Abstract
Beclin 1 (BECN1) is a key regulator of autophagy, a critical catabolic homeostasis pathway that involves sequestration of selected cytoplasmic components by multilayered vesicles called autophagosomes, followed by lysosomal fusion and degradation. BECN1 is a core component of class III phosphatidylinositol-3-kinase complexes responsible for autophagosome nucleation. Without heterologous binding partners, BECN1 forms an antiparallel homodimer via its coiled-coil domain (CCD). However, the last 16 CCD residues, composing an “overlap helix” (OH), have been crystallized in two mutually exclusive states: either as part of the CCD or packed against the C-terminal β-α repeated, autophagy-specific domain (BARAD). Here, using CD spectroscopy, isothermal titration calorimetry, and small-angle X-ray scattering, we show that in the homodimeric state, the OH transitions between these two different packing states, with the predominant state comprising the OH packed against the BARAD, contrary to expectations based on known BECN1 interactions with heterologous partners. We confirmed this observation by comparing the impact of mutating four residues that mediate packing of the OH against both the CCD and BARAD on structure and stability of the CCD, the OH+BARAD, and the two-domain CCD–BARAD. Last, we used cellular assays to demonstrate that mutation of these OH-interface residues abrogates starvation-induced up-regulation of autophagy but does not affect basal autophagy. In summary, we have identified a BECN1 helical region that transitions between packing as part of either one of two conserved domains (i.e. the CCD or the BARAD). Our findings have important implications for the relative stability of autophagy-inactive and autophagy-active BECN1 complexes.
Keywords: autophagy, Beclin-1 (BECN1), circular dichroism (CD), conformational change, isothermal titration calorimetry (ITC), protein stability, small-angle X-ray scattering (SAXS)
Introduction
Macroautophagy, hereafter called autophagy, is a conserved cellular homeostasis pathway responsible for engulfment of long-lived, misfolded, aggregated, damaged, or mutated proteins, organelles, and pathogens in multilayered vesicles called autophagosomes that fuse with lysosomes to enable degradation of the sequestered contents, facilitating nutrient recycling (1–5). BECN1/Beclin 1/ATG6/VPS30 homologs, first identified as a BCL2-interacting protein in humans (6) and among the first autophagy proteins to be identified in mammals (4), are highly conserved in eukaryotes. As core components of the class III phosphatidylinositol 3-kinase (PI3KC3) complexes essential for autophagosome nucleation and maturation (7–11), BECN1 homologs form a parallel heterodimer with either ATG14 or UVRAG/VPS38 via their coiled-coil domains (CCDs)3 (12–14). Defective BECN1 or deficient expression levels are linked to various diseases, including neurodegenerative disorders (15–17), cancers (4, 18–21), and infectious diseases (6, 22–24).
BECN1 up-regulates PI3KC3 activity and autophagy in response to diverse signals, although the exact mechanisms by which BECN1 performs this function remain unclear. BECN1 appears to be a major interaction hub for autophagy, interacting with at least 20 different cellular proteins as well as five virus (HIV, influenza virus, α-, β-, and γ-herpesviruses)-encoded proteins that inhibit autophagy to prevent virus degradation (24–26). Although the mechanism(s) by which BECN1 binds so many diverse partners is unknown, conformational flexibility often facilitates multivalent protein interactions, allowing one protein to interact with various binding partners to carry out diverse functions. Thus, understanding BECN1 conformational flexibility is key to understanding varied BECN1 interactions, which, in turn, will improve our understanding of the role of BECN1 in autophagy. Ultimately, this is essential to better understand how eukaryotic organisms respond to and survive cellular stressors.
BECN1 is a conformationally flexible protein that has at least four domains (27): an N-terminal intrinsically disordered region (26, 28), a flexible helical domain (29), a CCD (30, 31), and a β-α repeated, autophagy-specific domain (BARAD) (32, 33). The CCD (residues 175–265 in human BECN1) and the BARAD (residues 266–450 in human BECN1) are highly conserved, well-folded domains. The BARAD bears an aromatic finger composed of residues 359–361 (residues 430–432 of yeast VPS30) that is required for BARAD binding to membranes (33). Mass spectroscopy hydrogen–deuterium exchange experiments show that this aromatic finger probably interacts with membranes when part of the PI3KC3 complex (13).
Interestingly, residues 248–264 preceding the BARAD form a helical region that we have named the “overlap helix” (OH), because it has been crystallized in two differently packed states: (i) as part of the anti-parallel CCD homodimer (30, 31), constituting the C-terminal four turns of each CCD, or (ii) packed against the BARAD (33). Notably, in the structure of the quaternary complex of yeast VPS34/PI3KC3, VPS15, VPS30/BECN1, and VPS38/UVRAG, VPS30 residues 304–327 (equivalent to residues 248–271 of human BECN1) are part of the VPS30/BECN1 CCD and pack against the VPS38/UVRAG CCD, stabilizing the parallel CCD heterodimer (13). This led to the assumption that the OH is part of the CCD in the context of full-length BECN1. Notably, mass spectroscopy hydrogen–deuterium exchange experiments indicate that VPS30 residues 312–327 (equivalent to BECN1 residues 256–271), which compose the latter half of the OH, undergo membrane binding–induced conformational changes, although these residues do not directly bind membranes (13). Thus, different conformational states of the BECN1 OH probably play important roles in regulating protein–membrane or protein–protein interactions.
Co-immunoprecipitation assays show that the BECN1 homodimer exists in cells (34). Inclusion of the OH as part of the anti-parallel CCD homodimer would greatly increase stability of the autophagy-inactive homodimer. However, it is unknown whether the BECN1 OH is part of the CCD or the BARAD when BECN1 is not part of PI3KC3 complexes; nor have the factors that regulate this conformational change been established. Therefore, to determine the packing state of the OH in the absence of heterologous binding partners, we investigated the solution structure of a two-domain CCD–BARAD BECN1 fragment using CD spectroscopy and size-exclusion chromatography (SEC) in tandem with small angle X-ray scattering (SAXS). Further, we assessed the impact of mutating OH residues that pack against either the partner helix of the CCD or the BARAD on self-dissociation, using isothermal titration calorimetry (ITC); on structure and stability of the CCD, BARAD, and CCD–BARAD by monitoring thermal denaturation by CD and structural changes by SAXS; and last on cellular autophagy levels by evaluating the change in the number of autophagosomes per cell.
Results
The BECN1 OH cannot simultaneously pack against the CCD homodimer partner helix and the BARAD
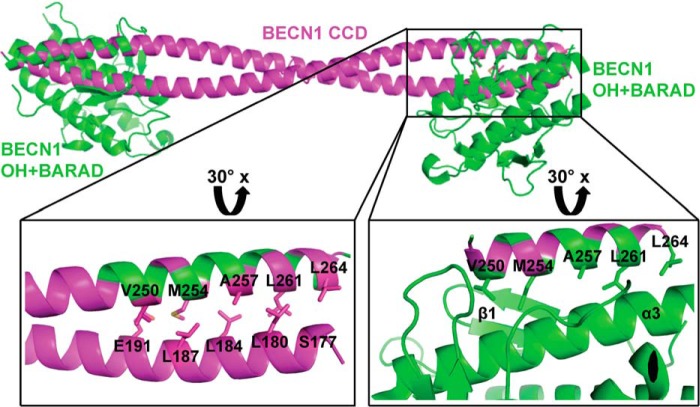
Superimposition of the human BECN1 OH of the OH+BARAD (PDB entry 4DDP) and one chain of the CCD homodimer (PDB entry 5HHE) reveals that the OH cannot simultaneously be within the CCD and packed against the BARAD, as there are extensive steric conflicts between the BARAD and the partner helix of the CCD (Fig. 1). Further, the same five OH residues (Val250, Met254, Ala257, Leu261, and Leu264) involved in the CCD homodimer interface also pack against the BARAD (Fig. 1). Thus, the OH in the CCD and OH+BARAD crystal structures represent mutually exclusive conformations of BECN1.
Figure 1.
The OH has two different packing states. The BECN1 CCD (magenta) and BARAD (green) are shown in ribbons. The OHs in the CCD and BARAD structures are superimposed. Boxes indicate regions that are rotated 30° about the x axis and enlarged to show the mutually exclusive packing arrangement of interacting OH side chains displayed as sticks. The first β-sheet and third helix of the BARAD, against which the OH side chains pack, are labeled β1 and α3, respectively. This and other molecular figures were made using PyMOL version 1.5.0.2 (Schrödinger, LLC, New York) (41).
Approximately 1359 Å2 of surface area are buried upon the OH packing against the BARAD, significantly more than the 937 Å2 buried when the OH packs within the CCD. Within the CCD, OH residues are only involved in paired interactions. However, OH residues pack against the first β-sheet and third α-helix of the BARAD (Fig. 1), resulting in more extensive packing.
Each of the OH interface residues is largely conserved from yeast to humans (supplemental Fig. S1A). The position equivalent to human Met254 is least well-conserved, although all examined vertebrates have a Met or Val at equivalent positions. In comparison with the OH, residues of the partner BECN1 CCD that pack against the OH are not as well-conserved (supplemental Fig. S1B). Based on this preliminary analysis, we hypothesized that simultaneous alanine mutagenesis of four of the common OH interface residues (V250A, M254A, L261A, and L264A (tetrad mutant)) would differently impact structure and stability of the CCD and BARAD. Structural and thermodynamic analyses of these differences would allow us to determine whether the OH is part of the CCD, packs against the BARAD, or transitions between the two states.
Aromatic finger mutation (AFM) increases solubility of BARAD-containing BECN1 fragments
We find that the WT OH+BARAD and CCD–BARAD proteins aggregate during purification. Analysis of the OH+BARAD crystal structure (PDB entry 4DDP) indicates that the OH+BARAD molecules are arranged in a head-to-tail manner in the crystal lattice, stabilized by the aromatic finger, consisting of Phe359, Phe360, and Trp361, of one OH+BARAD molecule being buried within a hydrophobic pocket formed partly by the OH in the next OH+BARAD molecule (33) (supplemental Fig. S2). We hypothesized that this interaction may be the cause of protein aggregation during purification of WT OH+BARAD–containing constructs.
The AFM, wherein the aromatic finger residues, Phe359, Phe360, and Trp361, are mutated to Asp, has no effect on the secondary structure content estimated from CD (supplemental Fig. S3 and Table S1) or the SEC-SAXS profiles (supplemental Fig. S4 and Table S2) of the CCD+BARAD constructs. This is consistent with previously published analyses that showed that the AFM does not change the overall secondary structure or thermal stability of the BECN1 OH+BARAD (33). However, we find that the AFM improves the yield of soluble homogeneous protein for both OH+BARAD and CCD+BARAD fragments. Because introduction of the AFM dramatically reduced aggregation and permitted large-scale purification of homogenous, high-quality protein without affecting structure, all BARAD-containing proteins used for subsequent analysis bear the AFM.
BECN1 CCD structure is destabilized by the OH tetrad mutation
The BECN1 CCD interface is composed of 26 interacting pairs comprising 13 unique pairs related by 2-fold homodimer symmetry. Only six of these 13 unique pairs constitute residue pairs that form good hydrophobic interactions, whereas for each of the remaining seven pairs, one residue has a polar and/or bulky side chain, resulting in non-ideal packing (30, 31). At the termini of each set of 13 interacting pairs within the CCD homodimer, each OH contributes to five pairs of interactions: Val250–Glu191, Met254–Leu187, Ala257–Leu184, Leu261–Leu180, and Leu264–Ser177 (supplemental Fig. S1). This includes three of the six good hydrophobic interacting pairs. Thus, the OH contributes half of the hydrophobic interactions that stabilize the BECN1 CCD homodimer.
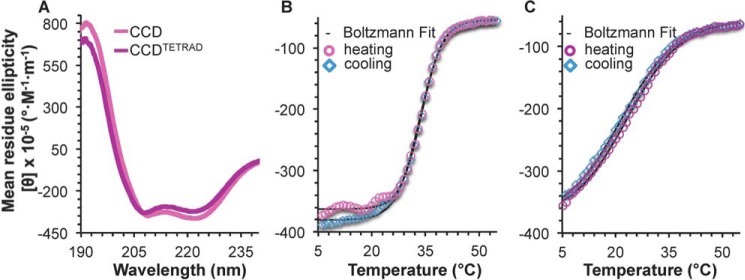
To assess the effect of the tetrad mutation on CCD homodimerization, we compared dimer self-dissociation constants (Kd) of the CCD and CCDTETRAD, as quantified by ITC. At 20 °C, self-association of the CCDTETRAD is ∼1.5-fold weaker than that of the WT CCD (Table 1). A comparison of the secondary structure content estimated by analysis of CD spectra for the CCD and CCDTETRAD indicates that the tetrad mutation reduces helicity by ∼9.5 residues with a concomitant ∼9-residue increase in coil content of the CCD but does not impact β-strand content (Fig. 2A and Table 2). Therefore, the tetrad mutation destabilizes CCD secondary structure. The cooperative, reversible melting curves indicate that the melting temperature (Tm) of ∼33.5 °C of the WT CCD (Fig. 2B and Table 2) decreases substantially to 22.5 °C for the CCDTETRAD (Fig. 2C and Table 2), which is also indicative of the tetrad mutation destabilizing the CCD structure.
Table 1.
Thermodynamics of self-dissociation of CCD-containing BECN1 fragments
| BECN1 fragment | Kd | ΔHa | ΔSb |
|---|---|---|---|
| μm | kJ/mol | J/K mol | |
| CCD | 44.8 ± 2.0 | 45.0 ± 10.5 | 236.6 ± 36.2 |
| CCDTETRAD | 70.7 ± 3.1 | 157.0 ± 8.8 | 615.1 ± 30.0 |
| CCD-BARADAFM | 61.0 ± 8.1 | 207.1 ± 21.5 | 787.1 ± 73.0 |
| CCD-BARADAFM,TETRAD | 134.2 ± 4.9 | 143.0 ± 10.3 | 561.9 ± 34.9 |
a ΔH = change in enthalpy.
b ΔS = change in entropy.
Figure 2.
BECN1 CCD CD spectra and melting curves. A, BECN1 CCD (magenta) and BECN1 CCDTETRAD (purple). B, BECN1 CCD melting curve. C, BECN1 CCDTETRAD melting curve. Boltzmann fits to the melting curves are shown in black.
Table 2.
CD analysis: Estimated secondary structure content and melting temperatures
| Protein | Average estimated secondary structure content |
Melting temperature |
|||
|---|---|---|---|---|---|
| Helix | Strand | Coil | Forward | Reverse | |
| % | % | % | °C | °C | |
| BECN1 CCD | 89.8 ± 6.1 | 1.9 ± 1.1 | 8.6 ± 6.4 | 33.9 ± 0.1 | 33.3 ± 0.1 |
| BECN1 CCDTETRAD | 80.4 ± 4.8 | 1.9 ± 1.6 | 17.6 ± 6.8 | 23.2 ± 0.2 | 21.6 ± 0.2 |
| BECN1 OH+BARADAFM | 46.1 ± 7.9 | 20.2 ± 3.0 | 34.2 ± 9.8 | 47.7 ± 0.1 | NAa |
| BECN1 OH+BARADAFM,TETRAD | 46.3 ± 3.8 | 10.6 ± 2.6 | 44.0 ± 3.2 | 41.4 ± 0.1 | NA |
| BECN1 CCD-BARADAFM | 65.2 ± 6.7 | 5.3 ± 1.6 | 29.9 ± 7.4 | 23.9 ± 0.6; 49.0 ± 0.3 | NA |
| BECN1 CCD-BARADAFM,TETRAD | 56.2 ± 4.1 | 7.5 ± 0.2 | 36.7 ± 3.7 | 22.1 ± 0.6; 45.1 ± 0.2 | NA |
a NA, not available; proteins containing the BARAD precipitated, even upon heating just to the melting temperature; therefore, reverse Tm could not be determined.
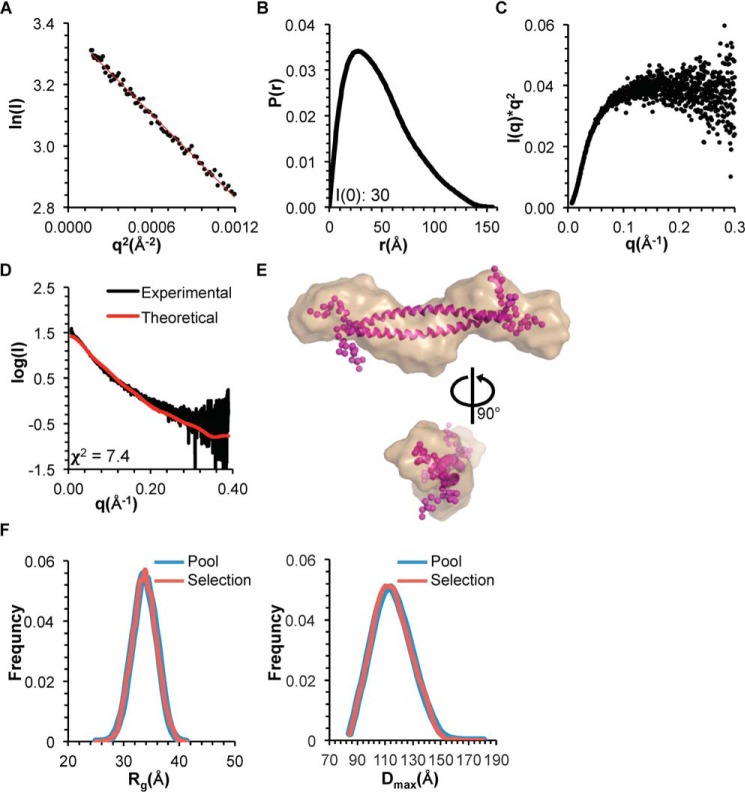
We used SEC-SAXS to investigate the impact of the tetrad mutation on size, shape, and structure of the CCD. The Rg of the CCDTETRAD, estimated from the Guinier plot and P(r) distribution, is ∼39 Å (Fig. 3 (A and B) and Table 3), which is similar to the Rg of ∼36 Å reported for the WT CCD (31). However, the Dmax of the CCDTETRAD obtained from the P(r) distribution is 155 Å, which is about 30% longer than the Dmax reported for the CCD (31) (Fig. 3B and Table 3). Further, the Kratky plot indicates that the CCDTETRAD is largely unfolded (Fig. 3C), in contrast to the CCD, which is well-folded (31). The increase in the Dmax observed in the CCDTETRAD is consistent with the destabilization of the CCD by the tetrad mutation, observed in secondary structure estimations and melting temperature calculations by CD, because increased disorder would result in polypeptides sampling larger volumes in solution.
Figure 3.
SAXS analysis of BECN1 CCDTETRAD. A, Guinier plot. B, P(r) pairwise distribution. C, Kratky plot. D, theoretical scattering curve calculated from the best fit EOM model of the BECN1 CCD with flexible OH fitted to the experimental data. E, best fit EOM model (magenta, flexible residues modeled as spheres) superimposed into the filtered SAXS envelope (sand). F, Rg (left) and Dmax (right) distribution of initial pool and selected ensemble of models.
Table 3.
SAXS data analysis
| BECN1 domain |
||||||
|---|---|---|---|---|---|---|
| CCD (31) | CCDTETRAD | OH+BARADAFM | OH+BARADAFM,TETRAD | CCD-BARADAFM | CCD-BARADAFM,TETRAD | |
| From sequence | ||||||
| Molecular mass (kDa) | 24.4 (dimer) | 21.8 (dimer) | 26.0 (monomer) | 26.7 (monomer) | 64.6 (dimer) | 64.8 (dimer) |
| SAXS parameters | ||||||
| Rg (Å) | 34 | 37 | 20 | 22 | 45 | 47 |
| P(r) Rg (Å) | 37 | 40 | 20 | 23 | 53 | 53 |
| Dmax (Å) | 121 | 155 | 77 | 92 | 195 | 229 |
The theoretical scattering data calculated from the best fit ensemble optimization method (EOM) (35) model of the CCDTETRAD fit the experimental SAXS data poorly with a χ2 of 7.4 (Fig. 3, D and E), which supports the conclusion that the CCDTETRAD is highly flexible, and, because the Kratky plot suggests that the CCDTETRAD is largely unfolded (Fig. 3C), this further suggests that the tetrad mutation destabilizes additional regions of the CCD, such as the N-terminal region of the CCD that it packs against. In contrast, the theoretical scattering calculated from the WT CCD crystal structure fits the WT SAXS data very well (31). Analysis using EOM further confirms that the CCDTETRAD is highly flexible. EOM generates a random pool of models and selects an ensemble of models from the pool that best fit the scattering data, providing an Rg and Dmax distribution for both the pool and selected ensemble of models (35). A narrower Rg and Dmax distribution for the selected ensemble compared with the random pool suggests that the molecule is rigid, whereas an Rg and Dmax distribution for the ensemble that is as broad as the pool suggests that the molecule is flexible (36). The Rg and Dmax distribution for the CCDTETRAD pool and selected ensemble are equally broad (Fig. 3F), suggesting that the CCDTETRAD is highly flexible.
Taken together, the secondary structure and melting temperature estimations by CD, homodimer association affinity quantified by ITC, solution size and shape parameter determinations by SEC-SAXS, and assessment of flexibility by the Kratky plot and EOM all indicate that CCD secondary, tertiary, and quaternary structure stability is significantly disrupted by the OH tetrad mutation.
The OH tetrad mutation impacts BECN1 OH+BARAD structure less than that of the CCD
The crystal structure of the human BARAD (33) includes the OH packed against the BARAD, but the OH was not included in the yeast VPS30 BARAD crystal structure (32). In each case, the crystal structure as well as solution data indicate that the BARAD is a monomer. Therefore, the OH does not mediate homodimerization of the BARAD; nor does it appear to be required for stability of the BARAD structure in all VPS30/BECN1 homologs.
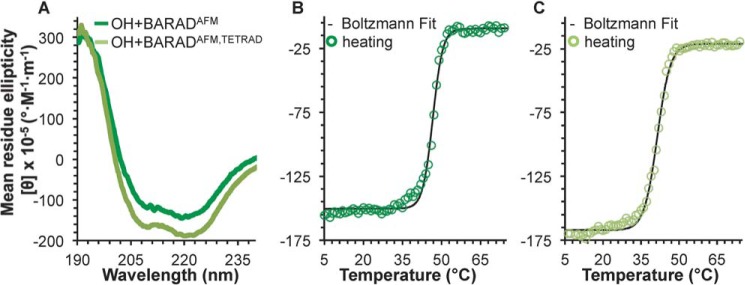
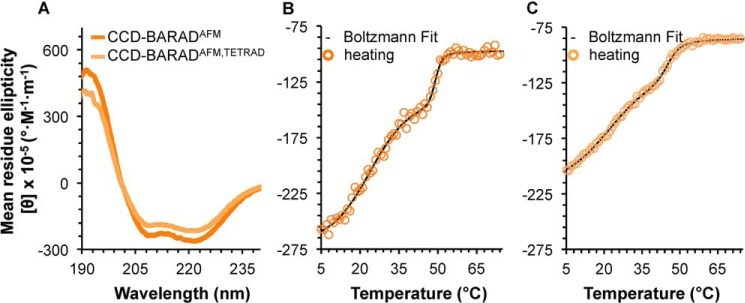
A comparison of the secondary structure content estimated from CD spectra analysis for the OH+BARADAFM and OH+BARADAFM,TETRAD indicates that the tetrad mutation reduces β-strand content by ∼21 residues with a concomitant increase of ∼25 residues in coil content of the OH+BARAD but does not impact α-helical content (Fig. 4A and Table 2). This suggests that although the tetrad mutation does not disrupt the OH itself, it does distort BARAD secondary structure. Reversible melting curves were not obtained for any construct containing the BARAD, as the proteins precipitated even when heated only to the Tm. However, melting is cooperative for the OH+BARADAFM and OH+BARADAFM,TETRAD (Fig. 4, B and C), enabling us to assess Tm. The tetrad mutation decreases the Tm of the BARAD by only 7 °C (Table 2). Thus, the BARAD is less affected by the tetrad mutation than the CCD.
Figure 4.
BECN1 OH+BARAD CD spectra and melting curves. A, BECN1 OH+BARADAFM (green) and BECN1 OH+BARADAFM,TETRAD (light green). B, BECN1 OH+BARADAFM melting curve. C, OH+BARADAFM,TETRAD melting curve. Boltzmann fits to the melting curves are shown in black.
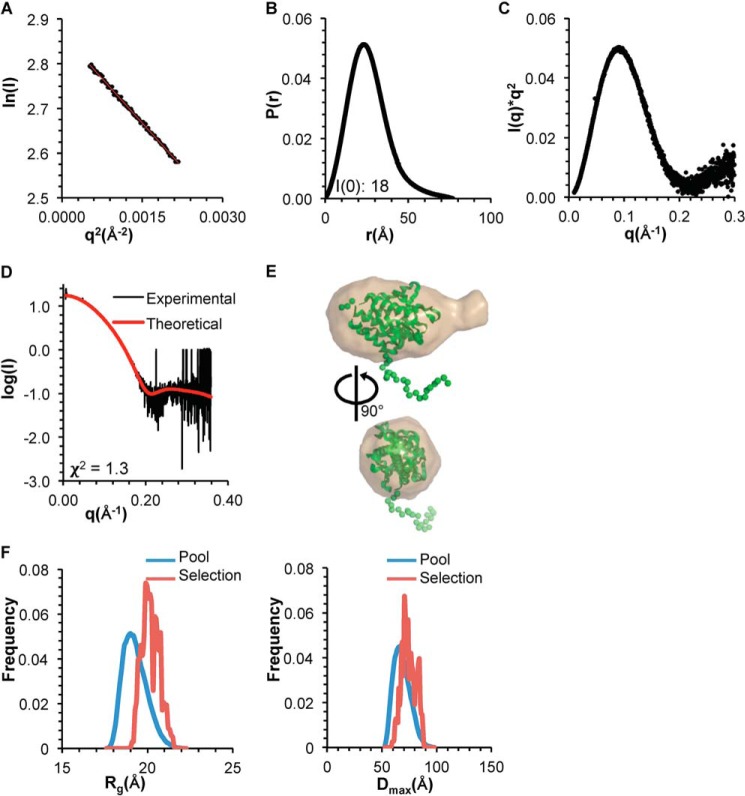
Next, we used SEC-SAXS to investigate the impact of the tetrad mutation on size, shape, and structure of the OH+BARAD. The Rg of the OH+BARADAFM estimated from the Guinier plot and P(r) distribution is ∼20 Å (Fig. 5 (A and B) and Table 3). Further, the shapes of the P(r) distribution plot and the Kratky plot indicate that the OH+BARADAFM is a well-folded, globular domain (Fig. 5, B and C). EOM was used to generate an ensemble of OH+BARAD models that would account for the flexible, N-terminal His6 tag. The best fit of the experimental SAXS data to the theoretical scattering data calculated from an EOM model has a χ2 of 1.3 (Fig. 5, D and E), indicative of a very good fit.
Figure 5.
SAXS analysis of BECN1 OH+BARADAFM. A, Guinier plot. B, P(r) pairwise distribution. C, Kratky plot. D, theoretical scattering curve calculated from the best fit EOM model of BECN1 OH+BARADAFM, with OH modeled against the BARAD and a flexible His6 tag, fitted to the experimental data. E, best fit EOM model (green, flexible residues modeled as spheres) superimposed into the filtered SAXS envelope (sand). F, Rg (left panel) and Dmax (right panel) distribution of initial pool and selected ensemble of models.
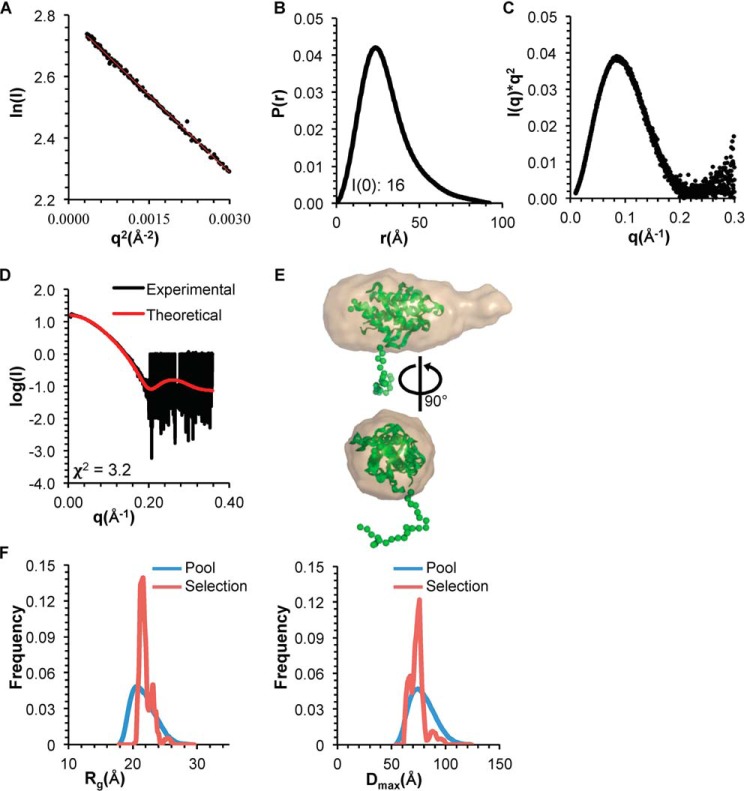
The Rg of the OH+BARADAFM,TETRAD estimated from the Guinier plot and P(r) distribution is ∼23 Å (Fig. 6 (A and B) and Table 3), which is similar to that of the OH+BARADAFM. Similarly, the shape of the P(r) distribution curve and the Kratky plot reveals that the OH+BARADAFM,TETRAD is also well-folded and globular (Fig. 6, B and C). However, the 15-Å increase in Dmax and the extended tail on the P(r) distribution of the OH+BARADAFM,TETRAD (Fig. 6B) relative to the OH+BARADAFM (Fig. 5B) suggest the presence of a more flexible region in the OH+BARADAFM,TETRAD. Indeed, the normalized Kratky (37) shows that although the OH+BARADAFM,TETRAD is well-folded, it is slightly more flexible than the OH+BARADAFM (data not shown). This is consistent with the increased coil content estimated by CD.
Figure 6.
SAXS analysis of BECN1 OH+BARADAFM,TETRAD. A, Guinier plot. B, P(r) pairwise distribution. C, Kratky plot. D, theoretical scattering curve calculated from the best fit EOM model of BECN1 OH+BARADAFM,TETRAD, with OH modeled against the BARAD and a flexible His6 tag, fitted to the experimental data. E, best fit EOM model (green, flexible residues modeled as spheres) superimposed onto the filtered SAXS envelope (sand). F, Rg (left) and Dmax (right) distribution of initial pool and selected ensemble of models.
EOM was used to generate an ensemble of OH+BARADAFM,TETRAD models to account for the flexible, N-terminal His6 tag of the OH+BARAD fragments. The best fit of the experimental SAXS data to the theoretical scattering data calculated from an EOM model has a χ2 of 3.2 (Fig. 6, D and E). Last, for both the OH+BARADAFM and OH+BARADAFM,TETRAD, EOM analyses show that the Rg and Dmax distribution for the selected ensemble is narrower than that of the initial pool of ensembles (Figs. 5F and 6F), indicating that both constructs are largely conformationally homogeneous in solution.
Taken together, the secondary structure and Tm estimations by CD, solution size and shape parameter determinations by SEC-SAXS analyses, and assessment of flexibility by the Kratky plot and EOM, all demonstrate that the OH tetrad mutation impacts OH+BARAD structure and stability significantly less than it does the CCD.
The OH preferentially packs against the BARAD rather than within the CCD homodimer but transiently samples both conformations
A comparison of the secondary structure content estimated from the CD spectra for the BECN1 CCD–BARADAFM and CCD–BARADAFM,TETRAD indicates that the tetrad mutation decreases helical content by 25 residues and increases strand and coil content by ∼4 and ∼9 residues, respectively (Fig. 7A and Table 2). Thus, secondary structure of the two-domain construct is impacted by the tetrad mutation.
Figure 7.
BECN1 CCD+BARAD CD spectra and melting curves. A, CCD–BARADAFM (orange) and CCD–BARADAFM,TETRAD (light orange). B, CCD–BARADAFM melting curve. C, CCD–BARADAFM,TETRAD melting curve. Boltzmann fits to the melting curves are shown in black.
To determine whether the CCD–BARAD also forms a homodimer like the CCD, we used ITC to quantify the dimer Kd. At 20 °C, self-association of the CCD–BARADAFM is 1.4-fold weaker than that of the WT CCD but slightly tighter than that of the CCDTETRAD (Table 1). Notably, self-association of the CCD–BARADAFM,TETRAD is ∼2-fold weaker than the CCD–BARADAFM (Table 1). Taken together, these ITC data indicate that the tetrad mutation destabilizes both the CCD and CCD–BARAD homodimer. Equally importantly, compared with the WT CCD homodimer, the CCD–BARAD homodimer is substantially dissociated, with a Kd comparable with the CCDTETRAD rather than the CCD, suggesting that the OH helix does not stabilize the homodimer as much in the two-domain fragment.
To assess the thermostability of the CCD–BARAD, we used CD to measure the melting temperature of the two-domain fragment. Although protein precipitation prevented acquisition of reversible curves, melting curves for both the CCD–BARADAFM and CCD–BARADAFM,TETRAD contain two well-separated transitions (Fig. 7, B and C), indicating that each construct contains two independently folded domains. The higher Tm of 49 °C for the CCD–BARADAFM corresponds well with that of the OH+BARADAFM (Tm = 48 °C), but notably, the lower Tm of 24 °C corresponds to that of the CCDTETRAD (Tm = 23 °C) rather than the WT CCD (Tm = 33 °C) (Table 2). This suggests that in the CCD–BARAD construct, the OH is not part of the CCD homodimer but rather packs against the BARAD.
This is further corroborated by the melting temperatures of 22 and 45 °C observed for the CCD–BARADAFM,TETRAD (Table 2), as the 22 °C transition temperature is comparable with that of the lower Tm of the CCD–BARADAFM as well as that of the CCDTETRAD, whereas the 45 °C transition is a little less than the second Tm of 49 °C for the CCD–BARADAFM or the Tm of 48 °C for the OH+BARADAFM, albeit slightly higher than the Tm of 41 °C for the OH+BARADAFM,TETRAD (Table 2). Therefore, the tetrad mutation does not impact stability of the CCD in the CCD–BARAD construct but rather appears to marginally destabilize the OH+BARAD, consistent with the disruption of packing of the OH against the BARAD in the two-domain construct.
Thus, the melting temperature analysis indicates that the OH packs against the BARAD in the BECN1 homodimer. However, self-dissociation measurements by ITC indicate that the CCD–BARAD homodimerizes slightly better than the CCDTETRAD and that disruption of the OH packing interface weakens CCD-mediated homodimerization of the CCD–BARAD, although not as much as in the CCD alone. Taken together, these data suggest that in solution, the OH transitions between packing as part of the CCD or against the BARAD, with the predominant state comprising the OH packed against the BARAD.
SAXS indicates the OH packs against the BARAD in the CCD–BARAD protein fragment
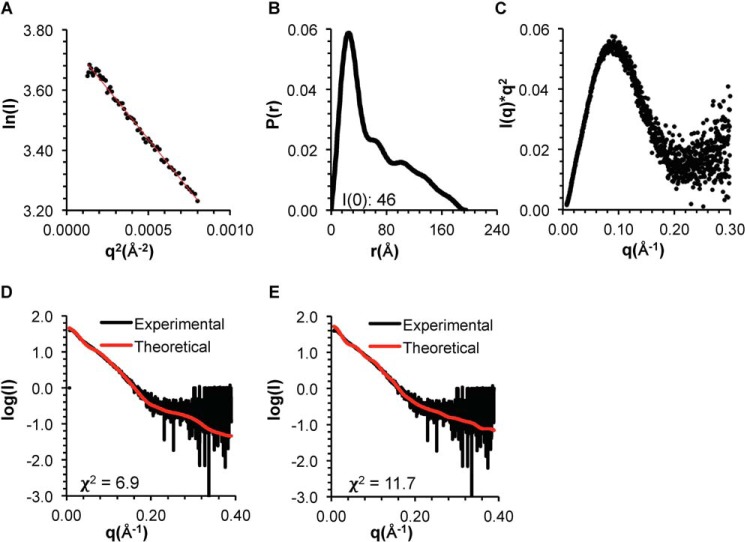
We used SEC-SAXS to further investigate the conformation of the OH in the CCD–BARADAFM and CCD–BARADAFM,TETRAD. In each case, the Rg estimated from the Guinier plot and P(r) distribution is ∼50 Å, (Figs. 8 (A and B) and 9 (A and B) and Table 3), and the shape of the P(r) distribution curves and the Kratky plots indicates that both are well-folded, dumbbell-shaped proteins (Figs. 8 (B and C) and 9 (B and C)).
Figure 8.
SAXS analysis of BECN1 CCD–BARADAFM. A, Guinier plot. B, P(r) pairwise distribution. C, Kratky plot. D, theoretical scattering data calculated from the best fit EOM model of the BECN1 CCD–BARADAFM, with the OH modeled as packed against the BARAD, fitted into the experimental data. E, theoretical scattering data calculated from the best fit EOM model of the BECN1 CCD–BARADAFM, with the OH modeled as part of the CCD, fitted into the experimental data.
Figure 9.
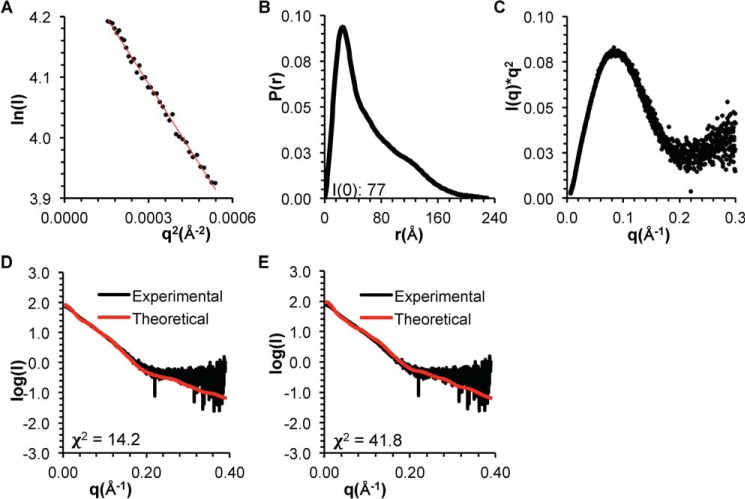
SAXS analysis of BECN1 CCD–BARADAFM,TETRAD. A, Guinier plot. B, P(r) pairwise distribution. C, Kratky plot. D, theoretical scattering data calculated from the best fit EOM model of the BECN1 CCD–BARADAFM,TETRAD, with the OH modeled as packed against the BARAD, fitted into the experimental data. E, theoretical scattering data calculated from the best fit EOM model of the BECN1 CCD–BARADAFM,TETRAD, with the overlap region as flexible loop, fitted into the experimental data.
EOM was used to generate the best ensemble of CCD–BARADAFM homodimer models. Two separate sets of ensembles were generated: one with the OH modeled within the CCD and the other with the OH modeled as packed against the BARAD. The average Rg of 53 Å for the ensemble wherein the OH is packed against the BARAD is more similar to the Rg of ∼50 Å estimated from SAXS data (Table 3) than the Rg of 65 Å for the ensemble wherein the OH is part of the CCD. Further, the experimental SAXS data fit better to the theoretical scattering curve calculated from the best fit EOM model wherein the OH is packed against the BARAD, as indicated by a χ2 of 6.9 (Fig. 8D), compared with the χ2 of 11.7 for the fit of the theoretical scattering curve calculated from the best fit EOM model wherein the OH is part of the CCD (Fig. 8E), although neither ensemble fits particularly well. Thus, whereas EOM analysis also suggests that the OH probably packs against the BARAD rather than within the CCD in the BECN1 CCD–BARAD homodimer, the poor fits of both modeled states suggest that different conformations are transiently occupied in solution.
EOM was also used to generate the best ensemble of CCD–BARADAFM,TETRAD homodimer models. Two separate sets of ensembles were generated: one with the OH modeled packed against the BARAD and the other with the OH modeled as coil. The fit of the experimental CCD–BARADAFM,TETRAD SAXS data was better to the theoretical scattering curve calculated from the best fit EOM model wherein the OH is packed against the BARAD, as indicated by a χ2 of 14.2 (Fig. 9D), compared with the χ2 of 41.8 for the fit of the theoretical scattering curves calculated from the best fit EOM model wherein the OH is a coil (Fig. 9E). However, all model fits to the CCD–BARADAFM,TETRAD SAXS data were worse than the fit of the best EOM model with the OH packed against the BARAD to the experimental CCD–BARADAFM SAXS data (χ2 = 6.9; Fig. 8D). Together, these results suggest that the tetrad mutation destabilizes OH packing in the CCD–BARAD homodimer.
The tetrad mutant decreases starvation-induced autophagy
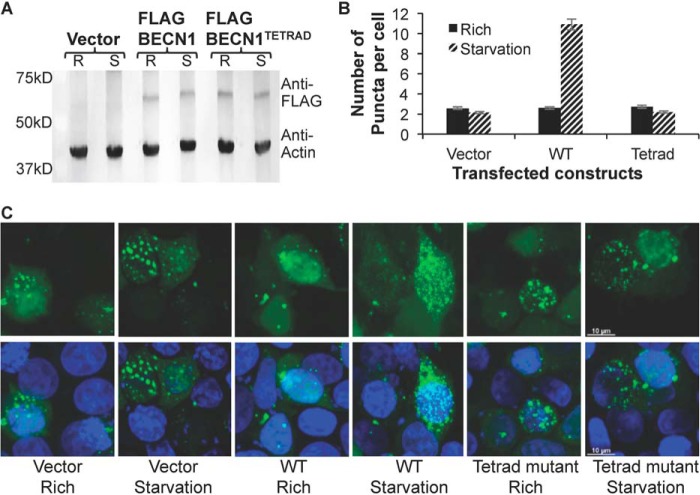
Last, we investigated the impact of destabilization of OH packing on cellular autophagy levels mediated by exogenously expressed full-length BECN1, by quantifying and comparing the impact of either WT or the tetrad mutant (V250A, M254A, L261A, and L264A) BECN1. Because BECN1 is known to be required for autophagosome nucleation, we evaluated cellular autophagy by monitoring and comparing levels of puncta labeled with GFP-tagged LC3, an autophagosome-specific marker, in cells grown in either nutrient-rich or starvation medium. Expression of WT and tetrad mutant BECN1 was comparable in starvation and nutrient-rich conditions (Fig. 10A). We used human breast adenocarcinoma MCF7 cells because they lack detectable endogenous expression of BECN1, resulting in very low basal levels of autophagy (4, 22, 38) even in starvation conditions unless BECN1 is ectopically expressed (Fig. 10, B and C). This allows the effect of BECN1 mutants to be assayed in the absence of endogenous BECN1.
Figure 10.
Effect of the OH tetrad mutation on autophagy. A, Western blotting of MCF7 cell extracts showing comparable expression levels of WT and mutant FLAG-BECN1 in nutrient-rich (R) and starvation (S) conditions, with actin as a loading control. B, light microscopy quantification of discrete GFP-LC3 puncta per cell in GFP-positive MCF7 cells co-transfected with GFP-LC3 and WT or mutant FLAG-BECN1 as indicated below the x axis. Bars, number of puncta per cell. Error bars, S.D. C, representative images of GFP-LC3 (green) and DAPI (blue) staining in cells grown in starvation or nutrient-rich medium and transfected with mutant FLAG-BECN1 as indicated.
Transient expression of BECN1 in MCF7 cells did not increase autophagy levels in nutrient-rich conditions (p = 0.951 for BECN1 expression versus no expression; Fig. 10B) but led to a marked increase in autophagy upon starvation (p = 0.005 for starved versus nutrient-rich cells; Fig. 10B). We find that the BECN1 tetrad mutation does not impact autophagy levels in nutrient-rich medium (p = 0.836 for mutant versus WT), consistent with the lack of impact of BECN1 expression on basal autophagy levels in nutrient-rich conditions. Strikingly, however, the BECN1 tetrad mutation decreases autophagy levels in starvation medium (p = 0.004 for mutant versus WT), indicating that these residues are essential for up-regulating starvation-induced autophagy. This is consistent with the established importance of other conserved regions of BECN1 in starvation-triggered autophagy rather than basal autophagy levels (29, 31).
Discussion
Based on the biophysical and structural analyses reported here, we conclude that the BECN1 C-terminal domains, the CCD and BARAD, are linked by a conformationally labile helix. Structural superimposition unambiguously demonstrates that the OH residues present in the crystal structure of both the CCD (30, 31) and OH+BARAD (33) cannot simultaneously pack as part of the CCD and against the BARAD. Our buried surface area calculations identify a larger interface between the OH and BARAD than between the OH and the partner helix within the CCD. Half of the hydrophobic interactions that stabilize the CCD homodimer map to the OH, and alanine mutagenesis of the four hydrophobic interacting OH residues, which are also key components of the interface with the BARAD, substantially weakens the CCD homodimer. Our results show that self-association of the CCD-BARAD is weaker than that of the WT CCD alone, but comparable with that of the tetrad mutant CCD, suggesting that in the CCD-BARAD, the OH primarily packs against the BARAD; however, tetrad mutation in the CCD-BARAD further weakens CCD-BARAD homodimerization, indicating that the OH transiently contributes to homodimerization. The comparative melting temperature and secondary structure content assessed by CD spectroscopy together with SEC-SAXS analyses also indicate that the tetrad mutation disrupts CCD structure more than the BARAD. Therefore, contrary to currently established ideas, the OH preferentially packs against the BARAD in the absence of heterologous binding partners but appears to also transiently sample conformations where it forms part of the CCD. Thus, relative to homodimer association reported for the CCD in this and previous reports (30, 31), full-length BECN1 probably homodimerizes more weakly, consistent with that reported here for the CCD–BARAD.
We further show that the BECN1 homodimer is unstable at physiological temperatures. Our melting temperature analyses show that nearly half of the CCD is unfolded in the two-domain CCD–BARAD fragment at 37 °C. Together, the weak self-association and instability of the CCD–BARAD homodimer suggest that BECN1 probably does not exist in a stable homodimeric state at physiological temperatures unless stabilized in that state by BECN1 domains other than the CCD or other protein partners, such as the BCL2 proteins. BECN1 homodimerization probably serves as an important means of regulating autophagy in cells. Notably, this may be a regulatory mechanism that evolved in higher eukaryotes such as vertebrates, as yeast do not encode BCL2 proteins, and it is unknown whether yeast VPS30 homodimerizes.
This is significant because up-regulation of autophagy by BECN1 requires either ATG14 or UVRAG to displace one molecule of the BECN1 homodimer, which would be facilitated by a weak BECN1 homodimer. The 4.4 Å crystal structure (PDB code 5DFZ) of full-length yeast VPS30/BECN1 in complex with VPS34/PI3KC3, VPS15/p150, and VPS38/UVRAG (13), shows that the OH is part of a parallel VPS30–VPS38 CCD heterodimer essential for stabilizing this complex. The 28 Å reconstruction of the quaternary complex of human PI3KC3, p150, BECN1, and ATG14 (12) indicates that BECN1 and ATG14 interact in an analogous manner. A SAXS data–constrained model of the BECN1–ATG14 heterodimer indicates that four of the OH interface residues identified in the homodimer also contribute to the heterodimer coiled-coil interface (Val250–Ile165, Met254–Asn169, Ala257–Leu172, and Leu261–Val176) (31). Notably, these OH residues are largely conserved, especially in vertebrates (supplemental Fig. S1A), and are predicted to pack against ATG14 residues that are also conserved. Indeed among these interacting pairs, the least conserved BECN1 residue, Met254, is paired with Asn169, the least conserved paired residue in ATG14 (31). Thus, the OH contributes critical binding determinants to the parallel CCD heterodimers required for formation of VPS34/PI3KC3 complexes essential for autophagy. Consistent with this, we show that the tetrad mutation of the OH interface residues abrogates the starvation-induced up-regulation of autophagy but has no effect on basal levels of autophagy.
Our experimental evidence showing that the OH preferentially packs against the BARAD in the BECN1 homodimer suggests a mechanism wherein homodimerization via the CCD results in BECN1 conformations that prevent the BARAD aromatic finger from associating with membranes, whereas BECN1 heterodimerization with ATG14 or UVRAG disrupts this inhibitory conformation. The crystal structure and mass spectroscopy hydrogen–deuterium exchange experiments of the yeast PI3KC3-p150-BECN1-UVRAG quaternary complex show that the aromatic finger interacts with membranes when the OH participates in the CCD interface of the heterodimer (13). However, formation of a BECN1 homodimer, wherein the OH preferentially packs against the BARAD, would shorten the CCD and involve reorientation of the BARAD relative to that observed in the yeast PI3KC3-p150-BECN1-UVRAG quaternary complex. This would reposition the BARAD such that the aromatic finger is no longer able to interact with membranes, thereby inhibiting BECN1-mediated autophagy.
Another important implication of the OH being preferentially packed against the BARAD in the homodimer is that this would release the N-terminal region of the partner helix within the anti-parallel CCD homodimer. The N-terminal region of the human BECN1 CCD (supplemental Fig. S1B) contains a leucine-rich nuclear export signal (NES) that binds the chromosomal protein 1 (CRM1) to enable nuclear export of BECN1 (39, 40). A functional NES is composed of the sequence motif LX(2–3)LX(2–3)LXL. NES residues Leu184 and Leu187 are essential for BECN1 nuclear export, BECN1-mediated autophagy, and tumor suppression (39, 40). In the CCD homodimer, NES residues Leu184 and Leu187 pack against OH interface residues, Ala257 and Met254, respectively (31). Therefore, to bind CRM1, the BECN1 NES cannot be packed against the BECN1 OH. Thus, our results showing that the OH preferentially packs against the BARAD in the homodimeric state provide a mechanism of how the NES may be exposed for CRM1 binding to enable nuclear export. Indeed, interaction with CRM1 may stabilize BECN1 conformations wherein the OH is packed against the BARAD. Interestingly, unlike vertebrates, lower eukaryotes do not have a NES located at the N terminus of the CCD (supplemental Fig. S1B). Therefore, this conformational switch is probably not needed for nuclear export in yeast, as the NES is located between yeast VPS30 residues 12 and 21 (39), which is part of the intrinsically disordered region, rather than the CCD.
In summary, previous studies have established the BECN1 CCD and BARAD as structurally well-defined domains, but here we show that these domains include a region, now termed the OH, that can adopt two distinct, mutually exclusive packing states, emphasizing overall BECN1 conformational flexibility. Further, in the absence of heterologous partners, the OH transitions between these states with a preference for packing against the BARAD, rather than the CCD, as has been commonly assumed. This region probably serves to regulate BECN1 function in autophagy and nuclear transport, and interactions with different partners probably stabilize one packing state or the other to promote these different functions. Understanding how different interactions stabilize different BECN1 conformational states is the next critical step toward understanding the regulation of BECN1-mediated autophagy.
Experimental procedures
Structural superimposition and buried surface area calculation
The crystal structures of the human BECN1 CCD homodimer (PDB entry 5HHE) and the human BECN1 BARAD containing the OH (OH+BARAD) (PDB entry 4DDP) were superimposed using the align function of the PyMOL Molecular Graphics System (Schrödinger, LLC) (41). The PDBePISA (42) server (http://www.ebi.ac.uk/pdbe/pisa/) was used to calculate interface buried surface areas.
Plasmid preparation
Human BECN1 residues 175–265 (CCD), residues 175–450 (CCD–BARAD), or residues 248–450 (OH+BARAD) were cloned between the NcoI and NotI restriction enzyme sites of the pMBP-Parallel-1 bacterial expression vector (43). The BECN1 OH+BARAD was also cloned between the XhoI and BamHI restriction enzyme sites of the pET-15b bacterial expression vector encoding an N-terminal His6 tag (supplemental Table S3). The OH tetrad mutation (V250A, M254A, L261A, and L264A) and AFM (F359D, F360D, and W361D) were generated via site-directed mutagenesis. A pcDNA3.1 mammalian expression vector encoding full-length, human WT BECN1 was subjected to site-directed mutagenesis to generate the OH tetrad mutation (V250A, M254A, L261A, and L264A).
Protein expression and purification
For the BECN1 CCD and CCDTETRAD, Escherichia coli BL21(DE3)·pLysS cells were transformed and grown in LB medium with 100 μg/ml ampicillin at 37 °C to an A600 of ∼0.8 before equilibrating the temperature to 20 °C. For WT and mutant BECN1 CCD–BARAD and His6-BECN1 OH+BARAD, E. coli Arctic Express cells were transformed and grown in LB medium with 100 μg/ml ampicillin at 30 °C to an A600 of ∼0.6 before equilibrating the temperature to 10 °C. In each case, cells were grown to an A600 of 0.8–1.2. Expression of the CCD and CCD–BARAD was induced by the addition of 0.5 mm isopropyl thio-β-d-galactoside. His6-OH+BARAD constructs were induced with 0.3 mm isopropyl thio-β-d-galactoside. All constructs were expressed overnight.
For all BECN1 CCD and BECN1 CCD–BARAD constructs, soluble maltose-binding protein–tagged fusion protein was purified from clarified crude cell lysate by amylose affinity chromatography (Wash Buffer 1: 25 mm Tris-HCl, pH 7.5, 300 mm NaCl, 1 mm EDTA, 2 mm DTT, 10% glycerol; Wash Buffer 2: 25 mm Tris-HCl, pH 7.5, 300 mm NaCl, 1 mm EDTA, 2 mm DTT) and subjected to overnight, on-column cleavage by GST-tobacco etch virus protease at 4 °C. Cleaved protein was washed from the column with Wash Buffer 2 and further purified by ion exchange chromatography on a 5/50 GL MonoQ (GE Healthcare) (MonoQ Buffer A: 50 mm Tris-HCl, pH 7.5, 2 mm DTT; MonoQ Buffer B: 50 mm Tris-HCl, pH 7.5, 2 mm DTT, 1 m NaCl). BECN1 protein fragments were purified to homogeneity by SEC, using a 10/30 Superdex 200 (GE Healthcare) column (SEC buffer: 25 mm Tris-HCl, pH 7.5, 300 mm NaCl, 2 mm DTT). The fusion proteins were concentrated in a 10,000 MWCO Amicon Ultra-0.5 centrifugal concentrator (EMD Millipore, Billerica, MA).
For His6-BECN1 OH+BARAD constructs, fusion protein was purified from clarified crude cell lysate by nickel immobilized ion affinity chromatography (Wash Buffer: 50 mm HEPES acid, pH 7.5, 300 mm NaCl, 10% (v/v) glycerol, 25 mm imidazole). Protein was eluted with Wash Buffer containing 350 mm imidazole. BECN1 OH+BARAD constructs were subsequently purified to homogeneity by SEC, using a 10/30 HR Superdex 200 column (GE Healthcare) (SEC buffer: 25 mm Tris-HCl, pH 7.5, 300 mm NaCl, 2 mm DTT) and concentrated to 5 mg/ml in a 10,000 molecular weight cut-off Amicon Ultra-0.5 centrifugal concentrator (EMD Millipore, Billerica, MA).
At each stage of purification, protein purity was evaluated by SDS-PAGE stained with Coomassie Blue. In each case, the final purified protein was estimated to be >90% pure by Coomassie Blue-stained SDS-PAGE.
CD spectroscopy
Continuous scanning CD spectra were recorded from 190 to 240 nm at 4 °C in a 300-μl quartz cell (0.1-cm path length) on a Jasco J-815 spectropolarimeter equipped with a Peltier thermoelectric temperature control for each protein sample at concentrations of 3–20 μm in 5 mm sodium phosphate dibasic, 5 mm sodium phosphate monobasic, pH 7.4, 100 mm (NH4)2SO4. Secondary structure content was estimated by analyzing CD data using the SDP48 reference protein database and three programs, SELCON3, CDSSTR, and CONTIN, from the CDpro suite within the Jasco software (44–46). For each spectrum, the average secondary structure was calculated by averaging estimated secondary structure content obtained from the three CD data analysis programs.
Variable temperature CD spectra were recorded at 222 nm at 1 °C intervals, with a ramp rate of 1 °C/min after dilution of protein samples to 3–20 μm in 5 mm sodium phosphate dibasic, 5 mm sodium phosphate monobasic, pH 7.4, 100 mm (NH4)2SO4. Spectra were recorded for BECN1 OH+BARAD and BECN1 CCD–BARAD proteins from 4 to 75 °C and for BECN1 CCD proteins from 4–55 or 55–4 °C for reverse measurements. Data were analyzed using OriginPro version 8 (OriginLab), where mean residue molar ellipticity was plotted against temperature, and the Tm was calculated by fitting the data to the Boltzmann or Double Boltzmann algorithm included in OriginPro version 8.
ITC
BECN1 CCD, CCDTETRAD, CCD–BARADAFM, and CCD–BARADAFM,TETRAD protein samples were dialyzed against 50 mm HEPES base, pH 8.0, 150 mm NaCl, and 2 mm β-mercaptoethanol. ITC experiments were performed in triplicate at 20 °C using a Low Volume Gold Nano isothermal titration calorimeter (TA Instruments, New Castle, DE). 65 μm CCD–BARADAFM, 100 μm CCD–BARADAFM,TETRAD, 350 μm CCD, and 100 μm CCDTETRAD were separately titrated into dialysis buffer using 20 injections of 2.5 μl each. Data were analyzed with the NanoAnalyze software (TA Instruments), using a dimer dissociation model to calculate the dimer dissociation constants.
SAXS data collection and analysis
SAXS data were recorded at the BioCAT beamline (ID18) (Advanced Photon Source, Argonne, IL) on a Pilatus 3 1M detector at a sample-to-detector distance of 3.5 meters at a wavelength of 1.03 Å. For all constructs, SEC was performed in tandem with SAXS to ensure that all SAXS data were collected from a homogenous sample eluting from the SEC column (10/300 GL Superdex 200 Increase). SEC-SAXS data were recorded by exposing the column eluate to the X-ray beam for 1 s with a periodicity of 3 s. Scattering data were normalized to the incident X-ray beam intensity, and scattering from buffer was subtracted using the BioCAT beamline pipeline based on the ATSAS suite of programs prior to further analysis. Data were analyzed with the ATSAS program suite (47), including PRIMUS (48) to calculate Guinier extrapolations to estimate the Rg and Kratky plots to evaluate disorder within the samples. The P(r) function plotted from the Fourier inversion of the scattering intensity, I(q), using GNOM (49) was used to calculate the Rg and Dmax and to reconstruct ab initio envelopes by applying 10 cycles in DAMMIF (50) with subsequent analysis by DAMSEL, DAMSUP, DAMAVER, and DAMFILT (51) to compare and identify the most probable model, align all models to the most probable model, average aligned models and compute a probability map, and filter the averaged model.
EOM 2.1 (35) was used to generate an ensemble of models for each protein fragment. For the CCDTETRAD, we modeled the OH and its partner helix (BECN1 residues 175–193) as flexible using PDB entry 5HHE, after removing OH and partner helix residues (ShortCCD), as a rigid body. For the BARADAFM and BARADAFM,TETRAD, we modeled the flexible His6 tag using PDB entry 4DDP (OH+BARAD) as a rigid body. For the CCD–BARADAFM, we generated two-domain models using (i) the ShortCCD and the OH+BARAD as rigid bodies or (ii) PDB entries 5HHE and 4DDP, after removing OH residues (BARAD), as rigid bodies. For the CCD–BARADAFM,TETRAD, we generated two-domain models using (i) the ShortCCD and the OH+BARAD as rigid bodies or (ii) the ShortCCD and BARAD as rigid bodies. Models were obtained using native-like chain options, and P2 symmetry was imposed on CCD-containing fragments by fixing the original coordinates of the PDB ID: 5HHE or ShortCCD. The theoretical scattering data calculated from each model in the ensemble were fitted to the averaged scattering data using CRYSOL (52). The EOM model that best agreed with the experimentally derived Rg and Dmax and best fit the averaged scattering curves was then fitted into the molecular envelopes using SUPCOMB (53).
Autophagy assays
Cellular autophagy levels were quantified by monitoring cellular localization of GFP-tagged LC3 protein (54). Each chamber of an 8-well culture slide (NuncTM Lab-TekTM chambered coverglass) was seeded with 3.0 × 105 MCF7 cells and cultured overnight in DMEM with 10% FBS until 80–90% confluent. The cells were co-transfected with 500 ng of total plasmid per well, comprising 200 ng of GFP-LC3 and 300 ng of FLAG-BECN1 WT or mutant expression plasmids, using Lipofectamine 2000 reagent (Invitrogen) according to the manufacturer's instructions. After transfection and incubation at 37 °C for 24 h, the cells were cultured in either rich (DMEM, 10% FBS, 2× essential amino acids, and 2× nonessential amino acids) or starvation (Earle's balanced salt solution) medium for 4 h. Cells were counterstained with DAPI to visualize nuclei and facilitate total cell counts, fixed to slides with 4% (v/v) paraformaldehyde, and then stored in 70% (v/v) glycerol. Cells were washed with PBS between the counterstaining, fixation, and storage stages. GFP-LC3-positive puncta were observed under a fluorescent microscope (Zeiss AxioObserver Z1) and quantified by counting a minimum of 50 cells for duplicate samples per condition using Imaris software (Bitplan AG, Zurich, Switzerland) in three independent experiments. The significance of alterations in autophagy levels was determined by a two-tailed, heteroscedastic Student's t test, wherein p ≤ 0.05 is considered significant.
Expression levels of WT FLAG-BECN1 and mutants were verified by Western blotting using commercial mouse monoclonal anti-FLAG M2-peroxidase antibody (Sigma). As a loading control, actin levels in MCF7 cell lysates were detected with mouse anti-actin (EMD-Millipore).
Author contributions
K. G. performed protein purification, biophysical experiments, and analysis; helped with cellular assays; and wrote and edited the manuscript. Y. L. performed protein purification, biophysical experiments, and analysis and helped edit the manuscript. S. M. performed the cellular assays. Z. L. assisted Y. L. with protein purification and mutagenesis. S. C. assisted with SEC-SAXS data collection. C. L. C. assisted with data analysis and edited the manuscript. S. C. S. directed the research, designed the experiments, and wrote and edited the manuscript. All authors analyzed the results and approved the final version of the manuscript.
Supplementary Material
Acknowledgments
Work performed at Bio-CAT was supported by NIH NIGMS Grant 9P41 GM103622 and and S10 OD018090. The use of the Pilatus 3 1M detector was supported by NIGMS, National Institutes of Health (NIH), Grant 1S10OD018090-01. This research used resources of the Advanced Photon Source, a United States Department of Energy Office of Science User Facility operated for the Department of Energy Office of Science by Argonne National Laboratory under Contract DE-AC02-06CH11357. We acknowledge the North Dakota State University (NDSU) Core Biology Facility (funded by NIH Grant P30 GM103332-01) for access to tissue culture facilities and the NDSU Advanced Imaging and Microscopy Core Laboratory for access to microscopy and imaging equipment. We also thank Dr. Pawel Borowicz for assistance with imaging and puncta quantification method development.
This work was supported by National Institutes of Health (NIH) Grants NINDS RO3 NS090939 and NIGMS R15 GM122035 (to S. C. S.) and R15 GM113227 (to C. L. C.), National Science Foundation Grant MCB-1413525 (to S. C. S.), and a North Dakota State University Graduate School doctoral dissertation award (to K. G.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article contains supplemental Tables S1–S3 and Figs. S1–S4.
- CCD
- coiled-coil domain
- AFM
- aromatic finger mutation
- BARAD
- β-α repeated, autophagy-specific domain
- Dmax
- maximum particle size
- EOM
- ensemble optimization method
- I(q)
- scattering intensity
- OH
- overlap helix
- PI3KC3
- class III phosphatidylinositol 3-kinase
- Rg
- radius of gyration
- SAXS
- small angle X-ray scattering
- SEC
- size exclusion chromatography
- ITC
- isothermal titration calorimetry
- NES
- nuclear export signal
- PDB
- Protein Data Bank.
References
- 1. De Duve C., and Wattiaux R. (1966) Functions of lysosomes. Annu. Rev. Physiol. 28, 435–492 [DOI] [PubMed] [Google Scholar]
- 2. Lawrence B. P., and Brown W. J. (1992) Autophagic vacuoles rapidly fuse with pre-existing lysosomes in cultured hepatocytes. J. Cell Sci. 102, 515–526 [DOI] [PubMed] [Google Scholar]
- 3. Baba M., Takeshige K., Baba N., and Ohsumi Y. (1994) Ultrastructural analysis of the autophagic process in yeast: detection of autophagosomes and their characterization. J. Cell Biol. 124, 903–913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Liang X. H., Jackson S., Seaman M., Brown K., Kempkes B., Hibshoosh H., and Levine B. (1999) Induction of autophagy and inhibition of tumorigenesis by Beclin 1. Nature 402, 672–676 [DOI] [PubMed] [Google Scholar]
- 5. Klionsky D. J. (2010) The autophagy connection. Dev. Cell 19, 11–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Liang X. H., Kleeman L. K., Jiang H. H., Gordon G., Goldman J. E., Berry G., Herman B., and Levine B. (1998) Protection against fatal Sindbis virus encephalitis by Beclin 1, a novel Bcl-2-interacting protein. J. Virol. 72, 8586–8596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kihara A., Noda T., Ishihara N., and Ohsumi Y. (2001) Two distinct Vps34 phosphatidylinositol 3-kinase complexes function in autophagy and carboxypeptidase Y sorting in Saccharomyces cerevisiae. J. Cell Biol. 152, 519–530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Itakura E., Kishi C., Inoue K., and Mizushima N. (2008) Beclin 1 forms two distinct phosphatidylinositol 3-kinase complexes with mammalian Atg14 and UVRAG. Mol. Biol. Cell 19, 5360–5372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Sun Q., Fan W., Chen K., Ding X., Chen S., and Zhong Q. (2008) Identification of Barkor as a mammalian autophagy-specific factor for Beclin 1 and class III phosphatidylinositol 3-kinase. Proc. Natl. Acad. Sci. U.S.A. 105, 19211–19216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Itakura E., and Mizushima N. (2009) Atg14 and UVRAG: mutually exclusive subunits of mammalian Beclin 1-PI3K complexes. Autophagy 5, 534–536 [DOI] [PubMed] [Google Scholar]
- 11. Matsunaga K., Saitoh T., Tabata K., Omori H., Satoh T., Kurotori N., Maejima I., Shirahama-Noda K., Ichimura T., Isobe T., Akira S., Noda T., and Yoshimori T. (2009) Two Beclin 1-binding proteins, Atg14L and Rubicon, reciprocally regulate autophagy at different stages. Nat. Cell Biol. 11, 385–396 [DOI] [PubMed] [Google Scholar]
- 12. Baskaran S., Carlson L.-A., Stjepanovic G., Young L. N., Kim D. J., Grob P., Stanley R. E., Nogales E., and Hurley J. H. (2014) Architecture and dynamics of the autophagic phosphatidylinositol 3-kinase complex. eLife 10.7554/eLife.05115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Rostislavleva K., Soler N., Ohashi Y., Zhang L., Pardon E., Burke J. E., Masson G. R., Johnson C., Steyaert J., Ktistakis N. T., and Williams R. L. (2015) Structure and flexibility of the endosomal Vps34 complex reveals the basis of its function on membranes. Science 350, aac7365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Su M., Li Y., Wyborny S., Neau D., Chakravarthy S., Levine B., Colbert C. L., and Sinha S. C. (2017) BECN2 interacts with ATG14 through a metastable coiled-coil to mediate autophagy. Protein Sci. 26, 972–984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Shibata M., Lu T., Furuya T., Degterev A., Mizushima N., Yoshimori T., MacDonald M., Yankner B., and Yuan J. (2006) Regulation of intracellular accumulation of mutant Huntingtin by Beclin 1. J. Biol. Chem. 281, 14474–14485 [DOI] [PubMed] [Google Scholar]
- 16. Pickford F., Masliah E., Britschgi M., Lucin K., Narasimhan R., Jaeger P. A., Small S., Spencer B., Rockenstein E., Levine B., and Wyss-Coray T. (2008) The autophagy-related protein Beclin 1 shows reduced expression in early Alzheimer disease and regulates amyloid beta accumulation in mice. J. Clin. Invest. 118, 2190–2199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Spencer B., Potkar R., Trejo M., Rockenstein E., Patrick C., Gindi R., Adame A., Wyss-Coray T., and Masliah E. (2009) Beclin 1 gene transfer activates autophagy and ameliorates the neurodegenerative pathology in α-synuclein models of Parkinson's and Lewy body diseases. J. Neurosci. 29, 13578–13588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Qu X., Yu J., Bhagat G., Furuya N., Hibshoosh H., Troxel A., Rosen J., Eskelinen E.-L., Mizushima N., Ohsumi Y., Cattoretti G., and Levine B. (2003) Promotion of tumorigenesis by heterozygous disruption of the beclin 1 autophagy gene. J. Clin. Invest. 112, 1809–1820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Koneri K., Goi T., Hirono Y., Katayama K., and Yamaguchi A. (2007) Beclin 1 gene inhibits tumor growth in colon cancer cell lines. Anticancer Res. 27, 1453–1457 [PubMed] [Google Scholar]
- 20. Miracco C., Cosci E., Oliveri G., Luzi P., Pacenti L., Monciatti I., Mannucci S., De Nisi M. C., Toscano M., Malagnino V., Falzarano S. M., Pirtoli L., and Tosi P. (2007) Protein and mRNA expression of autophagy gene Beclin 1 in human brain tumours. Int. J. Oncol. 30, 429–436 [PubMed] [Google Scholar]
- 21. Li Z., Chen B., Wu Y., Jin F., Xia Y., and Liu X. (2010) Genetic and epigenetic silencing of the beclin 1 gene in sporadic breast tumors. BMC Cancer 10, 98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Pattingre S., Tassa A., Qu X., Garuti R., Liang X. H., Mizushima N., Packer M., Schneider M. D., and Levine B. (2005) Bcl-2 antiapoptotic proteins inhibit Beclin 1-dependent autophagy. Cell 122, 927–939 [DOI] [PubMed] [Google Scholar]
- 23. Orvedahl A., Alexander D., Tallóczy Z., Sun Q., Wei Y., Zhang W., Burns D., Leib D. A., and Levine B. (2007) HSV-1 ICP34.5 confers neurovirulence by targeting the Beclin 1 autophagy protein. Cell Host Microbe 1, 23–35 [DOI] [PubMed] [Google Scholar]
- 24. Levine B., and Kroemer G. (2008) Autophagy in the pathogenesis of disease. Cell 132, 27–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. He C., and Levine B. (2010) The Beclin 1 interactome. Curr. Opin. Cell Biol. 22, 140–149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Mei Y., Su M., Soni G., Salem S., Colbert C. L., and Sinha S. C. (2014) Intrinsically disordered regions in autophagy proteins. Proteins 82, 565–578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Mei Y., Glover K., Su M., and Sinha S. C. (2016) Conformational flexibility of BECN1: essential to its key role in autophagy and beyond. Protein Sci. 25, 1767–1785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lee E. F., Perugini M. A., Pettikiriarachchi A., Evangelista M., Keizer D. W., Yao S., and Fairlie W. D. (2016) The BECN1 N-terminal domain is intrinsically disordered. Autophagy 12, 460–471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Mei Y., Ramanathan A., Glover K., Stanley C., Sanishvili R., Chakravarthy S., Yang Z., Colbert C. L., and Sinha S. C. (2016) Conformational flexibility enables function of a BECN1 region essential for starvation-mediated autophagy. Biochemistry 55, 1945–1958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Li X., He L., Che K. H., Funderburk S. F., Pan L., Pan N., Zhang M., Yue Z., and Zhao Y. (2012) Imperfect interface of Beclin1 coiled-coil domain regulates homodimer and heterodimer formation with Atg14L and UVRAG. Nat. Commun. 3, 662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Mei Y., Su M., Sanishvili R., Chakravarthy S., Colbert C. L., and Sinha S. C. (2016) Identification of BECN1 and ATG14 coiled-coil interface residues important for starvation-induced autophagy. Biochemistry 55, 4239–4253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Noda N. N., Kobayashi T., Adachi W., Fujioka Y., Ohsumi Y., and Inagaki F. (2012) Structure of the novel C-terminal domain of vacuolar protein sorting 30/autophagy-related protein 6 and its specific role in autophagy. J. Biol. Chem. 287, 16256–16266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Huang W., Choi W., Hu W., Mi N., Guo Q., Ma M., Liu M., Tian Y., Lu P., Wang F.-L., Deng H., Liu L., Gao N., Yu L., and Shi Y. (2012) Crystal structure and biochemical analyses reveal Beclin 1 as a novel membrane binding protein. Cell Res. 22, 473–489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Adi-Harel S., Erlich S., Schmukler E., Cohen-Kedar S., Segev O., Mizrachy L., Hirsch J. A., and Pinkas-Kramarski R. (2010) Beclin 1 self-association is independent of autophagy induction by amino acid deprivation and rapamycin treatment. J. Cell. Biochem. 110, 1262–1271 [DOI] [PubMed] [Google Scholar]
- 35. Tria G., Mertens H. D., Kachala M., and Svergun D. I. (2015) Advanced ensemble modelling of flexible macromolecules using X-ray solution scattering. IUCrJ 2, 207–217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Mertens H. D., and Svergun D. I. (2010) Structural characterization of proteins and complexes using small-angle X-ray solution scattering. J. Struct. Biol. 172, 128–141 [DOI] [PubMed] [Google Scholar]
- 37. Receveur-Brechot V., and Durand D. (2012) How random are intrinsically disordered proteins? A small angle scattering perspective. Curr. Protein Pept. Sci. 13, 55–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Sinha S., Colbert C. L., Becker N., Wei Y., and Levine B. (2008) Molecular basis of the regulation of Beclin 1-dependent autophagy by the γ-herpesvirus 68 Bcl-2 homolog M11. Autophagy 4, 989–997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Liang X. H., Yu J., Brown K., and Levine B. (2001) Beclin 1 contains a leucine-rich nuclear export signal that is required for its autophagy and tumor suppressor function. Cancer Res. 61, 3443–3449 [PubMed] [Google Scholar]
- 40. Zhang K., Chen Y., Zhang Y., Yao Q., and Zhuo H. (2014) Novel CRM1 inhibitor prevent the nuclear export of Beclin-1 blocks autophagy in mantle cell lymphoma. Blood 124 [Google Scholar]
- 41. DeLano W. L. (2015) The PyMOL Molecular Graphics System, version 1.8, Schrödinger, LLC, New York [Google Scholar]
- 42. Krissinel E., and Henrick K. (2007) Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 372, 774–797 [DOI] [PubMed] [Google Scholar]
- 43. Sheffield P., Garrard S., and Derewenda Z. (1999) Overcoming expression and purification problems of RhoGDI using a family of “Parallel” expression vectors. Protein Expr. Purif. 15, 34–39 [DOI] [PubMed] [Google Scholar]
- 44. Sreerama N., Venyaminov S. Y., and Woody R. W. (2000) Estimation of protein secondary structure from circular dichroism spectra: inclusion of denatured proteins with native proteins in the analysis. Anal. Biochem. 287, 243–251 [DOI] [PubMed] [Google Scholar]
- 45. Sreerama N., and Woody R. W. (2000) Estimation of protein secondary structure from circular dichroism spectra: comparison of CONTIN, SELCON, and CDSSTR methods with an expanded reference set. Anal. Biochem. 287, 252–260 [DOI] [PubMed] [Google Scholar]
- 46. Sreerama N., Venyaminov S. Y., and Woody R. W. (2001) Analysis of protein circular dichroism spectra based on the tertiary structure classification. Anal. Biochem. 299, 271–274 [DOI] [PubMed] [Google Scholar]
- 47. Petoukhov M. V., Franke D., Shkumatov A. V., Tria G., Kikhney A. G., Gajda M., Gorba C., Mertens H. D. T., Konarev P. V., and Svergun D. I. (2012) New developments in the ATSAS program package for small-angle scattering data analysis. J. Appl. Crystallogr. 45, 342–350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Konarev P. V., Volkov V. V., Sokolova A. V., Koch M. H. J., and Svergun D. I. (2003) PRIMUS: a Windows PC-based system for small-angle scattering data analysis. J. Appl. Crystallogr. 36, 1277–1282 [Google Scholar]
- 49. Svergun D. I. (1992) Determination of the regularization parameter in indirect-transform methods using perceptual criteria. J. Appl. Crystallogr. 25, 495–503 [Google Scholar]
- 50. Franke D., and Svergun D. I. (2009) DAMMIF, a program for rapid ab-initio shape determination in small-angle scattering. J. Appl. Crystallogr. 42, 342–346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Volkov V. V., and Svergun D. I. (2003) Uniqueness of ab initio shape determination in small-angle scattering. J. Appl. Crystallogr. 36, 860–864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Svergun D., Barberato C., and Koch M. H. J. (1995) CRYSOL: a program to evaluate x-ray solution scattering of biological macromolecules from atomic coordinates. J. Appl. Crystallogr. 28, 768–773 [Google Scholar]
- 53. Kozin M. B., and Svergun D. I. (2001) Automated matching of high- and low-resolution structural models. J. Appl. Crystallogr. 34, 33–41 [Google Scholar]
- 54. Kabeya Y., Mizushima N., Ueno T., Yamamoto A., Kirisako T., Noda T., Kominami E., Ohsumi Y., and Yoshimori T. (2000) LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J. 19, 5720–5728 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.