Abstract
Fibroblast Activation Protein (FAP) is a membrane-bound serine protease whose expression is often elevated in activated fibroblasts associated with tissue remodeling in various common diseases such as cancer, arthritis and fibrosis. Like the closely related dipeptidyl peptidase DPPIV, the extracellular domain of FAP can be released into circulation as a functional enzyme, and limited studies suggest that the circulating level of FAP correlates with the degree of tissue fibrosis. Here we describe a novel homogeneous fluorescence intensity assay for circulating FAP activity based on a recently identified natural substrate, FGF21. This assay is unique in that it can effectively distinguish endopeptidase activity of FAP from that of other related enzymes such as prolyl endopeptidase (PREP) and was validated using Fap-deficient mice. Structural modeling was used to elucidate the mechanistic basis for the observed specificity in substrate recognition by FAP, but not by DPPIV or PREP. Finally, the assay was used to detect elevated FAP activity in human patients diagnosed with liver cirrhosis and to determine the effectiveness of a chemical inhibitor for FAP in mice. We propose that the assay presented here could thus be utilized for diagnosis of FAP-related pathologies and for the therapeutic development of FAP inhibitors.
Introduction
Fibroblast activation protein, (FAP, also known as FAPα, seprase, or circulating antiplasmin-cleaving enzyme, (EC 3.4.21.B28) is a type II transmembrane serine protease and a member of the S9 family of proline-specific proteases which includes dipeptidyl peptidase IV (DPPIV), DPP8, DPP9, prolyl endopeptidase (PREP, also known as POP) (1–4). FAP was originally identified as a protein highly expressed on the surface of the activated stromal fibroblasts in tumors1–3. In adult mammals, elevated FAP expression in activated fibroblasts is associated with remodeling of tissues at sites of inflammation in disease, including several forms of cancer4–8, rheumatoid arthritis and osteoarthritis9,10, liver disease11–14, inflammatory bowel diseases15,16 and idiopathic pulmonary fibrosis17,18. Furthermore, stromal expression of FAP is linked to immunosuppressive tumor microenvironment and poor prognosis in various cancers, suggesting a role in cancer progression and anti-tumor immunity4–8,19. Although the contribution of FAP in the development of these diseases is not well understood, some preclinical studies support the role of FAP in determining disease severity8,20,21.
Despite intense studies over the past two decades, the identities of physiological substrates for FAP remain elusive. FAP, like its closest relative, DPPIV, exhibits dipeptidyl peptidase activity, cleaving after the proline present at the second position from the N-terminus in various secreted proteins and peptide hormones22. FAP, but not DPPIV, also possesses endopeptidase activity, preferentially cleaving carboxy-terminal to a Gly-Pro sequence23,24. The Gly-Pro requirement results from the unique property of glycine to adopt a positive phi dihedral angle, allowing the P3 residue to avoid steric clashes with the protease23,25. While collagens have repeating Gly-Pro motifs, they are resistant to degradation by FAP except in a denatured or partially processed form (e.g., gelatin)26. Consistent with the role of FAP in regulating collagen turnover, Fap KO mice show elevated collagen levels in pathogenic conditions20,21. FAP has also been proposed to cleave at a specific site near the N-terminus of α2-Antiplasmin (α2AP) to potentiate its activity27. Fibroblast growth factor 21 (FGF21) is a metabolic hormone that signals through the βKlotho and FGFR coreceptors and activation of this pathway is under investigation for the treatment of metabolic diseases, such as type 2 diabetes and non-alcoholic steatohepatitis28. We and others have recently found that FAP also cleaves FGF21 at a specific site proximal to the C-terminus, leading to its inactivation, as this region of the molecule is crucial for binding βKlotho29–31. In both α2AP and FGF21, the specific cleavage site targeted by FAP possesses the consensus Gly-Pro sequence at P2-P1 position, and these amino acid residues are essential for cleavage by FAP32.
Although FAP is produced as a membrane-bound protein, the extracellular domain encoding the active enzyme can be shed from the cell surface, and therefore soluble FAP protein is readily detectable in serum and plasma by a standard sandwich ELISA. The level of FAP protein has been shown to be elevated in patients with cirrhosis12,33,34, suggesting a diagnostic value to measuring circulating FAP levels in these and perhaps other diseases. Another approach to measure FAP protein levels is based on enzymatic activity, in contrast to the activity-independent ELISA. FAP protein can be isolated from tissue or blood samples by immunocapture with an FAP-specific antibody, followed by a general fluorescence intensity assay for dipeptidyl-peptidases using a peptide substrate attached to a chemically quenched dye, such as Z-Gly-Pro-7-amido-4-methylcoumarin (AMC) or Ala-Pro-7-amino-4-trifluoromethyl-coumarin (AFC)13,21,35,36. The immunocapture step is necessary to eliminate other related DPP enzymes from the reaction. Alternatively, endopeptidase substrates (e.g. Acetyl-Ala-Gly-Pro-AFC, MEPLGRQLTSGP-AMC, etc.) containing the consensus Gly-Pro dipeptide have been used without immunocapture37–39. These substrates are likely targeted also by other circulating proline-specific endopeptidases such as PREP that could be present in the reaction. Nonetheless, this type of assay could be appropriate for characterization of a broad S9 protease inhibitor such as Talabostat (also known as Val-boro-Pro or PT100)37,38. A fluorescence-based homogeneous assay reagent to specifically monitor FAP activity called 3144-AMC, ARI-3144 or “N-terminally blocked FAP specific substrate” has also been described by William Bachovchin and his collaborators11,14,29,40. However, the identity of this substrate remains unpublished and not readily available to a broader community. Here we describe a novel homogeneous fluorescence intensity assay for circulating FAP activity. This assay utilizes a modified peptide substrate based on the endopeptidase cleavage site of FGF21, a newly identified natural substrate for FAP, in a quenched dye format and is selective for FAP.
Results
An FGF21-based quenched-fluorescence peptide is cleaved by FAP and PREP
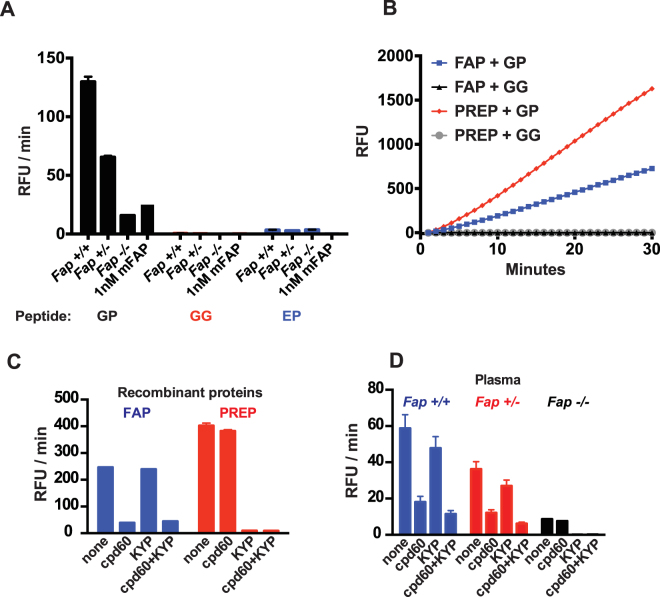
A peptide containing the six amino acid residues surrounding the FAP cleavage site near the C-terminus of human FGF21, termed the “GP” probe, (Fig. 1) was synthesized for the purpose of monitoring FAP endopeptidase activity. The peptide is flanked by a FRET-donor (HyLite Fluor 488) and a dark quencher (QXL 520). By design, fluorescence intensity is suppressed due to the close proximity of the quencher dye to the donor fluorophore, and it is liberated by protease-catalyzed cleavage of the peptide. As controls, variant peptides containing a substitution of the P1 proline with glycine (“GG” probe) or the homologous region of murine FGF21 (“EP” probe) were also generated. Both control probes lack the Gly-Pro consensus necessary for FAP-based cleavage, thus serve as negative controls (Fig. 1). These three peptides were used to evaluate the FAP endopeptidase activity in the plasma of wild type (WT), heterozygous and Fap-deficient (KO) mice. We have previously demonstrated by immunoblot analysis that FAP is present in the plasma from WT mice, to a lesser degree in heterozygotes and undetectable in KO mice29. When the GP probe was used, plasma from WT mice produced a strong signal, indicating efficient cleavage of the probe (Fig. 2A). As expected, plasma from Fap heterozygous mice exhibited approximately half the activity as that from WT mice, and that from homozygous Fap KO mice exhibited even lower, but significant activity. When plasma from WT mice was tested using GG and EP probes, no and a minimal signal was obtained respectively. Furthermore, purified recombinant mouse FAP protein cleaved the GP but not GG or EP probes (Fig. 2A).
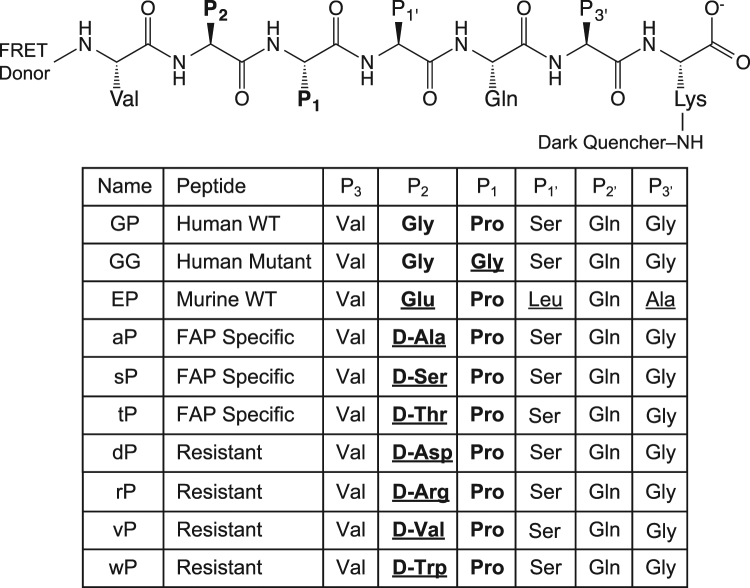
Figure 1.
Design of fluorescence-quenched peptides. Peptides contain an N-terminal fluorescent donor, followed by six amino acid residues of the region flanking the dominant FAP endopeptidase cleavage site of human FGF21, ending with an additional C-terminal lysine conjugated to a dark quencher. Variants include substitution of the P1 proline with glycine, the P2 glycine with D-alanine or the entire homologous region of murine FGF21.
Figure 2.
Plasma cleavage of fluorescence-quenched peptides. (A) Cleavage rate of human WT (GP), human mutant (GG) and murine (EP) FGF21-based peptides in plasma samples isolated from Fap +/+, Fap +/−, and Fap −/− mice, as indicated (N = 3 mice per genotype). As a control, the cleavage reaction was run with 1 nM recombinant mouse FAP. Concentration of peptide substrates in this experiment was 3 µM. (B) Cleavage rate of the GP peptide by recombinant human FAP or PREP proteins at 1 nM enzyme concentration. (C) Cleavage rate of the GP peptide by recombinant human FAP (left, blue) or PREP (right, red) in the presence of an FAP-specific inhibitor cpd60 and/or PREP-specific inhibitor KYP-2047 (KYP). (D) Cleavage rate of the GP peptide in plasma from Fap +/+ (blue), Fap +/− (red), and Fap −/− (black) mice in the presence of cpd60 and/or KYP (N = 3 per group).
The ability of Fap KO mouse plasma to cleave the GP probe and to a much lesser degree, the EP probe, but not the GG probe, suggested the presence of other proline-specific endopeptidases in plasma. Although prolyl endopeptidase (PREP) is predominantly a cytosolic enzyme, it is known to be present at variable levels in plasma41,42 and seemed a likely candidate for the source of residual probe cleavage by the KO plasma. Indeed, when 1 nM of recombinant human FAP or PREP was tested, both enzymes readily cleaved the GP peptide, while leaving the mutant GG peptide intact (Fig. 2B). To further investigate the involvement of PREP in the cleavage of the GP probe, we used an FAP-selective inhibitor, cpd6030,43, and a PREP-selective inhibitor KYP-204744,45 (Supplementary Fig. 1). Both inhibitors were tested at a concentration that provides selective inhibition of each enzyme (Fig. 2C). Using these inhibitors, we found that the residual GP probe cleaving activity in Fap KO sample was sensitive to KYP, but not cpd60 (Fig. 2D). At the same time, the GP probe activity in WT or heterozygous samples were clearly sensitive to cpd60. Taken together, we concluded that FAP is the major enzyme responsible for the cleavage of the GP probe in plasma, while PREP has a smaller but significant contribution.
Impact of P2 D-Ala substitution on FAP cleavage kinetics
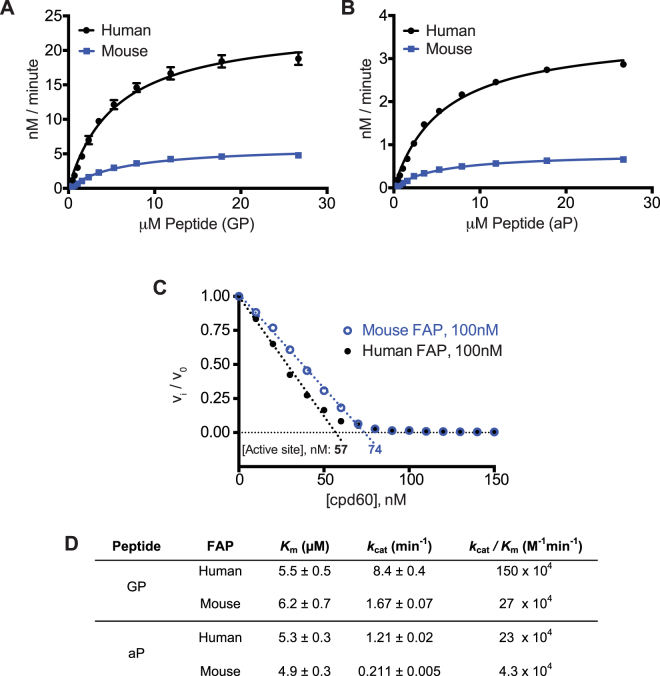
FAP has previously been shown to tolerate a change of the consensus glycine to a D-alanine at position P2 in a α2AP-based peptide substrate, although a weaker catalytic efficiency was observed relative to the parental peptide due to a decrease in k cat of approximately eight-fold23. In the context of FAP inhibitors, this substitution has been reported to impart specificity for FAP over PREP40. In an attempt to increase the specificity of our assay, we incorporated this substitution into the FGF21-based GP peptide to produce the “aP” probe (Fig. 1). For an activity probe to be of practical use for a diagnostic purpose, it is important that this change does not have a strong negative impact on the enzyme kinetics. We, therefore, compared the kinetics for both the GP and aP peptides, using both human and mouse FAP proteins (Fig. 3A,B & D).
Figure 3.
Cleavage kinetics of GP and aP substrates by recombinant FAP. (A and B) Michaelis-Menten saturation curves for human (black) or mouse (blue) recombinant FAP cleavage of (A) the GP or (B) aP peptides. (C) Plot of fractional velocity as a function of cpd60 concentration, for recombinant mouse or human FAP with cpd60, using GP peptide. Mouse and human FAP preparations were found to be 74% and 57% active, respectively. (D) Human and mouse FAP enzyme kinetics constants with the GP and aP substrates, corrected for experimentally determined active enzyme concentrations determined in (C). Error is SEM, determined from three experimental replicates. FAP molar concentrations are based on monomer for active site titration and kinetics calculations.
Interestingly, mouse FAP exhibited a lower catalytic efficiency than human FAP, mainly due to a reduced k cat (Fig. 3A & B). From the kinetics data alone, it was not clear if this is an innate efficiency difference between the orthologs or if it was due to differences in active enzyme concentration between the two preparations. To address this question, an active site titration was performed, using the tight binding inhibitor, cpd60 (Fig. 3C). We found that the preparation of mouse FAP had, in fact, a higher relative concentration of active enzyme, confirming its intrinsically weaker catalytic efficiency. The kinetics data shown in Fig. 3D are thus corrected for the experimentally determined active enzyme concentration. The K m value is essentially unchanged between the parental (GP) and aP peptides, and we observed the expected seven to eight-fold decrease in k cat for aP cleavage25.
P2 D-Ala substitution provides FAP specificity
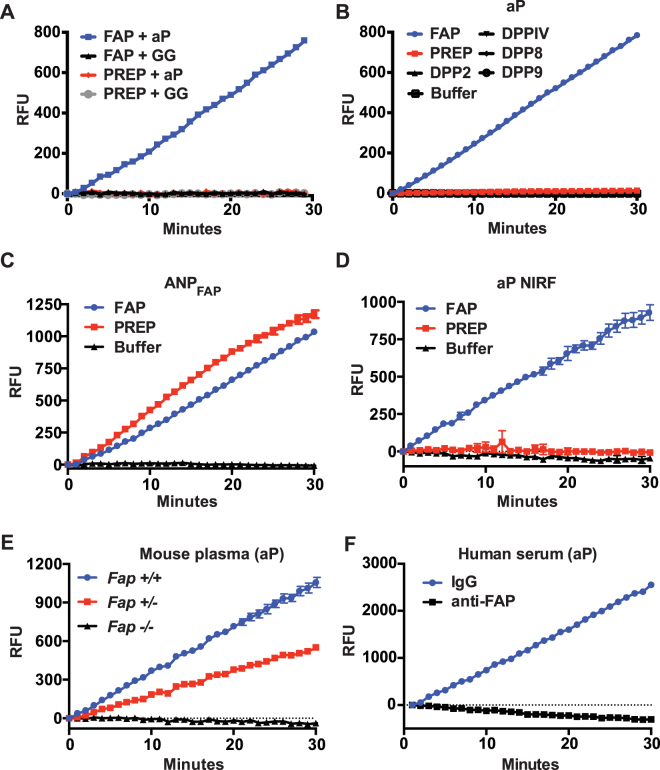
Encouraged by the kinetics data, we decided to test the specificity of the aP probe against PREP and a panel of related peptidases. Using a concentration of 1 nM enzyme, cleavage of the aP probe by human FAP is clearly detectable, but not the GG probe (Fig. 4A). In contrast, human PREP is unable to digest either probe. In addition, a broader set of human S9 and S28 peptidases, did not exhibit an ability to cleave the aP probe (Fig. 4B). This is consistent with the general lack of endopeptidase activity of these prolyl dipeptidyl peptidases.
Figure 4.
Specificity of aP peptide cleavage. (A) Cleavage rate of the aP or GG peptide by 1 nM recombinant, human FAP or PREP. (B) Cleavage rate of the aP peptide by a panel of recombinant, human prolyl peptidases. Each recombinant peptidase was used at 2.5 µg/ml. (C and D) Cleavage rate of ANPFAP (C) or aP NIRF (D) by 10 nM recombinant, human FAP or PREP. (E) Cleavage rate of the aP peptide in plasma from Fap +/+, Fap +/−, and Fap −/− mice. Plasma was diluted 10-fold. (F) Cleavage rate of the aP peptide in anti-FAP or control IgG-immunodepleted human serum. Serum was diluted 2.5-fold. Complete FAP removal following anti-FAP immunodepletion was shown previously by immunoblotting29.
Another internally-quenched FRET peptide substrate (ANPFAP) for FAP has recently been reported and demonstrated for use as an activity-based, in vivo imaging tool39. The peptide sequence contains two internal Gly-Pro dipeptide motifs, susceptible to FAP cleavage and a Cy5.5/QSY21, quenched-FRET pair. To evaluate specificity, the authors tested ANPFAP for cleavage in vitro by FAP, DPPIV and MMP-2, but only detected cleavage in the presence of FAP. Our experience with the GP probe led us to suspect that the Gly-Pro motifs in ANPFAP are also recognized by PREP. Therefore, ANPFAP was synthesized and tested for cleavage by recombinant human FAP and PREP (Fig. 4C). As suspected, both FAP and PREP efficiently cleave ANPFAP in vitro. While it is currently unclear how much of a liability PREP or other non-specific prolyl endopeptidase activity might be in the context of in vivo FAP imaging, we were interested to see if our aP probe would retain its specificity and FAP cleavability if the FRET pair was replaced with near infrared fluorophores (NIRF) more suitable to in vivo imaging. A new peptide was generated (aP NIRF), identical to aP, but the FRET donor and quencher were replaced with Cy5.5 and QSY21, respectively. In contrast to ANPFAP, we found that FAP, but not PREP, efficiently cleaves aP NIRF (Fig. 4D).
Finally, having demonstrated specificity using several purified recombinant enzymes, we returned to the analysis of the WT and Fap KO mice plasma as in Fig. 2A, but using the aP probe. As expected, the aP probe shows a clear signal with WT plasma, intermediate with heterozygous plasma and satisfyingly, the aP probe exhibits no cleavage with Fap KO plasma samples (Fig. 4E). Furthermore, the signal is lost after FAP immunodepletion from human serum, but not in control IgG-immunodepleted serum (Fig. 4F).
Molecular determinants for the substrate specificity of FAP
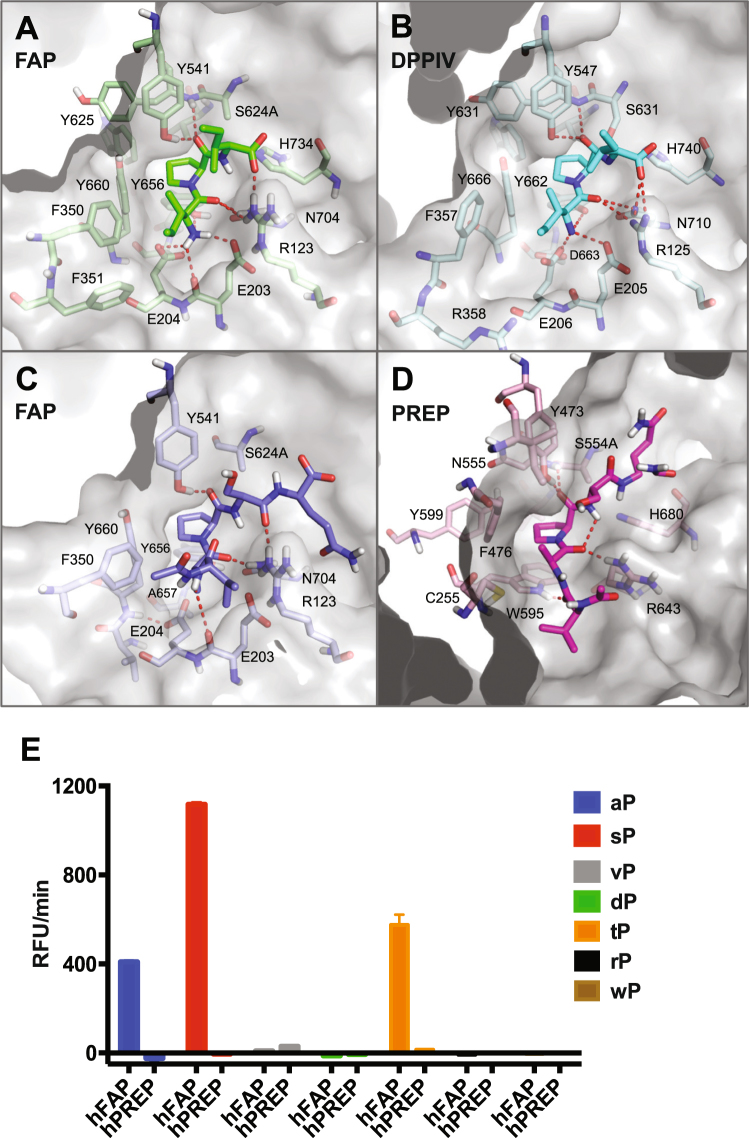
In order to elucidate the mechanistic basis for the observed aP probe recognition by FAP but not by DPP4 or PREP, a model of peptide-bound FAP was generated by superposition of the apo FAP coordinates onto diprotin A (Ile-Pro-Ile or IPI) tripeptide-bound DPPIV, with the active site side chain of S624 of FAP replaced by an alanine residue46,47. This model of FAP shows the coordination of the N-terminal amine at P2 (E203, E204 and Y656), coordination of the P2 carbonyl oxygen (R123 and N704), positioning of the P1 proline side chain in a hydrophobic pocket (Y625, V650, W653, Y656 and Y660), and coordination of the proline P1 carbonyl (Y541 and Y625 back bone NH) so as to position the scissile bond adjacent to the catalytic residue S624 (Fig. 5A). As a result of this binding mode, the side chain of the P2 residue is directed into the space proximal to F350 and F351. This is highly reminiscent of the substrate binding mode of DPPIV (Fig. 5B), and explains how both enzymes can act as dipeptidases for proteins terminating with an L-amino acid at P2 and a proline at P1.
Figure 5.
Binding mode of peptide substrates to FAP, DPPIV and PREP. (A) FAP active site (grey surface and pale green sticks) bound to modeled IPI substrate tripeptide (green sticks). (B) DPPIV active site (grey surface and pale cyan sticks) bound to IPI substrate tripeptide (cyan sticks). (C) FAP active site (grey surface and pale blue sticks) bound to modeled acetyl-VaPSQ substrate peptide (blue sticks). (D) Representative induced-fit docking model of PREP (grey surface and pale pink sticks) bound to acetyl-VAPSQ-amide substrate peptide (pink sticks). In all panels, red dashed lines indicate intermolecular and intra-substrate hydrogen bonds. (E) Comparison of cleavage rates by human FAP on VxPSQG peptide substrates containing various D-enantiomer residues at the P2 position (xP).
Induced fit docking was used to generate hypotheses for how FAP could also act as an endopeptidase. Residues within 5 Å of the IPI peptide were allowed to move during the docking calculation to accommodate the acetyl-VaPSQ substrate peptide. Three of the top 5 scoring ligand poses are able to accommodate the P2 side chain of the substrate by a modest reorientation of the E204 side chain towards A657 (χ1 changes from 180° to −60° and χ2 from +60° to 180°) with the concomitant formation of a hydrogen bond from the E204 carboxylate to the backbone NH of Y660 (Fig. 5C). These three ligand poses all have the substrate P2 residue with a positive backbone phi dihedral angle, the amide proton within hydrogen bonding distance of E203 (backbone NH or side chain carboxylate, depending on pose) and the alanine side chain in the pocket of space generated by relocation of the E204 side chain. Importantly, this binding mode readily accommodates acylation of the P3 amine with the additional N-terminal residues directed into the large space between the α/β hydrolase and β-propeller domains of FAP. This binding mode explains the ability of FAP to act as an endopeptidase for substrates with small D-amino acids at the P2 position. Moreover, such a binding mode would not be possible in DPP4 since A657 is replaced by the larger D663 and hence E206 (the equivalent of E204 in FAP) has no room to reorient, as has been suggested by others23,25,47. Indeed, a D663A substitution in DPPIV increases the endopeptidase activity of DPPIV while the corresponding A657D substitution in FAP decreases the endopeptidase activity of this enzyme47.
The substrate binding mode of PREP has been revealed in some co-crystal structures in which the active site serine residue has been mutated to alanine48,49. Conserved substrate interactions include a hydrogen bond from the P3 carbonyl to W595 indole, a hydrogen bond from the P2 carbonyl to R643 and an intra-substrate hydrogen bond from the P2 carbonyl to the P’1 amide NH (Fig. 5D). As a result of this hydrogen bond network, the substrate P2 residue is constrained to a have a negative phi backbone dihedral angle with the P2 side chain directed towards F476. A D-amino acid at P2 would not be accommodated due to the high energy of forcing such a residue to have a negative phi, and a steric clash since the P2 side chain would be orientated directly at the side chain of C255. Thus, the specific substrate-binding mode required by PREP prevents it from recognizing and cleaving VxPSQ or other substrates containing a D-amino acid residue at P2 position.
To validate the molecular modeling, relative cleavage rates of human FAP and PREP were experimentally determined using substrates containing a variety of D-residues at the P2 position (Fig. 5E). We found that FAP can cleave substrates containing D-Ser or D-Thr, in addition to D-Ala, at the P2 position. As predicted from the structural modeling, none of these D-residue containing peptides were cleaved by PREP to an appreciable degree.
The diagnostic utility of aP probe in analyzing FAP activity in blood
Previously, serum FAP levels and activity have been shown to correlate with liver fibrosis11. Therefore, we decided to determine if we can recapitulate these findings by the aP probe-based fluorescence assay. We obtained serum samples from healthy volunteers and from individuals who had been diagnosed with non-viral liver cirrhosis. The observed FAP activity correlated well with FAP protein level (R2 = 0.76, p values < 0.05) and using the activity assay, we were easily able to detect levels as low as 63 ng/ml (Fig. 6A). When the healthy and disease groups were compared, we found a measurable increase in FAP activity as determined using the aP probe, P < 0.05 (Fig. 6B) and in FAP levels determined by a conventional sandwich ELISA, P < 0.0001 (Fig. 6C).
Figure 6.
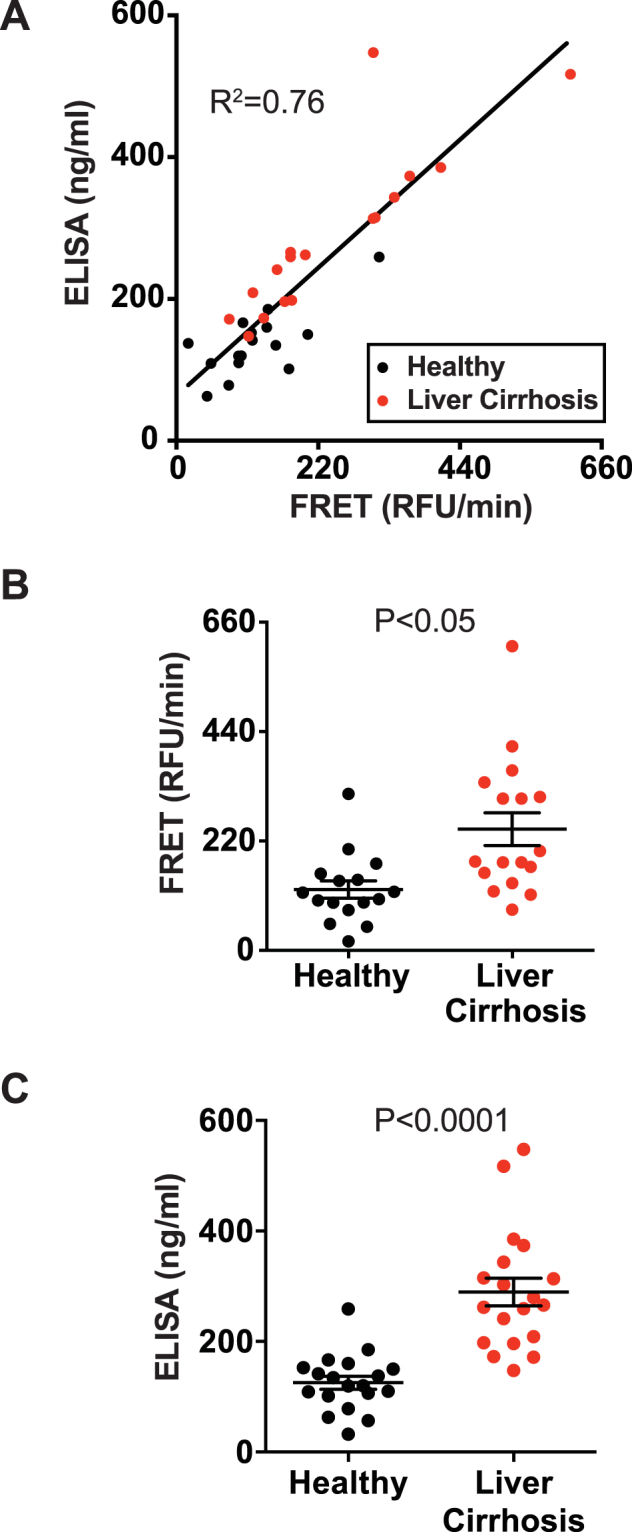
FAP activity in serum correlates with liver disease and FAP levels. (A) Scatter plot showing FAP activity levels determined by using the aP peptide (x axis) and FAP protein levels determined by ELISA in serum samples (y axis) from healthy individuals (N = 16, black circles) or individuals previously diagnosed with liver cirrhosis (N = 17, red circles). R2 value is indicated in the graph. (B and C) The same data as (A), with FRET and ELISA data presented separately, including mean ± SEM. P values indicated were calculated by student t-test.
We next wanted to determine whether the new aP-based assay can be used to determine the pharmacodynamic activity of FAP inhibitors in vivo. To do this, we fed mice with chow containing the FAP inhibitor cpd60 or a control diet without the inhibitor (Fig. 7). The FAP activity was determined in plasma isolated at days 3 and 7 after cpd60 feeding started. The results clearly showed that at 20 ppm, cpd60-feeding could strongly suppress circulating FAP activity in mice, while at 100 ppm suppression was complete. Therefore, we conclude that the aP-based assay can be used to evaluate the pharmacodynamics of FAP inhibitors selectively, without interference from PREP activity potentially present in plasma.
Figure 7.
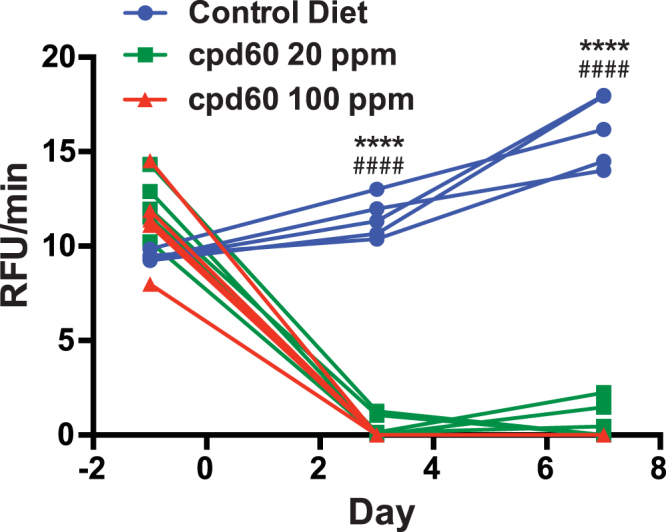
Plasma FAP activity is completely suppressed in mice fed chow with cpd60. Ten to eighteen-week old C57/BL6 background mice were fed ad libitum control chow (blue circles), diet containing 20 ppm of cpd60 (green squares), or diet containing 100 ppm of cpd60 (red triangles). Plasma was isolated from blood retrieved at the indicated time points and FAP activity levels determined using the aP peptide. N = 5 animals per group. P values indicated were calculated by student t-test (****P < 0.0001 for 20 ppm cpd60 vs control diet; ####P < 0.0001 for 100 ppm cpd60 vs control diet).
Discussion
Clinical development of an enzyme inhibitor as a therapeutic agent can be significantly facilitated by the ability to determine the activity of the enzyme in biological samples in a homogenous assay format. First, identification of patients with high levels of active enzyme may support the discovery of a patient sub-population who might benefit most from the use of an inhibitor or an enzyme-targeted anti-tumor agent, such as the recently described anti-DR5/FAP bispecific antibody50. This aspect of drug development has become ever more important in the era of precision medicine. In addition, the ability to measure the effect of an inhibitor in clinical trial subjects would enhance the understanding of the relationship between pharmacokinetics and pharmacodynamics; an aspect that is vital in selecting an appropriate dose regimen to balance drug efficacy and safety. In this regard, one major challenge in the clinical development of FAP inhibitors has been the lack of a selective homogenous assay for FAP activity.
The new FRET assay based on an FGF21 peptide containing the D-Ala-Pro dipeptide described here provides several notable advancements. First and foremost, the assay enables the measurement of FAP endopeptidase activity in serum or plasma samples without immunocapture or any other purification steps, even for samples that contain related S9 proteases, most notably PREP. Second, because of its strict specificity, the activity assay for FAP could substitute for the more cumbersome sandwich ELISA, at least for certain applications. Indeed, we observed a good correlation between FAP activity and FAP protein levels in human serum samples from both healthy individuals and patients with cirrhosis (Fig. 6). Thus, the FAP activity assay could save time and cost for investigators who are interested in determining FAP levels in samples that do not contain FAP inhibitors. Finally, the aP probe can be synthesized from commonly used fluorescent dyes and amino acids without any specialized methods. This means that the probe can be ordered from a peptide probe vendor through a custom synthesis request. This increases the accessibility, practicality and utility of the new assay method.
As a practical application of the new assay, we demonstrated that an FAP-selective inhibitor cpd6038 can be used in vivo to suppress the activity of circulating FAP in a sustained fashion in mice. We previously tested this compound in cynomolgus monkeys via bolus p.o. administration and achieved a transient decrease in FAP-related activity, consistent with the rapid clearance of this compound from circulation29. In this study, therefore, we tested the strategy of feeding cpd60-containing chow to mice and used the aP probe to track the resulting pharmacodynamics. The results indicated that cpd60 suppressed FAP activity in mice to undetectable levels. The ability of the aP probe to monitor changes in FAP activity without an influence of the residual PREP activity allowed us to demonstrate that cpd60 could completely suppress FAP at the dose we tested. We foresee that this feeding-based approach will promote pre-clinical investigation of the role of FAP to regulate endopeptidic substrates, such as α2AP and FGF21, as well as its role in diseases for which FAP has been implicated.
One of the most critical aspects of our new substrate probe described above is its ability to provide a strict FAP selectivity over highly related dipeptidyl peptidases such as DPPIV as well as more distally related endopeptidases such as PREP. To understand the mechanistic basis for the observed selectivity, we have compiled structural models of FAP, DPPIV and PREP with critical features of a peptide substrate included and experimentally substantiated these models. Our models highlight previously undescribed structural differences in substrate recognition by FAP and PREP, and the logic that enables recognition of peptides with a D-amino acid at P2 position by FAP but not by PREP.
In conclusion, we have developed a unique, accessible and specific homogenous assay for the endopeptidase activity of FAP. We propose that the assay presented here could guide preclinical investigations of FAP biology and the clinical development of FAP inhibitors or FAP-localized anti-tumor therapies in the future. As a complement to the in vitro assay, we are also investigating the potential utility of aP NIRF for in vivo imaging.
Methods
Ethical approval
All animal studies were conducted in accordance with the Guide for the Care and Use of Laboratory Animals, published by the National Institutes of Health (NIH) (NIH Publication 8523, revised 1985). The Institutional Animal Care and Use Committee (IACUC) at Genentech reviewed and approved all animal protocols. All the human samples were de-identified to investigators.
Recombinant proteins
Human prolyl oligopeptidase (PREP, 4308-SE), DPPIV (1180-SE), DPP9 (5419-SE), and DPPII/DPP7 (3438-SE) proteins were purchased from R&D Systems. Human DPP8 (BML-SE527) protein was purchased from Enzo Life Sciences. N-terminal octa-His-tagged human and mouse FAP proteins containing the extracellular domain (residues L26-D760 and L26-D761, respectively) were purified from the conditioned media of transiently-transfected CHO cells, by immobilized metal affinity chromatography (IMAC), followed by size exclusion chromatography (SEC).
Inhibitors
The FAP-specific inhibitor (S)-N-(2-cyano-4,4-difluoropyrrolidin-1-yl)-2-oxoethyl)-quinoline-4-carboxamide (cpd60) was synthesized according to the experimental procedure reported by Jansen et al.43. The PREP-specific inhibitor KYP-2047 was purchased from Sigma-Aldrich (St. Louis, MO).
In vivo studies
Mice were maintained in a pathogen-free animal facility at 21 °C under standard 12 h light/12 h dark cycle with access to normal chow (Labdiet 5010) and water ad libitum unless otherwise indicated. Generation of Fap KO mice was described previously30. For in vivo pharmacological FAP inhibition, C57BL/6 background mice were fed ad libitum control chow (TD.00588, Envigo, Madison, WI), or the same diet containing 20 or 100 ppm of cpd60.
Plasma and serum
Mouse plasma samples were prepared in K3-EDTA MiniCollect tubes (Greiner Bio-One, 450475) and frozen immediately at −80 °C before analysis. De-identified human serum samples were obtained from healthy donors through the Employee Donation Program at Genentech, Inc. The clinically diagnosed cirrhosis patient samples and healthy donor samples were purchased from Discovery Life Sciences (Los Osos, CA).
FAP ELISA
Serum FAP concentration was measured using the human FAP DuoSet ELISA kit (R&D Systems, DY3715), according to the manufacturer’s protocol.
Fluorescence resonance energy transfer (FRET)-quench assay
Peptides of the region flanking the C-terminal human FGF21 cleavage site (VGPSQG) or variants with D-amino acid at P2 were synthesized (Anaspec, Inc.), containing an amine-terminal donor (HyLite Fluor 488 or Cy5.5 for aP NIRF) and a dark quencher (QXL 520 or QSY21 for aP NIRF) conjugated to a supplementary C-terminal lysine. Additional peptides were generated as above, with the residue sequences shown in Fig. 1. ANPFAP 39 and aP NIRF were synthesized by CPC Scientific (Sunnyvale, CA). Assays were conducted at 37 °C in 50 mM HEPES (pH 7.2), 150 mM NaCl, 1 mM EDTA, 0.1 mg/ml BSA. The excitation/emission wavelengths of cleaved peptide are 490/520 nm or 675/695 nm for ANPFAP and aP NIRF. Fluorescence was monitored on a Tecan M1000 Pro plate reader in kinetic mode. Except where indicated otherwise, measurements of serum or plasma FAP activity were performed with the peptide substrate concentration at 6 µM and a 10-fold dilution of serum or plasma in assay buffer.
Enzyme kinetics
Kinetics measurements were carried out using 5 nM human or mouse recombinant FAP, with a series of peptide substrate titrations. The initial rate of substrate hydrolysis was determined using the Magellan software on a Tecan M1000 Pro plate reader and kinetic parameters were modeled using nonlinear regression analysis with GraphPad Prism software. Standard error was calculated from three experimental replicates.
Active site titration
The tight-binding FAP inhibitor, cpd60, was titrated into a GP peptide cleavage reaction with 100 nM of mouse or human recombinant FAP and 6 µM of GP. The IC50 of cpd60 for human and mouse FAP endopeptidase activity was previously determined to be <0.5 nM30. As described51, the active enzyme concentration was determined from the x-intercept.
Statistics
Unpaired student’s t-test (two-tailed) was used for statistical analysis to compare treatment groups. A p value < 0.05 was considered statistically significant. All the values were presented as means ± SEM unless otherwise noted.
Protein modeling
The Schrödinger Small-Molecule Drug Discovery Suite 2017-1, (Schrödinger, LLC, New York, NY, 2017) was used for modeling studies. Protein coordinate files were prepared using the default protein preparation workflow, including minimization of the coordinates using the OPLS3 force field52,53. Superposition of the IPI-peptide-bound coordinates of DPPIV (PDB Accession Number 1NU8) with those of apo FAP (PDB Accession Number 1Z68) allowed placement of the IPI peptide into the apo FAP structure. The FAP active site serine (S624) was replaced by alanine and the peptide-bound model of FAP was further refined by minimization. A model for PREP bound to an acetyl-VAPSQ-amide peptide was generated from peptide-bound PREP coordinates (PDB Accession Protein 1E8N); the peptide ligand was modified manually and the complex minimized. All structure figures were prepared using the PyMOL Molecular Graphics System, Version 1.8 (Schrödinger, LLC).
Ligand docking calculations
The Schrödinger Induced Fit docking workflow was used to generate models of the acetyl-VaPSQ peptide bound to FAP54. The docking protocol employed a core restraint to localize the position of the proline in the active site (to within 0.5 Å of its location in the IPI peptide model) and a hydrogen bond restraint to the phenolic hydroxyl proton of Y541. FAP residues within 5 Å of the IPI ligand were allowed to move during the docking protocol, with the exception of several key residues involve in coordinating the substrate in the active site (R123, E203, Y541, A624 and H734). The docking grid was centered on the IPI peptide; default settings were used for all other aspects of the protocol. 20 final poses were generated.
Data and materials availability
Datasets and materials generated during and/or analyzed during the current study are available from the corresponding author upon reasonable request.
Electronic supplementary material
Acknowledgements
We thank Genentech colleagues in Molecular Biology, Protein Chemistry, Bio-Molecular Engineering Group and Laboratory Animal Resources for technical assistance, and Suzette Weber for assistance formulating the cpd60 diet.
Author Contributions
T.W.B., D.R.D., J.S., J.A.E. conceived the project. D.R.D., T.W.B., N.M.K. performed assays. N.J.S. performed the computational studies. T.W.B. and J.A.E. contributed to purification of recombinant proteins. D.R.D. and N.M.K. performed in vivo experiments. T.W.B., D.R.D., N.J.S., J.S. and J.A.E. drafted the manuscript. All authors reviewed the results and approved the final version of the manuscript.
Competing Interests
This work was funded by Genentech, Inc. All the authors are present or former paid employees of Genentech/Roche. Genentech has filed patent applications related to this work.
Footnotes
Travis W. Bainbridge and Diana Ronai Dunshee contributed equally to this work.
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-017-12900-8.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Junichiro Sonoda, Email: sonoda.junichiro@gene.com.
James A. Ernst, Email: ernst.james@gene.com
References
- 1.Scanlan MJ, et al. Molecular cloning of fibroblast activation protein alpha, a member of the serine protease family selectively expressed in stromal fibroblasts of epithelial cancers. Proc Natl Acad Sci USA. 1994;91:5657–5661. doi: 10.1073/pnas.91.12.5657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rettig WJ, et al. Cell-surface glycoproteins of human sarcomas: differential expression in normal and malignant tissues and cultured cells. Proc Natl Acad Sci USA. 1988;85:3110–3114. doi: 10.1073/pnas.85.9.3110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rettig WJ, et al. Regulation and heteromeric structure of the fibroblast activation protein in normal and transformed cells of mesenchymal and neuroectodermal origin. Cancer Res. 1993;53:3327–3335. [PubMed] [Google Scholar]
- 4.Henry LR, et al. Clinical implications of fibroblast activation protein in patients with colon cancer. Clin Cancer Res. 2007;13:1736–1741. doi: 10.1158/1078-0432.CCR-06-1746. [DOI] [PubMed] [Google Scholar]
- 5.Shi M, et al. Expression of fibroblast activation protein in human pancreatic adenocarcinoma and its clinicopathological significance. World J Gastroenterol. 2012;18:840–846. doi: 10.3748/wjg.v18.i8.840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ha SY, Yeo SY, Xuan YH, Kim SH. The prognostic significance of cancer-associated fibroblasts in esophageal squamous cell carcinoma. PLoS One. 2014;9:e99955. doi: 10.1371/journal.pone.0099955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kim GJ, et al. Increased expression of CCN2, epithelial membrane antigen, and fibroblast activation protein in hepatocellular carcinoma with fibrous stroma showing aggressive behavior. PLoS One. 2014;9:e105094. doi: 10.1371/journal.pone.0105094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen, L., Qiu, X., Wang, X. & He, J. FAP positive fibroblasts induce immune checkpoint blockade resistance in colorectal cancer via promoting immunosuppression. Biochem Biophys Res Commun, 10.1016/j.bbrc.2017.03.039 (2017). [DOI] [PubMed]
- 9.Milner JM, et al. Fibroblast activation protein alpha is expressed by chondrocytes following a pro-inflammatory stimulus and is elevated in osteoarthritis. Arthritis Res Ther. 2006;8:R23. doi: 10.1186/ar1877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bauer S, et al. Fibroblast activation protein is expressed by rheumatoid myofibroblast-like synoviocytes. Arthritis Res Ther. 2006;8:R171. doi: 10.1186/ar2080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Williams KH, et al. Lower serum fibroblast activation protein shows promise in the exclusion of clinically significant liver fibrosis due to non-alcoholic fatty liver disease in diabetes and obesity. Diabetes Res Clin Pract. 2015;108:466–472. doi: 10.1016/j.diabres.2015.02.024. [DOI] [PubMed] [Google Scholar]
- 12.Wang XM, et al. Fibroblast activation protein and chronic liver disease. Front Biosci. 2008;13:3168–3180. doi: 10.2741/2918. [DOI] [PubMed] [Google Scholar]
- 13.Levy MT, et al. Fibroblast activation protein: a cell surface dipeptidyl peptidase and gelatinase expressed by stellate cells at the tissue remodelling interface in human cirrhosis. Hepatology. 1999;29:1768–1778. doi: 10.1002/hep.510290631. [DOI] [PubMed] [Google Scholar]
- 14.Keane FM, et al. Quantitation of fibroblast activation protein (FAP)-specific protease activity in mouse, baboon and human fluids and organs. FEBS Open Bio. 2013;4:43–54. doi: 10.1016/j.fob.2013.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Giuffrida P, Pinzani M, Corazza GR, Di Sabatino A. Biomarkers of intestinal fibrosis - one step towards clinical trials for stricturing inflammatory bowel disease. United European Gastroenterol J. 2016;4:523–530. doi: 10.1177/2050640616640160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rovedatti L, et al. Fibroblast activation protein expression in Crohn’s disease strictures. Inflamm Bowel Dis. 2011;17:1251–1253. doi: 10.1002/ibd.21446. [DOI] [PubMed] [Google Scholar]
- 17.Acharya PS, Zukas A, Chandan V, Katzenstein AL, Pure E. Fibroblast activation protein: a serine protease expressed at the remodeling interface in idiopathic pulmonary fibrosis. Hum Pathol. 2006;37:352–360. doi: 10.1016/j.humpath.2005.11.020. [DOI] [PubMed] [Google Scholar]
- 18.Egger C, et al. Effects of the fibroblast activation protein inhibitor, PT100, in a murine model of pulmonary fibrosis. Eur J Pharmacol. 2017;809:64–72. doi: 10.1016/j.ejphar.2017.05.022. [DOI] [PubMed] [Google Scholar]
- 19.Lopez JI, et al. Fibroblast activation protein predicts prognosis in clear cell renal cell carcinoma. Hum Pathol. 2016;54:100–105. doi: 10.1016/j.humpath.2016.03.009. [DOI] [PubMed] [Google Scholar]
- 20.Fan MH, et al. Fibroblast Activation Protein (FAP) Accelerates Collagen Degradation and Clearance from Lungs in Mice. J Biol Chem. 2016;291:8070–8089. doi: 10.1074/jbc.M115.701433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Santos AM, Jung J, Aziz N, Kissil JL, Pure E. Targeting fibroblast activation protein inhibits tumor stromagenesis and growth in mice. J Clin Invest. 2009;119:3613–3625. doi: 10.1172/JCI38988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gorrell, M. D. & Park, J. E. In Handbook of Proteolytic Enzymes Vol. 1 (eds Rawlings, N. D. & Salvesen, G. S.) Ch. 750, 3395–3402 (Academic Press, 2013).
- 23.Edosada CY, et al. Peptide substrate profiling defines fibroblast activation protein as an endopeptidase of strict Gly(2)-Pro(1)-cleaving specificity. FEBS Lett. 2006;580:1581–1586. doi: 10.1016/j.febslet.2006.01.087. [DOI] [PubMed] [Google Scholar]
- 24.Rosenblum JS, Kozarich JW. Proyl peptidases: a serine protease subfamily with high potential for drug discovery. Curr Opin Chem Biol. 2003;7:496–504. doi: 10.1016/S1367-5931(03)00084-X. [DOI] [PubMed] [Google Scholar]
- 25.Edosada CY, et al. Selective inhibition of fibroblast activation protein protease based on dipeptide substrate specificity. J Biol Chem. 2006;281:7437–7444. doi: 10.1074/jbc.M511112200. [DOI] [PubMed] [Google Scholar]
- 26.Pineiro-Sanchez ML, et al. Identification of the 170-kDa melanoma membrane-bound gelatinase (seprase) as a serine integral membrane protease. J Biol Chem. 1997;272:7595–7601. doi: 10.1074/jbc.272.12.7595. [DOI] [PubMed] [Google Scholar]
- 27.Lee KN, et al. Antiplasmin-cleaving enzyme is a soluble form of fibroblast activation protein. Blood. 2006;107:1397–1404. doi: 10.1182/blood-2005-08-3452. [DOI] [PubMed] [Google Scholar]
- 28.Sonoda, J., Chen, M. Z. & Baruch, A. FGF21-receptor agonists: an emerging therapeutic class for obesity-related diseases. Horm Mol Biol Clin Investig30, 10.1515/hmbci-2017-0002 (2017). [DOI] [PubMed]
- 29.Coppage AL, et al. Human FGF-21 Is a Substrate of Fibroblast Activation Protein. PLoS One. 2016;11:e0151269. doi: 10.1371/journal.pone.0151269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dunshee DR, et al. Fibroblast Activation Protein Cleaves and Inactivates Fibroblast Growth Factor 21. J Biol Chem. 2016;291:5986–5996. doi: 10.1074/jbc.M115.710582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhen EY, Jin Z, Ackermann BL, Thomas MK, Gutierrez JA. Circulating FGF21 proteolytic processing mediated by fibroblast activation protein. Biochem J. 2016;473:605–614. doi: 10.1042/BJ20151085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schechter I, Berger A. On the size of the active site in proteases. I. Papain. Biochem Biophys Res Commun. 1967;27:157–162. doi: 10.1016/S0006-291X(67)80055-X. [DOI] [PubMed] [Google Scholar]
- 33.Uitte de Willige S, Malfliet JJ, Janssen HL, Leebeek FW, Rijken DC. Increased N-terminal cleavage of alpha-2-antiplasmin in patients with liver cirrhosis. J Thromb Haemost. 2013;11:2029–2036. doi: 10.1111/jth.12396. [DOI] [PubMed] [Google Scholar]
- 34.Gorrell MD, et al. The Protease Fibroblast Activation Protein (FAP) in Liver Disease. Hepatology. 2014;60:592a–592a. [Google Scholar]
- 35.Park JE, et al. Fibroblast activation protein, a dual specificity serine protease expressed in reactive human tumor stromal fibroblasts. J Biol Chem. 1999;274:36505–36512. doi: 10.1074/jbc.274.51.36505. [DOI] [PubMed] [Google Scholar]
- 36.Niedermeyer J, et al. Targeted disruption of mouse fibroblast activation protein. Mol Cell Biol. 2000;20:1089–1094. doi: 10.1128/MCB.20.3.1089-1094.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Narra K, et al. Phase II trial of single agent Val-boroPro (Talabostat) inhibiting Fibroblast Activation Protein in patients with metastatic colorectal cancer. Cancer Biol Ther. 2007;6:1691–1699. doi: 10.4161/cbt.6.11.4874. [DOI] [PubMed] [Google Scholar]
- 38.Lee KN, Jackson KW, Christiansen VJ, Dolence EK, Mckee PA. Enhancement of fibrinolysis by inhibiting enzymatic cleavage of precursor alpha(2)-antiplasmin. Journal of Thrombosis and Haemostasis. 2011;9:987–996. doi: 10.1111/j.1538-7836.2011.04195.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li JB, et al. Activatable Near-Infrared Fluorescent Probe for In Vivo Imaging of Fibroblast Activation Protein-alpha. Bioconjugate Chem. 2012;23:1704–1711. doi: 10.1021/bc300278r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Poplawski SE, et al. Identification of Selective and Potent Inhibitors of Fibroblast Activation Protein and Prolyl Oligopeptidase. J Med Chem. 2013;56:3467–3477. doi: 10.1021/jm400351a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Maes M, et al. Components of biological variation in prolyl endopeptidase and dipeptidyl-peptidase IV activity in plasma of healthy subjects. Clin Chem. 1994;40:1686–1691. [PubMed] [Google Scholar]
- 42.Agusti-Cobos E, Tenorio-Laranga J. Enhancement of fibrinolysis by inhibiting enzymatic cleavage of precursor alpha(2)-antiplasmin: a rebuttal. Journal of Thrombosis and Haemostasis. 2011;9:1266–1267. doi: 10.1111/j.1538-7836.2011.04294.x. [DOI] [PubMed] [Google Scholar]
- 43.Jansen K, et al. Extended structure-activity relationship and pharmacokinetic investigation of (4-quinolinoyl)glycyl-2-cyanopyrrolidine inhibitors of fibroblast activation protein (FAP) J Med Chem. 2014;57:3053–3074. doi: 10.1021/jm500031w. [DOI] [PubMed] [Google Scholar]
- 44.Venalainen JI, et al. Binding kinetics and duration of in vivo action of novel prolyl oligopeptidase inhibitors. Biochemical Pharmacology. 2006;71:683–692. doi: 10.1016/j.bcp.2005.11.029. [DOI] [PubMed] [Google Scholar]
- 45.Jarho EM, et al. A cyclopent-2-enecarbonyl group mimics proline at the P2 position of prolyl oligopeptidase inhibitors. J Med Chem. 2004;47:5605–5607. doi: 10.1021/jm049503w. [DOI] [PubMed] [Google Scholar]
- 46.Thoma R, et al. Structural basis of proline-specific exopeptidase activity as observed in human dipeptidyl peptidase-IV. Structure. 2003;11:947–959. doi: 10.1016/S0969-2126(03)00160-6. [DOI] [PubMed] [Google Scholar]
- 47.Aertgeerts K, et al. Structural and kinetic analysis of the substrate specificity of human fibroblast activation protein alpha. J Biol Chem. 2005;280:19441–19444. doi: 10.1074/jbc.C500092200. [DOI] [PubMed] [Google Scholar]
- 48.Fulop V, Szeltner Z, Renner V, Polgar L. Structures of prolyl oligopeptidase substrate/inhibitor complexes. Use of inhibitor binding for titration of the catalytic histidine residue. J Biol Chem. 2001;276:1262–1266. doi: 10.1074/jbc.M007003200. [DOI] [PubMed] [Google Scholar]
- 49.Szeltner Z, et al. Electrostatic environment at the active site of prolyl oligopeptidase is highly influential during substrate binding. J Biol Chem. 2003;278:48786–48793. doi: 10.1074/jbc.M309555200. [DOI] [PubMed] [Google Scholar]
- 50.Brunker P, et al. RG7386, a Novel Tetravalent FAP-DR5 Antibody, Effectively Triggers FAP-Dependent, Avidity-Driven DR5 Hyperclustering and Tumor Cell Apoptosis. Mol Cancer Ther. 2016;15:946–957. doi: 10.1158/1535-7163.MCT-15-0647. [DOI] [PubMed] [Google Scholar]
- 51.Copeland, R. A. Enzymes: a practical introduction to structure, mechanism, and data analysis. 2nd edn, (J. Wiley, 2000).
- 52.Sastry GM, Adzhigirey M, Day T, Annabhimoju R, Sherman W. Protein and ligand preparation: parameters, protocols, and influence on virtual screening enrichments. J Comput Aided Mol Des. 2013;27:221–234. doi: 10.1007/s10822-013-9644-8. [DOI] [PubMed] [Google Scholar]
- 53.Harder E, et al. OPLS3: A Force Field Providing Broad Coverage of Drug-like Small Molecules and Proteins. J Chem Theory Comput. 2016;12:281–296. doi: 10.1021/acs.jctc.5b00864. [DOI] [PubMed] [Google Scholar]
- 54.Sherman W, Day T, Jacobson MP, Friesner RA, Farid R. Novel procedure for modeling ligand/receptor induced fit effects. J Med Chem. 2006;49:534–553. doi: 10.1021/jm050540c. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.