Abstract
ATP-binding cassette subfamily B member 10 (Abcb10) is a mitochondrial ATP-binding cassette (ABC) transporter that complexes with mitoferrin1 and ferrochelatase to enhance heme biosynthesis in developing red blood cells. Reductions in Abcb10 levels have been shown to reduce mitoferrin1 protein levels and iron import into mitochondria, resulting in reduced heme biosynthesis. As an ABC transporter, Abcb10 binds and hydrolyzes ATP, but its transported substrate is unknown. Here, we determined that decreases in Abcb10 did not result in protoporphyrin IX accumulation in morphant-treated zebrafish embryos or in differentiated Abcb10-specific shRNA murine Friend erythroleukemia (MEL) cells in which Abcb10 was specifically silenced with shRNA. We also found that the ATPase activity of Abcb10 is necessary for hemoglobinization in MEL cells, suggesting that the substrate transported by Abcb10 is important in mediating increased heme biosynthesis during erythroid development. Inhibition of 5-aminolevulinic acid dehydratase (EC 4.2.1.24) with succinylacetone resulted in both 5-aminolevulinic acid (ALA) accumulation in control and Abcb10-specific shRNA MEL cells, demonstrating that reductions in Abcb10 do not affect ALA export from mitochondria and indicating that Abcb10 does not transport ALA. Abcb10 silencing resulted in an alteration in the heme biosynthesis transcriptional profile due to repression by the transcriptional regulator Bach1, which could be partially rescued by overexpression of Alas2 or Gata1, providing a mechanistic explanation for why Abcb10 shRNA MEL cells exhibit reduced hemoglobinization. In conclusion, our findings rule out that Abcb10 transports ALA and indicate that Abcb10's ATP-hydrolysis activity is critical for hemoglobinization and that the substrate transported by Abcb10 provides a signal that optimizes hemoglobinization.
Keywords: ABC transporter, hemoglobin, mitochondria, porphyrin, transcription, ALA, abcb10, red cell
Introduction
ATP-binding cassette (ABC)3 proteins belong to one of the largest classes of transporters. They bind and hydrolyze ATP to translocate substrates across membranes and are involved in many biological processes. Several ABC transporters (Abcb6, Abcb7, Abcb8, and Abcb10) are localized to mitochondria and are involved in iron- and/or heme-related biological pathways (1). Heme is an essential co-factor and is involved in biological processes, including oxidative phosphorylation, oxygen transport (hemoglobin), metabolism, and detoxification (cytochromes P450). During erythropoiesis, heme biosynthesis increases and is concomitantly coordinated with iron uptake and globin synthesis through transcriptional regulators Gata1 and Bach1 (2–6). Mutations in the heme biosynthetic pathway give rise to human hematologic disorders, such as erythropoietic porphyria or sideroblastic anemia (7).
Abcb10 is localized to the inner mitochondrial membrane, and its expression is induced in developing erythroid cells by Gata1 (8). Deletion of Abcb10 in mice is embryonic lethal due to anemia, suggesting an essential role in erythropoiesis (9, 10). Characterization of the role of Abcb10 in erythropoiesis has determined that Abcb10 stabilizes mitoferrin1 (Mfrn1) and forms a complex with ferrochelatase (Fech) to enhance heme synthesis (11, 12). This interaction correlates with increased iron uptake into the mitochondria during hemoglobinization. It is interesting to note that Abcb10 is also expressed in other tissues, suggesting a role independent of erythroid differentiation. In support of this hypothesis, Abcb10 has been shown to have a protective effect against reactive oxygen species and plays a role in heme synthesis in cardiac cells (13). siRNA-mediated reductions in Abcb10 in cardiomyocytes showed decreased mitochondrial heme and decreased enzyme activity for heme-containing proteins but no accumulation of the heme precursor protoporphyrin IX (PPIX). Further, Bayeva et al. (13) showed that the addition of exogenous δ-aminolevulinic acid (ALA), the rate-limiting product in heme biosynthesis, to Friend murine erythroleukemic (MEL) cells rescued the heme defect associated with reductions in Abcb10 protein. Contrasting results were observed in a mouse hematopoietic-specific deletion of Abcb10, where PPIX accumulation was observed (14). More recently, Qiu et al. (15) have reported that Abcb10 does not play a role in ALA export from mitochondria. Because of these conflicting results, we examined the role of Abcb10 using the model organism Danio rerio and cultured murine MEL cells. We show that Abcb10 has a function in hemoglobinization independent of Mfrn1 and is not an ALA exporter. We observed significant down-regulation of the erythropoiesis transcriptional program in the absence of Abcb10, which can be ascribed to increased Bach1 occupancy on the β-Globin promoter.
Results
Loss of Abcb10 results in reduced heme levels without PPIX accumulation
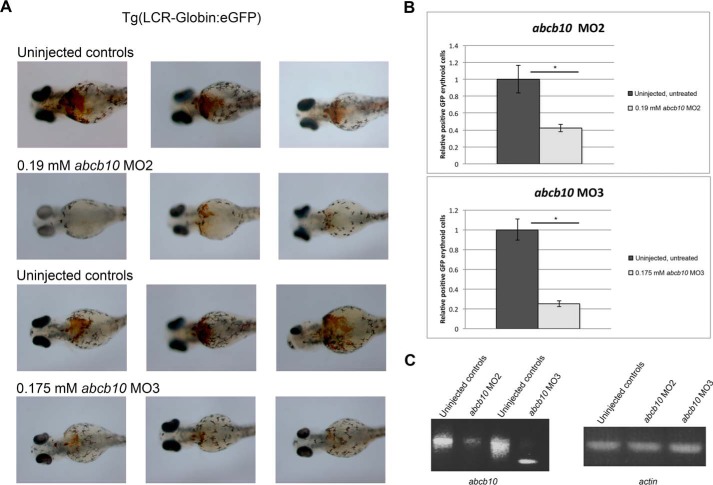
We utilized the D. rerio (zebrafish) model system for red blood cell development to determine whether the loss of abcb10 resulted in changes in PPIX, intermediate porphyrins, or heme. We employed morpholinos in the Tg(globin-LCR:eGFP) transgenic line of zebrafish, which expresses GFP under the globin locus control region (LCR) enhancer (16), and o-dianosidine staining to assess hemoglobinization and flow cytometry to assess changes in GFP+ red cell mass. Two different splice donor morpholinos (MOe2 and MOe3) were used, which are predicted to disrupt abcb10 mRNA formation. Embryos showed reductions in hemoglobinization with either abcb10-specific morpholino at 72 h postfertilization (hpf) (Fig. 1A). Flow cytometric analysis of GFP-positive cells showed marked reductions in erythrocytes (GFP-positive cells) compared with the uninjected controls (Fig. 1B). That both morpholinos gave rise to similar phenotypes and normal β-actin mRNA processing suggests that this is not an off-target or toxic effect of the morpholinos. Reductions of abcb10 mRNA in embryos was confirmed by PCR using primers spanning Abcb10 exon–intron junctions (Fig. 1C).
Figure 1.
Zebrafish abcb10 morpholinos reduce hemoglobinization. A, abcb10 splice-blocking morpholinos (MO2 and MO3) were microinjected into Tg(globin-LCR:eGFP) transgenic zebrafish embryos at the one-cell stage, and hemoglobinization was assessed at 72 hpf using o-dianisidine staining. Representative examples of uninjected and MO2- and MO3-injected embryos are shown. B, embryos as in A were examined for GFP-positive erythrocytes from a Tg(globin-LCR:eGFP) transgenic line at 72 hpf using flow cytometry. Error bars, S.E. C, semiquantitative RT-PCR analysis was performed on abcb10 from MO2 and MO3 zebrafish embryos as well as the efficacy of urod and fech MOs in zebrafish embryos (Table 3). *, p ≤ 0.05.
To confirm that the anemia resulted from a defect of heme production, heme and porphyrin levels were measured by HPLC in embryos at 72 hpf (Table 1). Morpholinos against uroporphyrinogen decarboxylase d (urod) and ferrochelatase (fech), the enzymes that catalyze the fifth and the last step of the heme biosynthetic pathway, respectively, were used as controls. Heme levels were severely decreased in both abcb10 morphants compared with the uninjected controls. As expected, heme levels were decreased in both urod and fech morpholino-treated embryos. Intermediate porphyrins accumulated in urod morphant-treated embryos and PPIX accumulated in fech morphant-treated embryos. In contrast, both abcb10 morphant-treated embryos (MO2 and MO3) did not accumulate either intermediate porphyrins or PPIX. The absence of abcb10 resulted in decreased heme levels with no accumulation of PPIX. These results are in agreement with previous studies in cardiac myocytes (13) but in contrast to the previous findings that PPIX accumulates in hematopoietic cells of a hematopoietic targeted Abcb10 knock-out mouse (14).
Table 1.
HPLC analysis of porphyrins from control zebrafish embryos and heme synthetic morphants
Pooled zebrafish embryos were analyzed as described under “Experimental procedures.” Significant changes (p ≤ 0.05) compared with uninjected controls are shown in boldface type and underlined.
| Hemin | S.D. hemin | PPIX | S.D. PPIX | ZnPPIX | S.D. ZnPPIX | Intermediate porphyrins | S.D. intermediate porphyrins | |
|---|---|---|---|---|---|---|---|---|
| pmol/mg | pmol/mg | pmol/mg | pmol/mg | |||||
| Uninjected controls | 648.0 | 36.0 | Trace | Trace | 1.4 | 0.3 | ||
| 0.19 mm abcb10 MO2 | 238.0 | 12.2 | Trace | Trace | 1.0 | 0.4 | ||
| Uninjected controls | 477.4 | 47.9 | 1.5 | 0.3 | Trace | Trace | ||
| 0.75 mm abcb10 MO3 | 77.5 | 30.5 | 1.9 | 0.6 | Trace | Trace | ||
| Uninjected controls | 237 | 11.4 | 1.2 | 0.3 | Trace | Trace | ||
| 0.175 mm urod MO | Trace | 14.5 | 2.1 | Trace | 51 | 24 | ||
| Uninjected controls | 726.5 | 60 | 2 | 0.9 | Trace | Trace | ||
| 0.75 mm fech MO | 102.3 | 17.1 | 518.4 | 98 | 32.4 | 3.8 | Trace |
Abcb10-silenced MEL cells show reduced hemoglobinization and decreased iron incorporation into heme
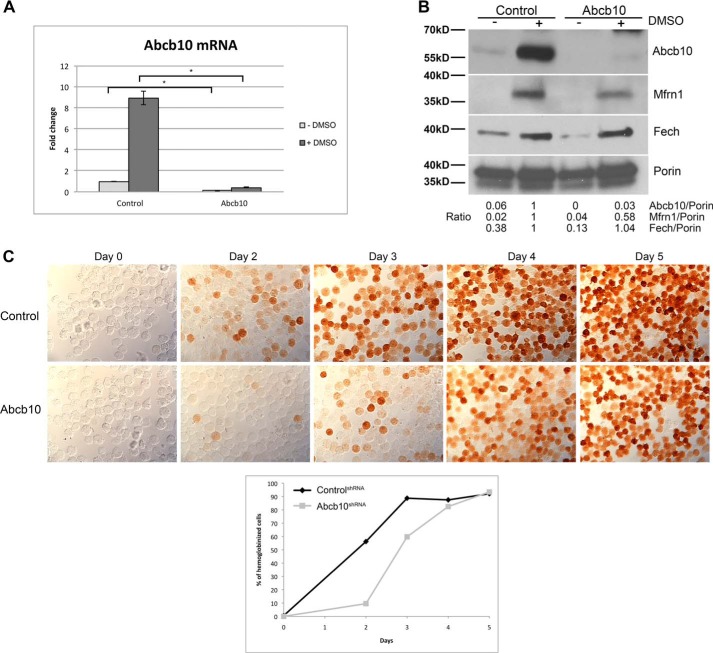
To better understand the role of Abcb10 in heme synthesis, we generated MEL cells containing stably expressed shRNA directed against Abcb10. We confirmed by qRT-PCR that Abcb10 mRNA levels were reduced (Fig. 2A). Western blot analysis showed reduced Abcb10 levels in differentiated MEL cells (Fig. 2B). Further, reductions in Abcb10 resulted in decreased levels of Mfrn1 as Abcb10 has been shown to interact with and stabilize Mfrn1 during red cell differentiation (11, 12, 17, 18). Abcb10 shRNA cells showed a delay in hemoglobinization as well as reduced amounts of hemoglobin in the majority of cells as assessed by o-dianisidine staining (Fig. 2C). As the MEL cell experiments are completely independent of the zebrafish experiments, these results validate the observations seen with morpholinos in zebrafish embryos.
Figure 2.
Abcb10 shRNA-stable MEL cells show decreased hemoglobinization. A, MEL cells stably expressing control shRNA or Abcb10-specific shRNA were treated with 1.5% DMSO for 3 days, and mRNA was isolated. qRT-PCR for Abcb10 and actin were performed. Error bars, S.E. B, mitochondria were isolated from cells as in A and lysed, and Western blot analysis was performed using rabbit anti-Abcb10, rabbit anti-Mfrn1, mouse anti-Fech, and mouse anti-Porin followed by HRP-conjugated goat anti-rabbit or anti-mouse IgG. Blots were quantified using NIH ImageJ with porin as a loading control and the ratios of Abcb10, Mfrn1, and Fech to Porin were determined with control shRNA MEL cells normalized to 1. An example blot with its quantification is shown. C, MEL cells stably expressing control shRNA or Abcb10-specific shRNA were treated with 1.5% DMSO for 0–5 days and stained for hemoglobin using o-dianisidine. Quantification of hemoglobinization of a representative experiment is shown (n = 100–200 cells/time). (The presence of a pink to red signal was considered positive in the quantification). *, p ≤ 0.05.
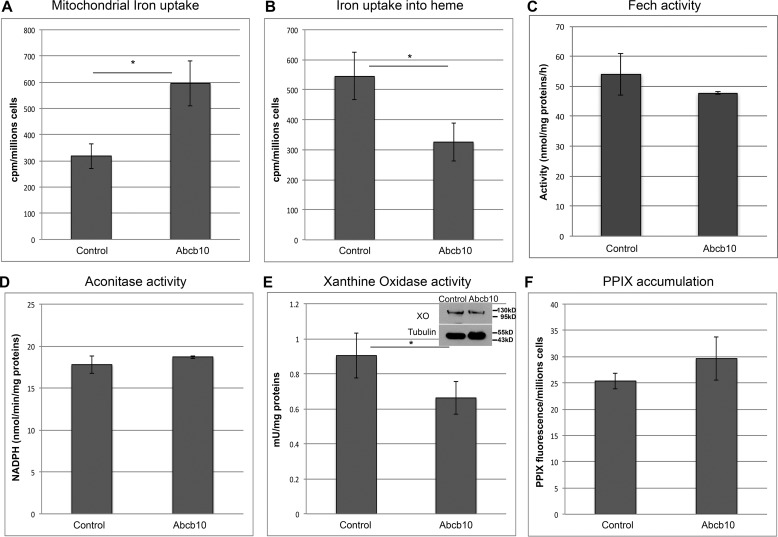
We utilized our Abcb10-specific shRNA MEL cells to examine mitochondrial iron uptake and iron incorporation into heme. Abcb10-specific shRNA MEL cells showed increased iron uptake into mitochondria (Fig. 3A) but reduced iron incorporation into heme (Fig. 3B). This suggests that there may be a defect in iron loading into the PPIX molecule. Iron incorporation into the PPIX molecule to generate heme requires the activity of the iron–sulfur (Fe-S) cluster protein Fech. Perhaps reductions in Abcb10 affect iron–sulfur cluster formation. We measured the activity of Fech (Fig. 3C), aconitase (Fig. 3D), and xanthine oxidase (Fig. 3E). Only xanthine oxidase showed a significant reduction in activity without a change in protein levels. These results demonstrate that the absence of Abcb10 does not affect mitochondrial Fe-S cluster proteins but does affect a cytosolic Fe-S cluster protein. Importantly, there was no increase in the levels of PPIX in Abcb10 shRNA MEL cells (Fig. 3F). Together, these results suggest that the absence of Abcb10 affects an early step in heme synthesis.
Figure 3.
Alterations in mitochondrial iron uptake and heme levels in Abcb10-silenced MEL cells. A, MEL cells stably expressing control shRNA or Abcb10-specific shRNA were treated with 1.5% DMSO for 60 h. Cells were incubated overnight with 200 μm BPS, followed by incubation with 1.2 mm ALA and Tf(59Fe)2 for 6–8 h. Mitochondria were isolated, and mitochondrial iron (59Fe) levels were measured. The data are expressed as cpm/million cells. Error bars, S.E. B, whole-cell heme levels were measured from cells treated as in A, and the data are expressed as cpm/million cells. Error bars, S.E. MEL cells as in A were lysed, and Fech (n = 2) (C) (error bars, S.D.), aconitase (D), and xanthine oxidase activity (E) (a minimum of 3) and protein levels were measured. Error bars, S.E. F, MEL cells stably expressing control shRNA or Abcb10-specific shRNA were treated with 1.5% DMSO for 60 h, followed by incubation with 1.2 mm ALA for 8 h, and protoporphyrin IX levels were measured. Data are expressed as arbitrary fluorescence units/million cells. *, p ≤ 0.05. Error bars, S.E.
Abcb10 signature and Walker B motifs are necessary for hemoglobinization
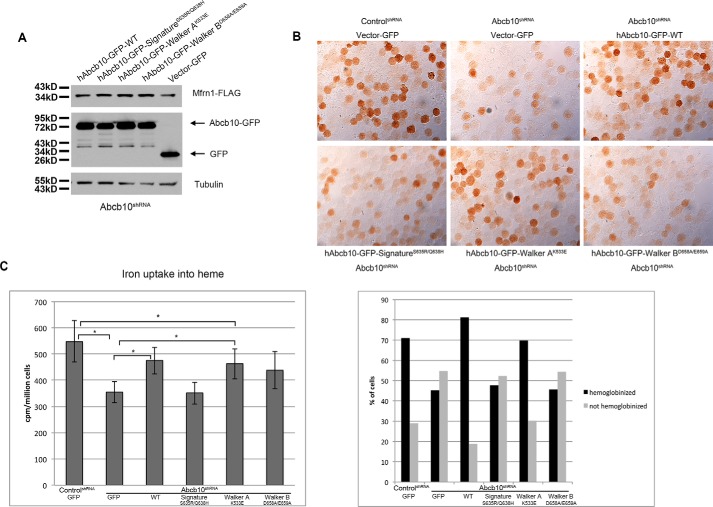
To determine whether the ATPase activity of Abcb10 is necessary for hemoglobinization, we generated plasmids containing either wild-type human Abcb10-GFP or human Abcb10-GFP with a mutated Walker A (K533E), Walker B (D658A/E659A) or signature (S635R/Q638H) motif as these motifs are responsible for ATP binding and hydrolysis (15, 19, 20). These constructs were transfected in Abcb10 shRNA MEL cells, and cells were differentiated for 3 days. All constructs were expressed and localized to mitochondria (supplemental Fig. 1). Western analysis showed that the ATP binding function of Abcb10 was not necessary for the stabilization of Mfrn1 (Fig. 4A). Abcb10 shRNA MEL cells expressing wild-type or Walker A mutant human Abcb10 showed increased hemoglobinization compared with Abcb10 shRNA MEL cells expressing control GFP, whereas Walker B and signature motif mutants were not able to rescue the hemoglobinization defect associated with loss of Abcb10 (Fig. 4B) or heme synthesis (Fig. 4C). These results demonstrate that ATP hydrolysis and subsequent substrate transport are necessary for hemoglobinization of MEL cells and suggest that Abcb10 has an additional role in heme synthesis besides the stabilization of Mfrn1.
Figure 4.
Complementation of the Abcb10-silenced phenotype by human Abcb10. MEL cells stably expressing control shRNA or Abcb10-specific shRNA were transfected with empty vector-GFP, wild-type Abcb10-GFP, or mutant Abcb10-GFP, and stable cells lines were selected by growth in 1.0 mg/ml G418. A, stable MEL cells were transfected with Mfrn1-FLAG, and Western blot analysis was performed. B, MEL cells as described in A were treated with 1.5% DMSO for 3 days, and hemoglobin was stained with o-dianisidine. Images were quantified, and the data are expressed as the percentage of cells hemoglobinized or not hemoglobinized. Representative images are shown and quantified (below image) as described in the legend to Fig. 2. C, cells treated as described for B were incubated with 1.2 mm ALA and Tf(59Fe)2 for 8 h, and iron (59Fe) incorporation into heme was measured and expressed as cpm/million cells. *, p ≤ 0.05. Error bars, S.E.
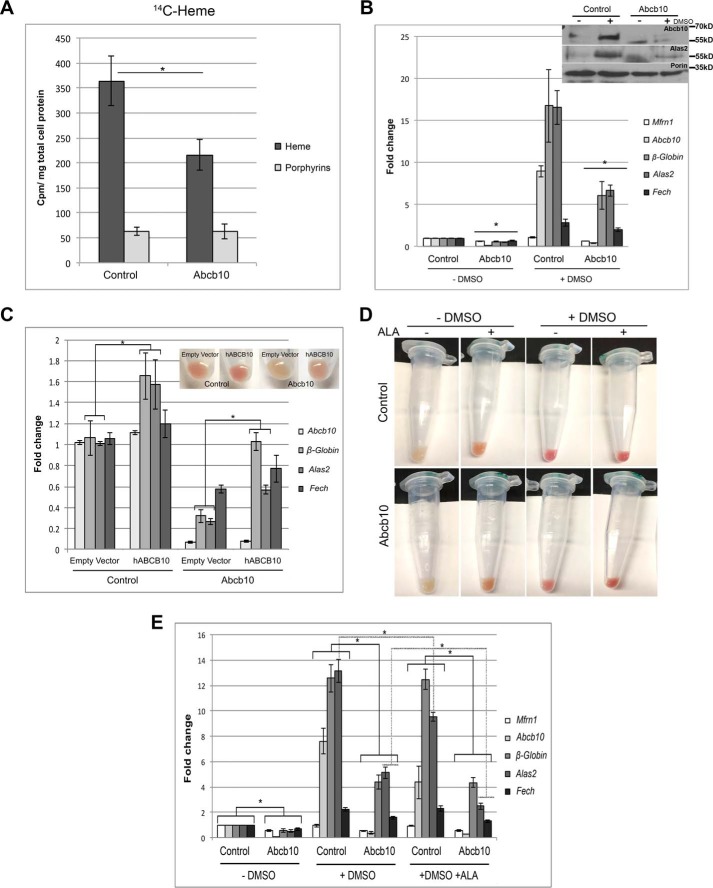
Abcb10 shRNA MEL cells show a marked reduction in Alas2 activity and a decrease in heme biosynthesis transcripts
To identify the site in the heme synthesis pathway that is disrupted by reductions in Abcb10, we fed cells [14C]glycine. Glycine and succinyl-CoA are the substrates for the rate-limiting step in heme synthesis, production of ALA. This step requires the enzyme 5′-aminolevulinate synthase 2 acid synthase 2 (Alas2) in red cells. Extracting 14C-heme and 14C-porphyrins provides a measure of Alas2 activity. Abcb10 shRNA MEL cells showed a marked reduction in 14C-heme compared with nonspecific shRNA control MEL cells (Fig. 5A). These results suggest that there is a defect in ALA synthesis. Because iron uptake into mitochondria is not limited in the Abcb10 shRNA MEL cells, we examined the mRNA levels of heme biosynthesis genes. As expected, heme biosynthesis genes are up-regulated in differentiated control cells as well as Abcb10 shRNA MEL cells, but the -fold increase in β-Globin, Alas2, and Fech was markedly reduced in Abcb10 shRNA MEL cells (Fig. 5B). Fech activity was not reduced in Abcb10 shRNA MEL (see Fig. 2C), but Alas2 protein levels were greatly reduced, suggesting reduced Alas2 activity. Heme biosynthesis transcripts for Pgbd, UroD, and Ppox were also significantly reduced even in undifferentiated Abcb10 shRNA MEL cells (supplemental Fig. 2A). Further, genes involved in erythroid differentiation (TfR1 and Bcl-xl) showed reduced transcripts, suggesting that the “tone” for erythroid differentiation is altered by reductions in Abcb10 levels. These reductions in transcripts are specific to the genes in the erythroid differentiation pathway because both heavy and light chain ferritin (Fth and Ftl) and superoxide dismutase 2 (Sod2) are unaffected in Abcb10 shRNA MEL cells (supplemental Fig. 2B). We confirmed that the hemoglobinization and transcript profile of Abcb10 shRNA MEL cells could be rescued by overexpression of human Abcb10 (Fig. 5C). That the overexpression of human Abcb10 did not fully complement transcripts back to control cell levels can be attributed to decreased efficiency of transduction in Abcb10 shRNA MEL cells compared with control shRNA MEL cells (supplemental Fig. 3).
Figure 5.
Differentiated Abcb10 shRNA MEL cells show reduced Alas2 mRNA levels. A, MEL cells stably expressing control shRNA or Abcb10-specific shRNA were treated with 1.5% DMSO for 3 days, and differentiated cells were shifted to growth in glycine-deficient medium (−glycine) and incubated with 1 μm Tf(Fe)2 for 30 min, followed by the addition of [14C]glycine for 1 h at 37 °C. Heme and porphyrins were extracted, and radioactivity was measured as described under “Experimental procedures.” The data are expressed as cpm/mg of total cell protein. Error bars, S.E. B, mRNA from MEL cells stably expressing control shRNA or Abcb10-specific shRNA was treated with 1.5% DMSO for 3 days, and qPCR for Mfrn1, Abcb10, β-Globin, Alas2, and Fech was performed using the primers listed in Table 3. Error bars, S.E. Western blot analysis of Abcb10, Alas2, and Porin protein levels in control and shRNA Abcb10 undifferentiated and differentiated MEL cells is shown. C, control shRNA or Abcb10-specific shRNA cells were transduced with a lentivirus containing either a control vector or human Abcb10-FLAG, and hemoglobinization and qPCR were performed. A representative experiment is shown. Error bars, S.E. D, hemoglobinization was assessed in cells as in B plus or minus 1.2 mm ALA. E, cells as in D were harvested, mRNA was isolated, and qPCR was performed. *, p ≤ 0.05. Error bars, S.E.
It has been proposed that Abcb10 is involved in the export of ALA from mitochondria to the cytosol, since the addition of ALA rescues defects in cardiomyocytes (13). To see whether ALA could rescue the hemoglobinization defect in Abcb10 shRNA MEL cells, ALA was added during differentiation. As shown in Fig. 5D, hemoglobinization was rescued in Abcb10 shRNA MEL cells by the addition of ALA. We noted that the levels of hemoglobinization were increased even in undifferentiated control cells grown with ALA. Similarly, Bayeva et al. (13) also saw increased mitochondrial heme levels in ALA-treated control cells, demonstrating that the presence of excess ALA affects heme accumulation independent of the presence of Abcb10. We examined whether the addition of ALA rescued the expression levels of heme biosynthesis genes. The addition of ALA to control cells slightly reduced the levels of Alas2, whereas β-Globin and Fech levels remained the same (Fig. 5E). The addition of ALA to Abcb10 shRNA MEL cells resulted in decreased Alas2 and Fech mRNA but did not increase of β-Globin transcripts. That β-Globin levels were still low in ALA-treated Abcb10 shRNA cells suggests that there may be a block in the ability to increase β-Globin transcription.
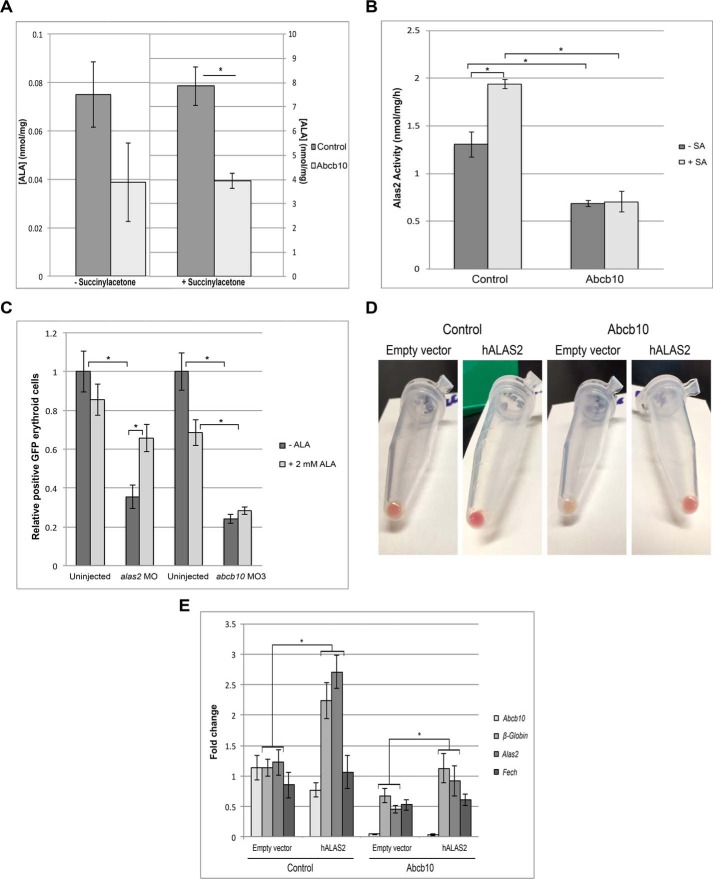
To further rule out the possibility that Abcb10 is an ALA exporter, we incubated cells with succinylacetone (SA), an inhibitor of the cytosolic enzyme ALA dehydratase, the second enzyme of the heme biosynthesis pathway that catalyzes the condensation of ALA to form porphobilinogen (21, 22). If Abcb10 is involved in the export of ALA, we would not expect to see an accumulation of ALA in Abcb10 shRNA MEL cells. Cells were differentiated for 3 days and incubated with SA for 3 h. ALA levels and Alas2 activity were measured. As expected, ALA levels increased by ∼100-fold in SA-treated differentiated control MEL cells but also increased 100-fold in Abcb10 shRNA MEL cells (Fig. 6A), demonstrating that ALA is exiting the mitochondria even in the absence of Abcb10. The levels of ALA, however, were reduced in Abcb10 shRNA MEL cells compared with control cells. Further, Alas2 activity was decreased by ∼2-fold in Abcb10 knockdown cells compared with control shRNA MEL cells (Fig. 6B), which confirms the reduced protein and transcript levels seen in Abcb10 shRNA MEL cells. As ALA did accumulate in SA-treated Abcb10 shRNA MEL cells, these experiments strongly suggest that Abcb10 does not play a role in ALA export from mitochondria. Further, the addition of SA did not alter Alas2 or β-Globin mRNA levels (supplemental Fig. 4). These results confirm those reported by the Shiriai group (15), which indicated that Abcb10 is not involved in the export of ALA from mitochondria. We also utilized the in vivo zebrafish model expressing GFP under the globin LCR enhancer as in Fig. 1C, where we could reduce the levels Abcb10 or Alas2, the rate-limiting enzyme in ALA synthesis. Treatment of zebrafish embryos with 2 mm ALA rescued hemogloblinization of alas2 morphants but did not rescue abcb10 morphants (Fig. 6C). That ALA did not rescue abcb10 morphants or change β-Globin transcript levels in Abcb10 shRNA MEL cells suggests that the levels of heme are insufficient to permit increased β-Globin expression. The addition of hemin to growth media increased β-Globin mRNA levels in control shRNA MEL cells; however, there were no significant changes in Abcb10 shRNA MEL cells (supplemental Fig. 5). Heme oxygenase-1 (HO-1) transcripts were only slightly increased in control and Abcb10 shRNA MEL cells. One possible explanation for this result is that MEL cells do not take up hemin well.
Figure 6.
Abcb10 shRNA MEL cells do not accumulate ALA. A, MEL cells stably expressing control shRNA or Abcb10-specific shRNA were treated with 1.5% DMSO for 3 days and incubated with 1 μm Tf(Fe)2 for 3 h, followed by a 3-h incubation in the presence or absence of 100 μm SA. Cells were harvested, and ALA was measured by HPLC as described under “Experimental procedures” (n = 2). Error bars, S.E. B, Alas2 activity in cells treated as in A was measured as described previously (33) (n = 2). Error bars, S.E. C, embryos from MO3 as in Fig. 1 and alas2 morphants were dechorionated at 24 hpf and then incubated with or without 2 mm ALA. At 72 hpf (48 h of ALA or vehicle exposure), the control and treated embryos were disaggregated and subjected to flow cytometry. The number of eGFP+ erythrocytes is a surrogate marker or index of red cell mass. Error bars, S.E. Control shRNA or Abcb10-specific shRNA cells were transduced with a lentivirus containing either a control vector or human Alas2-FLAG, and hemoglobinization was assessed (D) and qPCR was performed (E) using primers as in Table 3. *, p ≤ 0.05. Error bars, S.E.
We also tested whether the Alas2 cofactor pyridoxal 5′-phosphate was rate-limiting for ALA synthesis in Abcb10 shRNA MEL cells. The addition of pyridoxine or pyridoxal hydrochloride did not result in increased hemoglobinization of Abcb10 shRNA MEL cells (data not shown). The defect of hemoglobinization in Abcb10 knockdown cells could be due to the defect in Alas2, which is required to fully induce the hemoglobinization transcription program (5). We therefore overexpressed human Alas2 in control and Abcb10 shRNA MEL cells and measured hemoglobinization and heme biosynthesis transcripts. Overexpression of human Alas2 increased hemoglobinization in Abcb10 shRNA MEL cells (Fig. 6D) and a 2-fold increase in endogenous mouse Alas2 transcripts (Fig. 6E), suggesting that there might be a repression of mouse Alas2 expression in the absence of Abcb10 that can be partially rescued by overexpression of human Alas2.
Abcb10 shRNA MEL cells show increased levels of Bach1 on the β-Globin promoter
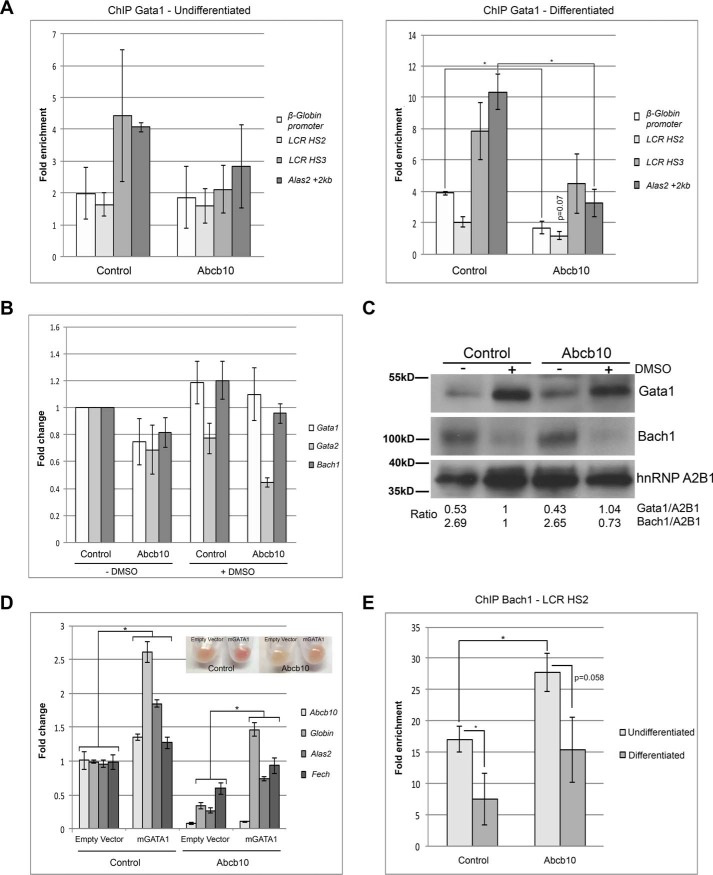
The transcription factor Gata1 is needed for proper erythropoiesis and is known to induce key erythroid genes during terminal differentiation, while simultaneously repressing non-erythroid genes (3). To determine the mechanism for reduced heme biosynthesis transcripts in Abcb10 shRNA MEL cells, we measured the Gata1 occupancy of β-Globin and Alas2 in undifferentiated and differentiated control and Abcb10 shRNA MEL cells. We examined both the LCR DNase I hypersensitive site (HS) HS2 and HS3 regions and the proximal region (Globin promoter) of the β-Globin promoter (23). ChIP analysis revealed no changes in promoter occupancy in undifferentiated Abcb10 shRNA MEL cells compared with control shRNA MEL cells, whereas in differentiated cells, there was less Gata1 present on the HS2 region and the proximal region of the β-Globin promoter and less Gata1 on the Alas2 promoter in Abcb10 shRNA MEL cells (Fig. 7A). We examined the levels of transcripts of several transcription factors involved in erythropoiesis. Gata1, Gata2, and Bach1 transcript levels were unchanged in differentiated Abcb10 shRNA MEL cells (Fig. 7B), and there were no changes in Gata1 or Bach1 protein compared with control MEL cells (Fig. 7C). These results show that Gata1 protein levels are similar in control shRNA and Abcb10 shRNA MEL cells but Gata1 is unable to occupy the β-Globin and Alas2 promoters efficiently in the absence of Abcb10. Overexpression of mouse Gata1 partially rescued the hemoglobinization defect and hemoglobinization transcripts in differentiated Abcb10 shRNA MEL cells (Fig. 7D). We note that the rescued hemoglobinization in Gata1 overexpression was not as robust as the rescues seen in cells overexpressing human Abcb10 or Alas2 (see Figs. 5C and 6E).
Figure 7.
Abcb10 shRNA MEL cells show increased Bach1 on the β-Globin promoter. A, Gata1 ChIP analysis was performed on Alas2 and β-Globin promoters in undifferentiated and differentiated control shRNA and Abcb10 shRNA MEL cells as described under “Experimental procedures.” Error bars, S.E. B, qPCR analysis of transcript levels of Gata1, Gata2, and Bach1 in undifferentiated and differentiated control shRNA and Abcb10 shRNA MEL cells. Error bars, S.E. C, undifferentiated and differentiated control shRNA and Abcb10 shRNA MEL cells were disrupted, and cytosol and membrane fractions were obtained. Membranes were lysed, and Western blot analysis was performed for Gata1 and Bach1 as described under “Experimental procedures.” D, control shRNA or Abcb10-specific shRNA cells were transduced with a lentivirus containing either a control vector or mouse Gata1, and hemoglobinization and qPCR were performed using primers as in Table 3. Error bars, S.E. E, Bach1 ChIP was performed on the β-Globin and actin promoters in undifferentiated and differentiated control shRNA and Abcb10 shRNA MEL cells as described under “Experimental procedures.” *, p ≤ 0.05. Error bars, S.D.
It is possible that there is a transcription factor repressing hemoglobinization and that excess Gata1 can partially overcome this repression. The transcriptional repressor Bach1 is known to interact with control elements, Maf antioxidant recognition elements, in the β-Globin promoter to coordinate transcription with the availability of heme (2, 24). To determine whether Bach1 is repressing hemoglobinization transcripts in Abcb10 shRNA MEL cells, we performed ChIP on the promoter regions of the β-Globin and β-actin (control) in undifferentiated and differentiated control shRNA and Abcb10 shRNA MEL cells. Bach1 was significantly enriched on the HS2 region of the β-Globin promoter in undifferentiated control cells and, as expected, was reduced upon differentiation (Fig. 7E). There was significantly more Bach1 present in the HS2 region of the β-Globin promoter in undifferentiated Abcb10 shRNA MEL cells. Upon differentiation, Bach1 levels on the β-Globin promoter were reduced but remained higher than that seen in differentiated control shRNA MEL cells. That there is increased Bach1 on the β-Globin promoter in Abcb10 shRNA MEL cells suggests that the levels of heme are not sufficient to release Bach1, and therefore the transcripts required for hemoglobinization are repressed even before cells are differentiated. These results provide a mechanistic explanation of why Abcb10 shRNA MEL cells show reduced hemoglobinization.
Discussion
Several studies have shown that Abcb10 is important in hemoglobinization (11, 12) and heme synthesis (13). It has been suggested that the role of Abcb10 in red cell hemoglobinization is to stabilize Mfrn1 (12), thus increasing mitochondrial iron import concomitant with the demand to make heme. It has also been suggested that Abcb10 may have other roles in protecting cells against oxidative damage independent of stabilizing Mfrn1 (8). This study shows that iron import into mitochondria is not decreased when Abcb10 levels are reduced, but rather mitochondrial iron is increased. One explanation for increased iron in mitochondria is that control MEL cells incorporate imported iron into PPIX and then export heme out of the mitochondria, resulting in less mitochondrial iron, whereas, in Abcb10 shRNA MEL cells, there is less Mfrn1 but also less heme is being made and thus less heme is exported out of mitochondria, giving rise to more mitochondrial iron accumulation. Increased iron without increased heme is also seen when cells overexpress both Mfrn1 and Abcb10 as porphyrin synthesis is still rate-limiting (12). Here we also show that there is no porphyrin accumulation, not PPIX or other intermediate porphyrins, in zebrafish treated with abcb10 morpholinos or Abcb10 shRNA-specific MEL cells, suggesting that although there is reduced flux through the porphyrin biosynthesis pathway, there is not a defect at one of the intermediate steps in heme biosynthesis. These results can be explained by the fact that any porphyrin made progresses to heme and is exported into the cytosol to combine with globin. That there is no PPIX accumulation in cells with reduced Abcb10 levels has been reported by others (12, 13), but Yamamoto et al. (14) showed increased PPIX in mice with a deletion of Abcb10 in hematopoietic tissue. Based upon our findings that abcb10 morphant zebrafish do not show PPIX accumulation and Abcb10 shRNA MEL cells do not show a defect in ferrocheletase activity, PPIX accumulation would not be predicted. It is unclear what the differences are between these studies. It may be that complete loss of Abcb10 in hematopoietic lineage cells has a more profound effect on the heme biosynthesis pathway in vivo.
So why is heme synthesis diminished, and what activity of Abcb10 is necessary for hemoglobinization? Our studies show that the ATP-hydrolysis activity of Abcb10 is important for hemoglobinization, and it is known that ATP hydrolysis is necessary for Abc substrate transport (1). It is interesting to note that the Walker A motif of Abcb10 is not necessary for hemoglobinization. One possibility is that there is still enough structural conservation with the intact Walker B and signature motifs that permits ATP binding in the Walker A K533E mutant. We have not been able to determine the substrate transported by Abcb10, but we have ruled out ALA as a substrate. Using the ALA dehydratase inhibitor SA, which acts in the cytosol, we showed that Abcb10 shRNA MEL cells accumulated ALA in the cytosol upon exposure to SA, but the amount of ALA was reduced compared with control MEL cells. The addition of ALA did not rescue our abcb10 zebrafish morphants, and it did not change the reduced β-Globin transcript levels seen in Abcb10 shRNA MEL cells, whereas overexpression of hAlas2 did increase β-Globin transcript levels in Abcb10 shRNA MEL cells. It is difficult to explain why ALA does not result in increased hemoglobinization transcripts, whereas overexpression of hAlas2 does increase β-Globin and mAlas2 transcripts. It may be that the amount of ALA taken up by MEL cells is not equivalent to that synthesized by overexpression of hAlas2 or that synthesis of ALA within the mitochondria is important.
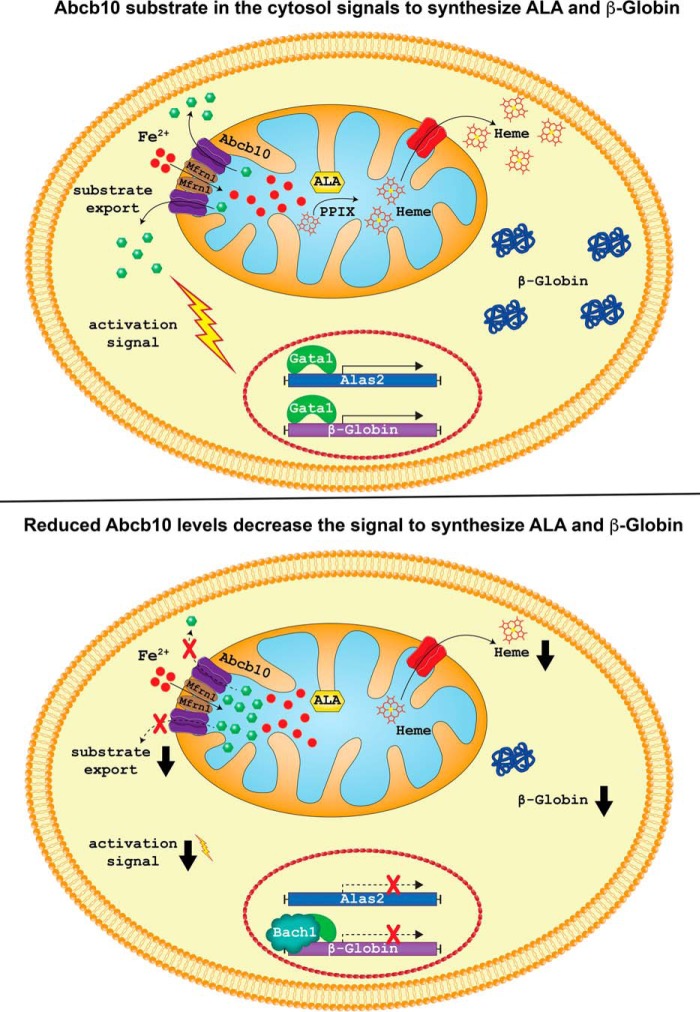
Our analysis of heme biosynthesis transcript changes in control and Abcb10 shRNA MEL cells revealed that even in undifferentiated Abcb10 shRNA MEL cells, Alas2 and β-Globin as well as other heme biosynthesis transcripts are decreased. Upon differentiation, those same transcripts were increased in control and Abcb10 shRNA MEL cells; however, the levels were greatly reduced in the absence of Abcb10. That Alas2 protein and activity is reduced in Abcb10 shRNA MEL cells provides an explanation for the reduced ALA levels. Previous studies by Bayeva et al. (13) also looked at heme biosynthesis transcripts in H9c2 rat embryonic heart cells but did not see changes in transcript levels. This difference can be explained by the fact that shRNA-mediated knockdown of Abcb10 was only 50% in H9c2 cells, whereas the knockdown seen in our MEL cells was >85%. Further, because MEL cells require significant up-regulation of heme biosynthesis genes, changes in transcript levels are probably amplified. These results demonstrate that the absence of Abcb10 affects heme production even in the absence of red cell differentiation and suggest that the transcriptional “tone” for precursor red cell hemoglobinization must be set early, even before differentiation. This tone in shRNA Abcb10 cells is set by significantly lower levels of heme. Reduced heme synthesis then allows for Bach1-mediated repression (5), which cannot be alleviated without either the substrate transported by Abcb10 or by bypassing Abcb10 with Alas2 overexpression. We show that there is increased Bach1 occupancy of the β-Globin promoter in Abcb10 shRNA MEL cells and that overexpression of Gata1 or Alas2 helps alleviate the repression, thus allowing for more heme synthesis. The increased heme then allows for some derepression and further hemoglobinization. It is interesting that hemin does not complement the hemoglobinization defect of Abcb10 shRNA MEL cells. Our model suggests that it is not just heme or ALA insufficiency, but that the substrate transported by Abcb10 provides an important condition or signal that optimizes hemoglobinization (Fig. 8). What the signal is and what Abcb10 transports remain to be determined.
Figure 8.
A model for hemoglobin synthesis under normal or reduced Abcb10 transport. Shown is a model for Abcb10 transporting a substrate out of the mitochondria (15) under normal conditions that provides a signal to permit β-Globin and Alas2 expression in a coordinated fashion, allowing for the production of hemoglobin (top) (modified from Ref. 12). When Abcb10 levels are reduced, there is decreased substrate export from the mitochondria, a reduced signal that then results in less β-Globin and Alas2 expression and less hemoglobin in the developing erythron (bottom).
Experimental procedures
Tissue culture, plasmids, and transfections
MEL cells (DS19 clone) were maintained in DMEM containing 10% FBS and penicillin and streptomycin. MEL cells stably expressing control shRNA or shRNA for mouse Abcb10 (Sigma-Aldrich) were selected in DMEM containing 10% FBS, penicillin, and streptomycin and 5 μg/ml puromycin. MEL cells stably expressing GFP, wild type human Abcb10-GFP, human mutant Abcb10-A (Walker A K533E)-GFP, human mutant Abcb10-B (Walker B D658A/E659A)-GFP, or human mutant Abcb10-S (signature S635R/Q638H)-GFP (Table 2) were grown in DMEM containing 10% FBS, 5 μg/ml puromycin, and 1 mg/ml G418. MEL cells were differentiated by incubating the cells in 1.5% DMSO for 3 or 5 days.
Table 2.
Plasmids used in this study
| Plasmid | Reference/Source |
|---|---|
| pEGFP-N1 | Clontech |
| pEGFP-N1-WT-hABCB10 | This study |
| pEGFP-N1-walker AK533E-hABCB10 | This study |
| pEGFP-N1-walker BD658A,E659A-hABCB10 | This study |
| pEGFP-N1-signatureS635R,Q638H-hABCB10 | This study |
| pEF1α-Mfrn1-FLAG | Ref. 12 |
| pFIN-EF1-GFP-2A- WPRE | Ref. 25 |
| pFIN-EF1-GFP-2A-hALAS2-FLAG -WPRE | This study |
| pFIN-EF1-GFP-2A-mGata1-WPRE | This study |
| pFIN-EF1-GFP-2A-hABCB10-FLAG -WPRE | This study |
Lentivirus production and transduction
Human ALAS2-FLAG, mouse Gata1, and human ABCB10-FLAG were cloned into a modified bicistronic lentiviral vector pFIN-EF1-GFP-2A-mCherry-HA-WPRE (25). The lentiviral vectors were packaged in HEK293T cells using a three-plasmid packaging system. The supernatant containing retroviruses was passed through a 0.45-μm filter and stored in aliquots at −80 °C. MEL cells were transduced with GFP, hALAS2, mGata1, or hABCB10 lentiviruses and 8 μg/ml Polybrene on day 0 and day 2 of differentiation.
o-Dianisidine staining
Staining solution (20 mg/ml o-dianisidine, 3% H2O2, and 1% acetic acid) was added to MEL cells resuspended in PBS for 30 min at room temperature. Cells were then centrifuged (2 min at 250 rpm) onto glass slides using a Shandon Cytospin2 centrifuge (41).
qRT-PCR
mRNA was extracted using the RNeasy kit from Qiagen. Two μg of total mRNA was used to synthesize cDNA using the High Capacity cDNA reverse transcription kit (AB Biosystems). Power SYBR Green Master Mix (Life Technologies) was used on a Realplex2 thermal cycler (Eppendorf). Actin was used as a control housekeeping gene. The ΔΔCt method was used to compare the variation of transcripts among samples. Specificity and efficiency were checked before using this method. Primers were validated by cloning and sequencing the PCR products. Primers used in this study are listed in Table 3.
Table 3.
Primers used for qPCR
| Gene | Primer sequence | Reference/Source |
|---|---|---|
| Mouse primers | ||
| β-Actin | ||
| Forward | GACGGCCAAGTCATCACTATTG | |
| Reverse | CCACAGGATTCCATACCCAAGA | |
| Abcb10 | ||
| Forward | ATGTACGCTTTCTGGGTTGG | |
| Reverse | TCCTGGAATACGGACACCTC | |
| Mfrn1 | ||
| Forward | TTGAATCCAGATCCCAAAGC | |
| Reverse | GTTTCCTTGGTGGCTGAAAA | |
| β-Globin | ||
| Forward | AGAAGGCTGCTGTCTCTTGC | |
| Reverse | CTGGGTCCAAGGGTAGACAA | |
| Alas2 | ||
| Forward | CTCCGAGGCATCTATGGCATC | Refs. 35 and 36 |
| Reverse | ACACGAGGGTGTCTGCTTATG | 156255174c3 |
| Fech | ||
| Forward | TCATCCAGTGCTTTGCAGAC | |
| Reverse | CAGTGGCTCCTACCTCTTGG | |
| Pgbd | ||
| Forward | TGCACGATCCTGAAACTCTG | |
| Reverse | TGCATGCTATCTGAGCCATC | |
| Urod | ||
| Forward | CTTGTTGTACCCCAGGCATT | |
| Reverse | TAAGGGTGATGGCTTGGAAC | |
| Ppox | ||
| Forward | GTCTGGAGGCTGACCACATT | |
| Reverse | ATGGCACCAAATGTCCAAAT | |
| Gata1 | ||
| Forward | GAAGCGAATGATTGTCAGCA | |
| Reverse | TTCCTCGTCTGGATTCCATC | |
| Gata2 | ||
| Forward | AGACGACAACCACCACCTTA | |
| Reverse | TCCTTCTTCATGGTCAGTGG | |
| Bach1 | ||
| Forward | CATGGGCCCTAAAGAAGACA | |
| Reverse | GCTGCAAATGTCACTCCAGA | |
| Bcl-xl | ||
| Forward | TGACCACCTAGAGCCTTGGA | Ref. 37 |
| Reverse | GCTGCATTGTTCCCGTAGA | |
| TfR1 | ||
| Forward | CCCAAGTATTCTCAGATATGATTTCA | Ref. 37 |
| Reverse | CAGTCCAGCTGGCAAAGATTAT | |
| HO-1 | ||
| Forward | ACATCGACAGCCCCACCAAGTTCAA | |
| Reverse | CTGACGAAGTGACGCCATCTGTGAG | |
| Fth | ||
| Forward | CTCCTACGTCTATCTGTCTATG | |
| Reverse | ATTCGGCCACCTCGCTGGTTCT | |
| Ftl | ||
| Forward | ATGACCTCTCAGATTCGTCAG | |
| Reverse | ATTCGCGGAAGAAGTGGCCTA | |
| Sod2 | ||
| Forward | GGCCAAGGGAGATGTTACAA | |
| Reverse | GCTTGATAGCCTCCAGCAAC | |
| Human/Zebrafish primers | ||
| h-ACTIN | ||
| Forward | ATGGCCACGGCTGCTTCCAGC | |
| Reverse | CATGGTGGTGCCGCCAGACAG | |
| h-ABCB10 | ||
| Forward | ATGACCGTGGGTGAACTCTC | |
| Reverse | CTCGTTAAAAGGCAGCTTGG | |
| h-ALAS2 | ||
| Forward | TGTCCGTCTGGTGTAGTAATGA | Refs. 35 and 36 |
| Reverse | GCTCAAGCTCCACATGAAACT | 195539358c3 |
| z-alas2r] | ||
| Forward | CCGAAATATCTCTGGGACGA | |
| Reverse | CATGATTGCCCATGTCTGAG | |
| abcb10 MO2 | ||
| Forward | TCCGCAGAGATGGAGACTGACAG | |
| Reverse | ACACGTATAACATCATGCCAACTC | |
| abcb10 MO3 | ||
| Forward | CGTGTCTACCTCATGCAAATCTCA | |
| Reverse | CCCAAAAGCTCGGACTGT | |
| actin | ||
| Forward | GTTGGTATGGGACAGAAAGACAG | |
| Reverse | ACCAGAGGCATACAGGGACAG | |
| ChIP primers | ||
| β-Globin prom | ||
| Forward | CAGGGAGAAATATGCTTGTCATCA | Ref. 38 |
| Reverse | GTGAGCAGATTGGCCCTTACC | |
| Alas2 + 2kb | ||
| Forward | AGGGCAGGACTTTGCCTCTAATCT | Ref. 38 |
| Reverse | AGATGTCCCAGTTCCTGCAGGTTT | |
| Zfpm1 + 2kb | ||
| Forward | CTTTTCTCCTGCCCAGTCG | Ref. 38 |
| Reverse | TGCTGTTGCCTCGAACC | |
| LCR HS2 | ||
| Forward | TGCAGTACCACTGTCCAAGG | Ref. 39 |
| Reverse | ATCTGGCCACACACCCTAAG | |
| LCR HS3 | ||
| Forward | CTAGGGACTGAGAGAGGCTGCTT | Ref. 38 |
| Reverse | ATGGGACCTCTGATAGACACATCT | |
| Actin b | ||
| Forward | TGTTACCAACTGGGACGACA | Ref. 40 |
| Reverse | CTATGGGAGAACGGCAGAAG | |
Heme assay
Iron in heme was measured as described previously (26). Briefly, cells were incubated with 200 μm bathophenonthroline disulfonate (BPS) overnight followed by incubation with 1.2 mm ALA and 200 nm Tf(59Fe)2 for 8 h. The cells were washed with PBS and lysed in lysis buffer (10 mm Tris-HCl, pH 7.2, 150 mm NaCl, 0.5 mm EDTA, 1% Triton X-100) containing protease inhibitor mixture (Roche Applied Science). One hundred μl of 0.1 n HCl was added to 500 μl of cell lysate and vortex for 1 min. Six hundred μl of ethyl acetate/acetic acid (3:1) was added and vortexed for 1 min. Samples were centrifuged at 15,000 rpm for 5 min, 250 μl of organic fraction was collected, and radioactivity was detected using a γ-counter.
Mitochondrial isolation and mitochondrial iron uptake assay
Mitochondria were isolated from cells as described previously (26). Cells were washed with PBS and homogenized in mitochondrial buffer (10 mm Tris-HCl, pH 7.8, 0.25 m sucrose, 0.2 mm EDTA), followed by centrifugation at 1000 × g for 10 min at 4 °C. The supernatant was centrifuged at 12,000 × g for 15 min at 4 °C to pellet the mitochondria. The crude mitochondria were resuspended in mitochondrial buffer and centrifuged at 12,000 × g for 15 min at 4 °C. The pellets were collected as mitochondrial fraction. For the mitochondrial iron uptake assay, cells were treated with 200 μm BPS overnight followed by incubation with 1.2 mm ALA and 200 nm Tf(59Fe)2 for 8 h. Mitochondrial fractions were collected, and radioactivity was measured using a γ-counter.
Protoporphyrin IX assay
Cells were treated with 1.2 mm ALA for 8 h and lysed in lysis buffer. Ethyl acetate/acetic acid (4:1) was added to 500 μl of lysate and vortexed, followed by centrifugation at 15,000 rpm for 5 min at room temperature. Three hundred μl of organic fraction was collected, and 300 μl of 1.5 n HCl was added to the fraction and vortexed, followed by centrifugation at 15,000 rpm for 5 min at room temperature. The organic fraction was removed, and fluorescence was measured using an excitation wavelength of λ405 nm and emission wavelength of λ600 nm on a PerkinElmer fluorescence spectrophotometer.
Enzyme assays
Cells were lysed in lysis buffer containing 1 mm PMSF. After incubation on ice for 30 min, the lysate was centrifuged, and supernatant was collected. For aconitase, 4 μl of lysate was incubated in assay buffer (100 mm Tris-HCl, pH 8.0, 1 mm MgCl2, 1 mm NADP, 1 mm citrate, 0.8 units/ml recombinant isocitrate dehydrogenase) (USB Corp.) at 37 °C, and UV absorbance at a wavelength of λ340 nm was measured (27). Xanthine oxidase activity was measured using the Amplex Red xanthine/xanthine oxidase assay kit (Invitrogen).
In vitro heme assay
Mitochondrial heme biosynthesis was assayed as described (28).
Zebrafish injection and porphyrin analysis
Morpholinos specific to abcb10 MOe2/i2 intron 2 splice donor (5′-GTTTCACCATCCATACTTCACCTGA-3′), MOe3/i3 intron 3 splice donor (5′-CTAAAGGTCAAGCATCTCACCATCA-3′), and control morpholino (5′-CCTCTTACCTCAGTTACAATTTATA) were synthesized by Gene Tools, LLC (Philomath, OR). Morpholinos were injected at a concentration of 0.19–0.4 and 0.175 mm for abcb10, respectively. urod (5′-GTCCTTATCCATCATGACCGGCTTC-3′), fech (5′-CCCATATTCAGCATCAGAATGCCTG-3′), and alas2 (5′-CAGTGATGCAGAAAAGCAGACATGA-3′) morpholinos were synthesized by Gene Tools and injected at a concentration of 0.175, 0.75, and 0.1 mm, respectively (29, 30). HPLC analysis of porphyrin intermediates and heme was conducted in zebrafish embryo pools (n = 50) as described (31). ALA rescue experiments were performed using the alas2 and abcb10 morpholinos (i.e. MO3 splice blocking abcb10 MO) in the Tg(globinLCR:eGFP) transgenic line. At 24 hpf, embryos were dechlorionated before the addition of 2 mm ALA or vehicle. At ∼72 hpf (48 h of ALA or vehicle exposure), the control and treated embryos were disaggregated and analyzed by flow cytometry. The number of eGFP+ cells is a surrogate marker or index of red cell mass (26, 32).
[14C]Glycine heme measurement
Cells were differentiated for 3 days and moved to a glycine-free medium the day of the experiment and incubated with human holo-transferrin at 37 °C for 30 min before the addition of 1.5 μCi of [14C]glycine for an additional 1 h at 37 °C. The cells were then washed twice with cold PBS and lysed on ice for 30 min in lysis buffer containing protease inhibitor mixture. Five hundred μl of ethyl acetate/acetic acid (3:1) was added to 500 μl of cell lysate and vortexed for 2 min. Two hundred fifty μl of organic fraction was collected after centrifugation at 15,000 rpm for 5 min, and the extraction was repeated one more time. Extracts were washed twice with 750 μl of 0.3% sodium acetate and vortexed for 1 min. Three hundred μl of organic fraction was added, followed by 600 μl of 1.5 n HCl, and vortexed for 2 min. The organic fraction was collected, and radioactivity was measured with a liquid scintillation counter.
Quantification of ALA and Alas2 activity
Alas2 activity and ALA content were measured as described (33) using differentiated MEL cells (with DMSO for 3 days) incubated with or without 100 μm succinylacetone (4,6-dioxoheptanoic acid) for 3 h at 37 °C.
Chromatin immunoprecipitation
MEL cells were grown in 100-mm dishes and differentiated for 3 days in 1.5% DMSO. Briefly, proteins and DNA were cross-linked with 1% formaldehyde in medium for 2 min at room temperature. The cells were treated with 125 mm glycine to stop fixation. Cells were washed three times with ice-cold PBS, collected by centrifugation, lysed in radioimmune precipitation buffer supplemented with protease inhibitors, and sonicated for 5 cycles (20-s pulse and 40-s rest on ice). A portion of sheared chromatin was treated with RNase A to reverse the formaldehyde cross-link and labeled as “input DNA”. Chromatin was immunoprecipitated with IgG (control), Gata1, or Bach1 antibody. The immunoprecipitates were washed sequentially for 5 min each. Protein-DNA complexes were eluted from the antibody with freshly prepared elution buffer (1% SDS, 0.1 mm NaHCO3). Formaldehyde cross-links were reversed by the addition of RNase A and heating at 65 °C overnight. DNA was treated with proteinase K and purified using the Qiagen PCR-purification kit according to the manufacturer's instructions. qPCR was performed using Power SYBR Green Master mix (Life Technologies, Inc.).
Other procedures
Immunofluorescence was done by cytospinning fixed (3.7% formaldehyde for 20 min) transfected MEL cells onto glass slides. Cells were then permeabilized with 0.01% saponin, PBS, 1% BSA for 20 min at room temperature and incubated overnight at 4 °C with mouse anti-FLAG (1:750; Sigma) and rabbit anti-GFP (1:1000). Cells were washed extensively and incubated with Alexa 594–conjugated goat anti-mouse IgG and Alexa 488–conjugated goat anti-rabbit IgG (1:750; Thermo Fisher Scientific). Images were captured on an Olympus BX51 microscope (60 × 1.3 objective) using Picture Framer software. Western blotting was performed using the following primary antibodies: α-GFP (rabbit, 1:1000; Abcam), α-Myc (rabbit, 1:1000; Abcam), α-FLAG (rabbit, 1:2000; Sigma), α-Abcb10 (rabbit, 1:1000; a generous gift from Dr. O. S. Shirihai) or α-Abcb10 (rabbit, 1:1000; Santa Cruz Biotechnology), α-Alas2 (rabbit, 1:2000; a generous gift from Dr. H. Dailey), α-ferrochelatase (mouse, 1:1000; Santa Cruz Biotechnology), α-Gata1 (rat, 1:1000; Santa Cruz Biotechnology), α-A2B1 (rabbit, 1:2000; GeneTex), α-BACH1 (A1–6 rabbit; 1:1000) (34), α-tubulin (mouse, 1:5000; GeneTex), and α-porin (mouse, 1:1000; Thermo Fisher Scientific) and peroxidase-conjugated donkey α-rat, donkey α-goat, and goat α-rabbit or mouse IgG as a secondary (Jackson Immunoresearch). Western blots were developed using PerkinElmer Life Sciences Western Lightening Reagent, and blots were quantified using Bio-Rad gel quantification software. All experiments were performed a minimum of three times unless otherwise stated. Statistical analyses were performed using two-tailed Student's t test unless otherwise stated with significance of p ≤ 0.05 shown as an asterisk.
Author contributions
J. K., J. D. P., B. H. P., and D. M. W conceived the study; A. S., J. K., and D. M. W wrote the paper; A. S., N. T.-M., N. C. H., Y. Y. Y, G. M., J. W., J. C. W., M. D. K., T. B., and H. B. performed the experiments; M. M. and K. I. provided reagents; and A. S., N. C. H., Y. Y. Y., and H. B. performed quantitative and statistical analyses and prepared figures.
Supplementary Material
Acknowledgments
We thank the Ward, Kaplan, and Paw laboratories for critically reading the manuscript. We thank Johannes G. Wittig for the model illustration in Fig. 8, Arthur Skoutchi (Albert Einstein College of Medicine, Bronx, NY) for the MEL DS19 clone, and Leonard I. Zon (Boston Children's Hospital, Boston, MA) for the Tg(globinLCR:eGFP) transgenic line.
This work is supported by National Institutes of Health Grants DK052380 (to D. M. W.), DK070838 and P01 HL032262 (to B. H. P.), and U54DK110858 (to J. P.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article contains supplemental Figs. 1–5.
- ABC
- ATP-binding cassette
- PPIX
- protoporphyrin IX
- ALA
- 5-aminolevulinic acid
- MEL
- murine Friend erythroleukemia
- LCR
- locus control region
- hpf
- hours postfertilization
- qPCR and qRT-PCR
- quantitative PCR and RT-PCR, respectively
- SA
- succinylacetone
- HS
- hypersensitive site
- BPS
- bathophenonthroline disulfonate.
References
- 1. Zutz A., Gompf S., Schägger H., and Tampé R. (2009) Mitochondrial ABC proteins in health and disease. Biochim. Biophys. Acta 1787, 681–690 [DOI] [PubMed] [Google Scholar]
- 2. Tahara T., Sun J., Nakanishi K., Yamamoto M., Mori H., Saito T., Fujita H., Igarashi K., and Taketani S. (2004) Heme positively regulates the expression of beta-globin at the locus control region via the transcriptional factor Bach1 in erythroid cells. J. Biol. Chem. 279, 5480–5487 [DOI] [PubMed] [Google Scholar]
- 3. Welch J. J., Watts J. A., Vakoc C. R., Yao Y., Wang H., Hardison R. C., Blobel G. A., Chodosh L. A., and Weiss M. J. (2004) Global regulation of erythroid gene expression by transcription factor GATA-1. Blood 104, 3136–3147 [DOI] [PubMed] [Google Scholar]
- 4. Igarashi K., and Sun J. (2006) The heme-Bach1 pathway in the regulation of oxidative stress response and erythroid differentiation. Antioxid. Redox Signal. 8, 107–118 [DOI] [PubMed] [Google Scholar]
- 5. Tanimura N., Miller E., Igarashi K., Yang D., Burstyn J. N., Dewey C. N., and Bresnick E. H. (2016) Mechanism governing heme synthesis reveals a GATA factor/heme circuit that controls differentiation. EMBO Rep. 17, 249–265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Doty R. T., Phelps S. R., Shadle C., Sanchez-Bonilla M., Keel S. B., and Abkowitz J. L. (2015) Coordinate expression of heme and globin is essential for effective erythropoiesis. J. Clin. Invest. 125, 4681–4691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Dailey H. A., and Meissner P. N. (2013) Erythroid heme biosynthesis and its disorders. Cold Spring Harb. Perspect. Med. 3, a011676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Shirihai O. S., Gregory T., Yu C., Orkin S. H., and Weiss M. J. (2000) ABC-me: a novel mitochondrial transporter induced by GATA-1 during erythroid differentiation. EMBO J. 19, 2492–2502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hyde B. B., Liesa M., Elorza A. A., Qiu W., Haigh S. E., Richey L., Mikkola H. K., Schlaeger T. M., and Shirihai O. S. (2012) The mitochondrial transporter ABC-me (ABCB10), a downstream target of GATA-1, is essential for erythropoiesis in vivo. Cell Death Differ. 19, 1117–1126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Tang L., Bergevoet S. M., Bakker-Verweij G., Harteveld C. L., Giordano P. C., Nijtmans L., de Witte T., Jansen J. H., Raymakers R. A., and van der Reijden B. A. (2012) Human mitochondrial ATP-binding cassette transporter ABCB10 is required for efficient red blood cell development. Br. J. Haematol. 157, 151–154 [DOI] [PubMed] [Google Scholar]
- 11. Chen W., Dailey H. A., and Paw B. H. (2010) Ferrochelatase forms an oligomeric complex with mitoferrin-1 and Abcb10 for erythroid heme biosynthesis. Blood 116, 628–630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Chen W., Paradkar P. N., Li L., Pierce E. L., Langer N. B., Takahashi-Makise N., Hyde B. B., Shirihai O. S., Ward D. M., Kaplan J., and Paw B. H. (2009) Abcb10 physically interacts with mitoferrin-1 (Slc25a37) to enhance its stability and function in the erythroid mitochondria. Proc. Natl. Acad. Sci. U.S.A. 106, 16263–16268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Bayeva M., Khechaduri A., Wu R., Burke M. A., Wasserstrom J. A., Singh N., Liesa M., Shirihai O. S., Langer N. B., Paw B. H., and Ardehali H. (2013) ATP-binding cassette B10 regulates early steps of heme synthesis. Circ. Res. 113, 279–287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Yamamoto M., Arimura H., Fukushige T., Minami K., Nishizawa Y., Tanimoto A., Kanekura T., Nakagawa M., Akiyama S., and Furukawa T. (2014) Abcb10 role in heme biosynthesis in vivo: Abcb10 knockout in mice causes anemia with protoporphyrin IX and iron accumulation. Mol. Cell. Biol. 34, 1077–1084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Qiu W., Liesa M., Carpenter E. P., and Shirihai O. S. (2015) ATP binding and hydrolysis properties of ABCB10 and their regulation by glutathione. PLoS One 10, e0129772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ganis J. J., Hsia N., Trompouki E., de Jong J. L., DiBiase A., Lambert J. S., Jia Z., Sabo P. J., Weaver M., Sandstrom R., Stamatoyannopoulos J. A., Zhou Y., and Zon L. I. (2012) Zebrafish globin switching occurs in two developmental stages and is controlled by the LCR. Dev. Biol. 366, 185–194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Medlock A. E., Shiferaw M. T., Marcero J. R., Vashisht A. A., Wohlschlegel J. A., Phillips J. D., and Dailey H. A. (2015) Identification of the mitochondrial heme metabolism complex. PLoS One 10, e0135896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Paradkar P. N., Zumbrennen K. B., Paw B. H., Ward D. M., and Kaplan J. (2009) Regulation of mitochondrial iron import through differential turnover of mitoferrin 1 and mitoferrin 2. Mol. Cell. Biol. 29, 1007–1016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hollenstein K., Dawson R. J., and Locher K. P. (2007) Structure and mechanism of ABC transporter proteins. Curr. Opin. Struct. Biol. 17, 412–418 [DOI] [PubMed] [Google Scholar]
- 20. Jones P. M., O'Mara M. L., and George A. M. (2009) ABC transporters: a riddle wrapped in a mystery inside an enigma. Trends Biochem. Sci. 34, 520–531 [DOI] [PubMed] [Google Scholar]
- 21. Ebert P. S., Hess R. A., Frykholm B. C., and Tschudy D. P. (1979) Succinylacetone, a potent inhibitor of heme biosynthesis: effect on cell growth, heme content and δ-aminolevulinic acid dehydratase activity of malignant murine erythroleukemia cells. Biochem. Biophys. Res. Commun. 88, 1382–1390 [DOI] [PubMed] [Google Scholar]
- 22. Tschudy D. P., Ebert P. S., Hess R. A., Frykholm B. C., and Weinbach E. C. (1980) Effect of heme depletion on growth, protein synthesis and respiration of murine erythroleukemia cells. Biochem. Pharmacol. 29, 1825–1831 [DOI] [PubMed] [Google Scholar]
- 23. Noordermeer D., and de Laat W. (2008) Joining the loops: β-globin gene regulation. IUBMB Life 60, 824–833 [DOI] [PubMed] [Google Scholar]
- 24. Ogawa K., Sun J., Taketani S., Nakajima O., Nishitani C., Sassa S., Hayashi N., Yamamoto M., Shibahara S., Fujita H., and Igarashi K. (2001) Heme mediates derepression of Maf recognition element through direct binding to transcription repressor Bach1. EMBO J. 20, 2835–2843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Verrier J. D., Madorsky I., Coggin W. E., Geesey M., Hochman M., Walling E., Daroszewski D., Eccles K. S., Ludlow R., and Semple-Rowland S. L. (2011) Bicistronic lentiviruses containing a viral 2A cleavage sequence reliably co-express two proteins and restore vision to an animal model of LCA1. PLoS One 6, e20553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Chen C., Garcia-Santos D., Ishikawa Y., Seguin A., Li L., Fegan K. H., Hildick-Smith G. J., Shah D. I., Cooney J. D., Chen W., King M. J., Yien Y. Y., Schultz I. J., Anderson H., Dalton A. J., et al. (2013) Snx3 regulates recycling of the transferrin receptor and iron assimilation. Cell Metab. 17, 343–352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Chen O. S., Schalinske K. L., and Eisenstein R. S. (1997) Dietary iron intake modulates the activity of iron regulatory proteins and the abundance of ferritin and mitochondrial aconitase in rat liver. J. Nutr. 127, 238–248 [DOI] [PubMed] [Google Scholar]
- 28. Shah D. I., Takahashi-Makise N., Cooney J. D., Li L., Schultz I. J., Pierce E. L., Narla A., Seguin A., Hattangadi S. M., Medlock A. E., Langer N. B., Dailey T. A., Hurst S. N., Faccenda D., Wiwczar J. M., et al. (2012) Mitochondrial Atpif1 regulates haem synthesis in developing erythroblasts. Nature 491, 608–612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Wingert R. A., Galloway J. L., Barut B., Foott H., Fraenkel P., Axe J. L., Weber G. J., Dooley K., Davidson A. J., Schmidt B., Paw B. H., Shaw G. C., Kingsley P., Palis J., Schubert H., et al. (2005) Deficiency of glutaredoxin 5 reveals Fe-S clusters are required for vertebrate haem synthesis. Nature 436, 1035–1039 [DOI] [PubMed] [Google Scholar]
- 30. Guernsey D. L., Jiang H., Campagna D. R., Evans S. C., Ferguson M., Kellogg M. D., Lachance M., Matsuoka M., Nightingale M., Rideout A., Saint-Amant L., Schmidt P. J., Orr A., Bottomley S. S., Fleming M. D., et al. (2009) Mutations in mitochondrial carrier family gene SLC25A38 cause nonsyndromic autosomal recessive congenital sideroblastic anemia. Nat. Genet. 41, 651–653 [DOI] [PubMed] [Google Scholar]
- 31. Yien Y. Y., Robledo R. F., Schultz I. J., Takahashi-Makise N., Gwynn B., Bauer D. E., Dass A., Yi G., Li L., Hildick-Smith G. J., Cooney J. D., Pierce E. L., Mohler K., Dailey T. A., Miyata N., et al. (2014) TMEM14C is required for erythroid mitochondrial heme metabolism. J. Clin. Invest. 124, 4294–4304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kardon J. R., Yien Y. Y., Huston N. C., Branco D. S., Hildick-Smith G. J., Rhee K. Y., Paw B. H., and Baker T. A. (2015) Mitochondrial ClpX activates a key enzyme for heme biosynthesis and erythropoiesis. Cell 161, 858–867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Bergonia H. A., Franklin M. R., Kushner J. P., and Phillips J. D. (2015) A method for determining δ-aminolevulinic acid synthase activity in homogenized cells and tissues. Clin. Biochem. 48, 788–795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Matsumoto M., Kondo K., Shiraki T., Brydun A., Funayama R., Nakayama K., Yaegashi N., Katagiri H., and Igarashi K. (2016) Genomewide approaches for BACH1 target genes in mouse embryonic fibroblasts showed BACH1-Pparg pathway in adipogenesis. Genes Cells 21, 553–567 [DOI] [PubMed] [Google Scholar]
- 35. Spandidos A., Wang X., Wang H., and Seed B. (2010) PrimerBank: a resource of human and mouse PCR primer pairs for gene expression detection and quantification. Nucleic Acids Res. 38, D792–D799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Wang X., Spandidos A., Wang H., and Seed B. (2012) PrimerBank: a PCR primer database for quantitative gene expression analysis, 2012 update. Nucleic Acids Res. 40, D1144–D1149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Nai A., Lidonnici M. R., Rausa M., Mandelli G., Pagani A., Silvestri L., Ferrari G., and Camaschella C. (2015) The second transferrin receptor regulates red blood cell production in mice. Blood 125, 1170–1179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Byrska-Bishop M., VanDorn D., Campbell A. E., Betensky M., Arca P. R., Yao Y., Gadue P., Costa F. F., Nemiroff R. L., Blobel G. A., French D. L., Hardison R. C., Weiss M. J., and Chou S. T. (2015) Pluripotent stem cells reveal erythroid-specific activities of the GATA1 N-terminus. J. Clin. Invest. 125, 993–1005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Crusselle-Davis V. J., Vieira K. F., Zhou Z., Anantharaman A., and Bungert J. (2006) Antagonistic regulation of β-globin gene expression by helix-loop-helix proteins USF and TFII-I. Mol. Cell. Biol. 26, 6832–6843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Shakya A., Callister C., Goren A., Yosef N., Garg N., Khoddami V., Nix D., Regev A., and Tantin D. (2015) Pluripotency transcription factor Oct4 mediates stepwise nucleosome demethylation and depletion. Mol. Cell. Biol. 35, 1014–1025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Huber T. L., Zhou Y., Mead P. E., and Zon L. I. (1998) Cooperative effects of growth factors involved in the induction of hematopoietic mesoderm. Blood 92, 4128–4137 [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.