Abstract
The development of Sjögren’s syndrome (SjS) is a dynamic and temporal process with a female predilection. Following the initial influx of immune cells, T cell clusters develop, accelerating the pathology in the salivary glands. Proinflammatory cytokines, IFN-γ and IL-17A, produced by T cells contribute synergistically to the disease. In this study, we examined the sexual dimorphism in cellular infiltrates of the salivary glands by using functional single-cell microengraving analysis. Using high-throughput sequencing, we investigated the clonal diversity of the T cell receptors (TCRs) of infiltrating IFN-γ and IL-17A-producing T cells in male and female SjS-susceptible (SjSs) C57BL/6.NOD-Aec1Aec2 mice. There were elevated frequencies of IFN-γ and IL-17A-producing effector T cell populations in female SjSS mice compared to male SjSS mice. MEME analysis shows high frequency and unique, sexually dimorphic motifs in the TCR hypervariable regions in the SjSS mice. Male mice selected for TRAV8/TRAJ52 (CATDLNTGANTGKLTFG) TCR genes in Th1 cells and TRBV16/(TRBD1/2)TRBJ1-7 (CGGKRRLESIFR) in Th1 and Th17 cells. Female SjSS mice selected for TRAV8/TRAJ52 (CATDLNTGANTGKLTFG), TRAV13D-2/TRAJ23 (CVYLEHHFE), and TRBV23/(TRBD2)TRBJ2-2 (CRKLHSCATCALNFL) in Th1 cells. These findings suggest that there is an elevated prevalence of pathogenic effector T cells in the glands with a sexually dimorphic selection bias of TCR repertoires.
Introduction
Sjögren’s syndrome (SjS) is an autoimmune disorder characterized by the inflammation of secretory glands, namely the lacrimal and salivary glands (SG) leading to keratoconjunctivitis and xerostomia i.e. dry eyes and dry mouth1. Immunoglobulins, including autoantibodies against ribonuclear proteins such as SSA/Ro and SSB/La, are highly elevated in SjS patients. The disease progression is marked by the unregulated infiltration of lymphocytes in which some foci organize into germinal center-like formations in the glands causing periductal aggregates, inflammation, and apoptosis; this leads to gland dysfunction and loss of tear and saliva production2–4. CD4+ T helper cells (Th) expressing the αβ T cell receptor (TCR) dominate the infiltrates5–8. SjS presents a signature of type 2 interferon, Interferon-γ (IFN-γ) as well as Interleukin (IL)-17A likely produced by Th1 and Th17 cells respectively9–11.
Human and animal models of SjS exhibit high levels of IFN-γ and IFN-responsive factors. The upregulation of the IFN pathway induces the activation of macrophages, natural killer (NK) cells, and CD8+ T cells. It also induces vascular adhesion molecule-1 (VCAM-1), L-selectin, lymphocyte function-associated antigen-1 (LFA-1), and other molecules that can trigger the homing of immune cells to the glands12. Previous data indicated that organogenesis in the SG is retarded in the SjS-susceptible (SjSS) NOD (non-obese diabetic) model and ablation of Ifnγ resulted in a return to normal gland development13. It is very likely that IFN-γ plays a role in early disease development as well as the symptoms by attracting invasive lymphocytes as well as hindering gland development, which in turn exacerbates glandular dysfunction. In addition to IFN-γ producing Th1 cells, we, and others, have shown that IL-17A producing Th17 cells contribute profoundly to the disease pathogenesis10,14–16. L-17 is expressed as one of six isotypes, two of which are defining to Th17 cells (IL-17A and IL-17F) with IL-17A being the more prevalent isotype17. In addition to production of IL-17, which recruits neutrophils, Th17 cells produces IL-21 and IL-22, which regulates B cell differentiation and immunoglobulin isotype switching, and induces proliferation in a STAT3-dependent manner, respectively18. Our recent studies have indicated that IL-17A plays a critical role in the strong sexual dimorphism in the SjSS mouse model where it affects sialadenitis, plasma cells, and germinal center B cell populations, moreso in females19,20. Furthermore, because IL-17’s roles in inducing isotypic switching, recruiting neutrophils, and inducing proliferation, it strongly suggests that Th17 cells may have an intrinsic role in both disease development and progression.
The essential role of Th1 and Th17 cells at the specific stages of the autoimmune process is widely studied. In addition to the Th subsets, cytotoxic T cells expressing IFN-γ or IL-17A (Tc1 and Tc17 respectively) have been identified as propagating autoimmunity in other diseases21–23. In context of the major histocompatibility complex (MHC) expressed on antigen presenting cells, the effector T cell recognizes a peptide antigen in a specific interaction via the TCR, a heterodimeric membrane protein that comes in two forms, αβ and γδ, the former of which is present in 70% of T cells infiltrating the SG24. The recognition of cognate antigen triggers activation, cytokine secretion, and proliferation25. During the immune response, antigen-specific interactions lead to proliferation of only reactive T cells, this, in turn, leads to clonotypic restriction and loss in diversity26–28. Diversity of the TCR is generated from the unique pairing of variable (V) and joining (J) gene segments for the α chain or V, diversity (D), and J in the case of the β chain, which generates on the order of 1014 possible combinations29,30. Each chain possesses a hypervariable region or complementarity determining region 3 (CDR3), which contains amino acids responsible for interaction with the antigen. Diversity is skewed toward CDR3 in αβ TCRs as opposed to CDR1 and CDR2 regions, which contact the cognate MHC. In response to antigen interaction, non-template nucleotides (randomly added in a terminal deoxynucleotidyl transferase dependent manner), N in the α chain or NDN in the β chain, are inserted between the V and J gene segments during recombination which leads to a change in the CDR3 region length31,32. In the TCR repertoire, each clone is represented by a single conserved TCR sequence; a robust T cell response is characterized by a reduction of heterogeneity and restriction of epitopes of antigen presentation, i.e. monoclonal expansions of antigen experienced T cells.
In this study, we investigated the comprehensive TCR repertoires of individual infiltrating T cells in the SGs by analyzing the TCRαβ transcripts of CD4+ and CD8+ T cells that produce either IFN-γ or IL-17A in a SjSS animal model. Our results indicate that the TCR repertoire of the Th1 and Th17 cells in the SG of SjSS mice exhibited reduced diversity in the CDR3 regions, which expressed predominantly TRAV8/TRAJ52 (Th1) and TRBV16/(TRBD1/2)TRBJ1-7 (Th1 and Th17) in male and female mice as well as TRAV13/TRAJ23 and TRBV23/(TRBD2)TRBJ2-2 (Th1) in female mice. These clones indicate antigen mediated maturation of effector T cells to a common autoantigen in the SjSS mouse model. This helps elucidate the underlying initiation of the autoimmune cascade in SjS.
Results
Isolation of helper and cytotoxic T cells by microengraving shows an increase in IFN-γ and IL-17A producing cells in SjSS glands
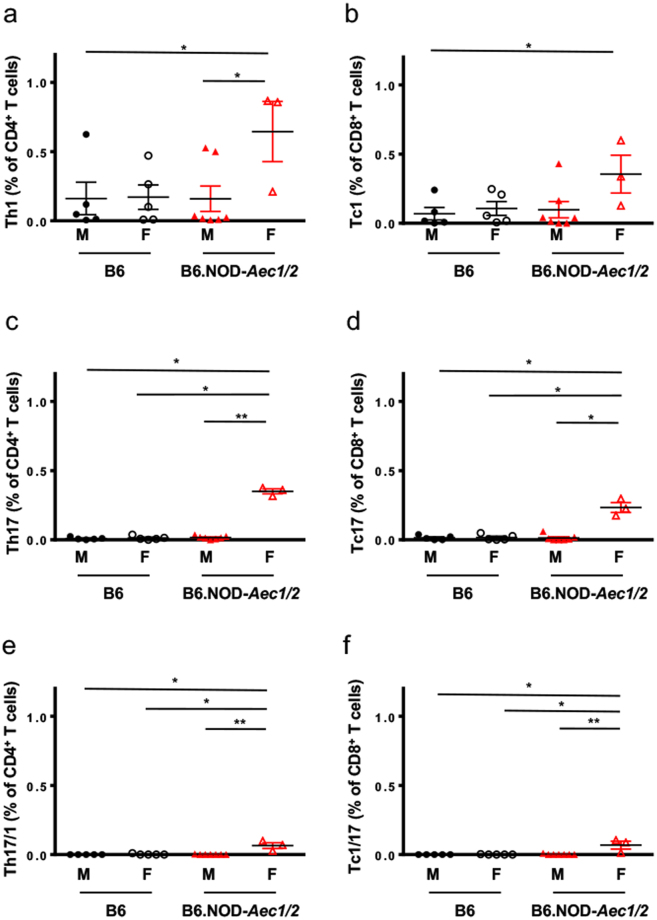
Both IFN-γ and IL-17A are essential for the development of the autoimmune cascade in both SjSS mice and patients13–15,20,33–35. Utilizing single-cell microengraving analysis with functional live cell characterization, we sought to isolate and characterize T helper (Th) and cytotoxic T (Tc) cells producing IL-17A and/or IFN-γ in the SG of SjSS mice. Stimulated lymphocytes were labeled and microengraved to identify the following subsets for isolation: CD4+IFN-γ+ (Th1), CD4+IL-17A+ (Th17), CD8+IFN-γ+ (Tc1), CD8+ IL-17A+ (Tc17), CD4+IL-17A+IFN-γ+ (Th17/1), and CD8+IL-17A+IFN-γ+ (Tc1/17) (Fig. 1). Due to the established sexually dimorphic function of IL-17A in SjS, it is pertinent to examine the phenotypic differences by sex in the mice. As expected, there were no significant differences between male and female B6 mice in regards to the frequencies of Th1, Tc1, Th17, Tc17, and Tc1/17 in the SG. However, female B6 mice showed an elevated, but statistically insignificant frequency of Th17/1 cells over their male counterparts. By comparison, the SjSS mice had a more than two-fold increase of infiltrating Th1, Tc1, Th17, Tc17, Th17/1, and Tc1/17 cells when compared to B6 mice (Fig. 2). The single-cell microengraving analysis did not alter the T cell subsets when compared to direct ex-vivo staining by flow cytometry (Supplementary Fig. S1 ). Interestingly, when the sexes were compared, it became apparent that there was a significant increase in these cell populations in female over male SjSS mice. As presented in Table 1 and Fig. 2, female SjSS B6.NOD-Aec1/2 exhibited 2- and 18-fold increases of Th17 and Tc17 (0.350 versus 0.161, and 0.233 versus 0.013, respectively) in comparison to males; there was also an approximately 4- and 3-fold increase in Th1 and Tc1 cells, respectively, in SjSS females versus males. Unexpectedly, there was a marked increase in the Th17-IFN-γ (65-fold) and Tc1-IL-17A (68-fold) populations in female SjSS mice, compared to males. This suggests that the infiltrating T cells in the SG of the SjSS mice are comprised of higher frequencies of effector T cells, particularly Th1, Tc1, Th17, Tc17, Th17-IFNγ, and Tc1-IL-17A cells. More importantly, female SjSS B6.NOD-Aec1/2 showed a significant increase in all the effector T cells compared to their male counterparts. These data clearly indicate that there is a significant infiltration of effector T cells in SjSS B6.NOD-Aec1/2 mice when compared to B6 mice, the most significant of which are those that produce IL-17A. Furthermore, these cell types are differentially detectable in males and females of B6.NOD-Aec1/2 mice.
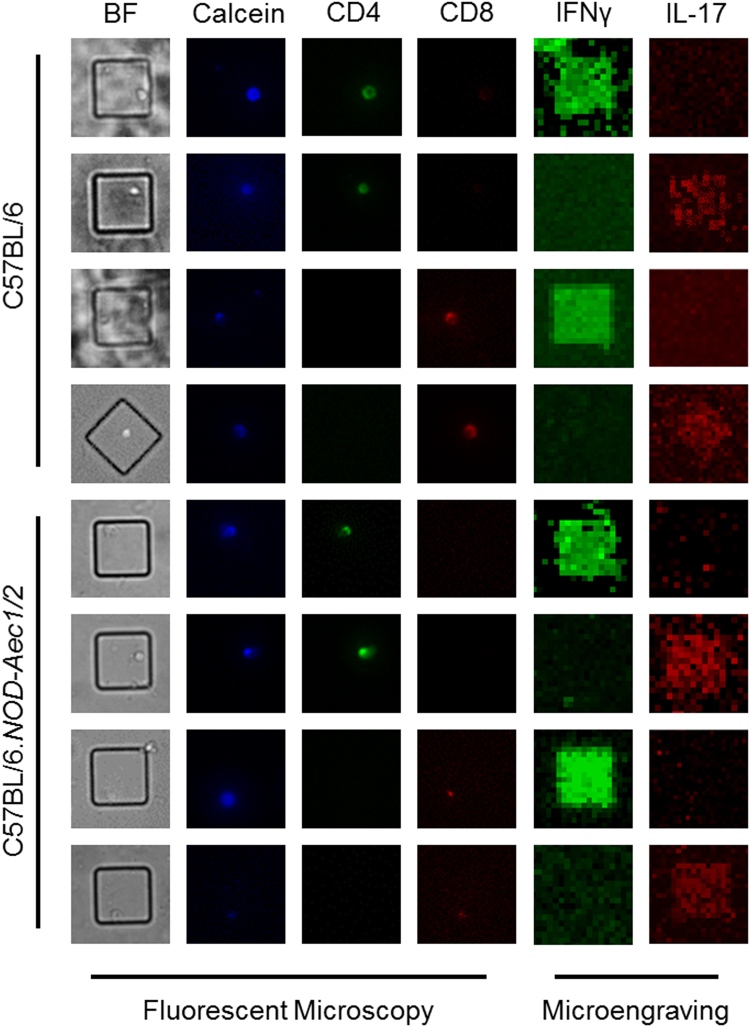
Figure 1.
Microengraving of T lymphocytes isolated from murine salivary glands. Representative fluorescent microscopy coupled with microengraving of secreted cytokines from isolated T-lymphocytes. Fluorescent antibody staining was performed with anti-CD4-FITC (green), anti-CD8-PE (Red), and Calcein violet-405 (blue), a marker of viable cells. Secreted cytokines were captured during microengraving and stained with anti-IFN-γ (green) and anti-IL-17A (red).
Figure 2.
Microengraving shows greater infiltration by IL-17A and IFN-γ producing cells in the salivary glands of SjSS mice. Quantification of T lymphocytes isolated from the major salivary gland of B6 male (●), B6 female (○), B6.NOD-Aec1Aec2 male ( ), and B6.NOD-Aec1Aec2 female (
), and B6.NOD-Aec1Aec2 female ( ) mice expressing (a) CD4+IFN-γ+, (b) CD8+IFN-γ+, (c) CD4+IL-17A+, (d) CD8+IL-17A+, (e) CD4+IFN-γ+IL-17A+, or (f) CD8+IFN-γ+IL-17A+. The frequency in percentage of was determined by using the percentage (multiplied by 100) of the total number of CD4+IFN-γ+ (Th1), CD4+IL-17A+ (Th17), CD8+IFN-γ+ (Tc1), CD8+ IL-17A+ (Tc17), CD4+IL-17A+IFN-γ+ (Th17/1), and CD8+IL-17A+IFN-γ+ (Tc1/17) cells from wells with single live cells among the total number of wells with single CD4+ or CD8+ cells. Group comparisons were made using a Kruskall-Wallis test with Dunn’s post hoc test. Statistical significance was defined as a p value < 0.05. All results are presented as means ± s.e.m. Significance was determined as *p < 0.05, **p < 0.01.
) mice expressing (a) CD4+IFN-γ+, (b) CD8+IFN-γ+, (c) CD4+IL-17A+, (d) CD8+IL-17A+, (e) CD4+IFN-γ+IL-17A+, or (f) CD8+IFN-γ+IL-17A+. The frequency in percentage of was determined by using the percentage (multiplied by 100) of the total number of CD4+IFN-γ+ (Th1), CD4+IL-17A+ (Th17), CD8+IFN-γ+ (Tc1), CD8+ IL-17A+ (Tc17), CD4+IL-17A+IFN-γ+ (Th17/1), and CD8+IL-17A+IFN-γ+ (Tc1/17) cells from wells with single live cells among the total number of wells with single CD4+ or CD8+ cells. Group comparisons were made using a Kruskall-Wallis test with Dunn’s post hoc test. Statistical significance was defined as a p value < 0.05. All results are presented as means ± s.e.m. Significance was determined as *p < 0.05, **p < 0.01.
Table 1.
Frequency of IFN-γ and IL-17A producing T cells in the salivary glands of male and female mice.
| B6M | B6F | B6.NOD-Aec1/2 M | B6.NOD-Aec1/2 F | |
|---|---|---|---|---|
| Th1 | 0.161 ± 0.117 | 0.171 ± 0.088 | 0.159 ± 0.092 | 0.644 ± 0.217 (4x)# |
| Th17 | 0.009 ± 0.004 | 0.013 ± 0.001 | 0.161 ± 0.004 | 0.350 ± 0.018 (2x)# |
| Th17/1 | 0.000 ± 0.000 | 0.002 ± 0.002 | 0.001 ± 0.001 | 0.065 ± 0.021† (65x)# |
| Tc1 | 0.069 ± 0.045 | 0.107 ± 0.050 | 0.098 ± 0.059 | 0.355 ± 0.136 (3x)# |
| Tc17 | 0.014 ± 0.007 | 0.017 ± 0.009 | 0.013 ± 0.008 | 0.233 ± 0.035 (18x)# |
| Tc1/17 | 0.000 ± 0.000 | 0.001 ± 0.001 | 0.001 ± 0.001 | 0.068 ± 0.028† (68x)# |
Summary of means with ± s.e.m. of infiltrating T cells. Mean is defined as the percentage of Th/CD4+ T cells or Tc/CD8+ T cells. Statistics were performed using a Kruskall-Wallis test with Dunn’s post hoc test. #Fold-change between female and male B6.NOD-Aec1/2. †Denotes significance of p < 0.05 or less between all other data sets in a row.
Loss of normal distribution in V/J pairing is associated with the SjSS B6.NOD-Aec1/2 mice
To explore the differences between individual effector T cells in the SG of males and females, functional live CD4+ and CD8+ T cells producing IFN-γ or IL-17A were examined for TCR gene rearrangement. After microengraving, each live cell that expressed IFN-γ or IL-17A, was isolated, then reverse transcription nested PCR was performed with two pairs of primers used in sequential reactions to target the amplification of the hypervariable, CDR3 region of the tcra and tcrb transcripts. Sequences were aligned to the International ImMunoGeneTics Information System (IMGT) using the IgBLAST tool to determine the V and J (and D) gene. Of the sequences recovered from B6 mice, 72% amplified the TCRα subunit gene and 40% amplified the TCRβ subunit gene, while in the B6.NOD-Aec1/2 mice 66% TCRα and 84% TCRβ were amplified. Of these, TCRα/β paired reads comprised only 12% and 15% for the B6 and the B6.NOD-Aec1/2 mice, respectively.
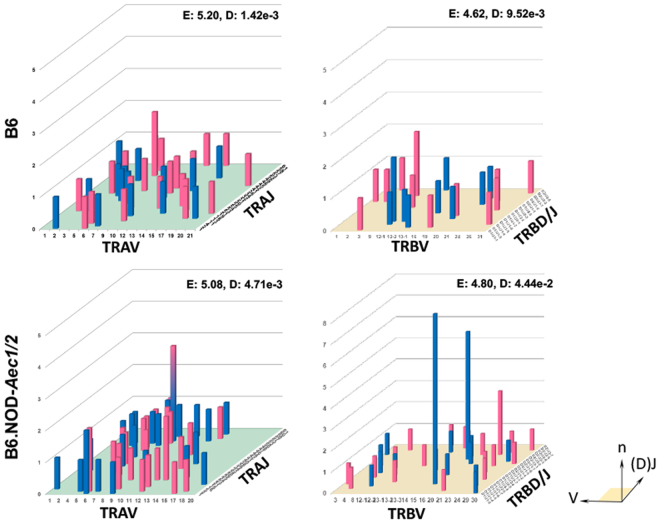
The diversity of the unique V/(D)J combinations in each group’s TCR gene pool reflects the progression of the immune response; a lower diversity indicates clonal expansion and positive selection of antigen experienced, autoimmune cells. Shannon’s Entropy depicts the diversity of a population by measuring uniformity; higher values are more uniform and thus normally distributed. In contrast, the Simpson’s index approximates diversity and indicates overlap, where a lower value indicates greater diversity and 0 indicates no clonal overlap. V/J combinations are catalogued in Fig. 3 as a representation of the repertoire of infiltrating T cells of male and female mice. A complete normal distribution would show a similar frequency of pairings of all of the V/J gene combinations, however it is only likely to see this distribution if all genes are used equally and if thousands of isolated cells are sequenced. B6 mice showed a more heterogeneous distribution of V/J parings across the TRα repertoire compared to B6.NOD-Aec1/2, which showed a higher Shannon’s entropy (E: 5.20 vs 5.08) but slightly lower Simpson’s diversity (1.42e-3 vs 4.71e-3). Interestingly, the B6.NOD-Aec1/2 mice showed a gene bias towards TRAV genes 1–13. There was only one high frequency pairing from the B6 mice, TRAV6/TRAJ52 observed in female mice, whereas the B6.NOD-Aec1/2 mice displayed pairing from: TRAV8/TRAJ52 (both males and females, indicated by the hybrid pink-blue bar), TRAV6/TRAJ1 (males) or TRAJ2 (females), and TRAV13/TRAJ23 (females). B6 mice showed a narrower distribution of TRβ (E: 4.62 vs 4.80) but had fewer high frequency pairings while B6.NOD-Aec1/2 mice had two high frequency pairs: TRBV16/(TRBD1/2)TRBJ1-7 from males and TRBV23/(TRBD2)TRBJ2-2 from females with potential TRBV bias toward TRBV genes 3–20 in males, while females showed an opposing bias toward later TRBV genes 12–30. The presence of high frequency V/J pairing in B6.NOD-Aec1/2 mice in combination with lower entropy, and higher diversity, indicated a restricted TCR repertoire in SjSS mice.
Figure 3.
Loss of normal distribution in V/J pairing is associated with the SjSS mouse. Total salivary gland T-lymphocyte three dimensional histograms of TCR gene V/(D)J combinations for B6 TRA (upper left) and B6 TRB (upper right) genes, as well as B6.NOD-Aec1/2 TRA (lower left) and TRB (lower right) genes. Blue bars indicate data from male mice, pink from female mice, and hybrid blue-pink bars for data shared between the sexes. The x-axis represents the analyzed Vα or Vβ gene segment, the z-axis is the analyzed Jα or D/Jβ gene segments, the height or y-axis indicates the total reads of specific V/(D)J combination. V and J genes were designated by the disambiguated IMGT nomenclature for each gene subgroup. TCR repertoire was quantified by entropy (E = Shannon’s Entropy) and diversity (D = Simpson’s Index).
Distribution V/J pairing contributed by effector Th1, Th17, Tc1 and Tc17 cells
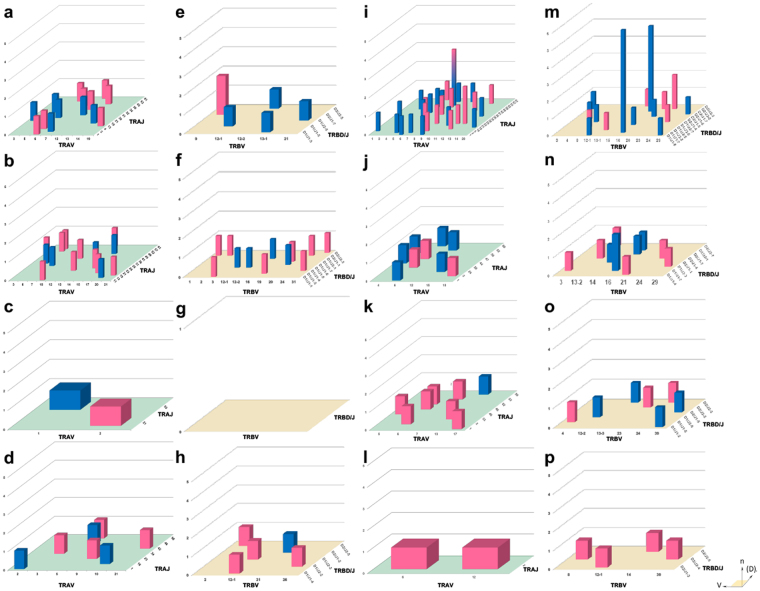
Next we looked at the contribution of Th1, Th17, Tc1 and Tc17 cells to the overall repertoires. In Fig. 4, B6 mice have an even distribution of TRAV/TRAJ pairings from Th1 and Th17 cells (Fig. 4a,b) with little contribution from Tc17 cells (Fig. 4d) and even less contribution from Tc1 cells (only 2 TRA pairs and 0 TRB pairs, Fig. 4c,g). The TRAV6/TRAJ52 pairing identified in Fig. 3 was evenly divided between Th1 and Th17 populations. SjSS B6.NOD-Aec1/2 mice have shown an extensive repertoire of V/J pairings from Th1 cells (Fig. 4I,m) and to a lesser extent Th17 (Fig. 4j,n) and Tc1 cells (Fig. 4k,o); the latter of which had a limited contribution from males. There was no contribution from Tc17 cells (Fig. 4l,p). The TRAV8/TRAJ52 pairing contributed solely to the repertoire of the SjSS B6.NOD-Aec1/2, Th1 cells and existed in both female and male mice. The TRAV6/TRAJ1 combination seen previously in Fig. 3 was divided between Th1 and Th17 repertoires of male mice and the TRAV6/TRAJ2 consisted of Tc1 and Tc17 in females. Another pairing unique to the SjSS B6.NOD-Aec1/2 mice, TRBV16/(TRBD1/2)TRBJ1-7 was present only in the male mice and contributed mostly to the Th1 (with pairings consisting of either TRBD gene) and in a minor extent the Th17 repertoires. The TRBV23/(TRBD2)TRBJ2-2 combination was comprised of Th1 and Tc1 cells derived from female SjSS B6.NOD-Aec1/2 mice. The SjSS B6.NOD-Aec1/2 mice display a pattern of expansion of Th1 and Th17 cells from TRAV8/TRAJ52 and TRBV16/(TRBD1/2)TRBJ2-2 pairings in male SjSS mice and Th1 cells from TRAV8/TRAJ52, TRAV13/TRAJ23, and TRBV23/(TRBD2)TRBJ2-2 pairings in female SjSS mice.
Figure 4.
T cells from the SjSS mouse show reduced diversity in V/J pairing. Salivary gland T-lymphocyte three dimensional histograms of TCR gene V/(D)J combinations derived from B6 Th1 cells (a) TRA and (e) TRB genes, Th17 cells (b) TRA and (f) TRB genes, Tc1 (c) TRA and (g) TRB, and Tc17 (d) TRA and (h) TRB genes as well as B6.NOD-Aec1/2 derived Th1 (i) TRA and (m) TRB genes, Th17 (j) TRA and (n) TRB genes, Tc1 (k) TRA and (o) TRB genes, and Tc17 (l) TRA and (p) TRB genes. Blue bars indicate data from male mice, pink from female mice, and hybrid blue-pink bars for data shared between the sexes. The x-axis represents the analyzed Vα or Vβ gene segment, the z-axis is the analyzed Jα or D/Jβ gene segments, the height indicates the total reads of specific V/(D)J combination. V and J genes were designated by the unambiguous IMGT nomenclature for each gene subgroup.
High frequency of CDR3 sequences were only found in T cells of the SjSS B6.NOD-Aec1/2 mice
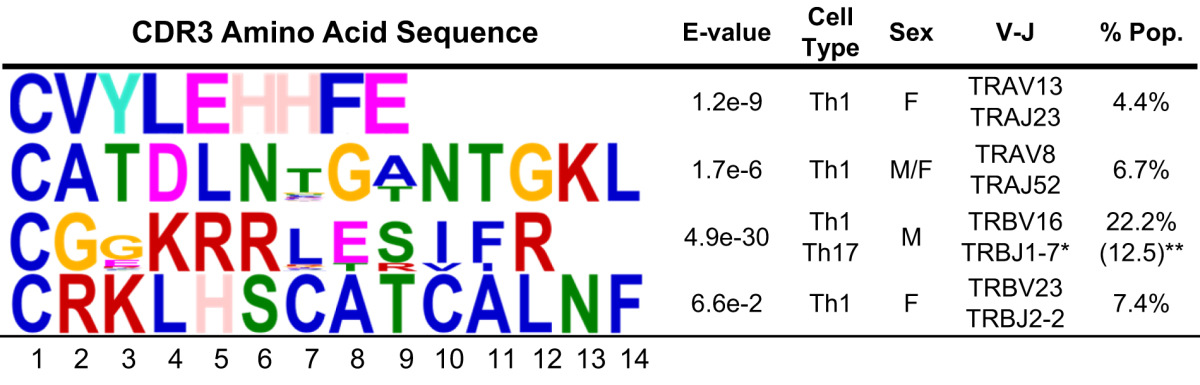
Since non-template N nucleotides yield higher diversity, it was utilized to examine the diversity within the CDR3 sequence and hence the TCR repertoires. B6 mice had no identical CDR3α/β clones (Supplementary Tables S1 and S2 ). B6 mice had only two premature stop codons (5.9% of CDR3α chain repertoire and 16.7% of the CDRβ chain repertoires) in the IFN-γ producing Th1 cells, whereas SjSS B6.NOD-Aec1/2 Th1 cells produced 13.2% premature stop codon for the CDR3α chain and 7.4% for the CDR3β chain while Th17 cells produced 20.1% for the CDRα chain and 12.6% for the CDRβ chain repertoire indicative of nonfunctional TCRs (Supplementary Tables S3 and S4). CDR3α and CDR3β of Th1 and Tc1 cells from B6 mice had an unusually low repertoire entropy (4.09 and 2.59, Supplementary Table S1) compared to the SjSS B6.NOD-Aec1/2 counterparts (5.34, 3.95, Supplementary Table S3) likely due to a smaller B6 repertoire. As expected, Th17 and Tc17 cells of B6 mice had greater entropy (α: 4.59, β: 4.09, Supplementary Table S2) compared to B6.NOD-Aec1/2 mice (CDR3α: 3.91, CDR3β: 3.88, Supplementary Table S4). There was no overlap in clones in the B6 Th1 and Tc1 sampling (Simpson’s Index values of 0 for both α and β chains) compared to SjSS B6.NOD-Aec1/2 (0.004, 0.063). Likewise, the Simpson’s Indices indicated no overlap in the CDR3α repertoire and little overlap in the CDR3β repertoire of SjSS Th17 and Tc17 cells (0.000 vs 0.008). In the B6.NOD-Aec1/2 mice, TRAV8/TRAJ52 (6.7%) and TRAV13/TRAJ23 (4.4%) pairings identified above were translated into CATDLNTGANTGKLTFG and CVYLEHHFE respectively, in which the former clone was derived from non-identical DNA sequences. CDR3β chain translation produced two clones of interest derived from combinations of TRBV16/(TRBD1/2)TRBJ1-7 (CGGKRRLESIFR, 22.2%) and TRBV23/(TRBD2)TRBJ2-2 (CRKLHSCATCALNFL, 7.4%). The former clone (CGGKRRLESIFR) was derived from multiple CDR3β sequences produced by Th1 and Th17 cells. A reduction in entropy with greater Simpson’s Indices, SjSS B6.NOD-Aec1/2 mice exhibit greater clonal expansion which is consistent with the disease process of SjS and other autoimmune diseases.
Motif analysis reveals high frequency and sexual dimorphism in the hypervariable CDR3 regions in the SjSS B6.NOD-Aec1/2 mice
MEME analysis (www.meme-suite.org) was utilized to determine the high-frequency motifs present in the CDR3 regions. Prior research has revealed that hydrophobic amino acids in the center of the CDR3 region have been linked to autoreactivity in T cells36. MEME analysis here was also utilized to determine which, if any, of the effector T cell clones of SjSS B6.NOD-Aec1/2 mice produced TCRs with this specific pattern. The core amino acids LEHH (positions 4–7) in the first motif from Table 2 have a hydrophilic amino acid, glutamate, flanked by leucine (hydrophobic) and two histidines (moderately hydrophobic). Likewise, the core amino acids from the last CDR3 region (SCAT in positions 6–9) contain two moderately hydrophobic amino acids (cysteine and alanine) flanked by two hydrophilic ones (serine and threonine). These two motifs, CVYLEHHFE and CRKLHSCATCALNF, were derived from clones with 100% DNA identity, which indicates that they are a product of clonal expansion. The second motif’s core is NTG (A or T) N in positions 6–10 yielding only one or two hydrophobic amino acids (glycine and alanine), however the glycine in position 8 is conserved among the clones analyzed. The final motif has a central KRRLE motif in positions 4–8, only one of these being hydrophobic (leucine), however this motif contains high confidence in lysine-arginine-arginine positively charge peptides. Three motifs analyzed in B6.NOD-Aec1/2 mice were present in female Th1 cells, where one (CATDLNTGANTGKLTFG) was also present in males and only one of these (CGGKRRLESIFR) came from a Th17 origin. Two CDR3 amino acid sequences derived from identical DNA sequences (CVYLEHHFE and CRKLHSCATCALNFL) and two derived from non-identical sequences (CATDLNTGANTGKLTFG and CGGKRRLESIFR) provide evidence for clonal expansion and clonal selection, in the SjS B6.NOD-Aec1/2 mouse autoimmune environment.
Table 2.
MEME analysis of high frequency CDR3 regions from SjSS mice. Amino acid motif analysis was performed on the high frequency CDR3 sequences from the B6.NOD-Aec1/2 mice. Bit height corresponds to the likelihood of the amino acid in each position. Blue – hydrophobic, neutral amino acids, Red – positively charge hydrophilic amino acids, Green – Neutral hydrophilic amino acids, Magenta – negatively charged hydrophilic amino acids, Orange-glycine, Teal-Tyrosine, Pink-Histidine (positively charged moderately hydrophobic). E-value indicates the model confidence for the amino acid in that position. Cytokine indicates the secreted molecule(s) associated with the motif, V/J indicates recombination gene segment pairings, and % pop. Indicates the percent of the motif present in the IFN-γ or the IL-17A producing population. *Most popular V/J combination. **Motif located in both IL-17A and IFN-γ producing populations in both TCRα and TCRβ chains.
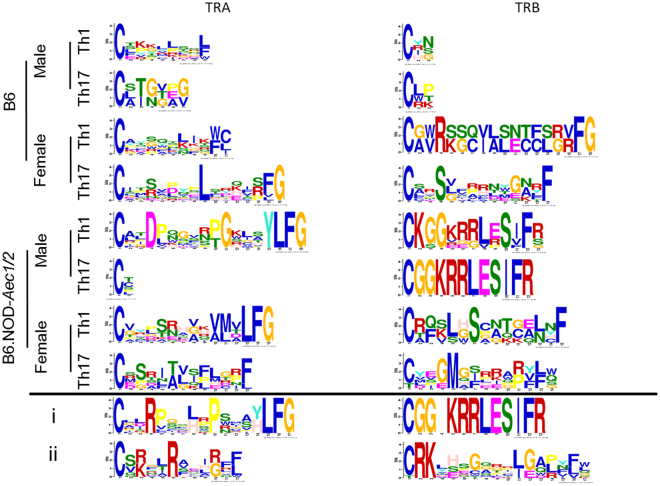
The GLAM2 algorithm (www.meme-suite.org) was employed to determine a full-length motif indicative of each sex and cell type derived from B6 and B6.NOD-Aec1/2 mice. We analyzed only the Th1 and Th17 repertoires because of the lack of the Tc1 and Tc17 repertoires in B6 mice and the presence of the high frequency CDR3 sequences in Th1 and Th17 cells (Table 2). Repertoires from female B6 mice had only a single leucine conserved in CDR3α peptides from Th1 and Th17 repertoires (Fig. 5), whereas β-chain motifs had an arginine or serine present in Th1 or Th17 repertoires. Several hydrophilic (serine, glutamine, asparagine, or threonine) amino acids are observed in the core of the CDR3α/β chains from B6 mice. Male B6 mice lacked conserved amino acids and produced short MEME analyses. Th1 cells of male SjSS B6.NOD-Aec1/2 mice contained a motif with an aspartate in the 4th position, either a proline or threonine in the 10th and 11th position of the CDR3α chain motif. Th17 cells from these mice produced abortive MEMEs in this motif analysis. β-chain of both Th1 and Th17 cells from male SjSS B6.NOD-Aec1/2 mice provided a highly-conserved motif of “CGGKRRLESIFR”. Female B6.NOD-Aec1/2 mice Th1 cells had either valine, glycine, or alanine in position 8 followed by either a valine or alanine in position 10 and then a methionine or leucine in position 11 of the CDR3α chain, all moderately hydrophobic amino acids. Th17 cells seemed to express serine, proline, or glycine in the 3rd position, threonine or alanine in the 6th position, valine, leucine or glycine in the 7 position and phenylalanine, proline or leucine in the 9 position of the CDR3α chain, again all moderately hydrophobic. Finally, the β-chain of Th1 cells in female SjSS mice showed a motif of leucine or serine in the 5th position and serine in the 7th position, whereas Th17 cells had a glycine or glutamate in the 4th position, methionine in the 5th position, and serine, isoleucine, or phenylalanine in the 7th position. Male B6.NOD-Aec1/2 mice displayed several cationic and hydrophobic amino acids in the core amino acids of the CDR3α/β chains for Th1 repertoires as well as Th17 CDR3β chain repertoire in contrast to male B6 mice which produced inconclusive motifs. B6.NOD-Aec1/2 produced an inconclusive motif for Th17 CDR3α chains implying a minimized role of the CDR3α chain selection, however a common motif for both Th1 and Th17 CDR3β chain implies selection. Female B6.NOD-Aec1/2 mice have a mix of hydrophobic and cationic amino acids (arginine, histidine, valine, and lysine) in the central positions of Th1 CDR3α chain and a mix of polar and nonpolar amino acids in the central positions in contrast with that of the female B6 mice with polar, hydrophilic amino acids. However, both B6 and B6.NOD-Aec1/2 female mice present similar hydrophilic amino acids (LSNT vs SCNT respectively) in the CDR3β chains of Th1 repertoire. The CDR3α chain of Th17 cells in B6.NOD-Aec1/2 female mice had motifs of hydrophilic and hydrophobic amino acids in contrast to one conserved leucine in B6 females. Because of contrasting motifs, T cells from male B6.NOD-Aec1/2 mice are being generated to different auto antigens as that of male B6 mice and female B6.NOD-Aec1/2 vis-à-vis female B6 mice.
Figure 5.
Motif analyses of SjSS mice show conserved amino acids in CDR3 regions of male and female T cells. Glam2 motif analyses were performed on B6 and B6.NOD-Aec1/2 Th1 and Th17 cell repertoires. Bit height corresponds to amino acid identity likelihood. Blue – hydrophobic, neutral amino acids, Red – positively charge hydrophilic amino acids, Green – Neutral hydrophilic amino acids, Magenta – negatively charged hydrophilic amino acids, Orange-glycine, Teal-Tyrosine, Pink-Histidine (positively charged moderately hydrophobic).
Two representative mice are presented to examine TCR repertoires in a single mouse. The first mouse, a male SjSS mouse (Fig. 5i), had several instances of the “CGGKRRLESIFR” CDR3β sequence (Table 2) as shown by their overall β-chain motif analysis. Motif analysis of the α-chain revealed arginine in the fourth position, leucine or histidine in the eighth position, proline in the tenth position, and either a tyrosine or a histidine in the fourteenth position. These were either positively charged or hydrophobic amino acids. This mouse had multiple clones of the “CATDLNTGANTGKLTFG” (Table 2), however this was obscured in the repertoire and not apparent in the overall CDR3α motif. The second mouse, a female SjSS mouse (Fig. 5ii), had a strong motif of arginine or lysine (cationic) in the 3rd position, arginine in the 6th position, and arginine or glycine in the 10th position while, the β-chain had a motif beginning with arginine-lysine in the 2nd and 3rd position, glycine, cysteine or tryptophan in the 7th position, leucine or threonine in the 11th position and glycine or glutamate in the 12th position. The arginine-lysine beginning of the motif matches the CDR3β sequence “CRKLHSCATCALNFL” (Table 2). Based on motif analyses, both the male and female mice selected for CDR3α/β amino acids that were hydrophobic or cationic in the SjS environment.
Discussion
The development of SjS is a dynamic and temporal process. Following the perturbation of the salivary glands, the influx of immune cells initially by T cell clusters followed by recruitment of B lymphocytes accelerates the gross pathology of the glands. Effector T cell-produced proinflammatory cytokines, IFN-γ and IL-17A, contribute synergistically to the disease process. T cells recognize autoantigens by their TCR, however, there remains speculation in regards to the clonal selection of these T cells based on their TCR repertoires. We sought to determine the clonal diversity and selection of the TCRs expressed by infiltrating IFN-γ and IL-17A-producing effector T cells, between male and female of SjSS B6.NOD-Aec1/2 mice. Compared to B6 mice, the SjSS mice show restricted Th1 and Th17 TCR repertoires in both males and females, with more highly limited repertoires in Th17 cells of females. Identified here were two TCR motifs present only in SjSS mice that arise from selection (CATDLNTGANTGKLTFG from Th1 cells and CGGKRRLESIFR in both Th1 and Th17 cells) as well as two TCR motifs that show evidence of expansion in Th1 cells (CVYLEHHFE and CRKLHSCATCALNFL).
IFN-γ induces adhesion molecules in the gland including: vascular cell adhesion molecule-1 (VCAM-1), α4β1 integrin, peripheral node addressing, L-selectin and LFA-1, which allows for the influx of inflammatory cells into salivary glands. Global transcription analyses have indicated that a number of chemokines like CCL5, CXCL16 (an IFN-γ regulated chemokine that attracts NK and memory T cells), CXCL9 and CXCL13 transcripts are up-regulated in the salivary glands of SjSS mice at the time of the disease onset37. IFN-γ activity is prominent in SG epithelial cells and biopsies from SjS patients38. IL-17A is involved in the proliferation, maturation and recruitment of neutrophils during the initial insult to the glands. Furthermore, IL-17A can function as a B cell helper by inducing a strong proliferative response of B cells and triggering antibody production with class switching and germinal center formation39,40. In various related autoimmune disorders, IL-17 can be directly correlated with disease severity41. Th17 (not Th1 or Th2) cells are highly prevalent in the SG of SjSS mice and human patients, with a strong sexual dimorphism in the mouse model20,21. Here we found a significant increase of Th17 cells in SjSS B6.NOD-Aec1/2 mice with a higher frequency in females versus males, while Th1, Tc1, and Tc17 cells were also expressed in female SjSS mice. These findings are consistent with research into other autoimmune diseases, including experimental autoimmune encephalitis (EAE), where it was found that females present a more robust immune response with elevated cell mediated responses including increased recruitment of Th1, Th17, Tc1, and Tc17 cells like in the glands of female SjSS mice42,43. Exogenous androgen treatment has been shown to reduce IL-12 and IFN-γ levels, lowering disease presentation of EAE44, due to the protective effect of androgens in certain genetic backgrounds towards inflammation19,20,45. Our data showed a disproportionately high amount of IL-17A produced by female SjSS mice. This upregulation could be attributed to the female sex hormone, 17β-estradiol, which reduces the transition of Th17 cells from the draining lymph node to the target organs as in rheumatoid arthritis46,47. SjS may bear sensitivity to estrogen by increasing the activation of Th17 cells via IL-23 causing regulatory dysfunction, allowing migration to the glands, and secretion of proinflammatory cytokines48.
The clinical manifestation of SjS in patients is attributed by the hyperactivity of B cells as evidenced by ectopic germinal center (GC) formation, hypergammaglobulinemia, and circulating autoantibodies49–51. The GC formation is mediated by the function of follicular helper T (Tfh) cells surrounding activated B cells in the follicles52. A study by Maehara et al.53 has indicated that Th2 and Tfh cells represented by their signature cytokines are positively correlated with increased lymphocytic foci and GC. In the mouse models, IL-4 of Th2 cells plays an important role during the clinical phase, while having little or no effect on the pathology associated with the preclinical disease state54,55. The mice examined in the study were at the advanced age, mimicking the clinical phase of human SjS which is predominated by Th2/Tfh response. The study did not examine the effector Th2/Tfh cells, but it could explain the low numbers of other effector T cells in the SG. One potential question which has not well understood or investigated in the SG is differentiation of recirculating memory T cells present in the vascular system of the SG and resident memory T cells in the parenchyma. As alluded to earlier, high numbers of effector T cells were identified in the glands of SjSS mice. As presented in Supplementary Fig. S2, the majority of the effector T cells are resident or gland-infiltrating T cells and not from circulation. Further work is needed to address the gland-specific pressure exerted on the enrichment for effector T cells and whether their expansion is initiated from the circulation or secondary lymphoid tissues.
The influx of inflammatory cells into the salivary glands raises concerns regarding the migratory nature as well as the selection process of infiltrating lymphocytes. The danger signals in the epithelium in conjunction with the activation of adhesion molecules and homing chemokines/chemokine receptors drive the flow of immune cells into the glands. There are two scenarios which could explain the migratory pattern of these infiltrating T cells and resulting TCR repertoires. First, there is a massive influx of various effector cell types with polyclonal TCR repertoires. The gland milieu and unique antigen presentation select for monoclonal TCR repertoires expressed on a specific subset of effector T cells, i.e. a selection for only SG antigen-specific T cells with a bias against T cells specific for other antigens. Alternatively, the initial infiltrates are composed of a few sporadic T cells with limited TCR repertoire. These early T cells proliferate and clonally expand, which maintains their monoclonal repertoires. There is no human data that examines the temporal influx of T cells into the SG, however using the SjSS mice, we have observed there is a sporadic infiltration of T cells in younger mice, followed by a massive occupation of T cell during the onset of the disease. This observation, in addition to studies that show reduced TCR diversity in pathogenic T cells in SjS and other autoimmune diseases56–59 (which even correlate disease severity with T cell expansion60) all appear to support the latter scenario. In some cases, restriction of the TCR diversity is necessary for rapid IL-17 secretion61. In the study, SjSS mice exhibited less normal distribution but a higher diversity in the CDR regions. Sequence analysis of the CDRs pointed to the presence of premature stop codons which were mapped to non-templated bases in the CDR regions, therefore they are more likely to represent real events. SjSS mice appeared to show an increase of the premature stop codons in TCRα and TCRβ chains by Th17 cells. As discussed, glandular Th17 cells are antigen-specific and hyperproliferative upon antigen-specific stimulation62,63. This finding suggests that hyperproliferation of Th17 generates more error-prone DNA rearrangements, which could generate more premature stop codons. It is possible that nonsense-mediated decay (NMD) that degrades aberrant transcripts harboring premature stop codons are not efficient or defective in SjS64, therefore truncated or pathogenic proteins from non-productively rearranged genes are being produced and possibly initiate glandular dysfunction.
The interaction of CDR3 with the SjS autoantigen has yet to be elucidated. However, hydrophobic amino acids in the N regions play a part in binding to autoantigens36,65. It is hypothesized that these hydrophobic amino acids promote additional interaction with the MHC-peptide complex to enforce a strong binding. In this manner, our data is consistent, in that hydrophobic and partially hydrophobic amino acids in these positions of the clonotypes were identified in this study; this supports that these CDR3 clonotypes were generated to an autoantigen. Although TCR repertoire analysis is being performed in other disease studies, is limited to CD4+ Th or CD8+ Tc cells and not effector subsets56,66 SjSS mice had a gene bias towards the selection of TRAV1-TRAV13. Though the significance of this bias is uncertain in SjS, the expression of the Th17 transcription regulator, RORγT, is associated with the development and survival of CD4/CD8 double positive thymocytes. RORγT skews TCRα selection towards distal Vα genes (TRAV1-TRAV6) in EAE67. Previous studies have identified certain V genes associated with SjS in the SG from humans. Sumida et al. identified the biased utilization of TRAV17-1, TRAV2, and TRAV11-1 quoting the use of GGPKT and VDxG motifs as well as TRBV2 and TRBV13 with the STxTLRNEQ motif. Motifs and pairings were unique to each SjS patient. Another study which was done on the NOD mouse concluded that 15% of their TCR repertoire was of TRBV8.1 and TRBV268. Another SjS patient study examining TCRβ observed that seven TCRβ genes were devoid in their set, two of which (TRBV16, and TRBV23) were enriched in our datasets69. The most prevalent CDR3 clonotype observed (CGGKRRLESIFR) was present in Th17 and Th1 cells while remaining exclusive to the SjSS male mice. This implies that both Th1 and Th17 cells may have a common selective pressure for autoantigen in the glands. With the divergence of results from the literature, we can see that the relationship between TCR diversity and SjS is still very complex.
This study is the first of its kind to identify sex differences in functionally distinct T cells of autoimmunity. We observed that males develop fewer Th1 and Th17 cells in the glands with unique TCR repertoires from female SjSS mice. Male Th1 and Th17 repertoires select for similar CDR3β amino acids with little selective pressure for Th17 CDR3α while female SjSS mice presented an alternative selection for hydrophobic and cationic amino acids. Furthermore, we have identified four CDR3 motifs which represent motifs associated with the SjSS mice. It is still unclear what causes the sexual bias in SjS, so it is necessary to interrogate the mechanism from which different sexes pose different degrees of selection. The temporal role of sex hormones, the balance of androgen and estrogen in different stages of life, may play a role in disease development and progression, and evident from this study, T cell selection. Lastly, the implications of the specific clonotypes and whether they inherently produce SjS or are simply indicative of the disease is still not understood.
Methods
Animals
SjSS C57BL/6 J.NOD/ShiLtJ-Aec1Aec2 (B6.NOD-Aec1/2) and non-SjSS C57BL/6 J (B6) control mice were bred and maintained under specific pathogen-free conditions in the animal facility of Animal Care Services at the University of Florida. The breeding and the use of animals described herein were approved by and carried out within accordance of the University of Florida’s Institutional Animal Care and Use Committee. All methods were performed in accordance with the relevant guidelines and regulations. Development of the B6.NOD-Aec1/2 mouse and its SjS-like disease phenotype are described elsewhere70,71. All animals were maintained on a 12-hour light-dark schedule and provided food and acidified water ad libitum. At times indicated in the study, mice were euthanized by cervical dislocation after deep anesthetization with isoflurane and their organs and tissues freshly harvested for analyses.
Single-cell microengraving process
Salivary gland lymphocytes of B6 and B6-NOD.Aec1/2 mice were isolated as previously described19. Lymphocytes were cultured in RPMI complete medium (12-167 F, Lonza) containing 10% fetal bovine serum (F4135, Sigma-Aldrich), 100 U/mL penicillin, 100 μg/mL streptomycin (SV30010, GE Life Science), 2 mM L-glutamine (25-005-Cl, Corning), 10 mM HEPES (SH30237.01, GE Life Science), 1X MEM-non essential amino acids (25-025-Cl, Corning), 1 mM pyruvate (SH30239.01,GE Life Science), and 0.0004% 2-mercaptoethanol (M3148, Sigma-Aldrich), under T-cell stimulation: 5 ug/mL anti-CD28 (553294, BD Biosciences) and 10 ug/mL anti-CD3 (145-2C11, BD Biosciences) for 64 hours at 37 °C. Following stimulation, lymphocytes were treated with 500 ng/mL Ionomycin sulfate (I3909-1ML, Sigma-Aldrich) and 50 ng/mL phorbol 12-myristate 13-acetate (PMA: P1585-1MG, Sigma-Aldrich), blocked using anti-CD16/32 TruStain FcX blocker (101320, Biolegend) for 5 minutes on ice, and then stained with cell surface markers, anti-CD3-Alexa Fluor 647 (HM3421, Invitrogen), anti-CD4-fluorescein isothiocyanate (FITC) (MCD0401, Invitrogen), anti-CD8-phycoerythrin (PE) (MCD0804, Invitrogen) as well as Calcein violet 405 nm Live/Dead marker (C4858,Invitrogen) for 30 minutes. Following surface staining, 500,000 cells were suspended in 300 μL complete medium were deposited onto the arrays of nanowells and imaged as previously described72. The nanowells were hybridized with the capture slide containing antibodies against IL-17A (AAM60, AbD Serotec) and IFN-γ (505827, Biolegend). After 2 hours, the capture slide was treated with anti-IL-17A conjugated with Dylight 594 (46413, Thermo Scientific) and anti-IFN-γ conjugated with Dylight 488 (46403, Thermo Scientific). The slide was imaged using the Genepix 4400 A scanner (Molecular Devices). Single-cell data analysis was performed as described previously72.
Sequencing of TCRs using nested polymerase chain reaction (PCR)
Cells expressing either CD4 or CD8 surface markers and either IFN-γ or IL-17A cytokines were collected using a Narashige micromanipulator loaded with a pulled glass capillary. Single, isolated cells were pipetted into PBS with 2% BSA and 200 U/mL Rnasin RNAse inhibitor (N251A, Promega) and frozen at −20 °C. Frozen single cells were directly used for reverse transcription (RT) reaction per manufacturer’s protocol (1708840, iScript cDNA Sythesis Kit, Biorad) supplemented with 0.1% Triton × 100 detergent to achieve cell lysis. Serial dilutions of about 5 cells per reaction were isolated from the spleen following a CD4+ enrichment via CD4+ untouched cell isolation kit (11141, Miltenyi Biotec) and used as positive controls. Following RT, PCR was performed using a protocol and primers described previously73. In short a 1:50 dilution of the cDNA produced from single-cell RT reaction was used as template for PCR using Primestar Taq (R010A, Clontech). Successful reactions were determined by gel electrophoresis and bands size confirmation.
Bioinformatic analysis of TRA and TRB Hypervariable Region and Motif analyses
Sanger DNA Sequencing was performed by Eton Bio (Raleigh, NC). T receptor gene and allele identification was determined using the IgBLAST tool (http://www.ncbi.nlm.nih.gov/igblast/). CDR3 sequences were determined using an in-house macro developed for use with Microsoft Excel. The macro was assembled to designate the sequence between the codon for cysteine (TGY) followed by the DNA sequence TTYGGX (where Y denotes either pyrimidine and X denotes any nucleic acid) as the CDR3 sequence. Only sequences less than 60 nucleotides were considered. This allowed for a level of stringency to identify CDR3 regions of high diversity. In some cases, unambiguous germline genes could not be defined due to insertions and deletions, however those sequences containing a canonical CDR3 sequence were retained for analysis. Motif analysis was performed using www.meme-suite.org website. Analyses of high frequency CDR3 amino acid sequences were performed using the MEME tool (http://meme-suite.org/tools/meme) to identify conserved amino acid motifs while the GLAM2 tool (http://meme-suite.org/tools/glam2) was used to identify gapped motifs among diverse datasets.
Statistical Analysis
Data was analyzed using a Kruskall-Wallis test (one-way ANOVA) with Dunn’s post hoc test. (GraphPad Prism, GraphPad Software Inc.) to determine to determine the statistical significance between female B6.NOD-Aec1/2 mice from each of the other groups. Significance was defined as a *p-value < 0.05, **p < 0.05, and ***p < 0.005. TCR Clonal diversity was determined with Shannon’s entropy as well as Simpson’s Diversity Index.
Electronic supplementary material
Acknowledgements
This study was supported financially in part by PHS grants DE023433 and AI122182 (CQN) from the National Institutes of Health (NIH), and funds from the University of Florida College of Veterinary Medicine Consolidated Faculty Research Award.
Author Contributions
Conceived and designed the experiments: A.W., C.Q.N., S.S., C.M.S., I.B., and A.V. Performed the experiments: A.W. Analyzed the data: A.W. Wrote the manuscript: A.W., C.Q.N., and A.V.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-017-12627-6.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Fox RI. Sjögren’s syndrome. The Lancet. 2005;366:321–331. doi: 10.1016/S0140-6736(05)66990-5. [DOI] [PubMed] [Google Scholar]
- 2.Chisholm DM, Mason DK. Labial salivary gland biopsy in Sjögren’s disease. Journal of Clinical Pathology. 1968;21:656–660. doi: 10.1136/jcp.21.5.656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Manganelli P, Fietta P. Apoptosis and Sjögren syndrome. Seminars in Arthritis and Rheumatism. 2003;33:49–65. doi: 10.1053/sarh.2003.50019. [DOI] [PubMed] [Google Scholar]
- 4.Mavragani CP, Moutsopoulos HM. Sjögren syndrome. CMAJ: Canadian Medical Association Journal. 2014;186:E579–E586. doi: 10.1503/cmaj.122037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Costa, S. et al. B-cell and T-cell quantification in minor salivary glands in primary Sjögren’s syndrome: development and validation of a pixel-based digital procedure. Arthritis Research & Therapy 18, 10.1186/s13075-016-0924-2 (2016). [DOI] [PMC free article] [PubMed]
- 6.Corsiero E, Sutcliffe N, Pitzalis C, Bombardieri M. Accumulation of Self-Reactive Naïve and Memory B Cell Reveals Sequential Defects in B Cell Tolerance Checkpoints in Sjögren’s Syndrome. PLoS ONE. 2014;9:e114575. doi: 10.1371/journal.pone.0114575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kapsogeorgou EK, et al. Minor Salivary Gland Inflammatory Lesions in Sjögren Syndrome: Do They Evolve? The Journal of Rheumatology. 2013;40:1566–1571. doi: 10.3899/jrheum.130256. [DOI] [PubMed] [Google Scholar]
- 8.Manoussakis MN, et al. Rates of infiltration by macrophages and dendritic cells and expression of interleukin-18 and interleukin-12 in the chronic inflammatory lesions of Sjögren’s syndrome: Correlation with certain features of immune hyperactivity and factors associated with high risk of lymphoma development. Arthritis & Rheumatism. 2007;56:3977–3988. doi: 10.1002/art.23073. [DOI] [PubMed] [Google Scholar]
- 9.Kyriakidis NC, Kapsogeorgou EK, Tzioufas AG. A comprehensive review of autoantibodies in primary Sjögren’s syndrome: Clinical phenotypes and regulatory mechanisms. Journal of Autoimmunity. 2014;51:67–74. doi: 10.1016/j.jaut.2013.11.001. [DOI] [PubMed] [Google Scholar]
- 10.Nguyen CQ, Hu MH, Li Y, Stewart C, Peck AB. Salivary Gland Tissue Expression of Interleukin-23 and Interleukin-17 in Sjögren’s Syndrome. Arthritis and rheumatism. 2008;58:734–743. doi: 10.1002/art.23214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nardi N, et al. Circulating auto-antibodies against nuclear and non-nuclear antigens in primary Sjögren’s syndrome. Clinical Rheumatology. 2005;25:341–346. doi: 10.1007/s10067-005-0059-3. [DOI] [PubMed] [Google Scholar]
- 12.Nguyen C, Peck AB. The interferon-signature of Sjögren’s syndrome: how unique biomarkers can identify underlying inflammatory and immunopathological mechanisms of specific diseases. Immunotherapies and Vaccines. 2013;4:142. doi: 10.3389/fimmu.2013.00142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cha S, et al. A Dual Role for Interferon-γ in the Pathogenesis of Sjögren’s Syndrome-Like Autoimmune Exocrinopathy in the Nonobese Diabetic Mouse. Scandinavian Journal of Immunology. 2004;60:552–565. doi: 10.1111/j.0300-9475.2004.01508.x. [DOI] [PubMed] [Google Scholar]
- 14.Lin X, et al. Th17 cells play a critical role in the development of experimental Sjogren’s syndrome. Ann Rheum Dis. 2015;74:1302–1310. doi: 10.1136/annrheumdis-2013-204584. [DOI] [PubMed] [Google Scholar]
- 15.Nguyen CQ, et al. Pathogenic effect of interleukin-17A in induction of Sjogren’s syndrome-like disease using adenovirus-mediated gene transfer. Arthritis Res Ther. 2010;12:R220. doi: 10.1186/ar3207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sakai A, Sugawara Y, Kuroishi T, Sasano T, Sugawara S. Identification of IL-18 and Th17 cells in salivary glands of patients with Sjogren’s syndrome, and amplification of IL-17-mediated secretion of inflammatory cytokines from salivary gland cells by IL-18. J Immunol. 2008;181:2898–2906. doi: 10.4049/jimmunol.181.4.2898. [DOI] [PubMed] [Google Scholar]
- 17.Weaver CT, Hatton RD, Mangan PR, Harrington LE. IL-17 Family Cytokines and the Expanding Diversity of Effector T Cell Lineages. Annual Review of Immunology. 2007;25:821–852. doi: 10.1146/annurev.immunol.25.022106.141557. [DOI] [PubMed] [Google Scholar]
- 18.Lavoie TN, et al. IL-22 regulation of functional gene expression in salivary gland cells. Genomics Data. 2015;7:178–184. doi: 10.1016/j.gdata.2015.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Voigt, A., Esfandiary, L. & Nguyen, C. Q. Sexual dimorphism in an animal model of Sjögren’s syndrome: a potential role for Th17 cells. Biology Open, bio.013771, 10.1242/bio.013771 (2015). [DOI] [PMC free article] [PubMed]
- 20.Voigt A, et al. Sexual dimorphic function of IL-17 in salivary gland dysfunction of the C57BL/6.NOD-Aec1Aec2 model of Sjögren’s syndrome. Scientific Reports. 2016;6:38717. doi: 10.1038/srep38717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mittrücker H-W, Visekruna A, Huber M. Heterogeneity in the Differentiation and Function of CD8+ T Cells. Archivum Immunologiae et Therapiae Experimentalis. 2014;62:449–458. doi: 10.1007/s00005-014-0293-y. [DOI] [PubMed] [Google Scholar]
- 22.Liang Y, Pan H-F, Ye D-Q. Tc17 Cells in Immunity and Systemic Autoimmunity. International Reviews of Immunology. 2015;34:318–331. doi: 10.3109/08830185.2014.954698. [DOI] [PubMed] [Google Scholar]
- 23.Huber M, et al. A Th17-like developmental process leads to CD8+ Tc17 cells with reduced cytotoxic activity. European Journal of Immunology. 2009;39:1716–1725. doi: 10.1002/eji.200939412. [DOI] [PubMed] [Google Scholar]
- 24.Zumla A, Mathur M, Stewart J, Wilkinson L, Isenberg D. T cell receptor expression in Sjögren’s syndrome. Annals of the Rheumatic Diseases. 1991;50:691–693. doi: 10.1136/ard.50.10.691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Garcia KC, Adams EJ. How the T Cell Receptor Sees Antigen—A Structural View. Cell. 2005;122:333–336. doi: 10.1016/j.cell.2005.07.015. [DOI] [PubMed] [Google Scholar]
- 26.Mora, T. & Walczak, A. Quantifying lymphocyte receptor diversity. (2016).
- 27.Eltahla AA, et al. Linking the T cell receptor to the single cell transcriptome in antigen-specific human T cells. Immunology and Cell Biology. 2016;94:604–611. doi: 10.1038/icb.2016.16. [DOI] [PubMed] [Google Scholar]
- 28.Kern J, Drutel R, Leanhart S, Bogacz M, Pacholczyk R. Reduction of T Cell Receptor Diversity in NOD Mice Prevents Development of Type 1 Diabetes but Not Sjögren’s Syndrome. PLOS ONE. 2014;9:e112467. doi: 10.1371/journal.pone.0112467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Murray, J. S. An old Twist in HLA-A: CDR3α Hook up at an R65-joint. Frontiers in Immunology6, 10.3389/fimmu.2015.00268 (2015). [DOI] [PMC free article] [PubMed]
- 30.Garcia KC, et al. An αβ T Cell Receptor Structure at 2.5 Å and Its Orientation in the TCR-MHC Complex. Science. 1996;274:209–219. doi: 10.1126/science.274.5285.209. [DOI] [PubMed] [Google Scholar]
- 31.Saada R, Weinberger M, Shahaf G, Mehr R. Models for antigen receptor gene rearrangement: CDR3 length. Immunology and Cell Biology. 2007;85:323–332. doi: 10.1038/sj.icb.7100055. [DOI] [PubMed] [Google Scholar]
- 32.Kepler TB, Borrero M, Rugerio B, McCray SK, Clarke SH. Interdependence of N nucleotide addition and recombination site choice in V(D)J rearrangement. The Journal of Immunology. 1996;157:4451–4457. [PubMed] [Google Scholar]
- 33.Katsifis, G. E., Rekka, S., Moutsopoulos, N. M., Pillemer, S. & Wahl, S. M. Systemic and local interleukin-17 and linked cytokines associated with Sjogren’s syndrome immunopathogenesis. Am J Pathol175, 1167–1177, doi:ajpath.2009.090319 (2009). [DOI] [PMC free article] [PubMed]
- 34.Hall JC, et al. Molecular Subsetting of Interferon Pathways in Sjogren’s Syndrome. Arthritis & rheumatology. 2015;67:2437–2446. doi: 10.1002/art.39204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nezos A, et al. Type I and II interferon signatures in Sjogren’s syndrome pathogenesis: Contributions in distinct clinical phenotypes and Sjogren’s related lymphomagenesis. Journal of autoimmunity. 2015;63:47–58. doi: 10.1016/j.jaut.2015.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stadinski BD, et al. Hydrophobic CDR3 residues promote the development of self-reactive T cells. Nature Immunology. 2016;17:946–955. doi: 10.1038/ni.3491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nguyen, C. Q. et al. Differential gene expression in the salivary gland during development and onset of xerostomia in Sjogren’s syndrome-like disease of the C57BL/6.NOD-Aec1Aec2 mouse. Arthritis Res Ther11, R56, doi:ar2676 (2009). [DOI] [PMC free article] [PubMed]
- 38.Hall JC, et al. Precise probes of type II interferon activity define the origin of interferon signatures in target tissues in rheumatic diseases. Proc Natl Acad Sci USA. 2012;109:17609–17614. doi: 10.1073/pnas.1209724109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mitsdoerffer, M. et al. Proinflammatory T helper type 17 cells are effective B-cell helpers. Proc Natl Acad Sci USA107, 14292–14297, doi:1009234107 (2010). [DOI] [PMC free article] [PubMed]
- 40.Pisitkun P, et al. Interleukin-17 cytokines are critical in development of fatal lupus glomerulonephritis. Immunity. 2012;37:1104–1115. doi: 10.1016/j.immuni.2012.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tabarkiewicz J, Pogoda K, Karczmarczyk A, Pozarowski P, Giannopoulos K. The Role of IL-17 and Th17 Lymphocytes in Autoimmune Diseases. Archivum Immunologiae et Therapiae Experimentalis. 2015;63:435–449. doi: 10.1007/s00005-015-0344-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Libert C, Dejager L, Pinheiro I. The X chromosome in immune functions: when a chromosome makes the difference. Nature Reviews Immunology. 2010;10:594–604. doi: 10.1038/nri2815. [DOI] [PubMed] [Google Scholar]
- 43.Whitacre CC, et al. A Gender Gap in Autoimmunity. Science. 1999;283:1277–1278. doi: 10.1126/science.283.5406.1277. [DOI] [PubMed] [Google Scholar]
- 44.Dalal M, Kim S, Voskuhl RR. Testosterone therapy ameliorates experimental autoimmune encephalomyelitis and induces a T helper 2 bias in the autoantigen-specific T lymphocyte response. The Journal of Immunology. 1997;159:3–6. [PubMed] [Google Scholar]
- 45.Voskuhl R. Sex differences in autoimmune diseases. Biology of Sex Differences. 2011;2:1. doi: 10.1186/2042-6410-2-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Andersson A, et al. IL-17-producing γδT cells are regulated by estrogen during development of experimental arthritis. Clinical Immunology. 2015;161:324–332. doi: 10.1016/j.clim.2015.09.014. [DOI] [PubMed] [Google Scholar]
- 47.Andersson A, et al. Estrogen regulates T helper 17 phenotype and localization in experimental autoimmune arthritis. Arthritis Research & Therapy. 2015;17:32. doi: 10.1186/s13075-015-0548-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Newcomb DC, et al. Estrogen and progesterone decrease Let-7f miRNA expression and increase IL-23/IL-23R signaling and IL-17A production in severe asthma. The Journal of allergy and clinical immunology. 2015;136:1025–1034.e1011. doi: 10.1016/j.jaci.2015.05.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Voulgarelis M, Skopouli FN. Clinical, immunologic, and molecular factors predicting lymphoma development in Sjogren’s syndrome patients. Clin Rev Allergy Immunol. 2007;32:265–274. doi: 10.1007/s12016-007-8001-x. [DOI] [PubMed] [Google Scholar]
- 50.Fox RI, Kang HI. Pathogenesis of Sjogren’s syndrome. Rheum. Dis. Clin. North Am. 1992;18:517–538. [PubMed] [Google Scholar]
- 51.Gordon TP, Bolstad AI, Rischmueller M, Jonsson R, Waterman SA. Autoantibodies in primary Sjogren’s syndrome: new insights into mechanisms of autoantibody diversification and disease pathogenesis. Autoimmunity. 2001;34:123–132. doi: 10.3109/08916930109001960. [DOI] [PubMed] [Google Scholar]
- 52.Vinuesa CG, Tangye SG, Moser B, Mackay CR. Follicular B helper T cells in antibody responses and autoimmunity. Nature reviews. Immunology. 2005;5:853–865. doi: 10.1038/nri1714. [DOI] [PubMed] [Google Scholar]
- 53.Maehara T, et al. Selective localization of T helper subsets in labial salivary glands from primary Sjogren’s syndrome patients. Clin Exp Immunol. 2012;169:89–99. doi: 10.1111/j.1365-2249.2012.04606.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Brayer JB, et al. IL-4-dependent effector phase in autoimmune exocrinopathy as defined by the NOD.IL-4-gene knockout mouse model of Sjogren’s syndrome. Scand. J. Immunol. 2001;54:133–140. doi: 10.1046/j.1365-3083.2001.00958.x. [DOI] [PubMed] [Google Scholar]
- 55.Gao J, et al. Sjogren’s syndrome in the NOD mouse model is an interleukin-4 time-dependent, antibody isotype-specific autoimmune disease. J. Autoimmun. 2006;26:90–103. doi: 10.1016/j.jaut.2005.11.004. [DOI] [PubMed] [Google Scholar]
- 56.Schneider-Hohendorf T, et al. CD8+ T-cell pathogenicity in Rasmussen encephalitis elucidated by large-scale T-cell receptor sequencing. Nature Communications. 2016;7:11153. doi: 10.1038/ncomms11153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Thapa, D. R. et al. Longitudinal analysis of peripheral blood T cell receptor diversity in patients with systemic lupus erythematosus by next-generation sequencing. Arthritis Research & Therapy 17, 10.1186/s13075-015-0655-9 (2015). [DOI] [PMC free article] [PubMed]
- 58.Imberti, L. et al. Reduced T-cell repertoire restrictions in abatacept-treated rheumatoid arthritis patients. Journal of Translational Medicine13, 10.1186/s12967-014-0363-2 (2015). [DOI] [PMC free article] [PubMed]
- 59.Okajima M, et al. Analysis of T cell receptor Vβ diversity in peripheral CD4+ and CD8+ T lymphocytes in patients with autoimmune thyroid diseases. Clinical and Experimental Immunology. 2009;155:166–172. doi: 10.1111/j.1365-2249.2008.03842.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Joachims, M. L. et al. Single-cell analysis of glandular T cell receptors in Sjögren’s syndrome. JCI Insight1, 10.1172/jci.insight.85609. [DOI] [PMC free article] [PubMed]
- 61.Wei Y-L, et al. A highly focused antigen receptor repertoire characterizes γδ T cells that are poised to make IL-17 rapidly in naive animals. T Cell Biology. 2015;6:118. doi: 10.3389/fimmu.2015.00118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Voigt A, Esfandiary L, Nguyen CQ. Sexual dimorphism in an animal model of Sjogren’s syndrome: a potential role for Th17 cells. Biol Open. 2015;4:1410–1419. doi: 10.1242/bio.013771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Voigt A, et al. Sexual dimorphic function of IL-17 in salivary gland dysfunction of the C57BL/6.NOD-Aec1Aec2 model of Sjogren’s syndrome. Scientific reports. 2016;6:38717. doi: 10.1038/srep38717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wang J, Vock VM, Li S, Olivas OR, Wilkinson MF. A quality control pathway that down-regulates aberrant T-cell receptor (TCR) transcripts by a mechanism requiring UPF2 and translation. J Biol Chem. 2002;277:18489–18493. doi: 10.1074/jbc.M111781200. [DOI] [PubMed] [Google Scholar]
- 65.Chowell D, et al. TCR contact residue hydrophobicity is a hallmark of immunogenic CD8+ T cell epitopes. Proceedings of the National Academy of Sciences of the United States of America. 2015;112:E1754–1762. doi: 10.1073/pnas.1500973112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Dong L, Li P, Oenema T, McClurkan CL, Koelle DM. Public TCR use by HSV-2-specific human CD8 CTL. Journal of immunology (Baltimore, Md. : 1950) 2010;184:3063–3071. doi: 10.4049/jimmunol.0903622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Guo Y, et al. Inhibition of RORgammaT Skews TCRalpha Gene Rearrangement and Limits T Cell Repertoire Diversity. Cell Rep. 2016;17:3206–3218. doi: 10.1016/j.celrep.2016.11.073. [DOI] [PubMed] [Google Scholar]
- 68.Skarstein K, Wahren M, Zaura E, Hattori M, Jonsson R. Characterization of T cell receptor repertoire and anti-Ro/SSA autoantibodies in relation to sialadenitis of NOD mice. Autoimmunity. 1995;22:9–16. doi: 10.3109/08916939508995294. [DOI] [PubMed] [Google Scholar]
- 69.Dwyer E, Itescu S, Winchester R. Characterization of the primary structure of T cell receptor beta chains in cells infiltrating the salivary gland in the sicca syndrome of HIV-1 infection. Evidence of antigen-driven clonal selection suggested by restricted combinations of V beta J beta gene segment usage and shared somatically encoded amino acid residues. J Clin Invest. 1993;92:495–502. doi: 10.1172/JCI116593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Brayer J, et al. Alleles from chromosomes 1 and 3 of NOD mice combine to influence Sjogren’s syndrome-like autoimmune exocrinopathy. J Rheumatol. 2000;27:1896–1904. [PubMed] [Google Scholar]
- 71.Cha S, Nagashima H, Peck AB, Humphreys-Beher MG. IDD3 and IDD5 alleles from nod mice mediate Sjogren’s syndrome-like autoimmunity. Advances in experimental medicine and biology. 2002;506:1035–1039. doi: 10.1007/978-1-4615-0717-8_146. [DOI] [PubMed] [Google Scholar]
- 72.Love JC, Ronan JL, Grotenbreg GM, van der Veen AG, Ploegh HL. A microengraving method for rapid selection of single cells producing antigen-specific antibodies. Nature Biotechnology. 2006;24:703–707. doi: 10.1038/nbt1210. [DOI] [PubMed] [Google Scholar]
- 73.Dash P, et al. Paired analysis of TCRα and TCRβ chains at the single-cell level in mice. The Journal of Clinical Investigation. 2011;121:288–295. doi: 10.1172/JCI44752. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.