Abstract
Metal-dependent lysine deacetylases (KDACs) are involved in regulation of numerous biological and disease processes through control of post-translational acetylation. Characterization of KDAC activity and substrate identification is complicated by inconsistent activity of prepared enzyme and a range of multi-step purifications. We describe a simplified protocol based on two-step affinity chromatography. The purification method is appropriate for use regardless of expression host, and we demonstrate purification of several representative members of the KDAC family as well as a selection of mutated variants. The purified proteins are highly active and consistent across preparations.
Keywords: hdac, histone deacetylase, lysine deacetylase
1. Introduction
Metal-dependent lysine deacetylases (KDACs, also known as histone deacetylases, EC 3.5.1.98) are enzymes that reverse the post-translational modification of lysine acetylation, by catalyzing the hydrolysis of ε-N-acetyllysine residues in proteins via a conserved mechanism [1–3]. Thousands of acetylated protein sequences have been identified in mammalian cells, and thus are subject to deacetylation by KDACs [4–9]. Properly regulated acetylation and deacetylation have been linked to many biological processes, while aberrant KDAC activity has also been linked to numerous diseases [10,11]. Based on the therapeutic potential of regulating KDACs in vivo, research efforts are focused on identifying molecules that inhibit or activate these enzymes [11–14], as well as identifying substrates of specific KDACs [15–19]. KDACs are commonly grouped into several classes, with class I, II, and IV KDACs being metal-dependent, and class III (sirtuins) being NAD-dependent. Metal-dependent KDACs require a divalent metal ion in the active site. While KDACs can utilize different metal ions, activity levels are partially dependent upon which metal is present. In addition, KDAC8 is inhibited by excess zinc which binds to a second site on the enzyme [3]. Over 1000 inhibitors for KDACs have been identified, and several are in clinical trials or have already been approved for therapeutic use [11,12]. Despite high interest in understanding KDAC function, relatively few substrates (i.e. acetylated proteins) have been definitively assigned to a particular KDAC. To accomplish this task, purified KDACs are required for in vitro activity assays.
Protocols for recombinant expression and purification of KDAC8 from E. coli have previously been reported, but are time-consuming and labor-intensive. Expression is generally done in BL21 E. coli or a BL21 derivative strain overnight at reduced temperature. Most of the purification protocols rely at least partially on immobilized metal affinity chromatography (IMAC) of TEV protease-cleavable His6-tagged KDAC8. This initial purification step is usually followed by removal of the tag and a secondary purification step, often involving anion exchange and/or size exclusion chromatography. These secondary purification steps result in a dilute enzyme prep, which must then be concentrated [3,20,21]. One frequently cited protocol then requires an additional step to chelate metals, resulting in a metal-free preparation of apo-KDAC8. Enzyme stored in this manner must be metalated before being used in experiments allowing control of which metal ion resides in the active site to ensure that activity between different preps are comparable [3]. Following purification, most protocols require storage at −80 °C in small aliquots to avoid freeze/thawing [3,21]. There is even greater variability when considering protocols for purifying the other KDACs, including varying the expression system, tags, and purification methods [18,22].
Critically, KDACs purified using different methods demonstrate differences in activity with the same substrate. A previous comparison of enzyme activity of KDAC8 purified using different metal affinity chromatography protocols resulted in a four-fold difference in activity [23]. In another report, the catalytic efficiency of KDAC8 purified from insect cells was reported to be 3–5 fold higher than the same enzyme purified from E. coli [15], although it is unclear whether this is due to a difference in the enzyme resulting from the two different cell types or an artifact of the different purification protocols. Nevertheless, these differences make it impossible to compare KDAC activity against different substrates across reports, as activity differences could be attributed to either the difference in substrate or the enzyme preparation. Here, we present a robust protocol for expression and purification of metal-dependent KDACs. It is applicable across KDACs and expression systems, and is simpler than previously reported protocols. Most importantly, KDAC activity is highly reproducible, even between preparations from different expression systems.
2. Materials and Methods
2.1 KDAC expression in E. coli
Expression of KDACs was modified from a previously reported procedure [24]. pJExpress401 vectors (DNA 2.0) containing codon-optimized genes were obtained to express human KDAC8 (pJExpress-KDAC8), KDAC4 (aa648-1057; pJExpress-KDAC4), and KDAC7 (aa521-942; pJExpress-KDAC7) fused to a tobacco-etch virus (TEV) protease cleavage site and His6 tag. pJExpress-KDAC4 and pJExpress-KDAC8 were subjected to site-directed mutagenesis to introduce the H976Y mutation in KDAC4 (KDAC4HY) and the H143A mutation in KDAC8 (KDAC8HA). For expression in E. coli, plasmids were introduced into BL21(DE3) cells for expression. Cells were grown in LB overnight at 37 °C with shaking at 250 rpm, then diluted 1:100 into 2X YT broth and grown under the same conditions. When cells reached an OD600=0.8–1.0, ZnCl2 was added to a final concentration of 50 μM and expression induced with 1 mM IPTG, followed by an additional 3.5 hr of growth at 37 °C. After induction, cells were harvested by centrifugation at 3500 rpm for 20 min at 4 °C. Cells pellets were stored at −20 °C until lysis.
2.2 KDAC expression in insect cells
KDAC6 (a gift from Eric Verdin, Addgene plasmid #13823)[25], KDAC7 (aa521-942), KDAC8, and KDAC8HA were cloned into pFastbac1 (Life Technologies) From pJExpress vectors with the TEV protease cleavage site and His6 tag. Constructs were transformed into DH10Bac E. coli cells to produce bacmids containing KDAC8 [26]. Bacmids were purified and transfected into Sf9 cells using Cellfectin II (Life Technologies) as described elsewhere [27]. Baculovirus from these transfections was amplified in Sf9 cells, then used to infect High Five insect cells in suspension in Express Five SFM (Gibco). At 72 hours post-infection, cells were pelleted and frozen at −20 °C until lysis.
2.3 KDAC purification
Similar to a previously reported protocol [24], cells were resuspended in either E. coli lysis buffer (30 mM MOPS pH 8.0, 150 mM KCl, 5% glycerol, 5 mM imidazole, 2 mM MgCl2, 0.5 mM CaCl2, 0.5 mg mL−1 egg white lysozyme, 2 U mL−1 DNaseI [New England Biolabs], 1X HALT protease inhibitor [Thermo Scientific]) and incubated with rocking for 30 min on ice or insect cell lysis buffer (30 mM MOPS pH 8.0, 150 mM KCl, 5% glycerol, 5 mM imidazole, 2 mM MgCl2, 1X HALT protease inhibitor [Thermo Scientific]). Typically 10 mL of lysis buffer was used per 1 L E. coli culture or 250 mL High Five insect cell culture harvested. Cell suspensions were sonicated five times at 50% amplitude for 10 s (Fisher Scientific Sonic Dismembrator Model 120, 1/8″ probe), followed by 30 s on ice. Lysates were clarified by centrifugation at 27,000 xg for 20 min at 4 °C.
Clarified lysate was added to TALON cobalt resin (Clontech) equilibrated with column buffer (30 mM MOPS pH 8.0, 150 mM KCl, 5% glycerol, 5 mM imidazole) and incubated on ice for 15 min with rocking (resin bed volume of 1 mL per 10 mL lysis buffer). Resin was pelleted by centrifugation at 700 xg for 5 min and washed twice with 10 bed volumes of column buffer each time. After final centrifugation, resin was transferred to column housing and washed with an additional 10 bed volumes of column buffer. KDACs were eluted (30 mM MOPS pH 8.0, 150 mM KCl, 5% glycerol, 150 mM imidazole) and collected in fractions. KDAC6 was dialyzed into storage buffer (30 mM MOPS pH 8.0, 150 mM KCl, 25% glycerol) overnight at 4 °C with one buffer change. Following dialysis, tris (2-carboxyethyl)phosphine (TCEP) was added to a final concentration of 1 mM. For other KDACs, TEV protease, expressed and purified as described previously [24], was added (1:25) to fractions containing protein, and the mixture was dialyzed in TEV cleavage buffer (30 mM MOPS pH 8.0, 150 mM KCl, 5% glycerol, 1 mM 2-mercaptoethanol, 0.3 mM EDTA pH 7.0) overnight at 4 °C with one buffer change. This was followed by dialysis into buffer containing 30 mM MOPS pH 8.0, 150 mM KCl, and 5% glycerol overnight at 4 °C with one buffer change. Protein was recovered from dialysis and flowed over TALON resin equilibrated with the final dialysis buffer for secondary purification. Purified KDAC (flow-through) was collected. Glycerol and TCEP were added to final concentrations of 25% and 1 mM, respectively.
Where noted, Ni Superflow resin (Clontech) was used for nickel-based purification instead of cobalt. For experiments using zinc-containing resin, TALON resin was stripped with five bed volumes of 0.2M EDTA pH 8.0 and washed with five bed volumes of dH2O. Then it was regenerated by flowing ten bed volumes of 50 mM ZnCl2 over the resin, followed by seven bed volumes of dH2O, three bed volumes of 300 mM NaCl, and seven bed volumes of dH2O.
2.4 SDS-PAGE analysis
Purified KDACs were loaded onto SDS-PAGE gels made from NEXT gel polyacrylamide solution (VWR Amresco) and run at 150 V for 90–120 min. Protein was visualized by staining with Gelcode blue stain reagent (Thermo Fisher).
2.5 Activity assays
{K-ac}-AMC was commercially obtained (Fluor-de-Lys; Enzo Life Sciences). All other peptide substrates were commercial custom peptide syntheses purified to > 95% (Genscript). Fluorescamine assays were performed in assay buffer (30 mM potassium phosphate pH 7.6, 100 mM KCl, 5% glycerol) as described previously [24]. Deacetylation reactions using the Fluor-de-Lys substrate were conducted in the same buffer as above and the assay was conducted as previously described [24]. 100 μM substrate was incubated with either KDAC8 (200 nM), KDAC6-His6 (50 nM), or KDAC6-GST (20 nM) for 1 hour at 25°C. When noted, excess Co2+ (ICP-MS standard quality; Ultra Scientific) was pre-incubated with enzyme prior to the addition of substrate. Commercially obtained KDAC8 was purchased from BPS Bioscience and Novus Biologicals, and used where indicated. KDAC6-GST was purchased from BPS Bioscience.
2.6 Circular Dichroism spectroscopy (CD)
Purified KDACs in storage buffer were diluted to 500 nM in assay buffer in a 2 mm cuvette and subjected to CD scans from 200–245 nm at 25 °C, with a data pitch of 1.0 nm, scan rate of 5 nm min−1, an integration time of 16 s, and 2 accumulated scans. A scan of buffer was performed under the same conditions in the same cuvette and the buffer signal at each wavelength was subtracted from the KDAC sample. The resulting values for wild-type KDACs and their variants were normalized based on the most negative value for each set (221 nm for KDAC8 and variants or 207 nm for KDAC4 and variants).
2.7 Detection of His6 tag by MALDI-TOF mass spectrometry
Approximately 2 μg of His6-tagged KDAC was incubated with 0.1 μg TEV protease in TEV cleavage buffer overnight at 4 °C. Reactions were diluted 1:10 in TA85 (85% acetonitrile, 15% water, 0.1% trifluoroacetic acid). 0.5 μl was spotted onto a MTP Anchorchip target plate (Bruker Daltonics) and allowed to dry. 0.5 μl matrix (1.4 mg/ml α-cyano-4-hydroxycinnamic acid in TA85) was spotted on top of each sample. Samples were analyzed by matrix assisted laser desorption/ionization (MALDI) time-of-flight (TOF) mass spectrometry (MS) on an Autoflex Speed MALDI TOF/TOF (Bruker Daltonics) in positive reflector mode and masses were assigned to peaks using flexanalysis software (Bruker Daltonics).
3. Results and Discussion
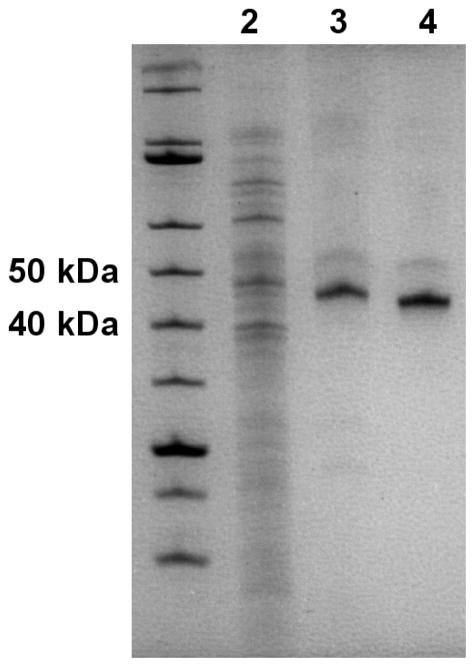
Several protocols have been previously published for the expression and purification of KDACs; however, they vary widely, are often considerably laborious, and the reported activity for the purified enzymes varies substantially between reports. We have developed a robust and broadly applicable expression and two-step affinity purification method, which relies on a TEV protease-cleavable His6 tag. Figure 1 shows a representative progression of this process. KDACs were purified from cell lysate by metal affinity chromatography utilizing a C-terminal His6 tag and cobalt resin. The pooled protein-rich fractions typically resulted in 2–10 mg of total protein with the KDAC as the major protein (Figure 1). Following the initial purification step, the tag was removed from the protein by incubating with TEV protease, which recognizes a TEV protease cleavage site that is positioned between the KDAC and the His6 tag. This mixture was subjected to a second purification step, again using cobalt-containing resin. In this step, the flow-through contained the KDAC of interest, while His6-tagged TEV protease and other contaminants from the lysate were retained by the column. Recovered total protein was typically 25–50% of the total sample loaded onto the second column. Specific activity of the KDACs was enhanced 1.2-fold to 2-fold compared to the His6-tagged proteins, indicating that the His6 tag has little to no effect on the enzymatic activity. While expression in E. coli was suitable for some KDACs, a subset of the wild-type enzymes (KDAC6 and KDAC7) and several variants did not express well in bacteria. Several parameters were varied to try to increase yield of these enzymes, including expression strain, growth in minimal media, lower expression temperature, and longer expression time; however, none of these adjustments to the protocol improved yields (data not shown). Instead, we found that switching to an insect cell expression system allowed us to express these proteins in usable quantities.
Figure 1. Representative purification progression.
Approximately 10 μg total E. coli lysate containing recombinant KDAC8-His6 (lane 2), 1.0 μg total protein following the first column (lane 3), and 1.0 μg total protein after the second column (lane 4) are shown on a 12.5% SDS-PAGE gel. KDAC8-His6 is apparent at 43.5 kDa in lane 3, and KDAC8 at 42.5 kDa in lane 4. KDACs were not obviously apparent in the cell lysate from E. coli or Sf9 cells.
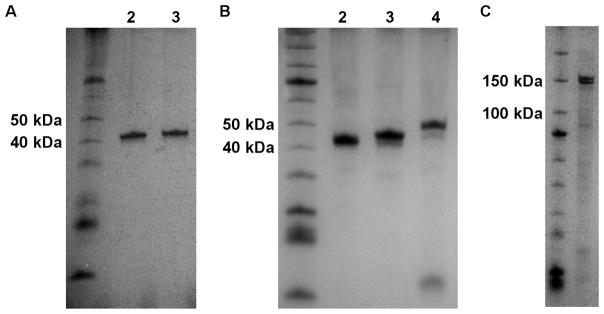
Ideally, we wanted to develop a single purification strategy for lysates obtained from multiple expression systems. KDAC8 expressed in both E. coli and insect cells, allowing a direct comparison of yield and purity. Indeed, we were able to purify KDAC8 from both lysates using the same purification method, resulting in 0.5–1 mg purified KDAC8 per liter of E. coli and approximately 5 mg from 250 mL insect cell culture. We obtained similar purity between KDAC8 purified from E. coli and insect cell lysates (Figure 2A). Furthermore, the purification method was successful for all KDACs tested (KDAC4, KDAC6, KDAC7, KDAC8, and several variants containing amino acid substitutions) and resulted in a purity ≥90% in most cases (Figure 2B and 2C). The one noted exception was KDAC6 which was recovered during the initial purification, but did not flow through the column during the secondary purification step. We hypothesized that the TEV protease cleavage site is not accessible to TEV protease in this particular protein, resulting in a failure to remove the His6 tag. Consistent with this hypothesis, we were clearly able to identify the cleaved His6 tag from KDAC8, but not KDAC6, using MALDI-TOF mass spectrometry following incubation of the KDAC with TEV protease (Figure S1).
Figure 2. Purified KDACs.
A. Approximately 0.5 μg KDAC8 (42.5 kDa) purified from either insect cells (lane 2) or E. coli (lane 3) was compared by gel electrophoresis on a 12.5% SDS-PAGE gel, indicating similar purity from both expression systems. B. Approximately 1.5 μg purified KDAC8 (lane 2), KDAC7 (lane 3, 46 kDa), and KDAC4 (Lane 4, 48 kDa) are shown on a 12.5% SDS-PAGE gel. C. Approximately 1 μg KDAC6-His6 (134 kDa) is shown on a 10% SDS-PAGE gel.
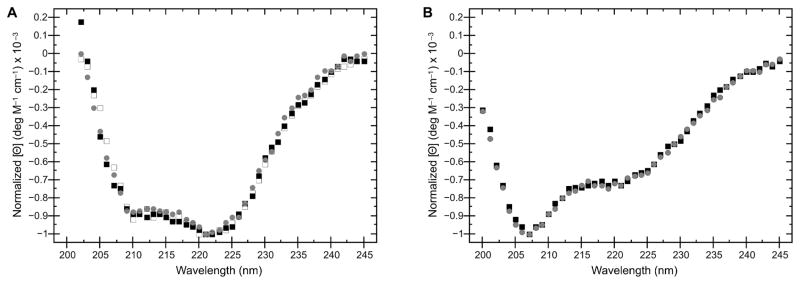
To determine whether we could successfully purify variants, we chose two previously characterized KDAC mutants, KDAC8HA and KDAC4HY. KDAC8HA has previously been reported as a loss-of-function variant, as the H143 residue is involved in catalysis [28]. Conversely, KDAC4HY is a gain-of-function mutation, and it is well-established that this variant demonstrates increased catalytic activity compared to wild-type KDAC4 [29,30]. Like their wild-type counterparts, we were able to successfully purify both variants (Figure S2). As previously reported, KDAC8HA demonstrated no measurable activity, while KDAC4HY showed activity with substrates for which KDAC4 was not active ([28–30]; data not shown). To determine whether our protocol resulted in purification of variants that were structurally similar to their wild-type counterparts, we performed CD spectroscopy on each protein. In both cases, there were no detectable differences in overall structure (Figure 3). This result implied that any differences in activity are attributable to previously suggested alterations in the reaction mechanism [30,31] and not simply a major structural change due to incorrect folding, which is especially important for variants that result in a loss of activity, such as KDAC8HA.
Figure 3. CD spectroscopy of KDAC variants.
A. Normalized CD spectra comparing wild-type KDAC8 purified from E. coli (black squares), as well as wild-type KDAC8 (grey circles) and KDAC8HA (white squares) purified from insect cells. B. Wild-type KDAC4 (black squares) compared to KDAC4HY (grey circles). Spectra indicate that there are no significant changes in secondary structure between wild-type KDACs and their respective variants, or due to the expression system used.
Metal affinity chromatography most commonly relies on nickel [32]; however, nickel in the active site of KDAC8 has been previously reported to result in reduced activity [3]. Resins containing cobalt are now available and avoid the risk of contaminating the enzyme with an inactive metal, as the cobalt-containing enzyme is active. The metal typically found in the active site of purified KDACs is zinc, and it is likely that zinc is a biologically active metal for some or all of the KDACs whereas cobalt is unlikely to be biologically relevant.[3,31] Therefore, we were interested in the possibility of using zinc-containing resin as this would presumably allow us to recover KDACs containing zinc in the active site. To compare purification using affinity for different metals, we attempted to purify KDAC8 using cobalt, nickel, and zinc resin. Although all three resins partially purified KDAC8 during the first purification step, KDAC8 was only present in the flow through of cobalt-containing resin (Figure S3). Thus, using cobalt-containing resin was critical to the success of our purification strategy. As previously reported, it is possible to elute the cleaved KDAC8 from a nickel resin using low amounts of imidazole [3]. However, collecting direct flow-through off a cobalt resin has the advantage that the protein is immediately ready for use and does not require additional cleanup steps, while avoiding the risk of introducing nickel into the enzyme active site.
We compared the activity of three preparations of KDAC8, two expressed in E. coli and one from insect cells, using a previously described fluorescence-based assay which relies on fluorescamine to react with deacetylated lysines on peptide substrates after incubation with KDACs [24]. There was no significant difference between preparations of KDAC8 with respect to activity against a previously identified peptide substrate, despite being expressed in different systems and purified independently at different times (Table 1). Importantly, we previously reported data using a different assay demonstrating that KDAC8 purified using our method was approximately ten-fold more active than a commercially obtained preparation of KDAC8 [23]. A second commercially obtained preparation of KDAC8 was significantly less pure and demonstrated no activity against the same substrate in our assay (Figure S4). There was no significant difference between the activity of KDAC6-His6 purified using our method and a commercially obtained GST-tagged preparation of KDAC6 with several peptide substrates (Table 2).
Table 1.
Comparison of purified KDAC8 activity against ac-FR{K-ac}RW-am
| KDAC8 prep | Activity (pmol min−1 μg−1) |
|---|---|
| E. coli 1a | 19 ± 4 |
| E. coli 2 | 21 ± 1 |
| Insect cells | 20 ± 1 |
Previously reported.[24]
Table 2.
Comparison of activity of purified KDAC6-His6 with commercially obtained KDAC6-GST with peptide substrates.
| Peptide sequence | KDAC6-His6 activity (pmol min−1 μg−1)a | KDAC6-GST activity (pmol min−1 μg−1) |
|---|---|---|
| ac-{K-ac}-am | 4.8 ± 1.1 | 8.9 ± 4.1 |
| ac-{K-ac}W-am | 33.2 ± 13.7 | 30.9 ± 7.6 |
| ac-RG{K-ac}-am | 9.7 ± 5.1 | 12.2 ± 2.7 |
| ac-RG{K-ac}W-am | 45.2 ± 13.5 | 40.1 ± 5.9 |
| ac-RH{K-ac}-{K-ac}-am | 11.4 ± 3.7 | 8.7 ± 2.5 |
| ac-RH{K-ac}-{K-ac}W-am | 39.5 ± 18.7 | 29.7 ± 3.6 |
Previously reported.[33]
Metal-dependent KDACs require a single divalent cation in the active site [3,31]. To determine whether our purified KDACs were saturated with metal at the active site, we titrated in cobalt and assayed the effect of the additional metal on activity. Adding cobalt up to a ratio of 100 mol Co/1 mol KDAC8 did not increase the activity of KDAC8 (Table S1). Based on this and the reproducibility between preps, we concluded that our purification and storage protocol reliably resulted in metal saturated KDACs. Unlike the commonly cited protocol for purifying KDACs [15], we did not demetalate the enzymes before storage. Even in the metalated state, the purified KDACs were stable for long periods of time stored at −20 °C, as we did not notice a significant decrease in activity over a period of greater than one year as long as the storage buffer did not freeze. While demetalating KDACs after purification is advantageous for any studies directly related to the biologically relevant active site metal, the simpler protocol presented here is suitable for general applications.
4. Conclusions
We have presented here a relatively simple and straightforward protocol for purification of KDACs. It is robust and can be used to purify several different KDACs from multiple expression systems, allowing for sufficient yields for all KDACs and variants tested. The resulting enzymes are highly-purified, stable for long periods of time, and demonstrate consistent activity between preparations, even when different recombinant expression systems are utilized. Using this protocol for KDAC expression and purification will not only improve the quality and efficiency of KDAC purifications, it will also allow for more direct comparison of KDAC activity across substrates, as variations in activity due to preparatory method will not be a confounding factor.
Supplementary Material
Highlights.
A relatively simple, straightforward, and robust protocol for KDAC purification.
Applicable to multiple expression hosts.
High yield of highly active, stable enzymes.
Enhanced preparation consistency for more reliable characterization.
Acknowledgments
Funding
This material is based upon work supported by, or in part by, the U. S. Army Research Laboratory and the U. S. Army Research Office [grant number W911NF1310129], the Louisiana Cancer Research Consortium, the National Institutes of Health [grant numbers 5G12MD007595, TL4GM118968, 5RL5GM118966, UL1GM118967], the National Science Foundation [grant number CHE 1625993], the Xavier University of Louisiana Center for Undergraduate Research, UNCF Stem Scholars Program, and the Xavier University of Louisiana Ronald E. McNair program. The contents are solely the responsibility of the authors and do not necessarily represent the official views of the Louisiana Cancer Research Consortium or the National Institutes of Health.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Finnin MS, Donigian JR, Cohen A, Richon VM, Rifkind RA, Marks PA, Breslow R, Pavletich NP. Structures of a histone deacetylase homologue bound to the TSA and SAHA inhibitors. Nature. 1999;401:188–193. doi: 10.1038/43710. [DOI] [PubMed] [Google Scholar]
- 2.Chen K, Zhang X, Wu YD, Wiest O. Inhibition and mechanism of HDAC8 revisited. J Am Chem Soc. 2014;136:11636–11643. doi: 10.1021/ja501548p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gantt SL, Gattis SG, Fierke CA. Catalytic activity and inhibition of human histone deacetylase 8 is dependent on the identity of the active site metal ion. Biochemistry. 2006;45:6170–6178. doi: 10.1021/bi060212u. [DOI] [PubMed] [Google Scholar]
- 4.Kim SC, Sprung R, Chen Y, Xu Y, Ball H, Pei J, Cheng T, Kho Y, Xiao H, Xiao L, Grishin NV, White M, Yang XJ, Zhao Y. Substrate and functional diversity of lysine acetylation revealed by a proteomics survey. Mol Cell. 2006;23:607–618. doi: 10.1016/j.molcel.2006.06.026. [DOI] [PubMed] [Google Scholar]
- 5.Basu A, Rose KL, Zhang J, Beavis RC, Ueberheide B, Garcia BA, Chait B, Zhao Y, Hunt DF, Segal E, Allis CD, Hake SB. Proteome-wide prediction of acetylation substrates. Proc Natl Acad Sci USA. 2009;106:13785–13790. doi: 10.1073/pnas.0906801106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Choudhary C, Kumar C, Gnad F, Nielsen ML, Rehman M, Walther TC, Olsen JV, Mann M. Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science. 2009;325:834–840. doi: 10.1126/science.1175371. [DOI] [PubMed] [Google Scholar]
- 7.Zhao S, Xu W, Jiang W, Yu W, Lin Y, Zhang T, Yao J, Zhou L, Zeng Y, Li H, Li Y, Shi J, An W, Hancock SM, He F, Qin L, Chin J, Yang P, Chen X, Lei Q, Xiong Y, Guan KL. Regulation of cellular metabolism by protein lysine acetylation. Science. 2010;327:1000–1004. doi: 10.1126/science.1179689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lundby A, Lage K, Weinert BT, Bekker-Jensen DB, Secher A, Skovgaard T, Kelstrup CD, Dmytriyev A, Choudhary C, Lundby C, Olsen JV. Proteomic analysis of lysine acetylation sites in rat tissues reveals organ specificity and subcellular patterns. Cell Rep. 2012;2:419–431. doi: 10.1016/j.celrep.2012.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Scholz C, Weinert BT, Wagner SA, Beli P, Miyake Y, Qi J, Jensen LJ, Streicher W, McCarthy AR, Westwood NJ, Lain S, Cox J, Matthias P, Mann M, Bradner JE, Choudhary C. Acetylation site specificities of lysine deacetylase inhibitors in human cells. Nat Biotechnol. 2015;33:415–423. doi: 10.1038/nbt.3130. [DOI] [PubMed] [Google Scholar]
- 10.Yao YL, Yang WM. Beyond histone and deacetylase: an overview of cytoplasmic histone deacetylases and their nonhistone substrates. J Biomed Biotechnol. 2011;2011:146493. doi: 10.1155/2011/146493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tang J, Yan H, Zhuang S. Histone deacetylases as targets for treatment of multiple diseases. Clin Sci. 2013;124:651–662. doi: 10.1042/CS20120504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Murugan K, Sangeetha S, Ranjitha S, Vimala A, Al-Sohaibani S, Rameshkumar G. HDACiDB: a database for histone deacetylase inhibitors. Drug Des Devel Ther. 2015;9:2257–2264. doi: 10.2147/DDDT.S78276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Howitz KT, Bitterman KJ, Cohen HY, Lamming DW, Lavu S, Wood JG, Zipkin RE, Chung P, Kisielewski A, Zhang LL, Scherer B, Sinclair DA. Small molecule activators of sirtuins extend Saccharomyces cerevisiae lifespan. Nature. 2003;425:191–196. doi: 10.1038/nature01960. [DOI] [PubMed] [Google Scholar]
- 14.Singh RK, Mandal T, Balsubramanian N, Viaene T, Leedahl T, Sule N, Cook G, Srivastava DK. Histone deacetylase activators: N-acetylthioureas serve as highly potent and isozyme selective activators for human histone deacetylase-8 on a fluorescent substrate. Bioorg Med Chem Lett. 2011;21:5920–5923. doi: 10.1016/j.bmcl.2011.07.080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Olson DE, Udeshi ND, Wolfson NA, Pitcairn CA, Sullivan ED, Jaffe JD, Svinkina T, Natoli T, Lu X, Paulk J, McCarren P, Wagner FF, Barker D, Howe E, Lazzaro F, Gale JP, Zhang YL, Subramanian A, Fierke CA, Carr SA, Holson EB. An unbiased approach to identify endogenous substrates of “histone” deacetylase 8. ACS Chem Biol. 2014;2014:2210–2216. doi: 10.1021/cb500492r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wegener D, Wirsching F, Riester D, Schwienhorst A. A fluorogenic histone deacetylase assay well suited for high-throughput activity screening. Chem Biol. 2003;10:61–68. doi: 10.1016/S1074-5521(02)00305-8. [DOI] [PubMed] [Google Scholar]
- 17.Gurard-Levin ZA, Kilian KA, Kim J, Bahr K, Mrksich M. Peptide arrays identify isoform-selective substrates for profiling endogenous lysine deacetylase activity. ACS Chem Biol. 2010;5:863–873. doi: 10.1021/cb100088g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Riester D, Hildmann C, Grunewald S, Beckers T, Schwienhorst A. Factors affecting the substrate specificity of histone deacetylases. Biochem Biophys Res Commun. 2007;357:439–445. doi: 10.1016/j.bbrc.2007.03.158. [DOI] [PubMed] [Google Scholar]
- 19.Gurard-Levin ZA, Kim J, Mrksich M. Combining mass spectrometry and peptide arrays to profile the specificities of histone deacetylases. Chembiochem. 2009;10:2159–2161. doi: 10.1002/cbic.200900417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Riester D, Wegener D, Hildmann C, Schwienhorst A. Members of the histone deacetylase superfamily differ in substrate specificity towards small synthetic substrates. Biochem Biophys Res Commun. 2004;324:1116–1123. doi: 10.1016/j.bbrc.2004.09.155. [DOI] [PubMed] [Google Scholar]
- 21.Aramsangtienchai P, Spiegelman NA, He B, Miller SP, Dai L, Zhao Y, Lin H. HDAC8 Catalyzes the Hydrolysis of Long Chain Fatty Acyl Lysine. ACS Chem Biol. 2016;11:2685–2692. doi: 10.1021/acschembio.6b00396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hai Y, Christianson DW. Histone deacetylase 6 structure and molecular basis of catalysis and inhibition. Nat Chem Biol. 2016;12:741–747. doi: 10.1038/nchembio.2134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Toro TB, Pingali S, Nguyen TP, Garrett DS, Dodson KA, Nichols KA, Haynes RA, Payton-Stewart F, Watt TJ. KDAC8 with high basal velocity is not activated by N-acetylthioureas. PLoS ONE. 2016;11:e0146900. doi: 10.1371/journal.pone.0146900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Toro TB, Watt TJ. KDAC8 substrate specificity quantified by a biologically-relevant, label-free deacetylation assay. Protein Sci. 2015;24:2020–2032. doi: 10.1002/pro.2813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fischle W, Emiliani S, Hendzel MJ, Nagase T, Nomura N, Voelter W, Verdin E. A new family of human histone deacetylases related to Saccharomyces cerevisiae HDA1p. J Biol Chem. 1999;274:11713–11720. doi: 10.1074/jbc.274.17.11713. [DOI] [PubMed] [Google Scholar]
- 26.Ciccarone VC, Polayes DA, Luckow VA. Molecular Diagnosis of Infectious Diseases. Humana Press; New Jersey: 1997. Generation of recombinant baculovirus DNA in E. coli using a baculovirus shuttle vector; pp. 213–236. [DOI] [PubMed] [Google Scholar]
- 27.Hawley-Nelson P, Ciccarone V, Moore ML. Transfection of cultured eukaryotic cells using cationic lipid reagents. In: Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K, editors. Current Protocols in Molecular Biology. John Wiley & Sons, Inc; Hoboken, NJ, USA: 2008. [DOI] [PubMed] [Google Scholar]
- 28.Dowling DP, Gantt SL, Gattis SG, Fierke CA, Christianson DW. Structural studies of human histone deacetylase 8 and its site-specific variants complexed with substrate and inhibitors. Biochemistry. 2008;47:13554–13563. doi: 10.1021/bi801610c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bottomley MJ, Lo Surdo P, Di Giovine P, Cirillo A, Scarpelli R, Ferrigno F, Jones P, Neddermann P, De Francesco R, Steinkuhler C, Gallinari P, Carfi A. Structural and functional analysis of the human HDAC4 catalytic domain reveals a regulatory structural zinc-binding domain. J Biol Chem. 2008;283:26694–26704. doi: 10.1074/jbc.M803514200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lahm A, Paolini C, Pallaoro M, Nardi MC, Jones P, Neddermann P, Sambucini S, Bottomley MJ, Lo Surdo P, Carfi A, Koch U, De Francesco R, Steinkuhler C, Gallinari P. Unraveling the hidden catalytic activity of vertebrate class IIa histone deacetylases. Proc Natl Acad Sci USA. 2007;104:17335–17340. doi: 10.1073/pnas.0706487104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gantt SML, Decroos C, Lee MS, Gullett LE, Bowman CM, Christianson DW, Fierke CA. General Base-General Acid Catalysis in Human Histone Deacetylase 8. Biochemistry. 2016;55:820–832. doi: 10.1021/acs.biochem.5b01327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Block H, Maertens B, Spriestersbach A, Brinker N, Kubicek J, Fabis R, Labahn J, Schafer F. Immobilized-metal affinity chromatography (IMAC): a review. Meth Enzymol. 2009;463:439–473. doi: 10.1016/S0076-6879(09)63027-5. [DOI] [PubMed] [Google Scholar]
- 33.Toro TB, Bryant JR, Watt TJ. Lysine deacetylases exhibit distinct changes in activity profiles due to fluorophore-conjugation of substrates. Biochemistry. 2017 doi: 10.1021/acs.biochem.7b00270. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.