Abstract
Polycyclic aromatic hydrocarbons (PAHs) pollutions often occur in marine and other saline environment, largely due to anthropogenic activities. However, study of the PAHs-degradation genotypes in halophiles is limited, compared with the mesophilic terrestrial PAHs degraders. In this study, a bacterial consortium (CY-1) was enriched from saline soil contaminated with crude oil using phenanthrene as the sole carbon source at 10% salinity. CY-1 was dominated by the moderate halophilic Marinobacter species, and its dominant PAHs ring-hydroxylating dioxygenase (RHD) genotypes shared high identity to the classic nah-related RHDs found in the mesophilic species. Further cloning of a 5.6-kb gene cluster from CY-1 unveiled the existence of a new type of PAHs degradation gene cluster (hpah), which most probably evolves from the nah-related gene clusters. Expression of the RHD in this gene cluster in E. coli lead to the discovery of its prominent salt-tolerant properties compared with two RHDs from mesophiles. As a common structural feature shared by all halophilic and halotolerant enzymes, higher abundance of acidic amino acids was also found on the surface of this RHD than its closest nah-related alleles. These results suggest evolution towards saline adaptation occurred after horizontal transfer of this hpah gene cluster into the halophiles.
Introduction
Polycyclic aromatic hydrocarbons (PAHs) are a group of pollutants that consist of two or more fused benzene rings and are prevailing in both terrestrial and marine environment mainly due to anthropogenic activities1,2. Microbial biodegradation is the main process for elimination of PAHs in the environment1,3. In bacteria, PAHs degradation is initialized via di-hydroxylation catalyzed by ring-hydroxylating dioxygenase (RHD)3, which is composed of four components, namely a large subunit (coded by pahAc), a small subunit (pahAd), a reductase (pahAa), and a ferredoxin (pahAb)4,5. RHD, the large subunit in particular, is often used as a marker to detect PAHs degraders6–8, and the phylogenies of RHD can reflect genotypes of PAHs degraders. So far, more than ten types of gene clusters (pah) that consist of PAHs degradation genes have been identified9–15, and most of them are from terrestrial PAHs degraders. Pah gene clusters of the same type are often found within the same genus, family, or order16–18.
Although PAHs pollutions occur frequently across marine and other saline environments, genotypes of PAHs degraders from saline environment are poorly characterized. So far, marine PAHs degraders within dozens of genera have been isolated1; however, only two genotypes with the archetype in Cycloclasticus sp. A510 and Alteromonas sp. SN213 have been reported. While strain A5′s the genotype was exclusively found within Cycloclasticus 19,20, strain SN2’s genotype was found across Alteromonas, Neptunomonas 21, and Pseudoalteromonas 22 genera. Both of these genotypes are distinct from their counterparts in terrestrial PAHs degraders. Indeed, this segregation of genotype is common for all terrestrial microorganisms and marine microorganisms, which was mainly caused by the molecular mechanisms to adapt the distinct salt concentrations23. Therefore, novel PAH-degrading genotypes are expected to be existed in saline environments and their molecular mechanisms to salt adaption remain to be investigated.
Noticeably, a PAHs-degrading Marinobacter strain (NCE312) was once reported to have a pahAc fragment similar to the dioxygenase of nah-related genotypes24. Nah-related gene clusters are a set of evolutionarily related pah gene clusters found within diverse Proteobacteria, including the nah type within Pseudomonas 9, nag type within Burkholderiales25, as well as the nitrotoluenes-degradation (NT) type within Burkholderiales26. Both nag and NT gene clusters are mainly detected in terrestrial PAHs degraders. In contrast, nah genotype is the only PAHs degradation genotype that is widely detected across terrestrial and coastal marine environments6,7,16,27. The presence of the nah gene clusters in the coastal marine environment makes it possible to study nah genes transfer between the terrestrial and marine lineages, as suggested by the pahAc fragment discovered in Marinobacter sp. NCE31224. Unfortunately, there is currently no evidence except the case of Marinobacter sp. NCE312 that allow us to address this issue. Therefore, it is worthy to further explore nah gene clusters in marine environment and determine whether the horizontal transfer of the nah gene clusters or other PAHs degradation genotypes between terrestrial and marine lineages occurs commonly.
In addition, halotolerant enzymes with biotechnological applications from halotolerant or halophilic microorganisms are drawing increasing attentions because of their impressive tolerance to a wide range of salinity28,29. Recently, several halotolerant enzymes, including two catechol dioxygenases from Marinobacter species, have been characterized30–32. However, the halotolerant properties and salt adaption mechanism of catabolic enzymes in PAHs degradation remain unknown although there was one has been cloned from halophilic PAHs degraders10.
In this study, we identified nah-related PAHs dioxygenases as the dominant genotype in a halophilic phenanthrene-degrading bacterial consortium (CY-1), which is dominated by Marinobacter species. PahAcs from this consortium form an independent clade in the phylogenetic tree, rather than clustering with any known nah-related pahAc sequences available in the Genbank. A 5.6 kb PAHs degradation gene cluster belonging to the dominant nah group is cloned and the dioxygenase part in this cluster is overexpressed in E. coli successfully. Further efforts were made to characterize the property of the PAHs dioxygenase in this halophilic consortium and explore the evolution scenario of the new pah gene cluster in this consortium. To date, this is the first report on cloning and overexpression of a halotolerant nah-like dioxygenase and prediction of horizontal transfer of nah-like gene clusters between halophilic and non-halotolerant bacteria.
Results and Disscussion
Microbial community structure in CY-1
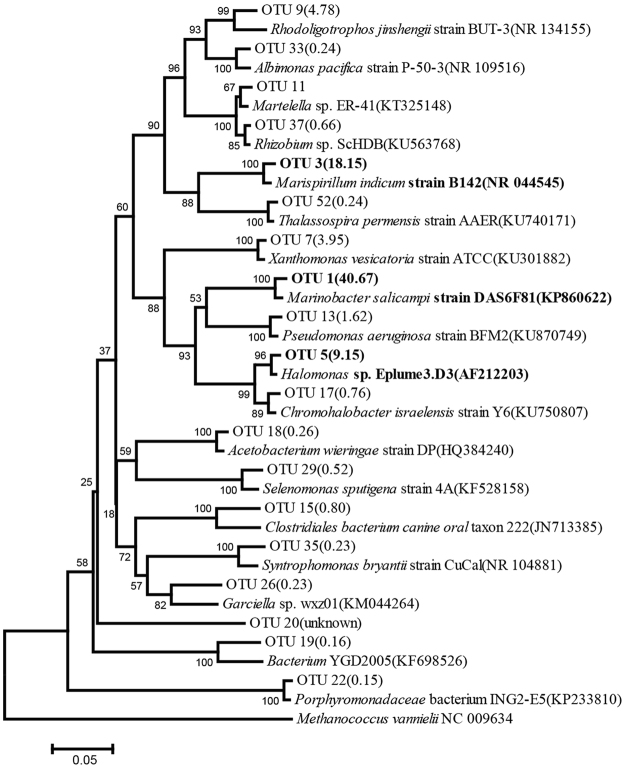
The halophilic bacterial consortium CY-1 was enriched by weekly transfers in sea salt-defined media (SSDM) medium30 with 10% salinity and phenanthrene (100 mg/L) for 3 months. It could completely degrade 100 mg/L phenanthrene in 6 days. The microbial community structure was determined through high-throughput sequencing (Fig. 1). Marinobacter was found to be the most abundant genus and account for 40.67% of the total 16 s rRNA gene sequences, followed by Marispirillum (18.15%), and Halomonas (9.15%). All of these abundant species were halophiles.
Figure 1.
Phylogenetic Neighbor-Joining tree showing the V1 to V3 region of the 16 S rRNA gene sequences of CY-1. The sequences data is separated into different OTUs with 97% identity. The proportions of different genera in CY-1 are shown following the OUT names, and the three most abundant OTUs are shown in bold. For each genus present in the consortium, a reference sequence is included in the phylogenetic tree, and the accession number is shown after the specie names. The percentages of replicate trees in which the associated taxa clustered together in the bootstrap test (1000 replicates) are shown next to the branches. 16 s rRNA gene sequence from Methanococcus Vannielii is used as the outgroup in the tree.
PAHs-degrading strains within genus Marinobacter had been isolated from coastal marine environment all around the world24,33,34. These Marinobacter strains could utilize naphthalene or phenanthrene as the sole carbon source. In a previous study, Marinobacter species were also the predominant PAHs degraders in a halophilic phenanthrene-degrading consortium enriched at 10% salinity35. The genus Marispirillum was identified recently and contained only one Marispirillum species, the type strain of which was isolated from an oil-degrading marine consortium but could not degrade PAHs36. Halomonas species were found in many oil or PAHs-degrading halophilic bacterial consortiums20,35; however, none of them were able to degrade PAHs. The consortium also contained several less abundant genera, within which PAHs-degrading strains have been reported previously, i.e. Pseudomonas (1.62%), and Thalassospira 37 (0.24%).
We also tried to isolate pure strains that can grow with phenanthrene as the sole carbon source from CY-1. However, this attempt ended in failure. We did achieve several Halomonas strains on the nutrient medium, but we failed to get strains within Marinobacter and other genera. In accordance with other previous reports20,35, these isolated Halomonas strains could not degrade PAHs. Based on the community structure and previous studies24,35, the main degraders in this phenanthrene-degrading consortium are most likely to be Marinobacter species, while Halomonas strains serve as utilizers of intermediates of degradation.
Profile of PAHs dioxygenase in CY-1
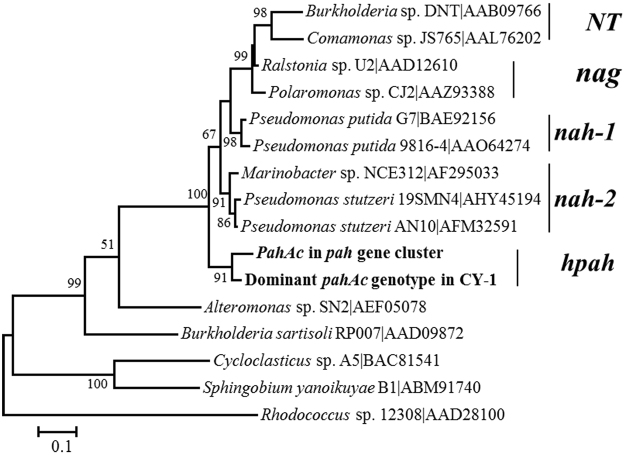
Using primers PAH-RHD-396F and PAH-RHD-696R as described previously7, the profile of the PAHs dioxygenase in this consortium was investigated. A total of 85 pahAc gene sequences were recovered, and corresponded to 7 unique sequences. All of them fell into 3 OTUs when 97% of the amino acid sequence was used as the dividing standard, or one OTU when 90% identity was used. A blast analysis of the representative pahAc sequences from CY-1 showed that they were mostly similar to the nah-related dioxygenase from Pseudomonas and Burkerholderiales. We further built a phylogenetic tree with the representative sequence in CY-1 and other well characterized nah-related dioxygenase (Fig. 2). PahAc gene sequences cloned from CY-1 formed a single clade, namely hpcah, in parallel with the nah and nag clades in the phylogenetic tree. We also analyzed the phylogeny of all available nah-related dioxygenase gene sequences from cultured strains in the Genbank. All previously-reported nah-related pahAc fell into the three clades of the well-characterized dioxygenases (Fig. S1). These results indicated the dominant dioxygenases in CY-1 represented a new type of nah-related PAHs dioxygenases.
Figure 2.
Maximum Likelihood tree of the representative pahAc of the dominant genotype in CY-1, the full length pahAc gene in the newly-cloned hpah gene cluster and other representative nah-related Ac genes, built with predicted amino acid sequences. Several characterized pahAcs from Alteromonas sp. SN2, Burkerholderia sp. RP007, Sphingomonas sp. B1 and Rhodococcus sp. NCIB12308 were used as outgroup. PahAc sequences cloned in this study are shown in bold. The five types of the nah-related pahAcs are labeled in the tree, with the group’s name shown in the right of the tree.
Cloning and sequence analysis of a PAHs-degrading gene cluster
Since different types of nah-related clusters vary in structures and PAHs degradation pathways25,38,39, it’s attractive to determine the structures of the gene clusters associated with the newly-identified pahAcs. After a subsequent gene walking strategy, a 5.6-kb PAH-degradation gene cluster (hpah, accession number: KM025345) containing pahAc of the dominant type in CY-1 was cloned. A RT-PCR experiment with the total RNA extracted from the phenanthrene-induced CY-1 consortium revealed that this gene cluster was actively transcribed during phenanthrene degradation as shown in Fig. S2.
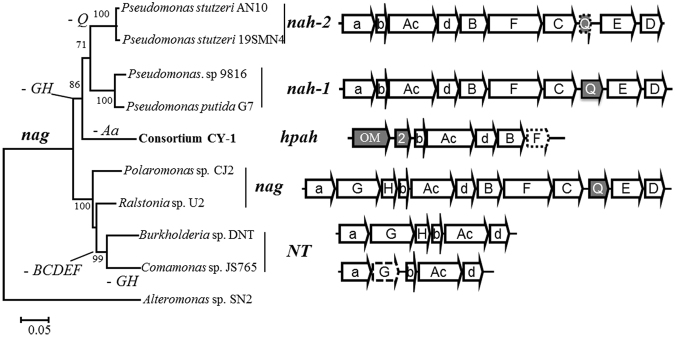
The whole gene cluster contains seven open reading frames (ORFs) (Fig. 3). All except orf2 are initiated by a canonical ATG start codon and are preceded by a putative ribosomal binding site. Through blast analysis, the five downstream ORFs were found to be most similar to the nah-related PAHs degradation genes, namely the nahAb, nahAc, nahAd, nahB, and nahF, with the highest identities ranging from 81–87% (Table 1). These five ORFs, designed as hpahAbAcAdBF, are arranged in the same way as the well-characterized nah and nag gene clusters25,38,39. The homologies of these ORFs and their arrangement in this newly-cloned hpah gene cluster suggest that it was evolutionarily-related to the nah-related gene clusters.
Figure 3.
Concatenated tree of nah-related AbAcAdB genes, organization of nah-related gene clusters and proposed evolutionary scenario. On the left is the concatenated tree of nah-related AbAcAdB genes deduced by both the Maximum Likelihood and Neighbor-Joining method; AcAdB genes from Alteromonas sp. SN2 were used as the outgroup. On the right are the archetypal structures of the five types of the nah-related gene clusters. Arrows in white represent PAHs degradation genes and their transcription directions. For clarity, nahAaAbAcAd are shown as a, b, Ac, d, etc. Arrows in gray represent genes encoding hypothetical proteins. Arrows with dashed borders represent interrupted genes. The proposed evolutionary events are shown next to the corresponding branches in the tree.
Table 1.
Annotation of genes in hpah gene cluster.
| Genes | Proposed function | Protein size | Protein with highest identity | Identity (aa)* | Accession no.a | Organism |
|---|---|---|---|---|---|---|
| OM1 | outer membrane protein | 461 | aromatic hydrocarbon degradation protein | 76% | AHI32768 | Marinobacter salaries R9SW1T |
| tbuX, toluene transporter | 54% | AAF03168 | Ralstonia pickettii PK01 | |||
| styE, styrene transporter | 45% | AAR24508 | Pseudomonas putida CA-3 | |||
| todX, toluene transporter | 38% | AAC43318 | Pseudomonas putida F1 | |||
| fadL, Long-chain fatty acid transport protein | 23% | P10384 | E. coli K12 | |||
| orf2 | 105 | |||||
| hpahAb | ferredoxin | 105 | naphthalene dioxygenase ferredoxin | 87% | AAD02135 | Pseudomonas stutzeri AN10 |
| hpahAc | RHDs α subunit | 449 | Naphthalene 1,2-dioxygenase subunit α | 83% | Q51494 | Pseudomonas aeruginosa PaK1 |
| hpahAd | RHDs β subunit | 194 | naphthalene 1,2-dioxygenase | 85% | EZQ14081 | Pseudomonas bauzanensis W13Z2 |
| hpahB | cis-naphthalene dihydrodiol dehydrogenase | 259 | 2,3-dihydroxy-2,3-dihydrophenylpropionate | 81% | EXF45161 | Pseudomonas sp. BAY1663 |
| hpahF | salicylaldehyde dehydrogenase | 452 | salicylaldehyde dehydrogenase | 86% | 4JZ6_A | Pseudomonas putida G7 |
aOnly sequences with the highest identity to orf1, hpahAb, hpahAc, hpahAd, hpahB, and hpahF from the blast results are shown in this table. OM1 shares the highest identity with a protein from Marinobacter salaries R9SW1T, and several other proteins with known functions are also listed.
The two upstream ORFs, however, exhibit no identities to the upstream genes of the nah-related gene clusters. For orf2, nothing was returned for the blast analysis with the default setting in GenBank, indicating its low similarity with the existing proteins. The orf1 (designed as OM1) was identified as a FadL-type outer membrane protein (OM), and exhibited the highest identity (75%) with an uncharacterized OM from Marinobacter salaries R9SW1T (Table 1). A closer examination of the OM gene in Marinobacter salaries R9SW1T unveils it is associated with a toluene degradation gene cluster in the genome. Several FadL-type OMs, i.e. TodX40,41, TbuX40,42, and styE43, have been reported to be involved in the degradation of toluene and other aromatics, and were proposed to be responsible for aromatics uptake from the environment. The OM1 ORF exhibited moderate identity with these proposed aromatics transporters (Table 1), thus was likely involved in PAHs uptake.
In addition, passive diffusion and active transport were two known mechanisms for PAHs uptake by bacteria44–47. Because PAHs are poorly soluble in the water, the association of a transport protein to facilitate active PAHs uptake could largely promote PAHs utilization efficiency for PAHs degraders46. However, in Pseudomonas strains of the nah genotype, no potential transporter genes are found in the surrounding regions of the nah gene clusters. Accordingly, they are reported to take in PAHs through passive diffusion44,47. Currently, the only reported PAHs transporter of all PAHs degraders was an ompW-type OM in a Pseudomonas strain, of which the PAHs degradation genotype was undetermined46. Thus, we cannot ascertain whether the FadL-type OM associated with this newly-cloned hpah gene cluster was actually a PAHs transporter. Anyway, this is the first time to observe a FadL-type OM associated with a nah-related gene cluster, which may evolved after horizontal transfer.
RHD cloning and substrate specificity
Using primer RHD-F and RHD-R, nahAb/Ac/Ad genes in this cluster was cloned into the expression vector pET28a (+) and was successfully overexpressed in E. coli BL21 (DE3). An analysis of crude extracts of IPTG-induced recombinant cells by SDS-PAGE revealed the over-production of two soluble recombinant polypeptides, shown as the first and third stripes (from the top) representing the product of nahAd and nahAc, respectively (Fig. S2). The nahAb product was not detected probably because the standard SDS-PAGE used here was improper for resolving polypeptides in the 10,000- to 15,000-Da range. These results indicated that the RHD were correctly assembled in E. coli, which made it possible to analyze its enzymatic characteristics.
We first determined the substrate specificity of this cloned RHD, using GC-MS. As expected, it could readily oxidize naphthalene and phenanthrene into their cis-dihydrodiol forms, with the highest activity towards phenanthrene (Table 2). This RHD could also slightly transform phenol to hydroquinone and oxidize biphenyl Biphenyl cis-2,3-dihydrodiol or n-hydroxy-biphenyl, indicating its monohydroxylation activity. No products were detected for dibenzothiophene and PAHs with four rings, i.e. fluoranthene, pyrene and benz[a]anthracene. Generally, the substrate specificity and the reaction type of this RHD were consistent with other nah-related RHDs, which are well known for their ability to oxidize low-molecular-weight PAHs, especially naphthalene and phenanthrene25,48,49.
Table 2.
Substrate specificity of RHD expressed in E. coli.
| Substrate | Degradation rate(%) | Products | RT(min) | µM Diol/h mgProta |
|---|---|---|---|---|
| Phenol | 8.31 | hydroquinone | 8.34c | 0.0045 |
| Biphenyl | 12.51 | Biphenyl cis-2,3-Dihydrodiol | 12.13c | 0.0043 |
| 4-hydroxy-Biphenyl | 9.59c | |||
| 2-hydroxy-Biphenyl | 11.32c | |||
| Dibenzothiophene | 6.50 | nd | 4.20b | 0.0014 |
| 4.70b | ||||
| Naphthalene | 27.44 | Naphthalene 1,2-dihydrodiol | 9.75c | 0.0113 |
| phenanthren | 49.93 | Phenanthrene cis-3,4-dihydrodiol | 14.46c | 0.0148 |
| Fluoranthene | 27.17 | nd | 5.43b | 0.0071 |
| Pyrene | 1.85 | nd | 7.92b | 0.0005 |
| benz[a]anthracene | 5.38 | nd | 5.56b | 0.0014 |
aCalculated from the HPLC peak areas of biphenyl degraded after 13 h of incubation. The values are averages of three separate determinations with the control data deduced. bHPLC retention time with those of the authentic sample. cGC-MS retention time nd: dibenzothiophene, fluoranthene, pyrene,benz[a]anthracene,benz[a]pyrene, did not give any detectable products.
Halotolerant properties of the newly-cloned RHD
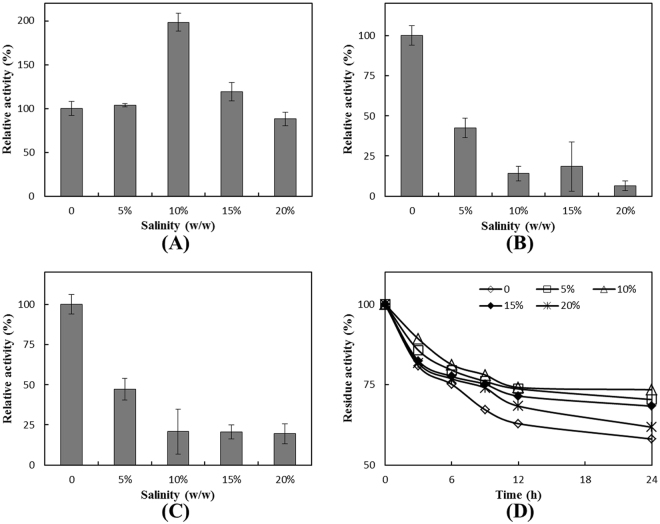
This RHD was active over a broad range of NaCl concentrations, as shown in Fig. 4A. It exhibited the highest activity in the PBS medium with 10% NaCl, and could still maintain nearly 100% activity with 20% NaCl, indicating a strong tolerance to NaCl. Furthermore, it could maintain 75% activity after 24 h with 10% NaCl, compared with the 60% activity with 0% NaCl, indicating a higher stability under saline conditions (Fig. 4D).The halotolerant property of this RHD was prominent, especially when compared with another nah type dioxygenase (84% identity) from a Pseudomonas strain (Fig. 4B) and the PAHs dioxygenase from Delftia sp. Cs1–4 (Fig. 4C).
Figure 4.
Effects of NaCl concentration on the activity (A) and salinity stability (D) of RHD obtained from CY-1 and the effect of NaCl concentration on the RHD activity from a non-halotolerant species, Pseudomonas (B) and Delftia sp. Cs1–4 (C). Effects of NaCl concentration (A, B and C) was determined at 4 °C in 50 mM PBS buffer (pH = 7.5). Activity was detected by the standard method. The value obtained without NaCl in the reaction mixture was taken as 100%.The enzyme was incubated in sodium phosphate buffers (pH 7.5) and PBS contain 5%, 10%, 15%, 20% NaCl respectively to detect the RHD salinity stability (D) and residual activity was determined at 3, 6, 9, 12, 24 h. The values shown represent averages from triplicate experiments. Error bars were not shown in this fig for better looking effect.
The halotolerant properties of this RHD are similar to that reported for two catechol 2,3-dioxygenases from Marinobacter species, all exhibiting higher activity and stability at high NaCl concentration30. However, the activity and stability of this RHD were the highest with 10% NaCl and dropped when the concentration of NaCl increased or decreased, while the activity and stability of the two catechol 2,3-dioxygenases remained almost unchanged when the concentration of NaCl fluctuated. In previous studies, several halotolerant enzymes cloned from halotolerant and halophilic bacteria were also reported to exhibit higher activity in saline environment than their non-halotolerant counterparts31,32. All of them can maintain activity across a wide range of salinity, from 0% up to 15% or 20%. However, the relationship between the enzymatic activity and salinity varied for different halotolerant enzymes including this RHD.
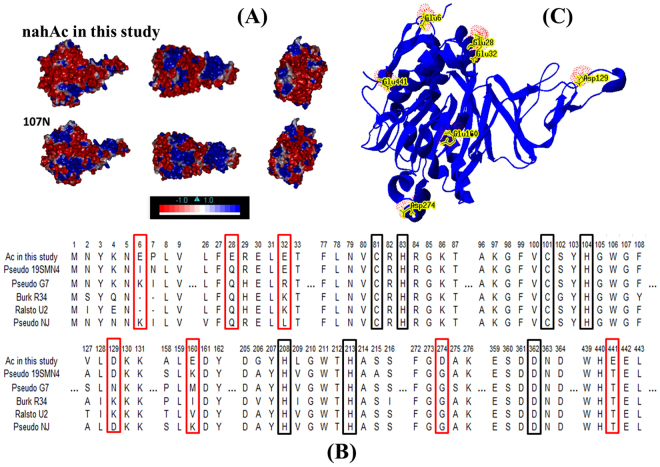
Amino acid composition, especially those on the protein surface of the newly-cloned RHD was calculated. Compared with other classic nah-related dioxygenase, this RHD had the highest content of acidic amino acids and the lowest content of alkaline amino acids in the non-conservative region (Table S1). In the large subunit, acidic animo acids at sites 6 (Glu), 28 (Glu), 32(Glu), 129 (Asp), 154 (Asp), 160 (Asp), 263 (Asp) and 274 (Glu) were exclusively found on the surface of this RHD (Fig. 5B,C). Correspondingly, the negative electrostatic surface potential of this RHD was predicted to be much lower than a mesophilic nah-related RHD (1O7N, with 83% identity to this RHD) (Fig. 5A). The excess of acidic amino acids were also found on the surface of the small subunit (Ad) of this RHD compared with other nah-related RHD (Table S1). Other fragments in this cluster also contained the higher proportion of acidic amino acids than their mesophilic alleles (Table S1). It might be the high acidic amino acids content at the protein surface that made the pahAc obtained from CY-1 present such a high halotolerant property29,50.
Figure 5.
Surface electrostatic potentials analysis. (A) Surface electrostatic potentials of nahA3, and Naphthalene 1,2-dioxygenase from Pseudomonas putida NCIB 9816–4 (PDB no. 1O7N) as obtained using Discovery Studio 2.5 software. The first, second and third line were seen from the front, top and left of the structures, respectively, with the red surface corresponding to negatively charged residues and the blue surface corresponding to positively charged residues (color figure online). (B) The key amino acid sites in nahAc. Fe2+ binding sites are labelled in black squares and the sequence near these active sites are also shown. Mutated amino acid sites are labelled in red frame. (C) The position of these mutated sites appear at the surface of the large subunit. This graph is illustrated by Swiss-pdb viewer.
Phylogenies of the hpah gene cluster
To further elucidate the evolutionary relationships between this hpah gene cluster and other nah-related gene clusters, we conducted phylogenetic analysis with the hpah genes and their alleles in the nah-related gene clusters.
In the phylogenetic trees of all genes, hpah gene formed an independent branch, in parallel with two nah clades (nah-1 and nah-2) and one nag clade (including the NT clade) (Figs 2, S4–S6). However, the topology of trees differed in the differentiation order of each clade for different genes. For Ac, and B genes, hpah clade first separated from other clades; in contrast, for Ab, Ad genes, the nag clade first separated from other clades. HpahAb gene from CY-1 diverged before the divergence of B genes in the nah-1 and nah-2 clades; in contrast, hpahAd gene from CY-1 clustered with the nah-2 clade.
The inconsistence of phylogenetic trees of different genes created an obstacle to infer the evolution process of hpah gene cluster and other nah-related gene clusters. Since genes in these gene clusters were likely to evolve and be transferred as a whole rather than separately, we further constructed a concatenated tree of AbAcAdB genes to track the evolution of the whole gene cluster. Interestingly, the topology of the concatenated tree was consistent with the tree of Ab genes, indicating nag gene clusters first separated from other nah-related gene clusters (Fig. 3), followed by the divergence of the newly-identified hpah clade and the two nah clades from Pseudomonas. The concatenated tree was consistent with the organization of the nah-related gene clusters (Figs 3, S7), indicating this hpah gene cluster represents a new type of nah-related gene cluster.
Evolutionary scenario of nah-related gene clusters
The nah gene clusters (nah_1 in Fig. 3) in Pseudomonas were the first PAHs-degradation gene clusters described and are still the best characterized38,49. Rafael Bosch et al.39 revealed the existence of a nah-related gene cluster (nah_2 type in Fig. 3), in which nahQ gene was absent, in Pseudomonas stutzeri AN10. In this study, we found the absence of nahQ gene was the common feature in all nah_2 type gene clusters (Fig. S7), and further analysis revealed a nahQ remnant between nahC and nahE in these gene clusters, suggesting the deletion of nahQ was probably the point at which the nah_2 gene cluster diverged from the nah_1 gene cluster. The nag gene cluster was first discovered in Ralstonia sp. U225, and was later found in Polaromonas 51 and Burkholderia 52. A comprehensive analysis in this study revealed that the nag gene clusters were widely distributed in PAH-degrading Burkholderiales (Figs S1, S7). The main difference between the nag and nah gene clusters was the existence of two genes (nagG and nagH) between the nagAa and nagAb genes. NT gene clusters in the nitrotoluenes degraders, which were also found mainly in Burkholderiales species, are another type of nah-related gene clusters (Figs S1, S6). It is generally accepted that the NT gene clusters were derived from the nag gene clusters through the elimination of the downstream nagBFCQED genes and further deletion of the nagGH genes26.
The newly-cloned hpah gene cluster was clustered with type nah−1 and nah−2 gene clusters in the phylogenetic tree and did not contain the G and H genes. It differed from the nag and nah gene clusters in the absence of pahAa gene and the presence of a potential PAHs transporter gene (OM1) in the upstream. Because Aa gene is present in both nag and nah gene clusters, Aa gene was probably present in the progenitor of hpah and nah gene clusters and was deleted in the subsequent evolution process of the hpah gene cluster. Because nah gene clusters have been identified within diverse Pseudomonas species and across diverse environments, it is likely that the hpah gene cluster was derived from nah gene clusters.
The first divergence of the nah-related gene clusters occurred between nag gene clusters and the progenitor of nah and hpah gene clusters (Fig. 3). Which is the initial progenitor was yet unsolved. The most obvious difference between these two groups is the nagGH genes. In the nag gene clusters, products of nagAa and nagAb genes served as the electron transport components (ETCs) both in salicylate-5-hydroxylating oxygenase coded by nagGH genes and in PAHs dioxygenase coded by nagAcAd genes53. ETC components have been thought to be one of the important selective forces in the evolution of oxygenase5. To determine whether nagAaAb are the inherent ETCs in salicylate-5-hydroxylating oxygenase or in PAHs dioxygenase may provide insight in the evolution of process the nag gene cluster.
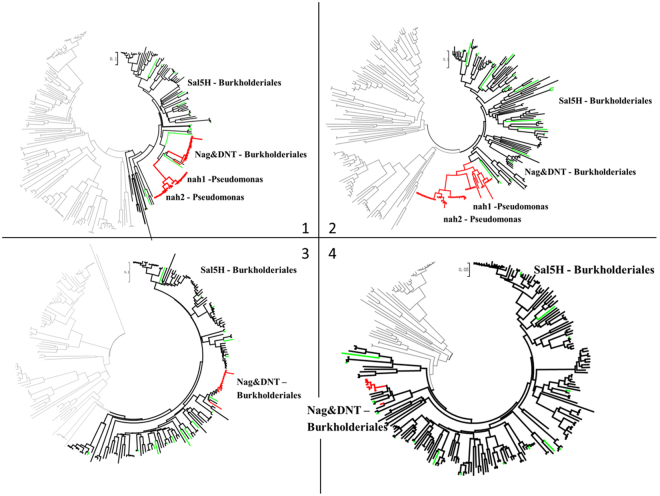
Comparison of the phylogenetic trees, built with Aa, Ab, Ac, Ad, G, and H genes and their homologous genes, revealed a similar topology for the trees of Aa, Ab, G, and H (Fig. 6). In each tree, genes from nag gene clusters formed a single clade surrounded by their homologous alleles mostly from Burkholderiales (Fig. 6). Most of the homologous alleles of Aa, Ab, G, and H genes were found to coexist in same strains (Fig. S8). Moreover, these homologous genes were adjacent to each other in the genome (of nearly twenty randomly-chosen strains), i.e., they belonged to the same operon (Fig. S8). These results indicated that Aa, Ab, G, and H genes have clustered and coevolved in the salicylate-degradation gene clusters (sal) long before they were integrated in the PAHs-degradation gene clusters, i.e. nagAaAb are the inherent ETCs in salicylate-5-hydroxylating oxygenase.
Figure 6.
Neighbour-Joining trees for nagA1 (1), nagA2 (2), nagG (3), nagH (4) and their homologous genes. Branches in red are from the nah-related gene clusters; branches in bold black are from the Sal5H gene clusters; branches in green are from some representative Sal5H gene clusters whose structures are present in Fig. S7.
Based on the above analysis, the progenitor of nah-related gene clusters can be proposed to be a nag type gene cluster. Furthermore, it can be concluded that the nag gene cluster initially arose in a PAHs-degrading strain within Burholderiales, because the sal gene cluster was widely distributed in Burkholderiales, but was rarely found in other bacteria (Figs 6, S8). The nag gene cluster was then rampantly transferred across species within Burkholderiales. The following evolutionary scenario of the nah-related gene clusters was proposed, as shown in Fig. 3. The nag gene clusters are the progenitor of other nah-related gene clusters and arose in PAHs-degrading bacteria within Burkerholderales. NT gene clusters were derived from the nag gene cluster after the deletion of nagBCDEF genes and the gradual deletion of the GH genes. The nag gene cluster was later transferred into other linkages and became the common ancestor of hpah gene cluster found within halophiles and of nah gene clusters found within Pseudomonas species. The nagGH genes were deleted subsequently, probably because the central one-ring aromatic intermediate is not gentisate in the new host. Hpah gene cluster then separated from nah gene clusters, and the Aa gene was probably deleted. In Pseudomonas strains, the initial nah gene cluster further diverged into two groups after the deletion of the useless Q gene in the nah−2 gene cluster. Although nah gene clusters are found mainly in Pseudomonas, several gene fragments similar to nahAc have been reported in other genera including Marinobacter (Fig. S1), suggesting that further inter-genera horizontal gene transfer had taken place. In a word, these nah-related gene clusters were evolving to become more compact through elimination of the redundant genes.
Back to hpah gene cluster in the halophiles, as mentioned above, it is derived from the nah-related gene clusters in the mesophiles via horizontal gene transfer. PAHs degraders with nag gene clusters are exclusively isolated from terrestrial environment. Although PAHs degraders with the nah gene clusters were found in saline environment, the nah type PAHs dioxygenase from Pseudomonas sp. strain HY-1 did not possess ability to adapt saline environment in our study. The halotolerant property and structural feature of the hpah RHD pointed to the evolutionary process towards adaptation of the saline environment after horizontal gene transfer. Many barriers that can affect the fate of the transferred gene in the new host have been identified or proposed54. It have been reported that further evolution process are required for proper work in the new host after horizontal gene transfer for some pathways55,56. Microorganisms in marine environment generally have different lifestyles in comparison to the terrestrial microorganisms due to the molecular mechanisms required for adaptation to distinct concentration of salt23. The distinct salt concentration can not only cause the segregation of the terrestrial and marine bacteria, but can also cause the inadaptation of gene clusters that undergo horizontal transfer between the terrestrial and marine bacteria. For PAHs degradation genes which are undergoing rampant horizontal transfer, the distinct genotypes present in terrestrial and marine environment just reflect such segregation effects by distinct salt concentration. The newly-discovered hpah gene cluster in our study provided the first example of an enzyme that evolved towards salt adaptation after a recent horizontal gene transfer event.
Still much work is required for further understanding this hpah gene cluster in CY-1. First, we cannot affirm whether the dominant pahAcs and hpah gene cluster were actually from Marinobacter or from other genera. Taking more efforts to isolate the PAHs-degrading pure strains or sequencing the metagenomics of the consortium are two feasible ways to address this question. Second, the PAHs dioxygenase cloned from this Marinobacter-predominant halophilic consortium exhibited remarkable halotolerant capability, similar properties were also reported for two Marinobacter-related catechol 2,3-dioxygenase. Whether halotolerant properties present in these enzymes are common feature for the majority of proteins in Marinobacter and other halophilic bacteria in the marine environment is an appealing question that requires far more efforts. Moreover, we proposed the newly-cloned hpah RHD have gone through an evolution process towards adaptation of saline environment after horizontal gene transfer. It will be attractive to explore whether this process is common for other HGT events between the mesophiles and halophiles. In addition, it is also attractive to further explore whether inadaptation to the distinct salt concentrations is a basic barrier for HGT between the mesophiles and the halophiles, which may serve as an explanation for the distinct PAHs-degradation and other functional genotypes between the terrestrial and marine environments.
Materials and Methods
Reagents, strains and plasmids
N-butylboronate (NBB) and antibiotics were obtained from Sigma-Aldrich (United States). PAHs were purchased from Alfa Aesar. Restriction endonucleases, Taq DNA polymerase, DNAiso Reagent, RNAiso Plus, DNase I, cDNA Synthesis Kit, SYBR® Premix Dimer Eraser™ and T4 DNA ligase were from TaKaRa (TAKARA, Dalian, China). pET28a (+) (Invitrogen, Shanghai, China) was used as an expression vector. E. coli strains TOP 10 and BL21 (DE3) (Qiagene, Beijing, China) were used for general cloning and protein expression.
Pseudomonas sp. strain HY-1 is isolated from oil-polluted soil and maintained using a mixture of phenanthrene and naphthalene as the sole carbon source in our lab; its 16 S gene sequence has been deposited in GenBank (accession number: MF689060). Delftia sp. strain Cs1–4 was kindly denoted by William J. Hickey’lab, and has been reported previously12.
Bacterial consortium and growth conditions
A halophilic bacteria consortium CY-1 capable of degrading phenanthrene under 10% salinity (w/w) was cultured using sea salt-defined media (SSDM) medium under 30 degrees, 150 rpm as described previously30.
PAH-degrading gene cluster amplification and sequencing analysis
DNA extraction, consortium structure and profile of PAHs dioxygenase
DNA extraction, consortium structure and profile of PAHs dioxygenase. The genome DNA from the enrichment was extracted by DNeasy Blood & Tissue Kit (Qiagen, Germany) with the process according to manufacturer’s instructions.
The consortium structure in CY-1 was investigated by 454-pyrosequencing following Polymerase Chain Reaction (PCR) strategy as previously described. The RHD diversity was investigated using primers PAH-RHD-396F (5′-ATTGCGCTTAYCAYGGBTGG-3′) and PAH-RHD-696R (5′-ATAGGTGTCTCCAACRAARTT-3′) as described previously7. A clone library of pahAcs was then constructed in E. coli. A total of one hundred clones were selected for sequencing, and 85 pahAc gene sequences were recovered.
Gene cluster amplification and annotation
The first fragment containing the naphthalene dioxygenase was achieved by PCR using the degenerate primers as described by Gomes57. The other fragments contained in the whole PAHs degradation cluster were obtained by thermal asymmetric interlaced PCR (Tail-PCR) according to the manufacturer’s instructions, using different sets of divergent primers designed on the basis of step by step sequences. The amplified gene cluster were completely sequenced and uploaded in GenBank (Accession no KM025345). Gene annotation and similarity analysis were carried out using the NCBI Web BLAST service58.
Phylogenetic analysis
To collect all the nah-related pahAc gene sequences and their corresponding gene clusters that are available in the GenBank database, amino acid sequence of the hpahAc cloned in this study is used to blast against the Non-redundant protein database using the NCBI Web BLAST service58, and 5000 sequences with the lowest e-values from cultured microorganisms are retrieved. With the retrieved sequences, a preliminary distance tree was created by using the NCBI tree viewer tool with the default parameters on the NCBI Web BLAST service58. All pahAc sequences that fell into the nah-related clade of the distance tree were collected for further analysis. The corresponding gene clusters of the nah-related pahAcs, if existing, are collected. Phylogenetic tree of each gene was then created by using the Maximum Likelihood method and the neighbor-joining method. Total bootstrap value was 100 in each tree. To construct the concatenated tree of hpahAbAcAdB genes, sequences of each gene were first aligned; and the aligned gene sequences from the same bacterium were then concatenated; the concatenated sequence for each bacterium was then used to construct the final phylogenetic tree in the same way as the individual genes. All phylogenetic analyses were conducted in MEGA659.
RHD cloning, overexpression and characterization
Cloning and overexpression
The nahAbAcAd fragment encoding RHD in the whole cluster was amplified by PCR using primers pairs: RHD-F (5′-GCGCATATGATGACAGAGAAGTGGATC-3′) and RHD-R (5′-CGGAGCTCTTACAGAAAGACCATTACG-3′), introducing the Nde I and Sac I (in underlined) at the end of primer. PCR products were ligated into pMD-19T, sequenced and then sub-cloned into the Nde I and Sac I sites of expression vector pET28a (+) (Novagen, USA). The construct was transformed into E. coli strain BL21 (DE3) for expression analysis.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE)
Bacterial cells were pelleted by centrifugation and washed with phosphate buffer (pH 7.5). One milliliter of phosphate buffer was added to the pellet and the suspension was then subjected to sonication on ice for 2 min (5 s pulse interval; 40% of maximum amplitude). After centrifugation, the supernatant and the pellet were detected by SDS-PAGE. After electrophoresis, the gel was stained with Coomassie Brilliant Blue. The molecular mass of the protein was determined by comparison of the control.
RT-PCR analysis of RHD gene
Real-time PCR analysis was performed using the SYBR Green-based detection system to monitor RHD gene expression. Cells were harvested at 0, 4, 8, 12, 16, 24, 36, 48, 72 and 96 h by centrifugation at 10,000 g at 4 °C for 1 min. Total DNA and RNA were extracted directly from each culture using the DNAiso Reagent and RNAiso Plus. The RNA was treated with RNase-free DNase according to the manufacturer’s instructions. Reverse transcription of RNA samples was performed using the Prime Script 1st strand cDNA synthesis Kit.
RHD genes and transcripts were quantified in an iCycleriQ (Bio-Rad, America) with the primer pairs as follow: nahAd-5′-GGTTGTGTCACGGGAACTG-3′, nahAd-5′-GATCAGGTTGGAGCGAACG-3′. At the end of the Real-Time PCR a melting curve analysis was performed by a final step that consisted of measurement of the SYBR Green I signal intensities during a 0.5 °C temperature increment every 10 s from 51 °C to 95 °C. The standard curve for gene quantification was performed with the nahAd PCR product cloned in the pMD-19T TA cloning kit. The gene expression level was calculated based on the transcript/gene ratio.
Substrates preference of the RHD
The activity of recombinant nahAb/Ac/Ad expressed in E. coli was assayed as follows. Strains BL21 (DE3) (pET28a) were grown in Luria-Bertani medium containing suitable antibiotics to an OD600 of 0.6 and then incubated at 30 °C with 0.5 mM IPTG. After 5 hours, cells were centrifuged, washed and resuspended to an OD600 of 1.2 in M9 minimal medium containing 0.2% glucose, and distributed as 5 ml aliquots into Erlenmeyer flasks. Fifty microliters of PAHs, including phenol, biphenyls, naphthalene, phenanthrene, dibenzothiophene, fluoranthene, pyrene, benz[a]anthracene and benzo[a]pyrene, dissolved in N,N-dimethyl formamide (stock 100 mg/ml) were added to the sterile liquid medium. The flasks were incubated at 150 rpm for 13 h at 30 °C. All the tests were performed in triplicates.
Water-soluble products resulting from PAHs oxidation were extracted with ethyl acetate, dried over anhydrous Na2SO4 and evaporated under nitrogen gas. The dried samples were dissolved in 500 μl acetonitrile for GC-MS (Agilent 7890 A GC, Inert MSD with TripleAxis Detector, Agilent, USA) analyses.
PAHs and their degradation products were analyzed by reversed phase HPLC with UV detection at 254 nm. The mobile phase, supplied at 1 ml min−1, was a 10 min linear gradient from 60% (v/v) to 90% (v/v) aqueous methanol with holding at 90% aqueous methanol for 10 min. The analytical column was a 4.6 × 250-mm, 5-μm C18 Inertsil ODS-3 column (Beijing Mairuida Technology, Beijing, China).
Before GC-MS analysis, the extracts were derivatized either with bis (trimethysilyl) trifluoroacetamide: trimethylchlorosilane (99:1) or NBB, according to the manufacturer′s instructions. An HP-5 capillary column (30 m × 0.25 mm I.D. × 0.25 µm film thickness) was used to separate the products. The column temperature was programmed as follows: started at 60 °C and held for 2 min, ramped at 10 °C/min to 280 °C, and then held for 10 min. Helium gas was used as the carrier gas at a flow rate of 1 ml/min. The mass spectrometer was operated in the mass scan mode.
Enzyme assays and effects of NaCl on RHD activity
Cells induced by 0.5 mM IPTG were disrupted by sonication, the cellular debris was removed by centrifugation at 14,000 g for 20 min, and the supernatant was used as the crude cell extract. Dioxygenase activity was assayed through the transform of naphthalene by HPLC.
RHD characteristics were measured after the enzyme incubated under different conditions such as a wide range of salinity and pH for 30 min. The RHD temperature stability was measured after incubation under correspond temperatures for 3 h, 6 h, 9 h, 12 h and 24 h. The RHD salinity stability was measured after incubation under correspond salinities for 3 h, 6 h, 9 h, 12 h and 24 h at 4 °C.
Surface electrostatic potential
The surface electrostatic potential of the RHD obtained in CY-1 and a mesophilic homolog obtained from protein database (PDB no. 1O7N) was calculated by Discovery Studio 2.5 software (Accelrys, SanDiego, CA, USA).
Electronic supplementary material
Acknowledgements
This work was supported by the National Science Foundation of China (41573065, 51138006), National 863 Program of China (2013AA06A210), and National major projects on water pollution control and management technology (2012ZX07301-001).
Author Contributions
C.W. and G.G. were the joint first authors as they contributed equally to the work. C.W. enriched the consortium, determined the community structure and RHD profile of this consortium, analyzed the amino acids sequence of the RHD, detected the halotolerant properties of the newly-cloned RHD and other two PAHs-RHDs, and prepared Figs 1, 2, 4, 5(b,c), S4~S8 and Table S1. G.G. cloned the gene cluster, expressed the RHD, characterized its substrate preference and prepared Figs 5(a), S2, S3 and Tables 1, 2. C.Y. and G.G. draft the manuscript text together. Y.H. analyze the data and write the final manuscript, and prepared Figs 3, 6, S1 and Table 1. H.W. modified the manuscript text; H.W. is the corresponding author. All authors revised the manuscript.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-017-12979-z.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Vila J, Tauler M, Grifoll M. Bacterial PAH degradation in marine and terrestrial habitats. Current opinion in biotechnology. 2015;33:95–102. doi: 10.1016/j.copbio.2015.01.006. [DOI] [PubMed] [Google Scholar]
- 2.Tingting, et al. Effect of salinity on community structure and naphthalene dioxygenase gene diversity of a halophilic bacterial consortium. Frontiers of Environmental Science & Engineering. 2016;10:16. [Google Scholar]
- 3.Peng R-H, et al. Microbial biodegradation of polyaromatic hydrocarbons. FEMS microbiology reviews. 2008;32:927–955. doi: 10.1111/j.1574-6976.2008.00127.x. [DOI] [PubMed] [Google Scholar]
- 4.Gibson DT, Parales RE. Aromatic hydrocarbon dioxygenases in environmental biotechnology. Current Opinion in Biotechnology. 2000;11:236–243. doi: 10.1016/S0958-1669(00)00090-2. [DOI] [PubMed] [Google Scholar]
- 5.Kweon O, et al. A new classification system for bacterial Rieske non-heme iron aromatic ring-hydroxylating oxygenases. BMC biochemistry. 2008;9:1. doi: 10.1186/1471-2091-9-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lozada M, et al. Novel aromatic ring-hydroxylating dioxygenase genes from coastal marine sediments of Patagonia. BMC microbiology. 2008;8:1. doi: 10.1186/1471-2180-8-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ding GC, et al. Soil type-dependent responses to phenanthrene as revealed by determining the diversity and abundance of polycyclic aromatic hydrocarbon ring-hydroxylating dioxygenase genes by using a novel PCR detection system. Applied & Environmental Microbiology. 2010;76:4765–4771. doi: 10.1128/AEM.00047-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Iwai S, Johnson TA, Chai BL, Hashsham SA, Tiedje JM. Comparison of the Specificities and Efficacies of Primers for Aromatic Dioxygenase Gene Analysis of Environmental Samples. Applied and Environmental Microbiology. 2011;77:3551–3557. doi: 10.1128/AEM.00331-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Habe H, Omori T. Genetics of polycyclic aromatic hydrocarbon metabolism in diverse aerobic bacteria. Bioscience, biotechnology, and biochemistry. 2003;67:225–243. doi: 10.1271/bbb.67.225. [DOI] [PubMed] [Google Scholar]
- 10.Kasai Y, Shindo K, Harayama S, Misawa N. Molecular characterization and substrate preference of a polycyclic aromatic hydrocarbon dioxygenase from Cycloclasticus sp. strain A5. Applied and environmental microbiology. 2003;69:6688–6697. doi: 10.1128/AEM.69.11.6688-6697.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pinyakong O, et al. Isolation and characterization of genes encoding polycyclic aromatic hydrocarbon dioxygenase from acenaphthene and acenaphthylene degrading Sphingomonas sp. strain A4. FEMS microbiology letters. 2004;238:297–305. doi: 10.1016/j.femsle.2004.07.048. [DOI] [PubMed] [Google Scholar]
- 12.Hickey WJ, Chen S, Zhao J. The phn island: a new genomic island encoding catabolism of polynuclear aromatic hydrocarbons. Frontiers in microbiology. 2012;3:125. doi: 10.3389/fmicb.2012.00125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Math RK, et al. Comparative genomics reveals adaptation by Alteromonas sp. SN2 to marine tidal-flat conditions: cold tolerance and aromatic hydrocarbon metabolism. PLoS One. 2012;7:e35784. doi: 10.1371/journal.pone.0035784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Singleton DR, Hu J, Aitken MD. Heterologous expression of polycyclic aromatic hydrocarbon ring-hydroxylating dioxygenase genes from a novel pyrene-degrading betaproteobacterium. Applied and environmental microbiology. 2012;78:3552–3559. doi: 10.1128/AEM.00173-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim S-J, Kweon O, Jones RC, Edmondson RD, Cerniglia CE. Genomic analysis of polycyclic aromatic hydrocarbon degradation in Mycobacterium vanbaalenii PYR-1. Biodegradation. 2008;19:859–881. doi: 10.1007/s10532-008-9189-z. [DOI] [PubMed] [Google Scholar]
- 16.Ferrero M, et al. Coexistence of two distinct copies of naphthalene degradation genes in Pseudomonas strains isolated from the western Mediterranean region. Applied and environmental microbiology. 2002;68:957–962. doi: 10.1128/AEM.68.2.957-962.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kulakov LA, Chen S, Allen CC, Larkin MJ. Web-type evolution of Rhodococcus gene clusters associated with utilization of naphthalene. Applied and environmental microbiology. 2005;71:1754–1764. doi: 10.1128/AEM.71.4.1754-1764.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.DeBruyn JM, Mead TJ, Sayler GS. Horizontal transfer of PAH catabolism genes in Mycobacterium: evidence from comparative genomics and isolated pyrene-degrading bacteria. Environmental science & technology. 2011;46:99–106. doi: 10.1021/es201607y. [DOI] [PubMed] [Google Scholar]
- 19.Cui Z, et al. Isolation and characterization of Cycloclasticus strains from Yellow Sea sediments and biodegradation of pyrene and fluoranthene by their syntrophic association with Marinobacter strains. International Biodeterioration & Biodegradation. 2014;91:45–51. doi: 10.1016/j.ibiod.2014.03.005. [DOI] [Google Scholar]
- 20.Wang B, Lai Q, Cui Z, Tan T, Shao Z. A pyrene‐degrading consortium from deep‐sea sediment of the West Pacific and its key member Cycloclasticus sp. P1. Environmental microbiology. 2008;10:1948–1963. doi: 10.1111/j.1462-2920.2008.01611.x. [DOI] [PubMed] [Google Scholar]
- 21.Hedlund BP, Geiselbrecht AD, Bair TJ, Staley JT. Polycyclic aromatic hydrocarbon degradation by a new marine bacterium, Neptunomonas naphthovorans gen. nov., sp. nov. Applied and Environmental Microbiology. 1999;65:251–259. doi: 10.1128/aem.65.1.251-259.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hedlund BP, Staley JT. Isolation and characterization of Pseudoalteromonas strains with divergent polycyclic aromatic hydrocarbon catabolic properties. Environmental Microbiology. 2006;8:178–182. doi: 10.1111/j.1462-2920.2005.00871.x. [DOI] [PubMed] [Google Scholar]
- 23.Lozupone CA, Knight R. Global patterns in bacterial diversity. Proceedings of the National Academy of Sciences. 2007;104:11436–11440. doi: 10.1073/pnas.0611525104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hedlund BP, Geiselbrecht AD, Staley JT. Marinobacter strain NCE312 has a Pseudomonas-like naphthalene dioxygenase. Fems Microbiology Letters. 2001;201:47–51. doi: 10.1111/j.1574-6968.2001.tb10731.x. [DOI] [PubMed] [Google Scholar]
- 25.Fuenmayor SL, Wild M, Boyes AL, Williams PA. A gene cluster encoding steps in conversion of naphthalene to gentisate in Pseudomonas sp. strain U2. Journal of Bacteriology. 1998;180:2522–2530. doi: 10.1128/jb.180.9.2522-2530.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Johnson GR, Jain RK, Spain JC. Origins of the 2, 4-dinitrotoluene pathway. Journal of bacteriology. 2002;184:4219–4232. doi: 10.1128/JB.184.15.4219-4232.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Flocco CG, Gomes N, Mac Cormack W, Smalla K. Occurrence and diversity of naphthalene dioxygenase genes in soil microbial communities from the Maritime Antarctic. Environmental microbiology. 2009;11:700–714. doi: 10.1111/j.1462-2920.2008.01858.x. [DOI] [PubMed] [Google Scholar]
- 28.Yin J, Chen JC, Wu Q, Chen GQ. Halophiles, Coming Stars for Industrial Biotechnology. Biotechnology Advances. 2014;33:1433–1442. doi: 10.1016/j.biotechadv.2014.10.008. [DOI] [PubMed] [Google Scholar]
- 29.Dassarma S, Dassarma P. Halophiles and their enzymes: Negativity put to good use. Current Opinion in Microbiology. 2015;25:120–126. doi: 10.1016/j.mib.2015.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Guo, G. et al. Isolation and characterization of two novel halotolerant Catechol 2, 3-dioxygenases from a halophilic bacterial consortium. Scientific reports5 (2015). [DOI] [PMC free article] [PubMed]
- 31.Wang C-Y, et al. Purification and characterization of a novel halostable cellulase from Salinivibrio sp. strain NTU-05. Enzyme and Microbial Technology. 2009;44:373–379. doi: 10.1016/j.enzmictec.2009.02.006. [DOI] [Google Scholar]
- 32.Jiang X, et al. Cloning, expression and characterization of a halotolerant esterase from a marine bacterium Pelagibacterium halotolerans B2T. Extremophiles. 2012;16:427–435. doi: 10.1007/s00792-012-0442-3. [DOI] [PubMed] [Google Scholar]
- 33.Gauthier MJ, et al. Marinobacter hydrocarbonoclasticus gen. nov., sp. nov., a new, extremely halotolerant, hydrocarbon-degrading marine bacterium. International Journal of Systematic and Evolutionary Microbiology. 1992;42:568–576. doi: 10.1099/00207713-42-4-568. [DOI] [PubMed] [Google Scholar]
- 34.Gao W, et al. Marinobacter nanhaiticus sp. nov., polycyclic aromatic hydrocarbon-degrading bacterium isolated from the sediment of the South China Sea. Antonie Van Leeuwenhoek. 2013;103:485–491. doi: 10.1007/s10482-012-9830-z. [DOI] [PubMed] [Google Scholar]
- 35.Dastgheib SMM, Amoozegar MA, Khajeh K, Shavandi M, Ventosa A. Biodegradation of polycyclic aromatic hydrocarbons by a halophilic microbial consortium. Applied Microbiology and Biotechnology. 2012;95:789–798. doi: 10.1007/s00253-011-3706-4. [DOI] [PubMed] [Google Scholar]
- 36.Lai Q, Yuan J, Gu L, Shao Z. Marispirillum indicum gen. nov., sp. nov., isolated from a deep-sea environment. International journal of systematic and evolutionary microbiology. 2009;59:1278–1281. doi: 10.1099/ijs.0.003889-0. [DOI] [PubMed] [Google Scholar]
- 37.Zhou H, Wang H, Huang Y, Fang T. Characterization of pyrene degradation by halophilic Thalassospira sp. strain TSL5-1 isolated from the coastal soil of Yellow Sea, China. International Biodeterioration & Biodegradation. 2016;107:62–69. doi: 10.1016/j.ibiod.2015.10.022. [DOI] [Google Scholar]
- 38.Eaton RW, Chapman PJ. Bacterial metabolism of naphthalene: construction and use of recombinant bacteria to study ring cleavage of 1, 2-dihydroxynaphthalene and subsequent reactions. Journal of bacteriology. 1992;174:7542–7554. doi: 10.1128/jb.174.23.7542-7554.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bosch R, García-Valdés E, Moore ER. Genetic characterization and evolutionary implications of a chromosomally encoded naphthalene-degradation upper pathway from Pseudomonas stutzeri AN10. Gene. 1999;236:149–157. doi: 10.1016/S0378-1119(99)00241-3. [DOI] [PubMed] [Google Scholar]
- 40.Hearn EM, Patel DR, Van den Berg B. Outer-membrane transport of aromatic hydrocarbons as a first step in biodegradation. Proceedings of the National Academy of Sciences. 2008;105:8601–8606. doi: 10.1073/pnas.0801264105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang Y, et al. Identification of a membrane protein and a truncated LysR-type regulator associated with the toluene degradation pathway in Pseudomonas putida F1. Molecular and General Genetics MGG. 1995;246:570–579. doi: 10.1007/BF00298963. [DOI] [PubMed] [Google Scholar]
- 42.Kahng H-Y, Byrne AM, Olsen RH, Kukor JJ. Characterization and role of tbuX in utilization of toluene by Ralstonia pickettii PKO1. Journal of bacteriology. 2000;182:1232–1242. doi: 10.1128/JB.182.5.1232-1242.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mooney A, O’Leary ND, Dobson AD. Cloning and functional characterization of the styE gene, involved in styrene transport in Pseudomonas putida CA-3. Applied and environmental microbiology. 2006;72:1302–1309. doi: 10.1128/AEM.72.2.1302-1309.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bateman JN, Speer B, Feduik L, Hartline RA. Naphthalene association and uptake in Pseudomonas putida. Journal of bacteriology. 1986;166:155–161. doi: 10.1128/jb.166.1.155-161.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bugg T, Foght JM, Pickard MA, Gray MR. Uptake and active efflux of polycyclic aromatic hydrocarbons by Pseudomonas fluorescens LP6a. Applied and environmental microbiology. 2000;66:5387–5392. doi: 10.1128/AEM.66.12.5387-5392.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Neher TM, Lueking DR. Pseudomonas fluorescens ompW: plasmid localization and requirement for naphthalene uptake. Canadian journal of microbiology. 2009;55:553–563. doi: 10.1139/W09-002. [DOI] [PubMed] [Google Scholar]
- 47.Miyata N, Iwahori K, Foght JM, Gray MR. Saturable, energy-dependent uptake of phenanthrene in aqueous phase by Mycobacterium sp. strain RJGII-135. Applied and environmental microbiology. 2004;70:363–369. doi: 10.1128/AEM.70.1.363-369.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Resnick S, Lee K, Gibson D. Diverse reactions catalyzed by naphthalene dioxygenase fromPseudomonas sp strain NCIB 9816. Journal of industrial microbiology. 1996;17:438–457. doi: 10.1007/BF01574775. [DOI] [Google Scholar]
- 49.Denome S, Stanley D, Olson E, Young K. Metabolism of dibenzothiophene and naphthalene in Pseudomonas strains: complete DNA sequence of an upper naphthalene catabolic pathway. Journal of bacteriology. 1993;175:6890–6901. doi: 10.1128/jb.175.21.6890-6901.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Siglioccolo A, Paiardini A, Piscitelli M, Pascarella S. Structural adaptation of extreme halophilic proteins through decrease of conserved hydrophobic contact surface. BMC structural biology. 2011;11:1. doi: 10.1186/1472-6807-11-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jeon CO, Park M, Ro H-S, Park W, Madsen EL. The naphthalene catabolic (nag) genes of Polaromonas naphthalenivorans CJ2: evolutionary implications for two gene clusters and novel regulatory control. Applied and environmental microbiology. 2006;72:1086–1095. doi: 10.1128/AEM.72.2.1086-1095.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tittabutr P, Cho IK, Li QX. Phn and Nag-like dioxygenases metabolize polycyclic aromatic hydrocarbons in Burkholderia sp. C3. Biodegradation. 2011;22:1119–1133. doi: 10.1007/s10532-011-9468-y. [DOI] [PubMed] [Google Scholar]
- 53.Zhou N-Y, Al-Dulayymi J, Baird MS, Williams PA. Salicylate 5-hydroxylase from Ralstonia sp. strain U2: a monooxygenase with close relationships to and shared electron transport proteins with naphthalene dioxygenase. Journal of bacteriology. 2002;184:1547–1555. doi: 10.1128/JB.184.6.1547-1555.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Thomas CM, Nielsen KM. Mechanisms of, and barriers to, horizontal gene transfer between bacteria. Nature reviews microbiology. 2005;3:711–721. doi: 10.1038/nrmicro1234. [DOI] [PubMed] [Google Scholar]
- 55.Michener JK, Neves AAC, Vuilleumier S, Bringel F, Marx CJ. Effective use of a horizontally-transferred pathway for dichloromethane catabolism requires post–transfer refinement. Elife. 2014;3:e04279. doi: 10.7554/eLife.04279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Michener JK, Nielsen J, Smolke CD. Identification and treatment of heme depletion attributed to overexpression of a lineage of evolved P450 monooxygenases. Proceedings of the National Academy of Sciences. 2012;109:19504–19509. doi: 10.1073/pnas.1212287109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nicholson CA, Fathepure BZ. Aerobic biodegradation of benzene and toluene under hypersaline conditions at the Great Salt Plains, Oklahoma. Fems Microbiology Letters. 2005;245:257–262. doi: 10.1016/j.femsle.2005.03.014. [DOI] [PubMed] [Google Scholar]
- 58.Johnson M, et al. NCBI BLAST: a better web interface. Nucleic acids research. 2008;36:W5–W9. doi: 10.1093/nar/gkn201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.