Abstract
The Wnt-signaling pathway is crucial to cell proliferation, differentiation, and migration. The secreted Frizzled-related proteins (sFRPs) represent the largest family of secreted Wnt inhibitors. However, their function in antagonizing Wnt signaling has remained somewhat controversial. Here, we report the crystal structure of Sizzled from Xenopus laevis, the first full-length structure of an sFRP. Tethered by an inter-domain disulfide bond and a linker, the N-terminal cysteine-rich domain (CRD) and the C-terminal netrin-like domain (NTR) of Sizzled are arranged in a tandem fashion, with the NTR domain occluding the groove of CRD for Wnt accessibility. A Dual-Luciferase assay demonstrated that removing the NTR domain and replacing the CRD groove residues His-116 and His-118 with aromatic residues may significantly enhance antagonistic function of Sizzled in inhibiting Wnt3A signaling. Sizzled is a monomer in solution, and Sizzled CRD exhibited different packing in the crystal, suggesting that sFRPs do not have a conserved CRD dimerization mode. Distinct from the canonical NTR domain, the Sizzled NTR adopts a novel α/β folding with two perpendicular helices facing the central mixed β-sheet. The subgroup of human sFRP1/2/5 and Sizzled should have a similar NTR domain that features a highly positively charged region, opposite the NTR–CRD interface, suggesting that the NTR domain in human sFRPs, at least sFRP1/2/5, is unlikely to bind to Wnt but is likely involved in biphasic Wnt signaling modulation. In summary, the Sizzled structure provides the first insights into how the CRD and the NTR domains relate to each other for modulating Wnt-antagonistic function of sFRPs.
Keywords: crystal structure, protein crystallization, recombinant protein expression, structure-function, Wnt signaling, Sizzled, antagonistic function, sFRPs
Introduction
The Wnt-signaling pathway plays a crucial role in mediating vertebrate and invertebrate development due to its contributions to cell proliferation, differentiation, and migration (1–4). Wnt signaling is also implicated in cancers and neurological disorders, and therefore it serves as potential targets for anti-cancer and pro-regeneration therapies (5–7). The Wnt genes encode secreted, cysteine-rich glycoproteins that activate receptors and co-receptors located on responder cell surface to initiate different signaling pathways, including the canonical β-catenin pathway, non-canonical planar cell polarity, and Ca2+ pathways (4, 8). The canonical Wnt pathway is activated by the binding of canonical Wnts (such as Wnt3A) to their receptors (like Frizzled7) and co-receptors, like LRP5/6. As a result, β-catenin accumulates in the cytosol and translocates to the nucleus for the activation of β-catenin–TCF transcriptional complexes (1). In contrast with Wnt3A, the non-canonical Wnts such as Wnt5A, simulate a non-canonical, β-catenin-independent pathway through binding to receptors like ROR2 (9).
The sFRPs4 represent the largest family of secreted Wnt inhibitors and have a region with high homology to the Wnt-binding CRD of the Frizzled family of Wnt receptors (10). In humans, the sFRP family includes five members, sFRP1 to sFRP5, that can be divided into two subgroups (sFRP1/2/5 versus sFRP3/4) based on phylogenetic analysis (11). A third subgroup of sFRPs, including Sizzled, Crescent, and Tlc, has been identified in Xenopus, zebrafish, and chick, and it shares sequence similarities with the sFRP1/2/5 subgroup (11). The sFRPs consist of 280–360 residues that fold into a CRD at the N terminus and a netrin-like domain (NTR) at the C-terminal end. The CRDs of sFRPs have relatively high sequence similarity and contain 10 conserved cysteine residues that form a pattern of five disulfide bonds as validated in the crystal structure of mouse sFRP3 CRD (12). The NTR domains of sFRPs were also found in the axon guidance protein netrin-1 and complement proteins (13). The sFRP1 NTR was claimed to feature with a heparin-binding motif (14) and three disulfide bridges (15). Different subgroups of sFRP appear to have alternative disulfide linkage patterns (15).
The CRD of sFRPs shares 30–50% sequence similarity with that of Wnt receptors, Frizzleds, which endows sFRPs with potentials to recognize Wnts. For example, either Wnt3A or Wnt5A binds to several sFRPs with affinities in the nanomolar range similar to that of Wnts for Frizzleds (16). Therefore, it is anticipated that sFRPs can effectively sequester Wnts away from active receptor complexes and thus inhibit both canonical and non-canonical Wnt signaling (10). In line with this standpoint, sFRP1 from vascular endothelium may specifically bind to Wnt-1 and inhibit its signaling (17), and deletion of the sFRP1 CRD may lead to the complete loss of Wnt antagonist function (18). sFRP2 may inhibit the ability of ectopic Wnt3A to stimulate proliferation in the developing chick neural tube (19) or reduce the dermomyotome-inducing activity of Wnt1 and Wnt4 in the somitic mesoderm (20). The biochemical and functional studies on Xenopus embryos and cultured cells demonstrated that sFRP3 binds to Wnt1 and XWnt8 and inhibits canonical Wnt signaling and that the CRD of sFRP3 is necessary and sufficient for both Wnt-binding and functional activities (21, 22). sFRP4 antagonizes the canonical Wnt-signaling pathway, resulting in decreased cellular proliferation and increased apoptosis of endothelial cells in angiogenesis (23). sFRP5 may coordinate foregut specification and morphogenesis by antagonizing both canonical and noncanonical Wnt11 signaling (24) or suppressing adipocyte mitochondrial metabolism through Wnt inhibition (25). Sizzled was also found to be a secreted XWnt8 antagonist expressed in the ventral marginal zone of Xenopus embryos (26). In addition to sFRPs, other endogenous CRD-harboring molecules, like V3Nter, bind to and inhibit Wnt signaling (27), which further demonstrates that the CRD seems to be determinant for Wnt-antagonistic function of sFRPs.
However, in contrast to the initial report of Sizzled's XWnt8 antagonistic function (26), subsequent studies showed that Sizzled was not able to antagonize Wnt signaling (28–30). A construct of sFRP1 with the CRD deleted retains the ability to bind to Wingless, the Drosophila Wnt homolog (14). As claimed by a study on sFRP1 in cell culture and medakafish embryos, the NTR domain mimics the function of the entire molecule in binding Wnt8 and inhibiting Wnt signaling (31). Furthermore, consistent with the previous report that sFRP1 is a biphasic modulator of Wnt signaling (14), recent biochemical and cellular studies suggested that sFRP1 may either inhibit or enhance canonical Wnt3A signaling in a concentration- and context-dependent way (32). More interestingly, sFRP1 interacts with Frizzled2 and regulates the growth of retinal ganglion cell axons through this receptor (33). sFRP2 may stabilize the Wnt5A-RoArg-2 complex at the membrane and promote the non-canonical Wnt5A signaling (34). All these data seemingly contradict the common function of sFRPs in antagonizing Wnt signaling by their CRDs that may prevent ligand-receptor interactions (10).
How to understand the above apparently conflicting results? One interpretation was that sFRPs might have multiple Wnt-binding sites, and the NTR might recognize Wnt (10). The other interpretation was that sFRPs may form homo- and heteromeric complexes through their CRDs with other CRD-containing Wnt receptors such as Frizzled (33) or ROR2 (34) so that sFRPs may modulate Wnt signaling in a receptor-dependent manner. Current structural knowledge of sFRPs is limited to the crystal structure of mouse sFRP3 CRD that implicated the potential for the CRDs to form a dimer (12). Nevertheless, whether the other sFRPs have the same dimerization mode as sFRP3 remains unknown. The recently published crystal structure of Xenopus Wnt8 (XWnt8) in complex with mouse Frizzled8 CRD revealed that the Frizzled CRD bears a deep groove and a hydrophobic depression, respectively, engaged by the thumb and index finger motifs of Wnt (4). However, there is lack of structural information for the NTR domain of sFRPs, regardless of the full-length sFRPs. Therefore, whether the NTR domain can recognize Wnt and how the two domains coordinate for Wnt binding and inhibition need more investigation at the structural level. Here, we present the crystal structure of the full-length Sizzled from Xenopus laevis, which, to our knowledge, is the first full-length structure of an sFRP. This structure not only provides the first insights into how the CRD and the NTR domains relate to each other, but it also shed lights on how to understand Wnt-antagonistic function of sFRPs.
Results
Structure determination by in-house sulfur-SAD phasing
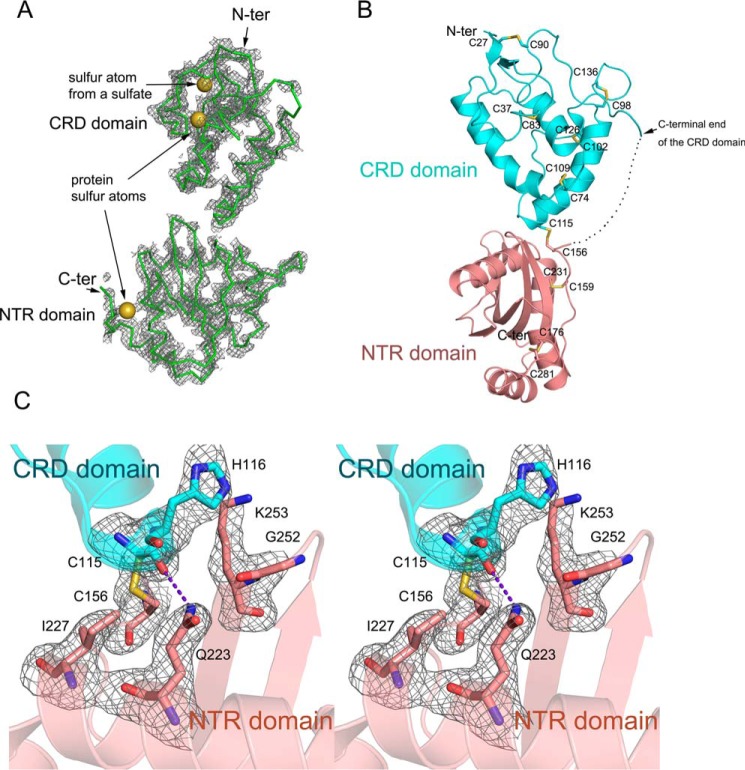
Sizzled (Uniprot code O73821) is a 281-residue secreted protein consisting of 16 cysteine residues. For guaranteeing its correct folding and disulfide-bond formation during recombinant expression, we utilized the BacMam system that facilitates post-translational modification for proteins (35). Sizzled was transiently expressed in the baculovirus-transduced mammalian cells and secreted into conditioned medium with a yield of ∼0.5 mg of protein per liter of medium. The purified Sizzled produced high-quality brick-like crystals that diffracted beyond 2.6-Å Bragg spacings on an in-house CuKα X-ray generator. Because of the high sulfur content of Sizzled, we used a SAD method for phasing. A dataset with a total oscillation range of 360° was acquired because high-redundancy data may increase signal-to-noise level for reflections and thereby anomalous scattering signals (36, 37). This dataset had a low Rmerge factor of 5.1% for all reflections, a relatively high data multiplicity of about 26, and a considerable mean anomalous difference of 3.5% (supplemental Table S1). Using AutoSol, three sulfurs were successfully located, and the initial density map calculated with experimental phases derived from the three sulfurs was clear enough for tracing ∼220 residues. It is worthy to mention that one sulfur atom was derived from solvent sulfate in reagents likely used for protein crystallization or cell culture (Fig. 1A). For refinement, a higher-resolution dataset was collected from synchrotron facility. The final model has Rwork/Rfree values of 21.3/23.0, respectively. Calculated with the phases from the final structure refined against the in-house data, an anomalous difference Fourier map clearly illustrates the sites of eight pairs of disulfide bonds (supplemental Fig. S1). The successful determination of Sizzled crystal structure sets good proof for the concept that the weak anomalous diffraction signals can be used for routine structure determination (38).
Figure 1.
Structure determination and overall structure of Sizzled. A, initial 2Fo − Fc electron density map (contour level, 1.2 σ; curve parameter, 1.6 Å) calculated with experimental phases derived from three sulfurs (yellow spheres). B, ribbon model of overall structure of Sizzled with the inter-domain linker (residues 143–154) represented by a dotted line and disulfide bonds by sticks. C, residues distributed on the CRD-NTR interface are shown by stick representation. The Fo − Fc electron density map (contour level, 3.0 σ) was calculated with phases derived from the final model except for these residues. The purple dashed line indicates the hydrogen bond.
Overall structure of Sizzled
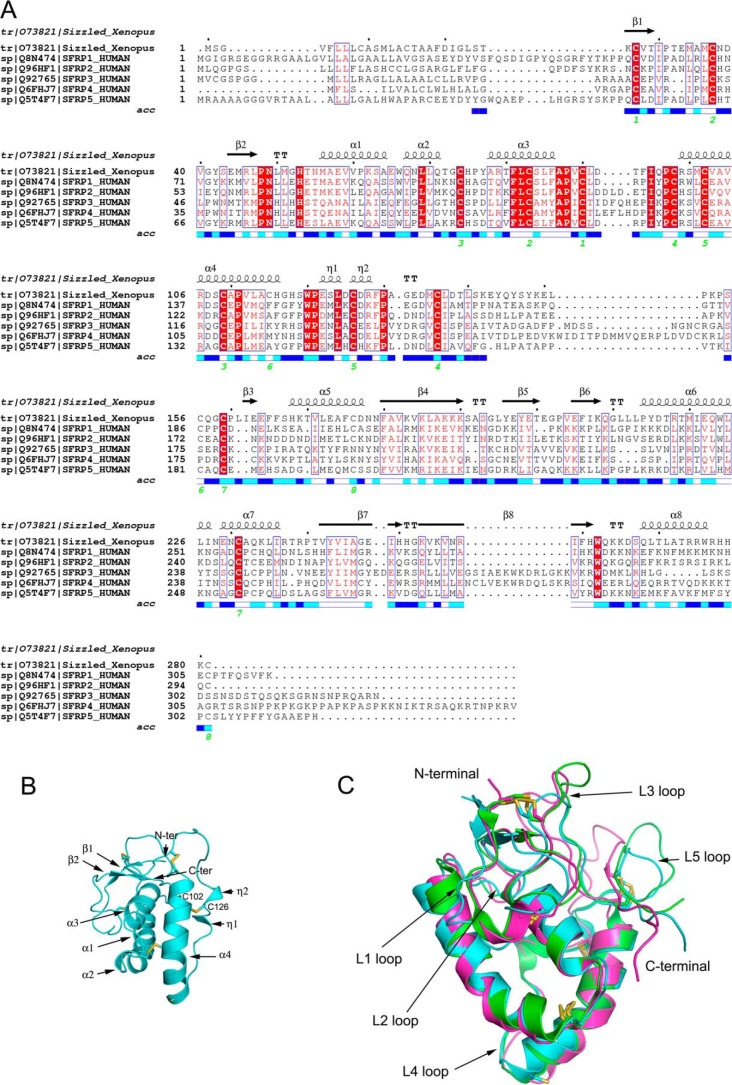
The overall structure of Sizzled resembles the arabic numeral “8,” and the elongated shape has a length of 75 Å and a maximal width of 50 Å (Fig. 1B). The N-terminal CRD (residues 24–142) packs onto the tip of the C-terminal NTR domain (residues 155–281) in a tandem fashion, only burying ∼444 Å2 total surface area at interface. Traversing the interfacial center is an inter-domain disulfide bond formed by Cys-115 of the CRD and Cys-156 of the NTR domain. Besides the two cysteines, His-116, Ile-227, Gln-223, and Lys-253 are involved in the interface and form a few van der Waals interactions and a hydrogen bond between the Cys-115 carbonyl oxygen and the amine nitrogen of Gln-223 (Fig. 1C). The linker between the two domains, including residues 143–154, was not visible in the density map, likely due to its high flexibility and multiple conformations. Although the linker may facilitate tethering the two domains together, it can be inferred that the Cys-115–Cys-156 disulfide bridge constitutes the main force for stabilizing the overall structure of Sizzled. In addition to the interdomain disulfide bridge Cys-115–Cys-156, Sizzled has seven more disulfide pairs. The five disulfide bonds Cys-27–Cys-90, Cys-37–Cys-83, Cys-74–Cys-109, Cys-98–Cys-136 and Cys-102–Cys-126 in the CRD are absolutely conserved among the human sFRP family members, whereas the other two, Cys-159–Cys-231 and Cys-176–Cys-281, are not strictly conserved (Fig. 2A). This suggests that the two domains of Sizzled have different extents of topological conservation in comparison with their counterparts in human sFRPs.
Figure 2.
Sequence alignment of sFRPs and structural mode of Sizzled CRD. A, sequence alignment of X. laevis Sizzled and human sFRPs based on Sizzled structure with paired cysteines labeled by green numbers. The solvent accessibility (acc) of each Sizzled residue is presented from white to blue. The deeper blue means higher solvent accessibility. B, ribbon model of Sizzled CRD with secondary structure elements labeled. C, superimposition of X. laevis Sizzled (cyan), mouse Frizzled 8 (green), and mouse sFRP3 (pink) CRD ribbon models. Inter-helical loops are shown.
CRD structure and its topological conservation
The Sizzled CRD is primarily α-helical with ∼50% residues assigned to four α-helices, α1–α4. Preceding the long helix α1, the N-terminal region folds into a minimal two-strand β-sheet with one strand passing through a cysteine knot founded by disulfide bonds Cys-27–Cys-90 and Cys-37–Cys-83. Following α4, the C-terminal fragment comprises two short 310 helices, η1 and η2 (Fig. 2B). The η1 helix is initiated by a conserved acidic residue Glu-122 and ends with a conserved hydrophobic residue Leu-124, whereas the η2 helix starts from the highly conserved residue Cys-126 that forms a disulfide bond with Cys-102 on the long helix α4. As introduced previously, all cysteines in the CRD except Cys-115, form five absolutely conserved disulfide bonds and cross-link the relatively conserved secondary structural elements (long helices α1–α4 and short helices η1–η2 and β-strands) of this domain into a compact structure. Therefore, it is not unexpected that the structure of Sizzled CRD is topologically similar to those of mouse sFRP3 (12) and mouse Frizzled-8 (4) with the root mean-square deviation (r.m.s.d.) of 1.32 and 1.35 Å for 90 and 97 aligned Cα atoms, respectively. In the meantime, it should be noted that their structural variation mainly lies in the loops L1 (connecting strands β1 and β2), L3 (between helices α3 and α4), and L5 (between 310 helix η2 and C136) as well as their C-terminal extremities (Fig. 2C).
NTR domain structure and its novel structural features
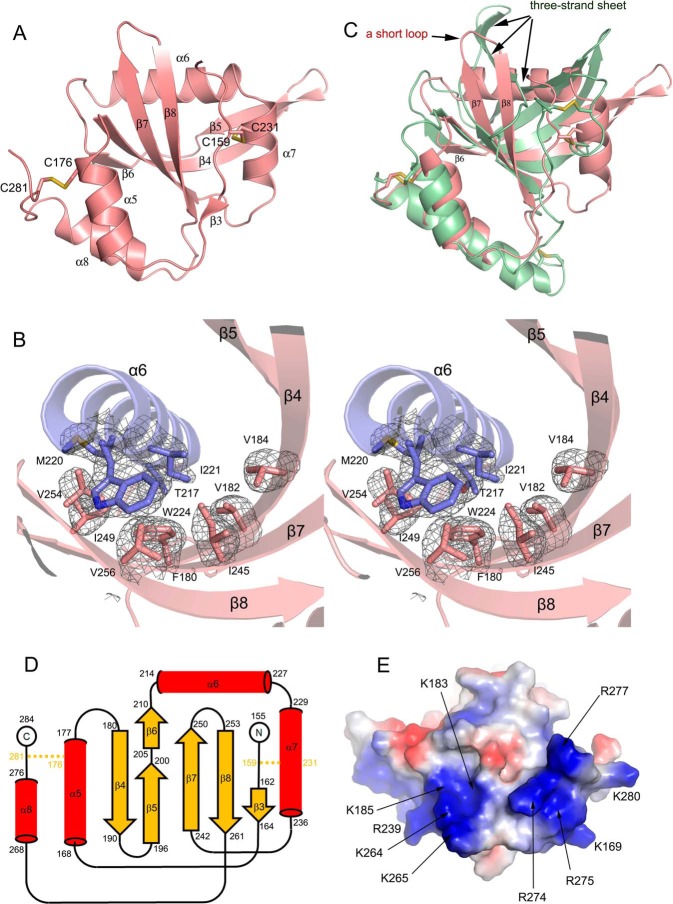
The NTR domain adopts a α/β fold with a mixed β-sheet flanked by helices. The β-sheet consists of six β-strands, β3–β8, with the longest strand β4 located in the center. The strand β4 is twisted and can be divided into two portions (Fig. 3A). At the N-terminal portion, β4 forms an antiparallel β-sheet with β6, β7, and β8, whereas its C-terminal portion is followed by β5 that is arranged with β4 also in an antiparallel fashion. The N-terminal segment of NTR is flexible except that it assumes a minimal β-strand at residues 162–164, named β3, which forms a short parallel sheet with the C-terminal residues of β8 (Fig. 3A).
Figure 3.
Structural novelties of Sizzled NTR domain. A, ribbon model of Sizzled NTR domain with disulfide bonds shown in stick representation. B, stereoview of hydrophobic residue contacts among α6 (slate) and the central mixed β-sheet (salmon). The side chains of the residues are shown as sticks. The Fo − Fc electron density map (gray mesh) for these residues were contoured at 3.0 σ. C, overlay of Sizzled NTR (salmon) with human complement C5 NTR domain (grass green). D, secondary-structure diagram of the Sizzled NTR fold. Disulfide bridges are indicated by orange lines. E, bottom surface of Sizzled NTR colored by electrostatic potential from red (negatively charged) to blue (positively charged). Conserved residues are labeled with arrows.
Because the twisted β4 interacts with different β-strands separately at its two termini, the total mixed β-sheet looks like a bent boat. Facing the concave side of the boat, two amphipathic helices α6 and α7 are orthogonally arranged and link β6 and β7 with the long axis of α6 being perpendicular to β7. The hydrophobic residues Met-220, Ile-221, Trp-224, and Leu-225 from α6 (covering residues 214–227) and Ala-232, Leu-235, and Ile-236 from α7 (residues 229–236) are packed against the inward-facing side chains of a constellation of hydrophobic residues Phe-180, Val-182, Val-184, Ile-245, Ile-249, Val-254, and Val-256 from the β-sheet, thereby forming a hydrophobic core for stabilizing the NTR configuration (Fig. 3B). In addition, α7 is cross-linked to the N-terminal flexible fragment of NTR by the disulfide bond Cys-159–Cys-231. Overhanging at the bottom of the boat are two helices. One is the helix α5 (including residues 168–177) that connects β3 and β4 with its long axis in parallel with β8. The other one is α8, covering residues 268–276, that follows β8 with five residues as a spacer. The residues Val-171, Ala-174, and Phe-175 on α5 and Leu-269, Ala-272, and Trp-276 on α8 form extensive van der Waals interactions with the hydrophobic residues distributed on the convex side of the sheet, such as Ala-181 of β4, Ile-207 of β6, Val-244 and Ala-246 of β7, and Phe-260 of β8. Moreover, the helix α5 is bridged to the C terminus of NTR by the disulfide bond Cys-176–Cys-281 that may facilitate tethering α5 and α8 together. In short, it is hydrophobic amino acid contacts and disulfide-bond bridges that maintain interactions between four helices and the mixed β-sheet.
As identified by the program DALI (39), the most significant structural homology with Sizzled NTR domain is the human complement C5 NTR domain (PDB code 5hce; residues 1532–1676) (40), with a relatively high score of 8.1. However, superimposition of the two structures leads to a large r.m.s.d. value of 3.7 Å for 105 aligned Cα atoms. As shown in Fig. 3C, in the core space facing the concave side of the central β-sheet, Sizzled adopts two perpendicular helices (α6 and α7), whereas human complement C5 assumes a twisted three-strand sheet. The strands β7 and β8 in Sizzled are interspaced by a short loop, whereas their counterparts in human complement C5 are linked by a long sequence that folds into two strands of the twisted three-strand sheet. More intriguingly, helix α6 of Sizzled is replaced by a strand in human complement C5. Such a striking difference in type and arrangement of secondary structural elements suggests that Sizzled NTR domain is a novel fold (Fig. 3D).
In addition, Sizzled NTR features a highly positively charged “bottom” surface, opposite the NTR-CRD interface. On the bottom surface, two adjacent positively charged regions are noteworthy. One region is composed of a spatial continuum of Lys-183, Lys-185, Arg-239, Lys-264, and Lys-265, whereas the other consists of Lys-169, Arg-274, Arg-275, Arg-277, and Lys-280 (Fig. 3E). Except for Lys-169, Arg-239, and Lys-280, other residues are highly conserved among sFRP1/2/5. In particular, the second region is mainly constituted by residues located on the C-terminal helix, α8, an absolutely conserved secondary structural element possessed by all sFRP family members. Several lines of evidence have shown that the C-terminal portion of sFRPs binds tightly to heparin, which may regulate Wnt binding and signaling (14, 21, 41–43). In terms of region location and residue conservation, it is conceivable that the two positively charged regions may confer heparin-binding properties.
Comparison of the structures of Sizzled and Fz8 CRD-XWnt8 complex
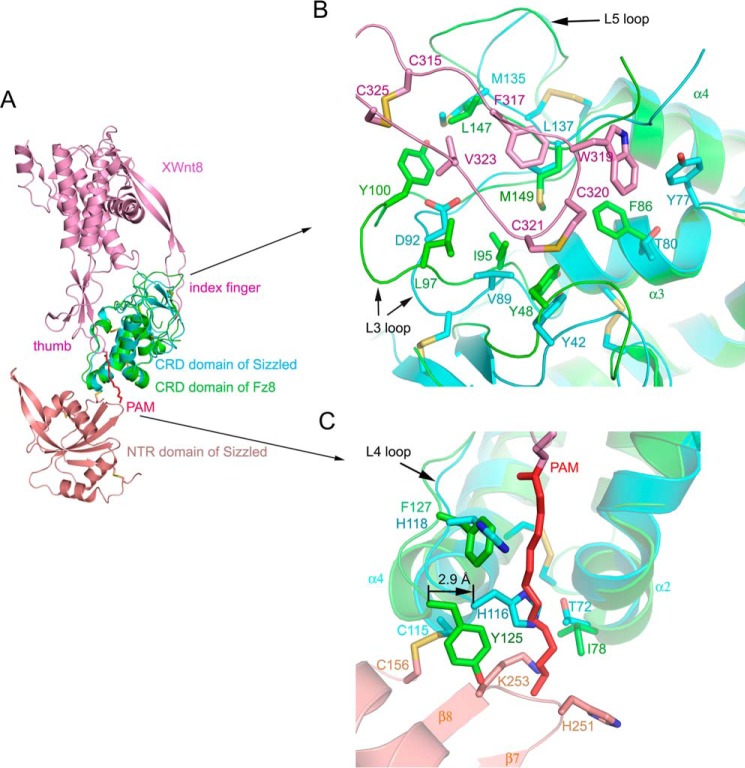
As mentioned above, the Sizzled CRD has a similar topological profile with mouse Fz8 CRD. Based on the CRD, the full-length Sizzled was superimposed with Fz8 CRD-XWnt8 complex (PDB code 4F0A (4)). As shown in Fig. 4A, XWnt8 would seemingly grasp Sizzled CRD at two opposing sites using the extended thumb and index fingers in a manner identical to that used for Fz8 CRD. In the index finger site, the hydrophobic residues Cys-315, Phe-317, Trp-319, Cys-320, Cys-321, Val-323, and Cys-325 of XWnt8 may engage in hydrophobic contacts with the depression between loops L3 and L5 of Sizzled that are mainly composed of hydrophobic residues Tyr-42, Tyr-77, Val-89, Met-135, and Leu-137 (Fig. 4B). This implicates a possibility that Sizzled would bind to XWnt8 for modulating its signaling. However, a careful view on the index finger site reveals a disadvantage in Sizzled for XWnt8 binding that lies in the L3 loop. Compared with the Sizzled L3 loop, the Fz8 CRD L3 loop is longer and extends up to the XWnt8 index finger so that residue Tyr-100 forms hydrophobic contacts with XWnt8 Val-323 and Cys-325 (Fig. 4B). Besides, in this loop, Fz8 Leu-97 is substituted with Asp-92 in Sizzled that may weaken hydrophobic interactions between XWnt8 index finger and the CRD depression.
Figure 4.
Structural comparison of Sizzled and Frizzled8 CRD-XWnt8 complex. A, superimposition of ribbon models of Sizzled and Frizzled8 CRD-XWnt8 complex (PDB code 4F0A). B, overlay of XWnt8 index finger (purple)-binding site on mouse Frizzled8 CRD (green) with Sizzled CRD (cyan). C, overlay of XWnt8 thumb (purple)-binding site on mouse Frizzled8 CRD (green) with Sizzled CRD (cyan). The XWnt8 lipid group, PAM, is shown as red stick representation.
Meanwhile, checking the superimposed structures around the thumb site by scrutiny, we further propose that Sizzled would not be able to bind to XWnt8. First, Sizzled NTR sterically collides with the tip of the extended palmitoleic acid (PAM) group, which is the key factor determining the interaction between CRD and Wnt. Tethered by the disulfide bond Cys-115–Cys-156 and the CRD-NTR linker, NTR interacts with CRD so that the PAM group bumps into the residues His-251, Gly-252, and Lys-253 on the short loop connecting β7 and β8. Second, in the Fz8-XWnt8 complex, the groove used for accommodating the PAM group is besieged by the secondary structural elements, Loop 4, and helices α2 and α4 (4). Relative to the case of Fz8 CRD, these elements of Sizzled are in a closer orientation relative to each other. For example, the Cα atom of Sizzled His-116 moves to the groove center by 2.9 Å, in comparison with its counterpart, Fz8 CRD Cys-115 Cα (Fig. 4C). Thus, the groove in Sizzled CRD is narrower, less solvent-exposed, and likely inaccessible for Wnt. Third, in the inner side of the groove, the lining residues, Ile-78, Tyr-125, and Phe-127, of Fz8 CRD are, respectively, replaced with Sizzled Thr-72, His-116, and His-118, three hydrophilic residues not compatible with the intrinsic hydrophobicity of PAM. Furthermore, the side chain of Sizzled His-116 points to the center of the groove and may directly occlude the access of PAM.
Because acylation is necessary for both Wnt intracellular trafficking and secretion for its activity and Wnts use the PAM group as a “hot spot” residue to engage its receptor (44, 45), it is reasonable to consider that, in comparison with the CRD depression site for binding to the Wnt index finger, the CRD groove (for Wnt PAM group binding) is more critical for Wnt-CRD interaction. Therefore, as shown below, we mutated residues in the Sizzled CRD groove to probe the Wnt-antagonizing function of Sizzled.
Wnt3A-antagonizing activity of Sizzled and its derivatives
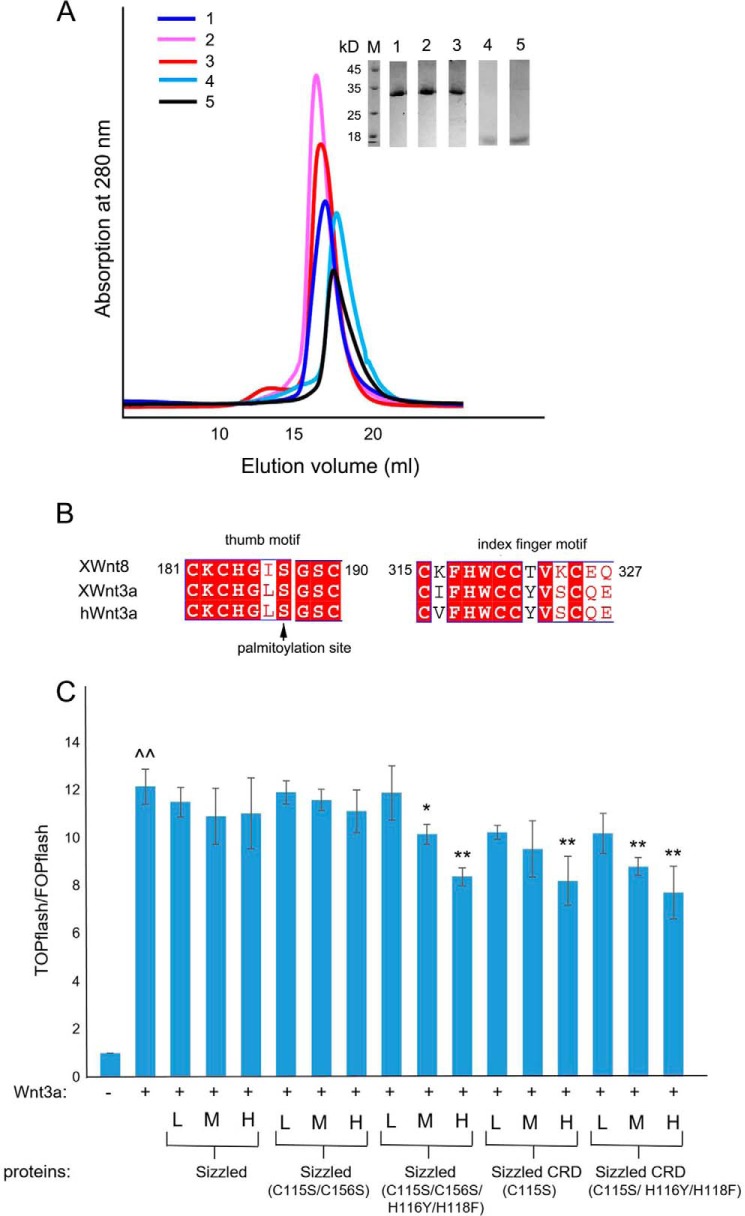
To validate whether Sizzled would inhibit Wnt signaling, we performed dual-luciferase reporter assays using Sizzled and its four derivatives: Sizzled with C115S/C156S; Sizzled with a quadruple mutation of C115S/C156S/H116Y/H118F (hereafter called “Sizzled quadruple mutant”); Sizzled CRD with a single mutation of C115S; and Sizzled CRD with a triple mutation of C115S/H116Y/H118F (hereafter called “Sizzled CRD triple mutant”). Judging from the crystal structure of Sizzled, the removal of NTR will expose Cys-115 in a free state; the C115S mutation was adopted to prevent CRD from forming oligomers through an intermolecular disulfide bond. Similar to the native Sizzled, all the derivatives were expressed well and purified into high purity and homogeneity by Ni2+-NTA affinity and gel filtration chromatography (Fig. 5A). Because X. laevis Wnt8 and human Wnt3A have the nearly identical thumb and index finger motif, which are critical for CRD recognition (Fig. 5B), the conditioned medium of L cells stably expressing human Wnt3A was used to stimulate HEK293A cells for inducing canonical Wnt signaling that is hallmarked by the stabilization and nuclear translocation of β-catenin. The nucleus-located β-catenin may associate with transcription factors of the T-cell factor family (TCF) and drive transcription of downstream genes, like TOPflash or FOPflash luciferase genes used in this assay (46, 47). Expectedly, the Sizzled derivatives that may bind to Wnt3A would decrease the ratio of TOPflash/FOPflash firefly luciferase activity.
Figure 5.
Preparation and Wnt3A antagonistic function assay of Sizzled and its derivatives. A, size-exclusion chromatography profiles of Sizzled (blue, 1), Sizzled C115S/C156S (pink, 2), Sizzled C115S/C156S/H116Y/H118F (red, 3), Sizzled CRD C115S (cyan, 4), and Sizzled CRD C115S/H116Y/H118F (black, 5) alongside an inset showing SDS-PAGE analysis of peak fractions. B, sequence alignment of thumb and index finger motifs of XWnt8, XWnt3a (X. laevis Wnt3A), and hWnt3a (human Wnt3A). C, fold changes of TOPflash/FOPflash firefly luciferase activity (each value normalized by Renilla luciferase activity) under different experimental conditions. For each protein, 3 columns from left to right, respectively, correspond to protein concentrations of 2.56, 25.6, and 256 nm (represented by L, M, and H, respectively). ∧∧, p value < 0.05, determined through comparing results in the presence of Wnt3A with the vehicle control; *, p value <0.05, and **, p value < 0.01, comparing results in the presence of the indicated protein sample with those obtained in the presence of Wnt3A alone.
As shown in Fig. 5C, the addition of Wnt3A alone increased the TCF-mediated transcriptional level of luciferase reporter gene in HEK293A cells by ∼12-fold. In the presence of Wnt3A, the addition of the full length of Sizzled or its mutant, C115S/C156S at the concentrations of 2.56, 25.6, and 256 nm did not significantly alter the luciferase gene transcription. Compared with Wnt3A alone, the supplement of the Sizzled quadruple mutant, Sizzled CRD C115S or Sizzled CRD triple mutant, at a high concentration of 256 nm decreased Wnt3A-induced luciferase gene transcription level by 30% (∼12-fold versus ∼8-fold) with a statistical significance. In the presence of Wnt3A, the addition of Sizzled quadruple mutant, Sizzled CRD, or Sizzled CRD triple mutant decreased the transcriptional activity of luciferase reporter gene in a dose-dependent manner: Sizzled quadruple mutant at 2.56, 25.6, and 256 nm, respectively, changed TOPflash/FOPflash ratio by 11.9-, 10.1-, and 8.4-fold; Sizzled CRD led to mean changes of 10.2-, 9.5-, and 8.2-fold; and Sizzled CRD triple mutant led to mean changes of 10.2-, 8.8-, and 7.7-fold, respectively. At the medium concentration, both Sizzled quadruple mutant and Sizzled CRD triple mutant significantly inhibited Wnt3A activity (p value < 0.05), whereas Sizzled CRD did without a statistical significance (p value = 0.1).
Crystal packing analysis of Sizzled
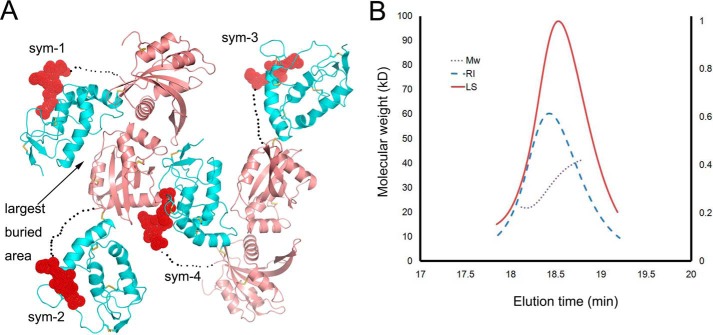
In the crystal, one Sizzled molecule is mainly packed against two symmetry mates. The largest buried surface area between two packed molecules (between sym-1 and sym-2, or between sym-3 and sym-4) can reach 1900 Å2 when one Sizzled NTR docks into the concave surface formed by the two domains of the other Sizzled molecule (Fig. 6A). Such a large buried surface area can not conclusively suggest that Sizzled would be a dimer. However, as shown in size-exclusion chromatography (Fig. 5A), the elution volume of Sizzled was 16.2 ml, a volume corresponding to an eluted protein with an apparent molecular mass of ∼35 kDa. Meanwhile, Sizzled migrated as a band on a SDS-polyacrylamide gel, equivalent to a mass of ∼33 kDa (Fig. 5A). Furthermore, as demonstrated in the Fig. 6B, Sizzled has an apparent molecular mass of 32.3 ± 0.4 kDa as measured by multi-angle light scattering (MALS), in agreement with the theoretical molecular mass of 31.5 kDa of a monomer. These results show that Sizzled exists as a monomer in the solution, whereas the crystallographic Sizzled dimer with a large buried surface area is only an artifact of crystallization.
Figure 6.
Crystal packing of Sizzled CRD and MALS analysis of Sizzled. A, crystal packing pattern of Sizzled (CRD in cyan, NTR in salmon, and linker in dashed line) with four symmetry mates presented. The NTR domain of sym-3 (or sym-2) docks into the concave surface formed by NTR and CRD of sym-1 (or sym-4), burying 1900 Å2 total surface area. The C-terminal portion of CRD is indicated by a red sphere. B, MALS analysis of Sizzled in solution. The brown dashed line across the refractive index peak (RI, blue dashed line) and the light scattering peak (LS, red line) indicates homogeneity of Sizzled. The calculated apparent molecular mass is 32.3 ± 0.4 kDa. The right vertical axis represents refractive index. For clarity, the vertical axis for light scattering intensity was not shown.
Discussion
The sFRPs represent the largest family of secreted Wnt inhibitors. Over the past 2 decades, it has remained controversial, to some extent, about their function in antagonizing Wnt signaling. Is Sizzled really able to antagonize Wnt signaling? How do sFRPs orchestrate their two domains when engaging in Wnt signaling? Is it possible for an NTR domain to mimic the full-length sFRPs in antagonizing Wnt? Do their CRDs dimerize following a conserved mode that suggests biological significance? Structural study will facilitate in elucidating these issues. However, structural information about this family was limited to mouse sFRP3 CRD (12). The crystal structure of Sizzled in this paper is the first full-length structure of the sFRP family member that may clarify these controversial issues to some extent.
Sizzled is unlikely able to antagonize Wnt signaling
It has been a long time since controversial reports on XWnt8 antagonistic function of Sizzled (26, 28–30). From the Sizzled structure, we deduced that Sizzled is unlikely able to bind to Wnt and antagonize its signaling because of the following: 1) the inter-domain disulfide bond Cys-115–Cys-156 and linker tether Sizzled CRD and NTR domains together so that the extended Wnt PAM group would collide with the NTR tip residues; 2) the narrow groove on Sizzled CRD is not appropriate for accommodating the PAM group; 3) the hydrophilic residues in the inner side of the Sizzled CRD groove is not compatible with the intrinsic hydrophobicity essence of the PAM group; and 4) the loops around the supposed index finger site on the Sizzled CRD expose less hydrophobic residues for docking Wnt than Fz8 CRD. To verify such a structure-based deduction, we constructed truncates or site-directed mutants of Sizzled for functional assays.
Because it has proven a challenge to determine binding affinities for Wnt and its binding partners due to the hydrophobic nature of the Wnt proteins and the concomitant difficulty of purifying them (48), we directly evaluated the Wnt-antagonizing function of Sizzled and its derivatives using the Dual-Luciferase assay in HEK293A cells stimulated with conditioned medium of human Wnt3A. As demonstrated in Fig. 5C, the full-length native Sizzled did not significantly alter Wnt3A signaling, suggesting that Sizzled is unlikely able to antagonize Wnt signaling because Wnt3A has basically an identical thumb and index finger motif with XWnt8A for binding CRD (Fig. 5B). For the full-length Sizzled mutants, the C115S/C156S mutant did not impair Wnt3A signaling, whereas the Sizzled quadruple mutant (C115S/C156S/H116Y/H118F) inhibited Wnt3A signaling by ∼30% (12.1-fold changes versus 8.4-fold changes) at 256 nm. These results have two implications. First, the disulfide bond Cys-115–Cys-156 is not an indispensable factor to restrain the interaction of Sizzled with Wnt. Removal of this interdomain disulfide bridge is not sufficient to release the blockage of the NTR domain for Wnt access to the CRD. Second, enhancing hydrophobicity of the lining residues in the CRD groove may increase Sizzled's binding to Wnt3A for antagonizing function.
Compared with the full-length native Sizzled, the Sizzled CRD C115S and Sizzled CRD triple mutant (C115S/H116Y/H118F) decreased Wnt3A-induced luciferase gene transcription level, and Sizzled CRD triple mutant presented higher potency in antagonizing Wnt signaling. These cellular assay results were consistent with structural implications. The removal of NTR may avoid steric hindrance between PAM and NTR residues and likely release the groove to a more PAM-accessible state. For Sizzled CRD triple mutant, the hydrophilic lining residues His-116 and His-118 in the groove were substituted with aromatic residues to increase groove hydrophobicity, which makes the CRD groove more compatible with the Wnt PAM group. This further supports that enhancing hydrophobicity of the lining residues in the CRD groove may increase the Wnt-binding and -antagonizing functions of Sizzled. Noteworthy, at the medium concentration, Sizzled CRD triple mutant exhibited the highest potency in inhibiting Wnt3A signaling, when compared with Sizzled CRD C115S and Sizzled quadruple mutant. This suggests that removal of the NTR domain and enhancing the CRD groove hydrophobicity may concertedly increase the Wnt-binding ability of Sizzled.
In short, based on the above structural analysis and cellular assay results, we propose that Sizzled is unlikely able to antagonize Wnt (including XWnt8) signaling. Also, we may conclude that the inter-domain disulfide bond is not an indispensable factor that restrains the NTR domain of Sizzled for Wnt binding to the CRD.
Sizzled structure provides a framework for understanding the full-length structure of sFRPs
As mentioned above, the two domains of Sizzled have different extents of topological conservation among sFRP family members. Structural comparison reveals that Sizzled CRD has a highly conserved topology due to five pairs of absolutely conserved disulfide bonds and relatively conserved secondary structure elements. In contrast, the Sizzled NTR domain is not topologically conserved among all sFRP proteins. Also, as seen in the sequence alignment (Fig. 2), the residues in Sizzled helices α6 and α7, two key secondary structure elements packing against the central β-sheet, are conserved in sFRP1/2/5 but not in sFRP3/4. Furthermore, secondary structure prediction by PepTool (49) suggests that the sFRP3/4 sequence equivalent to Sizzled helix α6 and α7 would fold into a β-strand (supplemental Table S2). Therefore, it can be implicated that sFRP3/4 NTR domain would have a fold similar to that of a canonical NTR domain and that sFRP1/2/5 and Sizzled would have a topologically similar NTR domain. Correspondingly, the sFRPs family members can be structurally classified into two subgroups, Sizzled/sFRP1/2/5 versus sFRP3/4. This structural classification is consistent with the sequence-based phylogenetic analysis that demonstrates that Sizzled is evolutionarily closer to sFRP1/2/5 than sFRP3/4 (supplemental Fig. S2).
In accordance with the previous mass spectrum assay (50), the Sizzled CRD and NTR domains are linked by the inter-domain disulfide bridge Cys-115–Cys-156. As shown in the sequence alignment (Fig. 2), this disulfide bond is not conserved among human sFRPs. Correspondingly, the cysteine in human sFRP1/2/5 equivalent to Sizzled Cys-156 should remain a free, reduced state. Therefore, sFRP1/2/5 should have only two pairs of disulfide bonds in their NTR domains, including one pair that is absolutely conserved among sFRPs and corresponds to the seventh pair in Sizzled, Cys-159–Cys-231. However, this is different from the previously predicted disulfide linkage pattern in sFRP1 NTR that was supposed to have three disulfide bridges Cys-185–Cys-255, Cys-188–Cys-257, and Cys-202–Cys-305 (15). Of note, Sizzled NTR was improperly considered to have a disulfide bond between Cys-156 and Cys-231, equivalent to sFRP1 NTR Cys-185–Cys-255 (15). Taking into account the fact that the disulfide bond Cys-159–Cys-231 and helices α6 and α7 of Sizzled are conserved in the subgroup of sFRP1/2/5, we speculate that human sFRP1 would assume a disulfide bond between Cys-188 and Cys-255 and remain both Cys-185 and Cys-257 in a free, unpaired state. Because of loop flexibility, the possibility could not be excluded that Cys-185 and Cys-257 in sFRP1 form a disulfide bond. Certainly, this speculation needs further validation at the structural level.
No matter which disulfide linkage pattern is adopted in human sFRP NTR, it is conclusive that human sFRPs lack an inter-domain disulfide bond. This implicates that, in contrast to Sizzled that tethers two domains together, human sFRPs have more freedom in locating NTR around CRD. Therefore, it is reasonable to conceive that NTR and CRD in human sFRPs are spatially independent on each other. That is likely why CRD of sFRP3 is necessary and sufficient for both Wnt-binding and functional activities (21, 22). As analyzed previously, Sizzled CRD has higher potency in antagonizing Wnt signaling than the full-length Sizzled, because NTR structurally occludes the CRD groove for PAM accessibility. However, this domain-to-domain occlusion in Sizzled was not the result of the inter-domain disulfide bond, but likely was by inter-domain interactions that need further investigation.
sFRPs do not have a conserved CRD dimerization mode
As elucidated previously, Sizzled exists as a monomer in the solution, whereas the observed Sizzled dimer in the crystal with a large buried surface area is only an artifact of crystallization. This is in sharp contrast with the case of both mouse sFRP3 and mouse Fz8 CRDs that exhibit the same dimer interface and suggest the potential for the different CRDs to homo- or heterodimerize (12). This potential has seemingly been convinced to be of biological significance by biochemical studies that sFRPs and/or Frizzleds could form homo- or hetero-complexes (33, 51, 52). Indeed, the dimerization of sFRP3 CRD is mainly mediated by hydrophobic residue contacts and an inter-molecular β-sheet formation engaged by the C-terminal fragment of this domain (supplemental Fig. S3). The hydrophobic residue interactions mainly occur among Trp-16, Met-18, Phe-54, Ile-63, Ile-66, Phe-68, and Val-114 (residues numbering as in PDB code 1ijx). These residues are not well-conserved in Sizzled and other sFRPs, even with some residues replaced by hydrophilic ones: Met-18 is replaced by Glu-44, and Ile-66 is replaced by Asp-92 in Sizzled. The residue Ser-149 of sFRP3 CRD, akin to the β-sheet formation, is replaced in sFRP2 with proline, a residue that generally disrupts β-strand formation. In contrast with the sFRP3 crystal, the carboxyl extremity of Sizzled in the crystal did not engage in β-sheet formation (Fig. 6A). All these demonstrate that sFRP family members do not have a conserved CRD dimerization mode. This means that the proposed potential for the different CRDs to homo- or heterodimerize (12) should be re-considered.
Actually, as shown in Fig. 2C, structural variation in the CRD of sFRPs mainly lies in the loops L1, L3, and L5 as well as the C-terminal portion. Correspondingly, the CRD of sFRPs has variable three-dimensional structural novelty and surface property for protein-protein interaction (including dimerization), although its topology is highly conserved. A piece of evidence for such a conception is that Sizzled is unique among sFRPs for its ability to specifically inhibit BMP-1 (50). Does sFRPs CRD really heterodimerize with Frizzled CRD for Wnt signaling? How does sFRPs CRD engage in Wnt interaction for Wnt signaling modulation? More structural information, especially for paired protein-protein complexes, is needed.
NTR domain of sFRPs, at least sFRP1/2/5, is unlikely to bind to Wnt but is likely involved in biphasic Wnt signaling modulation
Unexpectedly, there were reports that a construct of sFRP1 without the CRD retained the ability to bind to Wingless, the Drosophila Wnt homolog (14), and that the NTR domain mimicked the function of the full-length sFRP1 in binding Wnt8 and inhibiting Wnt signaling (31). However, from the structure of Sizzled NTR, we propose that the NTR domain of human sFRPs, at least of sFRP1/2/5, is unlikely to bind to Wnt. As analyzed above, the subgroup of sFRP1/2/5 may have an NTR domain resembling that of Sizzled, which lacks hydrophobic patches in its surface for Wnt recognition. On the contrary, the surface of NTR is characterized with two highly positively charged regions that are at the domain “bottom” and opposite the NTR-CRD interface. The residues lysine and arginine in the two positively charged regions are conserved in the subgroup of sFRP1/2/5, suggesting that sFRP1/2/5 may promote the electrostatic interactions through these regions with client proteins. Recent structural study shows that heparin may bind to a positively charged region formed by lysines and arginines (53). sFRP1/2/5 may also bind to heparin through the positively charged regions on NTR. This may explain the facts that the C-terminal portion of sFRPs binds tightly to heparin for Wnt binding and signaling modulation (14, 21, 41–43).
Recently, it was reported that sFRP1 may either inhibit or enhance signaling in the Wnt3a/β-catenin pathway, depending on its concentration and the cellular context (32). Interestingly, sFRPs were shown to enhance the diffusion and solubility of Wnts with the resulting enhancement of Wnt signaling in more distal cells (45, 54, 55). Rather presenting putative Wnt-antagonistic ability, sFRP2 was reported to augment Wnt signaling in cultured dermal papilla cells (56) or in tumor microenvironment (57). All these reports suggest that sFRPs may modulate Wnt signaling in a biphasic manner. Deducing from the crystal structure of Sizzled, we propose that the NTR domain of sFRPs is likely involved in biphasic Wnt signaling modulation (supplemental Fig. S4). Because the positively charged regions in the bottom of NTR are opposite and distal to the NTR-CRD interface, the event of client proteins or heparin binding to NTR may couple with Wnt binding to CRD. At a low concentration or under an appropriate condition, sFRPs would serve as a carrier for Wnt and be recruited to distal cells in a microenvironment containing client proteins or heparin, thus promoting Wnt signaling. When the local concentration of Wnt reached a critical point, sFRPs would potentially compete with Fzds for excess Wnt binding and serve a decoy to trap Wnt, thus inhibiting Wnt signaling. However, it will be necessary to perform a quantitative analysis of the pairwise Wnt/sFRP- and Fzd-binding affinities and find the physiological or pathological condition for sFRPs to switch from a Wnt carrier to a decoy. Given that Wnt signaling is implicated in cancers and neurological disorders (5–7), the proposed mechanism for biphasic modulation of Wnt signaling by sFRPs may give hints for designing anti-cancer and pro-regeneration therapies.
Experimental procedures
Cloning and recombinant baculovirus production
All proteins used for crystallization and Dual-Luciferase assay were produced using the BacMam system as reported previously (35, 58). A cDNA fragment encoding the full-length Sizzled (residues 18–281) from X. laevis was PCR-amplified from a synthetic DNA plasmid (SUNBIO, Beijing, China) and cloned to a modified BacMam vector with a C-terminal hexa-histidine tag (35). The constructed transfer plasmids and the BacVector-3000 baculovirus DNA (EMD Biosciences) were used to co-transfect Sf9 insect cells for generation of recombinant viruses. One week after transfection, the resulting low-titer virus stock was harvested and sequentially used to infect insect cells for amplification. When most insect cells presented cytopathic phenotypes, the amplified viruses were harvested and stored at 4 °C for usage.
Protein expression and purification
For large-scale protein expression, recombinant baculovirus was added to HEK293T cells at a density of 2 × 106 cells per ml, and culture flasks were shaken for 72 h at 37 °C in a large-capacity incubator filled with 5% CO2. Cells were removed by centrifugation, and the conditioned medium was concentrated and buffer-exchanged to HBS (10 mm HEPES, pH 7.5, 150 mm NaCl). The proteins were captured by addition of Ni2+-NTA-agarose (Invitrogen) and washed extensively with HBS containing different concentrations (from 20 to 300 mm) of imidazole in gradient. The eluted fractions were resolved by SDS-PAGE, and only those of high-purity interest protein were subjected to gel-filtration chromatography with a Superdex 200 Increase column pre-equilibrated and eluted with HBS. The fractions containing mono-dispersed proteins were pooled and concentrated to 8–10 mg/ml for crystallization.
Crystallization
Crystallization was performed at 20 °C using the sitting-drop vapor-diffusion method. The sitting drop was composed of 0.5 μl of reservoir solution and 0.5 μl of protein solution and equilibrated against 1 ml of reservoir solution. Over 1 week, brick-like Sizzled crystals grew from the reservoir solution containing 10% PEG8K, 0.1 m imidazole, pH 8.0, and 0.2 m lithium sulfate.
Diffraction data collection and processing
Crystals were flash-frozen in liquid nitrogen, followed by soaking in the reservoir solution supplemented with 24–26% ethylene glycol. For the Sizzled crystals, two data sets were collected: one data set of 1200 images was harvested in-house on a MicroMax-003 X-ray generator (Rigaku, Japan) equipped with a Saturn 944 detector; the other was collected on the beamline BL17U1 at Shanghai Synchrotron Radiation Facility (SSRF). All the data were collected at 100 K and processed with HKL2000 (59). The statistics are summarized in supplemental Table S1. Three parameters in this table are defined as follows: Rpim, precision-indicating merging R-factor (60); Rmeas, multiplicity-corrected merging R-factor (61); CC½ and CC*, Pearson correlation coefficients (62).
Structure determination, refinement, and validation
The high-redundancy in-house dataset was used to calculate the phases for the Sizzled crystal using the single-wavelength anomalous diffraction (SAD) method by the program Autosol in PHENIX (63). Three sulfur atoms per asymmetric unit were located, and initial SAD phases with an overall figure of merit of 0.41 for the reflections in the resolution range of 50–2.9 Å were obtained for model auto-building. The model was improved by iterative manual building in COOT (64) and refinement in CNS (65) against the higher resolution data collected at SSRF. Water molecules were automatically introduced using CNS and manually edited. Final refinement was performed PHENIX and validation with PROCHECK (66) and Molprobity (67).
Multiple-sequence alignment of sFRPs
Structure-based sequence alignment was executed using ClustalW (68) and mapped using ESPRIPT (69). Conservation of residues was based on chemical character: aromatic (Phe, Tyr, and Trp), hydrophobic (Leu, Ile, Val, and Met), acidic (Glu and Asp), basic (Lys, Arg, and His), polar (Ser and Thr), tiny (Gly and Ala), and amide (Asn and Gln).
Mutagenesis
A mutant of full-length Sizzled with C115S/C156S was created by a two-step PCR with overlapping primers. Using the constructed full-length Sizzled C115S/C156S mutant plasmid as a template, two mutants of the Sizzled CRD (residues 18–143), C115S and C115S/H116Y/H118F, as well as the quadruple mutant of the full-length Sizzled, C115S/C156S/H116Y/H118F, were produced either by a single-step PCR or by a two-step overlapping PCR. Subcloning, baculovirus recombination, and protein preparation of these mutants were executed following the same strategy as used for the wide-type construct.
MALS
MALS experiments were performed during size-exclusion chromatography on a WTC-015S5 (Wyatt Technology) with on-line static light-scattering (DAWN HELEOS II, Wyatt Technology) and differential refractive index (Optilab T-rEX, Wyatt Technology) detectors. Protein samples applied to MALS had previously been purified by size-exclusion chromatography with a Superdex 200 Increase column. Data were analyzed using the ASTRA software package (Wyatt Technology).
Dual-Luciferase reporter assay
Luciferase assays were carried out using the Dual-Luciferase Reporter Assay System (Promega). HEK293A cells, L cells (ATCC No. CRL-2648; generously given by the Prof. Ye-Guang Chen from Tsinghua University), and L-Wnt3A cells (ATCC No. CRL-2647; also from Prof. Chen) were maintained in DMEM (Invitrogen) supplemented with 10% FBS (Invitrogen), 100 IU/ml penicillin, and 100 μg/ml streptomycin and incubated in a humid incubator with 5% CO2 at 37 °C. For the L-Wnt3A cells, the culture was supplemented with 0.4 mg/ml G418 (Invitrogen). 5000 trypsinized HEK293A cells per well were seeded into 96-well plates for overnight incubation. In the presence of Lipofectamine 3000 in Opti-MEM medium (Invitrogen), cells were co-transfected with 50 ng of either TOPflash or FOPflash vector (kindly provided by Prof. Xiaoguang Liu from Peking University Third Hospital) and 1 ng of Renilla luciferase reporter vector pRL-TK (Promega) as the control for transfection efficiency. 6 h after transfection, the media of cells were changed to the complete DMEM containing FBS. After 18 h, cells were switched to FBS-free DMEM in addition to 40% of the conditioned medium of L cells or L-Wnt3A cells and immediately treated with Sizzled or its derivatives at three different concentrations 2.56, 25.6, and 256 nm. 24 h after protein treatment, the cells were lysed with PLB from the assay kit for 15 min and sequentially treated with firefly luciferase reagent LARII (for firefly luciferase activity) and Stop & Glo Reagent (for Renilla luciferase activity) according to manufacturer's protocol. Luminescent signals were recorded using a Centro XS3 LB960 microplate luminometer (Berthold Technology). TCF-mediated transcriptional activity was determined by the ratio of TOPflash/FOPflash firefly luciferase activity, each normalized to the Renilla luciferase activity of the pRL-TK reporter. All reporter assays were performed in triplicate. The data were presented as mean ± S.D. (error bars). The differences between groups were calculated by Student's t tests, and a value of p < 0.05 was considered statistically significant.
Author contributions
H. L. conceived the project, coordinated the study, analyzed the data, and wrote the manuscript. Q. B., Z. L., J. Z., F. X., and J. L. performed the experiments, analyzed the data, and contributed to manuscript preparation.
Supplementary Material
This work was supported by the China “Thousand Talents Program” and National Natural Science Foundation of China Grants 81322047 and 81673525 (to H. L.). The authors declare that they have no conflicts of interest with the contents of this article.
The atomic coordinates and structure factors (code 5XGP) have been deposited in the Protein Data Bank (http://wwpdb.org/).
This article contains supplemental Tables S1–S2 and Figs. S1–S4.
- sFRP
- secreted Frizzled-related protein
- CRD
- cysteine-rich domain
- NTR
- netrin-like domain
- HBS
- HEPES-buffered saline
- PAM
- palmitoleic acid
- SAD
- single-wavelength anomalous diffraction
- XWnt8
- X. laevis Wnt8
- Ni2+-NTA
- nickel-nitrilotriacetic acid
- r.m.s.d.
- root mean square deviation
- PDB
- Protein Data Bank
- MALS
- multi-angle light scattering
- SSRF
- Shanghai Synchrotron Radiation Facility
- Fzd
- Frizzled.
References
- 1. Logan C. Y., and Nusse R. (2004) The Wnt-signaling pathway in development and disease. Annu. Rev. Cell Dev. Biol. 20, 781–810 [DOI] [PubMed] [Google Scholar]
- 2. MacDonald B. T., Tamai K., and He X. (2009) Wnt/β-catenin signaling: components, mechanisms, and diseases. Dev. Cell 17, 9–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Angers S., and Moon R. T. (2009) Proximal events in Wnt signal transduction. Nat. Rev. Mol. Cell Biol. 10, 468–477 [DOI] [PubMed] [Google Scholar]
- 4. Janda C. Y., Waghray D., Levin A. M., Thomas C., and Garcia K. C. (2012) Structural basis of Wnt recognition by Frizzled. Science 337, 59–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Polakis P. (2012) Wnt signaling in cancer. Cold Spring Harb. Perspect. Biol. 4, a008052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Blagodatski A., Poteryaev D., and Katanaev V. L. (2014) Targeting the Wnt pathways for therapies. Mol. Cell Ther. 2, 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Driehuis E., and Clevers H. (2017) WNT signalling events near the cell membrane and their pharmacological targeting for the treatment of cancer. Br. J. Pharmacol., doi: 10.1111/bph.13758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Niehrs C. (2012) The complex world of WNT receptor signalling. Nat. Rev. Mol. Cell Biol. 13, 767–779 [DOI] [PubMed] [Google Scholar]
- 9. Endo M., Nishita M., Fujii M., and Minami Y. (2015) Insight into the role of Wnt5a-induced signaling in normal and cancer cells. Int. Rev. Cell Mol. Biol. 314, 117–148 [DOI] [PubMed] [Google Scholar]
- 10. Cruciat C. M., and Niehrs C. (2013) Secreted and transmembrane wnt inhibitors and activators. Cold Spring Harb. Perspect. Biol. 5, a015081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bovolenta P., Esteve P., Ruiz J. M., Cisneros E., and Lopez-Rios J. (2008) Beyond Wnt inhibition: new functions of secreted Frizzled-related proteins in development and disease. J. Cell Sci. 121, 737–746 [DOI] [PubMed] [Google Scholar]
- 12. Dann C. E., Hsieh J. C., Rattner A., Sharma D., Nathans J., and Leahy D. J. (2001) Insights into Wnt binding and signalling from the structures of two Frizzled cysteine-rich domains. Nature 412, 86–90 [DOI] [PubMed] [Google Scholar]
- 13. Bányai L., and Patthy L. (1999) The NTR module: domains of netrins, secreted frizzled-related proteins, and type I procollagen C-proteinase enhancer protein are homologous with tissue inhibitors of metalloproteases. Protein Sci. 8, 1636–1642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Uren A., Reichsman F., Anest V., Taylor W. G., Muraiso K., Bottaro D. P., Cumberledge S., and Rubin J. S. (2000) Secreted frizzled-related protein-1 binds directly to wingless and is a biphasic modulator of Wnt signaling. J. Biol. Chem. 275, 4374–4382 [DOI] [PubMed] [Google Scholar]
- 15. Chong J. M., Uren A., Rubin J. S., and Speicher D. W. (2002) Disulfide bond assignments of secreted frizzled-related protein-1 provide insights about frizzled homology and netrin modules. J. Biol. Chem. 277, 5134–5144 [DOI] [PubMed] [Google Scholar]
- 16. Wawrzak D., Métioui M., Willems E., Hendrickx M., de Genst E., and Leyns L. (2007) Wnt3a binds to several sFRPs in the nanomolar range. Biochem. Biophys. Res. Commun. 357, 1119–1123 [DOI] [PubMed] [Google Scholar]
- 17. Dennis S., Aikawa M., Szeto W., d'Amore P. A., and Papkoff J. (1999) A secreted frizzled-related protein, FrzA, selectively associates with Wnt-1 protein and regulates wnt-1 signaling. J. Cell Sci. 112, 3815–3820 [DOI] [PubMed] [Google Scholar]
- 18. Bhat R. A., Stauffer B., Komm B. S., and Bodine P. V. (2007) Structure-function analysis of secreted frizzled-related protein-1 for its Wnt antagonist function. J. Cell Biochem. 102, 1519–1528 [DOI] [PubMed] [Google Scholar]
- 19. Galli L. M., Barnes T., Cheng T., Acosta L., Anglade A., Willert K., Nusse R., and Burrus L. W. (2006) Differential inhibition of Wnt-3a by Sfrp-1, Sfrp-2, and Sfrp-3. Dev. Dyn. 235, 681–690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lee C. S., Buttitta L. A., May N. R., Kispert A., and Fan C. M. (2000) SHH-N upregulates Sfrp2 to mediate its competitive interaction with WNT1 and WNT4 in the somitic mesoderm. Development 127, 109–118 [DOI] [PubMed] [Google Scholar]
- 21. Leyns L., Bouwmeester T., Kim S. H., Piccolo S., and De Robertis E. M. (1997) Frzb-1 is a secreted antagonist of Wnt signaling expressed in the Spemann organizer. Cell 88, 747–756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lin K., Wang S., Julius M. A., Kitajewski J., Moos M. Jr., and Luyten F. P. (1997) The cysteine-rich frizzled domain of Frzb-1 is required and sufficient for modulation of Wnt signaling. Proc. Natl. Acad. Sci. U.S.A. 94, 11196–11200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Longman D., Arfuso F., Viola H. M., Hool L. C., and Dharmarajan A. M. (2012) The role of the cysteine-rich domain and netrin-like domain of secreted frizzled-related protein 4 in angiogenesis inhibition in vitro. Oncol. Res. 20, 1–6 [DOI] [PubMed] [Google Scholar]
- 24. Li Y., Rankin S. A., Sinner D., Kenny A. P., Krieg P. A., and Zorn A. M. (2008) Sfrp5 coordinates foregut specification and morphogenesis by antagonizing both canonical and noncanonical Wnt11 signaling. Genes Dev. 22, 3050–3063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Mori H., Prestwich T. C., Reid M. A., Longo K. A., Gerin I., Cawthorn W. P., Susulic V. S., Krishnan V., Greenfield A., and Macdougald O. A. (2012) Secreted frizzled-related protein 5 suppresses adipocyte mitochondrial metabolism through WNT inhibition. J. Clin. Invest. 122, 2405–2416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Salic A. N., Kroll K. L., Evans L. M., and Kirschner M. W. (1997) Sizzled: a secreted Xwnt8 antagonist expressed in the ventral marginal zone of Xenopus embryos. Development 124, 4739–4748 [DOI] [PubMed] [Google Scholar]
- 27. Lavergne E., Hendaoui I., Coulouarn C., Ribault C., Leseur J., Eliat P. A., Mebarki S., Corlu A., Clément B., and Musso O. (2011) Blocking Wnt signaling by SFRP-like molecules inhibits in vivo cell proliferation and tumor growth in cells carrying active β-catenin. Oncogene 30, 423–433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Bradley L., Sun B., Collins-Racie L., LaVallie E., McCoy J., and Sive H. (2000) Different activities of the frizzled-related proteins frzb2 and sizzled2 during Xenopus anteroposterior patterning. Dev. Biol. 227, 118–132 [DOI] [PubMed] [Google Scholar]
- 29. Collavin L., and Kirschner M. W. (2003) The secreted Frizzled-related protein Sizzled functions as a negative feedback regulator of extreme ventral mesoderm. Development 130, 805–816 [DOI] [PubMed] [Google Scholar]
- 30. Yabe T., Shimizu T., Muraoka O., Bae Y. K., Hirata T., Nojima H., Kawakami A., Hirano T., and Hibi M. (2003) Ogon/Secreted Frizzled functions as a negative feedback regulator of Bmp signaling. Development 130, 2705–2716 [DOI] [PubMed] [Google Scholar]
- 31. Lopez-Rios J., Esteve P., Ruiz J. M., and Bovolenta P. (2008) The Netrin-related domain of Sfrp1 interacts with Wnt ligands and antagonizes their activity in the anterior neural plate. Neural Dev. 3, 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Xavier C. P., Melikova M., Chuman Y., Üren A., Baljinnyam B., and Rubin J. S. (2014) Secreted Frizzled-related protein potentiation versus inhibition of Wnt3a/β-catenin signaling. Cell. Signal. 26, 94–101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Rodriguez J., Esteve P., Weinl C., Ruiz J. M., Fermin Y., Trousse F., Dwivedy A., Holt C., and Bovolenta P. (2005) SFRP1 regulates the growth of retinal ganglion cell axons through the Fz2 receptor. Nat. Neurosci. 8, 1301–1309 [DOI] [PubMed] [Google Scholar]
- 34. Brinkmann E. M., Mattes B., Kumar R., Hagemann A. I., Gradl D., Scholpp S., Steinbeisser H., Kaufmann L. T., and Özbek S. (2016) Secreted Frizzled-related protein 2 (sFRP2) redirects non-canonical Wnt signaling from Fz7 to Ror2 during vertebrate gastrulation. J. Biol. Chem. 291, 13730–13742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Dukkipati A., Park H. H., Waghray D., Fischer S., and Garcia K. C. (2008) BacMam system for high-level expression of recombinant soluble and membrane glycoproteins for structural studies. Protein Expr. Purif. 62, 160–170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Sarma G. N., and Karplus P. A. (2006) In-house sulfur SAD phasing: a case study of the effects of data quality and resolution cutoffs. Acta Crystallogr. D Biol. Crystallogr. 62, 707–716 [DOI] [PubMed] [Google Scholar]
- 37. Huet J., Teinkela Mbosso E. J., Soror S., Meyer F., Looze Y., Wintjens R., and Wohlkönig A. (2013) High-resolution structure of a papaya plant-defense barwin-like protein solved by in-house sulfur-SAD phasing. Acta Crystallogr. D Biol. Crystallogr. 69, 2017–2026 [DOI] [PubMed] [Google Scholar]
- 38. Liu Q., and Hendrickson W. A. (2015) Crystallographic phasing from weak anomalous signals. Curr. Opin. Struct. Biol. 34, 99–107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Holm L., and Sander C. (1995) Dali: a network tool for protein structure comparison. Trends Biochem. Sci. 20, 478–480 [DOI] [PubMed] [Google Scholar]
- 40. Jore M. M., Johnson S., Sheppard D., Barber N. M., Li Y. I., Nunn M. A., Elmlund H., and Lea S. M. (2016) Structural basis for therapeutic inhibition of complement C5. Nat. Struct. Mol. Biol. 23, 378–386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Finch P. W., He X., Kelley M. J., Uren A., Schaudies R. P., Popescu N. C., Rudikoff S., Aaronson S. A., Varmus H. E., and Rubin J. S. (1997) Purification and molecular cloning of a secreted, Frizzled-related antagonist of Wnt action. Proc. Natl. Acad. Sci. U.S.A. 94, 6770–6775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Wang S., Krinks M., Lin K., Luyten F. P., and Moos M. Jr. (1997) Frzb, a secreted protein expressed in the Spemann organizer, binds and inhibits Wnt-8. Cell 88, 757–766 [DOI] [PubMed] [Google Scholar]
- 43. Zhong X., Desilva T., Lin L., Bodine P., Bhat R. A., Presman E., Pocas J., Stahl M., and Kriz R. (2007) Regulation of secreted Frizzled-related protein-1 by heparin. J. Biol. Chem. 282, 20523–20533 [DOI] [PubMed] [Google Scholar]
- 44. Willert K., Brown J. D., Danenberg E., Duncan A. W., Weissman I. L., Reya T., Yates J. R. 3rd., and Nusse R. (2003) Wnt proteins are lipid-modified and can act as stem cell growth factors. Nature 423, 448–452 [DOI] [PubMed] [Google Scholar]
- 45. Janda C. Y., and Garcia K. C. (2015) Wnt acylation and its functional implication in Wnt signalling regulation. Biochem. Soc. Trans. 43, 211–216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. von Marschall Z., and Fisher L. W. (2010) Secreted Frizzled-related protein-2 (sFRP2) augments canonical Wnt3a-induced signaling. Biochem. Biophys. Res. Commun. 400, 299–304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Barker N., and Clevers H. (2006) Mining the Wnt pathway for cancer therapeutics. Nat. Rev. Drug Discov. 5, 997–1014 [DOI] [PubMed] [Google Scholar]
- 48. van Amerongen R., Fuerer C., Mizutani M., and Nusse R. (2012) Wnt5a can both activate and repress Wnt/β-catenin signaling during mouse embryonic development. Dev. Biol. 369, 101–114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Wishart D. S., and Fortin S. (2001) The BioTools Suite. A comprehensive suite of platform-independent bioinformatics tools. Mol. Biotechnol. 19, 59–77 [DOI] [PubMed] [Google Scholar]
- 50. Bijakowski C., Vadon-Le Goff S., Delolme F., Bourhis J. M., Lécorché P., Ruggiero F., Becker-Pauly C., Yiallouros I., Stöcker W., Dive V., Hulmes D. J., and Moali C. (2012) Sizzled is unique among secreted Frizzled-related proteins for its ability to specifically inhibit bone morphogenetic protein-1 (BMP-1)/Tolloid-like proteinases. J. Biol. Chem. 287, 33581–33593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Bafico A., Gazit A., Pramila T., Finch P. W., Yaniv A., and Aaronson S. A. (1999) Interaction of frizzled-related protein (FRP) with Wnt ligands and the frizzled receptor suggests alternative mechanisms for FRP inhibition of Wnt signaling. J. Biol. Chem. 274, 16180–16187 [DOI] [PubMed] [Google Scholar]
- 52. Carron C., Pascal A., Djiane A., Boucaut J. C., Shi D. L., and Umbhauer M. (2003) Frizzled receptor dimerization is sufficient to activate the Wnt/β-catenin pathway. J. Cell Sci. 116, 2541–2550 [DOI] [PubMed] [Google Scholar]
- 53. Liang W. G., Triandafillou C. G., Huang T. Y., Zulueta M. M., Banerjee S., Dinner A. R., Hung S. C., and Tang W. J. (2016) Structural basis for oligomerization and glycosaminoglycan binding of CCL5 and CCL3. Proc. Natl. Acad. Sci. U.S.A. 113, 5000–5005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Mii Y., and Taira M. (2009) Secreted Frizzled-related proteins enhance the diffusion of Wnt ligands and expand their signalling range. Development 136, 4083–4088 [DOI] [PubMed] [Google Scholar]
- 55. Kumar S., Žigman M., Patel T. R., Trageser B., Gross J. C., Rahm K., Boutros M., Gradl D., Steinbeisser H., Holstein T., Stetefeld J., and Özbek S. (2014) Molecular dissection of Wnt3a-Frizzled8 interaction reveals essential and modulatory determinants of Wnt signaling activity. BMC Biol. 12, 44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Kwack M. H., Ahn J. S., Jang J. H., Kim J. C., Sung Y. K., and Kim M. K. (2016) SFRP2 augments Wnt/β-catenin signalling in cultured dermal papilla cells. Exp. Dermatol. 25, 813–815 [DOI] [PubMed] [Google Scholar]
- 57. Sun Y., Zhu D., Chen F., Qian M., Wei H., Chen W., and Xu J. (2016) SFRP2 augments WNT16B signaling to promote therapeutic resistance in the damaged tumor microenvironment. Oncogene 35, 4321–4334 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 58. Liao M., Cao E., Julius D., and Cheng Y. (2013) Structure of the TRPV1 ion channel determined by electron cryo-microscopy. Nature 504, 107–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Otwinowski Z., and Minor W. (1997) Macromolecular crystallography part A. Methods Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 60. Weiss M. S., and Hilgenfeld R. (1997) On the use of the merging R factor as a quality indicator for X-ray data. J. Appl. Crystallogr. 30, 203–205 [Google Scholar]
- 61. Diederichs K., and Karplus P. A. (1997) Improved R-factors for diffraction data analysis in macromolecular crystallography. Nat. Struct. Biol. 4, 269–275 [DOI] [PubMed] [Google Scholar]
- 62. Karplus P. A., and Diederichs K. (2012) Linking crystallographic model and data quality. Science 336, 1030–1033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L. W., Kapral G. J., Grosse-Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R., Read R. J., Richardson D. C., et al. (2010) PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Emsley P., and Cowtan K. (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 65. Brünger A. T., Adams P. D., Clore G. M., DeLano W. L., Gros P., Grosse-Kunstleve R. W., Jiang J. S., Kuszewski J., Nilges M., Pannu N. S., Read R. J., Rice L. M., Simonson T., and Warren G. L. (1998) Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 54, 905–921 [DOI] [PubMed] [Google Scholar]
- 66. Laskowski R. A., Macarthur M. W., Moss D. S., and Thornton J. M. (1993) PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 26, 283–291 [Google Scholar]
- 67. Chen V. B., Arendall W. B. 3rd., Headd J. J., Keedy D. A., Immormino R. M., Kapral G. J., Murray L. W., Richardson J. S., and Richardson D. C. (2010) MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 66, 12–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Larkin M. A., Blackshields G., Brown N. P., Chenna R., McGettigan P. A., McWilliam H., Valentin F., Wallace I. M., Wilm A., Lopez R., Thompson J. D., Gibson T. J., and Higgins D. G. (2007) Clustal W and Clustal X version 2.0. Bioinformatics 23, 2947–2948 [DOI] [PubMed] [Google Scholar]
- 69. Gouet P., Courcelle E., Stuart D. I., and Metoz F. (1999) ESPript: analysis of multiple sequence alignments in PostScript. Bioinformatics 15, 305–308 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.