependent signaling can occur [32, 33]. Several regulators of the unique signaling pathway activated by CD14 have been defined [26, 32, 34–37], but the molecular details of this new pathway are unclear. The third consequence of LPS-CD14 interactions is the rapid modification of membrane phosphoinositides, in particular leading to the generation of PI(4,5)P2 [38, 39]. LPS detection by CD14 leads to the activation of the cytosolic phosphoinositide kinases PIP5K Iα and PIP5K Iγ, which phosphorylate the 5′ phosphate present in plasma membrane-localized PI(4)P to generate PI(4,5)P2 [38, 39]. This process (which is independent of TLR4 signaling) is perhaps most notable, as PI(4,5)P2 is a central regulator of TIRAP localization and the process of endocytosis (and phagocytosis) [19, 20, 40]. In this regard, one can consider CD14 to be orchestrating all cellular responses to LPS that are induced by TLR4. CD14 acts upstream of TLR4 to deliver LPS to TLR4 to promote dimerization, stimulate PI(4,5)P2 synthesis to promote myddosome formation and NF-κB/AP-1 dependent inflammatory responses, and stimulate TLR4 internalization, which consequently induces IRF3-dependent type I IFN expression.
There is also evidence that an intracellular pool of TLR4 is present on recycling endosomes in human and murine phagocytes [41, 42]. This pool is mobilized to early endosomes containing TLR4 that was internalized from the plasma membrane, and appears to amplify innate immune responses from this location [41]. Ultimately, endosomal TLR4 is sorted into the lumen of multivesicular bodies by a process dependent on the HRS trafficking adaptor [43]. This sorting event is important to inactivate TLR4 signal transduction [43].
While studies of CD14-dependent TLR4 endocytosis provided the first evidence that TLR4 does not control all cellular responses to LPS [26], work in recent years has revealed additional TLR4-independent LPS responses, and these are not regulated by CD14. For example, LPS delivery into the cytosol induces a potent pyroptotic cell death response [44] [45]. This response is important to prevent mammalian cells from being used as a growth substrate by intracellular bacteria [46]. LPS-induced pyroptosis is not dependent on CD14 or TLR4, but is rather mediated by the cytosolic LPS receptor caspase-11 (or caspase-4 and -5 in humans) [44, 45, 47]. LPS binding to the N-terminal caspase activation and recruitment domain (CARD) of caspase-11 promotes the oligomerization of this protein, and the activation of its intrinsic protease activity [47]. Active caspase-11 then cleaves its cytosolic substrate gasdermin D [48, 49], releasing its N-terminal domain to oligomerize into a ring-shaped pore that interacts with acidic lipids in the plasma membrane, such as PI(4,5)P2 [50, 51]. Formation of the gasdermin D pore in the plasma membrane disrupts the osmotic balance in the cell and ultimately causes swelling and disruption of the plasma membrane (i.e. pyroptosis).
While the process by which LPS promotes caspase-11 dependent pyroptosis is becoming more clear, the means by which LPS accesses the cytosol remains obscure. During bacterial infections, LPS accesses the cytosol through membrane damage inflicted in the host cells by virulence-associated secretion systems or pore-forming toxins [45]. However, outer membrane vesicles derived from non-pathogenic bacteria can also induce caspase-11 dependent responses [52], and injection of free LPS into mice induces caspase-11 dependent responses [53]. Under these conditions, where no virulent bacteria are present, we have minimal understanding of how LPS gains access to the cytosol.
If one considers the spectrum of LPS interactions that occur in mammalian cells, a remarkable diversity of mechanisms of interaction exist, with distinct domains of distinct proteins operating in distinct locations inside and outside the cell. Indeed, of the known proteins important for LPS-induced cellular responses, none are required for all. However, one factor is commonly implicated in the regulation of LPS responses—the plasma membrane localized lipid PI(4,5)P2. This phosphoinositide regulates 1) the localization of the sorting adaptor TIRAP, and its ability to induce myddosome formation, 2) the endocytosis of CD14 and TLR4, and 3) the pore forming activity of gasdermin D. As such, PI(4,5)P2 acts to initiate TLR4 signaling and execute caspase-11 activities. It is unknown whether PI(4,5)P2 regulates other proteins that act in the LPS response pathways, but the studies described above provide a mandate to consider this possibility.
LPS detection by plants
The extensive analysis of the LPS response systems in humans and mice has provided an increasingly wide view of the natural means by which these systems operate. For example, the diversity of known LPS receptors has increased to the point that one cannot simply perform a bioinformatic search for TLR4 to identify LPS response systems in non-mammals. Or, in other words, the lack of TLR4 in any given organism does not necessary indicate a lack of LPS responsiveness. Direct experimental support for this idea derives from the study of plants, which like all multicellular organisms, face the threat of bacterial infections [54]. Recent work has identified an LPS response protein in plants that has no similarity to any mammalian receptor [55]. LPS responses in the model organism Arabidopsis thaliana include the rapid increase in the concentration of cytosolic calcium, the induction of antibacterial oxygen species and the MAP kinase-dependent expression of antimicrobial peptides [56, 57]. However, the molecular basis of LPS responses in Arabidopsis thaliana (or any other plant) has been unclear.
In 2015, a genetic screen to identify A. thaliana mutants that are defective for LPS-induced calcium fluxes revealed a protein called LipoOligosaccharide-specific Reduced Elicitation (LORE) [55]. LORE is a member of a plant-specific group of transmembrane proteins called bulb-type lectin S-domain-1 kinases (SD-RLKs). Loss-of-function LORE mutants are unable to flux calcium in response to LPS, and are unable to restrict the replication of various bacterial pathogens [55]. Interestingly, LORE transgenes are sufficient to confer LPS-induced calcium fluxes to tobacco, which do not naturally encode a LORE orthologue. Based on these data, it has been suggested that LORE is a receptor for LPS [55, 58].
While LORE homologues are only present in the plant family Brassicaceae [55, 58], a comparison of its functions reveals similarities to mammalian LPS receptors. For example, LORE and TLR4 are genetically required for the restriction of bacterial replication, and both receptors induce the MAP kinase dependent expression of antibacterial genes [15, 55, 59]. Similarities also exist in the features of LPS that promote LORE-mediated responses in plants and inflammatory responses in mammals. The lipid A region of LPS activates CD14-dependent endocytosis, TLR4-dependent inflammatory gene expression, and caspase-11 dependent pyroptosis [7]. In all of these instances, the hexa-acylated form of lipid A (which is typically found in E. coli) can stimulate the activities of these receptors. Interestingly, the lipid A region of LPS is also sufficient to activate LORE-dependent responses [55]. Thus, while the aforementioned LPS receptors contain different structures, and are present in different kingdoms of life, they all recognize the lipid A domain of LPS. Structure-function analysis has revealed one notable distinction between the LPS recognition mechanisms in plants and animals, in that hexa-acylated LPS is unable to induce LORE dependent responses [55]. Rather, the preferred structure of LPS that is detected by LORE appears to be penta-acylated lipid A. This finding is interesting for two reasons. First, penta-acylated LPS is unable to induce robust TLR4 or caspase-11 dependent responses in mice [7]. Prior to the identification of LORE, the common preference of mammalian receptors for hexa-acylated LPS suggested that this structural feature may be a fundamental driving force that directed the evolution of all LPS receptors. While this statement may be correct in the case of TLR4 and caspase-11, the studies of LORE suggest a more diverse mechanism of LPS detection may occur in nature, at the level of the structural features of the receptor and ligand. Recent studies of CD14-induced endocytosis revealed that penta-acylated LPS was also capable of activating this process [32]. Thus, even within mammals, different LPS receptors can interact with different regions of this microbial product. The second reason why the activation of LORE by penta-acylated LPS is notable is based on the fact that hexa-acylated LPS is often used as a tool to induce inflammatory responses in mammals. This practice has also been taken during searches to identify LPS responses in non-mammals [60]. A lack of response to the presence of hexa-acylated LPS often results in the conclusion that the organism in question does not respond to LPS [60]. However, the preference of LORE for penta-acylated LPS raises the possibility that organisms that have been previously characterized as unresponsive to LPS may actually recognize a variant of this bacterial product.
Despite being recognized as the first-identified LPS receptor in plants, direct interactions between LPS and LORE have yet to be identified. However, it is worth noting that several years passed after the identification of TLR4 as an LPS receptor before definitive evidence of interactions between these molecules was presented. Indeed, the most definitive evidence of LPS-TLR4 interactions was the crystal structure of this complex [14]. It is therefore possible that crystallographic evidence will be necessary to firmly establish direct LORE-LPS interactions. In the interim, the fact that LORE expression is necessary and sufficient to confer LPS responsiveness to diverse plants suggests a central role of this factor in the LPS response pathway in a specific kingdom of life.
LPS responses in primitive organisms
Based on the presence of LPS receptors across the multicellular kingdoms of life, it would seem reasonable that LPS receptors would exist in all organisms in those kingdoms. However, experimental support for this suggestion is limited, as most of our knowledge of LPS responses derives from studies of a select set of organisms. Advances in genome sequencing have suggested that what has been learned from the study of mice and humans likely applies to all land mammals. For example, genes encoding LBP, CD14, MD-2 and TLR4 can be found in the genomes of numerous and diverse animal representatives of the terrestrial family tree [61]. The restriction of LORE and TLRs to distinct kingdoms of life suggests parallel evolutionary processes occurred after these kingdoms separated. However, genomics, combined with experimental evidence, suggests that we have not identified all LPS receptors in existence. Studies from a primitive eukaryote, the slime mold Dyctiostelium discoideum, illustrate this point. D. discoideum is an unusual organism, in that it spends part of its life in a single amoeboid state, and part as a multicellular organism [62]. Remarkably, this amoeba can respond to LPS by the induction of autophagy and the production of reactive oxygen species [63, 64]. These responses, which also occur in mammalian cells, are important for D. discoideum to survive bacterial infections. Thus, analogous cell-autonomous antibacterial responses are present in D. discoideum and mammalian cells. However, while these responses are mediated by TLRs in mammals, no TLRs are present in the genome of D. discoideum. Interestingly, D. discoideum produce a protein called TirA, which is necessary for LPS-induced antibacterial responses in this organism [65, 66]. TirA may have some relation to mammalian TLRs, as it contains a TIR domain. However, this protein contains no transmembrane domain and is not predicted to operate as a receptor. As such, it remains unclear how D. discoideum elicits LPS responses. The absence of any known LPS response regulators present in the genomic sequence of D. discoideum argues that a novel LPS response pathway exists in this organism.
Concluding Remarks
In this Review, I highlighted several LPS response systems that exist in nature. The observation that distinct LPS receptors exists in plants and terrestrial animals suggests independent evolutionary events occurred during the development of these systems. Additionally, the presence of an LPS response in the primitive eukaryote D. discoideum, coupled to the absence of a recognizable signaling network in its genome, suggests a novel LPS sensory system exists. Because the known LPS receptors do not contain a common fold or domain, it is difficult to determine how many other receptors exist in any organism. Indeed, even in mammals, new LPS receptors are still be identified and characterized. For example, the integrin CD11b has long-been recognized as an LPS receptor, and recent work highlighted its importance in controlling TLR4 endocytosis in cells that express low levels of CD14 [37]. Additionally, the G-protein coupled receptor (GPCR) BAI1 has been recently identified as a receptor for LPS, which promotes phagocytosis and killing of gram negative bacteria [67, 68]. This receptor is unusual, in that it does not interact with the lipid A portion of LPS, but rather binds the core oligosaccharide of this bacterial product [68]. As discussed above, most other LPS receptors in mammals recognize the lipid A region. Thus, a wide range of structurally distinct LPS binding proteins exists in mammals, which includes proteins from the TLR, caspase, integrin and GPCR families, among others (Figure 1). While much has been learned of how LPS is detected by eukaryotes, many unknowns remain (see Outstanding Questions). For example, we have a limited understanding of how the mammalian LPS response pathways operate on their own, and in collaboration with other pathways. Cell-type specific and species-specific questions in this area are also numerous. For example, while TLR4 and caspase-11 are expressed by many cell types, much of our knowledge of these receptors derives from studies of phagocytes. Studies of B cells provide what is perhaps the most compelling evidence for cell type-specific LPS responses. Exposure of these cells to LPS prompts TLR4-dependent cell division, whereas TLR4 signaling does not promote mitosis in phagocytes [69]. Caspase-11 has also been reported to have functions in disease models where no apparent source of LPS is present, such as endoplasmic reticulum stress responses and in neuronal disorders [70–72]. Much additional work is required to better explain these cell type-specific LPS responses.
Outstanding Questions Box.
How are the actions of mammalian LPS receptors coordinated during bacterial encounters?
Does the plant LPS receptor LORE bind directly to LPS, and do other plant-specific LPS receptors exist?
Does the central role of PI(4,5)P2 in LPS detection in mammals extend to plants?
What is the receptor and signaling pathway that controls LPS responses in single celled eukaryotes?
Can our knowledge of LPS response systems be leveraged to treat human diseases, and diseases of other organisms that occupy our ecosystem.
In plants, future work may further expand the repertoire of LPS receptors in this kingdom, as well as define the mechanisms and pathways that regulate LORE-LPS interactions. Finally, based on the diversity of organisms that encode LPS receptors, it may be worth reconsidering the long-held believe that most marine organisms, in particular sharks and fish, are LPS unresponsive [73]. While the elephant shark genome lacks any gene homologous to CD14 and MD-2, and the TLR4 homologue is riddled with stop codons [6], it is possible that unique receptors for LPS exist in the aquatic world. Indeed, there is evidence that LPS responses exist in at least some marine animals, as various species of horseshoe crab have long-been recognized to encode the LPS-binding protein Factor C [74–77]. Factor C is a protease that is highly expressed by phagocytic haemocytes present in the hemolymph of the horseshoe crab, and binds to the lipid A component of LPS. LPS interactions with Factor C activate a proteolytic cascade that prompts antibacterial clotting responses. These findings are notable, when considering the interactions between LPS and caspase-4/5/11 in mammals, as both systems utilize proteases as LPS receptors to induce host defense. Thus, just as the study of innate immunity in the invertebrate Drosophila melanogaster led to the identification of the immune functions of the mammalian TLRs [78], the study of LPS-induced clotting in the invertebrate horseshoe crab provided precedent for the idea that proteases can act as LPS receptors in mammals. It is therefore possible that other conserved or unique LPS response systems may be revealed by diversifying the LPS-host interactions that we study. Addressing these possibilities likely awaits the development of additional experimental animal models, which is a daunting (but worthy) challenge to overcome.
Trends Box.
Receptors for bacterial LPS are diverse, with representatives found in various families of proteins.
Different LPS receptors exist in the plant and animal kingdoms.
Distinct structures within LPS are detected by plant and animal receptors.
Increasing evidence suggests that unknown LPS response systems exist.
Acknowledgments
I would like to thank members of the Kagan lab for helpful discussions. This work was supported by NIH grants AI093589, AI116550 and P30 DK34854 to J.C.K. and an unrestricted gift from Mead Johnson & Company. J.C.K. holds an Investigators in the Pathogenesis of Infectious Disease Award from the Burroughs Wellcome Fund.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Rappuoli R, et al. Vaccines, new opportunities for a new society. Proc Natl Acad Sci U S A. 2014;111(34):12288–93. doi: 10.1073/pnas.1402981111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Crimmins EM. Lifespan and Healthspan: Past, Present, and Promise. Gerontologist. 2015;55(6):901–11. doi: 10.1093/geront/gnv130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wiethoelter AK, et al. Global trends in infectious diseases at the wildlife-livestock interface. Proc Natl Acad Sci U S A. 2015;112(31):9662–7. doi: 10.1073/pnas.1422741112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wielinga PR, Schlundt J. Food Safety: at the center of a One Health approach for combating zoonoses. Curr Top Microbiol Immunol. 2013;366:3–17. doi: 10.1007/82_2012_238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Eichinger L, et al. The genome of the social amoeba Dictyostelium discoideum. Nature. 2005;435(7038):43–57. doi: 10.1038/nature03481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Venkatesh B, et al. Elephant shark genome provides unique insights into gnathostome evolution. Nature. 2014;505(7482):174–9. doi: 10.1038/nature12826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tan Y, Kagan JC. A cross-disciplinary perspective on the innate immune responses to bacterial lipopolysaccharide. Mol Cell. 2014;54(2):212–23. doi: 10.1016/j.molcel.2014.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.O’Neill LA. Therapeutic targeting of Toll-like receptors for inflammatory and infectious diseases. Curr Opin Pharmacol. 2003;3(4):396–403. doi: 10.1016/s1471-4892(03)00080-8. [DOI] [PubMed] [Google Scholar]
- 9.Tobias PS, et al. Isolation of a lipopolysaccharide-binding acute phase reactant from rabbit serum. J Exp Med. 1986;164(3):777–93. doi: 10.1084/jem.164.3.777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gioannini TL, et al. Isolation of an endotoxin-MD-2 complex that produces Toll-like receptor 4-dependent cell activation at picomolar concentrations. Proc Natl Acad Sci U S A. 2004;101(12):4186–91. doi: 10.1073/pnas.0306906101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wright SD, et al. CD14, a receptor for complexes of lipopolysaccharide (LPS) and LPS binding protein. Science. 1990;249(4975):1431–3. doi: 10.1126/science.1698311. [DOI] [PubMed] [Google Scholar]
- 12.Ulevitch RJ, Tobias PS. Recognition of endotoxin by cells leading to transmembrane signaling. Curr Opin Immunol. 1994;6(1):125–30. doi: 10.1016/0952-7915(94)90043-4. [DOI] [PubMed] [Google Scholar]
- 13.Akashi S, et al. Lipopolysaccharide interaction with cell surface Toll-like receptor 4-MD-2: higher affinity than that with MD-2 or CD14. J Exp Med. 2003;198(7):1035–42. doi: 10.1084/jem.20031076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Park BS, et al. The structural basis of lipopolysaccharide recognition by the TLR4-MD-2 complex. Nature. 2009;458(7242):1191–5. doi: 10.1038/nature07830. [DOI] [PubMed] [Google Scholar]
- 15.Poltorak A, et al. Defective LPS signaling in C3H/HeJ and C57BL/10ScCr mice: mutations in Tlr4 gene. Science. 1998;282(5396):2085–8. doi: 10.1126/science.282.5396.2085. [DOI] [PubMed] [Google Scholar]
- 16.Triantafilou M, et al. Mediators of innate immune recognition of bacteria concentrate in lipid rafts and facilitate lipopolysaccharide-induced cell activation. J Cell Sci. 2002;115(Pt 12):2603–11. doi: 10.1242/jcs.115.12.2603. [DOI] [PubMed] [Google Scholar]
- 17.Fitzgerald KA, et al. Mal (MyD88-adapter-like) is required for Toll-like receptor-4 signal transduction. Nature. 2001;413(6851):78–83. doi: 10.1038/35092578. [DOI] [PubMed] [Google Scholar]
- 18.Horng T, et al. TIRAP: an adapter molecule in the Toll signaling pathway. Nat Immunol. 2001;2(9):835–41. doi: 10.1038/ni0901-835. [DOI] [PubMed] [Google Scholar]
- 19.Bonham KS, et al. A promiscuous lipid-binding protein diversifies the subcellular sites of toll-like receptor signal transduction. Cell. 2014;156(4):705–16. doi: 10.1016/j.cell.2014.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kagan JC, Medzhitov R. Phosphoinositide-mediated adaptor recruitment controls Toll-like receptor signaling. Cell. 2006;125(5):943–55. doi: 10.1016/j.cell.2006.03.047. [DOI] [PubMed] [Google Scholar]
- 21.Kagan JC, et al. SMOCs: supramolecular organizing centres that control innate immunity. Nat Rev Immunol. 2014 doi: 10.1038/nri3757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gay NJ, et al. What the Myddosome structure tells us about the initiation of innate immunity. Trends Immunol. 2011;32(3):104–9. doi: 10.1016/j.it.2010.12.005. [DOI] [PubMed] [Google Scholar]
- 23.Lin SC, et al. Helical assembly in the MyD88-IRAK4-IRAK2 complex in TLR/IL-1R signalling. Nature. 2010;465(7300):885–90. doi: 10.1038/nature09121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Motshwene PG, et al. An oligomeric signaling platform formed by the Toll-like receptor signal transducers MyD88 and IRAK-4. J Biol Chem. 2009;284(37):25404–11. doi: 10.1074/jbc.M109.022392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bryant CE, et al. Toll-like receptor signalling through macromolecular protein complexes. Mol Immunol. 2014 doi: 10.1016/j.molimm.2014.06.033. [DOI] [PubMed] [Google Scholar]
- 26.Zanoni I, et al. CD14 controls the LPS-induced endocytosis of Toll-like receptor 4. Cell. 2011;147(4):868–80. doi: 10.1016/j.cell.2011.09.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yamamoto M, et al. Role of adaptor TRIF in the MyD88-independent toll-like receptor signaling pathway. Science. 2003;301(5633):640–3. doi: 10.1126/science.1087262. [DOI] [PubMed] [Google Scholar]
- 28.Gutierrez MG, et al. Autophagy is a defense mechanism inhibiting BCG and Mycobacterium tuberculosis survival in infected macrophages. Cell. 2004;119(6):753–66. doi: 10.1016/j.cell.2004.11.038. [DOI] [PubMed] [Google Scholar]
- 29.Rathinam VA, et al. TRIF licenses caspase-11-dependent NLRP3 inflammasome activation by gram-negative bacteria. Cell. 2012;150(3):606–19. doi: 10.1016/j.cell.2012.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rosadini CV, et al. A Single Bacterial Immune Evasion Strategy Dismantles Both MyD88 and TRIF Signaling Pathways Downstream of TLR4. Cell Host Microbe. 2015;18(6):682–93. doi: 10.1016/j.chom.2015.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zanoni I, et al. IL-15 cis presentation is required for optimal NK cell activation in lipopolysaccharide-mediated inflammatory conditions. Cell Rep. 2013;4(6):1235–49. doi: 10.1016/j.celrep.2013.08.021. [DOI] [PubMed] [Google Scholar]
- 32.Tan Y, et al. Mechanisms of Toll-like Receptor 4 Endocytosis Reveal a Common Immune-Evasion Strategy Used by Pathogenic and Commensal Bacteria. Immunity. 2015;43(5):909–22. doi: 10.1016/j.immuni.2015.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zanoni I, et al. CD14 regulates the dendritic cell life cycle after LPS exposure through NFAT activation. Nature. 2009;460(7252):264–8. doi: 10.1038/nature08118. [DOI] [PubMed] [Google Scholar]
- 34.Chiang CY, et al. Phospholipase Cgamma-2 and intracellular calcium are required for lipopolysaccharide-induced Toll-like receptor 4 (TLR4) endocytosis and interferon regulatory factor 3 (IRF3) activation. J Biol Chem. 2012;287(6):3704–9. doi: 10.1074/jbc.C111.328559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kagan JC, et al. TRAM couples endocytosis of Toll-like receptor 4 to the induction of interferon-beta. Nat Immunol. 2008;9(4):361–8. doi: 10.1038/ni1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lin YC, et al. The tyrosine kinase Syk differentially regulates Toll-like receptor signaling downstream of the adaptor molecules TRAF6 and TRAF3. Sci Signal. 2013;6(289):ra71. doi: 10.1126/scisignal.2003973. [DOI] [PubMed] [Google Scholar]
- 37.Ling GS, et al. Integrin CD11b positively regulates TLR4-induced signalling pathways in dendritic cells but not in macrophages. Nat Commun. 2014;5:3039. doi: 10.1038/ncomms4039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Plociennikowska A, et al. Contribution of CD14 and TLR4 to changes of the PI(4,5)P2 level in LPS-stimulated cells. J Leukoc Biol. 2016 doi: 10.1189/jlb.2VMA1215-577R. [DOI] [PubMed] [Google Scholar]
- 39.Plociennikowska A, et al. LPS-induced clustering of CD14 triggers generation of PI(4,5)P2. J Cell Sci. 2015;128(22):4096–111. doi: 10.1242/jcs.173104. [DOI] [PubMed] [Google Scholar]
- 40.Botelho RJ, et al. Localized biphasic changes in phosphatidylinositol-4,5-bisphosphate at sites of phagocytosis. J Cell Biol. 2000;151(7):1353–68. doi: 10.1083/jcb.151.7.1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Husebye H, et al. The Rab11a GTPase controls Toll-like receptor 4-induced activation of interferon regulatory factor-3 on phagosomes. Immunity. 2010;33(4):583–96. doi: 10.1016/j.immuni.2010.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nair-Gupta P, et al. TLR signals induce phagosomal MHC-I delivery from the endosomal recycling compartment to allow cross-presentation. Cell. 2014;158(3):506–21. doi: 10.1016/j.cell.2014.04.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Husebye H, et al. Endocytic pathways regulate Toll-like receptor 4 signaling and link innate and adaptive immunity. Embo J. 2006;25(4):683–92. doi: 10.1038/sj.emboj.7600991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kayagaki N, et al. Noncanonical inflammasome activation by intracellular LPS independent of TLR4. Science. 2013;341(6151):1246–9. doi: 10.1126/science.1240248. [DOI] [PubMed] [Google Scholar]
- 45.Hagar JA, et al. Cytoplasmic LPS activates caspase-11: implications in TLR4-independent endotoxic shock. Science. 2013;341(6151):1250–3. doi: 10.1126/science.1240988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jorgensen I, et al. Programmed cell death as a defence against infection. Nat Rev Immunol. 2017;17(3):151–164. doi: 10.1038/nri.2016.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shi J, et al. Inflammatory caspases are innate immune receptors for intracellular LPS. Nature. 2014;514(7521):187–92. doi: 10.1038/nature13683. [DOI] [PubMed] [Google Scholar]
- 48.Kayagaki N, et al. Caspase-11 cleaves gasdermin D for non-canonical inflammasome signalling. Nature. 2015;526(7575):666–71. doi: 10.1038/nature15541. [DOI] [PubMed] [Google Scholar]
- 49.Shi J, et al. Cleavage of GSDMD by inflammatory caspases determines pyroptotic cell death. Nature. 2015;526(7575):660–5. doi: 10.1038/nature15514. [DOI] [PubMed] [Google Scholar]
- 50.Ding J, et al. Pore-forming activity and structural autoinhibition of the gasdermin family. Nature. 2016;535(7610):111–6. doi: 10.1038/nature18590. [DOI] [PubMed] [Google Scholar]
- 51.Liu X, et al. Inflammasome-activated gasdermin D causes pyroptosis by forming membrane pores. Nature. 2016;535(7610):153–8. doi: 10.1038/nature18629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vanaja SK, et al. Bacterial Outer Membrane Vesicles Mediate Cytosolic Localization of LPS and Caspase-11 Activation. Cell. 2016;165(5):1106–19. doi: 10.1016/j.cell.2016.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang S, et al. Murine caspase-11, an ICE-interacting protease, is essential for the activation of ICE. Cell. 1998;92(4):501–9. doi: 10.1016/s0092-8674(00)80943-5. [DOI] [PubMed] [Google Scholar]
- 54.Staskawicz BJ, et al. Common and contrasting themes of plant and animal diseases. Science. 2001;292(5525):2285–9. doi: 10.1126/science.1062013. [DOI] [PubMed] [Google Scholar]
- 55.Ranf S, et al. A lectin S-domain receptor kinase mediates lipopolysaccharide sensing in Arabidopsis thaliana. Nat Immunol. 2015;16(4):426–33. doi: 10.1038/ni.3124. [DOI] [PubMed] [Google Scholar]
- 56.Newman MA, et al. MAMP (microbe-associated molecular pattern) triggered immunity in plants. Front Plant Sci. 2013;4:139. doi: 10.3389/fpls.2013.00139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zeidler D, et al. Innate immunity in Arabidopsis thaliana: lipopolysaccharides activate nitric oxide synthase (NOS) and induce defense genes. Proc Natl Acad Sci U S A. 2004;101(44):15811–6. doi: 10.1073/pnas.0404536101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zipfel C. A new receptor for LPS. Nat Immunol. 2015;16(4):340–1. doi: 10.1038/ni.3127. [DOI] [PubMed] [Google Scholar]
- 59.Takeuchi O, et al. Differential roles of TLR2 and TLR4 in recognition of gram-negative and gram-positive bacterial cell wall components. Immunity. 1999;11(4):443–51. doi: 10.1016/s1074-7613(00)80119-3. [DOI] [PubMed] [Google Scholar]
- 60.Berczi I, et al. Comparative studies on the toxicity of Escherichia coli lipopolysaccharide endotoxin in various animal species. Can J Microbiol. 1966;12(5):1070–1. doi: 10.1139/m66-143. [DOI] [PubMed] [Google Scholar]
- 61.Neyen C, Lemaitre B. Sensing Gram-negative bacteria: a phylogenetic perspective. Curr Opin Immunol. 2016;38:8–17. doi: 10.1016/j.coi.2015.10.007. [DOI] [PubMed] [Google Scholar]
- 62.Schaap P. Evolution of developmental signalling in Dictyostelid social amoebas. Curr Opin Genet Dev. 2016;39:29–34. doi: 10.1016/j.gde.2016.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Pflaum K, et al. Lipopolysaccharide induction of autophagy is associated with enhanced bactericidal activity in Dictyostelium discoideum. Biochem Biophys Res Commun. 2012;422(3):417–22. doi: 10.1016/j.bbrc.2012.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Walk A, et al. Lipopolysaccharide enhances bactericidal activity in Dictyostelium discoideum cells. Dev Comp Immunol. 2011;35(8):850–6. doi: 10.1016/j.dci.2011.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chen G, et al. Immune-like phagocyte activity in the social amoeba. Science. 2007;317(5838):678–81. doi: 10.1126/science.1143991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhang X, et al. Social amoebae trap and kill bacteria by casting DNA nets. Nat Commun. 2016;7:10938. doi: 10.1038/ncomms10938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Billings EA, et al. The adhesion GPCR BAI1 mediates macrophage ROS production and microbicidal activity against Gram-negative bacteria. Sci Signal. 2016;9(413):ra14. doi: 10.1126/scisignal.aac6250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Das S, et al. Brain angiogenesis inhibitor 1 (BAI1) is a pattern recognition receptor that mediates macrophage binding and engulfment of Gram-negative bacteria. Proc Natl Acad Sci U S A. 2011;108(5):2136–41. doi: 10.1073/pnas.1014775108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hoshino K, et al. Cutting edge: Toll-like receptor 4 (TLR4)-deficient mice are hyporesponsive to lipopolysaccharide: evidence for TLR4 as the Lps gene product. J Immunol. 1999;162(7):3749–52. [PubMed] [Google Scholar]
- 70.Shibata M, et al. Caspases determine the vulnerability of oligodendrocytes in the ischemic brain. J Clin Invest. 2000;106(5):643–53. doi: 10.1172/JCI10203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Furuya T, et al. Caspase-11 mediates inflammatory dopaminergic cell death in the 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine mouse model of Parkinson’s disease. J Neurosci. 2004;24(8):1865–72. doi: 10.1523/JNEUROSCI.3309-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Fradejas N, et al. Caspase-11 mediates ischemia-induced astrocyte death: involvement of endoplasmic reticulum stress and C/EBP homologous protein. J Neurosci Res. 2010;88(5):1094–105. doi: 10.1002/jnr.22280. [DOI] [PubMed] [Google Scholar]
- 73.Iliev DB, et al. Endotoxin recognition: in fish or not in fish? FEBS Lett. 2005;579(29):6519–28. doi: 10.1016/j.febslet.2005.10.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ariki S, et al. Factor C acts as a lipopolysaccharide-responsive C3 convertase in horseshoe crab complement activation. J Immunol. 2008;181(11):7994–8001. doi: 10.4049/jimmunol.181.11.7994. [DOI] [PubMed] [Google Scholar]
- 75.Kawabata S, Muta T. Sadaaki Iwanaga: Discovery of the lipopolysaccharide- and beta-1,3-D-glucan-mediated proteolytic cascade and unique proteins in invertebrate immunity. J Biochem. 2010;147(5):611–8. doi: 10.1093/jb/mvq026. [DOI] [PubMed] [Google Scholar]
- 76.Koshiba T, et al. A structural perspective on the interaction between lipopolysaccharide and factor C, a receptor involved in recognition of Gram-negative bacteria. J Biol Chem. 2007;282(6):3962–7. doi: 10.1074/jbc.M609198200. [DOI] [PubMed] [Google Scholar]
- 77.Nakamura T, et al. Interaction between lipopolysaccharide and intracellular serine protease zymogen, factor C, from horseshoe crab (Tachypleus tridentatus) hemocytes. J Biochem. 1988;103(2):370–4. doi: 10.1093/oxfordjournals.jbchem.a122276. [DOI] [PubMed] [Google Scholar]
- 78.Hoffmann JA. The immune response of Drosophila. Nature. 2003;426(6962):33–8. doi: 10.1038/nature02021. [DOI] [PubMed] [Google Scholar]